Abstract
A two-stage strategy was used to identify and confirm quantitative trait loci (QTLs) associated with the changes in locomotor activity induced by a 1.5 gm/kg ethanol challenge. For stage 1, putative QTLs were identified by analysis of the strain means for 25 strains of the BXD recombinant inbred (RI) series (males only). QTLs were identified on chromosomes 1, 2, 4, and 6. The activity response to chlordiazepoxide generated similar QTLs on chromosomes 2 and 6. None of the QTLs were similar to those generated from analysis of the saline response data. For stage 2, 900 male C57BL/6J (B6) × DBA/2J (D2) F2 intercross animals were phenotyped for ethanol response, and the phenotypic extremes (those animals > and <1 SD from the mean) were identified. These extremes differed by >10,000 cm/15 min in their response to ethanol. The extreme progeny were used for a genome-wide scan both to confirm the putative RI-generated QTLs and to detect new QTLs. The F2 analysis generated no new QTLs with logarithm of the likelihood for linkage (LOD) scores >3. For RI-generated QTLs, only the QTL on chromosome 2 was confirmed (LOD = 5.3). The position of the peak LOD was estimated to be 47 cM with a 20 cM 1 LOD support interval; this QTL accounted for 6% of the phenotypic variance. The 1 LOD support interval overlaps with QTLs previously identified for alcohol preference and acute ethanol withdrawal (Melo et al., 1996; Buck et al., 1997; Phillips et al., 1999).
Keywords: ethanol, mouse, genes, recombinant inbred, QTL, activity
The current study asks the question, “What genes contribute to the variability in the ethanol-induced locomotor response?” The study builds on previous observations of genetics and ethanol locomotor responses. (1) Among inbred strains of mice it is now clearly established that moderate doses of ethanol (1–2 gm/kg) markedly stimulate locomotor activity in some strains, e.g., DBA/2J (D2), but only mildly affect activity in other strains, e.g., C57BL/6J (B6) (Crabbe et al., 1982, 1983; Crabbe, 1986; Dudek and Phillips, 1990; Dudek et al., 1991; Dudek and Tritto, 1994). (2) Dudek and Tritto (1994) completed a full Mendelian analysis of the locomotor response for the B6D2 genotypes. These authors found an additive mode of inheritance for the total distance measure (of activity) and determined that there was no evidence of sex linkage or sex effects. Narrow sense heritability was estimated at 0.35. (3) The locomotor response to ethanol has been subject to three different quantitative trait locus (QTL) analyses involving the BXD recombinant inbred (RI) series. Crabbe et al. (1983) (but analyzed by Gora-Maslak et al., 1991) reported significant QTLs on chromosomes 2, 4, 9, and 13. Cunningham (1995) studied a different phenotype from that of Crabbe et al. (1983), in that the focus was on the initial ethanol response (first 5 min after injection) as opposed to beginning measurements 10 min after injection. Cunningham (1995) detected two QTLs at p < 0.01 on chromosomes 7 and 18. Phillips et al. (1995), using a yet different behavioral paradigm but still focusing on the initial ethanol response, confirmed the presence of a significant (p < 0.01) QTL on the distal part of chromosome 18 but also found a significant (p < 0.01) QTL on chromosome 3. For all QTLs significant at p < 0.05,Cunningham (1995) and Phillips et al. (1995) agreed on four (chromosomes 3, 12, 17, and 18).
Because the BXD RI series contains only 26 strains, the smallest QTL effect size that can be detected at p < 0.01 is associated with 25% of the genotypic variance. At this level of significance it is necessary to use an additional strategy for verification (Belknap et al., 1996). In the current study we have phenotyped the BXD RI strains for their locomotor response to a 1.5 gm/kg ethanol challenge; a B6D2 F2 intercross genome-wide scan was used both to confirm the RI generated QTLs and to detect additional QTLs. Of the three RI studies noted above, the behavioral paradigm was closest to that of Crabbe et al. (1983) and was patterned after the testing procedure initially developed for the selection of the FAST and SLOW selected lines (Crabbe et al., 1987) and subsequently used in the biometric analysis of Dudek and Tritto (1994). To compare across RI studies (see above), data for ethanol response were collected over a broad time interval. A secondary goal of the current study was to use the RI strains to test the hypothesis that a genetic correlation exists between the ethanol- and benzodiazepine-induced effects on locomotor activity.
MATERIALS AND METHODS
Animals and sample sizes. Male and female C57BL/6 (B6), DBA/2 (D2), and B6D2 F1 mice and male BXD RI mice (25 strains) were obtained from The Jackson Laboratory (Bar Harbor, ME). All of the RI strains were obtained over a 3 month period from December 1997 to March 1998. We attempted to obtain an N of 15 per strain, although this was not always possible. Mice were housed two to four per cage in a constant-temperature colony room with a 12 hr light/dark cycle. Food and water were provided ad libitumthroughout the study. All testing was conducted during the light cycle and between 9 A.M. and 3:30 P.M. All mice were allowed a minimum of 10 d to acclimatize to the colony conditions before testing. All animal care and testing protocols were approved by the Laboratory Animal Users Committee at the State University of New York at Stony Brook and conformed to National Institutes of Health Guidelines for Using Animals in Intramural Research.
For production of the F2 mice, D2B6 F1 mice were bred locally from the Jackson parental lines. Reciprocal F1 crosses were used to generate a total of 900 male F2 mice from June 1, 1996 to August 1, 1997. Sample size for the genome-wide scan was estimated as described by Soller et al. (1976) and Lander and Botstein (1989) for an F2 population from n = (Zα +Zβ)2/(sQTL2/sRES2), where Zα and Zβ are the normal variates for the desired values of α and β, sQTL2 is the variance associated with or explained by the QTL, andsRES2 is the residual unexplained variance. The minimum hQTL2 was arbitrarily set at 0.05. It was recognized that the effect size for most QTLs is <0.05; however, the data from other behavioral phenotypes, e.g., Kanes et al. (1996), suggested that some QTLs of this size or larger were likely to be present. For (sQTL2/sRES2) = 0.053 (from 0.05/0.95), α = 0.0001 [Zα = 3.89 (two-tailed)], and β = 0.05 (Zβ = 1.64), the estimated sample size was 575. However, because only the extreme phenotypes (> and <1 SD from the mean) would be genotyped, to maintain power the sample size must be increased by 1/0.81 or 1.23 (Lander and Botstein, 1989). This correction is relatively small because most of the genetic information is found in the extreme phenotypes. We also included an additional correction of 1.15 to account for nonadditive genetic effects. Sample size estimates were based on the assumption that all QTLs have equal and additive effects (Fisher, 1918; Barton and Turelli, 1989). Because this assumption most certainly was violated, some correction was necessary. Finally, a correction of 1.11 was included to account for the loss of animals from data censoring. Overall, these correction factors caused the sample size to increase from 575 to 900.
Measurement of ethanol and chlordiazepoxide effects on locomotor activity. Mice were removed from the home cage and placed individually in the testing arena; the arena floor was covered with standard laboratory bedding. Thirty minutes later, the mice were administered saline and returned to the testing arena, and activity was monitored for 20 min. On the next day, the procedure was repeated, except that the animals were administered 1.5 gm/kg ethanol dissolved in saline (20% v/v). One week later the saline/ethanol days were repeated for the F2 mice only. For the RI strains, the second week of testing included saline on day 1 and chlordiazepoxide (10 mg/kg) on day 2. Locomotor activity was assessed in a San Diego Instruments Flex Field locomotor system. The apparatus comprised a four-by-eight array of photo cells mounted in a 25 × 47 cm metal frame, situated 1 cm off the floor, and surrounding a 22 × 42 × 20-cm-high plastic arena. Activity was recorded over four 5 min blocks. The distance traveled during each block was used as the measure of activity.
For the F2 sample, ethanol response (day2 − day1) data were obtained from two trials, and these data were used to identify the 10% of individuals showing the poorest test–retest reliability. These individuals, with a test–retest reliability of < 0.1, were censored from the data set. The remaining individuals were ranked from least to greatest ethanol-induced activation, and the phenotypic extremes (those individuals > and <1 SD from the mean) were identified. The extremes were denoted for historical reasons (Demarest et al., 1997) as very responsive (RR) and very nonresponsive (NN), although the NN group actually showed a marked inhibition of locomotor activity.
The phenotypic data generated from the RI strains and the F2 sample were analyzed using standard ANOVA techniques; the Student–Neuman–Keuls test was used for the post hocanalysis. Because of the multiple tests being performed for each phenotype, the threshold for significance was set at p< 0.01.
DNA isolation. High molecular weight genomic DNA was isolated from liver samples as follows: 250–500 mg of liver tissue was minced with a sterile razor blade, transferred to a 15 ml polypropylene Falcon tube with 5 ml of lysis buffer (100 mm Tris-HCl, pH 8.0, 5 mm EDTA, 100 μg/ml proteinase K, and 200 mm NaCl), and incubated with rocking at 55°C overnight. After incubation, 20 μl/ml 5 m NaCl was added with gentle inversion. The tissue digest was extracted twice with equilibrated phenol, once with equal volumes of phenol and chloroform/isoamyl alcohol (chisam, 24:1) and once with chisam alone. DNA was precipitated with 0.5 vol of 7.5 m ammonium acetate and 2 vol of ice cold ethanol. Dried DNA pellets were resuspended in double distilled water. Purity and concentration of the final samples were evaluated by UV spectroscopy, and only samples with a 260:280 ratio >1.4 were used for genotyping.
Genotyping the microsatellite polymorphisms. All of the genotyping involved the -(CA)n- repeating microsatellites first described by Dietrich et al. (1992). The PCR primer sets were obtained from the Massachusetts Institute of Technology–Whitehead catalog (Research Genetics). One to 5 ng of genomic DNA was amplified with an 18 pm concentration of each primer, 0.5 U ofTaq polymerase (AmpliTaq, Perkin-Elmer, Emeryville, CA; orTaq DNA polymerase, Boehringer Mannheim, Indianapolis, IN), and 100 nm dNTPs in a 20 μl reaction under the standard conditions recommended by the manufacturer. All reactions were amplified in a Perkin-Elmer thermal cycler. Products were visualized by electrophoresis in 1× Tris borate-EDTA buffer on a 3% agarose gel (3:1 Metaphor agarose; FMC Bioproducts, Rockland, ME). Bands were visualized by ethidium bromide staining.
Detection and mapping of QTLs. Candidate QTLs were generated from the BXD RI data essentially as described elsewhere (Kanes et al., 1996). Briefly, the analysis was performed by correlating the strain means with the strain distribution patterns of 1500 marker gene loci, each mapped to a chromosome region and showing allelic differences between the B6 and D2 strains. By convention, the B6 allele is scored as 0, and the D2 allele is scored as 1; the correlation coefficient has the same meaning as a t test for significant differences between strains with the B6 and D2 alleles. Because of linkage disequilibrium, the actual number of independent tests is <1500 but >100 (Belknap, 1992). Thus, although the threshold for acceptance of a candidate QTL was set at p < 0.01, some of these QTLs will be significant because of chance. Belknap et al. (1996) have estimated the false-positive rate to be 50%. We recognize that it is common, even in our own work (Kanes et al., 1996), to report QTL data generated from RI analyses that are significant at p < 0.05 but not p < 0.01. However, because no behavioral QTL detected in this range has been independently confirmed using a non-RI strategy, we feel there is little value in reporting these data. For the interested reader, the strain means reported in Tables 1-3 are sufficient to conduct such analyses.
Table 1.
Saline-induced locomotor response in the BXD RI series
| Strain | N | Time interval: distance traveled (cm) | ||||
|---|---|---|---|---|---|---|
| 0–5 min | 5–10 min | 10–15 min | 15–20 min | 5–20 min | ||
| 1 | 15 | 4018 ± 428 | 2908 ± 266 | 2391 ± 183 | 1964 ± 123 | 7263 ± 572 |
| 5 | 15 | 1876 ± 235 | 1767 ± 243 | 1442 ± 250 | 1420 ± 226 | 4629 ± 718 |
| 6 | 15 | 2893 ± 263 | 2469 ± 243 | 2302 ± 157 | 2272 ± 196 | 7043 ± 596 |
| 8 | 15 | 1784 ± 156 | 1807 ± 158 | 1753 ± 162 | 1656 ± 158 | 5216 ± 477 |
| 9 | 15 | 3817 ± 249 | 3533 ± 220 | 2865 ± 137 | 2531 ± 175 | 8929 ± 531 |
| 11 | 11 | 1647 ± 198 | 1333 ± 147 | 1166 ± 166 | 1268 ± 209 | 3767 ± 522 |
| 12 | 4 | 3998 ± 847 | 2730 ± 662 | 2391 ± 448 | 2084 ± 584 | 7204 ± 1694 |
| 13 | 15 | 2876 ± 146 | 2356 ± 149 | 1806 ± 145 | 2148 ± 218 | 6311 ± 512 |
| 14 | 15 | 3481 ± 261 | 2640 ± 158 | 2433 ± 179 | 2658 ± 205 | 7731 ± 541 |
| 15 | 9 | 1086 ± 174 | 892 ± 106 | 865 ± 106 | 954 ± 80 | 2710 ± 293 |
| 16 | 15 | 1603 ± 225 | 1752 ± 159 | 1612 ± 146 | 1383 ± 171 | 4747 ± 477 |
| 18 | 15 | 3117 ± 356 | 3032 ± 222 | 2775 ± 211 | 2642 ± 227 | 8449 ± 660 |
| 19 | 15 | 2888 ± 241 | 2900 ± 204 | 2625 ± 185 | 2442 ± 165 | 7966 ± 553 |
| 20 | 15 | 2713 ± 237 | 2367 ± 223 | 1868 ± 217 | 1794 ± 187 | 6029 ± 627 |
| 21 | 15 | 1929 ± 99 | 2200 ± 91 | 1852 ± 147 | 1781 ± 182 | 5833 ± 419 |
| 22 | 15 | 1926 ± 226 | 1859 ± 247 | 1430 ± 168 | 1347 ± 115 | 4636 ± 530 |
| 23 | 15 | 2511 ± 305 | 1864 ± 244 | 1573 ± 217 | 1351 ± 154 | 4787 ± 615 |
| 24 | 14 | 2317 ± 245 | 1854 ± 214 | 1418 ± 195 | 1340 ± 233 | 4612 ± 641 |
| 25 | 15 | 2391 ± 199 | 2316 ± 220 | 1752 ± 214 | 1594 ± 169 | 5663 ± 603 |
| 27 | 15 | 2194 ± 233 | 1925 ± 184 | 1691 ± 188 | 1797 ± 254 | 5413 ± 615 |
| 28 | 13 | 1449 ± 177 | 1645 ± 140 | 1476 ± 173 | 1519 ± 129 | 4640 ± 443 |
| 29 | 15 | 4543 ± 298 | 4018 ± 343 | 3395 ± 286 | 3083 ± 280 | 10496 ± 909 |
| 30 | 15 | 2128 ± 184 | 2143 ± 171 | 1850 ± 226 | 1598 ± 204 | 5590 ± 601 |
| 31 | 15 | 2099 ± 251 | 2266 ± 186 | 2013 ± 185 | 1775 ± 161 | 6055 ± 531 |
| 32 | 15 | 1559 ± 131 | 1789 ± 149 | 1657 ± 127 | 1770 ± 162 | 5215 ± 438 |
| B6 | 18 | 5131 ± 284 | 3595 ± 248 | 2783 ± 173 | 2580 ± 186 | 8958 ± 607 |
| D2 | 18 | 2170 ± 173 | 1894 ± 136 | 1582 ± 138 | 1317 ± 109 | 4793 ± 383 |
| F1 | 12 | 2811 ± 144 | 2046 ± 158 | 1845 ± 174 | 1554 ± 166 | 5445 ± 498 |
| Avg. RI1-a | 2514 ± 255 | 2255 ± 214 | 1936 ± 193 | 1847 ± 199 | 6037 ± 605 | |
Animals were administered normal saline (20 mg/kg, i.p.) and placed in the testing arena (San Diego Instruments Flex Field System), and activity was monitored for 20 min; data were collected in four 5 min blocks, and the metric used for the measurement of locomotion was the distance traveled in centimeters. For each interval data are presented as the mean ± SE. Data are also presented for the collapsed 5–20 min interval. Correlation matrix among each of the 5 min activity intervals: Time (min) 0–5 5–10 10–15 15–20 0–5 1.00 0.91 0.87 0.82 5–10 1.00 0.97 0.91 10–15 1.00 0.95 15–20 1.00.
Average locomotor response in the BXD RI strains, calculated from the strain means.
Table 2.
Ethanol-induced locomotor response (difference score) in the BXD RI series
| Strain | N | Time interval: distance traveled (cm) | ||||
|---|---|---|---|---|---|---|
| 0–5 min | 5–10 min | 10–15 min | 15–20 min | 5–20 min | ||
| 1 | 15 | 1274 ± 516 | 533 ± 283 | 182 ± 265 | 639 ± 336 | 1354 ± 884 |
| 5 | 15 | 323 ± 413 | −144 ± 293 | −824 ± 280 | −936 ± 270 | −1905 ± 843 |
| 6 | 15 | −208 ± 364 | −504 ± 420 | −1115 ± 348 | −1376 ± 316 | −2995 ± 1083 |
| 8 | 15 | 1182 ± 324 | 1387 ± 322 | 1344 ± 318 | 1147 ± 309 | 3879 ± 950 |
| 9 | 15 | 289 ± 370 | −2024 ± 460 | −2458 ± 169 | −2107 ± 151 | −6588 ± 779 |
| 11 | 11 | 308 ± 265 | 459 ± 213 | 599 ± 249 | 578 ± 280 | 1636 ± 743 |
| 12 | 4 | 1819 ± 347 | 2298 ± 279 | 1244 ± 141 | 1495 ± 188 | 5037 ± 608 |
| 13 | 15 | 925 ± 291 | 392 ± 306 | 282 ± 199 | −308 ± 296 | 366 ± 800 |
| 14 | 15 | 146 ± 375 | 288 ± 224 | −354 ± 416 | −857 ± 304 | −923 ± 944 |
| 15 | 9 | 943 ± 341 | −119 ± 132 | −324 ± 158 | −420 ± 133 | −864 ± 423 |
| 16 | 12 | 1845 ± 410 | 1884 ± 351 | 1555 ± 427 | 1416 ± 422 | 4855 ± 1200 |
| 18 | 15 | 2042 ± 447 | 2243 ± 436 | 2235 ± 367 | 1799 ± 299 | 6277 ± 1102 |
| 19 | 15 | 1786 ± 338 | 264 ± 402 | −357 ± 444 | −606 ± 424 | −717 ± 1271 |
| 20 | 15 | 1423 ± 389 | 2291 ± 437 | 1742 ± 498 | 1234 ± 241 | 5267 ± 1175 |
| 21 | 15 | 1228 ± 250 | 1904 ± 361 | 1693 ± 339 | 1406 ± 323 | 5003 ± 1023 |
| 22 | 15 | 481 ± 269 | −1116 ± 257 | −591 ± 233 | −528 ± 233 | −2235 ± 724 |
| 23 | 15 | 109 ± 420 | −138 ± 318 | −450 ± 260 | −173 ± 248 | −761 ± 826 |
| 24 | 12 | 683 ± 560 | −281 ± 444 | −319 ± 336 | −380 ± 293 | −979 ± 1073 |
| 25 | 15 | 698 ± 403 | −378 ± 558 | −634 ± 355 | −711 ± 375 | −1722 ± 1288 |
| 27 | 15 | 447 ± 266 | 1689 ± 528 | 800 ± 368 | 319 ± 396 | 2808 ± 1293 |
| 28 | 13 | 486 ± 126 | 566 ± 142 | 749 ± 203 | 515 ± 269 | 1829 ± 614 |
| 29 | 13 | 155 ± 600 | 1038 ± 253 | 799 ± 325 | 142 ± 369 | 1980 ± 947 |
| 30 | 15 | 983 ± 495 | 902 ± 424 | 758 ± 315 | 1065 ± 211 | 2725 ± 951 |
| 31 | 15 | 414 ± 325 | 671 ± 216 | 908 ± 301 | 1225 ± 200 | 2805 ± 716 |
| 32 | 15 | 1073 ± 289 | 515 ± 296 | 766 ± 391 | 851 ± 238 | 2132 ± 924 |
| B6 | 18 | 208 ± 321 | 536 ± 330 | 356 ± 331 | 215 ± 232 | 1107 ± 893 |
| D2 | 18 | 2284 ± 261 | 1689 ± 353 | 1094 ± 233 | 739 ± 207 | 3522 ± 793 |
| F1 | 12 | 2227 ± 410 | 721 ± 325 | 271 ± 296 | 117 ± 256 | 1109 ± 877 |
| Avg. RI2-a | 834 ± 368 | 585 ± 334 | 336 ± 309 | 217 ± 285 | 1137 ± 928 | |
Twenty-four hours after the saline trial (Table 1), animals were administered 1.5 gm/kg ethanol dissolved in normal saline (20% v/v, i.p.) and placed in the testing arena (San Diego Instruments Flex Field System), and activity was monitored for 20 min; data were collected in four 5 min blocks, and the metric used for the measurement of locomotion was the distance traveled in centimeters. The ethanol response was calculated as the difference in activity between day2 (ethanol treatment) and day1 (saline treatment). For each interval data are presented as the mean ± SE. Data are also presented for the collapsed 5–20 min interval. Correlation matrix among each of the 5 min activity intervals: Time (min) 0–5 5–10 10–15 15–20 0–5 1.00 0.63 0.61 0.63 5–10 1.00 0.94 0.88 10–15 1.00 0.95 15–20 1.00.
The values in bold are significantly different (p < 0.01) from the relative values in the 0–5 min column.
Average locomotor response in the BXD RI strains, calculated from the strain means.
Table 3.
CDP-induced locomotor response (difference score) in the BXD RI series
| Strain | N | Time interval: distance traveled (cm) | ||||
|---|---|---|---|---|---|---|
| 0–5 min | 5–10 min | 10–15 min | 15–20 min | 5–20 min | ||
| 1 | 15 | −716 ± 282 | −248 ± 283 | −462 ± 245 | −817 ± 248 | −1526 ± 776 |
| 5 | 15 | −40 ± 182 | −487 ± 209 | −846 ± 231 | −705 ± 170 | −2039 ± 610 |
| 6 | 15 | −9 ± 448 | −36 ± 410 | −137 ± 413 | −358 ± 318 | −531 ± 1141 |
| 8 | 15 | −284 ± 391 | −18 ± 446 | −285 ± 269 | −425 ± 269 | −727 ± 985 |
| 9 | 15 | 197 ± 163 | −1623 ± 286 | −2600 ± 262 | −2564 ± 356 | −6786 ± 903 |
| 11 | 9 | 919 ± 668 | 1166 ± 246 | 518 ± 381 | 319 ± 284 | 2002 ± 911 |
| 12 | 3 | 112 ± 156 | 1306 ± 119 | 477 ± 773 | −383 ± 707 | 1401 ± 1600 |
| 13 | 15 | −509 ± 310 | −451 ± 365 | −642 ± 239 | −888 ± 242 | −1981 ± 846 |
| 14 | 15 | −594 ± 250 | 132 ± 370 | 443 ± 414 | 268 ± 390 | 843 ± 1174 |
| 15 | 7 | 216 ± 233 | −1104 ± 304 | −1036 ± 171 | −835 ± 359 | −2974 ± 833 |
| 16 | 12 | 7 ± 239 | 1377 ± 407 | 1013 ± 337 | 814 ± 387 | 3204 ± 1131 |
| 18 | 15 | 668 ± 276 | 10 ± 431 | 266 ± 372 | 367 ± 347 | 643 ± 1149 |
| 19 | 15 | 335 ± 299 | −39 ± 228 | −403 ± 306 | −828 ± 309 | −1270 ± 843 |
| 20 | 15 | 744 ± 295 | 510 ± 505 | 453 ± 487 | −40 ± 420 | 923 ± 1412 |
| 21 | 15 | 76 ± 115 | −246 ± 299 | −520 ± 255 | −354 ± 319 | −1119 ± 873 |
| 22 | 15 | 787 ± 282 | −448 ± 295 | −743 ± 297 | −622 ± 335 | −1812 ± 926 |
| 23 | 8 | −113 ± 208 | 256 ± 545 | 104 ± 378 | 825 ± 552 | 1185 ± 1475 |
| 24 | 7 | 739 ± 317 | 174 ± 393 | −390 ± 370 | −351 ± 206 | −567 ± 969 |
| 25 | 14 | −39 ± 304 | −86 ± 294 | −110 ± 345 | −132 ± 365 | −327 ± 1005 |
| 27 | 6 | −413 ± 337 | −497 ± 574 | −555 ± 658 | −1058 ± 181 | −2110 ± 1413 |
| 28 | 10 | 387 ± 280 | 409 ± 278 | 9 ± 170 | −340 ± 206 | 78 ± 653 |
| 29 | 8 | 742 ± 291 | 1909 ± 316 | 903 ± 362 | 1059 ± 399 | 3870 ± 1077 |
| 30 | 15 | 204 ± 328 | −590 ± 283 | −28 ± 281 | −312 ± 160 | −930 ± 724 |
| 31 | 15 | 328 ± 268 | 527 ± 331 | 774 ± 305 | 233 ± 371 | 1534 ± 1007 |
| 32 | 15 | 299 ± 200 | 500 ± 370 | 419 ± 345 | 504 ± 314 | 1423 ± 1030 |
| B6 | 6 | −696 ± 292 | −459 ± 253 | −69 ± 352 | 297 ± 156 | −231 ± 761 |
| D2 | 6 | −81 ± 123 | 863 ± 307 | 875 ± 219 | 857 ± 293 | 2595 ± 819 |
| Avg. RI3-a | 162 ± 285 | 96 ± 344 | −135 ± 347 | −265 ± 329 | −304 ± 1019 | |
Twenty-four hours after a saline trial, animals were administered 10 mg/kg CDP, i.p., and placed in the testing arena (San Diego Instruments Flex Field System), and activity was monitored for 20 min; data were collected in four 5 min blocks, and the metric used for the measurement of locomotion was the distance traveled in centimeters. The data for the saline trial are not presented, but the correlation between the ethanol saline trial and the CDP saline trial was at 0.80 or better for all intervals. The CDP response was calculated as the difference in activity between day2 (CDP treatment) and day1 (saline treatment). For each interval data are presented as the mean ± SE. Data are also presented for the collapsed 5–20 min interval. Correlation matrix among each of the 5 min activity intervals: Time (min) 0–5 5–10 10–15 15–20 0–5 1.00 0.32 0.19 0.24 5–10 1.00 0.880.79 10–15 1.00 0.91 15–20 1.00.
The values in bold are significantly different (p < 0.001) from the relative values in the 0–5 min column.
Average locomotor response in the BXD RI strains, calculated from the strain means.
For analysis of the F2 intercross, 25 each of the extreme phenotypes (approximately one-fifth of each extreme sample) wererandomly selected for the genome-wide scan, except in the regions of the RI-generated QTLs, where a minimum N of 100 per extreme was used. For QTL detection, markers were spaced at ∼20 cM (Darvasi et al., 1993). The screening threshold for a significant segregation of the alleles was set at p < 0.05. For markers meeting the screening threshold, 25 each of the extreme phenotypes were again randomly selected and genotyped to confirm a significant effect. For markers meeting the threshold of p < 0.01, the entire sample of phenotypic extremes was genotyped, and additional markers were added in the region of interest. The data from this more dense map were then subjected to analysis using the MAPMAKER-QTL program (Lander and Botstein, 1989) to determine the position of the peak logarithm of the likelihood for linkage (LOD) and 1 LOD support intervals.
RESULTS
RI strains: saline response
The saline-induced locomotor response among 25 BXD RI strains is summarized in Table 1 for the four 5 min intervals and the collapsed 5–20 min interval. Data are also provided for the B6 and D2 inbred strains and the B6D2 F1 hybrids (but not the reciprocal D2B6 cross). The correlation matrix for the activity intervals is presented in the legend to Table 1. All of the intervals were highly correlated; the correlation coefficients ranged from 0.82 to 0.97. The ANOVA for both the 0–5 and 5–20 min intervals revealed highly significant (p < 0.00001) strain effects (F(24,295) = 10 and 8.4, respectively). The split-halves reliabilities (Blizard, 1992) for the RI strain means were 0.94 (0–5 min) and 0.93 (5–20 min). The narrow-sense heritability [DeFries et al., 1989;h2 = 0.5VA/(0.5VA +VW)] was 0.59 for the 0–5 min interval and 0.38 for the 5–20 min interval. The data in Table 1 illustrate that the average activity in the RI strains was similar to that seen in the D2 strain and markedly less than the activity of the B6 strain. Activity in the F1 cross was similar to that of the D2 strain.
RI strains: ethanol response (difference score)
The ethanol response was calculated as the difference in activity between day2 [ethanol (1.5 gm/kg) challenge] and day1 (saline challenge). The ethanol locomotor responses among the 25 BXD RI strains, the B6 and D2 inbred strains, and the B6D2 F1 hybrids are summarized in Table2 and presented graphically in Figure1 for the 0–5 min interval and the collapsed 5–20 min interval. The correlation matrix for the activity intervals is presented in the legend to Table 2. All of the intervals were highly correlated; the correlation coefficients ranged from 0.61 to 0.95. However, the correlations among the 5–10, 10–15, and 15–20 min intervals were stronger and in two cases significantly (p < 0.01) so when compared with the correlations between the 0–5 min interval and the subsequent intervals. The ANOVA for both the 0–5 and 5–20 min intervals revealed highly significant strain effects: 0–5 min (F(23,288) = 2.4; p < 0.0003) and 5–20 min (F(24,288) = 9.0;p < 0.00001). Post hoc analysis revealed numerous significant differences among the strain means, some of which are illustrated in Figure 1.
Fig. 1.
Strain means for the activity response to ethanol (1.5 gm/kg) among the BXD RI series (n = 25 strains). On the day after a saline challenge, animals were administered 1.5 gm/kg ethanol intraperitoneally, and activity was monitored for the next 20 min. Data in this figure are expressed as the difference score (day2 − day1) and reported for the 0–5 and 5–20 min intervals after ethanol administration. In addition to data for the RI strains, data are also reported for the B6 and D2 inbred strains and the B6D2 F1hybrids. Black bars illustrate the range of significant (p < 0.01) steps (of difference) for the RI strains as determined by the Student–Neuman–Keuls test. Thus for the 5–20 min data, strains 24 and 8 are significantly different, but strains 14 and 27 are not. All data are reported as the mean ± SE for the distance traveled.
The split-halves reliabilities for the RI strain means were 0.65 (0–5 min) and 0.88 (5–20 min). The narrow-sense heritabilities were 0.19 (0–5 min) and 0.32 (5–20 min). The data in Table 2 and Figure 1illustrate that the average ethanol response in the RI strains for the 0–5 min interval was intermediate to that seen in the D2 and B6 strains; in contrast, the response in the F1 cross was identical to that found in the D2 strain (marked activation). A different pattern of ethanol responses was observed for the 5–20 min interval. Over this time frame, ethanol responses in the RI strains (average response), in the B6 strain and in the F1 cross were identical and markedly less than the response found in the D2 strain. The 5–20 min interval was characterized by several RI strains that showed a marked decrease in activity after the ethanol challenge.
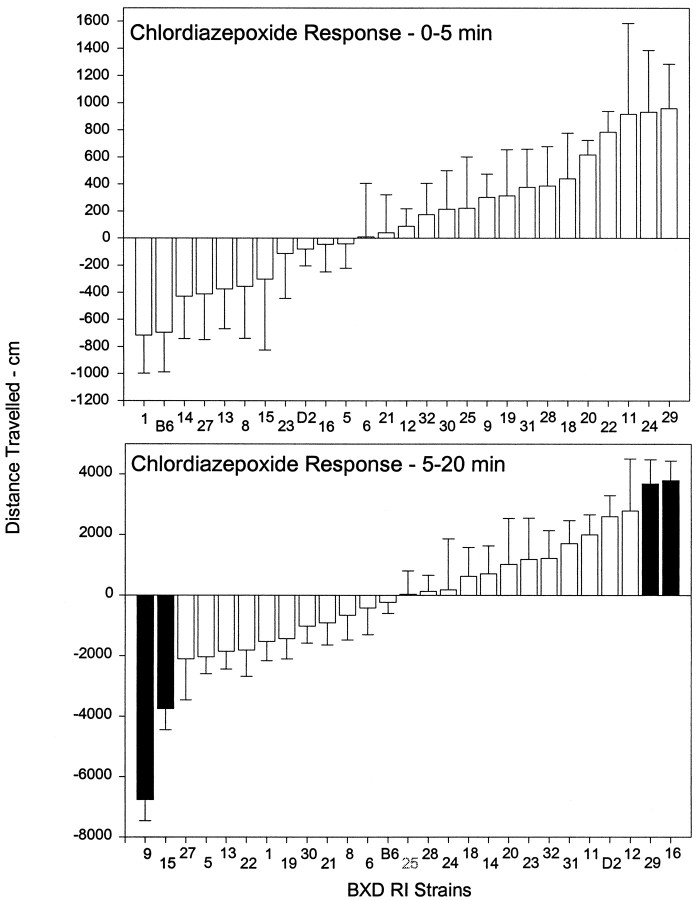
RI strains: chlordiazepoxide response
In a series of preliminary studies, B6 and D2 mice (n = 6–8 per strain per dose) were challenged with saline (day1) and 3, 10, or 30 mg/kg of chlordiazepoxide (CDP) (day2) and the difference scores calculated. The 10 mg/kg dose produced the maximum activation in the D2 strain (Table 3), which was observed over the 5–20 min interval. In contrast, no significant effect was observed in the B6 strain. At 30 mg/kg, CDP significantly decreased activity in both the B6 and D2 strains (data not shown). Thus, the 10 mg/kg CDP dose was used to challenge the RI strains.
The CDP locomotor responses among the 25 BXD RI strains and the B6 and D2 inbred strains are summarized in Table 3 for the four 5 min blocks and presented graphically in Figure 2 for the 0–5 min interval and the collapsed 5–20 min interval. The correlation matrix for the activity blocks is presented in the legend to Table 3. There were no significant correlations between the 0–5 min interval and the subsequent intervals. However, the 5–10, 10–15, and 15–20 min intervals were all highly correlated (range of rvalues, 0.79–0.91). The ANOVA for the 0–5 min interval did not meet the study criterion (p < 0.01) for a significant strain effect (F(24,283) = 1.7;p < 0.028) and was not considered for further analysis. In contrast, the ANOVA for the 5–20 min interval was highly significant (F(24,283) = 3.8; p< 0.0001); the post hoc analysis detected strain mean differences significant at p < 0.01 or better (see Fig. 2). The split-halves reliability for the RI strain means (5–20 min) was 0.91, and the narrow-sense heritability was 0.14. The data in Table 3 and Figure 2 illustrate that the average CDP response in the RI strains for 5–20 min interval was similar to that seen in the B6 strain. Overall, the RI strain means showed a trend toward expansion in the direction of an inhibitory effect on activity.
Fig. 2.
Strain means for the activity response to chlordiazepoxide (10 mg/kg) among the BXD RI series (n = 25 strains). Seven to 14 d after the ethanol challenge, animals were challenged with saline (day1) and chlordiazepoxide (day2). The data presented are for the difference score (day2 − day1). All other details are the same as in the legend to Figure 1.
RI strains: correlations among phenotypes
For the 5–20 min data, baseline (saline response) locomotor activity was not significantly (p > 0.2) correlated with either the ethanol or the CDP response. However, it was observed that there was a significant correlation between the ethanol and CDP response for this interval (r = 0.60;p < 0.01).
Phenotypic characterization of the F2 sample
A total of 900 male B6D2 F2 mice were phenotyped for their response to saline (day1) and ethanol (1.5 gm/kg; day2). As with the RI strains, the ethanol response was defined as the difference in activity between day2 and day1. The entire procedure was repeated one week later (trial2). The results obtained are summarized in Table 4. For the saline response it is useful to compare the results in Table 4 with those in Table 1. The data illustrate that the average activity in the RI strains and the F2 sample were quite similar: the saline response phenotype is similar to that seen in the D2 strain. As expected (see Markel et al., 1995), the average activity obtained from the test–retest design significantly reduced the sample variance without affecting activity. For both the 0–5 and 5–20 min intervals, the F test was 1.3 (p < 0.01). The test–retest reliability from trial1 to trial2was 0.54 (p < 0.0001) for both the 0–5 and 5–20 min intervals. The correlation matrix for each of the 5 min intervals (average data) is presented in Table 4; all of the intervals were highly correlated (range of r values, 0.63–0.84). Animals were rank-ordered on the basis of the test–retest reliability. The effect of censoring the data set of the bottom 10% (those individuals showing the poorest reliability) is also illustrated in Table 4. Censoring significantly improved the test–retest reliability of the remaining sample for both the 0–5 and 5–20 min intervals from 0.54 to 0.71 (p < 0.01) without affecting the sample means or variances.
Table 4.
Saline and ethanol (difference score) locomotor responses in male B6D2 F2 intercross animals (N = 900)
| Treatment | Trial/analysis | 0–5 min | 5–10 min | 10–15 min | 15–20 min | 5–20 min |
|---|---|---|---|---|---|---|
| Saline | 1 | 2052 ± 930 | 1772 ± 810 | 1645 ± 750 | 1602 ± 827 | 5017 ± 2141 |
| 2 | 2095 ± 1013 | 1855 ± 931 | 1718 ± 876 | 1655 ± 910 | 5212 ± 2487 | |
| Average4-a | 2068 ± 854 | 1809 ± 749 | 1679 ± 695 | 1626 ± 738 | 5115 ± 2033 | |
| Average-C | 2009 ± 823 | 4985 ± 1968 | ||||
| F test | 1.3 | 1.36 | 1.38 | 1.39 | 1.3 | |
| Test–retest | 0.54 | 0.47 | 0.45 | 0.44 | 0.54 | |
| Test–retest-C | 0.71 | 0.71 | ||||
| Ethanol | 1 | 970 ± 1460 | 410 ± 1610 | 48 ± 1430 | −4 ± 1340 | 450 ± 4050 |
| 2 | 1180 ± 1490 | 910 ± 1580 | 480 ± 1430 | 370 ± 1330 | 1760 ± 3990 | |
| Average4-b | 1080 ± 1120 | 660 ± 1240 | 270 ± 1140 | 180 ± 1030 | 1110 ± 3170 | |
| Average-C | 1090 ± 1100 | 1090 ± 3160 | ||||
| F test | 1.73 | 1.65 | 1.57 | 1.68 | 1.6 | |
| Test/retest | 0.15 | 0.21 | 0.22 | 0.22 | 0.25* | |
| Test/retest-C | 0.38 | 0.51* |
The saline and ethanol responses were determined as described in the legends to Tables 1 and 2. The saline–ethanol challenge was repeated in all animals 7–10 d after the first trial. For each interval data are also presented for the average of trial1and trial2. The F test compared the average of the variances from trials 1 and 2 to the variance associated with the average activity. Test–retest data were calculated as the correlation between trial1 and trial2. The absolute value of the difference between trial1 and trial2 was calculated and used to rank order the animals. The 10% of the animals showing the poorest test–retest reliability were censored from the data set. The censored (C) values for the average response and the test–retest reliability are also presented (0–5 and 5–20 min intervals, only). All data are presented as the mean ± SD.
*Significantly different from the 0–5 min value; p < 0.01.
Correlation matrix for the average saline response: Time (min) 0–5 5–10 10–15 15–20 0–5 1.00 0.78 0.71 0.63 5–10 1.00 0.84 0.75 10–15 1.00 0.81 15–20 1.00.
Correlation matrix for average ethanol response: Time (min) 0–5 5–10 10–15 15–20 0–5 1.00 0.63 0.50 0.41 5–20 1.00 0.840.73 10–15 1.00 0.81 15–20 1.00.
The values in bold are significantly different (p < 0.01) from the relative value in the 0–5 min column.
The ethanol response data for the F2 sample are also found in Table 4. The average response in the F2 sample was similar to the average response in the RI strains (Table 2). A potential confound in using the test–retest design was the sensitization effect from trial1 to trial2; i.e., on average there was a greater activation response in trial2 compared with trial1. However, sensitization and locomotor activity appear to be phenotypically independent. No significant relationship was detected between the average ethanol effect and the difference in the ethanol effect from trial1 to trial2; for the 0–5 min interval, r was 0.017 and for the 5–20 min interval, r was 0.08. For all intervals, the test–retest design significantly reduced the sample variance (F > 1.6; p < 0.0001). The test–retest reliability of the uncensored data were poor and ranged from 0.15 for the 0–5 min interval to 0.25 for the 5–20 min interval; this difference was significant (p < 0.01). The correlation matrix for each of the 5 min intervals (average data) is presented in Table 4; all of the intervals were significantly correlated (range of r values, 0.41–0.84). However, the correlations among the 5–10, 10–15, and 15–20 min intervals were significantly (p < 0.01) stronger when compared with the correlations between the 0–5 min interval and the subsequent intervals. The effect of censoring the data set of the bottom 10% is also illustrated in Table 4. Censoring significantly improved the test–retest reliability of the remaining sample for the 0–5 min sample from 0.15 to 0.38 (p < 0.001) and for 5–20 min intervals from 0.25 to 0.51 (p < 0.01) without affecting the sample means or variances. For the censored data, the reliability of the 5–20 min interval remained significantly (p < 0.01) better than the reliability of the 0–5 min data.
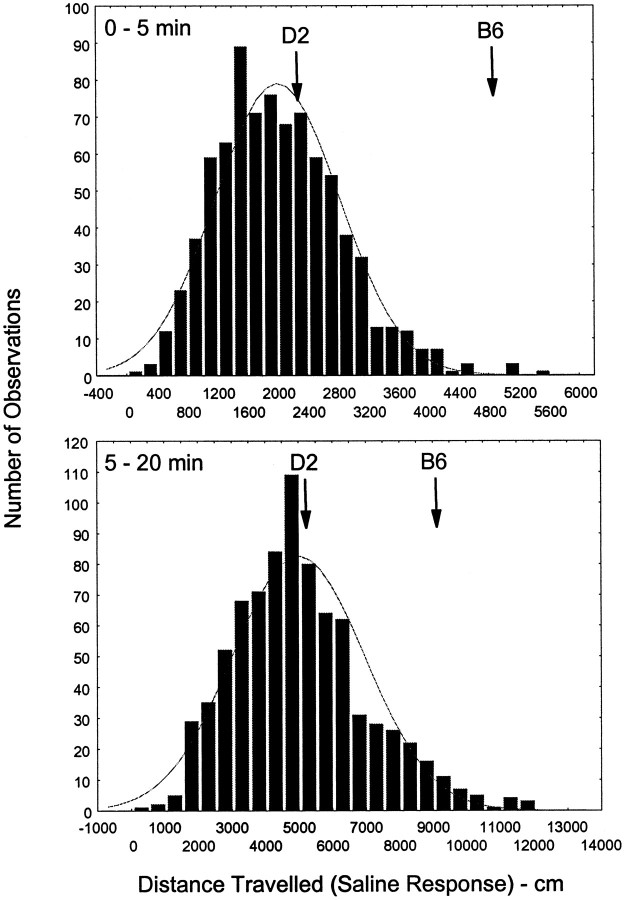
The distributions of the censored data sets for the 0–5 min and 5–20 min intervals are shown in Figures 3(saline response) and 4 (ethanol response). The data in Figure 3 illustrate that the saline responses were generally normally distributed, despite the floor effect (activity cannot be <0). For the parental strains, there was no significant change in activity from trial1 to trial2; the average (of trial1 and trial2) D2 and B6 data are shown in Figure 3. For the F2 sample, only a few animals showed a saline response that exceeded that seen in the B6 strain, but numerous animals were less active than the D2 strain. The data in Figure 4 illustrate that the F2 sample ethanol responses were also approximately normally distributed. For the parental D2 strain, there was no significant change in the ethanol response from trial1 to trial2; however, there was a significant sensitization in the B6 strain (activity increased from 1107 to 1940 cm/15 min).The average (of trial1 and trial2) D2 and B6 data are shown in Figure 4 for comparison with the F2 sample. The F2 extreme phenotypes were defined as those individuals > and <1 SD from the mean and were denoted RR and NN. For the 0–5 min interval, activity in the RR and NN extremes was 3100 ± 60 and −400 ± 60 cm, respectively. For the 5–20 min interval, activity in the RR and NN extremes was 7310 ± 190 and −2910 ± 110 cm, respectively. A comparison of these data for the 5–20 min interval with the mean ± SE values for the D2 and B6 strains (3850 ± 225 and 1940 ± 175 cm, respectively) illustrates the expansion in the F2 sample of the ethanol response phenotype beyond the parental boundaries. For both the 0–5 min interval (r= 0.09) and the 5–20 min interval (r = 0.01), there was no significant relationship between the average saline response and the average ethanol response.
Fig. 3.
Distribution of the average saline response in the B6D2 F2 intercross. Data are presented for the average from trials 1 and 2 (censored data set). In addition, the mean activity in the parental B6 and D2 strains (n = 18 per strain) are also illustrated. For both the 0–5 and 5–20 min intervals, the average intercross activity was similar to that seen in the D2 strain. Both distributions were not significantly different from normal.n = 810.
Fig. 4.
Distribution of the average ethanol response in the B6D2 F2 intercross. Difference score data are presented for the average from trials 1 and 2 (censored data set). In addition, the mean activities in the parental B6 and D2 strains (n = 18 per strain) are also illustrated. For both the 0–5 and 5–20 min intervals, a significant proportion of the animals showed an ethanol response below that seen in the B6 strain; for the 5–20 min interval, significant inhibition of activity was observed. Both distributions were not significantly different from normal. n = 810.
QTL analysis of the RI strain means
Table 5 lists the QTLs detected from analysis of the RI strain means (Tables 1-3) and found to be significant at p < 0.01 or better. For the saline response, a total of four QTLs were identified on chromosomes 1, 3, 5, 10, and 18. With the possible exception of the QTL on chromosome 1, these QTLs appear to be distinct from those detected for the ethanol or CDP responses. The QTL on chromosome 5 appears to be the same as a baseline activity QTL identified by Phillips et al. (1995, 1998) for the BXD RI series and also by K. Buck, T. Lisehka, J. Dorow, and J. C. Crabbe (unpublished observations).
Table 5.
QTL analysis of the BXD RI strain means for the saline-, ethanol-, and CDP-induced responses
| Chr | Marker | cM | Saline | Ethanol | CDP 5–20 min | ||
|---|---|---|---|---|---|---|---|
| 0–5 min | 5–20 min | 0–5 min | 5–20 min | ||||
| 1 | Ugta1 | 52 | −0.51 | ||||
| 1 | D1Mit87 | 62 | −0.55 | ||||
| 1 | DiRik88 | 80–90 | −0.66 | −0.56 | |||
| 2 | D2Mit17 | 53–72 | 0.69 | ||||
| 2 | Iapls2.4 | 86 | 0.53 | 0.55 | |||
| 2 | Pmv33 | 98 | 0.56 | ||||
| 3 | D3Byu3 | 12 | 0.53 | ||||
| 3 | D3Mit12 | 49 | −0.57 | ||||
| 4 | D4Mc2 | 35–40 | −0.67 | ||||
| 4 | Ly31 | 66–78 | 0.56 | 0.52 | |||
| 5 | D5Byu1 | 24–38 | −0.55 | ||||
| 6 | D6Rik59 | 36–48 | −0.59 | ||||
| 6 | Ly49c | 62 | −0.55 | ||||
| 9 | D9Lws1 | 48 | 0.58 | ||||
| 10 | D10Bir1 | 55 | −0.51 | ||||
| 13 | Tcrg | 10 | 0.52 | ||||
| 15 | D15Mit2 | 43–56 | −0.63 | ||||
| 18 | D18Mit111 | 2–11 | −0.53 | −0.57 | |||
The data from Tables 1-3 were subjected to a QTL analysis as described in Materials and Methods. Candidate QTLs reported are those that are significant at p < 0.01 or better. The markers reported are those that showed the most significant effect. The range of the QTL effect was estimated from correlations significant atp < 0.05.
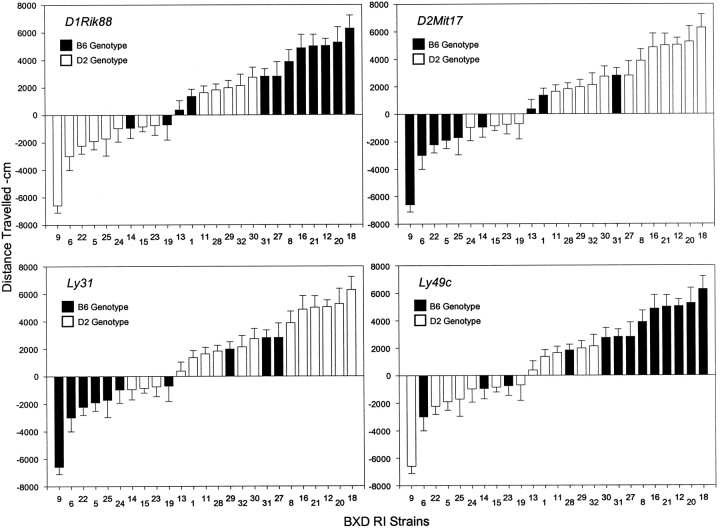
Significant QTLs for ethanol response were identified on chromosomes 1, 2, 4, and 6; the relationships between RI strain genotype and phenotype (5–20 min data) are illustrated in Figure5. These data demonstrate both the genetic homogeneity of the extreme phenotypes and for some QTLs the reversal of genotype based on the parental phenotype. Thus, on chromosomes 1 and 6, it is the B6 allele that contributes to increased locomotor activity and the D2 allele that contributes to decreased activity.
Fig. 5.
Relationship between phenotype and genotype for the activity response QTL (5–20 min) data. Data are presented for the four QTLs described in Table 5. For each marker the graph illustrates whether a particular RI strain carried the B6 or D2 genotype.
The QTLs on chromosomes 1 and 2 were detected for both the 0–5 min and the 5–20 min intervals. The data for chromosome 2 suggest that the QTL interval stretches from ∼50 to 100 cM. Four significant QTLs were detected for the CDP response (chromosomes 2, 4, 6, and 15). The QTLs on chromosomes 2 and 6 appear to be similar to those found for the ethanol response.
QTL confirmation and detection of additional QTLs
Approximately 100 each of the F2 ethanol response phenotypic extremes from the 0–5 and 5–20 min intervals were used to confirm the RI-generated ethanol response QTLs described in Table 5. Given the expansion of the genetic map in the RI strains and the associated difficulty in directly comparing map distances in the RI and F2 samples, summary data are provided for chromosomes 1, 2, 4, and 6 (Fig. 6). The QTLs on chromosomes 1, 4, and 6 were not confirmed in this analysis, although there was a trend (LOD = 1.6) for confirmation of the 0–5 min QTL on chromosome 1. On chromosome 2, the data illustrate that the QTLs for the 0–5 and 5–20 min data were probably identical, but the effect sizes for the 5–20 min data were greater. For the 0–5 min data, the peak LOD was 3.0, which is below the suggested criteria for acceptance (Lander and Kruglyak, 1995); however, combining the RI and F2 data would yield an LOD > 4. For the 5–20 min data, additional markers were added for fine mapping, and the data were analyzed used the MAPMAKER/QTL program. Because there was generally good agreement between the primary linkage map and the consensus map for chromosome 2 (Peters et al., 1998), the consensus map was used to place the markers in Figure 7. This figure illustrates that a peak LOD of 5.3 was obtained at ∼48 cM with either the free or additive QTL model. The 1 LOD support interval was 20 cM, and the peak LOD was associated with 6% of the phenotypic variance.
Fig. 6.

Summary of the screening results for ethanol response QTLs on chromosomes 1, 2, 4, and 6. The 0–5 and 5–20 min phenotypic extremes of the F2 intercross were genotyped for microsatellite markers on each of the four chromosomes identified from analysis of the RI strain means to contain ethanol response QTLs. The relative placement of each marker (based on the consensus map) is indicated. Each marker was analyzed independently (Kanes et al., 1996) and the χ2 value obtained was transformed to an LOD score (Lynch and Walsh, 1998).
Fig. 7.
LOD plot of chromosome 2 for the 5–20 min ethanol response data based as determined by an interval mapping approach (MAPMAKER/QTL). The markers used to generate the plot are found on thex-axis. Data were obtained by genotyping the NN and RR phenotypic extremes from the F2 intercross and ∼100 of the intermediate phenotypes. Curves are presented for the free, additive, dominant, and recessive models. The schematic chromosome indicates the location of some potential candidate genes.
The remainder of the genome in the F2 sample was screened for additional QTLs using the strategy outlined in Materials and Methods. The distribution of the markers used is given in Figure8. No QTLs with LOD scores > 3 were detected. However, it is important to note that the screening strategy used was biased to detecting additive genetic effects. To detect dominant genetic effects, which are common for quantitative traits (Lynch and Walsh, 1998), would require that a substantially greater proportion of the total F2 sample be used for screening.
Fig. 8.
Location of the microsatellite markers used in the genome-wide scan for ethanol response QTLs.
DISCUSSION
It is estimated that 40–60% of the risk for developing alcoholism is explained by genetic factors (Schuckit, 1994). However, neither the genes involved nor the mode of inheritance have been characterized. There are several reasons why progress has been slow. (1) Although the diagnosis of alcoholism is highly reliable, it is quite clear that one is dealing with a heterogeneous phenotype, which markedly reduces the power to detect specific gene effects. (2) Alcoholism is clearly a polygenic disorder and, thus, will require large samples to detect relatively small gene effects. For complex phenotypes under the control of multiple genes (or QTLs), gene effect sizes of 5–10% are generally considered large (Paterson et al., 1991). (3) Although recent reports show promise (Camp and Bansal, 1997;Gu and Rao, 1997; Hoeschele et al., 1997; Iyengar et al., 1997;Martin et al., 1997), there has been a lag in the execution of clinical QTL studies (in which one must deal with outbred populations and it is not possible to manipulate either the genome or the environment).
An alternative approach to finding clinically relevant QTLs is to first detect and map the QTLs using highly informative animal models and then look for the QTL in the homologous region of the human genome. Because the homologous regions of the mouse and human chromosomes are so well defined, it is possible to identify the chromosomal location of a gene in humans by mapping it in mice, often with far greater precision than human mapping studies (Copeland et al., 1993). Validation of this approach is seen in the recent report of Lembertas et al. (1997), who identified a putative QTL for human obesity on chromosome 20 based on homology with mouse chromosome 2, in which a significant obesity QTL had been mapped. In the application of the animal-to-human approach, alcoholism has a significant advantage over other areas of substance abuse. Studies on the genetics of alcohol-related behaviors have covered a wide variety of phenotypes and experimental strategies. Most investigations have focused on ethanol preference and choice, ethanol withdrawal, and/or acute ethanol sensitivity. Several recent studies have illustrated that QTL analysis can be successfully applied to such phenotypes, and importantly, some QTLs have been independently confirmed (Phillips et al., 1994; Rodriguez et al., 1995; Melo et al., 1996; Belknap et al., 1997). The work of the current study falls in the category of phenotypes related to acute ethanol sensitivity. QTL data are available for such diverse phenotypes as loss of the righting reflex (LORR), hypothermia, and locomotor activity (Crabbe et al., 1994; Cunningham, 1995; Phillips et al., 1995; Markel et al., 1997). These phenotypes may be relevant to alcoholism because one of the factors that appears to be inherited in alcoholic families is a difference in alcohol sensitivity (Schuckit et al., 1994); family history-positive subjects are generally less sensitive than family history-negative subjects to the effects of alcohol.
The data presented here confirm and extend the QTL analysis of theCrabbe et al. (1983) data set (Gora-Maslak et al., 1991). In this analysis, four provisional QTLs were detected on chromosomes 2, 4, 9, and 13; only the QTL on chromosome 4 was significant atp < 0.01. The analysis of the BXD RI data in Table 2confirmed the QTLs on chromosomes 2 and 4 and detected two additional QTLs on chromosomes 1 and 6. However, only the QTL on chromosome 2 was confirmed in the F2 analysis. Although the false-positive rate for the RI analysis was high (75%), it was not unexpected given the numerous comparisons being made (Belknap et al., 1996) and was similar to previous results from this laboratory (Kanes et al., 1996). None of the QTLs detected at p < 0.01 in the studies of Cunningham (1995) and Phillips et al. (1995) (chromosomes 3, 17 and 18) was confirmed in the present study. In this regard, it is important to note that these authors used a higher dose of ethanol (2 gm/kg) and different apparatus for measuring activity. Erwin et al. (1997b)examined the locomotor response to various doses of ethanol in the LS × SS RI series (24 strains). These authors found there was only one common QTL between those calculated for the ethanol activation slope and 2.0 gm/kg activity; the activation slope was calculated from the dose–response data obtained at 1, 1.25 and 1.5 gm/kg ethanol.Erwin et al. (1997b) concluded that for the LS × SS RI series, the 1.5 gm/kg dose of ethanol demarcates the end of the mainly stimulatory effects of ethanol and that the 2 gm/kg dose involves both the stimulatory and inhibitory influences of ethanol, making it a mixture of two different quantitative traits, which could be difficult to disentangle. This conclusion is supported in part by the observations of Dudek et al. (1991) that the stimulatory and inhibitory effects of ethanol are genetically independent. The exact dose conclusions of Erwin et al. (1997b) do not seem applicable to the BXD RI strains, because numerous strains show inhibition of activity over a time course very similar to that used to characterize the LS × SS RI series. However, it is reasonable to assume that for any particular dose of ethanol, one is observing (from a locomotor perspective) a mixture of quantitative traits, and this mixture is dose-dependent. Thus, the behavior seen at 2 gm/kg ethanol probably differs substantially from that found at lower doses. The argument that one is studying a mixture of quantitative traits has important implications for the QTL analysis. If there are separate QTLs for the activating and inhibitory effects of ethanol on locomotor activity, and assuming that one always has some mixture of these influences, then the power of both the RI and F2 analyses will be substantially reduced. Furthermore, in a genome-wide F2 analyses we could expect to find QTLs where the genetic segregation is asymmetrical; i.e., the responders but not the nonresponders (or vice versa) show a significant deviation from the expected 1:2:1 ratio of genotypes. However, the QTL on chromosome 2 was reasonably symmetrical; the genotypic ratios for the RR and NN individuals at D2Mit94were 2.1:1.4:0.5 and 0.8:1.7:1.5, respectively. These data strongly suggest that this marker is linked to a gene or genes in which the D2 alleles cause activation, whereas the B6 alleles cause inhibition.
The activity QTL on chromosome 2 is in the same general region previously identified to contain QTLs associated with ethanol preference and consumption (Melo et al., 1996; Phillips et al., 1999) and acute ethanol withdrawal (Buck et al., 1997) and appears to be significantly proximal from the loss of righting reflex QTL (Lorr2) identified by Markel et al. (1997). The observation that the ethanol and CDP responses in the RI strains are highly correlated and both map to the same region of chromosome 2, suggests the presence of a gene or genes that could provide a link between the ethanol response and ethanol effects at the GABA-benzodiazepine receptor arrays (see Deitrich et al., 1989). A potential GABA-related candidate gene that is also likely to affect ethanol preference and withdrawal is glutamic acid decarboxylase 1 (Gad1) (see Fig. 7). GAD activity among inbred mouse strains, including the B6 and D2 strains, has been reviewed (Hitzemann et al., 1995), and in general there is no substantial evidence for a functional polymorphism; although see Buck (1996). However, it is of interest to note that stress has been reported to increase GABA levels in the amygdala of the D2 but not B6 strain (Simler et al., 1982); recent evidence from this laboratory has suggested that the amydala and in particular the central nucleus are important in the regulation of the ethanol activity response (Hitzemann and Hitzemann, 1997; Demarest et al., 1998, 1999).
In conclusion, the data presented here are the first to confirm a QTL for the locomotor response to ethanol. This QTL joins a growing list of alcohol-related QTLs that have met or exceeded the generally accepted criteria for acceptance of LOD > 4.4 (Lander and Schork, 1994;Lander and Kruglyak, 1995). This list includes QTLs for alcohol preference (Melo et al., 1996; Phillips et al., 1998), acute ethanol withdrawal (Buck et al., 1997), and loss of the righting reflex (Markel et al., 1997). Although QTL detection for ethanol phenotypes has generally been quite successful, it is but the first step in the lengthy process of gene identification. The next step will be the fine mapping of the QTLs to intervals of ≤1 cM; for this step it will be necessary to use strategies such as advanced intercross lines or interval-specific congenic strains (Darvasi, 1998). The formation of congenic strains will also allow for a direct test of whether the QTLs, such as those identified on chromosome 2, are indeed pleitropic.
Footnotes
These studies were supported in part by US Public Health Service Grant AA 11043.
Correspondence should be addressed to Dr. Robert Hitzemann, Department of Psychiatry, State University of New York at Stony Brook, Stony Brook, NY 11794-8101.
REFERENCES
- 1.Barton NH, Turelli M. Evolutionary quantitative genetics: how little do we know? Annu Rev Genet. 1989;23:337–370. doi: 10.1146/annurev.ge.23.120189.002005. [DOI] [PubMed] [Google Scholar]
- 2.Belknap JK. Empirical estimates of Bonferroni corrections for use in chromosome mapping studies with the BXD recombinant inbred strains. Behav Genet. 1992;22:677–684. doi: 10.1007/BF01066638. [DOI] [PubMed] [Google Scholar]
- 3.Belknap JK, Mitchell SR, O’Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombination inbred mouse strains. Behav Genet. 1996;26:149–160. doi: 10.1007/BF02359892. [DOI] [PubMed] [Google Scholar]
- 4.Belknap JK, Richards SP, O’Toole LA, Helms ML, Philips TJ. Short-term selective breeding as a tool for QTL mapping: ethanol preference drinking in mice. Behav Genet. 1997;27:27–55. doi: 10.1023/a:1025615409383. [DOI] [PubMed] [Google Scholar]
- 5.Blizard DA. Recombinant-inbred strains: general methodological considerations relevant to the study of complex characters. Behav Genet. 1992;22:621–633. doi: 10.1007/BF01066634. [DOI] [PubMed] [Google Scholar]
- 6.Buck KJ. Molecular genetic analysis of the role of GABAergic systems in the behavioral and cellular actions of alcohol. Behav Genet. 1996;26:313–326. doi: 10.1007/BF02359387. [DOI] [PubMed] [Google Scholar]
- 7.Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. J Neurosci. 1997;17:3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camp NJ, Bansal A. The effect of selective sampling on mapping quantitative trait loci. Gen Epidemiol. 1997;14:767–772. doi: 10.1002/(SICI)1098-2272(1997)14:6<767::AID-GEPI34>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 9.Copeland NG, Jenkins NA, Gilbert DJ, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Weaver A, Lincoln SE, Steen RG, Stein LD, Nadeau JH, Lander ES. A genetic linkage map of the mouse: current applications and future prospects. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- 10.Crabbe JC. Genetic differences in locomotor activation in mice. Pharmacol Biochem Behav. 1986;25:289–292. doi: 10.1016/0091-3057(86)90267-4. [DOI] [PubMed] [Google Scholar]
- 11.Crabbe JC, Johnson NA, Gray DK, Kosobud A, Young ER. Biphasic effects of ethanol on open field activity: sensitivity and tolerance to C57BL/6N and DBA/2N mice. J Comp Physiol Psychol. 1982;96:440–451. doi: 10.1037/h0077898. [DOI] [PubMed] [Google Scholar]
- 12.Crabbe JC, Kosobud A, Young ER, Janowsky JS. Polygenic and single-gene determination for responses to ethanol in BXD/Ty recombinant inbred mouse strains. Neurobehav Toxicol Teratol. 1983;5:181–187. [PubMed] [Google Scholar]
- 13.Crabbe JC, Young ER, Deutsch CM, Tam BR, Kosobud A. Mice genetically selected for differences in open-field activity after ethanol. Pharmacol Biochem Behav. 1987;27:577–581. doi: 10.1016/0091-3057(87)90371-6. [DOI] [PubMed] [Google Scholar]
- 14.Crabbe JC, Belknap JK, Buck KJ. Genetic animal models of alcohol and drug abuse. Science. 1994;264:1715–1723. doi: 10.1126/science.8209252. [DOI] [PubMed] [Google Scholar]
- 15.Cunningham CL. Localization of genes influencing ethanol-induced conditioned place preference and locomotor activity in BXD recombinant inbred mice. Psychopharmacology. 1995;120:28–41. doi: 10.1007/BF02246142. [DOI] [PubMed] [Google Scholar]
- 16.Darvasi A. Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet. 1998;18:19–24. doi: 10.1038/ng0198-19. [DOI] [PubMed] [Google Scholar]
- 17.Darvasi A, Weinreb A, Minke V, Weller JI, Soller M. Detecting marker-QTL linkage and estimating QTL gene effect and map location using a saturated genetic map. Genetics. 1993;134:943–951. doi: 10.1093/genetics/134.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeFries JC, Wilson JR, Erwin VG, Petersen DR. LS × SS recombinant inbred strains of mice: initial characterization. Alcohol Clin Exp Res. 1989;13:196–200. doi: 10.1111/j.1530-0277.1989.tb00310.x. [DOI] [PubMed] [Google Scholar]
- 19.Deitrich RA, Dunwiddie TV, Harris RA, Erwin VG. Mechanism of action of ethanol: initial central nervous system actions. Pharmacol Rev. 1989;41:489–537. [PubMed] [Google Scholar]
- 20.Demarest K, Hitzemann B, McCaughran J, Mahjubi E, Hitzemann R. Ethanol, the Fos response and the central nucleus of the amygdala. Soc Neurosci Abstr. 1997;23:551.11. [Google Scholar]
- 21.Demarest K, Hitzemann B, Hitzemann R. Further evidence that the central nucleus of the amygdala is associated with ethanol-induced activation. Alcohol Clin Exp Res. 1998;22:1531–1537. [PubMed] [Google Scholar]
- 22.Demarest K, Hitzemann B, Phillips T, Hitzemann R (1999b) Ethanol-induced expression of c-Fos differentiates the FAST and SLOW selected lines of mice. Alcohol Clin Exp Res, in press. [PubMed]
- 23.Dietrich W, Katz H, Lincoln SE, Shin H, Friedman J, Dracopoli N, Lander ES. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudek BC, Phillips TJ. Distinctions among sedative, disinhibitory, and ataxic properties of ethanol in inbred and selectively bred mice. Psychopharmacology. 1990;101:93–99. doi: 10.1007/BF02253724. [DOI] [PubMed] [Google Scholar]
- 25.Dudek BC, Tritto T. Biometrical genetic analysis of ethanol’s psychomotor stimulant effect. Alcohol Clin Exp Res. 1994;18:956–963. doi: 10.1111/j.1530-0277.1994.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 26.Dudek BC, Phillips TJ, Hahn ME. Genetic analyses of the biphasic nature of the alcohol dose-response curve. Alcohol Clin Exp Res. 1991;15:262–269. doi: 10.1111/j.1530-0277.1991.tb01867.x. [DOI] [PubMed] [Google Scholar]
- 27.Erwin VG, Markel PD, Johnson TE, Gehle VM, Jones BC. Common quantitative trait loci for alcohol-related behaviors and central nervous system neurotensin measures—hypnotic and hypothermic effects. J Pharmacol Exp Ther. 1997a;280:911–918. [PubMed] [Google Scholar]
- 28.Erwin VG, Radcliffe RA, Gehle VM, Jones BC. Common quantitative trait loci for alcohol-related behaviors and central nervous system neurotensin measures: locomotor activation. J Pharmacol Exp Ther. 1997b;280:919–926. [PubMed] [Google Scholar]
- 29.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinb. 1918;52:399–433. [Google Scholar]
- 30.Gora-Maslak G, McClearn GE, Crabbe JC, Phillips TJ, Belknap JK, Plomin R. Use of recombinant inbred strains to identify quantitative trait loci in psychopharmacology. Psychopharmacology. 1991;104:413–424. doi: 10.1007/BF02245643. [DOI] [PubMed] [Google Scholar]
- 31.Gu C, Rao DC. A linkage strategy for detection of human quantitative-trait loci. I. Generalized relative risk ratios and power of sib pairs with extreme trait values. Am J Hum Genet. 1997;61:200–210. doi: 10.1086/513908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hitzemann B, Hitzemann R. Genetics, ethanol and the Fos response: a comparison of the C57BL/6J and DBA/2J inbred mouse strains. Alcohol Clin Exp Res. 1997;21:1497–1507. [PubMed] [Google Scholar]
- 33.Hitzemann R, Hitzemann B, Kanes S, Dains K. Genetics and organization of the basal ganglia. Int Rev Neurobiol. 1995;38:43–94. doi: 10.1016/s0074-7742(08)60524-3. [DOI] [PubMed] [Google Scholar]
- 34.Hoeschele I, Uimari P, Grignola FE, Zhang O, Gage KM. Advances in statistical methods to map quantitative trait loci in outbred populations. Genetics. 1997;147:1445–1457. doi: 10.1093/genetics/147.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyengar S, Calafell F, Kidd KK. Detection of major genes underlying several quantitative traits associated with a common disease using different ascertainment schemes. Genet Epidemiol. 1997;14:809–814. doi: 10.1002/(SICI)1098-2272(1997)14:6<809::AID-GEPI41>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 36.Kanes S, Hitzemann B, Hitzemann R. Mapping the genes for haloperidol-induced catalepsy. J Pharmacol Exp Ther. 1996;277:1016–1025. [PubMed] [Google Scholar]
- 37.Lander ES, Botstein E. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 39.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 40.Lembertas AV, Perusse L, Chagnon YC, Fisler JS, Warden GH, Purcell-Huynh DA, Dionne FT, Gagnon J, Nadeau A, Lusis AJ, Bouchard C. Identification of an obesity quantitative trait locus on mouse chromosome 2 and evidence of linkage of body fat and insulin on the human homologous region 20q. J Clin Invest. 1997;100:1240–1247. doi: 10.1172/JCI119637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch M, Walsh B. Genetics and analysis of quantitative traits. Sinauer; Sunderland, MA: 1998. [Google Scholar]
- 42.Markel PD, DeFries JC, Johnson TE. Ethanol-induced anaesthesia in inbred strains of long-sleep and short-sleep mice: a genetic analysis of repeated measures using censored data. Behav Genet. 1995;25:67–73. doi: 10.1007/BF02197243. [DOI] [PubMed] [Google Scholar]
- 43.Markel PD, Bennett B, Beeson M, Gordon L, Johnson TE. Confirmation of quantitative trait loci for ethanol sensitivity in long-sleep and short-sleep mice. Genome Res. 1997;7:92–99. doi: 10.1101/gr.7.2.92. [DOI] [PubMed] [Google Scholar]
- 44.Martin N, Boomsma D, Machin G. A twin-pronged attack on complex traits. Nat Genet. 1997;17:387–392. doi: 10.1038/ng1297-387. [DOI] [PubMed] [Google Scholar]
- 45.Melo JA, Shendure J, Pociask K, Silver LM. Identification of sex-specific quantitative trait loci controlling alcohol preference in C57BL/6 mice. Nat Genet. 1996;13:147–153. doi: 10.1038/ng0696-147. [DOI] [PubMed] [Google Scholar]
- 46.Paterson AH, Damon S, Hewitt JD, Zamir D, Rabinowitch HD, Lincoln SE, Lander ES, Tanksley SD. Mendelian factors underlying quantitative traits in tomato: comparison across species, generations, and environments. Genetics. 1991;127:181–197. doi: 10.1093/genetics/127.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters J, Siracusa LD, Pomp D, Zuberi AR, Church D, Koratkar R, Abbott C. Mouse chromosome 2. Mamm Genome. 1998;8:S27–S49. doi: 10.1007/s003359900647. [DOI] [PubMed] [Google Scholar]
- 48.Phillips TJ, Crabbe JC, Metten P, Belknap JK. Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res. 1994;18:931–941. doi: 10.1111/j.1530-0277.1994.tb00062.x. [DOI] [PubMed] [Google Scholar]
- 49.Phillips TJ, Huson M, Gwiazdon C, Burkhart-Kasch S, Shen EH. Effects of acute and repeated ethanol exposures on the locomotor activity of BXD recombinant inbred mice. Alcohol Clin Exp Res. 1995;19:1–10. doi: 10.1111/j.1530-0277.1995.tb01502.x. [DOI] [PubMed] [Google Scholar]
- 50.Phillips TJ, Huson MG, McKinnon CS. Localization of genes mediating acute and sensitized locomotor responses to cocaine in BXD/Ty recombinant inbred mice. J Neurosci. 1998;18:3023–3034. doi: 10.1523/JNEUROSCI.18-08-03023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips TJ, Belknap JK, Buck KJ, Cunningham CL (1999) Genes on mouse chromosomes 2 and 9 determine variation in ethanol consumption. Mamm Genome, in press. [DOI] [PubMed]
- 52.Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE. Alcohol acceptance, preference, and sensitivity in mice. II. Quantitative trait loci mapping analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res. 1995;19:367–373. doi: 10.1111/j.1530-0277.1995.tb01517.x. [DOI] [PubMed] [Google Scholar]
- 53.Schuckit MA. A clinical model of genetic influences in alcohol dependence. J Stud Alcohol. 1994;55:5–17. doi: 10.15288/jsa.1994.55.5. [DOI] [PubMed] [Google Scholar]
- 54.Simler S, Puglisi-allegra S, Mandel P. Gamma-aminobutyric acid in brain areas of isolated aggressive or non-aggressive inbred strains of mice. Pharmacol Biochem Behav. 1982;16:57–61. doi: 10.1016/0091-3057(82)90013-2. [DOI] [PubMed] [Google Scholar]
- 55.Soller M, Brody T, Genizi A. On the power of experimental designs for the direction of linkage between marker loci and quantitative loci in crosses between inbred lines. Theor Appl Genet. 1976;47:35–39. doi: 10.1007/BF00277402. [DOI] [PubMed] [Google Scholar]