Abstract
The Drosophila larva modulates its pattern of locomotion when exposed to light. Modulation of locomotion can be measured as a reduction in the distance traveled and by a sharp change of direction when the light is turned on. When the light is turned off this change of direction, albeit significantly smaller than when the light is turned on, is still significantly larger than in the absence of light transition. Mutations that disrupt adult phototransduction disrupt a subset of these responses. In larvae carrying these mutations the magnitude of change of direction when the light is turned on is reduced to levels indistinguishable from that recorded when the light is turned off, but it is still significantly higher than in the absence of any light transition. Similar results were obtained when these responses were measured in strains where the larval photoreceptor neurons were ablated by mutations in the glass(gl) gene or by the targeted expression of the cell death gene head involution defective(hid). A mutation in the homeobox gene sine oculis (so) that ablates the larval visual system, or the targeted expression of the reaper(rpr) cell death gene, abolishes all responses to light detected as a change of direction. We propose the existence of an extraocular light perception that does not use the same phototransduction cascade as the adult photoreceptors. Our results indicate that this novel visual function depends on the blue-absorbing rhodopsin Rh1 and is specified by the so gene.
Keywords: insect, larval photobehavior, locomotion, Drosophila, photoreceptor, Bolwig’s organ
The Drosophila melanogaster larva spends most of its life foraging, burrowed in the food substrate. Consistent with this general behavior pattern, theD. melanogaster larva is repelled by light (Lilly and Carlson, 1990; Gordesky-Gold et al., 1995; Sawin-McCormack et al., 1995). In the middle of the third instar the larva ceases foraging and leaves the food substrate in search of an adequate site in which to undergo metamorphosis. This behavior is referred to as wandering (Sokolowski et al., 1984). Modulation of larval photobehavior has been reported to occur during this transition from foraging to wandering (Sawin-McCormack et al., 1995). Interestingly, it coincides with the contact of the larval optic nerve by a serotonergic arborization (Mukhopadhyay and Campos, 1995), suggesting extrinsic modulation of this sensory pathway by 5-HT as demonstrated in other systems (Katz, 1995).
The larval visual system was first described in the house flyMusca domestica by Bolwig (1946) and henceforth was named the Bolwig’s organ. Similarly, in D. melanogaster, the larval visual system is composed of two bilateral groups of 12 photoreceptor cells located anteriorly and juxtaposed to the mouth hooks (Steller et al., 1987). The axons of the photoreceptor cells form the larval optic nerve that innervates the optic lobe primordium area of the brain lobes. The early development and the establishment of connectivity in this system has been described previously (Green et al., 1993; Campos et al., 1995).
We report that in the Drosophila larva a light stimulus modulates the direction of movement as well as quantitative aspects of locomotion such as path length and frequency of turning. Mutations that disrupt phototransduction in the adult eye disrupt aspects of the larval response to light measured in our assay. These results suggest that the larval and adult visual systems are similar from the functional point of view. These mutations, however, fail to abolish all perception of light, suggesting the existence of a light detection mechanism that does not require these gene products. The analysis of developmental mutants and of strains where the cell death genesreaper (rpr) and head involution defective (hid) are ectopically expressed suggests that this novel light detection mechanism is not located in the Bolwig’s organ.
MATERIALS AND METHODS
Fly stocks
Fly strains were grown at 25°C in 12 hr light/dark cycles on standard medium containing inactivated yeast, sucrose, and agar supplemented with fresh active yeast. Tegosept in ethanol and propionic acid were used to prevent mold growth. Strains used in addition to wild types Canton-S (CS) and Oregon-R(OR) are listed below:
glass. The glass(gl) gene encodes a zinc finger transcription factor required for the development of photoreceptor cells (Moses et al., 1989): gl60j is a severe allele that contains a 30 kb insertion (Moses et al., 1989);gl1 is a moderate allele; andgl+ contains a wild-type glgene in a gl60j background.
glass multimer reporter-head involution defective. This strain contains a fusion vector in which the cell death gene hid is expressed under the control of thegl promoter (Grether et al., 1995).
glass multimer reporter-reaper. This strain contains a fusion vector in which the cell death gene rpr is expressed under the control of the gl promoter (White et al., 1996).
neither inactivation nor afterpotential C. The neither inactivation nor afterpotential C (ninaC) gene encodes two isoforms (3.6 and 4.8 kb RNA) of adult photoreceptor specific cytoskeleton proteins consisting of a protein kinase and a myosin head domain (Montell and Rubin, 1988). ninaC5is a null mutant that has reduced levels of both the 3.6 and 4.8 kb RNA and leads to abnormal ERG, light and age-dependent retina degeneration (Porter and Montell, 1993; Hofstee et al., 1996) as well as a defect in response termination (Porter et al., 1992).ninaC2 is a mutant that has reduced levels of the 4.8 Kb RNA (Montell and Rubin, 1988).
neither inactivation nor afterpotential E. The neither inactivation nor afterpotential E (ninaE) gene encodes the opsin moiety of the Rh1 rhodopsin and is expressed in the adult photoreceptors R1–R6 (O’Tousa et al., 1985) as well as the larval visual system (Zuker et al., 1985; Pollock and Benzer, 1988).ninaE17 contains a 1.6 kb deletion. Flies have very low rhodopsin levels and respond poorly to light stimulus (O’Tousa et al., 1989). ninaE8 contains three missense mutations within the sixth transmembrane domain, T283m, W289R, C297S (Washburn and O’Tousa, 1989). Flies have <1% normal rhodopsin levels (O’Tousa et al., 1989).
no-receptor potential A. The no-receptor potential A (norpA) gene encodes a phospholipase C, which in null mutants, leads to a complete block of the phosphoinositide cascade mediating phototransduction (Hardie and Minke, 1995). Adult flies lack light elicited receptor potentials in the compound eyes and ocelli (Pak et al., 1970). norpAP24 contains a 28 base pair deletion in the norpA gene which produces a premature termination codon (Pearn et al., 1996).norpAP12 contains a nucleotide substitution in the norpA gene which produces a premature termination codon (Pearn et al., 1996).
sine oculis. The sine oculis (so) gene encodes a homeobox containing protein required for visual system determination (Fischbach and Technau, 1984).somda exhibits absence of larval photoreceptors and target area (Serikaku and O’Tousa, 1994).
Harvest of synchronized larvae
Adult flies aged from 1–7 d were allowed to lay eggs in a fresh food plate (100 mm × 15 mm; Fisher Scientific, Houston, TX) supplemented with vitamin A (Jamisons β carotene, 1.25 gm/l) and coated with yeast paste. After a minimum of two 2-hr precollections, a 1 hr egg collection was incubated at 25°C. At 20–22 hr after egg lay (AEL) all newly hatched first instar larvae were removed under a dissection microscope. After a 1 hr incubation period, ∼70 newly hatched first instar larvae were collected and transferred to a fresh food plate coated with yeast paste. Third instar larvae were tested for photobehavior between 84 and 90 hr AEL.
Photobehavior assays
Measurements of larval photobehavior were made in the ON/OFF assay. This consists of a plastic Petri dish (100 mm × 15 mm; Fisher Scientific) containing 15 ml of 1% agarose cooled to room temperature. Drosophila larvae prefer to remain in crevices. For this reason test plates need to be free of depressions (agar bubbles), and the test cannot be performed near the edge of the plate where the agar touches the side of the plate. Thus a circular 1 cm boundary from the plate edge was established beyond which the the collected data were discarded.
Manipulation of the larvae before the test was conducted using a darkroom light (20 W lamp with Kodak GBX-2 filter), and testing was conducted using a cool white bulb with a spectrum of 400–650 nm with peaks at 440 and 560 nm (20 W Cool White, Philips) and with a throughput of ∼320 microwatts/cm2. The darkroom light (20 W lamp with Kodak GBX-2 filter) used in this assay is the same used to record circadian-regulated locomotory behavior ofDrosophila in free-running conditions (“constant darkness”) (Sehgal et al., 1992). Larval photobehavior assays (Lilly and Carlson, 1990; M. Busto, J. Hassan, B. Iyengar, and A. R. Campos, unpublished observations) conducted using the darkroom light as the sole light source yielded response indices close to zero, confirming previous reports that Drosophila does not respond to light stimulus above the 650 nm range (Ashburner, 1989).
With use of a moist paintbrush, individual larva were removed from the culture dish. Each larva was carefully rinsed with distilled water to remove any excess food particles. They were removed from the distilled water, using a flathead paintbrush, and placed on a pre-test plate for a period of 1 min to allow them to acclimatize to the agar surface. Each larva was then positioned in the center of the test plate and allowed to move.
Individual plates were placed on a dark background and illuminated from above [20 W cool white bulb (Phillips) in a Rapid Start mechanism (Thomas Lighting)] in intermittent 10 sec pulses of light and dark. Throughout the duration of the assay the darkroom light (20 W lamp with Kodak GBX-2 filter) was on to allow recording of larval behavior. To estimate the influence of different light sources, controls using different kinds of light bulbs (incandescent, daylight, and cool white) were performed. Current oscillation causes the light sources to flicker at 60 Hz frequency. The amplitude of this oscillation varies according to the light source (i.e., fluorescent or incandescent). Response indices (RIs) obtained using various light sources were not significantly different (F(5,40) = 1.47,p > 0.05). In addition, RIs derived for wild-type larvae over the course of the 100 sec did not vary (F(4,87) = 1.77, p > 0.05).
Temperature
Surface temperature recordings were taken in 25 sec intervals for 200 sec during the course of the ON/OFF using a 21× Micrologger (Campbell Scientific). Temperature readings in the ON/OFF assay or under safelight conditions were 21.5 ± 0.5°C.
Data collection and analysis
Larval movement was visualized through a Fujinon TV·Z zoom lens (Fuji Optical) attached to a CCD TV camera (Elmo) and recorded on videotape (Fuji HQ-120, RCA VCR). Larvae were recorded either until they reached the 1 cm boundary or total test time (100 sec) had elapsed. Data derived for each of the strains were obtained from two to three sets of samples in which 10 larvae were tested.
Paths in the ON/OFF assay were first traced from a video monitor (8 inch × 10 inch Hitachi 1-chrome) onto acetate sheets and digitized using an Apple One Scanner at 72 dpi. Path length and the angle between path direction before and after the light switch were analyzed using public domain NIH Image software (developed at National Institutes of Health and available athttp://rsb.info.nih.gov/nih-image/) on a Macintosh Performa 5200CD computer. Response indices [(path length in dark-path length in light)/total path length per cycle] were calculated on a per larva basis, and a mean average of these individual indices was taken.
The data are depicted as means ± SEM. Transformation of the data were not necessary because variances did not differ significantly (Fmax test). ANOVAs and Tukey-Kramer multiple comparison tests (a = 0.05) were performed on the raw data using SAS-Jmp and Minitab software for MacIntosh (Sokal and Rohlf, 1995).
RESULTS
Light modulates the larval pattern of locomotion
The larval response to light has been measured in assays where opposing light conditions are presented to the animal at the same time (Lilly and Carlson, 1990; Gordesky-Gold et al., 1995; Sawin-McCormack et al., 1995; Busto, Hassan, Iyengar, and Campos, unpublished observations). In these assays locomotion is apparently a reflection of the larva’s attempt to remain in the dark environment (i.e., light avoidance when confronted with a dark/light boundary) combined with a direct effect of light on locomotion. Thus the behavior measured in these assays is in fact the composite of various responses.
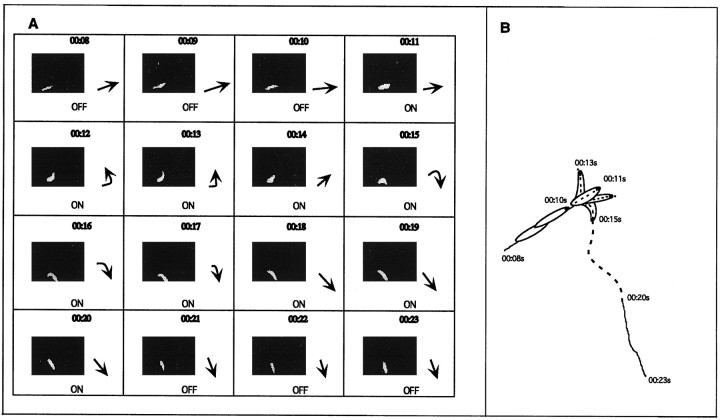
We hypothesized that single gene mutations can be used to dissect the network of cell types and molecules required for specific aspects of the Drosophila larva response to light. To test this hypothesis it was necessary to design a new assay that measures discrete aspects of the individual larva’s response to light. To that end an assay was designed (ON/OFF) in which the larva is subjected to intermittent pulses of light (10 sec each), and its locomotion is recorded. Visual inspection of the recorded larval behavior under the conditions of this assay suggest that the distance traveled in the presence of light is considerably shorter than in the absence of light. Likewise, head swinging and change of direction of the larval path are apparently triggered by light (Fig.1A,B).
Fig. 1.
Larval behavior during the ON/OFF assay.A, Videotape of a single CS larva tested in the ON/OFF assay was used to generate frame-by-frame photographs depicting 16 consecutive seconds. To the right of each panel is a schematic diagram of the larva representing the relative position of the head (arrowhead) and body (line). The first three frames (seconds00.08–00.10) show a larva immediately before a lightsOFF to ON transition. Lights are turned ON in the eleventh second, and head swinging is observed (00.12–00.14) followed by a change in direction (00.15–00.18). The final three framesshow a larva during lights OFF immediately after the lights ON to OFF transition (00.20–00.21). B, Line drawing of larval path shown in A. The solid lines represent the larval path during a portion of the dark pulse (00.08–00.10 and 00.21–00.23). The broken line represents larval path during the light pulse (00.11–00.20). The larval outlinedepicts the larval head swinging that occurs soon after the lights are turned on. During this time (00.12–00.15) the larva is stationary. This behavior is followed by a sharp change in the direction of the larval path.
These phenomena were quantified by analyzing the path tracings derived from the recordings using an image analysis software (NIH image). The effect of light on the distance traveled is represented by an RI derived from the resulting path length difference between light and dark (distance traveled in dark-distance traveled in light/total distance traveled in light and dark). An RI of ∼0.3 reflects a 50% reduction in path length when the light is turned on. To quantify head swinging behavior under the two light conditions, path tracings were drawn following the position of the mouth hooks such that head movements as well as the direction of the path were recorded (Fig.1B).
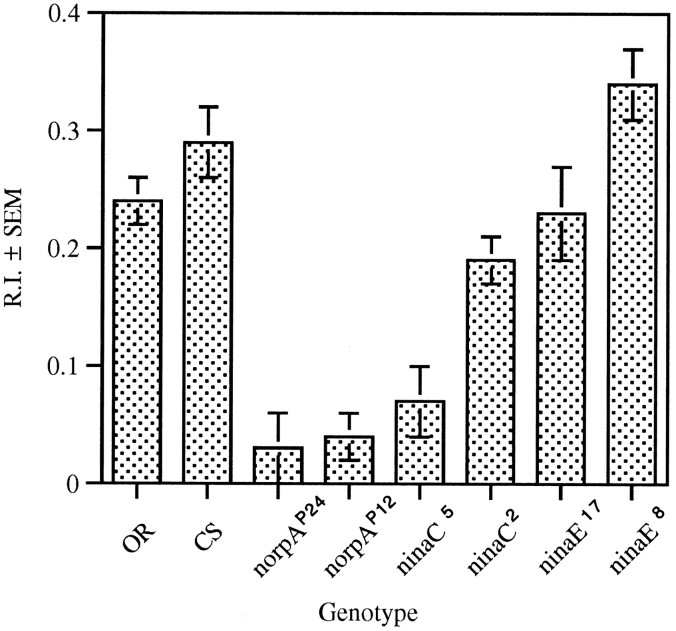
The wild-type strains tested reduce their path lengths when exposed to light as determined by the RI (Fig. 2). This response was abolished by mutations in genes that disrupt the phototransduction cascade (norpA and ninaC) but not by mutations in the blue-absorbing rhodopsin gene ninaE(Rh1) (Fig. 2). The two ninaC mutants tested (ninaC5 andninaC2) yielded opposite results. TheninaC5 mutants exhibited a severely reduced RI, whereas ninaC2 mutants behaved as wild type. Larvae homozygous for two mutant alleles of theninaE (ninaE17 andninaE8) gene displayed RIs in this assay that were indistinguishable from wild type (Fig. 2).
Fig. 2.
Response in the ON/OFF assay of wild type and larvae with mutations in genes involved in phototransduction. A response index (R.I.) was derived per larva, and a genotype average was calculated. The RIs for the strains are significantly different (ANOVA F(1,181) = 16.90, p < 0.001). Post hocanalysis of paired mean comparisons reveals no differences between the wild-type strains (OR, n = 30;CS, n = 30) and ninaE[ninaE17 (n = 20);ninaE8 (n = 20)], but a significant reduction in the larval response to light of thenorpA [norpAP24(n = 30); norpAP12(n = 20)] and ninaC[ninaC5 (n = 20);ninaC2 (n = 19)] mutants.
The ninaC gene encodes two retina-specific chimeric proteins consisting of a protein kinase and a myosin head domain (Montell and Rubin, 1988). One of these, a 132 kDa protein (p132), is expressed primarily in the cytoplasm. The other, a 174 kDa protein (p174), is localized predominantly in the rhabdomere (Hicks and Williams, 1992;Porter et al., 1992). Although ninaC5 has reduced levels of both p132 and p174,ninaC2 has reduced levels of only p174. Therefore, the wild-type response seen inninaC2 mutant larvae but notninaC5 larvae indicates that p132, not p174, is required for the larval response to light as measured by RI.
The norpA gene encodes a phospholipase C, an essential component of the phototransduction signaling cascade in the adult eye (Bloomquist et al., 1988; Ranganathan et al., 1995). ThenorpA gene is expressed as two developmentally regulated transcripts (subtypes I and II) generated by alternative splicing (Kim et al., 1995). Subtype I is specific to the adult eye, whereas subtype II is found in the CNS of adults and larvae (Kim et al., 1995). Therefore, disruption in the response to light in larvae carrying a null allele of the norpA gene may be caused by lack of this gene’s function in the CNS and/or larval visual system.
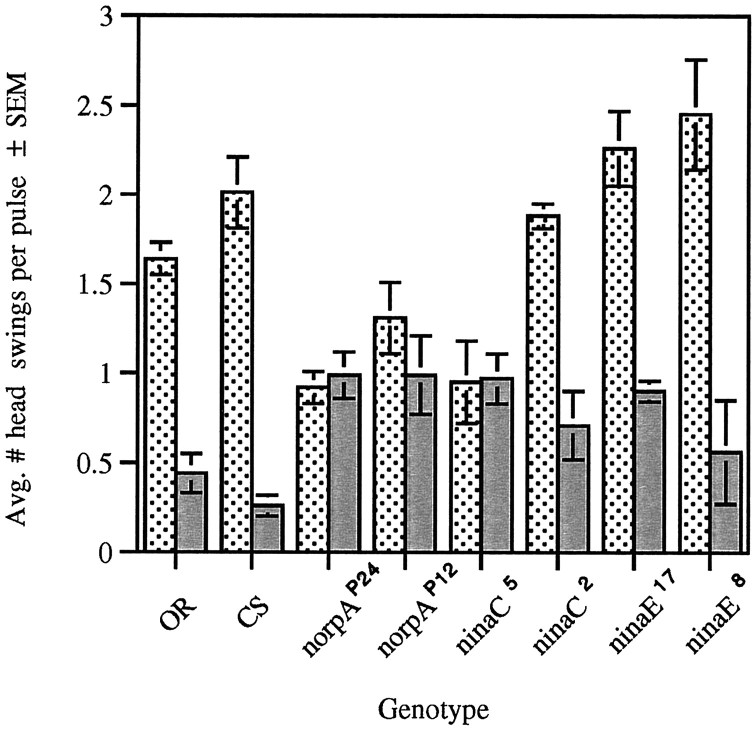
In addition to reduction in path length during the light pulse measured as an RI, wild-type larvae also exhibited a significant increase in head swinging when the light was turned on (Fig.3). This response was also abolished in the norpAP24,norpAP12, andninaC5 mutants but not in theninaC2 mutants (Fig. 3). These results suggest that p132 is required for this behavior also. Wild-type responses were also seen in larvae with severely reduced levels of Rh1 (ninaE17 andninaE8 mutants) (Fig. 3).
Fig. 3.
Head swinging behavior of wild-type strains and larvae with mutations in genes involved in phototransduction during the ON/OFF assay. Head swings, defined as an abrupt movement of the anterior portion of the larva away from original path choice, were counted in light (stippled bar) and dark (gray bar) pulses on a per larva basis, and an average for each genotype was derived. There is a significant increase in head swinging by wild-type larvae (CS,n = 30; OR, n = 30) during light pulses, relative to that during dark (ANOVA:CS, F(1,58) = 15.69,p < 0.001; OR,F(1,58) = 20.51, p < 0.001). This difference is abolished in the phototransduction mutants norpAP24 (n = 30), norpAP12 (n = 20), and ninaC5 (n = 20) but not in theninaC2 (n = 18),ninaE17 (n = 20), and ninaE8 (n = 20) mutants (ANOVA:norpAP24,F(1,58) = 0.09, NS;norpAP12,F(1,38) = 2.58, NS;ninaC5,F(1,38) = 0.05, NS;ninaC2,F(1,34) = 11.53, p < 0.001; ninaE17,F(1,38) = 30.82, p < 0.001; ninaE8,F(1,38) = 29.81, p < 0.001).
Taken together these results suggest that reduction in path length is attributable, at least in part, to immobilization of the larva while it swings its head in an apparent search for a dark environment. These responses are performed by a similar phototransduction cascade described for the adult visual system. Additionally, these results demonstrate that light-induced path length reduction and head swinging can be mediated by photoreceptors expressing rhodopsins other than Rh1.
Change of direction in larval path in different light conditions reveals a genetically distinct visual system function
Change of direction in the larval path was quantified by measuring the angle formed by the path tracing at the dark-to-light and light-to-dark boundaries. The magnitude of the angle formed by the two paths reflects the magnitude of the change in the direction of the larval path at the time of transition. Controls are represented by similar calculations performed at 10 sec intervals in path tracings derived from recordings performed in the absence of a light stimulus (Figs. 1B, 4).
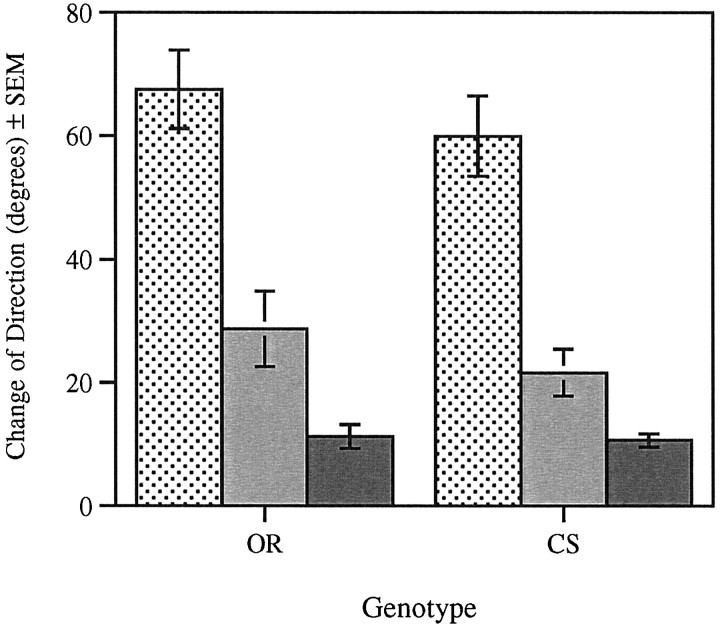
Fig. 4.
Change of direction in wild-type strains during the ON/OFF assay. Change of direction (in degrees) was measured at the dark to light (stippled bar), light to dark (gray bar), and in the absence of light transitions (solid bar). OR larvae display a significant difference between each of the light conditions (n = 30, F(1,87) = 33.89, p < 0.001). CS larvae display a significant difference between the dark to light and light to dark transitions only (n = 30,F(1,87) = 42.49, p < 0.001).
In wild-type strains (CS and OR), direction changes significantly more when the light is turned on [dark to light (D to L)] than when it is turned off (Fig. 4). Furthermore, comparison of paired means within genotypes demonstrates that in OR, change of direction when the lights are turned off [light to dark (L to D)] is significantly above that recorded in the absence of light transition (D to D). That is, D to L > L to D > D to D. In the wild-type strain CS, the change of direction when the lights are turned off (L to D) is considerably higher than that recorded in control conditions (absence of light transitions). Statistical analysis (comparison of paired means) indicates that this difference is not significant.
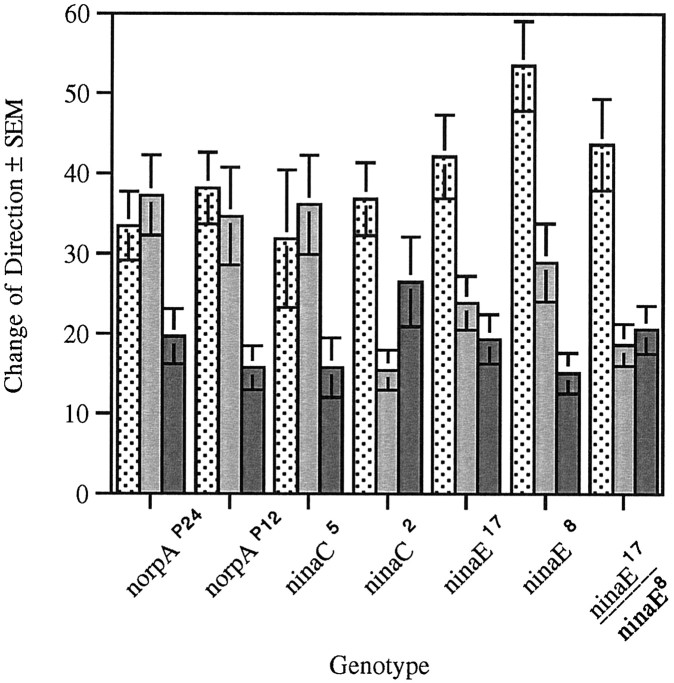
Similar to what is found for the other larval responses to light, two mutations in the norpA gene (norpAP24 andnorpAP12) and theninaC5 mutation abolish the light-induced difference in the amplitude of change of direction at the transitions D to L and L to D. Interestingly, these norpA andninaC mutations did not affect the difference between the change of direction found at L to D and that recorded during the absence of light pulses (D to D) (Fig.5). In contrast to the previous measured responses (RI and head swings), theninaC2 mutants did not respond like wild type (Fig. 5). Although light had a significant effect on direction change, the correlation seen in wild type was not exhibited by these larvae. Instead, the only statistically significant difference was that the change of direction at D to L was greater than that at the L to D transition.
Fig. 5.
Change of direction in strains with mutations in genes involved in adult phototransduction during the ON/OFF assay. Change of direction (in degrees) was measured at the dark to light (stippled bar), light to dark (gray bar), and in the absence of light transitions (solid bar). norpAP24 (n = 30, F(2,87) = 10.12, p < 0.001),norpAP12 (n = 20,F(2,57) = 6.21, p < 0.005), and ninaC5 (n= 20, F(2,57) = 5.17, p< 0.006) mutant larvae exhibit changes of direction at the dark to light and light to dark transitions that are not different from each other but are different from change of direction in the absence of light. ninaC2 mutant larvae (n = 20, F(2,57) = 5.64,p < 0.008) exhibit a significant difference at the dark to light and light to dark transition changes that in turn is not significantly different from that measured in the absence of light transition. ninaE17 mutant larvae also exhibit a significant difference between dark to light and light to dark, but the difference between the light to dark and absence of light transitions has been abolished (n = 20,F(2,57) = 8.93, p < 0.001). The same is true of larvae that are heterozygous [ninaE17/ninaE8(n = 20, F(2,57) = 12.2,p < 0.001)]. ninaE8(n = 20, F(2,57) = 12.21, p < 0.001) larvae display a significant difference between each of the transitions.
In contrast, the ninaE17 mutation reduces the change of direction at the L to D transition to levels indistinguishable from that recorded in the absence of light transition (D to D). These mutant larvae do exhibit a D to L change of direction that is greater than both the L to D change of direction and direction change in the absence of light transition. Although theninaE8 homozygous larvae behave as wild type at all transitions, in the heterozygous flies (ninaE8/ninaE17) the difference in change of direction at L to D and D to D transitions is abolished. Our interpretation of these results is that Rh1 expression in the ninaE8 strain is lower than wild type but still above the threshold for the performance of this particular behavior. This level of Rh1 expression, however, is not sufficient to overcome the deficit caused by theninaE17 mutation.
These results suggest the existence of a visual system function(s) that distinguishes between lights being turned on (D to L), lights being turned off (L to D), and no light transition (D to D). The distinction between lights being turned on and off requires the same phototransduction cascade as that described for RI and head swings; that is, it is abolished by mutations in the norpA andninaC genes. The results described above indicate that this light perception is not mediated by Rh1. However, Rh1-mediated phototransduction is required to distinguish presence from absence of light transitions.
Ablation of the Bolwig’s organ disrupts only a subset of the larval responses to light
In D. melanogaster, the larval visual system (Bolwig’s organ) is composed of two bilateral groups of 12 photoreceptor cells located anteriorly and juxtaposed to the mouth hooks, similar to what is found in larger flies (Steller et al., 1987). These photoreceptors project posteriorly and ventrally around the brain hemispheres and terminate in the optic lobe primordium (Schmucker et al., 1992, 1997;Green et al., 1993; Campos et al., 1995). To further dissect larval visual system requirements, so and gl, two genes directly involved in visual system specification and development, were studied.
The so gene encodes a homeodomain protein expressed in a number of places during embryogenesis (Cheyette et al., 1994; Serikaku and O’Tousa, 1994). In the visual system, so functions during embryogenesis in the regulation of genes necessary for proper optic lobe invagination and Bolwig’s organ formation (Serikaku and O’Tousa, 1994). Here, we used the somdaallele, the only allele that specifically disrupts the development of the larval visual system.
The gl gene, which encodes a transcription factor essential for photoreceptor development, is expressed in a more spatially restricted manner and acts downstream of so (Moses et al., 1989; Serikaku and O’Tousa, 1994). gl is expressed in the larval and adult photoreceptor neurons as well as in two groups of ∼21 neurons in each brain hemisphere (Moses et al., 1989). The effect of gl mutations in the development of thegl-expressing central neurons is not known. This is attributable to the absence of markers, besides gl gene expression itself, that allow the visualization of these neurons.
To determine whether the photoreceptors in Bolwig’s organ mediate the various responses to light measured in the ON/OFF assay, larvae carrying mutations in the so and gl gene were assayed. In addition, a gl mutant strain displaying wild-type adult phenotype, attributable to the expression of a wild-type gl gene present in a P element transposon, was tested (Moses et al., 1989). Two strains in which a cell death gene (hid or rpr) is under the control of thegl promoter were similarly analyzed (Grether et al., 1995;White et al., 1996).
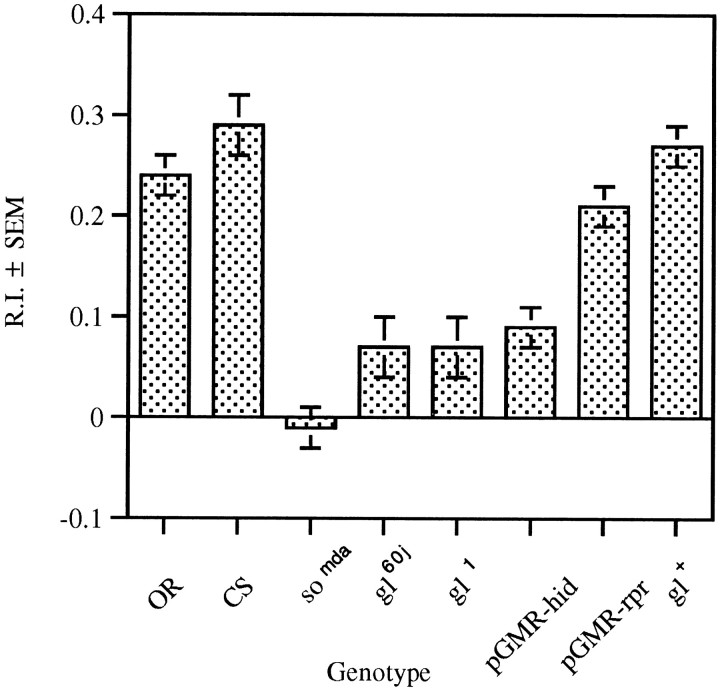
No significant difference between the RIs obtained for the wild-type strains and gl+ or glassmultimer reporter-reaper (pGMR-rpr) was detected (Fig. 6). A significant reduction in the RI was observed in the somda,gl60j, gl1, and glass multimer reporter-head involution defective-hid (pGMR-hid) mutant strains. Similar results were found when the frequency of head swinging was calculated during light and dark pulses (Fig. 7). The significant increase in head swinging frequency during the light pulse displayed by wild-type larvae is abolished by mutations in both the so and gl genes. This differential head swinging was restored by thegl+-containing transposon. Again, although the increase in head swinging during the light pulse was abolished in larvae carrying the pGMR-hid fusion, larvae containing the pGMR-rpr fusion were not affected in this manner.
Fig. 6.
RI in the ON/OFF assay of larvae with mutations in the so and gl genes. The RIs for the strains are significantly different (F(7,178) = 15.55, p < 0.001). Post hoc analysis of paired means reveals no difference between the wild-type strains (OR,n = 30; CS, n = 30) and the pGMR-rpr (n = 20) andgl+ (n = 16). A significant reduction is observed in the larval response to light of the somda (n = 20),gl60j (n = 20),gl1 (n = 20), andpGMR-hid (n = 30) mutants.
Fig. 7.
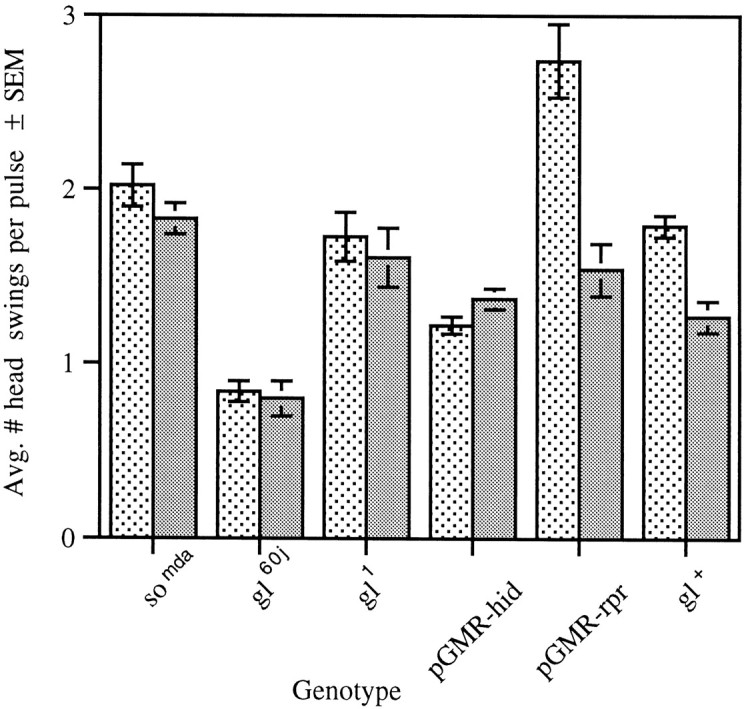
Head swinging behavior in the ON/OFF assay of larvae with mutations in the so and glgenes. The increase in head swinging behavior seen during the light pulses (stippled bar) over that seen during the dark pulses (gray bar) is abolished in thesomda mutant (n = 20,F(1,38) = 0.76, p > 0.05) as well as in the gl mutantsgl60j (n = 20,F(1,38) = 0.03, p > 0.05) and gl1 (n = 20, F(1,38) = 0.03, p > 0.05) and in the pGMR-hid strain (n= 30, F(1,58) = 0.03, p> 0.05), which lacks larval photoreceptor cells. A light pulse elicits differential head swinging behavior in pGMR-rpr(n = 20, F(1,38) = 15.33, p < 0.001), which exhibits a less severe adult phenotype than pGMR-hid, and ingl+ (n = 16,F(1,30) = 9.44, p < 0.005), which is the gl rescue line.
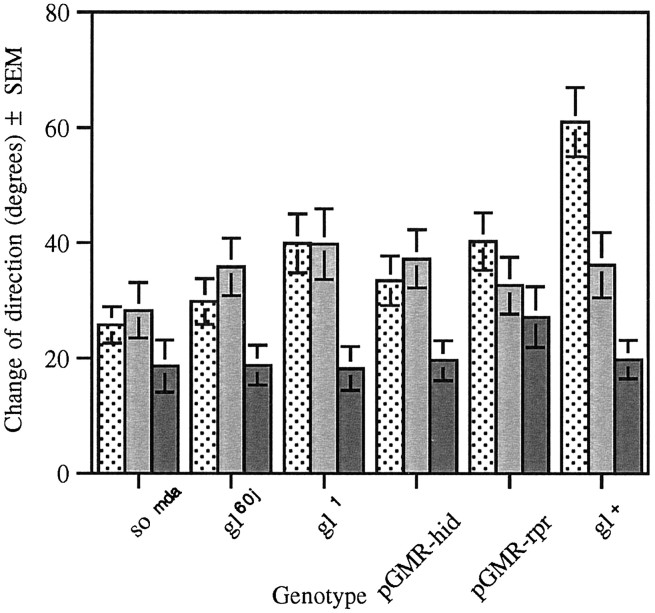
Disruption in the development of the larval visual system, caused bygl mutations or expression of the hid gene, abolished the difference in the magnitude in the change of direction at the D to L and L to D transitions (Fig.8). However, change of direction in the absence of a light transition is still significantly lower than either of the test conditions (Fig. 8). In contrast, thesomda mutation or the expression of the cell death gene rpr under the gl promoter (pGMR-rpr) abolished the difference in the magnitude of change of direction at all transitions (Fig. 8).
Fig. 8.
Change of direction in the ON/OFF assay of larvae with mutations in the so and gl genes. Change of direction (in degrees) was measured at the dark to light (stippled bar), light to dark (gray bar), and in the absence of light transitions (solid bar). Light has a significant effect on path direction in each of the strains tested, with the exception of pGMR-rpr(n = 20, F(1,57) = 0.98,p > 0.05) and somda(n = 20, F(1,57) = 1.79,p > 0.05), in which the presence or absence of light had no effect. The gl mutant strainsgl60j (n = 20,F(1,57) = 4.42, p < 0.02) and gl1 (n = 20, F(1,57) = 6.23, p < 0.005) and pGMR-hid (n = 30,F(1,87) = 4.57, p < 0.01) show no difference between degree of direction change at the light transitions. However, change of direction in the absence of light transitions is significantly lower than either of the test conditions. The gl+ strain displays a degree of direction change in the dark to light transition that is significantly higher than the other test conditions (n = 16,F(2,45) = 16.23, p < 0.001).
The apparent contradiction in the results obtained with pGMR-hid and pGMR-rpr strains can be attributed to different sensitivity of diverse cell types to the ectopic expression of these cell death genes. In fact, developing adult photoreceptors are more sensitive to the ectopic expression ofhid than rpr (H. Steller, personal communication). These observations lend further support to the proposal that these various responses to light are mediated by genetically distinct cell types.
These results demonstrate that the larval visual function, which is dependent on a phototransduction cascade similar to that described for the adult stage, requires at least the proper development of the larval photoreceptors. Although a mutation in the so gene abolishes all responses to light, as measured in this assay, mutations in thegl gene appear to disrupt only a subset of these responses. These larvae, at the L to D transition, exhibit changes of direction greater than at the D to D transition. Thus, these results demonstrate that a larval visual function exists that is not dependent on an adult-like phototransduction cascade. The cells that mediate this proposed visual function are not housed in gl-dependent neurons but in neurons dependent on the function of the homeobox-containing transcription factor so. Our results indicate that these neurons, although not dependent on glgene function, do express this transcription factor.
DISCUSSION
The Drosophila larval response to light represents a quantifiable behavior likely to include components of more complex behaviors executed by higher organisms. As a model system,Drosophila provides high-resolution genetic and molecular biology tools to dissect the components, molecular and cellular, required for the larval response to light (Miklos and Rubin, 1996).
The Drosophila larva response to light can be defined as klinokinesis and orthokinesis
The locomotory reaction of organisms to biotic or abiotic factors has been traditionally defined relative to the source of stimulus (Fraenkel and Gunn, 1961). In a directed reaction (taxis), the movement is modulated to position the long axis of the organism toward or away from the source of stimulation. In undirected locomotory reactions (kinesis), quantitative aspects of locomotion such as speed and frequency of turning are modulated by the stimulus. These definitions can be further refined when the stimulus is varied temporally and quantitatively. In klinotaxis, orientation is achieved by comparison of the stimulus intensity over time, whereas in tropotaxis the differential stimulation of paired receptors in space orients the animal relative to the stimulus source.
Kineses are similarly distinguished as klinokinesis, where the path shape (frequency of turning) is modulated by the differential intensity of stimulation over time, or orthokinesis, where quantitative aspects of locomotion (speed or frequency of locomotion) are affected by the intensity of the stimulation (Fraenkel and Gunn, 1961). Our results demonstrate that the Drosophila larva displays kinesis. In our assay, frequency of turning (change of direction) and frequency of locomotion (path length) are affected by alternating pulses of light and dark over time, suggesting that the Drosophila larva is able to compare light intensity over time. These observations suggest that the ON/OFF assay is assessing behaviors previously described as klinokinesis and orthokinesis (Fraenkel and Gunn, 1961).
The reduction in path length seen when the light turns on can be caused by different factors. The larva may stop more often as it searches for a preferred dark environment (head swinging). Alternatively, or additionally, the presence of light may change fundamental aspects of locomotion such as frequency of the peristaltic contractions that constitutes the larval stride or the amplitude of these contractions. The current assay does not have the level of resolution that distinguishes between these two alternatives.
Klinokinesis and orthokinesis in the ON/OFF assay are dependent on an adult-like phototransductions cascade
We demonstrate that in the wild-type strains tested, the change of direction of the larval path is significantly greater when the light is turned on than when it is turned off. This directionality in the temporal perception of the light stimulus is abolished by null mutations in the norpA and ninaC genes but not by mutations in the ninaE gene, suggesting that this light perception is performed by a phototransduction cascade similar to that described for the adult visual system but is not mediated by the blue-absorbing rhodopsin Rh1. Moreover, the absence of this response in strains that lack the Bolwig’s organ (gl andsomda mutants) further confirms this structure as the Drosophila larva’s main photosensory organ. Locomotion in the absence of a light stimulus was not significantly affected by these mutations (data not shown), demonstrating that intact phototransduction is not required for basic aspects of larval locomotion. These observations also demonstrate that these mutations do not have a pleiotropic effect on larval behavior.
The ON/OFF assay defines a novel extraocular light perception function
In the wild-type strains tested, change of direction when the light is turned off is greater than in the absence of light transitions, suggesting that turning off the light is a transition perceived by the animal. This observation supports the notion that a simple mechanism for the perception of light exists in the D. melanogaster larva that distinguishes changes in light conditions from absence of light transitions but is unable to distinguish whether the light is being turned on or off. This light response is mediated by the blue-absorbing rhodopsin (Rh1) because it is abolished in part by mutations in the ninaE gene. Interestingly, it does not rely on the same phototransduction pathway as that of the adult visual system as seen by the wild-type response of norpA andninaC mutant larvae.
Our results indicate that these hypothetical photoreceptors are not housed within the Bolwig’s organ, defined as the larval photoreceptors that depend on the gl gene function for differentiation. However, the observation that the function of this visual system is impaired in larvae where the cell death gene rpr is expressed under the control of the gl promoter demonstrates that these are cells in which the gl transcription factor is functional. Thus it is possible that this novel function is performed by a small number of cells that express Rh1 and the gl gene product but whose differentiation and Rh1 expression are not under the control of the gl gene.
Two different groups of cells are likely to be involved in this novel light perception. The observation that it is dependent on theso gene function but not gl suggests that these cells are included in the optic lobe primordium. The ablation of this proposed function by expression of the cell death gene rprunder the gl promoter suggests that the central brain neurons that express the gl gene are also involved in this behavior.
A precedent for a light detector that does not rely on known elements of the phototransduction machinery in adults is the photic input pathway required for the entrainment of the circadian rhythm (Wheeler et al., 1993). The novel visual system function proposed in this paper presents other parallels with cells involved in the control and generation of circadian rhythms. Mutations in the gl gene do not abolish circadian rhythms. However, the expression of theperiod (per) gene under the control of thegl promoter is sufficient to restore circadian rhythmicity in per mutant flies (Vosshall and Young, 1995). These results strongly suggest that the gl-expressing cells that are not the photoreceptors house the circadian pacemaker. It is possible that this novel visual function that distinguishes changes in light condition from absence of light transitions but is unable to distinguish whether light is being turned on or off is also involved in the control of pacemaker oscillation.
Footnotes
This work was supported by an operating grant to A.R.C by the Medical Research Council (MRC) of Canada. We are indebted to W. J. Bell, M. B. Sokolowski, and T. Tully for discussions on behavior, and to Roger Jacobs, Colin Nurse, and Andre Bedard for comments on this manuscript. We thank the generosity of the following fly workers: Joe O’Tousa, Randall Shortridge, and Kathy Matthews for the prompt donation of stocks.
M.B. and B.I. contributed equally to this work.
Correspondence should be addressed to Ana Regina Campos, Department of Biology, McMaster University, 1280 Main Street West, Hamilton, Ontario, Canada L8S 4K1.
REFERENCES
- 1.Ashburner M. Drosophila: A laboratory handbook, pp 139–298. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. Developmental biology. [Google Scholar]
- 2.Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- 3.Bolwig N. Sense and sense organs of the anterior end of the house fly larvae. Vidensk Medd Dan Naturhist Foren. 1946;109:81–217. [Google Scholar]
- 4.Campos AR, Lee KJ, Steller H. Establishment of neuronal connectivity during development of the Drosophila visual system. J Neurobiol. 1995;28:313–329. doi: 10.1002/neu.480280305. [DOI] [PubMed] [Google Scholar]
- 5.Cheyette BNR, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 6.Fischbach KF, Technau G. Cell degeneration in the developing optic lobes of the sine oculis and small-optic-lobes mutants of Drosophila melanogaster. Dev Biol. 1984;104:219–239. doi: 10.1016/0012-1606(84)90050-2. [DOI] [PubMed] [Google Scholar]
- 7.Fraenkel GS, Gunn DL. The orientation of animals. Dover; New York: 1961. [Google Scholar]
- 8.Gordesky-Gold B, Warrick JM, Bixler A, Beasley JE, Tompkins L. Hypomorphic mutations in the larval photokinesis A (lphA) gene have stage-specific effects on visual system function in Drosophila melanogaster. Genetics. 1995;139:1623–1629. doi: 10.1093/genetics/139.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green P, Hartenstein AY, Hartenstein V. The embryonic development of the Drosophila visual system. Cell Tissue Res. 1993;273:583–598. doi: 10.1007/BF00333712. [DOI] [PubMed] [Google Scholar]
- 10.Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 11.Hardie RC, Minke B. Phosphoinositide-mediated phototransduction in Drosophila photoreceptors: the role of Ca2+ and trp. Cell Calcium. 1995;18:256–274. doi: 10.1016/0143-4160(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 12.Hicks JL, Williams DS. Distribution of the myosin I-like ninaC proteins in the Drosophila retina and ultrastructural analysis of mutant phenotypes. J Cell Sci. 1992;101:247–254. doi: 10.1242/jcs.101.1.247. [DOI] [PubMed] [Google Scholar]
- 13.Hofstee CA, Henderson S, Hardie RC, Stavenga DG. Differential effects of ninaC proteins (p132 and p174) on light-activated currents and pupil mechanisms in Drosophila photoreceptors. Vis Neurosci. 1996;13:897–906. doi: 10.1017/s0952523800009147. [DOI] [PubMed] [Google Scholar]
- 14.Katz PS. Intrinsic and extrinsic neuromodulation of motor circuits. Curr Opin Neurobiol. 1995;5:799–808. doi: 10.1016/0959-4388(95)80109-x. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, McKay RR, Millern K, Shortridge RD. Multiple subtypes of phospholipase C are encoded by the norpA gene of Drosophila melanogaster. J Biol Chem. 1995;270:14376–14382. doi: 10.1074/jbc.270.24.14376. [DOI] [PubMed] [Google Scholar]
- 16.Lilly M, Carlson JR. smellblind: a gene required for Drosophila olfaction. Genetics. 1990;124:293–302. doi: 10.1093/genetics/124.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miklos GLG, Rubin GM. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 18.Montell C, Rubin GM. The Drosophila ninaC locus encodes two photoreceptor cell specific proteins with domains homologous to protein kinases and the myosin heavy chain head. Cell. 1988;52:757–772. doi: 10.1016/0092-8674(88)90413-8. [DOI] [PubMed] [Google Scholar]
- 19.Moses K, Ellis MC, Rubin GM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay M, Campos AR. The larval optic nerve is required for the development of an identified serotonergic arborization in Drosophila melanogaster. Dev Biol. 1995;169:629–643. doi: 10.1006/dbio.1995.1175. [DOI] [PubMed] [Google Scholar]
- 21.O’Tousa JE, Baehr W, Martin RL, Hirsh J, Pak WL, Applebury ML. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 22.O’Tousa JE, Leonard DS, Pak WL. Morphological defects in oraJK84 photoreceptors caused by mutation in R1–6 opsin gene of Drosophila. J Neurogenet. 1989;6:41–52. doi: 10.3109/01677068909107099. [DOI] [PubMed] [Google Scholar]
- 23.Pak WL, Grossfield J, Arnold KS. Mutants of the visual pathway of Drosophila melanogaster. Nature. 1970;227:518–520. doi: 10.1038/227518b0. [DOI] [PubMed] [Google Scholar]
- 24.Pearn MT, Randall LL, Shortridge RD, Burg MG, Pak WL. Molecular, biochemical, and electrophysiological characterization of Drosophila norpA mutants. J Biol Chem. 1996;271:4937–4945. doi: 10.1074/jbc.271.9.4937. [DOI] [PubMed] [Google Scholar]
- 25.Pollock JA, Benzer S. Transcript localization of four opsin genes in the three visual organs of Drosophila; RH2 is ocellus specific. Nature. 1988;333:779–782. doi: 10.1038/333779a0. [DOI] [PubMed] [Google Scholar]
- 26.Porter JA, Montell C. Distinct roles of the Drosophila ninaC kinase and myosin domains revealed by systematic mutagenesis. J Cell Biol. 1993;122:601–612. doi: 10.1083/jcb.122.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter JA, Hicks JL, Williams DS, Montell C. Differential localizations of and requirements for the two Drosophila ninaC kinase/myosins in photoreceptor cells. J Cell Biol. 1992;116:683–693. doi: 10.1083/jcb.116.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranganathan R, Malicki DM, Zuker CS. Signal transduction in Drosophila photoreceptors. Annu Rev Neurosci. 1995;18:283–317. doi: 10.1146/annurev.ne.18.030195.001435. [DOI] [PubMed] [Google Scholar]
- 29.Sawin-McCormack E, Sokolowski MB, Campos AR. Characterization and genetic analysis of Drosophila melanogaster photobehaviour during larval development. J Neurogenet. 1995;10:119–135. doi: 10.3109/01677069509083459. [DOI] [PubMed] [Google Scholar]
- 30.Schmucker D, Taubert H, Jaeckle H. Formation of the Drosophila larval photoreceptor organ and its neuronal differentiation require continuous Kruppel gene activity. Neuron. 1992;9:1025–1039. doi: 10.1016/0896-6273(92)90063-j. [DOI] [PubMed] [Google Scholar]
- 31.Schmucker D, Jaeckle H, Gaul U. Genetic analysis of the larval optic nerve projection in Drosophila. Development. 1997;124:937–948. doi: 10.1242/dev.124.5.937. [DOI] [PubMed] [Google Scholar]
- 32.Serikaku MA, O’Tousa JE. Sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sehgal A, Price J, Young MW. Ontogeny of a biological clock in Drosophila melanogaster. Proc Natl Acad Sci USA. 1992;89:1423–1427. doi: 10.1073/pnas.89.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokal RR, Rohlf FJ. Biometry, Ed 3. W.H. Freeman and Company; New York: 1995. [Google Scholar]
- 35.Sokolowski MB, Kent C, Wong J. Drosophila larval foraging behaviour: developmental stages. Anim Behav. 1984;32:645–651. [Google Scholar]
- 36.Steller H, Fischbach KF, Rubin G. disconnected: a locus required for neuronal pathway function in the visual system of Drosophila. Cell. 1987;50:1139–1153. doi: 10.1016/0092-8674(87)90180-2. [DOI] [PubMed] [Google Scholar]
- 37.Vosshall LB, Young MW. Circadian rhythms in Drosophila can be driven by period expression in a restricted group of central brain cells. Neuron. 1995;15:345–360. doi: 10.1016/0896-6273(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 38.Washburn T, O’Tousa JE. Molecular defects in Drosophila rhodopsin mutants. J Biol Chem. 1989;264:15464–15466. [PubMed] [Google Scholar]
- 39.Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. Behavior in light:dark cycles of Drosophila mutants that are arrhythmic, blind or both. J Biol Rhythms. 1993;8:67–94. doi: 10.1177/074873049300800106. [DOI] [PubMed] [Google Scholar]
- 40.White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- 41.Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from Drosophila melanogaster. Cell. 1985;40:851–858. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]