Abstract
Cyclic nucleotide-gated (CNG) channels play central roles in visual and olfactory signal transduction. In the retina, rod photoreceptors express the subunits CNCα1 and CNCβ1a. In cone photoreceptors, only CNCα2 expression has been demonstrated so far. Rat olfactory sensory neurons (OSNs) express two homologous subunits, here designated CNCα3 and CNCα4. This paper describes the characterization of CNCβ1b, a third subunit expressed in OSNs and establishes it as a component of the native channel. CNCβ1b is an alternate splice form of the rod photoreceptor CNCβ1a subunit. Analysis of mRNA and protein expression together suggest co-expression of all three subunits in sensory cilia of OSNs. From single-channel analyses of native rat olfactory channels and of channels expressed heterologously from all possible combinations of the CNCα3, -α4, and -β1b subunits, we conclude that the native CNG channel in OSNs is composed of all three subunits. Thus, CNG channels in both rod photoreceptors and olfactory sensory neurons result from coassembly of specific α subunits with various forms of an alternatively spliced β subunit.
Keywords: cAMP, cGMP, sensory transduction, olfaction, ion channels, channel structure, subunit
Sensory transduction in retinal photoreceptors and olfactory sensory neurons (OSNs) is mediated by ion channels that are gated open by the intracellular messengers cGMP and cAMP, respectively. Cyclic nucleotide-gated (CNG) channels can be composed of several distinct subunits in unknown stoichiometry (for review, see Kaupp, 1995; Finn et al., 1996; Zagotta and Siegelbaum, 1996). Rod and cone photoreceptors, as well as OSNs, each express a specific principal subunit, commonly referred to as α subunit, which is responsible for many of the key channel characteristics. When heterologously expressed, α subunits form functional channels, probably by assembling into homotetrameric complexes (Gordon et al., 1996; Liu et al., 1996). We have adopted the following nomenclature for the vertebrate CNG channel α subunits expressed in sensory tissue: CNCα1 in rod photoreceptors (Kaupp et al., 1989), CNCα2 in cone photoreceptors (Bönigk et al., 1993; Weyand et al., 1994), and CNCα3 in OSNs (Dhallan et al., 1990; Ludwig et al., 1990). These homologous α subunits share common structural features such as an S4-like voltage-sensor motif (Jan and Jan, 1990), a pore region (Goulding et al., 1992; Bönigk et al., 1993), a cyclic nucleotide-binding fold (Kaupp et al., 1989; Dhallan et al., 1990;Ludwig et al., 1990), and a similar transmembrane topology (Henn et al., 1995).
Additional subunits have been discovered for the rod (Chen et al., 1994; Körschen et al., 1995) and olfactory (Bradley et al., 1994;Liman and Buck, 1994; Sautter et al., 1998) CNG channels. These subunits have the same structural features as described for the α subunits, with some added diversity. The second subunit of rod photoreceptors, CNCβ1a, does not form functional channels on its own, but when coexpressed with CNCα1 it bestows the hetero-oligomeric channel with properties characteristic of the native rod channel: in particular, flickery gating, sensitivity to blockage by l-cis-diltiazem (Chen et al., 1993), and modulation by Ca2+/calmodulin (CaM) (Hsu and Molday, 1993; Chen et al., 1994; Körschen et al., 1995). Electrophysiological analyses along with extensive biochemical characterization (Cook et al., 1987;Molday et al., 1990; Hsu and Molday, 1993; Körschen et al., 1995) suggest that the native rod channel is built exclusively from these two subunits.
The second subunit of the olfactory CNG channel has a much shorter N terminus than rod CNCβ1a (Bradley et al., 1994; Liman and Buck, 1994) and phylogenetically is more closely related to α subunits than to CNCβ1a (Kaupp, 1995). We therefore refer to this subunit as CNCα4. Like CNCβ1a from rod photoreceptors, CNCα4 does not form functional CNG channels on its own, and when coexpressed with the CNCα3 subunit it imparts flickery gating and increased cAMP sensitivity to the resultant hetero-oligomeric channel (Bradley et al., 1994; Liman and Buck, 1994). One caveat made clear in these papers concerns the cAMP sensitivity of the native olfactory channel (Frings et al., 1992), which still is more than twofold higher than that of CNCα3/CNCα4 heteromers.
In a recent report, another subunit mRNA was identified that is expressed in rat olfactory epithelium (Sautter et al., 1998). This subunit represents an alternative splice form of the rod CNCβ1a subunit and is referred to here as CNCβ1b. Coexpression of the CNCβ1b-, -α3, and -α4 subunits produced channels that display aK1/2 value for activation by cAMP similar to that of the native channel. Despite this similarity, it could not be concluded unequivocally that these subunits form the native channel in sensory cilia. For example, some subunits might be differentially targeted to the axon terminals in the olfactory bulb and may not be present in sensory cilia. Moreover, information on the properties of the native rat channel is sparse; in particular, no single-channel recordings are available. Here we examine the expression patterns of subunit mRNAs as well as the localization and association of the channel polypeptides in the rat olfactory epithelium. We find that all three subunit mRNAs are expressed in OSNs and that the three channel polypeptides are colocalized in the sensory cilia. For a functional assay of subunit composition, we performed a detailed single-channel analysis of the native CNG channel in rat OSNs. We then coexpressed the CNCα3-, -α4, and -β1b subunits in all possible combinations, looking for one combination that would resemble the native channel in all experimental parameters. The best match with respect to gating kinetics, single-channel conductance, ion selectivity, and cAMP sensitivity was observed with channels containing all three subunits. Together these findings suggest that the native CNG channel in mature olfactory neurons is built from the CNCα3-, -α4, and -β1b subunits.
MATERIALS AND METHODS
Reverse transcription PCR and construction of complete cDNA clone. Cloning of cDNA encoding a CNG channel β subunit from olfactory neurons was accomplished in two steps. Degenerate PCR primers were designed corresponding to segments in the N-terminal region, the transmembrane regions (S1–S6), and the cyclic nucleotide-binding region of the β subunit from bovine rod photoreceptors. These primers were used in PCR reactions together with first-strand cDNA transcribed from poly(A+) RNA from rat olfactory epithelium (OE). Primer pair 476/477 (AARTAYATGGCNTTYTT, positions 1147–1163; CATRTCRAADATCATYTG, positions 1750–1767) amplified a segment between S4 and the beginning of the cGMP-binding site; primer pair 1451/1452 (AARGARCGNACNGARAARGT, positions 571–590; CCARCARTTCCARTTCCA, positions 844–862) amplified a segment between a putative CaM-binding site in the N-terminal region and S1; and primer pair 1477/1476 (TGACGTCACCTCCGATGAGG, positions 612–631; GTAGGCTTTGCTGAGGATGG, positions 1187–1206) amplified a segment between the CaM-binding site and S5. The 5′ and 3′ ends of the cDNA were obtained by the rapid amplification of cDNA ends (RACE) technique (Frohman et al., 1988) using gene-specific primers 1489 and 1490 (TTCTACAAGATCCCCCAGGTC, positions 1556–1580, and TGGTTCAGCTTCCGGACAAGATGCG, positions 1655–1689) for the 3′ extension, and primers 1494 and 1495 (ACCACGAAGAACAGCCACAGGATG, positions 813–836, and TGAGGTTGGTCAGTGGGTCGATGC, positions 785–808) for the 5′ extension. The final clone, CNCβ1b, was constructed from overlapping PCR fragments.
To distinguish in retinal cDNA between the photoreceptor and olfactory splice variants CNCβ1a and CNCβ1b, we used two primer pairs corresponding to sequences directly 5′ (primer pair 2066/1526: GCTCCATCCGTCGCCTG, positions 206–222 and 813–829) and directly 3′ (primer pair 2067/1526: GTACCAGCCACGAAAGAG, positions 223–240; AGAACAGCCACAGGATG, positions 813–829) to the splice site.
In situ hybridization. In situ hybridization was performed with sections from 3-week-old Sprague Dawley rats. The procedure, a modification of the protocol of Schaeren-Wiemers and Gerfin-Moser (1993), as well as the RNA probes against CNCα1, CNCα3, CNCα4, SCG10, and I7, have been described previously (Bradley et al., 1997). The probe directed against the 3′ region in common between retinal CNCβ1a and olfactory CNCβ1b (608 nucleotides), contains both 3′ untranslated sequences and sequences encoding the 129 C-terminal residues of the channel. The probe specific for olfactory CNCβ1b corresponds to the first 362 5′ untranslated nucleotides of the short form of the CNCβ1b cDNA. There is no significant homology between any of the channel subunit probes used. We showed directly by in situ hybridization to HEK 293 cells, transiently expressing high levels of each of the channel mRNAs, that the probes did not cross-hybridize under our hybridization conditions (data not shown). A positive signal is indicated by the purple enzymatic reaction product of the alkaline phosphatase (AP) reaction on the substrate nitro-blue tetrazolium. Development times were kept equal for all the probes relative to a given tissue (30 hr for olfactory epithelium and 4 hr for retina). To ascertain that the patterns observed for retinal and olfactory channel mRNAs are bona fide, we used, in parallel, antisense probes directed against the transcription factor SCG10 and the olfactory receptor I7, respectively. The SCG10 probe specifically labeled ganglion cells, whereas the I7 signal was restricted to a subset of OSNs in a discrete ventral zone of the sensory epithelium (data not shown) (cf. Vasser et al., 1993), confirming the validity of our in situ hybridization procedure.
RNase protection assay. Total RNA was isolated from freshly dissected olfactory turbinates or eyes (postnatal day 18 male Wistar rat) by extraction with TRIzol (Life Technologies/BRL). The levels of the various CNC mRNA transcripts were determined by RNase protection (Ambion, HybSpeed RPA) according to the manufacturer’s protocol. Briefly, 32P-labeled antisense RNA probes were synthesized in the presence of [α-32P]-UTP by transcriptionin vitro, from subcloned PCR products (Invitrogen). Mixtures of ∼90 ng of a CNC probe (∼30,000 cpm, specific activity 3.5 × 108 cpm/μg) and ∼300 ng of glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) probe (∼7500 cpm, specific activity 2.5 × 107 cpm/μg) were hybridized to yeast tRNA (50 μg), with or without added olfactory or eye target RNA (15 μg). After hybridization, the unhybridized sequences were digested with a mixture of RNases A and T1, then separated by electrophoresis on a 5% polyacrylamide/7 murea sequencing gel. For the controls of yeast RNA alone and no RNase digestion (lanes 11–15), only one-tenth of the total reaction was loaded. The gel was then dried and exposed to x-ray film without an intensifying screen. The probes used for detection of the various CNC subunit mRNAs were as follows. The 195 nucleotide CNCα4 probe protects a band of 126 nucleotides and spans base pairs 2127–2253 (GenBank U12623); the 282 nucleotide CNCβ1b probe protects a band of 213 nucleotides and spans base pairs 642–855 (GenBank AF068572); the 271 nucleotide CNCα1 probe protects a band of 200 nucleotides and spans base pairs 2415–2577 (GenBank X55519); the 328 nucleotide pan-β probe protects a band of 260 nucleotides and spans base pairs 3066–3326 (GenBank AF068572); and the 102 nucleotide rat GAPDH probe protects a band of 68 nucleotides and spans base pairs 575–644 (GenBank M17701). For quantifying the bands, the dried gel was exposed to a phosphorimager plate and analyzed with the TINA program package (v2.09) for the BAS-Reader series of Fuji Image Plate scanners (BAS1000). After quantifying, the bands representing protection of channel probes were corrected for uridine content and normalized to the loading control, GAPDH.
Membrane protein preparation. Rat olfactory epithelium was homogenized in a glass/Teflon homogenizer in ice-cold 10 mmHEPES, 0.1 mm EGTA, 1 mm DTT, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 1 μg/ml pepstatin, 500 μg/ml Pefabloc SC (Boehringer Mannheim, Mannheim, Germany), 10 mmbenzamidine, pH 7.5. The suspension was washed twice by centrifugation at 100 × g for 7 min (4°C) to separate the membranes from nuclei. Membranes were collected by centrifugation at 21,000 × g for 30 min (4°C). The membrane pellet was resuspended in 10 mm HEPES, 500 mm NaCl, 0.1 mmEGTA, 1 mm DTT, pH 7.5 (plus protease inhibitors), washed by centrifugation, and resuspended in cold 10 mm HEPES, 100 mm NaCl, 0.1 mm EGTA, 1 mm DTT, pH 7.5. Membrane proteins from transfected and nontransfected HEK 293 cells were isolated by the same procedure.
Isolation of olfactory cilia. Olfactory cilia preparations were obtained using the calcium-shock method (Anholt et al., 1986; Chen et al., 1986). Briefly, after a short wash of the olfactory epithelium in ice-cold saline solution (120 mm NaCl, 5 mmKCl, 1.6 mm K2HPO, 25 mmNaHCO3, 7.5 mm glucose, pH 7.4), the tissue was subjected to Ringer’s solution containing 10 mmcalcium and gently stirred for 5 min at 4°C. Detached cilia were isolated by three sequential centrifugation steps for 5 min at 7700 × g. The supernatants containing the cilia were collected, and pellets were resuspended in Ringer’s solution containing 10 mm CaCl2 as described above. The cilia preparation was obtained after a final centrifugation step of all the pooled supernatants for 15 min at 27,000 × g. The pellet containing the cilia was resuspended in hypotonic buffer (10 mm Tris, 3 mm MgCl2, 2 mm EGTA, pH 7.4) and stored at −70°C. The yield of cilia was ∼0.5 mg per rat.
Western blot analysis. Protein amounts for ciliary and epithelial preparations were determined in parallel on one filter according to Schaffner and Weissmann (1973). From each preparation, 15 μg were separated by SDS-PAGE, transferred to Immobilon-P membrane (Millipore), which was blocked with 0.5% milk powder in PBS, and sequentially probed with the following purified primary antibodies: mouse anti-α4 1:30 [mAB7B11, directed against residues 392–575 of CNCα4 (Bradley et al., 1997)]; rabbit anti-β1a 1:500 [polyclonal FPc21K, directed against residues 574–763 representing the N-terminal domain of the β′ part of retinal CNCβ1a (Körschen et al., 1995; Wiesner et al., 1998)]; rabbit anti-α3 1:150 [polyclonal, directed against residues 559–664 of CNCα3 (Bradley et al., 1997)]; and rabbit anti-ACIII 1:500 (Santa Cruz Biotechnology) directed against the olfactory adenylyl cyclase type III. Appropriate secondary antibodies were HRP-coupled and detected by enhanced chemiluminescence (Amersham).
For deglycosylation, membrane proteins were first denatured in the presence of 0.5% SDS/1% 2-mercaptoethanol for 10 min at room temperature and then incubated in 50 mm PBS/1% NP-40 and 500 U of peptide/N-glycosidase F (NEB) for 2 hr at 37°C.
Immunohistochemistry. Three-week-old rats (Sprague Dawley) were anesthetized with fluothane and quickly decapitated. The rostral part of the skull, containing the nasal cavity, was dissected and immersed in 4% paraformaldehyde/0.1 m phosphate buffer (PB), pH 7.4, for 1 hr. To enable access of the fixative to the olfactory epithelium, the air was evacuated from the nasal cavity in a low-pressure chamber for a few minutes. After several rinses in PB, tissue was cryoprotected in 30% sucrose in PB overnight. The next day, tissue was embedded in OCT compound (Miles Scientific) and frozen onto the cryostat stage. Coronal sections (16 μm thick) were cut on a cryostat (Reichert & Jung) and collected on gelatin-coated slides. Sections were air-dried, post-fixed in 4% paraformaldehyde for 5 min, washed in PB, and incubated in 10% normal goat serum (NGS), 0.5% Triton X-100 in PB for 1 hr. Primary antibodies were diluted in 5% NGS, 0.5% Triton X-100, 0.05% NaN3 in PB. Purified antibody directed against CNCα3 was diluted 1:500; purified antibody FPc21K directed against CNCβ1a was diluted 1:1000; monoclonal antibody mAB7B11 (Bradley et al., 1997) was diluted 1:5; anti-ACIII was diluted 1:1000. Sections were incubated with primary antibodies for several hours or overnight at room temperature. After several rinses in PB, sections were incubated with biotinylated secondary antibodies (Sigma; anti-rabbit-biotin 1:1000 or anti-mouse-biotin 1:80) diluted in 5% NGS, 0.5% Triton X-100 for 1.5 hr. Sections were rinsed in PB and subsequently incubated with Extravidin-HRP (Sigma; 1:300 dilution in PB) for 1.5 hr. After several rinses in PB, immunoreactivity was visualized using diaminobenzidine (DAB) as chromogen (0.05% DAB, 0.01% H2O2 in PB). Sections were coverslipped with Mowiol solution (Hoechst) and photographed using differential interference contrast optics.
Electrophysiological experiments. Rat OSNs were isolated from 3- to 6-week-old Sprague Dawley rats as described previously (Frings et al., 1992). After isolation the olfactory epithelium was washed in a solution containing (in mm): 145 NaCl, 5 KCl, 10 HEPES, 10 glucose, adjusted to pH 7.4 with NaOH. After dissection into small pieces, the tissue was incubated for 40 min in the same solution containing 0.2 mg/ml trypsin at 37°C. After trituration, the cell suspension was transferred to the recording chamber. Isolated OSNs were identified by their characteristic morphology and investigated with the patch-clamp technique (Hamill et al., 1981) using a LIST PC amplifier (LIST, Darmstadt, Germany). Macroscopic recordings were obtained from membrane patches excised from dendritic knobs, and single-channel recordings were obtained from the membrane of somata and dendrites (Frings et al., 1992). Expression of cloned cDNAs encoding the CNCα3, CNCα4, and CNCβ1b subunits in HEK 293 cells was performed as described previously (Bönigk et al., 1993;Baumann et al., 1994). Transient expression was driven by insertion of cDNAs into pCIS (Genentech; CNCα3 and CNCα4,) or pcDNAIamp (Invitrogen; CNCβ1b).
The solutions for cotransfection contained the following approximate molar ratios of plasmids: CNCα3/CNCα4 = 4:1; CNCα3/CNCα4/ CNCβ1b = 2:1:2. The combination CNCα3/CNCβ1b was tested at the ratios 3:7, 1:1, and 7:3. CNCα4 and CNCβ1b were tested with 1:1 and 7:3 mixtures. We did not observe any difference in the expression pattern of α3β1b channels using these three different plasmid ratios. Single-channel currents were recorded from inside-out patches excised from the membrane of cells expressing the respective subunits. The solution in the recording pipette contained (in mm): 120 NaCl, 3 KCl, 10 HEPES, 10 EGTA, adjusted to pH 7.4 with NaOH. For experiments under symmetrical ionic conditions, the same solution was used on both sides of the patch. The bath solution for bi-ionic experiments contained (in mm): 120 KCl, 5 NaCl, 10 HEPES, 10 EGTA, adjusted to pH 7.2 with KOH. cAMP concentrations were determined spectrophotometrically. Steady-state macroscopic currents were digitized and recorded at 50 Hz (PhoCal, Life Science Resources, Cambridge, UK); MacroscopicI–Vm relations were sampled at 1 kHz (PhoClamp, Life Science Resources). After currents were measured at various cAMP concentrations, leak currents were subtracted. Dose–response relations were constructed for each patch by fitting to the data a Hill-type function, I/Imax =cn/[cn +K1/2n], whereImax is the current at saturating concentrations of the ligand, c is the ligand concentration, nis the Hill coefficient, and K1/2 is the concentration for half-maximal channel activation. The mean values forK1/2 and n from all patches were used to construct the solid lines in the dose–response plots. The Figures also show the mean values of I/Imaxfor each concentration with SDs. Results in the text, are given as means ± SD, with numbers of experiments in parentheses. Single-channel currents were recorded with a filter frequency of 5 kHz (eight-pole Bessel filter) on a DAT recorder (DTR-1202, Biological). The data were digitized at 3 kHz and filtered at 1 kHz for analysis (PAT, Life Science Resources). Single-channel current and open probability were determined from all-point amplitude histograms obtained from single-channel recordings of 20–50 sec duration. Single-channel currents were obtained from the difference in open and closed peaks, whereas open probability was derived from the fractional area of the open peak. Amplitude histograms in the Figures display on the ordinate the percentage of the total recorded time spent in each current level indicated on the abscissa.
RESULTS
Cloning of CNCβ1b, a CNG channel β subunit from rat olfactory neurons
The cDNA encoding rat CNCβ1b was cloned by RT-PCR using degenerate primers derived from sequences of CNCβ1a, the β subunit of rod photoreceptors (Chen et al., 1994; Körschen et al., 1995). The 5′ and 3′ ends of the cDNA were obtained by the RACE technique. Overlapping PCR fragments were used to generate the final recombinant clone encoding the rat olfactory β subunit, termed CNCβ1b. As shown in Figure 1, CNCβ1b codes for a protein of 858 amino acid residues with a calculated molecular mass of 96.4 kDa and high sequence similarity to the β subunit from rod photoreceptors (CNCβ1a). The CNCβ1b sequence from residues 75–858 shares 87.5% amino acid identity with bovine rod CNCβ1a and 100% with rat CNCβ1a, whereas the N-terminal sequence from residues 1–74 is entirely different. One obvious implication of such a large domain of identity is that olfactory CNCβ1b and retinal CNCβ1a are derived from the same gene by alternative splicing.
Fig. 1.
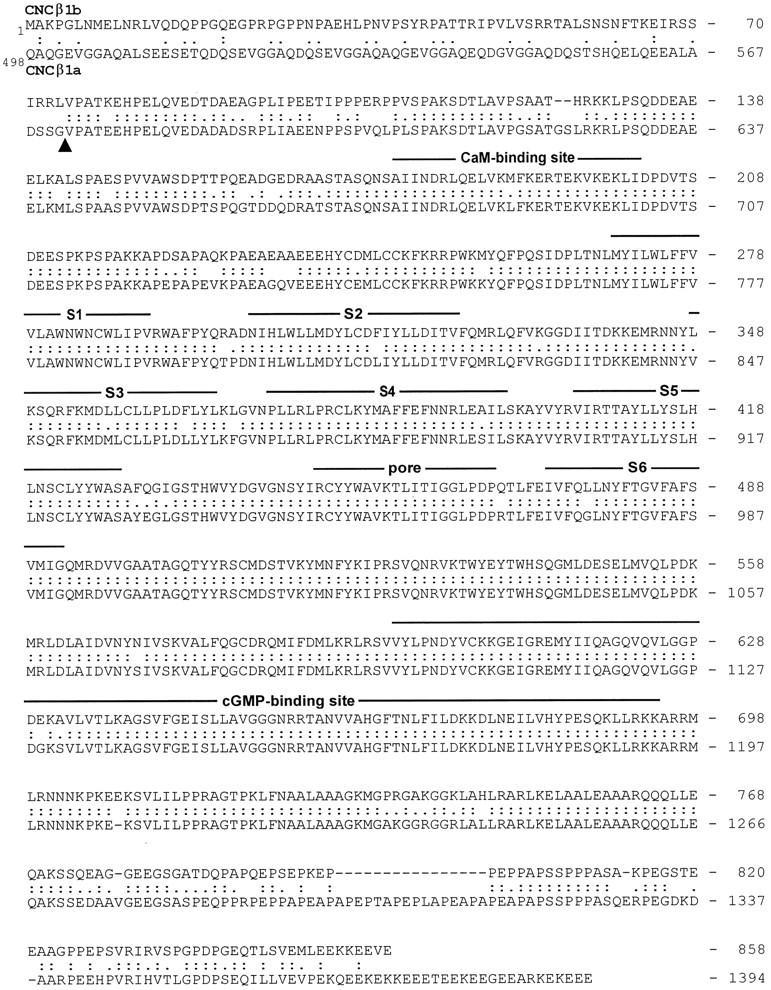
Alignment of the deduced amino acid sequence of rat olfactory CNCβ1b with bovine rod CNCβ1a. Numbersindicate positions of amino acid residues in the polypeptide. The sequence of CNCβ1a is presented starting at residue 498.Colons and periods between the two sequences indicate identical residues and conservative substitutions, respectively. Structural features similar to those of α subunits are represented by lines above the sequence.S1–S6, Membrane-spanning segments; S4, voltage sensor-like motif; P, the pore motif that lines the cavity of the channel; CaM, a nonconventional calmodulin-binding site (Weitz et al., 1998). Arrowheadindicates an exon boundary identified in human rod CNCβ1a (Ardell et al., 1996).
The CNCβ1a polypeptide is much larger than CNCβ1b (calculated molecular mass: 155 kDa vs 96.4 kDa) and is characterized by an unusual bipartite structure (Körschen et al., 1995). The C-terminal half (referred to as the β′ part) shows significant structural similarity to α subunits, whereas the large N-terminal half is nearly identical to a glutamic acid-rich protein (referred to as the GARP part). It has been shown that residues G571/V572 (corresponding to V75 in CNCβ1b) mark the boundary between the GARP part and the β′ part of bovine CNCβ1a (Fig. 1, arrowhead) (Körschen et al., 1995). In addition, an intron interrupts the genomic sequence in the human CNCβ gene at this site (Ardell et al., 1996). In accordance with the gene-structure analysis of CNCβ1 presented by Sautter et al. (1998)and our own RNase protection assays (see Fig. 3), we conclude that olfactory CNCβ1b and rod CNCβ1a are derived from alternatively spliced CNCβ transcripts.
Fig. 3.
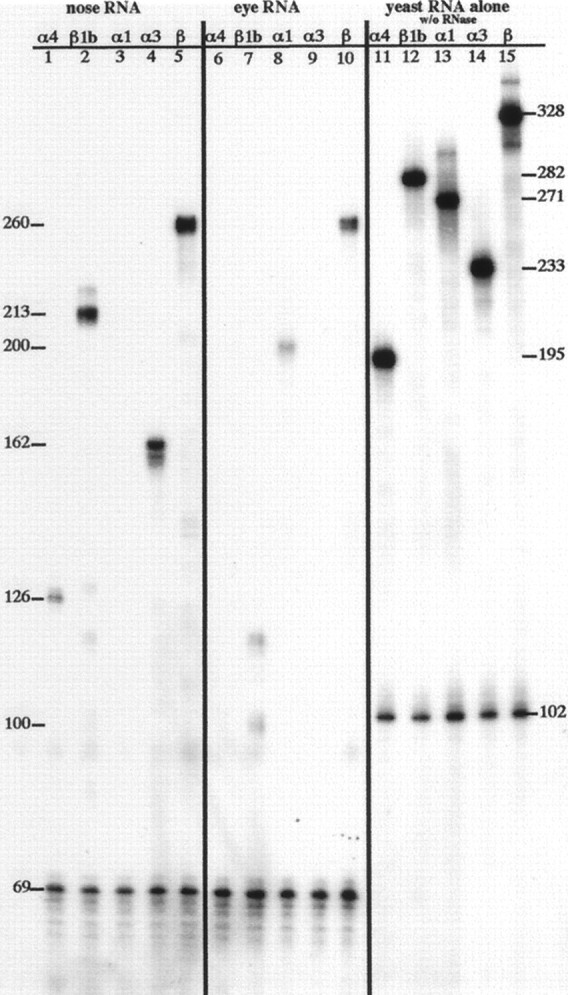
Analysis of CNC α and β mRNA expression by RNase protection. 32P-labeled antisense RNA probes were hybridized to total RNA from olfactory or eye tissue, subjected to RNase digestion, and resolved on a sequencing gel. Left panel, lanes 1–5, Protection of probes for GAPDH and CNC subunits α4, β1b, α1, α3, and β by olfactory RNA. Center panel, lanes 6–10, Protection of probes for GAPDH and CNC subunits α4, β1b, α1, α3, and β by eye RNA. Right panel, lanes 11–15, Probes for GAPDH and CNC subunits α4, β1b, α1, α3, and β. Numbers along right sideindicate length of undigested probe. Numbers alongleft side indicate length of protected products obtained with olfactory or eye RNA. See Materials and Methods for complete description of probes and protected products. Exposure shown was for 70 hr at room temperature.
Detection of CNCα3, -α4, and -β1b mRNA in olfactory neurons
We performed in situ hybridization of digoxigenin-labeled antisense RNA probes to 20 μm cryostat sections. Probes were directed against nonconserved regions (mostly nontranslated sequences) of the subunits and detected with an AP-conjugated anti-digoxigenin antibody. Expression of the olfactory CNCβ1b subunit message in sections of olfactory epithelium is clearly detected in the sensory neurons using a probe directed against the olfactory-specific 5′ region of CNCβ1b mRNA (Fig.2A), and by a probe that hybridizes to the common 3′ region of olfactory and retinal β messages (Fig. 2B). The AP reaction product respects the respiratory/sensory epithelial border (Fig. 2A,arrow), indicating specific expression in OSNs. As a control, expression of CNCα1 mRNA was not detected in OSNs, but in the retina (Fig. 2, compare C, H). As judged by the intensity of the AP product, the level of expression of CNCβ1b in OSNs is similar to that of CNCα3 (Figs. 2, compareA, B, and E). The expression levels of CNCα3 and CNCβ1b message appeared to be higher than that of CNCα4 (Figs. 2, compare E, A, and D) (cf.Bradley et al., 1994). For direct comparison, the color reactions in Figure 2A–E have been developed for the same time. Five times longer development time of the CNCα4 signal resulted in a staining intensity similar to that seen with CNCβ1b- or CNCα3-specific probes (data not shown). The very weak signal of the CNCα4 probe possibly reflects a lower expression level of CNCα4 mRNA. We have confirmed this observation by performing quantitative RNase protection assays (see below).
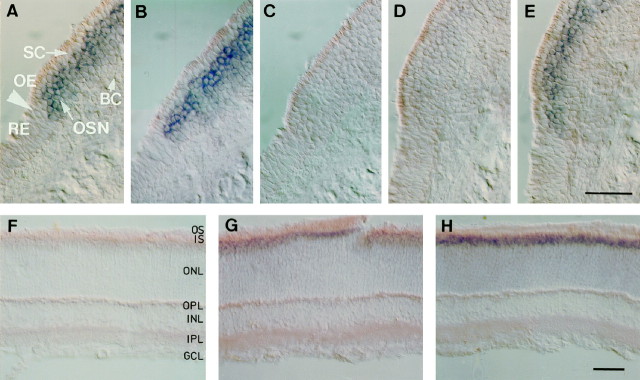
Fig. 2.
Cell type-specific expression of CNCα3, -α4, and -β1b mRNAs in the olfactory epithelium. In situhybridization analysis of olfactory channel subunits expressed in rat olfactory epithelium (A–E) and retina (F–H). Expression of channel mRNA was examined in 20-μm-thick sections using digoxigenin-labeled antisense RNA probes against nonconserved regions of each subunit. Visualization was achieved with an AP-conjugated anti-digoxigenin antibody. A positive signal is indicated by a purple AP reaction product. Shown are signals with probes against (A, F) the olfactory specific 5′ region of CNCβ1b, (B, G) the 3′ region in common with CNCβ1a and CNCβ1b, (C, H) CNCα1, (D) CNCα4, and (E) CNCα3. Arrows inA mark the transition zone between olfactory epithelium (OE) and respiratory epithelium (RE). Positive signals correspond to expression in olfactory sensory neurons (OSN), not supporting cells (SC) or basal cells (BC). In F–H, positive signals are restricted to the inner segment (IS) layer of photoreceptors; other layers are outer segments (OS), outer nuclear layer (ONL), outer plexiform layer (OPL), inner nuclear layer (INL), inner plexiform layer (IPL), and ganglion cell layer (GCL). Scale bars, 50 μm.
To distinguish between olfactory and retinal splice variants of β subunit mRNA, we performed hybridizations to sections of rat retina with both the olfactory CNCβ1b-specific 5′ probe, and the probe directed against the common 3′ region of CNCβ1a and CNCβ1b. As expected, the common 3′ probe, detected expression of CNCβ1a in retinal photoreceptors as indicated by the reaction product localized to the inner segments of these cells (Fig. 2G). The 5′ probe did not produce any signal in the retina (Fig. 2F), confirming that the β subunit mRNA exists in at least two splice forms, one in retinal rod photoreceptors (CNCβ1a) and one in olfactory sensory neurons (CNCβ1b).
Quantitative analysis of CNC α and β mRNA expression
The expression levels of the various subunit transcripts were analyzed by quantitative RNase protection assay to answer the following questions. We sought to clarify the nature of the weak CNCα4 signal seen in our in situ hybridization, distinguishing whether—relative to the other CNC subunits—this signal represents a lower level of expression of CNCα4 mRNA or is just an example of the nonquantitative nature of in situ hybridizations. We also wished to analyze the olfactory-specific use of the exon encoding the 74 amino acids unique to the N terminus of CNCβ1b. Primers were designed for amplification of five short amplicons (126–260 bp in length) from the cDNA clones for CNCα1, -α3, -α4, and -β1b (see Materials and Methods). These were subcloned and used as templates for transcribing [α-32P]-UTP-labeled antisense probes against the CNC mRNA transcripts. For CNCβ1b, two probe templates were amplified. One amplicon was directed against the 3′ end of the cDNA to generate a pan-β probe, which should detect all β gene transcripts. The second amplicon spans the position of divergence between CNCβ1b and CNCβ1a in their respective 5′ coding regions. Therefore, the antisense probe generated from this second β1b template has 100 of 213 complementary bases in common with CNCβ1a and is entirely complementary to CNCβ1b.
The result of an RNase protection assay with these probes is shown in Figure 3. Each reaction contained two labeled probes, one complementary to a CNC subunit and a second used as a loading control complementary to GAPDH. The signals in lanes 11–15 (each one-tenth of a reaction with no RNase digestion) are stronger than or equal to those in lanes 1–10. Thus, we can be assured that the probes were in excess during the hybridization and that our results were quantitative. In lane 1, the band migrating at 126 nucleotides represents the expression level of CNCα4 mRNA in olfactory turbinates. The results confirm that the expression level of CNCα4 mRNA in the olfactory turbinates is 6- to 14-fold lower than those of either CNCα3 or CNCβ1b (compare lane 1 with lanes 4 and 5). This result may explain the weak signal seen for CNCα4 mRNA expression in the in situ hybridization (Fig. 2D). Corroborative support for a lower level of CNCα4 mRNA expression comes from results of screening an olfactory cDNA library for CNCα3 and CNCα4. The abundance of CNCα3 clones was found to be ∼1 in 104, whereas CNCα4 clones were 1 in 105 (Bradley, 1996).
Turning to CNCβ expression, lane 2 shows that 213 nucleotides of the CNCβ1b probe are protected when hybridized to the olfactory RNA. When hybridized to RNA from the eye (lane 7), the same probe protected a band of 100 nucleotides, as expected for detection of β1a expression in this tissue. Protection is also observed for a band of 120 nucleotides, suggesting that perhaps a short (∼20 bp) sequence of CNCβ1b (5′ of the divergent point between CNCβ1b and CNCβ1a) is expressed as part of an mRNA in the eye. If both the 100 and 120 nucleotide bands in lane 7 represent β gene mRNAs expressed in the eye, then their signals should sum to the signal of the 260 nucleotide band seen with the pan-β probe (lane 10). After correction for the amount loaded (GAPDH signal) and the difference in length (i.e., uridine content), the signal of the 100 nucleotide band alone accounts for the 260 nucleotide pan-β probe signal in lane 10. Furthermore, in RT-PCR experiments with primers against the CNCβ1b sequence—5′ primer either just upstream or just downstream of the CNCβ1b/β1a divergence point, and a 3′ primer downstream in CNCβ1b—we were never able to detect expression of the CNCβ1b-specific sequences in the eye (data not shown; and see Materials and Methods). Therefore, we conclude that the 120 nucleotide band protected with the CNCβ1b probe in lanes 7 and 2 is an artifact of the assay. Lane 3 shows that there is no expression of CNCα1 in the olfactory turbinates. Likewise, lanes 6 and 9 show that there is no expression of CNCα3 or CNCα4 in the eye. These reactions also serve as negative controls for our demonstration of β expression in both the eye and olfactory tissue (lanes 5 and 10, respectively).
Expression of all three subunits in sensory cilia
The presence in OSNs of transcripts encoding all three channel subunits provides suggestive evidence but does not prove that the subunit polypeptides are expressed in sensory cilia. In particular, the low expression level of CNCα4 message in OSNs raises the question to what extent the CNCα4 protein contributes to the native channel in the sensory cilia. To examine this, we compared membrane protein extracts from whole OE (including cilia) to preparations of isolated cilia. Using the same amount of membrane protein from each preparation, proteins were separated by SDS-PAGE and analyzed by Western blotting and probing with subunit-specific antibodies. As control for the purity of our cilia preparation, we used an antibody directed against adenylyl cyclase type III (ACIII). ACIII has been proposed to mediate the odorant-induced rise of cAMP in OSNs and is localized to cilia (Pace et al., 1985; Pfeuffer et al., 1989; Bakalyar and Reed, 1990; Menco et al., 1992). Figure4A demonstrates that ACIII (∼230 kDa), in fact, is highly enriched in purified cilia (lane 2) compared with whole OE (lane 1). In membranes from whole OE, a polyclonal antibody raised against the rat CNCα3 subunit (Bradley et al., 1997) recognized a faint band of ∼75 kDa and a diffuse “smear” at 110–145 kDa (Fig. 4B, lane 1). In purified cilia, both the smear and the 75 kDa band were greatly enriched (Fig. 4B, lane 2). The CNCα3 polypeptide, expressed in HEK 293 cells, had almost the same size as the 75 kDa signal in cilia (Fig. 4B, comparelanes 2 and 5), suggesting that this band indeed represents the CNCα3 subunit. With some cilia preparations, however, the 75 kDa band was not enriched or was even less intensely labeled compared with whole OE preparations. On the other hand, the “smear” was always enriched in cilia membranes over whole OE membranes. We were concerned about this result because (1) it would indicate that the majority of the CNCα3 protein is not in cilia, and (2) the pronounced smear, if caused by unspecific cross-reactivity, would render suspect any immunohistochemical localization with anti-α3 antibody. We tested whether glycosylation of CNCα3 or formation of a stable complex with other ciliary components explains the fuzzy 110–145 kDa signal. Several treatments known to dissociate protein complexes were ineffective in reducing the smear signal (data not shown). In contrast, treatment of whole OE (lane 3) and cilia preparations (lane 4) with glycosidase abolished the diffuse smear entirely and strongly enhanced the 75 kDa signal. Before glycosidase treatment, the weak 75 kDa band in cilia consistently exhibited a slightly lower electrophoretic mobility than the 75 kDa band after deglycosylation (Fig.4B, compare lane 2 with lanes 3, 4, 5). This minor difference may indicate that even the ∼75 kDa form is glycosylated, although to a much lower degree. These results demonstrate that the vast majority of the CNCα3 polypeptide exists in a highly glycosylated form. In our cilia preparations, we observed batch to batch variation in the intensity of the 75 kDa band relative to preparations from whole OE, whereas the smear was always enhanced. It therefore seems likely that the glycosylated form is specifically targeted to cilia, whereas the nonglycosylated form might be expressed elsewhere in OSNs, for example the soma.
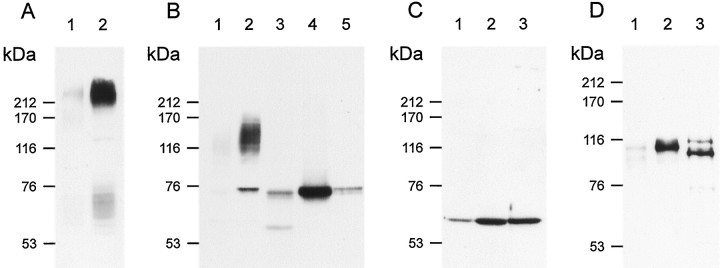
Fig. 4.
Analysis by Western blot of CNCα3, -α4 and -β1b expression in sensory cilia of rat olfactory epithelium.A, Western blot of equal amounts of membrane protein prepared from either whole olfactory epithelium (OE) (lane 1) or from isolated olfactory cilia (lane 2) probed with an anti-ACIII-specific antibody produced a much stronger ∼230 kDa signal with the cilia preparations. B, Western blot probed with polyclonal anti-α3 antibody. The antibody recognized a ∼75 kDa band and a “fuzzy” smear between ∼110 and 145 kDa in the preparations from both whole OE (lane 1) and cilia (lane 2). The CNCα3 subunit expressed in HEK 293 cells displays an apparent molecular mass of ∼75 kDa (lane 5, 10 μg protein), suggesting that the 75 kDa band recognized in olfactory tissue, in fact, represents the CNCα3 subunit. Treatment of membrane proteins from whole OE (lane 3) and cilia (lane 4) with N-glycosidase F abolished the smear entirely and correspondingly increased the intensity of the 75 kDa band. C, Western blot of membranes derived from whole OE (lane 1), cilia (lane 2), and HEK 293 cells expressing CNCα4 (lane 3) probed with monoclonal antibody mAB7B11 against the CNCα4 subunit. D, Western blot as in C, probed with polyclonal antibody FP21K against CNCβ1b subunit.Lane 3 is membrane derived from HEK 293 cells expressing CNCβ1b. As seen in B with CNCα3, both the CNCα4- and -β1b antibodies produced much stronger signals with preparations of cilia membranes, indicating that the molar concentration of the respective polypeptide (relative to the total protein content) is higher in ciliary-enriched than in whole OE membranes.
Enrichment of subunit polypeptide is also observed using monoclonal antibody mAB7B11 directed against the CNCα4 protein (Fig.4C) (Bradley et al., 1997). We observed a much stronger signal with isolated cilia than with whole OE membranes (Fig.4C, lanes 1 and 2). The apparent molecular mass of CNCα4 (∼ 62 kDa) in cilia (lane 2) is identical with the apparent molecular mass of the subunit expressed in HEK 293 cells (lane 3) and matches well the calculated molecular mass (65.7 kDa). No other proteins were stained by this antibody. These data show that CNCα4 is expressed in olfactory cilia and that the molar concentration of this subunit (relative to the total protein content) is higher in cilia than in nonciliary membranes.
Polyclonal antibody FPc21K, directed against CNCβ1b, also demonstrated enrichment of the β subunit in ciliary membranes (Fig.4D, lanes 2 and 3). The apparent molecular mass of CNCβ1b is significantly larger than predicted from the amino acid sequence (∼116 kDa vs 96.4 kDa). A similar difference was reported for the β′ part of bovine CNCβ1a, which comprises the region common to both CNCβ1a and CNCβ1b (apparent: 110 kDa; calculated: 92.7 kDa) as well as for the complete retinal CNCβ1a subunit (apparent: 240 kDa; calculated: 155 kDa) (Körschen et al., 1995). The olfactory CNCβ1b, like the retinal CNCβ1a subunit, is susceptible to proteolytic degradation (W. Bönigk, unpublished observations). The smaller size of CNCβ1b expressed in HEK 293 cells is probably caused by proteolysis (Fig.4D, lane 3). Glycosidase treatment had no effect on the electrophoretic mobility of CNCα4 and CNCβ1b, demonstrating that these subunits are not glycosylated (data not shown).
We used immunohistochemistry to localize CNG channel subunits in the olfactory tissue. Figure 5 shows coronal sections through the olfactory epithelium. In A, the upper part of the nasal cavity is shown. The septum on the right side can be recognized, and several turbinates are covered with olfactory epithelium. The section is stained with an antibody against ACIII. A thin dark line on the surface of the epithelium is stained, representing the cilia of OSNs. In this low-power magnification and wide-field illumination, the cartilage within the turbinates and septum appears dark when photographed in black and white, but it is not stained by the antibodies. In B, a section stained with the anti-α3 antibody is shown. Again, the ciliary layer is homogeneously stained. The prominent staining of the cilia is shown at higher magnification in Figure 5C. At anti-α3 dilutions ≤1:100, weak labeling was observed in the somata of OSNs, which appeared mostly of cytoplasmic origin (data not shown). In D, a section stained with the antibody mAB7B11 against the CNCα4 subunit is shown. In the ciliary layer, no staining above background is found. The staining in the submucosal layer reflects unspecific binding of the secondary antibody. We were concerned about the negative result with anti-α4, all the more because Western blot and electrophysiological analyses unequivocally show that CNCα4 is enriched in cilia and is present in native channels, respectively (Fig. 4 and see below). Therefore, we repeated the stainings with mAB7B11 on tissue specimens that were fixed in different ways (conditions included fixation in 2% or 4% paraformaldehyde for 20 or 60 min, or in 1% 1-ethyl-3-(3-dimethylamino-propyl)carbodiimid, or in methanol at −20°C; some specimens were sectioned unfixed, and sections were slightly post-fixed with paraformaldehyde, paraformaldehyde/picric acid mixtures, or methanol). In none of these cases was staining of olfactory cilia observed (F. Müller, data not shown). These negative results do not necessarily indicate the absence of the CNCα4 subunit from cilia membranes. It is known that some antibodies reliably detect an antigen in Western blots, but fail to stain the fixed antigen in the native tissue because of epitope masking (Kaprielian et al., 1995). Although mAB7B11 did not stain cilia in sections, it unequivocally demonstrated the enrichment of CNCα4 in cilia preparations by Western blot (Fig. 4). The antiserum FPc21K against the β subunit strongly stained cilia of OSNs (Fig. 5E).
Fig. 5.
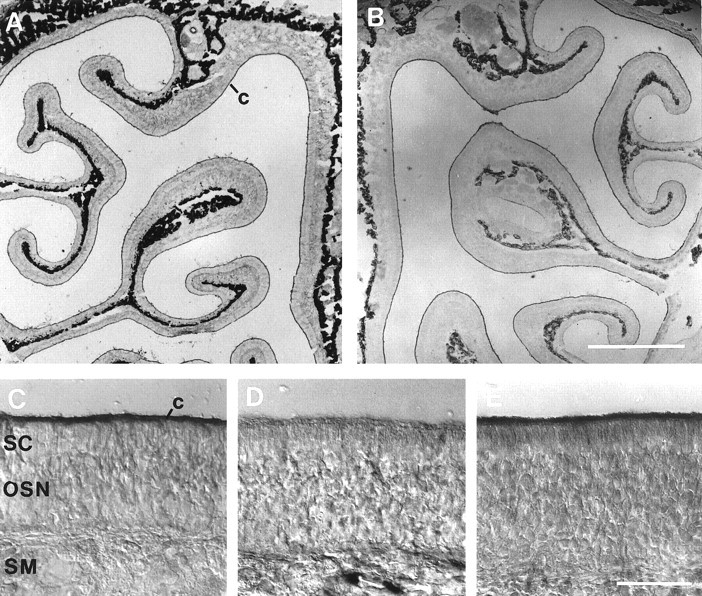
Immunohistochemical staining of olfactory epithelium. A, Upper part of the nasal cavity, stained with an antibody against ACIII. Strong staining is found in the thin ciliary layer (c) covering the epithelium on the septum (right border) and turbinates. The cartilage appears dark because of an artifact of the low-power optics but is not stained. B, The anti-α3 antibody homogeneously stained the ciliary layer. C, Higher magnification of the field shown in B. D, Antibody mAB7B11 against the CNCα4 subunit. No staining above background is found in the cilia. E, Purified antiserum FPc21K against the β subunit strongly stained the cilia of OSNs. Scale bars: A, B, 1 mm; C–E, 50 μm.
To investigate which of the three channel polypeptides co-assemble in the ciliary membrane to form a protein complex, we performed immunoprecipitation experiments under nondenaturing conditions. Using any of the three subunit-specific antibodies to precipitate the native channel protein from a solubilized cilia preparation, we found that each of the three subunits could be identified after SDS-PAGE and Western blotting (data not shown). In light of these results it is plausible that all three subunits co-assemble in one protein complex, probably the native CNG channel of the sensory cilia. As we will show below, however, CNG channel subunits can co-assemble into several different channel species. If different channel species do coexist, then it is not possible to distinguish among CNCα3β1b, CNCα4β1b, and CNCα3α4β1b by immunoprecipitation unless performed in a quantitative and subtractive manner (Tretter et al., 1997; Shamotienko et al., 1997; Jechlinger et al., 1998). Therefore, we have instead investigated the functional properties of the various subunit combinations expressed heterologously, while in parallel characterizing the functional properties of native channels from OSNs.
Functional analysis of subunit composition: macroscopic currents
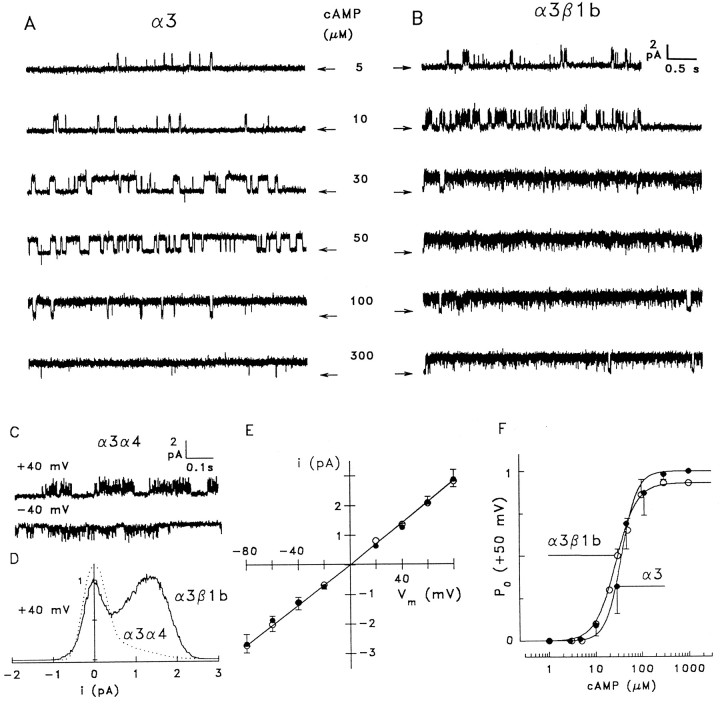
As a further analysis of subunit composition, we compared the electrophysiological properties of native channels with channels composed of the cloned subunits. Activity of the native channel was recorded in inside-out patches excised from somatic and dendritic membranes of rat olfactory neurons. Cloned channel subunits were studied in excised inside-out patches of HEK 293 cells transfected with all possible combinations of the cDNAs encoding CNCα3, CNCα4, or CNCβ1b. Transfection with the cDNA encoding CNCα3 alone, as well as the subunit combinations CNCα3/CNCβ1b, CNCα3/CNCα4, and CNCα3/CNCα4/CNCβ1b, produced functional channels (for simplicity referred to in the following as α3, α3β1b, α3α4, and α3α4β1b channels, respectively). Expression of CNCα4 or CNCβ1b either alone or co-transfected did not produce functional CNG channels.
Ligand sensitivity was determined by recording macroscopic currents at different concentrations of cAMP. Figure6A shows data for the two extreme cases, α3 and native channels. Figure6B compares the cAMP dose–response relations derived from these patches. Fitting the Hill equation to the data from each experiment gave values for the concentration of half-maximal activation (K1/2) and the Hill coefficient (n) listed in Table 1. In agreement with Sautter et al. (1998), the K1/2values of native and α3α4β1b channels are similar (4.1 and 4.8 μm, respectively), whereas theK1/2 values of all other channels are significantly larger, indicating that all three subunits are necessary for the high cAMP sensitivity of native channels.
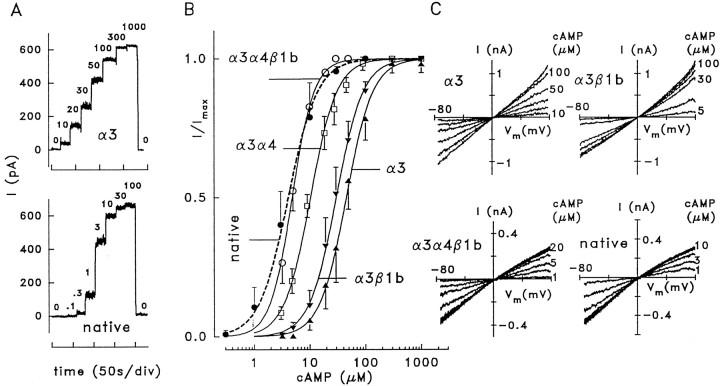
Fig. 6.
Ligand sensitivity and ion selectivity of native and heterologously expressed olfactory CNG channels. A, Macroscopic current recordings from inside-out patches of HEK 293 cells transfected with CNCα3 and from a patch excised from a dendritic knob of a rat OSN containing native CNG channels. Current was recorded at +40 mV and the indicated cAMP concentrations. B, Dose–response relations for the activation of macroscopic currents by cAMP at +40 mV. Lines were constructed by fitting a Hill-type function to the normalized current (see Materials and Methods). Fitting parameters are given in Table 1. C, Macroscopic currents recorded from inside-out patches with the indicated cAMP concentrations. The main permeable ions were Na+(extracellular) and K+ (intracellular). The current shows inward rectification only with α3α4β1b and native channels, indicating that these channels conduct Na+ better than K+.
Table 1.
Activation of macroscopic currents by cAMP
| Channel | K1/2(μm) | n | Patches |
|---|---|---|---|
| α3 | 45.3 ± 10.6 | 1.8 ± 0.2 | 11 |
| α3β1b | 29.9 ± 6.0 | 1.8 ± 0.1 | 33 |
| α3α4 | 10.3 ± 1.0 | 1.8 ± 0.2 | 6 |
| α3α4β1b | 4.8 ± 0.6 | 2.2 ± 0.3 | 8 |
| Native | 4.1 ± 1.9 | 1.7 ± 0.1 | 4 |
Concentrations for half-maximal activation,K1/2, of macroscopic currents at +40 mV, together with Hill coefficients, n, and numbers of patches.
K1/2 values derived from macroscopic currents are only meaningful if the channel population in the patch is homogeneous. If different kinds of channels are expressed by a cell, the measured K1/2 value represents the arithmetic mean of the weighted contributions from each channel population. Two observations demonstrate that mixed populations rarely occur with olfactory subunits. First, single-channel recordings show that each combination of olfactory subunits produced a single type of channel in almost all experiments (see below). Second, the cAMP sensitivities derived from macroscopic currents for each channel type (Table 1) and those obtained from open probability analysis of single-channel recordings (Table 2) do not differ significantly, indicating virtually homogeneous channel populations (see below). In conclusion, the co-expression of three subunits produces channels with cAMP sensitivity similar to the native olfactory channel, whereas the α3β1b and α3α4 combinations result in channels with significantly lower cAMP sensitivity.
Table 2.
Single-channel parameters of olfactory CNG channels
| Channel | K1/2/μm | n | Rectification ratio | |
|---|---|---|---|---|
| Symmetrical | Bi-ionic | |||
| α3 | 40 (4) | 2.5 (4) | 1.05 (15) | 1.03 (12) |
| α3β1b | 28 (3) | 1.9 (3) | 1.0 (12) | 1.03 (9) |
| α3α4β1b | 4.0 (3) | 2.0 (3) | 1.28 (13) | 0.57 (13) |
| Native | 3.4 (4) | 2.3 (4) | 1.30 (14) | 0.55 (9) |
Concentrations of cAMP, K1/2, that induced an open probability of 0.5 at +50 mV (native at −50 mV), together with Hill coefficients, n, and numbers of experiments in parentheses. Rectification ratios represent i (+50 mV)/i (−50 mV) with symmetrical Na+ or with extracellular Na+ and intracellular K+(bi-ionic condition).
The native channel from rat OSNs conducts Na+ better than K+. In macroscopic current recordings from the apical membrane of rat OSNs, K+ ions produced only half the current amplitude carried by Na+ ions (IK/INa ≅ 0.5) (Frings et al., 1992). We used this characteristic feature as an additional indicator of subunit composition of the native olfactory CNG channel. Figure 6C compares the Na+ and K+ ion selectivity of the native channel with that of several subunit combinations. With 140 mm extracellular Na+ and 140 mm K+ on the cytosolic side of inside-out patches, theI–Vm relations of α3 and α3β1b channels are linear or even slightly outwardly rectifying (Fig. 6C,top panels). In contrast, under identical conditions theI–Vm relations of native and α3α4β1b channels become slightly inwardly rectifying (Fig. 6C,bottom panels). The rectification ratios under bi-ionic conditions (I+50/I−50) of 0.66 ± 0.05 (3) for native and 0.73 ± 0.03 (2) for α3α4β1b channels indicate a similar degree of current rectification, distinctly more pronounced than for α3 [1.05 ± 0.02 (2)] and α3β1b [1.13 ± 0.03 (2)] channels. Thus, current rectification, as displayed by the native channel with Na+ on the outside and K+ on the inside of the membrane, is only observed with the combination of all three subunits.
Functional analysis of subunit composition: single-channel recordings
A detailed analysis of channel properties requires single-channel recording. Channel conductance, open probability, and kinetics of gating transitions represent characteristic features that distinguish ion channels from each other. Single-channel recordings can be obtained from the membranes of dendrites and somata of OSNs where the channel density is much lower than in the ciliary membrane. This raises the concern that channels investigated in the soma membrane may be different from channels expressed in sensory cilia. However, we will present data indicating that rat OSNs express only a single type of cAMP-gated channel that is expressed both in cilia and somata.
Native channels of rat OSNs
In all 46 single-channel patches excised from somata, we observed a channel described in Figure 7, suggesting that all cAMP-gated channels in the soma membrane are of the same type. Figure 7A shows current recordings obtained from a native channel activated with 1 μm cAMP at various membrane voltages in symmetrical Na+ solutions. The channel shows a sizeable voltage-dependence ofPo, displaying prolonged dwell periods in the open state at positive potentials and only brief openings at negative potentials. This voltage dependence is more pronounced at low compared with high cAMP concentrations. The ratio of open probabilities [Po(−60 mV)/Po(+60 mV)] is 0.17 ± 0.06 (3) at 1 μm cAMP and increases to 0.76 ± 0.13 (3) at 3 μm cAMP. At 3 μm cAMP (Fig. 7B), the channel is mostly open at positive Vm, whereas negativeVm induces frequent transitions between open and closed states. Single-channel currents were derived from amplitude histograms; the two distinct peaks at 0 pA and −1.95 pA represent the closed and open states of the channel, respectively, at −70 mV (Fig.7D). Single-channel currents were measured between −80 and +80 mV (Fig. 7E); the apparent slope conductance is 27 pS at negative voltages for both symmetrical (Na+) and bi-ionic (Na+/K+) conditions (14 patches). At positive Vm, the conductance is 35 pS (five patches) in symmetrical Na+ and 14.5 pS (nine patches) in bi-ionic conditions. These values represent an underestimate of the channel conductance because brief opening and closing events are only partially resolved under our recording conditions (see also Torre et al., 1992; Sesti et al., 1994; Bucossi et al., 1997). The particularly high frequency of brief opening and closing events at negative voltages may fully account for the slight outward rectification of single-channel current at symmetrical solution. Rectification ratios under bi-ionic conditions are consistent between single-channel data from the soma (i+50/i−50 = 0.55) and macroscopic recordings from the apical membrane (I+50/I−50 = 0.66), indicating that CNG channels with similar Na+/K+ selectivity are expressed in apical and basolateral membranes of OSNs.
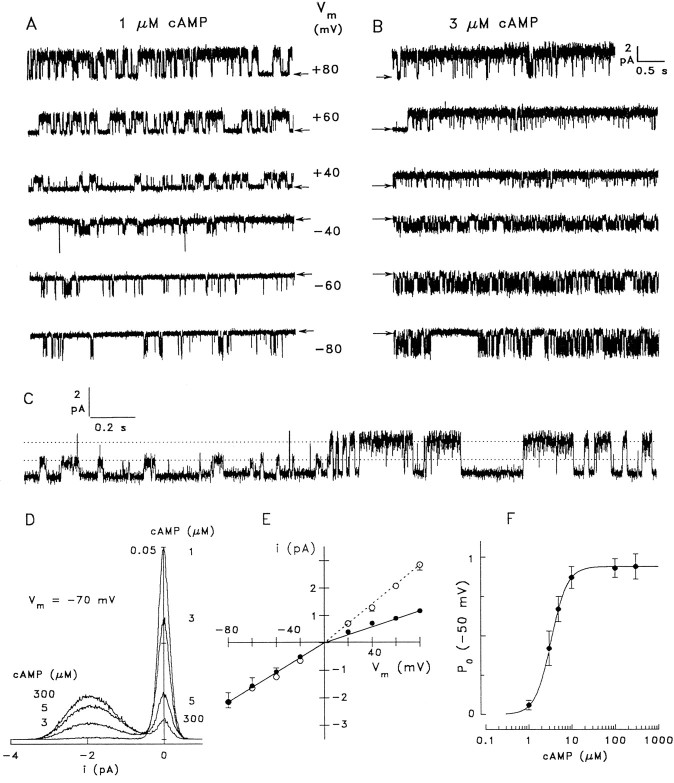
Fig. 7.
Single-channel analysis of native CNG channels from rat OSNs. A, Recording from an inside-out patch with 1 μm cAMP at the indicated membrane voltages.Arrows indicate the closed state. The channel shows a significant voltage dependence of open probability. B, The same channel as in A at 3 μm cAMP. Positive voltages favor the open state, whereas negative voltages induce continuous flickering. Single-channel recordings were obtained in symmetrical Na+ solutions, sampled at 3 kHz and filtered at 1 kHz. C, Transition of the native channel from a state of low conductance (14–16 pS) to the conductance level that is observed most of the time (35 pS); 1 μm cAMP, +60 mV. D, All-point amplitude histograms of four 40 sec recordings from a native channel at −70 mV and the indicated cAMP concentrations. Although brief, unresolved closing events result in a comparably broad current distribution in the open state (approximately −1.9 pA), the two peaks reflecting open and closed states can be clearly distinguished. The ordinate indicates the percentage of total time spent at each current level plotted. E, Voltage dependence of the single-channel current recorded in symmetrical (○, means of 5 channels), and bi-ionic conditions (●, means of 9 channels). The conductance at negative voltages (fitted to the mean values) was 27 pS in both symmetrical and bi-ionic (intracellular K+, extracellular Na+) conditions. At positive voltages the conductance was 35 pS in symmetrical and 14.5 pS in bi-ionic conditions. F, Dependence of open probability on cAMP concentration. Single-channel analysis from four patches at −50 mV yieldedK1/2 = 3.4 μm,n = 2.3.
Occasionally, the native channel displayed a state of reduced conductance. In this state, channels conduct ∼45% of the current in the main open state (Fig. 7C). This substate was observed infrequently, could not be correlated with any particular cAMP concentration or membrane voltage, and was typically stable for 10 sec to well over 30 sec.
Ligand sensitivity of native channels was analyzed by measuringPo at various cAMP concentrations (using amplitude histograms derived from recordings of 20–50 sec duration; −50 mV) and yielded a K1/2 of 3.4 μm (n = 2.3; four patches) (Fig.7F). Thus, the single-channel analysis is in good agreement with the values obtained from macroscopic recordings (4.1 ± 1.9 μm; +40 mV; Table 1), although they were recorded at different voltages. The small difference between activation constants could be explained by the voltage dependence ofPo illustrated above. ForPo analysis of native channels, it was necessary to record at negative potentials to obtain sufficiently stable patches. This almost perfect match of cAMP sensitivity between channels in the apical membrane and the membrane of somata strongly suggests that the same cAMP-gated channels are expressed in both membranes, probably with the same subunit composition.
α3, α3β1b, and α3α4 channels
We performed a similar single-channel analysis of α3 homomeric channels recorded under bi-ionic conditions. The channel opened for relatively long durations (Fig.8A); however, it required higher cAMP concentrations than the native channel. Amplitude histograms yielded a linear i–Vmrelation (Fig. 8E, ●) from which a mean conductance of 34.3 ± 1.4 pS (nine patches) was calculated. Outward (K+) conductance was not significantly different from the inward (Na+) conductance (Table 2). A plot of the open probability versus cAMP concentration, using data obtained from four single-channel recordings at +50 mV (Fig.8F, ●), was fitted withK1/2 = 40 μm andn = 2.5 (four patches). Po was weakly voltage dependent, with Po(−60 mV)/Po(+60 mV) = 0.66 ± 0.16 (two patches) at 30 μm cAMP.
Fig. 8.
Analysis of channels consisting of CNCα3 plus the CNCβ1b or CNCα4 subunit. A, Single-channel recording from a homomeric α3 channel at +50 mV and the indicated concentrations of cAMP. The channel shows a lower ligand sensitivity than native channels and long uninterrupted dwell periods in the open state. B, Single-channel recording from a α3β1b channel. Coexpression of the CNCβ1b subunit slightly increases ligand sensitivity and causes rapid open–closed transitions at both positive and negative membrane voltages. C, Recordings from α3α4 channels at 10 μm cAMP display extremely rapid flickering. D, All-point histograms from single-channel recordings from α3α4 channels (10 μm cAMP, +40 mV,dotted line) and α3β1b channels (30 μmcAMP, +40 mV, solid line). The skewed histogram obtained for α3α4 channels did not permit determination of single-channel current and open probability. E, Voltage dependence of single-channel current yielded an apparent conductance of 33.7 ± 2.7 pS (12 patches) for α3 (●) and 34.3 ± 1.4 pS (9 patches) for α3β1b channels (○). F, cAMP dependence of open probability measured at +50 mV. Solid lines were fitted for α3 channels (●, 4 patches) withK1/2 = 40 μm,n = 2.5, and for α3β1b channels (○, 3 patches) with K1/2 = 28 μm, n = 1.9.
Co-expression of CNCα3 with CNCβ1b increased the cAMP affinity and dramatically altered the gating properties of the α3 channel. As illustrated in Figure 8B, the α3β1b channel displayed a slightly increased cAMP affinity (K1/2 = 28 μm,n = 1.9) (Fig. 8F, ○) but lost the ability to adopt an open state for prolonged periods. Even at the saturating cAMP concentration of 300 μm, the channel showed frequent brief closing events, something rarely seen in α3 or native channels at saturating ligand concentrations. Amplitude histograms of the α3β1b channel produced two partially overlapping peaks (Fig. 8D, solid line) that could be used to calculate the single-channel current andPo. Thus, similar to the heteromeric rod photoreceptor channel (CNCα1/CNCβ1a) (Körschen et al., 1995;Bucossi et al., 1997), the olfactory β subunit imparts flickery gating to the heteromeric channel. The α3β1b channel, like the α3 channel described above, conducts K+ just as well as Na+ (Fig. 8E, ○; 12 patches) (Table 2). Analysis of Po revealed a very weak voltage dependence with Po(−60 mV)/Po(+60 mV) = 0.87 ± 0.09 (2) at 30 μm cAMP. Macroscopic and single-channel data are consistent, indicating that macroscopic α3β1b currents shown in Figure 6C are determined almost exclusively by the 34 pS channel species. Taken together, we have defined three distinguishing characteristics that set the α3 and α3β1b channels apart from the native channel. These include cAMP sensitivity, cation selectivity, and gating kinetics.
Channels consisting of CNCα3 and CNCα4 subunits were readily identified by extremely rapid flickering (Fig. 8C) that produced a single skewed peak in the amplitude histogram (Fig. 8D, dotted line), as reported previously (Bradley et al., 1994; Liman and Buck, 1994; Broillet and Firestein, 1997). These rapid kinetics set the α3α4 channels apart from the native channel and all other subunit combinations. Opening events were so brief that we could not measure the open-channel current; consequently, single-channel analysis was not feasible with α3α4 channels.
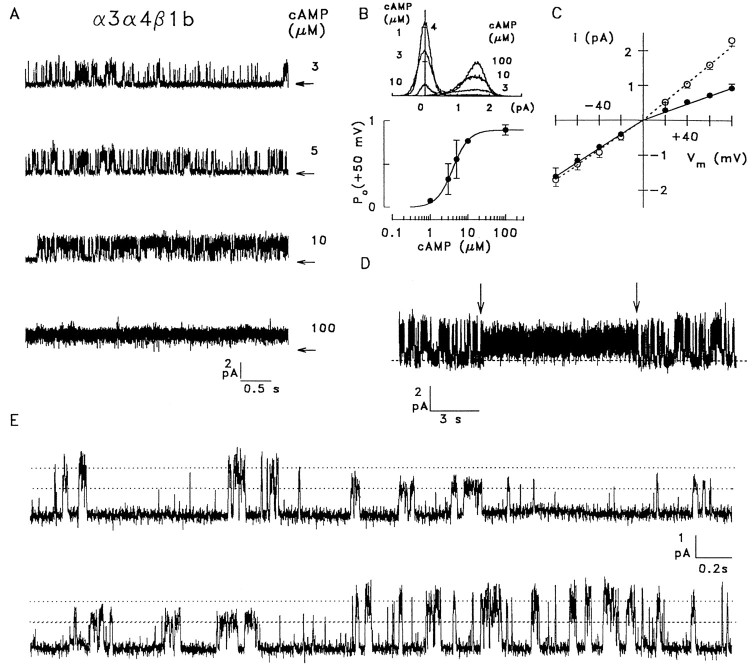
Channels containing all three subunits
When all three subunits were coexpressed, one distinct channel type was observed in almost all of the 54 patches that were analyzed (Fig. 9A), and three single-channel patches permitted analysis of Po. The resulting K1/2 for activation by cAMP was 4.0 μm, with a Hill coefficient of 2.0 (three patches) (Fig. 9B), not significantly different from the native channel. The voltage dependence of single-channel currents is shown in Figure 9C. While the i–Vmrelation was almost linear in symmetrical Na+solutions, replacement of Na+ by K+ on the cytosolic side of the patches decreased the apparent single-channel conductance for outward currents from 27 pS (Na+) to 12 pS (K+) (13 patches). Similar to the native and α3β1b channels, these values are underestimates of the channel conductance because of the brief, unresolved opening and closing events. Thus, the apparent conductance of the α3α4β1b channel is slightly smaller than in native channels, but both channels discriminate between Na+and K+ to a similar extent, in contrast to the other heterologously expressed combinations of subunits. A further similarity to native channels is the ability of α3α4β1b to adopt a subconductance state for extended periods of time. Figure 9Dshows a 20 sec record of an α3α4β1b channel at 3 μmcAMP that switches for 9.5 sec (arrows) from the predominant gating mode into a state that displays a smaller conductance, and a more vigorous gating activity. Transitions to and from the subconductance state are shown with better resolution for another α3α4β1b channel in Figure 9E.
Fig. 9.
Single-channel analysis of α3α4β1b channels.A, Single-channel recording from an α3α4β1b channel at +50 mV and the indicated concentrations of cAMP.B, Dependence of the open probability on cAMP concentration. Top panel, All-point amplitude histogram of 19–23 sec recordings at 1, 3, 10, and 100 μm cAMP and +50 mV. Bottom panel, Mean values obtained from four single-channel recordings were fitted with a Hill-type equation usingK1/2 = 4 μm,n = 2, and a maximal P0of 0.9. C, Voltage dependence of channel current for symmetrical Na+ solutions (21 pS inward, 27 pS outward) and for bi-ionic solutions (21 pS for Na+inward, 12 pS for K+ outward currents).D, Recording from an α3α4β1b channel at +60 mV with 3 μm cAMP, showing a 9.5 sec episode during which the channel switched from the main gating state into a substate with reduced conductance and different gating kinetics (arrows). E, Transition of an α3α4β1b channel from the main conductance state (27 pS) to a subconductance state (15 pS, top trace) and back to the main state (bottom trace); 5 μm cAMP, −50 mV; symmetrical Na+ solutions.
Thus, strong similarities are observed between native and α3α4β1b channels. Closer inspection of single-channel traces, however, do reveal subtle differences in the kinetics of open-closed transitions. Furthermore, the apparent single-channel conductance of the α3α4β1b channel is slightly smaller then observed in the native channel. However, all other subunit combinations result in channels that show profound functional differences compared with the native olfactory channel. Although we did not find a perfect match between native and α3α4β1b channels, the similarity of cAMP sensitivity and relative Na+/K+ conductance and the ability to adopt a subconductance state, together with the evidence of colocalization and co-assembly of all three subunits in the sensory cilia, strongly suggest that the native channel is composed of the CNCα3, CNCα4, and CNCβ1b subunits.
DISCUSSION
In this study we report the molecular identification of a CNG channel β subunit, CNCβ1b, which represents an alternatively spliced variant of the previously described CNCβ1a subunit expressed in rod photoreceptors. In accordance with a previous report (Sautter et al., 1998), we find that 74 amino acid residues in the CNCβ1b subunit replace the large, glutamic acid-rich intracellular domain (GARP) that forms the N-terminal part of CNCβ1a (Körschen et al., 1995). Because CNCβ1b is the third CNG channel polypeptide identified in rat olfactory epithelium (CNCα3, CNCα4, and CNCβ1b), we examined which of the three subunits contribute to the formation of the native CNG channel that mediates olfactory transduction.
Subunit composition of the olfactory CNG channel
We find expression of mRNA coding for all three subunits in OSNs, although the message for CNCα4 is less abundant than for CNCα3 and CNCβ1b. Using subunit-specific antibodies, we find expression of the three channel polypeptides in the sensory cilia, the site of chemoelectrical signal transduction in OSNs. Furthermore, immunoprecipitation experiments are consistent with the notion that the three polypeptides form a common protein complex in the ciliary membrane. Because the CNCα4 protein appears to be expressed at a level comparable to CNCα3 and CNCβ1b, the much lower level of CNCα4 mRNA perhaps indicates differential regulation of translation of these channel-encoding messages.
To functionally assess CNG channel subunit composition, we performed a biophysical analysis of the native rat olfactory CNG channel. Five functional criteria were used to compare native channels with heterologously expressed channels of various subunit composition: (1) the sensitivity for activation by cAMP, (2) the discrimination between Na+ and K+, (3) the single-channel conductance, (4) the kinetics of open–closed transitions, and (5) the ability to adopt a subconductance state. Homomeric α3 channels as well as α3α4 and α3β1b channels display clear differences in all of these criteria relative to the native channel, ruling out that these combinations represent the native subunit composition. In contrast, α3α4β1b channels resemble the native channel in each of the five criteria tested, strongly supporting the idea that the native olfactory channel consists of the CNCα3, CNCα4, and CNCβ1b subunits. The subtle differences between native and α3α4β1b channels with respect to the apparent single-channel conductance and the kinetics of open–closed transitions may be the consequence of expression in OSNs versus HEK 293 cells, respectively. In fact, previous studies have shown that the kinetic properties of α3α4 channels are influenced by the expression system used. The channels display desensitization when expressed in Xenopusoocytes (Liman and Buck, 1994) but not in HEK 293 cells (Bradley et al., 1994). Even more important, glycosylation of the α3 subunit may affect gating and permeation of the native channel in subtle ways that are absent in the nonglycosylated recombinant channels.
Because of the small size of cilia and the large channel density (Kurahashi and Kaneko, 1991; Kleene et al., 1994; Kleene, 1997), it is not feasible to obtain single-channel data from patches of olfactory cilia or the dendritic knob; instead we resorted to properties of single channels in membranes of the soma and dendrite. This inherent difficulty provokes the question regarding whether somatic and ciliary CNG channels are the same. For example, Torre and coworkers (Torre et al., 1992; Sesti et al., 1994) observed a subpopulation of CNG channels in the inner segment of rod photoreceptors that differs from the channel in the outer segment. However, we consistently observed only a single type of channel in membrane patches of the soma or dendrite of OSNs. Moreover, the cAMP sensitivity and the conductance ratio for Na+ and K+ measured in channels from the soma of OSNs, and from recombinant α3α4β1b channels, are very similar to the respective channel properties measured in the membrane patches from the dendritic knob. Because dendritic knob and sensory cilia together form the apical membrane of OSNs, it is reasonable to assume that they contain the same membrane proteins and have the same CNG channels. Although these observations do not entirely rule out the existence of mixed channel populations, they strongly suggest that α3α4β1b is the predominant channel species in cilia, the dendrite, and the soma.
Implication for olfactory transduction
The specific contributions of CNCα3, CNCα4, and CNCβ1b to the properties of the native channel have important ramifications for several functional aspects of olfactory transduction. This is because certain key structural features are distinctively different between the three subunits (cAMP-binding site, pore motif, and N terminus). Some of the implications of channel subunit composition are discussed below.
Olfactory CNG channels are highly permeable to Ca2+(Frings et al., 1995; Dzeja et al., 1999). A pronounced Ca2+ influx through the CNG channels during odor stimulation causes a rapid increase of the Ca2+concentration in the lumen of sensory cilia (Leinders-Zufall et al., 1997, 1998). This Ca2+ signal controls both excitation and adaptation of the sensory neuron. Ciliary Ca2+-activated Cl− channels conduct a depolarizing Cl− current that probably represents a large fraction of the receptor current (Kleene and Gesteland, 1991; Kurahashi and Yau, 1993; Lowe and Gold, 1993; Hallani et al., 1998; Reuter et al., 1998). The Ca2+ signal also terminates the sensory response through a CaM-mediated decrease of CNG channel activity (Chen and Yau, 1994; Kurahashi and Menini, 1997). Because of the prominent role of Ca2+ in olfactory signal transduction, it is important to understand how the CNCα3, -α4, and -β1b subunits contribute to the Ca2+conductance of the native channel. All known α subunits contain a negatively charged residue in the pore motif (Glu342in CNCα3, Asp234 in CNCα4). These residues form an intrapore Ca2+-binding site of high affinity (Frings et al., 1995; Seifert et al., 1999), which is crucial for ion selectivity, gating, and blockage by extracellular Ca2+ ions (Root and MacKinnon, 1993; Eismann et al., 1994; Sesti et al., 1995; Bucossi et al., 1996). The olfactory CNCβ1b subunit carries an uncharged glycine residue (Gly463) at the homologous position of its pore motif. As discussed in Dzeja et al. (1999) and Seifert et al. (1999), pores containing β subunit(s) are less “sticky” than pores consisting only of α subunits. Consequently, Ca2+is expected to pass more readily through heterooligomeric (α + β) compared with homomeric (only α) channels. Therefore, the heterooligomeric channels containing this β subunit should have different Ca2+ permeability from that of channels consisting of α subunits alone.
Adaptation in OSNs is a Ca2+-dependent process, driven by the binding of Ca2+/CaM to the channel. A CaM-binding site in the N-terminal region of the CNCα3 subunit (Liu et al., 1994) binds CaM with high affinity in a Ca2+-dependent manner, thereby reducing the apparent ligand sensitivity. The N terminus of CNCα4 is shorter than that of CNCα3 by 108 amino acid residues, and it lacks the CaM-binding site. Recently, a CaM-binding site was identified in the N-terminal region of the retinal CNCβ1a subunit and was shown to bestow CaM sensitivity to heterologously expressed α1β1a channels (Grunwald et al., 1998;Weitz et al., 1998). This site is also present in the olfactory CNCβ1b subunit. Recent studies have shown that native olfactory CNG channels show characteristic differences in response to CaM compared with α3 homomeric channels. The K1/2 for cAMP of the native channel can increase by 20- to 60-fold (compared with only tenfold in CNCα3 homomers), and in contrast to CNCα3 homomers, the open probability at saturating cAMP concentrations is not suppressed (Chen and Yau, 1994; Balasubramanian et al., 1996). These differences may indicate that the CNCα4 and CNCβ1b subunits participate in the CaM effect on the native channel. Therefore, investigation of α3α4β1b channels should further our understanding of the negative feedback regulation that leads to adaptation in OSNs.
Finally, we have recently shown that the ligand sensitivity of α3 channels can be increased tenfold by protein phosphorylation (Müller et al., 1998). When a single serine residue in the N terminus (Ser95) is phosphorylated by protein kinase C (PKC), the channel is activated at lower cAMP concentrations. This serine residue is conserved in all CNCα3 orthologs but is missing in CNCα4. The N terminus of CNCβ1b contains several consensus sequences for PKC, but any functional roles of these sites await assessment. Future coexpression studies with the CNCα3, -α4, and -β1b subunits should yield important insights into the regulation of the native olfactory CNG channel. This will promote our understanding not only of olfactory signal transduction, but also of other sensory and nonsensory signal transduction systems that use these CNG channel subunits.
Footnotes
This work was supported by grants from the European Community (CHRX-CT94-0543) and the Ministerium für Wissenschaft und Forschung des Landes Nordrhein-Westfalen (IVA6-10201095) to U.B.K, by the Human Frontiers Science Program (LT-448-96) to J.B., by the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm “Molekulare Sinnesphysiologie”) to S.F., and by the Develbiss Fund, the McKnight Foundation, and National Institute on Deafness and Other Communication Disorders/National Institutes of Health Grant RO1DC02979 to G.V.R. We thank Dr. Philippe Ascher for valuable comments on this manuscript. We gratefully acknowledge the assistance of Kerstin Bach, Mechthilde Bruns, and Helga Vent. We thank Dr. Heinz Körschen for the anti-CNCβ1a antibody FPc21K.
The CNCβ1b sequence has been deposited in GenBank (accession numberAF068572).
Correspondence should be addressed to Professor U. B. Kaupp, Forschungszentrum Jülich, IBI-1, 52425 Jülich, Germany.
Dr. Sesti’s present address: Yale University, School of Medicine, BCMM, 295 Congress Avenue, New Haven, CT 06536.
REFERENCES
- 1.Anholt RRH, Aebi U, Snyder SH. A partially purified preparation of isolated chemosensory cilia from the olfactory epithelium of the bullfrog, Rana catesbeiana. J Neurosci. 1986;6:1962–1965. doi: 10.1523/JNEUROSCI.06-07-01962.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardell MD, Aragon I, Oliveira L, Porche GE, Burke E, Pittler SJ. The β subunit of human rod photoreceptor cGMP-gated cation channel is generated from a complex transcription unit. FEBS Lett. 1996;389:213–218. doi: 10.1016/0014-5793(96)00588-1. [DOI] [PubMed] [Google Scholar]
- 3.Bakalyar HA, Reed RR. Identification of a specialized adenylyl cyclase that may mediate odorant detection. Science. 1990;250:1403–1406. doi: 10.1126/science.2255909. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramanian S, Lynch JW, Barry PH. Calcium-dependent modulation of the agonist affinity of the mammalian olfactory cyclic nucleotide-gated channel by calmodulin and a novel endogenous factor. J Membr Biol. 1996;152:13–23. doi: 10.1007/s002329900081. [DOI] [PubMed] [Google Scholar]
- 5.Baumann A, Frings S, Godde M, Seifert R, Kaupp UB. Primary structure and functional expression of a Drosophila cyclic nucleotide-gated channel present in eyes and antennae. EMBO J. 1994;13:5040–5050. doi: 10.1002/j.1460-2075.1994.tb06833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bönigk W, Altenhofen W, Müller F, Dose A, Illing M, Molday RS, Kaupp UB. Rod and cone photoreceptor cells express distinct genes for cGMP-gated channels. Neuron. 1993;10:865–877. doi: 10.1016/0896-6273(93)90202-3. [DOI] [PubMed] [Google Scholar]
- 7.Bradley J, Li J, Davidson N, Lester HA, Zinn K. Heteromeric olfactory cyclic nucleotide-gated channels: a new subunit that confers increased sensitivity to cAMP. Proc Natl Acad Sci USA. 1994;91:8890–8894. doi: 10.1073/pnas.91.19.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley J. PhD thesis. Caltech; 1996. Molecular analysis of olfactory signal transduction. [Google Scholar]
- 9.Bradley J, Zhang Y, Bakin R, Lester HA, Ronnett GV, Zinn K. Functional expression of the heteromeric “olfactory” cyclic nucleotide-gated channel in the hippocampus: a potential effector of synaptic plasticity in brain neurons. J Neurosci. 1997;17:1993–2005. doi: 10.1523/JNEUROSCI.17-06-01993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broillet M-C, Firestein S. β subunits of the olfactory cyclic nucleotide-gated channel form a nitric oxide activated Ca2+ channel. Neuron. 1997;18:951–958. doi: 10.1016/s0896-6273(00)80334-7. [DOI] [PubMed] [Google Scholar]
- 11.Bucossi G, Eismann E, Sesti F, Nizzari M, Seri M, Kaupp UB, Torre V. Time-dependent current decline in cyclic GMP-gated bovine channels caused by point mutations in the pore region expressed in Xenopus oocytes. J Physiol (Lond) 1996;493:409–418. doi: 10.1113/jphysiol.1996.sp021392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bucossi G, Nizzari M, Torre V. Single-channel properties of ionic channels gated by cyclic nucleotides. Biophys J. 1997;72:1165–1181. doi: 10.1016/S0006-3495(97)78765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T-Y, Yau K-W. Direct modulation by Ca2+-calmodulin of cyclic nucleotide-activated channel of rat olfactory receptor neurons. Nature. 1994;368:545–548. doi: 10.1038/368545a0. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, Pace U, Heldman J, Shapira A, Lancet D. Isolated frog olfactory cilia: a preparation of dendritic membranes from chemosensory neurons. J Neurosci. 1986;6:2146–2154. doi: 10.1523/JNEUROSCI.06-08-02146.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T-Y, Peng Y-W, Dhallan RS, Ahamed B, Reed RR, Yau K-W. A new subunit of the cyclic nucleotide-gated cation channel in retinal rods. Nature. 1993;362:764–767. doi: 10.1038/362764a0. [DOI] [PubMed] [Google Scholar]
- 16.Chen T-Y, Illing M, Molday LL, Hsu Y-T, Yau K-W, Molday RS. Subunit 2 (or β) of retinal rod cGMP-gated cation channel is a component of the 240-kDa channel-associated protein and mediates Ca2+-calmodulin modulation. Proc Natl Acad Sci USA. 1994;91:11757–11761. doi: 10.1073/pnas.91.24.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook NJ, Hanke W, Kaupp UB. Identification, purification, and functional reconstitution of the cyclic GMP-dependent channel from rod photoreceptors. Proc Natl Acad Sci USA. 1987;84:585–589. doi: 10.1073/pnas.84.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhallan RS, Yau K-W, Schrader KA, Reed RR. Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature. 1990;347:184–187. doi: 10.1038/347184a0. [DOI] [PubMed] [Google Scholar]
- 19.Dzeja C, Hagen V, Kaupp UB, Frings S. Ca2+ permeation in cyclic nucleotide-gated channels. EMBO J. 1999;18:131–144. doi: 10.1093/emboj/18.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eismann E, Müller F, Heinemann SH, Kaupp UB. A single negative charge within the pore region of a cGMP-gated channel controls rectification, Ca2+ blockage and ionic selectivity. Proc Natl Acad Sci USA. 1994;91:1109–1113. doi: 10.1073/pnas.91.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn JT, Grunwald ME, Yau K-W. Cyclic nucleotide-gated ion channels: an extended family with diverse functions. Annu Rev Physiol. 1996;58:395–426. doi: 10.1146/annurev.ph.58.030196.002143. [DOI] [PubMed] [Google Scholar]
- 22.Frings S, Lynch JW, Lindemann B. Properties of cyclic nucleotide-gated channels mediating olfactory transduction: activation, selectivity, and blockage. J Gen Physiol. 1992;100:45–67. doi: 10.1085/jgp.100.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frings S, Seifert R, Godde M, Kaupp UB. Profoundly different calcium permeation and blockage determine the specific function of distinct cyclic nucleotide-gated channels. Neuron. 1995;15:169–179. doi: 10.1016/0896-6273(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 24.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon SE, Oakley JC, Varnum MD, Zagotta WN. Altered ligand specificity by protonation in the ligand binding domain of cyclic nucleotide-gated channels. Biochemistry. 1996;35:3994–4001. doi: 10.1021/bi952607b. [DOI] [PubMed] [Google Scholar]
- 26.Goulding EH, Ngai J, Kramer RH, Colicos S, Axel R, Siegelbaum SA, Chess A. Molecular cloning and single-channel properties of the cyclic nucleotide-gated channel from catfish olfactory neurons. Neuron. 1992;8:45–58. doi: 10.1016/0896-6273(92)90107-o. [DOI] [PubMed] [Google Scholar]
- 27.Grunwald ME, Yu W-P, Yu H-H, Yau K-W. Identification of a domain on the β-subunit of the rod cGMP-gated cation channel that mediates inhibition by calcium-calmodulin. J Biol Chem. 1998;273:9148–9157. doi: 10.1074/jbc.273.15.9148. [DOI] [PubMed] [Google Scholar]
- 28.Hallani M, Lynch JW, Barry PH. Characterization of calcium-activated chloride channels in patches excised from the dendritic knob of mammalian olfactory receptor neurons. J Membr Biol. 1998;161:163–171. doi: 10.1007/s002329900323. [DOI] [PubMed] [Google Scholar]
- 29.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 30.Henn DK, Baumann A, Kaupp UB. Probing the transmembrane topology of cyclic nucleotide-gated ion channels with a gene fusion approach. Proc Natl Acad Sci USA. 1995;92:7425–7429. doi: 10.1073/pnas.92.16.7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu Y-T, Molday RS. Modulation of the cGMP-gated channel of rod photoreceptor cells by calmodulin. Nature. 1993;361:76–79. doi: 10.1038/361076a0. [DOI] [PubMed] [Google Scholar]
- 32.Jan LY, Jan YN. A superfamily of ion channels. Nature. 1990;345:672. doi: 10.1038/345672a0. [DOI] [PubMed] [Google Scholar]
- 33.Jechlinger M, Pelz R, Tretter V, Klausberger T, Sieghart W. Subunit composition and quantitative importance of hetero-oligomeric receptors: GABAA receptors containing α6 subunits. J Neurosci. 1998;18:2449–2457. doi: 10.1523/JNEUROSCI.18-07-02449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaprielian Z, Cho KO, Hadjargyrou M, Patterson PH. CD9, a major platelet cell surface glycoprotein, is a ROCA antigen and is expressed in the nervous system. J Neurosci. 1995;15:565–573. doi: 10.1523/JNEUROSCI.15-01-00562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaupp UB. Family of cyclic nucleotide gated ion channels. Curr Opin Neurobiol. 1995;5:434–442. doi: 10.1016/0959-4388(95)80002-6. [DOI] [PubMed] [Google Scholar]
- 36.Kaupp UB, Niidome T, Tanabe T, Terada S, Bönigk W, Stühmer W, Cook NJ, Kangawa K, Matsuo H, Hirose T, Miyata T, Numa S. Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP-gated channel. Nature. 1989;342:762–766. doi: 10.1038/342762a0. [DOI] [PubMed] [Google Scholar]
- 37.Kleene SJ. High-gain, low-noise amplification in olfactory transduction. Biophys J. 1997;73:1110–1117. doi: 10.1016/S0006-3495(97)78143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleene SJ, Gesteland RC. Calcium-activated chloride conductance in frog olfactory cilia. J Neurosci. 1991;11:3624–3629. doi: 10.1523/JNEUROSCI.11-11-03624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleene SJ, Gesteland RC, Bryant SH. An electrophysiological survey of frog olfactory cilia. J Exp Biol. 1994;195:307–328. doi: 10.1242/jeb.195.1.307. [DOI] [PubMed] [Google Scholar]
- 40.Körschen HG, Illing M, Seifert R, Sesti F, Williams A, Gotzes S, Colville C, Müller F, Dosé A, Godde M, Molday L, Kaupp UB, Molday RS. A 240 kDa protein represents the complete β subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron. 1995;15:627–636. doi: 10.1016/0896-6273(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 41.Kurahashi T, Kaneko A. High density cAMP-gated channels at the ciliary membrane in the olfactory receptor cell. NeuroReport. 1991;2:5–8. doi: 10.1097/00001756-199101000-00001. [DOI] [PubMed] [Google Scholar]
- 42.Kurahashi T, Menini A. Mechanism of odorant adaptation in the olfactory receptor cell. Nature. 1997;385:725–729. doi: 10.1038/385725a0. [DOI] [PubMed] [Google Scholar]
- 43.Kurahashi T, Yau K-W. Co-existence of cationic and chloride components in odorant-induced current of vertebrate olfactory receptor cells. Nature. 1993;363:71–74. doi: 10.1038/363071a0. [DOI] [PubMed] [Google Scholar]
- 44.Leinders-Zufall T, Rand MN, Shepherd GM, Greer CA, Zufall F. Calcium entry through cyclic nucleotide-gated channels in individual cilia of olfactory receptor cells: spatiotemporal dynamics. J Neurosci. 1997;17:4136–4148. doi: 10.1523/JNEUROSCI.17-11-04136.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leinders-Zufall T, Greer CA, Shepherd GM, Zufall F. Imaging odor-induced calcium transients in single olfactory cilia: specificity of activation and role in transduction. J Neurosci. 1998;18:5630–5639. doi: 10.1523/JNEUROSCI.18-15-05630.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liman ER, Buck LB. A second subunit of the olfactory cyclic nucleotide-gated channel confers high sensitivity to cAMP. Neuron. 1994;13:611–621. doi: 10.1016/0896-6273(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 47.Liu DT, Tibbs GR, Siegelbaum SA. Subunit stoichiometry of cyclic nucleotide-gated channels and effects of subunit order on channel function. Neuron. 1996;16:983–990. doi: 10.1016/s0896-6273(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 48.Liu M, Chen T-Y, Ahamed B, Li J, Yau K-W. Calcium-calmodulin modulation of the olfactory cyclic nucleotide-gated cation channel. Science. 1994;266:1348–1354. doi: 10.1126/science.266.5189.1348. [DOI] [PubMed] [Google Scholar]
- 49.Lowe G, Gold GH. Nonlinear amplification by calcium-dependent chloride channels in olfactory receptor cells. Nature. 1993;366:283–286. doi: 10.1038/366283a0. [DOI] [PubMed] [Google Scholar]
- 50.Ludwig J, Margalit T, Eismann E, Lancet D, Kaupp UB. Primary structure of cAMP-gated channel from bovine olfactory epithelium. FEBS Lett. 1990;270:24–29. doi: 10.1016/0014-5793(90)81226-e. [DOI] [PubMed] [Google Scholar]
- 51.Menco BPM, Bruch RC, Dau B, Danho W. Ultrastructural localization of olfactory transduction components: the G protein subunit Golfα and type III adenylyl cyclase. Neuron. 1992;8:441–453. doi: 10.1016/0896-6273(92)90272-f. [DOI] [PubMed] [Google Scholar]
- 52.Molday LL, Cook NJ, Kaupp UB, Molday RS. The cGMP-gated cation channel of bovine rod photoreceptor cells is associated with a 240-kDa protein exhibiting immunochemical cross-reactivity with spectrin. J Biol Chem. 1990;265:18690–18695. [PubMed] [Google Scholar]
- 53.Müller F, Bönigk W, Sesti F, Frings S. Phosphorylation of mammalian olfactory cyclic nucleotide-gated channels increases ligand sensitivity. J Neurosci. 1998;18:164–173. doi: 10.1523/JNEUROSCI.18-01-00164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pace U, Hanski E, Salomon Y, Lancet D. Odorant-sensitive adenylate cyclase may mediate olfactory reception. Nature. 1985;316:255–258. doi: 10.1038/316255a0. [DOI] [PubMed] [Google Scholar]
- 55.Pfeuffer E, Mollner S, Lancet D, Pfeuffer T. Olfactory adenylyl cyclase. J Biol Chem. 1989;264:18803–18807. [PubMed] [Google Scholar]
- 56.Reuter D, Zierold K, Schröder W, Frings S. A depolarizing chloride current contributes to chemo-electrical transduction in olfactory sensory neurons in situ. J Neurosci. 1998;18:6623–6630. doi: 10.1523/JNEUROSCI.18-17-06623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Root MJ, MacKinnon R. Identification of an external divalent cation-binding site in the pore of a cGMP-activated channel. Neuron. 1993;11:459–466. doi: 10.1016/0896-6273(93)90150-p. [DOI] [PubMed] [Google Scholar]
- 58.Sautter A, Zong X, Hofmann F, Biel M. An isoform of the rod photoreceptor cyclic nucleotide-gated channel β subunit expressed in olfactory neurons. Proc Natl Acad Sci USA. 1998;95:4696–4701. doi: 10.1073/pnas.95.8.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissues and cultured cells: in situ hybridization using digoxigenin-labeled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 60.Schaffner W, Weissmann C. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- 61.Seifert R, Eismann E, Ludwig J, Baumann A, Kaupp UB. Molecular determinants of a Ca2+-binding site in the pore of cyclic nucleotide-gated channels: S5/S6 segments control affinity of intrapore glutamates. EMBO J. 1999;18:119–130. doi: 10.1093/emboj/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sesti F, Straforini M, Lamb TD, Torre V. Gating, selectivity and blockage of single channels activated by cyclic GMP in retinal rods of the tiger salamander. J Physiol (Lond) 1994;474:203–222. doi: 10.1113/jphysiol.1994.sp020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sesti F, Eismann E, Kaupp UB, Nizzari M, Torre V. The multi-ion nature of cGMP-gated channel from vertebrate rods. J Physiol (Lond) 1995;487:17–36. doi: 10.1113/jphysiol.1995.sp020858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shamotienko OG, Parcej DN, Dolly JO. Subunit combinations defined for K+ channel Kv1 subtypes in synaptic membranes from bovine brain. Biochemistry. 1997;36:8195–8201. doi: 10.1021/bi970237g. [DOI] [PubMed] [Google Scholar]
- 65.Torre V, Straforini M, Sesti F, Lamb TD. Different channel-gating properties of two classes of cyclic GMP-activated channel in vertebrate photoreceptors. Proc R Soc Lond B Biol Sci. 1992;250:209–215. doi: 10.1098/rspb.1992.0151. [DOI] [PubMed] [Google Scholar]
- 66.Tretter V, Ehya N, Fuchs K, Sieghart W. Stoichiometry and assembly of a recombinant GABAA receptor subtype. J Neurosci. 1997;17:2728–2737. doi: 10.1523/JNEUROSCI.17-08-02728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vasser R, Ngai J, Axel R. Spatial segregation of odorant-receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 68.Weitz D, Zoche M, Müller F, Beyermann M, Körschen HG, Kaupp UB, Koch K-W. Calmodulin controls the rod photoreceptor CNG channel through an unconventional binding site in the N-terminus of the β-subunit. EMBO J. 1998;17:2273–2284. doi: 10.1093/emboj/17.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weyand I, Godde M, Frings S, Weiner J, Müller F, Altenhofen W, Hatt H, Kaupp UB. Cloning and functional expression of a cyclic-nucleotide-gated channel from mammalian sperm. Nature. 1994;368:859–863. doi: 10.1038/368859a0. [DOI] [PubMed] [Google Scholar]
- 70.Wiesner BWJ, Middendorf R, Hagen V, Kaupp UB, Weyand I. Cyclic nucleotide-gated channels on the flagellum control Ca2+ entry into sperm. J Cell Biol. 1998;142:473–484. doi: 10.1083/jcb.142.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zagotta WN, Siegelbaum SA. Structure and function of cyclic nucleotide-gated channels. Annu Rev Neurosci. 1996;19:235–263. doi: 10.1146/annurev.ne.19.030196.001315. [DOI] [PubMed] [Google Scholar]