Abstract
A mathematical model for the intracellular circadian rhythm generator has been studied, based on a negative feedback of protein products on the transcription rate of their genes. The study is an attempt at examining minimal but biologically realistic requirements for a negative molecular feedback loop involving considerably faster reactions, to produce (slow) circadian oscillations. The model included mRNA and protein production and degradation, along with a negative feedback of the proteins upon mRNA production. The protein production process was described solely by its total duration and a nonlinear term, whereas also the feedback included nonlinear interactions among protein molecules. This system was found to produce robust oscillations in protein and mRNA levels over a wide range of parameter values. Oscillations were slow, with periods much longer than the time constants of any of the individual system parameters. Circadian oscillations were obtained for realistic values of the parameters. The system was readily entrainable to external periodic perturbations. Two distinct classes of phase response curves were found, viz. with or without a time domain within the circadian cycle in which external perturbations fail to induce a phase shift (“dead zone”). The delay and nonlinearity in the protein production and the cooperativity in the negative feedback (Hill coefficient) were for this model found to be necessary and sufficient to generate robust circadian oscillations. The similarities between model outcomes and empirical findings establish that circadian rhythmicity at the cellular level can plausibly emerge from interactions among molecular systems which are not in themselves rhythmic.
Keywords: SCN, circadian rhythm, molecular clock, entrainment, phase–response curves, models
Free-running behavioral rhythms with a period close to 24 hr (“circadian”) are found in many species and constitute a fundamental mechanism for entrainment to environmental day–night cycles. These rhythms are further characterized by typical shifting of phase after external (light) stimulation, being dependent on the phase point of application. Experimental evidence suggests an intracellular origin for the generation of such rhythms, with a critical involvement of negative feedback in the protein synthesis system (Hardin et al., 1990, 1992; Aronson et al., 1994; Sassone-Corsi, 1994).
The protein synthesis cascade involves much faster reactions than the circadian period, making it not easily understood what are the critical conditions for such highly stable and slow periodic protein oscillations to occur. Mathematical models have been used by Goldbeter (1995), Lewis et al. (1997), and L. F. Abbott (personal communication), to study circadian rhythm generation inDrosophila, whereas Ruoff and Rensing (1996) concentrated on temperature compensation using Goodwin’s (1965) model. Different mechanisms were proposed by these authors to obtain slow oscillations.Goldbeter (1995) included protein phosphorylation (twofold) and nuclear entry reactions of a bidirectional (equilibrium) type as well as a Hill coefficient of n = 4 to account for cooperativity in the negative feedback. Abbott (personal communication) did not include a Hill coefficient but assumed up to 11 unidirectional phosphorylation reactions to get a long enough delay in nuclear entry to generate a 24 hr rhythm. Ruoff and Rensing (1996) included a single after-processing step only, but needed to assume an unrealistically high Hill coefficient of n = 9 to obtain circadian oscillations. In a modified Goodwin model, Griffith (1968) found stable limit cycles only with a Hill coefficient of eight or more. Lewis et al. (1997) assumed a threshold type of reaction in the feedback loop and included a delay of 8 hr for phosphorylation of period protein (PER) and transport of the PER/TIM (timeless protein) complex into the nucleus. Clearly, all these approaches were concerned primarily with the question of how to generate slow oscillations from a feedback loop consisting only of reactions with much faster kinetics. The models differ among themselves in the selection of reaction steps in the protein synthesis system that are modeled explicitly. When the reaction constants are used as optimization parameters, however, their outcomes will necessarily strongly depend on the completeness and the precise implementation of all reaction steps. Additionally, these approaches require the availability of empirical data about the kinetics of each of these reaction steps.
Our model avoids the strong constraint of specifying all the processes involved in the production of the “effective protein” (mRNA translation, protein postprocessing, transport, and nuclear entry) by characterizing the full chain of reactions solely in terms of (1) the total duration and (2) the nonlinear relationship between input and (delayed) output of the reaction chain. Effective protein refers to the molecular state directly capable of inhibiting mRNA production. We show that these properties are sufficient for generating stable and robust circadian oscillations for biologically realistic parameter values. Additionally, the model shows realistic entrainment properties and phase–response curves when subjected to periodic or single external stimulation, respectively.
MATERIALS AND METHODS
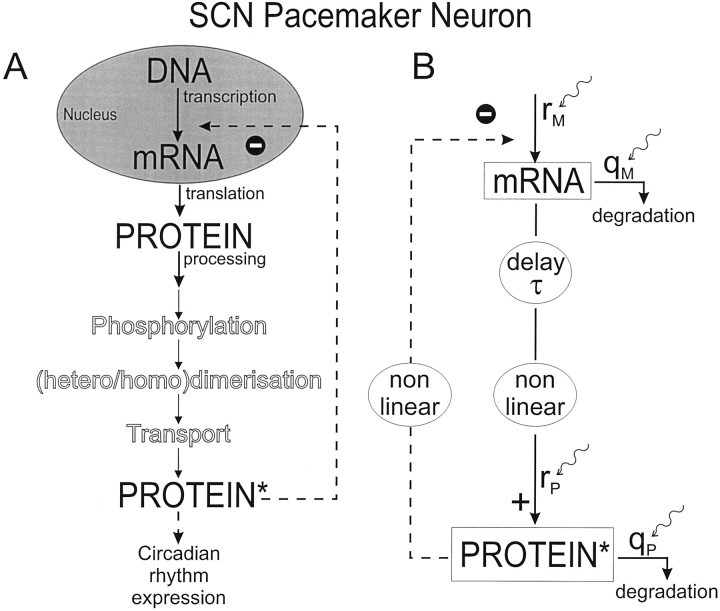
Structure of the model. The essential components of the reaction loop assumed to be involved in the generation of the circadian rhythm are depicted in Figure1A. The reaction loop consists of a cascade for the production of the effective protein from its mRNA and a negative feedback from the effective protein on the production of its mRNA. The protein production cascade involves the translation and subsequent processing steps, such as phosphorylation (Edery et al., 1994b), dimerization, transport, and nuclear entry (Zerr et al., 1990; Young, 1996; Saez and Young, 1996). Figure1B depicts a functional abstraction of the reaction loop emphasizing that the protein production cascade and the negative feedback are assumed to be nonlinear processes, whereas the total time involved in the protein production and subsequent processing (up to its negative effect on its own mRNA) is represented by a single delay term. It is assumed that these nonlinearities and the delay term are the critical parameters in the feedback loop that determine the free-running periodicity.
Fig. 1.
A, Schematic representation of the biological elements of the protein synthesis cascade, assumed to be elementary to the circadian rhythm generator. These include the auto inhibition of the protein at translational or transcriptional level and posttranslational processing such as phosphorylation, dimerization, and transport. Protein* denotes the effective protein, being in the molecular state capable of inhibiting mRNA production, as well as expressing the circadian rhythm. B, Model interpretation of A, emphasizing the delay (τ) and nonlinearity in the protein production cascade, the nonlinear negative feedback, as well as the mRNA and protein production and degradation. The mRNA and protein production (rM,rP) and degradation (qM,qP) rate constants, respectively, are also used as targets for external stimulation (curly arrows).
To analyze the circadian behavior of the intracellular circadian oscillator, the model is defined as follows:
| Equation 1 |
| Equation 2 |
with M and P denoting relative concentrations of mRNA and the effective protein, respectively,rM the scaled mRNA production rate constant,rP the protein production rate constant,qM and qP the mRNA and protein degradation rate constants, respectively, n the Hill coefficient, the exponent m the nonlinearity in the protein production cascade, the delay τ the total duration of protein production from mRNA, and k a scaling constant. A derivation of these scaled equations is given in the . The formulation in Equations 1 and 2 builds on the work of Goodwin (1965), who was the first to study theoretically the negative feedback loop in protein synthesis.
The values for the seven parameters in the model have been chosen according to the following considerations (Table1). Based on data in Kornhauser et al. (1990) and Zhang et al. (1996), a delay of 4 hr has been chosen. The exponent m implements the nonlinearity in the protein production term [i.e., a value of m different from one indicates that the (delayed) protein production does not follow the mRNA levels in a linear manner]. This occurs, for instance, when multiple mRNA molecules are the substrate in the production of a protein or when further processing steps introduce nonlinear interactions. The Hill coefficient n was given a value of 2 in view of the high probability of protein–protein interactions during the circadian oscillation and the putative role of dimerization in negative feedback (Young, 1996; Zeng et al., 1996; King et al., 1997). This value is lower than used by Goldbeter (1995) and Ruoff and Rensing (1996). Degradation rate constants were chosen from biological half-lives, for mRNA in the range of 0.07 and 1.39 hr−1 and for rapid degradation proteins from 0.35 to 4.16 hr−1 (Alberts et al., 1989). We chose a degradation rate constant of 0.21 hr−1, and a production rate constant of 1 hr−1 for both mRNA and protein. The scaling constant k was given the value of 1 throughout.
Table 1.
Parameter values for the unperturbed model
| Parameter | Values resulting in a free-running rhythm of 24.6 hr | Transition from steady state to limit cycle at values |
|---|---|---|
| rM, mRNA production rate constant | 1.0 hr−1 | 0.27 hr−1 |
| rP, protein production rate constant | 1.0 hr−1 | 0.02 hr−1 |
| qM, mRNA degradation rate constant | 0.21 hr−1 | 0.08 hr−1 |
| qP, protein degradation rate constant | 0.21 hr−1 | 0.08 hr−1 |
| n, Hill coefficient | 2.0 | 1.27 |
| m, nonlinearity in protein synthesis cascade | 3.0 | 1.81 |
| τ, duration of protein synthesis cascade | 4.0 hr | 2.21 hr |
| k, scaling constant | 1 |
Method for solving the nonlinear delay equations. Delay equations are notoriously difficult to solve analytically (Gumowski, 1981; Murray, 1989). The model was therefore analyzed by numerical integration of the equations, Equation 1 and Equation 2, with a Runge–Kutta fourth-order differential integrator with step sizes of 0.01–1 hr. A step size of 0.1 hr proved to be the most efficient. Periods were calculated by determining the periodogram by means of power spectrum estimation (Press et al., 1992).
Analysis of the unperturbed (free-running) system. Because the system is defined by seven parameters {n,m, τ, rM,rP, qM,qP} it is not feasible to explore the behavior of the system throughout the full seven-dimensional parameter space. We have therefore chosen to investigate the behavior along single dimensions around the set point (Table 1), by changing only one parameter at a time while keeping the others at their set point value. Additionally, the {n, m, τ} subspace was investigated more extensively to assess the critical role of the nonlinearity and delay parameters. It was especially important to find those areas in parameter space in which the system has stable limit cycles and those where the system has stable steady states. At the boundaries between these areas, transitions in the qualitative behavior of the system occur (bifurcations).
Analysis of the perturbed system. The effect of external input was studied by briefly changing the production and the degradation rate constants in the model. This choice was based on the assumption that production and degradation rates are more sensitive to external interference than the parameters n, m, and τ, which reflect more structural than dynamic aspects of the chemical reactions involved. Examples are light-induced TIM degradation (Hunter-Ensor et al., 1996; Myers et al., 1996) whereas PER or TIM induction shifts the phase of the clock (Edery et al., 1994a). To this end, Equations 1 and 2 were extended with perturbation termsSrM, SrP,SqM, and SqP for the parameters rM,rP, qM, andqP, respectively:
| Equation 3 |
| Equation 4 |
The perturbation terms SrM,SqM, SrP, and SqP were kept at zero value in the unperturbed model, but assumed either a positive or a negative value during stimulation in the perturbed model.
“Entrainment” was studied by investigating the oscillatory behavior of the system under conditions of periodic stimulation, which was simulated by switching on at regular intervals the perturbation term for 1 hr. “Entrainment windows”, (i.e., range of stimulus periods resulting in entrainment) were determined by changing the stimulus period and monitoring for which range of stimulus periods the system remained phase-locked to the stimulus. “Phase responses” have been determined by applying a brief single stimulus of 1 hr to the system at different points during the free-running cycle, and quantifying the induced phase shift in the oscillation. This phase shift has been determined from the time difference in peak values between the free-running and the perturbed oscillation after the transient effect of the stimulus had damped out. Both the mRNA and protein production and degradation rates were subjected to a single pulse stimulation or inhibition. “Phase–response curves” (PRCs) were obtained by plotting the phase shift as a function of the time point of application during the cycle.
RESULTS
Unperturbed (free-running) system
Dynamic states
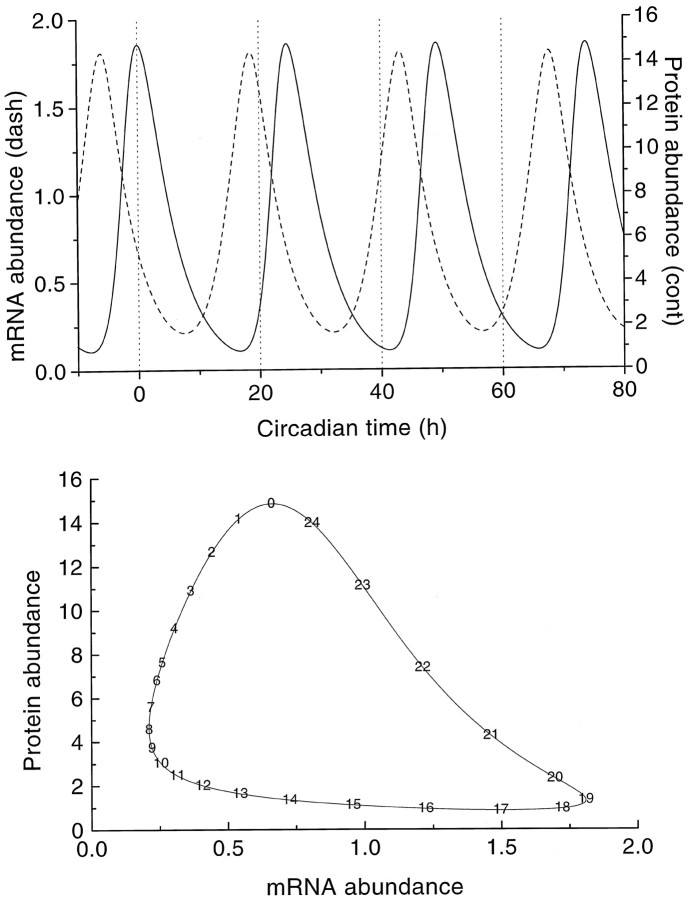
Stable oscillations with a period of 24.6 hr were obtained for the parameter values given in Table 1, with mRNA and protein concentrations fluctuating over a wide range (Fig.2A). The lag time between the peak concentrations of mRNA and protein was 6 hr. The time course of both oscillations deviated significantly from sinusoidal as is also shown by the limit cycle contour (Fig. 2B) with the mRNA concentration plotted versus the protein concentration. Nonsinusoidal behavior can be expected because the set of equations, Equation 1 and Equation 2, differs from that for a harmonic oscillator. The limit cycle contour is annotated with the time points in hours of circadian model time (CMT), with the zero time point (0 hr CMT) set arbitrarily at the maximum protein concentration. The mRNA concentration fluctuated with a rising phase of ≈11.1 hr and a falling phase of ≈13.5 hr, whereas the protein concentration oscillated with a rising phase of ≈7.8 hr and a falling phase of ≈16.8 hr. The dynamic behavior of the system (Eqs. 1 and 2) was studied by an extensive search in parameter space for possible bifurcations. Apart from stable points and stable limit cycles, no other behavior was encountered. Nevertheless, the possibility of chaotic behavior cannot be excluded for remote areas of the parameter space. Additionally, subharmonic oscillations might possibly be found close to the bifurcation points.
Fig. 2.
Free-running oscillation for the parameter values given in Table 1. Top panel, Time plot of the oscillating protein concentration (continuous line) and the oscillating mRNA concentration (dashed line).Bottom panel, Corresponding limit cycle contour, annotated with the time points within the 24.6 hr circadian cycle. The zero time point (0 hr CMT) is taken at maximum protein concentration.
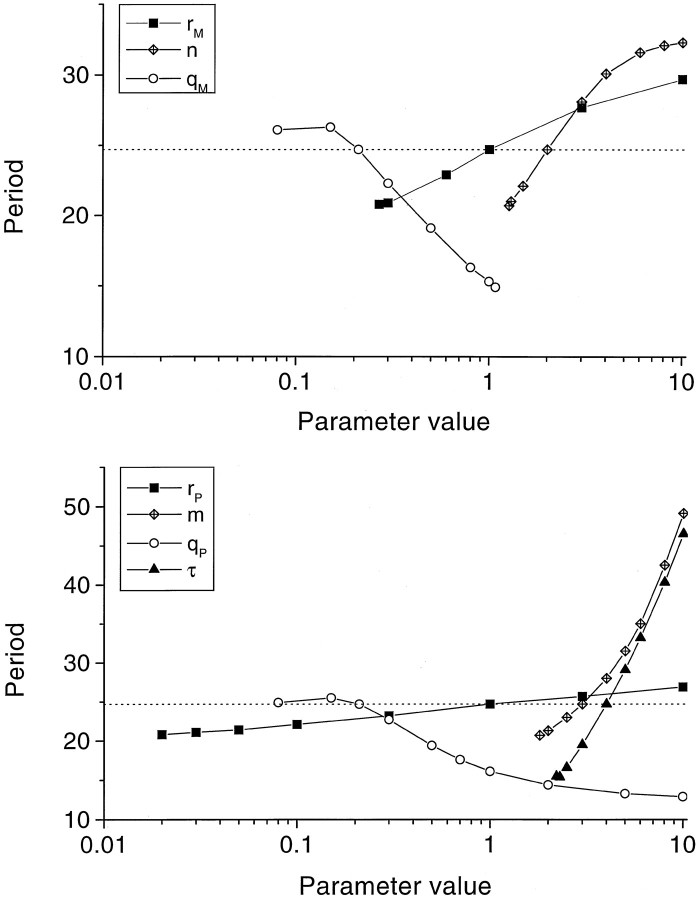
Boundaries in parameter space for limit cycle behavior, and the resulting periods
Figure 3 shows how the period of a free-running oscillation changes when one of the parameters is changed with the other parameters remaining constant. The lowest value for a given parameter in Figure 3 marks the bifurcation from a stable steady state to a limit cycle, see Table 1. For all parameters, exceptqM, the limit cycle behavior continued up to the largest value indicated (10), and an even further increase (up to 30) failed to change this pattern. For parameterqM, on the other hand, a bifurcation from limit cycle to stable steady state appeared at an upper value of 1.08. For all parameter values studied, the period of oscillation did not become shorter than ≈13 hr. A more extensive exploration of the {n, m, τ} subspace showed that at the {n = 1, m = 1, τ = 4} point the system always converged to a stable steady state for any choice of the other parameters. For {n = 1, τ = 4} a bifurcation was found at m ≈ 4, whereas the limit cycles for m > 4 attain circadian periods atm ≈ 6. For {m = 1, τ = 4} a bifurcation was found at n ≈ 4, whereas the limit cycles for n > 4 attain circadian periods atn ≈ 5.5. Smaller values of the delay parameter τ required substantially larger values for n or m, to obtain circadian oscillations. For instance, with τ = 3, such oscillations were found at {n = 1, m= 9} or at {n = 14, m = 1}. For larger values of τ, the parameter area for circadian oscillations encounters the bifurcation boundary. For instance, with τ = 5, the system is stable (for small values of n or m) or oscillates with periods larger than 24 hr (for larger values ofn or m).
Fig. 3.
Period of oscillation plotted against the value of (top panel) the mRNA production rate constantrM, mRNA degradation rate constantqM, and Hill coefficientn, and (bottom panel) the protein production rate constant rP, protein degradation rate constant qP, and the nonlinearity m and duration τ of the protein production cascade. The intersections with the dotted line indicate the set of parameter values for which the system oscillates with a (free-running) period of 24.6 hr. Note, that each parameter was varied while keeping the other parameters at their original value.
Sensitivity of periodicity to model parameters
The slope of each line in Figure 3 indicates how sensitively the period depends on the parameter in question. Especially the nonlinearity m and the duration τ of the protein production sequence strongly influence the period. Thus, changing the protein production rate constant rP over almost three orders of magnitude allowed the period increasing from ≈21 to ≈27 hr, whereas an increase of the delay term τ from 2 to 8 hr caused the period to increase more than threefold. The curves for the degradation rate constants qM andqP were the only ones with negative slopes, i.e., increased protein or mRNA degradation result in both cases in shorter free-running periods.
Perturbed system
Entrainment to periodic external stimulation
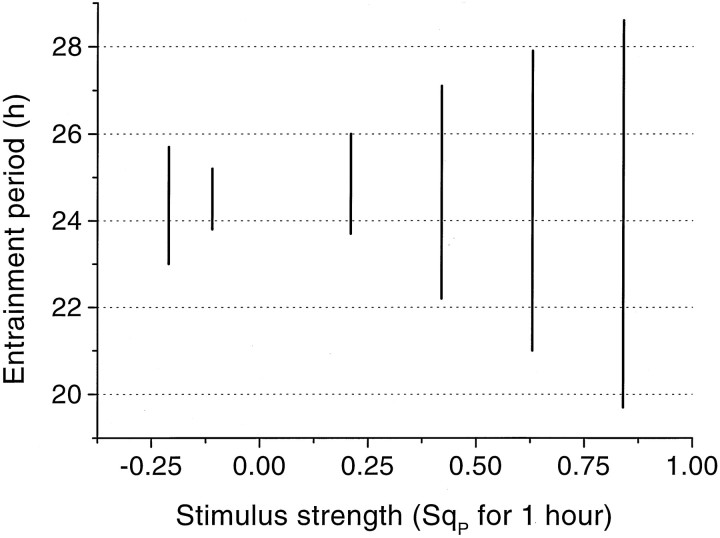
Periodic external stimulation was studied by periodically switching on for 1 hr one of the parametersSrM, SqM,SrP, SqP in Equations 3 and 4. For all four parameters entrainment of the oscillator was indeed found to occur to both shorter and longer cycles than the free-running one (Fig. 4). At the onset of periodic stimulation, the system went through a transient episode lasting approximately one or two periods until it became phase-locked. Entrainment appeared to depend on the stimulus strength: small input strengths had little effect, whereas large input strengths made the model to become a slave oscillator of the external signal. Then, the stimulus cannot be considered anymore as a perturbation but gets full control over the behavior of the system. The stimulus strength also determines the entrainment window, viz., the maximal deviation of the stimulus period from the free-running period for which entrainment still occurs. An example is given in Figure5 in which the entrainment window is shown for different perturbations of the protein production rate constant qP. For instance, with a perturbation of SqP = 0.2 on the value ofqP of 0.21 hr−1, the system becomes phase-locked only between imposed cycles of ≈23.6 hr and ≈26.1 hr.
Fig. 4.
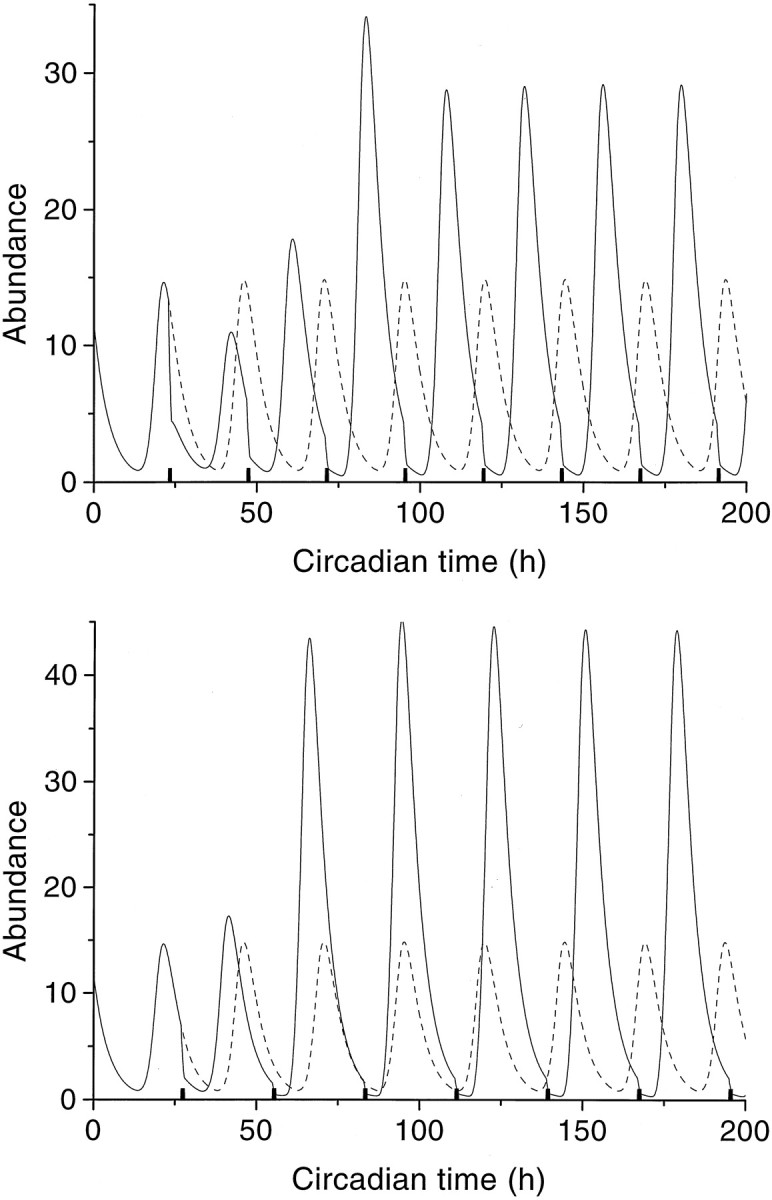
Entrainment of a free-running oscillator (Tfr = 24.6 hr) to an external periodic stimulation of the protein degradation rateqP from 0.21 into 0.42 for 1 hr (black bars) with stimulus periodTstim = 24.0 (top panel) or Tstim = 28.0 hr (bottom panel). Note the onsets of entrainment of the protein oscillation (continuous line). The unperturbed free-running oscillations (24.6 hr) are included for reference (dashed line).
Fig. 5.
Entrainment windows versus the strength of the periodic 1 hr perturbation in the protein production rateSqP. The entrainment window indicates the range of stimulus periods for which entrainment occurs.
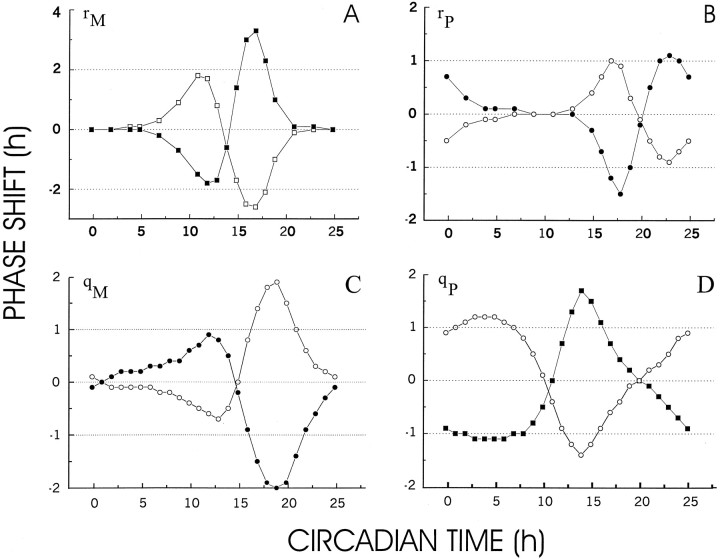
Phase–response curves
Phase–response curves were obtained by perturbing for 1 hr the mRNA production rate constant rM (Fig.6A), the protein production rate constant rP (Fig.6B), the mRNA degradation rate constantqM (Fig. 6C), and the protein degradation rate constant qP (Fig.6D). The PRCs demonstrate that each type of perturbation can result in a positive (phase advance) or in a negative phase shift (phase delay). Additionally, stimulatory and inhibitory perturbations had opposing effects on the phase shifts. For instance, whereas mRNA production stimulation of 1 hr at 17 hr CMT resulted in a phase delay of ≈2.5 hr, mRNA production inhibition at 17 hr CMT results in a phase advance of ≈3 hr. Characteristic for the mRNA and protein production PRCs (Fig.6A,B), is that during some periods in the circadian cycle perturbations fail to produce any phase shift (dead zone). For instance, perturbing the mRNA production rate constantrM results in clear phase shifts when applied between 5 and 23 hr CMT but had little or no effect when applied at the other time points during the circadian cycle. The dead zone for the protein production perturbation occurs between ∼6 and 12 hr CMT. The degradation PRCs (Fig. 6C,D) show patterns quite different from the production PRCs, in that they have no dead zones. The PRCs for the four rate constants differ considerably in the phase point of maximal phase shift as well as in the phase advance and delay time ranges. Investigating the PRCs at two other points in parameter space {n = 1.5, m = 4, τ = 4} and {n = 4, m = 1.5, τ = 4} yielded similar results, with in the latter case only a shorter dead zone in the rP PRC.
Fig. 6.
A, Phase–response curve for a single pulse of 1 hr duration of mRNA production stimulation (open squares) and inhibition (filled squares). During stimulation, parameterrM was changed from a value of 1 into a value of 2, whereas during inhibition the value was set at zero.B, Phase–response curve for a single pulse of 1 hr duration of protein production stimulation (open squares) and inhibition (filled squares). During stimulation, parameter rP was changed from a value of 1 into a value of 2, whereas during inhibition the value was set at 0. C, Phase–response curve for a single pulse of 1 hr duration of mRNA degradation stimulation (open squares) and inhibition (filled squares). During stimulation, parameterqM was changed from a value of 0.21 into a value of 0.42 (SqM = 0.21), whereas during inhibition the value was set at 0 (SqM = −0.21). D, Phase–response curve for a single pulse of 1 hr duration of protein degradation stimulation (open squares) and inhibition (filled squares). During stimulation, parameter qP was changed from a value of 0.21 into a value of 0.42 (SqP = 0.21), whereas during inhibition the value was set at 0 (SqP = 0.21).
DISCUSSION
A minimal intracellular model for circadian rhythm generation is shown in the present study to produce stable and robust circadian oscillations with realistic entrainment and phase–response properties. Characteristic for this model is that the complexity of the reaction cascade, involving protein synthesis, phosphorylation, transport, and nuclear entry, is reduced to simply the overall duration τ of the cascade along with a nonlinearity exponent m. By this lumping, the strong constraint for a complete description of all the reactions involved in the cascade and their respective kinetics is avoided. The model thus distinguishes itself from other recent modeling studies, all of which included more detailed descriptions of the cascade of reactions which constitute the negative feedback loop of proteins after the expression of their respective genes (Goldbeter, 1995; Lewis et al., 1997) (Abbott, personal communication).
Robustness of oscillation
At the chosen set point in parameter space, the system shows a robust and stable free-running oscillation with a period of 24.6 hr. Robust, because the precise parameter values are not critical for oscillations to occur. Limit-cycle behavior was found for a large area in parameter space (explored around the set point by changing only one parameter at a time, and more extensively in the {n,m, τ} subspace). Along the main axes this area is bounded by lower limit values, all greater than one, where bifurcations are found, and below which the system has only stable steady states. More extensive investigation of the {n, m, τ} subspace (by changing more than one parameter at the same time) demonstrated that elimination of both nonlinearities, i.e., {n = 1, m = 1}, results in a system having only stable steady states. Elimination of only one of the nonlinearities requires substantially stronger nonlinearities for the other to let the system oscillate. For instance, the conditionm = 1 required a Hill coefficient of at leastn ≈ 6, and the condition n = 1 required a protein production nonlinearity of at leastm ≈ 6. Such large values for the nonlinearities seem biologically unrealistic. Smaller values of the delay τ (e.g., τ = 3 hr) require even larger values for n or m, whereas larger values (i.e., τ > 5 hr) made it impossible for the system to oscillate with circadian periods (i.e., the system went to a steady state or oscillated with larger periods). These findings demonstrate that the duration τ, the nonlinearity m of the protein synthesis cascade, and the protein–protein interaction in the feedback loop (Hill coefficient n) are essential for the emergence of oscillations in the system, with lower limit values along the main axes of n = 1.27, τ = 2.21 hr, andm = 1.81. The positive lower limits for the other parameters further underscore that none of them can be excluded from the description, making the present model sufficient as well as necessary for the emergence of circadian rhythms in the molecular system.
Period of oscillation
For the area in parameter space where oscillations occur, the period of oscillation was always longer than ≈13 hr. Clearly, this period is much longer than the duration (τ = 4 hr) of the protein synthesis cascade and the protein and mRNA production and degradation rates. The slow and robust oscillation must therefore be considered to be an emergent property of the system, i.e., not arising simply from the properties of any of its components. A rather strong dependence of the period of oscillation was found for the delay parameter τ and the nonlinearity parameter m of the protein synthesis cascade, constituting the essential structure of this cascade. Much less dependence was found for the protein production and degradation rate constants. In other words, small changes in τ and m result in large changes in the period. For survival an organism needs both a robust circadian rhythm and the capacity to adapt and fine-tune its intrinsic rhythm to environmental conditions. Because of the discrete nature of reaction cascades, alterations in the core structure of the feedback loop result in discrete and possibly large changes in the period of oscillation. It is therefore plausible to assume that the fine-tuning capacity is provided by graded adaptations in the production and degradation rates, whereas the required robustness is based on a stable core structure of the feedback loop (represented by the delay and nonlinearity parameters). From an evolutionary point of view, the structure of the feedback loop must have been a principle target for natural selection as the period of circadian oscillation is an “all-pervading characteristic of living organisms, conserved throughout evolution and provided by highly stable and protected regions of the clock genome” (Marques and Waterhouse, 1994). Because of the sensitivity of the period for changes in the reaction cascade, large and discrete differences in period are expected to occur by mutations in the clock genes (Konopka and Benzer, 1971), suggesting that this property has been an important factor in the natural selection process.
Protein and mRNA oscillation
Although the duration τ chosen for the protein synthesis cascade was only 4 hr, the peak in the simulated protein oscillation curve occurred 6 hr later than the mRNA peak. This theoretical finding is in excellent agreement with experimental data, which show either a lag of 6 hr between these two peak levels (Takahashi, 1991, 1995;Aréchiga, 1993; Dunlap, 1996; King et al., 1997) or a lag of at least 4 hr (Zeng et al., 1994). The time between the protein peak (and, thus, the maximal negative feedback on mRNA production) and the subsequent mRNA peak (∼18 hr) is in good agreement with experimental findings of Merrow et al. (1997) in which the elapsed time between induced repression of frq transcript levels inNeurospora by FRQ and full recovery was found to be 14–18 hr.
Entrainment to external periodic stimuli
The entrainment properties of the system have been studied by periodically altering one of the production or degradation rate constants. These parameters were selected as targets for stimulation, assuming that light, too, interferes at one of these points in the feedback loop. In all cases entrainment occurred to both shorter and longer external periods but with a dependence on the intensity of the stimulus and the difference between the stimulus and the free-running period. For instance, sufficiently strong stimulation of the protein degradation rate resulted in an entrainment window between 20 and 28 hr (Fig. 5). This compares favorably with the empirical entrainment window observed in the melatonin rhythm of rams under symmetrical light/dark cycles, with a lower limit close to 20 hr cycles and an upper limit of at least 28 hr (Ravault et al., 1989).
Phase–response curves
Phase shifting was studied by changing either the production or the degradation rate constant of mRNA or protein during 1 hr. Both facilitatory and inhibitory perturbations were given, either of which could result in phase advance or phase delay, depending on the phase point of application during the free-running cycle. A remarkable observation was that almost all PRCs, obtained by perturbing the mRNA or protein production rate, contained dead zones, i.e., ranges in the free-running cycle in which perturbation failed to induce any phase shift. The mRNA and protein degradation rate PRCs, on the contrary, did not show dead zones, with phase shifts occurring at all points in the free-running cycle except at zero crossings. The PRCs for the four rate constants also turned out to differ considerably in the phase point of maximal phase shift. Phase–response curves have been calculated for three different points in parameter space, all yielding similar results and underscoring the robustness of the PRCs for parameter changes and the significance of their differences. Experimental PRCs are usually derived from the observation of phase shifts in locomotor or other activity brought about by light pulses. On the basis of our model findings, the observation that the PRC for Drosophila (Myers et al., 1996) lacks a dead zone suggests that light has targeted the degradation rather than the production of mRNA or protein. This conclusion is consistent with the finding that, inDrosophila, light pulses indeed cause enhanced degradation of TIM (one of the clock proteins) (Lee et al., 1996; Young, 1996; Zeng et al., 1996; Dembinska et al., 1997).
Expression of circadian rhythms
The expression of molecular circadian rhythm generators varies among species, and even tissues, taking the form of oscillating neuronal firing frequencies, hormonal levels, or behavioral expression, etc. Although one may assume that the period of oscillation is preserved in such expression, both the shape and the phase of the rhythms at supracellular organizational levels can differ considerably from the molecular “driver” rhythm. This consideration needs to be taken into account when comparing the molecular simulations with empirical findings.
Comparison with other modeling studies
The objective of the present study was to pinpoint those components of intracellular feedback systems that are essential for producing circadian rhythmicity. The nonlinearities and the delay, identified as essential components in this model, find their intracellular implementation in (complex) schemes of chemical reactions, the precise nature of which is still unknown. One of the mechanisms underlying the delay could be protein postprocessing and transport, as studied by Goldbeter (1995), Abbott (personal communication), and Lewis et al. (1997) by means of multiple PER phosphorylation steps and nuclear entry, but also the time required for protein synthesis itself contributes to the delay. With a value of 2 for the Hill coefficient, the present model needs only a moderate cooperativity (e.g., dimerization) in the feedback loop. In this sense, it distinguishes itself from other theoretical studies, all of which required stronger cooperativity (viz., higher values for the Hill coefficient) in order for the studied systems to generate stable circadian oscillations (Griffith, 1968; Goldbeter, 1995; Ruoff and Rensing, 1996). The present model predicts a shorter period of oscillation when the protein degradation rate increases (Fig. 3), whereas Goldbeter’s (1995) model shows a period increase under those conditions. It is not clear whether this difference reflects fundamental differences in the assumptions underlying the two models or originates merely from different model implementations of the reaction schemes.
Conclusion
Nonlinearities and delay in the protein synthesis negative feedback loop have been shown to be essential features in our model for the generation of robust circadian oscillations. Although the model was constructed on the basis of minimal requirements, it displays a rich and realistic repertoire of circadian rhythm behavior with respect to period, entrainment, and phase responses. Further outcomes of the present study are: (1) the prediction of dead zones in the phase–response curves for perturbations in the production rate but not in those for degradation rate perturbations, and (2) quantitative predictions of the dependence of the period of oscillation on the parameters of the system. These findings not only contribute to a better understanding of the putative molecular system underlying circadian rhythm generation but also serve as a basis for interpreting experimental findings as well as for formulating critical experiments for validation.
The model of the intracellular circadian oscillator has been defined as:
| Equation A1 |
| Equation A2 |
with M*(t) and P*(t) denoting the concentrations of mRNA and the effective protein,rM* the maximal mRNA production rate, rP the protein production rate constant,qM and qP the mRNA and protein degradation rate constants, respectively, n the Hill coefficient, the exponent m the nonlinearity in the protein production cascade, the delay τ the total duration of protein production from mRNA, and k* a scaling constant. Note, that the maximal production rate r*Mcarries the dimension [concentration]/[t], whereas the production and degradation rate constantsrP, qM, andqP carry the dimension 1/[t]. Introducing dimensionless quantities M =M*/M0, p =P*/M0, and k= k*/M0, and the quantity rM =rM*/M0with dimension 1/[t], with M0denoting the maximal mRNA concentration produced per unit of time, and dividing both equations by M0 finally result in Equations 1 and 2.
Footnotes
We thank Dr. Michael A. Corner for his critical comments and valuable suggestions regarding this manuscript
Correspondence should be addressed to Dr. Jaap van Pelt, Netherlands Institute for Brain Research, Meibergdreef 33, 1105 AZ Amsterdam, The Netherlands.
REFERENCES
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular biology of the cell, Ed 2. Garland Publishing; New York: 1989. [Google Scholar]
- 2.Aréchiga H. Circadian rhythms. Curr Opin Neurobiol. 1993;3:1005–1010. doi: 10.1016/0959-4388(93)90174-w. [DOI] [PubMed] [Google Scholar]
- 3.Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1583. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 4.Dembinska ME, Stanewsky R, Hall JC, Rosbash M. Circadian cycling of a PERIOD-β-galactosidase fusion protein in Drosophila: evidence for cyclical degradation. J Biol Rhythms. 1997;12:157–172. doi: 10.1177/074873049701200207. [DOI] [PubMed] [Google Scholar]
- 5.Dunlap JC. Genetic and molecular analysis of circadian rhythms. Annu Rev Genet. 1996;30:579–601. doi: 10.1146/annurev.genet.30.1.579. [DOI] [PubMed] [Google Scholar]
- 6.Edery I, Rutila JE, Rosbash M. Phase shifting of the circadian clock by induction of the Drosophila period protein. Science. 1994a;263:237–240. doi: 10.1126/science.8284676. [DOI] [PubMed] [Google Scholar]
- 7.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994b;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldbeter A. A model for circadian oscillations in the Drosophila period protein (PER). Proc R Soc Lond B Biol Sci. 1995;261:319–324. doi: 10.1098/rspb.1995.0153. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin BC. Oscillatory behaviour in enzymatic control processes. Adv Enzyme Regul. 1965;3:425–438. doi: 10.1016/0065-2571(65)90067-1. [DOI] [PubMed] [Google Scholar]
- 10.Griffith JS. Mathematics of cellular control processes. I. Negative feedback to one gene. II. Positive feedback to one gene. J Theor Biol. 1968;20:202–216. doi: 10.1016/0022-5193(68)90189-6. [DOI] [PubMed] [Google Scholar]
- 11.Gumowski I. Qualitative properties of some dynamic systems with pure delay. Lect Notes Biomath. 1981;41:139–152. [Google Scholar]
- 12.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 13.Hardin PE, Hall JC, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci USA. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 15.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TDL, Hotz Vitaterna M, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS. Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron. 1990;5:127–134. doi: 10.1016/0896-6273(90)90303-w. [DOI] [PubMed] [Google Scholar]
- 18.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 19.Lewis RD, Warman GR, Saunders DS. Simulations of free-running rhythms, light entrainment and the light-pulse phase response curves for the locomotor activity rhythm in period mutant of Drosophila melanogaster. J Theor Biol. 1997;185:503–510. [Google Scholar]
- 20.Marques MD, Waterhouse JM. Masking and the evolution of circadian rhythmicity. Chronobiol Int. 1994;11:146–155. doi: 10.3109/07420529409057234. [DOI] [PubMed] [Google Scholar]
- 21.Merrow MW, Garceau NY, Dunlap JC. Dissection of a circadian oscillation into discrete domains. Proc Natl Acad Sci USA. 1997;94:3877–3882. doi: 10.1073/pnas.94.8.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray JD. Mathematical biology. Springer; Berlin: 1989. [Google Scholar]
- 23.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 24.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes in C, Ed 2. Cambridge UP; Cambridge, UK: 1992. [Google Scholar]
- 25.Ravault JP, Arendt J, Tobler I, Chesneau D, Maulin O. Entrainment of melatonin rhythms in rams by symmetrical light-dark cycles of different period length. Chronobiol Int. 1989;6:329–339. doi: 10.3109/07420528909056939. [DOI] [PubMed] [Google Scholar]
- 26.Ruoff P, Rensing L. The temperature compensated Goodwin model simulates many circadian clock properties. J Theor Biol. 1996;179:275–285. [Google Scholar]
- 27.Saez L, Young MW. Regulation of nuclear entry of the Drosophila clock proteins PERIOD and TIMELESS. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 28.Sassone-Corsi P. Rhythmic transcription and autoregulatory loops: winding up the biological clock. Cell. 1994;78:361–364. doi: 10.1016/0092-8674(94)90415-4. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi JS. Circadian rhythms: from gene expression to behaviour. Curr Opin Neurobiol. 1991;1:556–561. doi: 10.1016/s0959-4388(05)80028-5. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi JS. Molecular neurobiology and genetics of circadian rhythms in mammals. Annu Rev Neurosci. 1995;18:531–553. doi: 10.1146/annurev.ne.18.030195.002531. [DOI] [PubMed] [Google Scholar]
- 31.Young MW. The Drosophila genes timeless and period collaborate to promote cycles of gene expression composing a circadian pacemaker. In: Buijs RM, Kalsbeek A, Romijn HJ, Pennartz CMA, Mirmiran M, editors. Progress in brain research, Vol 111. Elsevier Science; Amsterdam: 1996. pp. 29–39. [DOI] [PubMed] [Google Scholar]
- 32.Zeng H, Hardin PE, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 34.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Takahashi JS, Turek FW. Critical period for cycloheximide blockade of light induced phase advances of the circadian locomotor rhythm in golden hamsters. Brain Res. 1996;740:285–290. doi: 10.1016/s0006-8993(96)00900-6. [DOI] [PubMed] [Google Scholar]