Abstract
Transgenic technology, single-cell RT-PCR, and immunocytochemistry were combined to investigate the composition of the GABAA receptors of dopaminergic (interplexiform) amacrine (DA) cells. A mouse line was used in which these neurons were labeled with human placental alkaline phosphatase and could therefore be identified in vitro after dissociation of the retina. We performed single-cell RT-PCR on the isolated cells and showed that (1) DA cells contained the messages for α1, α3, α4, β1, β3, γ1, γ2S, and γ2L subunits; (2) this transcript repertory did not change on dissociation of the retina and throughout the time required for cell harvesting; and (3) all DA cells contained the entire transcript repertory. Immunocytochemistry with subunit-specific antibodies showed that all subunits were expressed and appeared homogeneously distributed throughout the cell membrane at a low concentration. In addition, with the exception of α4, the subunits formed clusters at the surface of the dendrites and on the inner pole of the cell body. Because of their size, shape, and topographic coincidence with GABAergic endings, the clusters were interpreted as postsynaptic active zones containing GABAAreceptors. The composition of the synaptic receptors was not uniform: clusters distributed throughout the dendritic tree contained α3, β3, and, less frequently, β1 subunits, whereas clusters containing the α1 subunit were confined to large dendrites. Therefore, DA cells possess at least two types of GABAA receptors localized in different synapses. Furthermore, they exhibit multiple extrasynaptic GABAA receptors.
Keywords: GABAA receptors, dopamine, retina, amacrine cell, single-cell RT-PCR, immunocytochemistry
The inhibitory neurotransmitter GABA acts on a large repertory of bicuculline-sensitive receptors that consist of various combinations of at least 14 subunits (Sieghart, 1995; Barnard et al., 1998). In situ hybridization and immunocytochemical studies demonstrated that each subunit has a unique distribution in the CNS (Fritschy et al., 1992; Laurie et al., 1992; Wisden et al., 1992); furthermore, transfection experiments proved that the various combinations of subunits found in specific regions of the brain have different pharmacological properties (Rabow et al., 1995). With in situ hybridization, however, it is often impossible to pinpoint the types of neurons that contain the subunit transcripts. On the other hand, pharmacological results on recombinant receptors may not always apply to native assemblies because there is no certainty that some of the transfected combinations of subunits exist in vivo on the surface of specific cell types (McKernan and Whiting, 1996;Sieghart et al., 1999). It is therefore crucial to identify the composition of the subunit assemblies that are expressed at the surface of individual neurons to understand their function in the computations performed by specific neural networks. So far, comprehensive information exists only for the cerebellar cortex (Santi et al., 1994;Wisden, 1995; Nusser et al., 1996a, 1998) and hippocampus (Pearce, 1993; Nusser et al., 1995, 1996b) where the various neuronal types have highly characteristic shape and distribution. The challenge is much greater for rare cell types, which cannot be easily identified on the basis of morphological criteria, either in tissue slices or after dissociation.
In this paper, we report the subunit composition of the GABAA receptors in the dopaminergic (interplexiform) amacrine (DA) cells of the mouse retina. Dopamine is in large measure responsible for neural adaptation to light (Witkovsky and Dearry, 1991; Djamgoz and Wagner, 1992). In turn, GABA inhibits dopamine release when the light is turned off by acting primarily on bicuculline-sensitive GABAAreceptors deployed at the surface of DA cells (Morgan and Kamp, 1980;Kamp and Morgan, 1981; O’Connor et al., 1986; Ishita et al., 1988;Kirsch and Wagner, 1989; Critz and Marc, 1992; Kolbinger and Weiler, 1993).
Because there are only 450 DA cells in each mouse retina and they cannot be distinguished from neighboring cells on the basis of their morphology, we labeled DA cells with human placental alkaline phosphatase (PLAP) by introducing into the mouse genome PLAP cDNA under the control of the promoter of the gene for tyrosine hydroxylase (TH), the rate-limiting enzyme for dopamine biosynthesis (Gustincich et al., 1997). Because PLAP is an enzyme that resides on the outer surface of the cell membrane, we could identify DA cells after dissociation of the retina by immunocytochemistry in the living state. Then, we analyzed their repertory of GABAA receptor subunit mRNAs by single-cell, RT-PCR. We finally confirmed by immunocytochemistry that the messages were translated into proteins, and we studied receptor composition and localization in the intact retina.
The pharmacological properties of the DA cell GABAA receptors have been published previously (Feigenspan et al., 1998).
MATERIALS AND METHODS
Harvesting of DA cells
Anesthetized, 1- to 6-month-old mice homozygous for the PLAP transgene were used. Dissociation of the retina by enzymatic digestion and mechanical trituration has been described elsewhere (Gustincich et al., 1997). The cell suspension was allowed to sediment on a concavalin A (1 mg/ml)-coated coverslip at the bottom of a recording chamber, and DA cells were identified in epifluorescence after staining with a monoclonal antibody to PLAP (E6) (De Waele et al., 1982) conjugated to the fluorochrome Cy3 (E6-Cy3) (Gustincich et al., 1997). Labeled cells were harvested after patch clamping. Patch pipettes were constructed from borosilicate glass (1.65 mm outer diameter, 1.2 mm inner diameter; A-M Systems, Everett, WA) using a horizontal two-stage electrode puller (BB-CH, Mecanex, Geneva, Switzerland); the electrode resistance ranged from 5 to 7 mΩ. Electrodes were connected to the amplifier via an Ag/AgCl wire. The electrode holder and the headstage were mounted on a piezoelectric, remote-controlled device attached to a tridimensional micromanipulator (Burleigh Instruments, Fishers, NY). The intracellular solution contained 130 mm KCl and 0.5 mm EGTA in 10 mm HEPES, pH 7.4. When the patch pipette was immersed into the bath, positive pressure was continuously applied to the fluid within. After seal formation on the surface of DA cells and disruption of the patch membrane, the cellular contents were aspirated into the pipette by gently applying negative pressure until the residual cell ghost became stuck to the pipette tip. The electrode was then lifted from the bath under constant visual control and removed from the holder. The tip of the pipette was finally broken into an Eppendorf tube containing 20 U RNase inhibitor (RNasin ; Boehringer Mannheim, Indianapolis, IN) and, after brief centrifugation, the tube was frozen on dry ice and stored at −80°C. The harvesting of DA cells lasted from 30 to 90 min from the time of plating.
Single-cell RT-PCR
After RT, each experiment comprised two successive rounds of amplification. The first one used forward (F) and reverse (R) primers, chosen according to the criteria specified below. To increase specificity, the second round used either an F or an R primer in combination with a primer internal to the first amplification product [nested (N)] or two N primers (FN and RN).
TH transcript
For amplification of TH cDNA, first-round F primer was CTGGCCTTCCGTGTGTTTCAGTG, which hybridizes with TH cDNA at nucleotide (nt) 915 (GenBank M69200), and the corresponding R primer was CCGGCTGGTAGGTTTGATCTTGG, which hybridizes with TH cDNA at nt 1296. The conditions of the amplification reactions are specified below. The first-round RT-PCR product was 382 bp long. The second-round FN primer was AGTGCACACAGTACATCCGTCAT, which hybridizes with TH cDNA at nt 934, and the corresponding RN primer was GCTGGTAGGTTTGATCTTGGTA, which hybridizes with TH cDNA at nt 1293. The final nested RT-PCR product was 360 bp long and contained a unique SacI site. The product was successfully amplified only from DA cells labeled by E6-Cy3. Unstained cells and control reactions without cells or with culture medium were negative. An aliquot of the reaction products from labeled cells was purified and cut with SacI to show the specificity of the amplification. As expected, the digestion with this restriction enzyme produced fragments of 271 and 89 bp.
GABAA receptor transcripts
Primers. Primer sequences are shown in Table 1. Primer pairs were chosen that annealed at 55 or 60°C so that all subunit cDNAs could be amplified simultaneously in a single multiplex experiment. Primers were designed in regions of the cDNA that are characterized by lack of homology among the various subunits, and their sequences were located in different exons to avoid amplification of genomic DNA.
Table 1.
Sequences of the PCR primers for GABAA receptor subunits
| α1 | X61430 | F/R 576 bp | MspI | α1F | ATCTTTGGGCCTGGACCCTCATTCT |
| N/R 526 bp | α1R | CTGGCTCTCTGGTCCACTCATAG | |||
| α1N | GGACAGCCCTCCCAAGATGAACT | ||||
| α2 | M86567 | F/R 445 cp | XbaI | α2F | TCTTCTGGTGTGGGACCCAGTCA |
| F/N 326 bp | α2R | GGCTTGGACTGTAAGCCTCATGG | |||
| α2N | CAGGAGTCCAGATTTTGCTGGCC | ||||
| α3 | M86568 | F/R 363 bp | HincII | α3F | TGGGCTTGGGAAGGCAAGAAGGT |
| N/R 219 bp | α3R | GCCCTTGATAGCTGATTCCCGGT | |||
| α3N | ATCTCCAAGTCTGCTGCTGCTCC | ||||
| α4 | New | F/R 207 bp | HaeIII | α4F | TGTCACCAGCTTTGGGCCCGTTT |
| F/N 152 bp | α4R | GGAGCTGTCATGTTATGTGAGAC | |||
| α4N | GGGTCCAAACTTTGGTGACCATC | ||||
| α5 | New | F/R 161 bp | HaeIII | α5F | GACGAGACCAATGACAACATCAC |
| F/N 121 bp | α5R | TCCATTTCCGTGTCGGACACTGG | |||
| α5N | AGATGTCTGTTCGCACCTGCGTG | ||||
| β1 | U14418 | F/R 334 bp | HindIII | β1F | GGGGCTTCTCTCTTTTCCCGTGA |
| F/N 260 bp | β1R | GGTGTCTGGTACCCAGAGTTGGT | |||
| β1N | GCCTCTTGTCCTTCCAAGACTGC | ||||
| β2 | U14419 | F/R 495 bp | HindIII | β2F | CAACTCTGGGTGCCTGACACCTA |
| N/R 350 bp | β2R | TCCTAATGCAACCCGTGCAGCAG | |||
| β2N | TGGACCTAAGGCGGTATCCACTG | ||||
| β3 | U14420 | F/R 390 bp | BamHI | β3F | GGTTTGCTGCGCTCAGAGCGTAA |
| N/R 339 bp | β3R | TACAGCACTGTCCCATCAGGGT | |||
| β3N | GGAGACGGTCGACAAGCTGTTGA | ||||
| γ1 | New | F/R 165 bp | RcaI | γ1F | CAGTTTGCATTTGTAGGGTTACG |
| N/R 135 bp | γ1R | AGACACCCAGGAAAGAACCACTG | |||
| γ1N | CTGAAATCTCTCACACAATCTCT | ||||
| γ2 | M62374 | F/R 322 bp | EarI | γ2F | GGTGGAGTATGGCACCCTGCATT |
| N/R 304 bp | γ2R | AGGCGGTAGGGAAGAAGATCCGA | |||
| γ2N | GTCAGCAACCGGAAGCCAAGCAA | ||||
| γ3 | X59300 | F/R 592 bp | HindII | γ3F | TGCTCGGTCCAGGAGGGTAGA |
| F/N 382 bp | γ3R | CTGATCAGCTGCCTCAACTGAATTTTT | |||
| γ3N | CCAGTGAGCCTCCGCTGTTTTAG | ||||
| δ | S42882 | F/R 398 bp | HindIII | δF | GACTACGTGGGCTCCAACCTGGA |
| N/R 283 bp | δR | ACTGTGGAGGTGATGCGGATGCT | |||
| δN | TTGCCCTAGAGGTGGCCAGCATT | ||||
| ε | New | F/R 225 bp | RcaI | εF | CAATGCGAAGAACACTTGGAAGC |
| F/N 202 bp | εR | CTGGCAGCAGCAGCTTCTATCTT | |||
| εN | GATCCAGAAGGAGACCCAGGAGA |
Sequences of the primers (forward, reverse, nested) used in the RT-PCR reactions for GABAA receptor subunit mRNAs, databank accession numbers of the mouse cDNA sequences, lengths of the PCR products, and restriction enzymes used to establish specificity of the reactions.
RT and cDNA amplification. Tubes containing single cells were incubated for 1.5 min at 65°C (Ghia et al., 1996). After cooling in ice, reverse transcriptase was added to each tube [75 mm KCl, 3 mmMgCl2, 10 mm DTT, 40 U RNasin (Boehringer Mannheim), 4 ng Pd(N)6(Pharmacia, Piscataway, NJ), 1 mm each dNTPs (Pharmacia), 200 U RT Superscript II H− (Life Technologies, Gaithersburg, MD) in 19 μl 50 mm Tris-HCl, pH 8.3], and first-strand cDNA was synthesized at 37°C for 1 hr. At the end of this reaction, first-round PCR mixture [35 mmKCl, 0.9 mm MgCl2, 40 pm each F and R primers, 1 U Taq polymerase (Boehringer Mannheim) in 80 μl 10 mm Tris-HCl, pH 8.3] was added to the tubes. First-round amplification comprised 2.5 min at 94°C; 10 cycles, each consisting of 30 sec at 94°C, 30 sec at 55°C, and 1 min at 72°C; 18 cycles, each consisting of 30 sec at 94°C, 30 sec at 55°C, and 1 min at 72°C, with a 5 sec extension time added to the final step of each cycle beginning from the second cycle; 10 min at 72°C. Aliquots (1 μl) of the first-round PCR mixtures were used as a template for second-round PCR reactions. To each tube, a solution was added containing 50 mm KCl, 1.5 mm MgCl2, 200 mm dNTPs, 20 pm each N and F or R primers, 1 U Taq polymerase in 49 μl 10 mm Tris-HCl, pH 8.3. The amplification was performed as follows: 2.5 min at 94°C; 10 cycles, each consisting of 30 sec at 94°C, 30 sec at 60°C, and 1 min at 72°C; 20 cycles, each consisting of 30 sec at 94°C, 30 sec at 60°C, and 1 min at 72°C, with a 5 sec extension time added to the final step of each cycle beginning from the second cycle. Aliquots (10 μl) of the second-round PCR mixtures were analyzed by electrophoresis in 1% Seakem and 1% Nusieve GTG agarose (FMC, Rockland, ME) containing 0.5 μg/ml ethidium bromide.
To confirm that each DA cell contained the entire repertory of subunit mRNAs, a multiplex semi-nested single-cell RT-PCR was performed. In this experiment, first-strand cDNA was synthesized as described above. The first-round PCR solution contained 40 pm F and R primers for all seven subunit cDNAs (α1, α3, α4, β1, β3, γ1, and γ2). The 30-cycle amplification was the same as described above. Aliquots (1 μl) of the first-round PCR reaction product were distributed to seven tubes, each containing N and F or R primers for a single subunit. Seven separate second-round PCR reactions were performed according to the protocol described above. The products were finally analyzed by agarose gel electrophoresis.
Precautions and controls. Our main concern was the preservation of the integrity of the RNA and the elimination of contamination. Recording chambers were washed with hydrogen peroxide; cotton-plug pipette tips, tubes, and glass for electrodes were autoclaved; reagents were stored at −20°C in single-use aliquots. Cells were harvested and solutions were prepared in rooms that were different from the laboratory in which PCR reactions and agarose gel electrophoresis were performed. DA cells, unlabeled neurons, and controls were examined simultaneously in each experiment, and six was the highest number of cells analyzed at any given time. Samples of the supernatant in the recording chamber were examined by PCR to rule out amplification of mRNA from floating or dead cells. Each couple of primers was tested first in an RT-PCR experiment from 0.5 μg of total brain RNA. These amplifications were performed for 40 cycles at the specific annealing temperatures. The RT-PCR products were digested with the appropriate restriction enzymes to confirm specificity. RNA dependency of the amplification was established by omitting RT during first-strand cDNA synthesis. First-round PCR products from total brain RNA, obtained using F and R primers for each subunit, were used as templates for second-round reactions with primers specific for other members of the same subunit family (e.g., the first-round RT-PCR product for the γ1 subunit was amplified in second-round reactions driven by the primers for γ2 and γ3 subunits). No difference in the repertory of PCR products from single cells was observed when the number of cycles was increased from 28+30 to 40+35 cycles. However, the number of cycles was kept as low as possible to avoid contamination and increase in nonspecific signals. When the experiments summarized in Table 2 were completed, a single-cell RT-PCR product for each subunit was cloned into the pCR-II vector by using the Original TA cloning Kit (Invitrogen, Carlsbad, CA). Both strands were sequenced using the dideoxy-chain termination method. DNA similarities were examined, and identity scores were generated by matching the query sequence to database entries using the BLAST and BESTFIT algorithms. All of the fragments were identified correctly.
Table 2.
Results of single-cell RT-PCR of GABAA receptor subunit subtype mRNAs in DA cells
| Subtype | No. of positive cells | Positive cells (%) |
|---|---|---|
| α1 | 8 /10 | 80 |
| α2 | 0 /10 | 0 |
| α3 | 12 /16 | 75 |
| α4 | 7 /10 | 70 |
| α5 | 0 /10 | 0 |
| β1 | 16 /18 | 88 |
| β2 | 0 /10 | 0 |
| β3 | 7 /10 | 70 |
| γ1 | 9 /12 | 75 |
| γ2 | 8 /10 | 80 |
| γ3 | 0 /10 | 0 |
| δ | 0 /10 | 0 |
| ε | 0 /10 | 0 |
cDNA cloning of mouse GABAA receptor subunits.Because sequences for the mouse subunits α4, α5, γ1, and ε were not available, we cloned and sequenced a small cDNA fragment of each subunit to design new primers for single-cell RT-PCR experiments from mouse cells. Total RNA from mouse retina, cerebrum, cerebellum, and amygdala was purified according to the acid guanidinium thiocyanate-phenol-chloroform method (Chomczynski and Sacchi, 1987) and used as a template in an RT-PCR reaction. Total RNA (0.5 μg) was incubated in the presence of 4 ng P(dN)6 for 5 min at 65°C. After cooling in ice for 2 min, the following mixture was added to the denatured RNA to a final volume of 19 μl: 75 mm KCl, 3 mm MgCl2, 10 mmdtt, 10 U RNasin, 1 mm each dNTPs in 50 mm Tris-HCl, pH 8.3. After 5 min at 37°C, 200 U RT Superscript II H− (Life Technologies) was added to the samples and incubated at the same temperature for 1 hr. For amplification, we used a high-fidelity Pwo polymerase (Boehringer Mannheim) with proofreading activity. The PCR reaction took place in a 100 μl volume containing 40 mm KCl, 0.6 mmMgCl2, 1.4 mm MgSO4, 5 mm(NH4)2SO4, 2 mm DTT, 200 μm each dNTPs, 40 pm F and R primers, 1 U Pwo polymerase, 20 mm Tris-HCl, pH 8.8. The amplification protocol was the following: 2.5 min at 95°C; 10 cycles, each consisting of 30 sec at 95°C, annealing for 30 sec, and 1 min at 72°C; 30 cycles, each consisting of 30 sec at 95°C, and 1 min at 72°C, with a 5 sec extension time added to the final step of each cycle starting from the second cycle; 10 min at 72°C (annealing temperatures are specified below). We choose primers specific for sequences identical in the rat and human genes. From rat α4 (L08493), ratα4F CCTGGATTTGGGGGTCCTGTTAC (nt 284) and ratα4R CATGGGACACTCCGCACTTATGG (nt 613) amplified a fragment 330 bp long. Annealing took place at 55°C. From rat α5 (L08494), ratα5F CTTATCCAGTCACTTTGGCTTTT (nt 350) and ratα5R TTTCATCTTTCCAGCTTTGTCGG (nt 603) amplified a fragment 254 bp long. Annealing took place at 45°C. From rat γ1 (X57514), ratγ1F AGCCCTCAGTGGAAGTGGCTGAT (nt 764) and ratγ1R CCCAGGGATGTTCTAGCAGGTAC (nt 1016) amplified a fragment 253 bp long. Annealing took place at 60°C. From rat ε (U92284), ratεF TTTCCAATGGATTCTCACTCTTG (nt 241) and ratεR GGGAAATTCTTACGAGAAAAGGT (nt 632) amplified a fragment 392 bp long. Annealing took place at 45°C. For each of the mouse subunits, two fragments from two different RT-PCR reactions were gel-purified, incubated with Taq polymerase for 10 min at 72°C, cloned, and sequenced as described above. The sequences of the mouse cDNA fragments are available at EMBL databank with the following accession numbers: α4 AF090373, α5 AF090374, γ1 AF090375, and εAF090376.
Immunocytochemistry
Fixation
GABAA receptor subunits are very sensitive to chemical fixation; thus, satisfactory preservation of antibody binding sites was obtained by fast chemical fixation under microwave irradiation. Eyecups were immersed in Ames medium (Sigma, St. Louis, MO) containing 40 mm glucose, and the retinas were separated from choroid and sclera. Immediately after immersion in 2% formaldehyde in PBS at 25°C (5 ml in 35 mm Falcon Petri dishes), specimens were irradiated for 10 sec in a microwave oven (Pelco; Ted Pella, Redding, CA) and rapidly returned to Ames medium. At the end of irradiation, the temperature of the fixative solution had increased by ∼20°C. After cryoprotection in 20% sucrose, retinas were frozen in partially solidified dichlorodifluoromethane; 5–8 μm horizontal sections through the retina were obtained in a cryostat and mounted on gelatinized slides.
Antibodies
Rabbit polyclonal antibodies to the α1, α3, α4, β1, β3, and γ1 subunits were described elsewhere (Sperk et al., 1997); they were used at the following protein concentrations (in μg/ml): α1 1.8, α3 5, α4L 2.6, α4N 6, β1 2, β3 3.4, γ1 4. Antibody to γ2 (Ebert et al., 1999) was diluted 1:100. Guinea pig antibody to the α3 subunit (Gao et al., 1993), a generous gift from J.-M. Fritschy (Institute of Pharmacology, University of Zürich, Switzerland), was diluted 1:3000. Rabbit polyclonal antibodies to the vescicular GABA transporter (VGAT) (Chaudhry et al., 1998), a generous gift from R. H. Edwards (Department of Neurology, University of California, San Francisco), was diluted 1:2000. DA cells were identified by staining with a monoclonal antibody to TH (Incstar, Stillwater, MN; 1:100).
Double- and triple-labeling studies
Slides were rinsed in PBS for 20 min, blocked in 10% normal goat serum (NGS) (Vector Laboratories, Burlingame, CA), 0.2% bovine serum albumin (BSA) (Sigma) in PBS for 1 hr, followed by 2% fish gelatin (Goldmark Biologicals, Phillipsburg, NJ) in PBS for 30 min. They were incubated overnight in a mixture of the primary antibodies diluted with 0.2% BSA in PBS, rinsed with PBS for 20 min, and incubated for 3 hr in a mixture of the secondary antibodies diluted with PBS containing 0.1% NGS, 0.2% fish gelatin, and 0.2% BSA. Slides were finally rinsed in PBS and coverslipped with Vectashield. For double-labeling experiments, the mixture of primary antibodies contained a rabbit polyclonal to a GABAA subunit and the monoclonal to TH. Secondary antibodies were goat FITC-conjugated anti-rabbit (Boehringer Mannheim; 1:500) and donkey Texas Red-conjugated anti-mouse (Jackson ImmunoResearch, West Grove, PA; 1:100). For triple-labeling experiments, the mixture of primary antibodies contained the guinea pig polyclonal to the α3 subunit, a rabbit polyclonal to one of the other subunits, and the monoclonal to TH. Secondary antibodies were the same as above with the addition of donkey Cy5-conjugated anti-guinea pig (Jackson; 1:100). Double- and triple-label immunocytochemistry did not change the pattern of staining when compared with labeling with a single antibody. Staining was absent when the antibodies to the GABAA subunits were omitted. In another experiment, the mixture of primary antibodies contained the guinea pig polyclonal to the α3 subunit, the rabbit polyclonal to VGAT, and the monoclonal to TH. Secondary antibodies were the same as above. Fluorescence was detected using a Bio-Rad (Bio-Rad Laboratories, Hercules, CA) MRC-1024 confocal imaging system equipped with an argon-krypton laser and a Zeiss Axiophot microscope. Sections were viewed with a 100× 1.4 NA plan apochromat, and the minimal thickness of the confocal image was adopted that was compatible with adequate emission. Images (1024 × 1024 pixels) were obtained sequentially from two or three channels by averaging seven scans; they were stored as TIFF files and processed by Adobe Photoshop (Adobe Systems, Mountain View, CA). Colocalization of α3 with other subunits in TH-positive cells was evaluated by overlaying the three stainings in three different color channels and labeling with text symbols the site of putative synapses. For double staining, data from one channel (TH) are represented in red and those from the other channel (GABAA receptor subunits) are represented in green; yellow indicates the postsynaptic active zones on DA cells that contain a GABAA receptor subunit. For triple staining, TH is red, the α3 subunit is blue, and the GABAA receptor subunits or VGAT is green; white indicates synapses overlaying DA cells that contain both α3 and another GABAA receptor subunit or aggregates of α3 subunits in register with a presynaptic cluster of GABA-containing vesicles. Retinal slices were obtained with a tissue chopper and incubated in the living state with antibody to γ2, followed by FITC-conjugated anti-rabbit antibody, both diluted 1:100 in Ames medium. Slices were subsequently fixed with 2% formaldehyde in PBS, blocked as above with the addition of 0.5% Triton X-100, and immunostained for TH as above.
RESULTS
Single-cell RT-PCR
After enzymatic digestion and mechanical trituration of the retina, DA cells were identified by labeling of their membrane with the fluorescent monoclonal antibody to PLAP E6-Cy3. DA and large unlabeled cells, possibly ganglion cells, were patch-clamped in the whole-cell configuration and individually harvested between 30 and 90 min from the moment when the retina was dissociated. This time interval was chosen to minimize changes in gene expression induced by cell damage during trituration, absence of synaptic inputs, or exposure to culture medium (see below).
Because we know from immunocytochemistry that in the intact retina all PLAP-expressing neurons contain TH (Gustincich et al., 1997), we used single-cell nested RT-PCR to detect the presence of TH mRNA in the fluorescent cells and thus prove beyond doubt that the neurons stained by E6-Cy3 were DA cells. In 23 of 28 labeled cells (82%) we amplified a 360 bp product specific for TH transcript (data not shown), whereas unlabeled neurons were consistently negative. With a technique based on processing individual cells and in the absence of false positive results, we have adopted a rate of success higher than 70% as a satisfactory test of the reproducibility and specificity of our experimental protocol. Negative outcomes are easily explained by loss of the cell, deterioration of the message, or inefficient amplification.
Single-cell RT-PCR technique was then applied to detect mRNAs for 13 subunits of the GABAA receptor: α6 was not studied because of its limited distribution in the CNS (Varecka et al., 1994; Jones et al., 1997). Table 1illustrates the sequences of the three primers (F, R, N) for each reaction, databank accession number of the mouse cDNA sequences, lengths of the PCR products, and restriction enzymes used to establish specificity of the 13 reactions. The primers for the γ2 subunit were selected in a region of the gene that is identical for its large (L) and short (S) variants. These differ for a 24-bp-long exon that codes for a PKC phosphorylation site (Whiting et al., 1990). Because mouse sequences were not available for α4, α5, γ1, and ε, we cloned and sequenced partial cDNAs from mouse brain to design sets of primers specific for this species.
In a two-rounds RT-PCR, one can analyze more than one transcript at a time from the same cell, provided that the messages are present in equivalent amounts. Because TH mRNA is abundantly expressed, we consistently failed in our efforts to demonstrate in the same cell the presence of the messages for both TH and one of the subunits of the GABAA receptor. Probably, amplification of the receptor transcript was competitively suppressed by the reaction for TH mRNA. Thus, because the relative amounts of the various receptor transcripts were not known, we studied first the expression of one subunit mRNA at a time in a total of 161 DA cells.
As shown in Figure 1 and summarized in Table 2, DA cells contained mRNAs for the α1, α3, α4, β1, β3, γ1, and γ2 subunits. Both the S and L spliced forms of the γ2 subunit transcript were present. We analyzed at least 10 DA cells for each of the subunits, and the rate of success was from 70 to 88% of the cells tested. The specificity of the amplification was established by restriction enzyme digestion of the PCR products. Furthermore, their identity was confirmed beyond doubt by cloning and sequencing; this control was performed after completion of all the experiments to avoid contamination. Several large, unlabeled neurons, probably ganglion cells, expressed some of the subunit transcripts that were also present in DA cells, whereas specific bands were consistently absent when the reaction was performed either on the supernatant or in absence of a cell. It is significant that anomalous reaction products were rarely seen when the specific message was present in the reaction mixture. Transcripts for the α2, α5, β2, γ3, δ, and ε subunits were absent in DA cells; to prove that this failure was not caused by insufficient sensitivity of our technique, we repeated single-cell RT-PCR reactions on unlabeled neurons until we observed expression of all the above transcripts (data not shown).
Fig. 1.
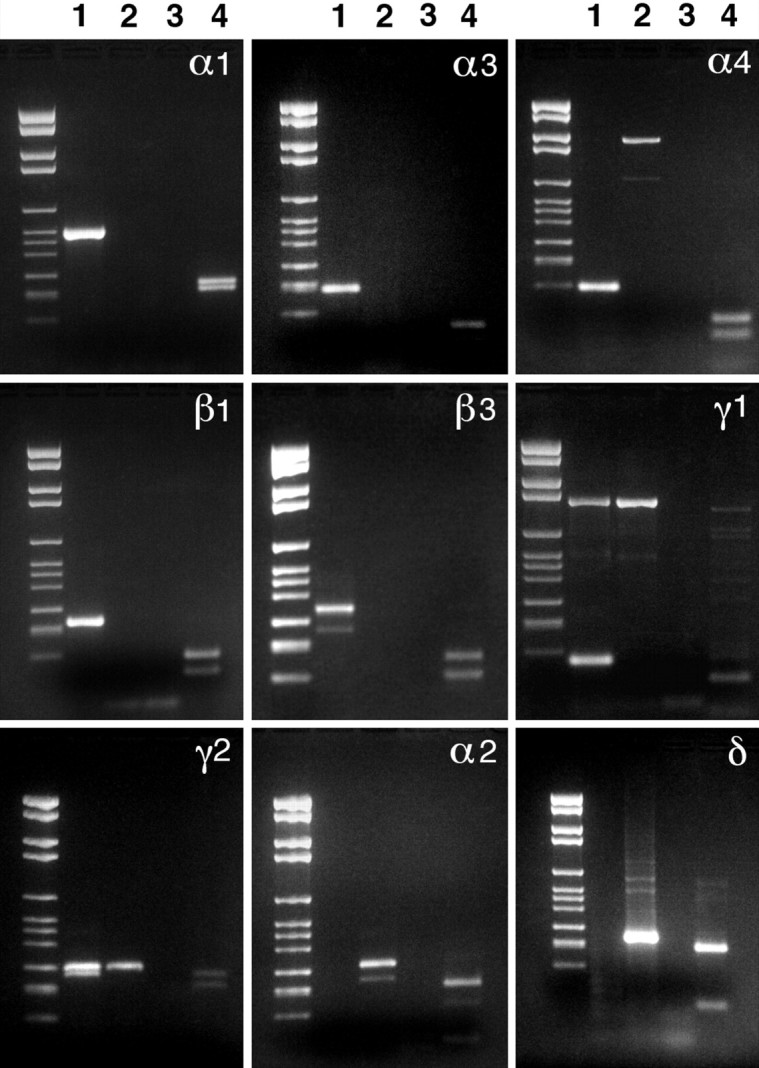
Agarose gel electrophoresis of the products of semi-nested single-cell RT-PCR for the transcripts of the subunits of the GABAA receptor in DA cells. In lane 1 is the product from a DA cell; in lane 2 is the product from a large cell that does not express PLAP; in lane 3is a control reaction in which the cell was omitted; and in lane 4 is the result of restriction enzyme digestion of the product of lane 1 to establish specificity. Bands inlanes 1 are the products of the reactions for the α1, α3, α4, β1, β3, γ1, γ2S, and γ2L subunits. Messages for α2 and δ subunits are absent in DA cells. The specific reaction products can be identified by comparison with Table 1, which lists their lengths and the restriction enzymes used to establish specificity. The molecular weights of the restriction products are those expected for each subunit cDNA. A nonspecific band of lower molecular weight is present in the reaction for β3. On the other hand, the reaction for γ1 consistently yielded a nonspecific band at higher molecular weights.
Next, we examined whether all DA cells contained seven GABAA receptor subunit transcripts or could be assigned to subpopulations each expressing a different repertory of GABAA receptor mRNAs. To this purpose, we resorted to multiplex semi-nested RT-PCR: in this method, first-strand cDNA was synthesized from single cells and amplified in the presence of primers for all subunit transcripts detected previously. Then, the product of the first-round amplification was distributed among seven different second-round reactions, each specific for a single subunit cDNA. Seven DA cells were examined, and four of them contained the transcripts of all seven subunits (Fig.2). In three cells the reaction failed, and we did not obtain any PCR product. Large, unlabeled neurons, possibly ganglion cells, expressed different repertories of subunit mRNAs.
Fig. 2.
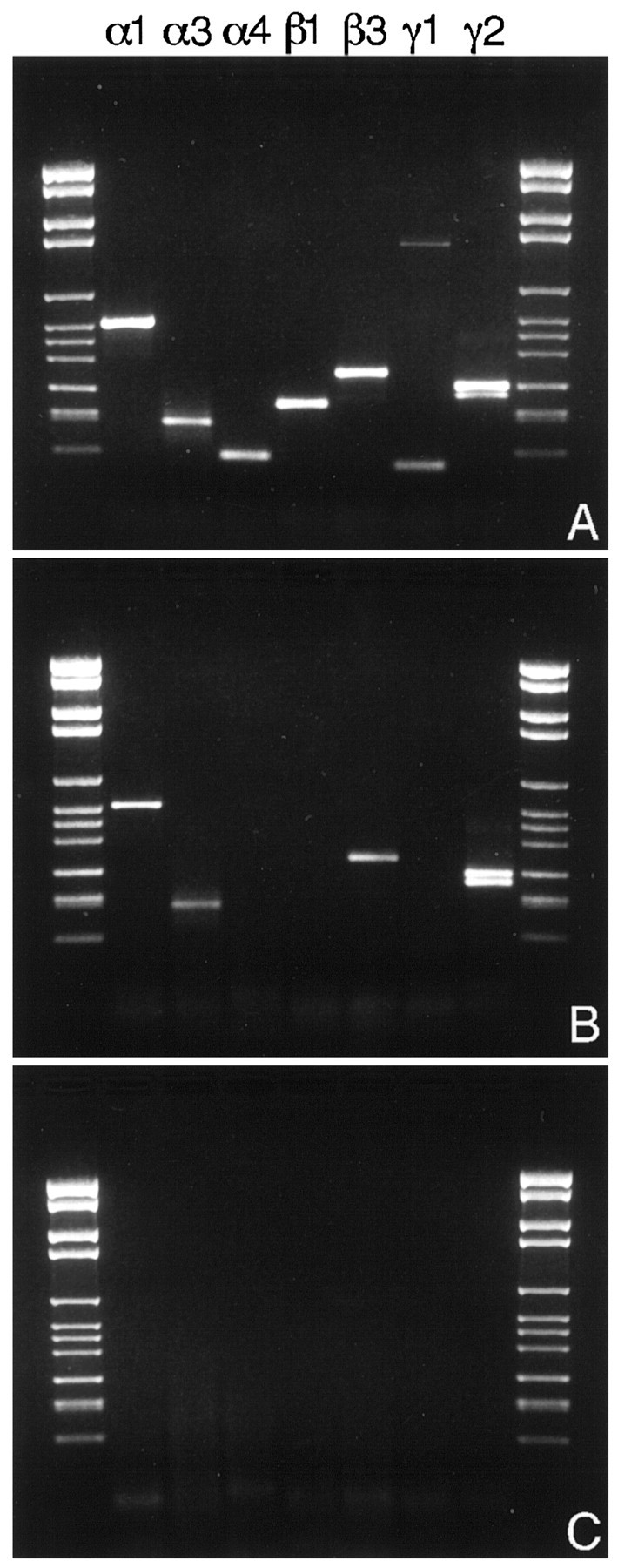
A, Agarose gel electrophoresis of the products of a multiplex semi-nested single-cell RT-PCR for the mRNAs of the α1, α3, α4, β1, β3, γ1, and γ2 subunits of the GABAA receptor in a DA cell. All seven subunits subtypes are expressed in the same cell. B, Pattern of expression of the seven subunits in a large bystander neuron, possibly a ganglion cell. Messages for α1, α3, β3, and γ2 are present.C, Control reaction in absence of the cell.
Because DA cells were isolated from the retina by enzymatic digestion and mechanical trituration and were exposed to abnormal culture stimuli before harvesting, we were concerned that their repertory of GABAA receptor subunit mRNAs did not reflect the true situation in the intact retina. Especially worrisome in this respect was the suppression of synaptic inputs that could alter the expression of postsynaptic receptor genes. Our approach to this problem was twofold. First, we investigated whether the repertory of transcripts varied over time: we began to observe loss of some of the messengers at 4.5 hr. As a result, cell harvesting was limited to the first 90 min after dissociation. To test whether experimental conditions induced a new pattern of gene expression, we shut offde novo transcription immediately after trituration by blocking the activity of RNA Polymerase II with α-amanitin (2 μg/ml) (Montarolo et al., 1986). Multiplex semi-nested RT-PCR analysis proved that the pattern of GABAAreceptor subunit mRNAs remained the same after inhibition of mRNA synthesis (data not shown). These finding confirmed that we were detecting transcripts that were present in the intact retina before dissociation.
Evidence was now sought to confirm that the subunit mRNAs were translated into proteins that were expressed at the surface of DA cells in the intact retina.
Immunocytochemistry
Vertical and horizontal sections of the retina were double-immunolabeled for TH and one of the seven subunits of the GABAA receptor whose mRNA had been identified in DA cells by single-cell RT-PCR. A 10 sec formaldehyde fixation under microwave irradiation was used to limit denaturation of the subunits. DA cells were readily recognized because they were the only retinal neurons stained by anti-TH antibody (Versaux-Botteri et al., 1984); furthermore, the entire neuron was immunoreactive, and this facilitated the analysis of the distribution of the subunits at the cell surface. As reported previously (Gustincich et al., 1997), mouse DA cells had a perikaryon 15 μm in diameter situated in the most vitreal tier of the inner nuclear layer. Large dendrites originated from the inner surface of the cell body and immediately spread horizontally in the inner plexiform layer (IPL). Here they branched repeatedly, giving rise to a dense plexus of varicose processes, rigorously confined to the most scleral stratum of the IPL. From this plexus, thinner, beaded processes occasionally descended into the center of the IPL and, after a long horizontal course, returned to the overlying dendritic plexus. DA cells also gave rise to vertically ascending processes. Most of them ended with a swelling in the outer plexiform layer (OPL); a few bent at a right angle and ran a long course in the OPL, where they formed a sparse horizontal plexus. Thus, some or all of DA cells belong to the interplexiform variety. DA cells conform to the general rule that most of the surface of the perikaryon of amacrine cells is devoid of synapses, except for the pole directed toward the IPL (data not shown).
Immunoreactivity for the GABAA receptor subunits was noted in three distinct locations: (1) as a diffuse staining of the entire cell surface; (2) in a small number of cytoplasmic organelles on the vitreal aspect of the nucleus, possibly endoplasmic reticulum and/or Golgi elements; and (3) as discrete clusters throughout the dendritic tree and vitreal pole of the perikaryon. The staining of the cell surface was distinct but weak. It was observed with the antibodies to all seven subunits (Fig.3A–H). To confirm that this staining resided in the cell membrane rather than in the underlying endoplasmic reticulum (Connolly et al., 1996) or Golgi complex, we resorted to an in vivo experiment of antibody-induced endocytosis. We treated slices of living retina with the polyclonal antibody to the γ2 subunit. This is the only subunit-specific antibody that recognizes the extracellular domain of the receptor (Ebert et al., 1999). After a rinse with Ames medium and incubation in fluorescein-conjugated anti-rabbit antibody, the entire surface of numerous cell bodies in the inner nuclear layer became fluorescent, showing that the γ2 subunit resided in the cell membrane. Within a matter of minutes, however, the label formed patches and was internalized. After fixation and permeabilization, DA cells were stained with the monoclonal anti-TH, followed by Texas Red-conjugated anti-mouse antibody. Newly formed, large endosomes containing the internalized subunit were present in the cytoplasm of DA cells (Fig. 3I). This experiment confirmed that DA cells express extrasynaptic GABAA receptors at the cell surface.
Fig. 3.
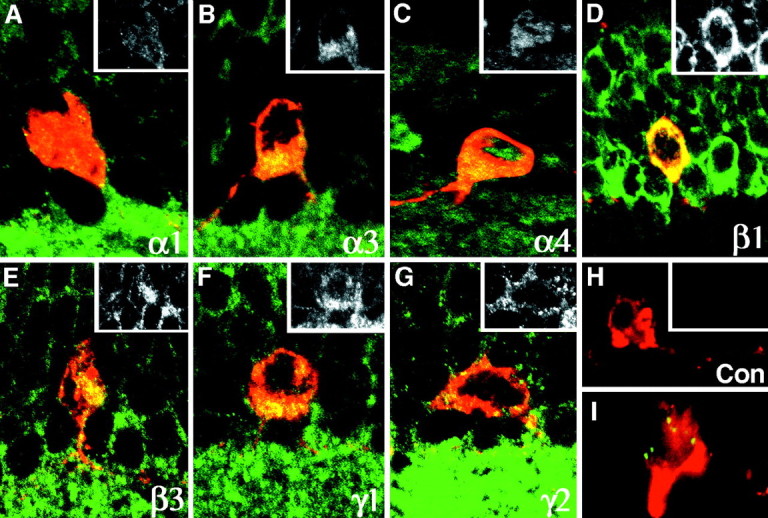
A–H, Vertical sections of the retina double-immunolabeled for TH (red) and one of the seven subunits of the GABAA receptor (green) whose mRNA had been identified in DA cells by single-cell RT-PCR. The cytoplasm of DA cells is stained by the TH antibody. The entire cell surface is weakly positive for all seven GABAA receptor subunits whose mRNA was identified by single-cell RT-PCR; this is confirmed by the insets that illustrate the staining with the subunit antibodies alone. In addition, cytoplasmic organelles are stained. Control experiments (Con) in which only the primary antibody to the subunits was omitted confirm the specificity of the reaction. I, In a DA cell, fluorescent endosomes contain the γ2 subunits that were swept from the cell surface by antibody-mediated endocytosis (for details see Results). Magnification 750×.
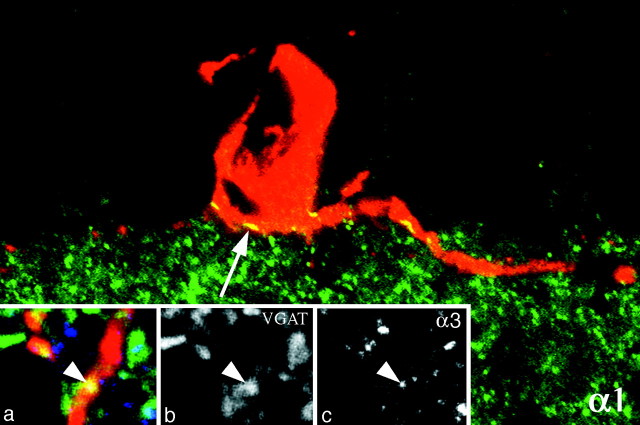
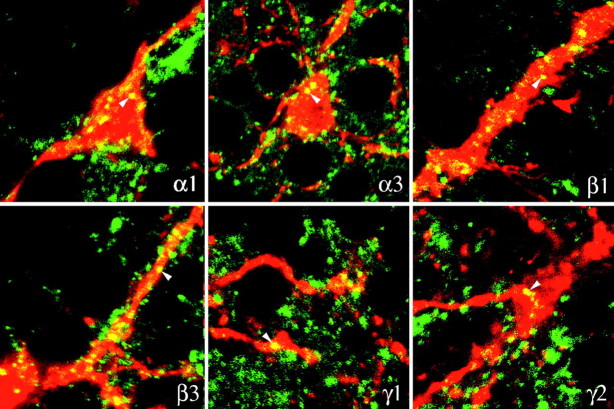
With the exception of α4, the six remaining subunits formed intensely fluorescent, compact clusters on the surface of the dendrites of DA cells and at the vitreal pole of their perikaryon (Figs.4, 5). At high magnification, the clusters appeared as round or elliptical plaques 0.2–0.5 μm in diameter when seen en face, and as short, thin lines when seen in profile along the edge of the dendrites (Fig. 4). Because of their size and shape, they were identified as postsynaptic specializations; this identification was supported by the fact that different subunits were localized in the same cluster (see below). Furthermore, after triple immunolabeling for TH, α3 subunit of the GABAA receptor, and VGAT, clusters of α3 subunits were seen in register with presynaptic GABAergic endings (Fig.4, insets). In places, the synaptic clusters were more irregular or appeared as loose aggregates of fluorescent puncta; these images were probably the result of inadequate stabilization of the synaptic membrane, because fixation was so brief.
Fig. 4.
In a vertical section of the retina stained for the α1 subunit, clusters of receptors appear as short, thin lines when seen in profile along the edge of dendrites (arrow). Because of their size and shape, these clusters correspond to postsynaptic active zones containing GABAAreceptors. This interpretation is confirmed by theinsets, in which triple-labeling for TH (a), VGAT (b), and α3 (c) shows that a cluster of α3 subunits is in register with a presynaptic GABAergic ending. Magnification 2500×.
Fig. 5.
Horizontal sections of the retina double-immunolabeled for TH (red) and six of the subunits of the GABAA receptor (green) whose mRNA had been identified in DA cells by single-cell RT-PCR. Results with α4 are absent, because no synaptic localization was observed for this subunit. Clusters intensely positive for the GABAA receptor subunits indicated in thebottom right corner of each micrograph are present at the surface of DA cells (arrowheads). The micrographs illustrating the results with the antibodies to α1, α3, and γ2 are horizontal sections through the vitreal pole of the perikaryon. Magnification 1000×.
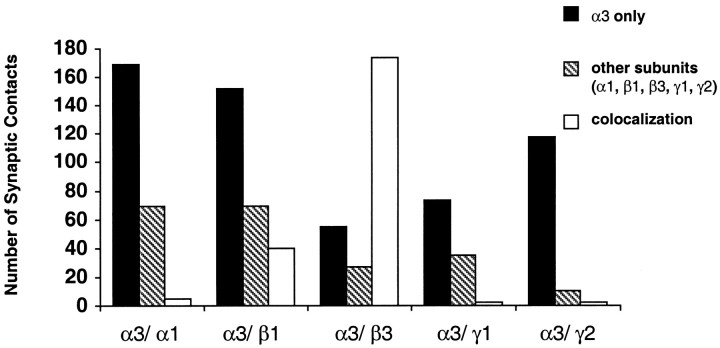
To establish whether the distribution of the subunits varied in different synapses, we resorted to triple-labeling with mouse antibody to TH, guinea pig antibody to α3 (Gao et al., 1993), and rabbit antibody to one of the remaining subunits (with the exception of α4), followed by secondary antibodies conjugated to different fluorophores (Fig. 6). The three high-magnification confocal images were finally stored sequentially in different color channels of a computer. A total of 1000 receptor clusters were examined by comparing their images in the different channels. We could therefore establish their position at the cell surface and their subunit composition. This examination also confirmed the total absence of cross-reactivity between secondary antibodies and of bleeding between different channels, because in absence of colocalization the stained clusters were consistently seen in a single channel. The histogram in Figure 7 illustrates our results in comparing the synaptic clusters stained by the guinea pig antibody to α3 with those stained by the rabbit antibodies to the remaining five synaptic subunits. We found that (1) α3 and α1 subunits were localized in different synapses (synapses with α3 alone = 73%, with α1 alone = 28%, with both subunits = 2%; 243 synapses examined). Furthermore, synapses containing α3 were distributed throughout the dendritic arborization, whereas those containing α1 had a preference for larger dendrites. (2) α3 was associated most commonly with β3 (synapses with α3 alone = 21%, with β3 alone = 11%, with both = 68%; 256 synapses examined) and more rarely with β1 (synapses with α3 alone = 58%, with β1 alone = 26%, with both = 15%; 261 synapses examined). (3) α3 did not colocalize to an appreciable extent with either γ1 (synapses with α3 alone = 66%, with γ1 alone = 32%, with both = 1.8%; 110 synapses examined) or γ2 (synapses with α3 alone = 91%, with γ2 alone = 8%, with both = 2%; 130 synapses examined). It should be noted that fewer synapses were stained by the antibodies to γ1 or γ2.
Fig. 6.
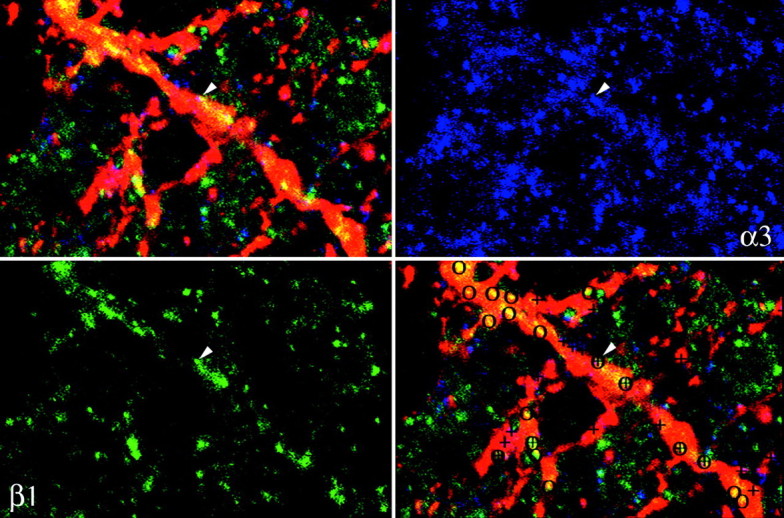
Horizontal sections of the retina triple-immunolabeled for TH (red), α3 (blue), and β1 (green). Subunit clusters on the dendrites of DA cells were labeled at high magnification by comparing images in different channels: circles indicate synapses that contain β1, and crosses indicate synapses that contain α3. The results of the counts are illustrated in Figure7. Magnification 1500×.
Fig. 7.
Synaptic colocalization of α3 with the other subunits present in DA cells. For details, see Results.
DISCUSSION
Combination of transgenic technology with single-cell RT-PCR and immunocytochemistry proved to be a powerful tool to identify the composition of the GABAA receptors in DA cells. First and foremost, we could obtain precise data on a very small neuronal population. This approach could be very useful for studying receptor gene expression in other neurons that are sparsely represented in the nervous system and do not possess a distinctive shape. Secondly, when cells are harvested individually after patch clamping and care is taken to avoid contamination, highly reproducible results were obtained with medium abundance transcripts, such as those for the subunits of this neurotransmitter receptor. Negative results, on the other hand, should be considered with caution, because the sensitivity of the technique remains to be determined.
Because little is known of the turnover of mRNAs in intact organs or after dissociation, we were concerned that our findings in the isolated cells did not reflect the true situation in the intact retina. Messages for the GABAA receptor, however, were stable for a long enough interval after dissociation that harvesting of a rare cell type could take place within a comfortable period of time. Our confidence that we had obtained a reliable representation of thein vivo transcript repertory was confirmed when mRNA synthesis was blocked and was reinforced by the finding that this repertory was remarkably consistent among different DA cells. In the past, only one report examined this important point: apparently, α1 and α6 subunit transcripts were not uniformly distributed among cerebellar granule cells (Santi et al., 1994). Our result that the subunit repertory is the same for all DA cells indicates that rules about an entire cell population can be extrapolated from findings obtained in individual cells. Furthermore, this observation could only be obtained by single-cell RT-PCR, because a quantitative evaluation of the electrophysiological properties of an entire neuronal population is prevented by variations in cell viability. Immunocytochemistry, on the other hand, is too much dependent on the quality of the antibody used and severely limited by the vagaries of the interaction between antigen epitopes and fixation. For instance, our results do not support previous observations in the rat retina that DA cells do not express α1 and β1 subunits (Greferath et al., 1995).
DA cells contain transcripts for seven subunits of the GABAA receptor, namely α1, α3, α4, β1, β3, γ1, γ2S, and γ2L; furthermore, all these messages are translated and their products are transported to the cell surface. Such a complexity in GABAA receptor composition is not new, for three other types of neurons, hippocampal pyramidal cells (Nusser et al., 1996b), and granule cells of both cerebellum and dentate gyrus (Laurie et al., 1992; Nusser et al., 1995, 1998), exhibit a similar richness in subunit subtypes.
Previous experiments showed that DA cells express bicuculline-sensitive GABAA receptors at their surface (Gustincich et al., 1997). More recently, we have demonstrated that most of the subunits are assembled into functional receptors. In fact, pharmacological dissection of the benzodiazepine sensitivity of DA cells’ responses to GABA suggested the presence of assemblies containing α1, one β, and γ2 subunits (Feigenspan et al., 1998). Furthermore, the GABA response was potentiated by loreclezole, which confirms the existence of the β3 subunit (Wafford et al., 1994;Wingrove et al., 1994). Further evidence that multiple GABAA receptors exist at the surface of DA cells emerges from previous electrophysiological observations after transfection of different subunit combinations: α4-containing receptors are insensitive to benzodiazepines (Benke et al., 1997), and the γ1 subunit does not co-assemble with γ2 (Mossier et al., 1994;Quirk et al., 1994).
With the exception of α4, six of the subunits formed clusters on the surface of DA cells. Because of their size, shape, and location opposite GABAergic endings, these clusters were interpreted as postsynaptic active zones containing GABAAreceptors. The postsynaptic clusters did not exhibit homogeneous composition. Clusters distributed throughout the dendritic tree contained the α3 subunit associated with β3 and, less frequently, β1, whereas clusters containing the α1 subunit were confined to large dendrites. Both types of clusters were found on the vitreal pole of the perikaryon. By exclusion, the β1 subunit had to be prevalently associated with α1. We were surprised that γ subunits were not colocalized with the α3 subunit, but the antibodies to γ1 and γ2 stained fewer synapses. It is possible that the antibodies to the γ subunits are less sensitive or recognize epitopes more easily denatured by the fixative fluid.
It is therefore tempting to speculate that a receptor consisting of α1, β1, and γ2 subunits is localized at postsynaptic active zones on large dendrites, whereas a second type of receptor containing α3 and β3 subunits is postsynaptic at contacts distributed throughout the dendritic tree. Both types of synapses would be present on the vitreal aspect of the perikaryon. Most likely, two or more varieties of GABAergic amacrines synapse with DA cells and inhibit dopamine release with different physiological actions at the postsynaptic membrane. This is not surprising if one considers that GABA is the major transmitter that controls dopamine release (Morgan and Kamp, 1980; Kirsch and Wagner, 1989; Gustincich et al., 1997). In fact, GABA suppresses light-evoked release in the intact retina, and GABA receptor antagonists induce dopamine release in the dark (Kamp and Morgan, 1981;O’Connor et al., 1986; Ishita et al., 1988; Critz and Marc, 1992;Kolbinger and Weiler, 1993; Gustincich et al., 1997). Multiple types of GABAergic amacrines would therefore be responsible for subtle regulations of DA cell activity, perhaps in different conditions of illumination.
In agreement with findings reported elsewhere in the nervous system (Fritschy et al., 1992; Nusser et al., 1995, 1996a), a large repertory of subunits is also present throughout the neuronal surface, suggesting the existence of multiple types of extrasynaptic receptors. It is interesting that only the α4 subunit is exclusively extrasynaptic. The function of α4 is poorly understood. It is commonly colocalized with the δ subunit (Benke et al., 1997), which in turn is exclusively extrasynaptic in cerebellar granule cells (Nusser et al., 1998).
The presence of extrasynaptic GABAA receptors suggests that the activity of a neuron responsible for neural adaptation to light must constantly integrate excitation and inhibition over its entire surface and accordingly regulate dopamine release. After all, the composition of the chemical soup in the intercellular spaces of the inner plexiform layer mirrors the diversity of the visual scene presented to the retina.
Footnotes
This research was supported by National Institutes of Health Grant EY-01344. We thank J.-M. Fritschy for providing the anti-α3 subunit antibody and R. H. Edwards for the anti-vesicular GABA transporter (VGAT) antibody. We are grateful to Heather Regan and Jennifer J. Wilson for assistance.
Correspondence should be addressed to Elio Raviola, Department of Neurobiology, Harvard Medical School, 220 Longwood Avenue, Boston, MA 02115.
REFERENCES
- 1.Barnard EA, Skolnick P, Olsen RW, Mohler H, Sieghart W, Biggio G, Braestrup C, Bateson AN, Langer SZ. International Union of Pharmacology. XV. Subtypes of γ-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol Rev. 1998;50:291–313. [PubMed] [Google Scholar]
- 2.Benke D, Michel C, Mohler H. GABAA receptors containing the α4-subunit: prevalence, distribution, pharmacology, and subunit architecture in situ. J Neurochem. 1997;69:806–814. doi: 10.1046/j.1471-4159.1997.69020806.x. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhry FA, Reimer RJ, Bellocchio EE, Danbolt NC, Osen KK, Edwards RH, Storm-Mathisen J. The vesicular GABA transporter, VGAT, localizes to synaptic vesicles in sets of glycinergic as well as GABAergic neurons. J Neurosci. 1998;18:9733–9750. doi: 10.1523/JNEUROSCI.18-23-09733.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Connolly CN, Krishek BJ, McDonald BJ, Smart TG, Moss SJ. Assembly and cell surface expression of heteromeric and homomeric γ-aminobutyric acid type A receptors. J Biol Chem. 1996;271:89–96. doi: 10.1074/jbc.271.1.89. [DOI] [PubMed] [Google Scholar]
- 6.Critz SD, Marc RE. Glutamate antagonists that block hyperpolarizing bipolar cells increase the release of dopamine from turtle retina. Vis Neurosci. 1992;9:271–278. doi: 10.1017/s0952523800010683. [DOI] [PubMed] [Google Scholar]
- 7.De Waele P, De Groote G, Van De Voorde A, Fiers W, Franssen J-D, Herion P, Urbain J. Isolation and identification of monoclonal antibodies directed against human placental alkaline phosphatase. Arch Int Physiol Biochim Biophys. 1982;90:B21. [Google Scholar]
- 8.Djamgoz MB, Wagner HJ. Localization and function of dopamine in the adult vertebrate retina. Neurochem Int. 1992;20:139–191. doi: 10.1016/0197-0186(92)90166-o. [DOI] [PubMed] [Google Scholar]
- 9.Ebert V, Scholze P, Fuchs K, Sieghart W. Identification of subunits mediating clustering of GABAA receptors by rapsyn. Neurochem Int. 1999;34:453–463. doi: 10.1016/s0197-0186(99)00039-x. [DOI] [PubMed] [Google Scholar]
- 10.Feigenspan A, Gustincich S, Raviola E. Electrophysiology of dopaminergic amacrine cells GABAA receptors. Exp Eye Res [Suppl] 1998;67:178. [Google Scholar]
- 11.Fritschy J-M, Benke D, Mertens S, Oertel WH, Bachi T, Möhler H. Five subtypes of type A γ-aminobutyric acid receptors identified in neurons by double and triple immunofluorescence staining with subunit-specific antibodies. Proc Natl Acad Sci USA. 1992;89:6726–6730. doi: 10.1073/pnas.89.15.6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao B, Fritschy JM, Benke D, Mohler H. Neuron-specific expression of GABAA-receptor subtypes: differential association of the α1- and α3-subunits with serotonergic and GABAergic neurons. Neuroscience. 1993;54:881–892. doi: 10.1016/0306-4522(93)90582-z. [DOI] [PubMed] [Google Scholar]
- 13.Ghia P, ten Boekel E, Sanz E, de la Hera A, Rolink A, Melchers F. Ordering of human bone marrow B lymphocyte precursors by single-cell polymerase chain reaction analyses of the rearrangement status of the immunoglobulin H and L chain gene loci. J Exp Med. 1996;184:2217–2229. doi: 10.1084/jem.184.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greferath U, Grünert U, Fritschy JM, Stephenson A, Möhler H, Wässle H. GABAA receptor subunits have differential distributions in the rat retina: in situ hybridization and immunohistochemistry. J Comp Neurol. 1995;353:553–571. doi: 10.1002/cne.903530407. [DOI] [PubMed] [Google Scholar]
- 15.Gustincich S, Feigenspan A, Wu DK, Koopman LJ, Raviola E. Control of dopamine release in the retina: a transgenic approach to neural networks. Neuron. 1997;18:723–736. doi: 10.1016/s0896-6273(00)80313-x. [DOI] [PubMed] [Google Scholar]
- 16.Ishita S, Negishi K, Teranishi T, Shimada Y, Kato S. GABAergic inhibition on dopamine cells of the fish retina: a [3H]dopamine release study with isolated fractions. J Neurochem. 1988;50:1–6. doi: 10.1111/j.1471-4159.1988.tb13221.x. [DOI] [PubMed] [Google Scholar]
- 17.Jones A, Korpi ER, McKernan RM, Pelz R, Nusser Z, Mäkelä R, Mellor JR, Pollard S, Bahn S, Stephenson FA, Randall AD, Sieghart W, Somogyi P, Smith AJH, Wisden W. Ligand-gated ion channel subunit partnerships: GABAA receptor α6 subunit gene inactivation inhibits δ subunit expression. J Neurosci. 1997;17:1350–1362. doi: 10.1523/JNEUROSCI.17-04-01350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamp CW, Morgan WW. GABA antagonists enhance dopamine turnover in the rat retina in vivo. Eur J Pharmacol. 1981;69:273–279. doi: 10.1016/0014-2999(81)90473-8. [DOI] [PubMed] [Google Scholar]
- 19.Kirsch M, Wagner HJ. Release pattern of endogenous dopamine in teleost retinae during light adaptation and pharmacological stimulation. Vision Res. 1989;29:147–154. doi: 10.1016/0042-6989(89)90120-x. [DOI] [PubMed] [Google Scholar]
- 20.Kolbinger W, Weiler R. Modulation of endogenous dopamine release in the turtle retina: effects of light, calcium, and neurotransmitters. Vis Neurosci. 1993;10:1035–1041. doi: 10.1017/s0952523800010142. [DOI] [PubMed] [Google Scholar]
- 21.Laurie DJ, Seeburg PH, Wisden W. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. II. Olfactory bulb and cerebellum. J Neurosci. 1992;12:1063–1076. doi: 10.1523/JNEUROSCI.12-03-01063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKernan RM, Whiting PJ. Which GABAA-receptor subtypes really occur in the brain? Trends Neurosci. 1996;19:139–143. doi: 10.1016/s0166-2236(96)80023-3. [DOI] [PubMed] [Google Scholar]
- 23.Montarolo PG, Goelet P, Castellucci VF, Morgan J, Kandel ER, Schacher S. A critical period for macromolecular synthesis in long-term heterosynaptic facilitation in Aplysia. Science. 1986;234:1249–1254. doi: 10.1126/science.3775383. [DOI] [PubMed] [Google Scholar]
- 24.Morgan WW, Kamp CW. A GABAergic influence on the light-induced increase in dopamine turnover in the dark-adapted rat retina in vivo. J Neurochem. 1980;34:1082–1086. doi: 10.1111/j.1471-4159.1980.tb09943.x. [DOI] [PubMed] [Google Scholar]
- 25.Mossier B, Tögel M, Fuchs K, Sieghart W. Immunoaffinity purification of γ-aminobutyric acidA (GABAA) receptors containing γ1-subunits: evidence for the presence of a single type of γ-subunit in GABAA receptors. J Biol Chem. 1994;269:25777–25782. [PubMed] [Google Scholar]
- 26.Nusser Z, Roberts JDB, Baude A, Richards JG, Sieghart W, Somogyi P. Immunocytochemical localization of the α1 and β2/3 subunits of the GABAA receptor in relation to specific GABAergic synapses in the dentate gyrus. Eur J Neurosci. 1995;7:630–646. doi: 10.1111/j.1460-9568.1995.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 27.Nusser Z, Sieghart W, Stephenson FA, Somogyi P. The α6 subunit of the GABAA receptor is concentrated in both inhibitory and excitatory synapses on cerebellar granule cells. J Neurosci. 1996a;16:103–114. doi: 10.1523/JNEUROSCI.16-01-00103.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nusser Z, Sieghart W, Benke D, Fritschy J-M, Somogyi P. Differential synaptic localization of two major γ-aminobutyric acid type A receptor α subunits on hippocampal pyramidal cells. Proc Natl Acad Sci USA. 1996b;93:11939–11944. doi: 10.1073/pnas.93.21.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nusser Z, Sieghart W, Somogyi P. Segregation of different GABAA receptors to synaptic and extrasynaptic membranes of cerebellar granule cells. J Neurosci. 1998;18:1693–1703. doi: 10.1523/JNEUROSCI.18-05-01693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connor P, Dorison SJ, Watling KJ, Dowling JE. Factors affecting release of 3H-dopamine from perfused carp retina. J Neurosci. 1986;6:1857–1865. doi: 10.1523/JNEUROSCI.06-07-01857.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearce RA. Physiological evidence for two distinct GABAA responses in rat hippocampus. Neuron. 1993;10:189–200. doi: 10.1016/0896-6273(93)90310-n. [DOI] [PubMed] [Google Scholar]
- 32.Quirk K, Gillard NP, Ragan CI, Whiting PJ, McKernan RM. γ-aminobutyric acid type A receptors in the rat brain can contain both γ2 and γ3 subunits, but γ1 does not exist in combination with another γ subunit. Mol Pharmacol. 1994;45:1061–1070. [PubMed] [Google Scholar]
- 33.Rabow LE, Russek SJ, Farb DH. From ion currents to genomic analysis: recent advances in GABAA receptor research. Synapse. 1995;21:189–274. doi: 10.1002/syn.890210302. [DOI] [PubMed] [Google Scholar]
- 34.Santi MR, Vicini S, Eldadah B, Neale JH. Analysis by polymerase chain reaction of α1 and α6 GABAA receptor subunit mRNAs in individual cerebellar neurons after whole-cell recordings. J Neurochem. 1994;63:2357–2360. doi: 10.1046/j.1471-4159.1994.63062357.x. [DOI] [PubMed] [Google Scholar]
- 35.Sieghart W. Structure and pharmacology of γ-aminobutyric acidA receptor subtypes. Pharmacol Rev. 1995;47:181–234. [PubMed] [Google Scholar]
- 36.Sieghart W, Fuchs K, Tretter V, Ebert V, Jechlinger M, Höger H, Adamiker D. Structure and subunit composition of GABAA receptors. Neurochem Int. 1999;34:379–385. doi: 10.1016/s0197-0186(99)00045-5. [DOI] [PubMed] [Google Scholar]
- 37.Sperk G, Schwarzer C, Tsunashima K, Fuchs K, Sieghart W. GABAA receptor subunits in the rat hippocampus I: immunocytochemical distribution of 13 subunits. Neuroscience. 1997;80:987–1000. doi: 10.1016/s0306-4522(97)00146-2. [DOI] [PubMed] [Google Scholar]
- 38.Varecka L, Wu C-H, Rotter A, Frostholm A. GABAA/Benzodiazepine receptor α6 subunit mRNA in granule cells of the cerebellar cortex and cochlear nuclei: expression in developing and mutant mice. J Comp Neurol. 1994;339:341–352. doi: 10.1002/cne.903390304. [DOI] [PubMed] [Google Scholar]
- 39.Versaux-Botteri C, Nguyen-Legros J, Vigny A, Raoux N. Morphology, density and distribution of tyrosine hydroxylase-like immunoreactive cells in the retina of mice. Brain Res. 1984;301:192–197. doi: 10.1016/0006-8993(84)90423-2. [DOI] [PubMed] [Google Scholar]
- 40.Wafford KA, Bain CJ, Quirk K, McKernan RM, Wingrove PB, Whiting PJ, Kemp JA. A novel allosteric modulatory site on the GABAA receptor β subunit. Neuron. 1994;12:775–782. doi: 10.1016/0896-6273(94)90330-1. [DOI] [PubMed] [Google Scholar]
- 41.Whiting P, McKernan RM, Iversen LL. Another mechanism for creating diversity in γ-aminobutyrate type A receptors: RNA splicing directs expression of two forms of γ2 subunit, one of which contains a protein kinase C phosphorylation site. Proc Natl Acad Sci USA. 1990;87:9966–9970. doi: 10.1073/pnas.87.24.9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wingrove PB, Wafford KA, Bain C, Whiting PJ. The modulatory action of loreclezole at the γ-aminobutyric acid type A receptor is determined by a single amino acid in the β2 and β3 subunit. Proc Natl Acad Sci USA. 1994;91:4569–4573. doi: 10.1073/pnas.91.10.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wisden W. Structure and distribution of multiple GABAA receptor subunits with special reference to the cerebellum. Ann NY Acad Sci. 1995;757:506–515. doi: 10.1111/j.1749-6632.1995.tb17510.x. [DOI] [PubMed] [Google Scholar]
- 44.Wisden W, Laurie DJ, Monyer H, Seeburg PH. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. I. Telencephalon, diencephalon, mesencephalon. J Neurosci. 1992;12:1040–1062. doi: 10.1523/JNEUROSCI.12-03-01040.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witkovsky P, Dearry A. Functional roles of dopamine in the vertebrate retina. Prog Retinal Res. 1991;11:247–292. [Google Scholar]