Abstract
Here, we show that a nitric oxide synthase (NOS) pseudogene is expressed in the CNS of the snail Lymnaea stagnalis. The pseudo-NOS transcript includes a region of significant antisense homology to a previously reported neuronal NOS (nNOS)-encoding mRNA. This suggested that the pseudo-NOS transcript acts as a natural antisense regulator of nNOS protein synthesis. In support of this, we show that both the nNOS-encoding and the pseudo-NOS transcripts are coexpressed in giant identified neurons (the cerebral giant cells) in the cerebral ganglion. Moreover, reverse transcription-PCR experiments on RNA isolated from the CNS establish that stable RNA–RNA duplex molecules do form between the two transcripts in vivo. Using an in vitro translation assay, we further demonstrate that the antisense region of the pseudogene transcript prevents the translation of nNOS protein from the nNOS-encoding mRNA. By analyzing NOS RNA and nNOS protein expression in two different identified neurons, we find that when both the nNOS-encoding and the pseudo-NOS transcripts are present in the same neuron, nNOS enzyme activity is substantially suppressed. Importantly, these results show that a natural antisense mechanism can mediate the translational control of nNOS expression in the Lymnaea CNS. Our findings also suggest that transcribed pseudogenes are not entirely without purpose and are a potential source of a new class of regulatory gene in the nervous system.
Keywords: nNOS, antisense, pseudogene, translational regulation, dsRNA, RNA interface
Nitric oxide (NO) is now recognized as an intercellular signaling molecule in the nervous system (Garthwaite et al., 1988). It is generated from l-arginine and molecular oxygen by the enzyme NO synthase (NOS), the neuronal isoform of which (nNOS) is activated by calcium/calmodulin (Bredt and Snyder, 1990). The neuronal expression of nNOS protein was first confirmed immunocytochemically in the rat brain (Bredt et al., 1990), and many subsequent studies show that NO is a neurotransmitter, although perhaps an enigmatic one (for review, see Bredt and Snyder, 1992; Dawson and Snyder, 1994; Hölscher, 1997).
To understand at cellular, molecular, and behavioral levels precisely how NO functions as a neurotransmitter, we have exploited the advantages of the presence of identifiable neurons in the nervous system of the mollusc Lymnaea stagnalis. Building on the finding that NO mediates the activation of feeding behavior inLymnaea (Elphick et al., 1995), we have now identified a number of nNOS-expressing neurons that are involved in feeding (Korneev et al., 1998; Park et al., 1998). These include two large and readily identifiable neurons known as the B2 motoneuron located in the buccal ganglion and the cerebral giant cell (CGC) of the cerebral ganglion. The existence of large identifiable nNOS-expressing neurons, together with our molecular characterization of NOS-encoding transcripts (Korneev et al., 1998), now allows us to investigate the expression of different NOS transcripts at the single-cell level. Our work has revealed a neuron-specific expression pattern for an unusual RNA molecule that cannot be translated into nNOS protein because it is transcribed from a NOS-related pseudogene. We show that this pseudo-NOS transcript has a crucial role in the regulation of nNOS protein synthesis through a natural antisense mechanism.
Pseudogenes are common in all eukaryotic genomes and are defined as inactive versions of currently functional genes (Lewin, 1990). They have been rendered inactive by the accumulation of deleterious mutations that prevent any or all of the stages of gene expression, and they are regarded as evolutionary dead ends. The pseudogene we have identified is a member of the NOS gene family and is actively transcribed in the Lymnaea CNS. Importantly the pseudo-NOS RNA contains a region that is antisense to the functional nNOS mRNA, and it was this that suggested a role for the pseudotranscript in the regulation of nNOS protein synthesis.
Here, we show that both pseudo-NOS and nNOS-encoding transcripts are coexpressed in identified neurons, that stable RNA–RNA duplexes form between them, and that the antisense region of the pseudo-NOS RNA specifically suppresses the synthesis of the nNOS protein. This represents a novel mechanism for regulating the expression of an important neuronal signaling pathway in the brain. Also, our findings suggest the NOS pseudogene is a member of a new class of regulatory genes that are derived from the pool of “nonfunctional” pseudogenes. Pseudogenes therefore are not all relics of evolution and entirely without purpose as the classical picture of them might suggest.
MATERIALS AND METHODS
cDNA library construction. Approximately 3 μg of poly(A+) RNA isolated from totalLymnaea CNS RNA by means of Dynabeads oligo-dT25 (Dynal, Great Neck, NY) were used to construct a cDNA library according to the manufacturer’s protocol for SuperScript Choice System (Life Technologies, Gaithersburg, MD).
Northern hybridization.Poly(A+) RNA (3–5 μg) isolated from the CNS using Oligotex mRNA Mini kit (Qiagen, Hilden, Germany) were resolved in 1% denaturing formaldehyde-containing agarose gel and transferred onto NYTRAN-N membrane (Schleicher & Schuell, Keene, NH).
Two different 32P-labeled probes were used, one corresponding to the 3′ untranslated region ofLymnaea nNOS cDNA. The other probe was generated by asymmetric PCR and represents a single-stranded DNA complementary to the antisense region of the pseudo-NOS RNA and cannot therefore recognize functional nNOS mRNA. Hybridizations were performed at 45°C in a buffer containing 10% dextran sulfate, 5× SSPE, 5× Denhardt’s solution, 50% formamide, 0.5% SDS, and 100 μg/ml denatured and sheared salmon sperm DNA.
Ribonuclease protection assay in vitro. Approximately 1 μg of linearized plasmid DNA containing functional nNOS cDNA was used inin vitro transcription reaction in the presence of digoxigenin (DIG)-UTP and T7 RNA polymerase (Boehringer Mannheim, Indianapolis, IN). A mixture of the DIG-labeled nNOS mRNA (200 ng) and a synthetic pseudo-NOS RNA (2 μg) was incubated for 24 hr at 50°C in a buffer containing 20 mm HEPES, pH 7.0, 100 mm KCl, and 1 mm EDTA and then treated with ribonuclease A (RNase A) under standard conditions (Shayiq, 1997). The products of the digestion were resolved in an agarose gel and blotted onto a positively charged nylon membrane. This was then subjected to an immunological detection procedure according to the manufacturer’s protocol. Crucially, a single band of ∼150 nucleotides (nt) corresponding to the expected size of the duplex was revealed, and no signal was detected in a control lane in which a sample containing RNase A-digested nNOS mRNA without the pseudo-NOS RNA was analyzed.
Detection of RNA duplexes in vivo. Cytoplasmic RNA was isolated from Lymnaea CNS under nondenaturing conditions. Briefly, the CNS was homogenized in a buffer containing 10 mm Tris HCl, pH 7.4, 3 mmCaCl2, 2 mmMgCl2, and 0.5% Nonidet P-40. The nuclei were then removed by centrifugation at 500 × g and 4°C for 10 min. The supernatant was treated with proteinase K and then extracted with phenol-chloroform. After precipitation with isopropanol, the RNA preparation was divided into two fractions; one was treated with an excess of RNase A and RQ1 DNase (Promega, Madison, WI), and the other was treated with RQ1 DNase (Shayiq, 1997). The two fractions of RNA were then reverse transcribed using SuperScript II (Life Technologies) and either P1 (5′-GCATGTTGAGATGGAAGAAC-3′) or P2 (5′AAAGACTGGTTTGAAAATCTC-3′) primers and amplified by means of Taq Supreme DNA polymerase (IGi) in the presence of P3 primer (5′-CAGAGCTGTGGAGTTCTC-3′) and either P1 or P2. Products of the PCR were then resolved in 3% MetaPhor agarose gel (FMC Bioproducts, Rockland, ME), cloned, and sequenced.
Reverse transcription-PCR on isolated identified neurons.The cell bodies of ∼20 CGCs and 20 B2 neurons were identified and then individually dissected from the CNS. Total RNA was extracted from each pool of neurons using the guanidine thiocyanate method (Chomczynski and Sacchi, 1987) and used as a template in a reverse transcription reaction in the presence of random primers and SuperScript II (Life Technologies). The cDNA generated was then subjected to 35 cycles of PCR amplification using the following parameters: denaturation, 94°C, 20 sec; annealing, 55°C, 30 sec; extension, 68°C, 90 sec. For detection of functional NOS mRNA, the PCR primers were as follows: #5, 5′-TGTGATCCTCACCGCTACAA-3′; and #8, 5′-GACTGTTGAGATGGAAGAAC-3′. For the detection of pseudo-NOS RNA, the primers were: #1, 5′-ATCTTCCTGTCTCCGAGGC-3′; and #4, 5′-TGTGGAAATGTGTTGCCCTT-3′. Nested PCRs were then performed under the same cycling parameters. The primers used for the nested PCR were as follows: #6, 5′-GCTCAACACCGAAACTGCGT-3′; and #7, 5′-GAAGGTACTAGTGATTACCA-3′ for detection of functional NOS mRNA. For detection of pseudo-NOS RNA, we used the following: #3, 5′-GCTAGTAGCCCAAGTCTCTT-3′; and #2, 5′-CACTATGGCATCTAAATGTTAAG-3′. For location of the primers, see Figure1B.
Fig. 1.
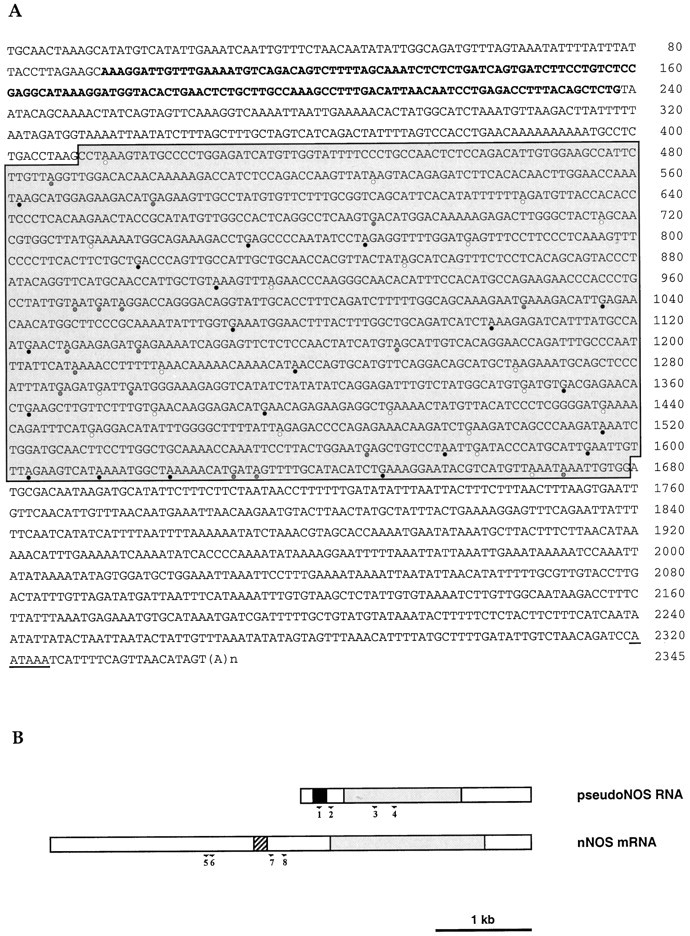
Molecular cloning of the pseudo-NOS transcript.A, Sequence of a full-length cDNA clone isolated from aLymnaea CNS cDNA library. The antisense region from 93 to 238 bp is indicated in bold type. The core region of high homology (>80%) to the nNOS-encoding transcript isshaded. A polyadenylation signal isunderlined. Stop codons within the core region are marked by circles: white, frame 1;shaded, frame 2; and black, frame 3.B, Schematic representation of the pseudo-NOS and nNOS-encoding transcripts. The antisense region in the pseudo-NOS transcript and its complementary counterpart are shown byblack and hatched boxes, respectively. Regions of high homology are shaded, and the unfilled areas have no significant homology to one another. The positions of the numbered primers used in RT-PCR experiments on isolated identified neurons are shown by arrows.
Hybrid arrest of translation. Lymnaea NOS cDNA cloned into pcDNA 3.1 was used as a template for an in vitrotranscription reaction to produce NOS cRNA, which was then capped according to the manufacturer’s protocol (Boehringer Mannheim). Similarly, cRNAs corresponding to the antisense region of the pseudo-NOS transcript were synthesized in either the sense or the antisense orientation with respect to the functional NOS cRNA, but they were not capped. Two hybridization reactions were performed: the first contained 0.5 μg of NOS cRNA and 0.2 μg of either sense or antisense pseudo-NOS cRNAs, whereas in the second, these values were 0.2 and 2 μg, respectively. Both mixtures were then incubated for 1 hr at 50°C in a buffer containing 20 mm HEPES, pH 7, 100 mm KCl, and 1 mmEDTA. The products of the hybridization were then translated in vitro using a wheat germ cell-free translational system (Promega) in the presence of 35S-methionine following the procedures of Schulz-Aellen et al. (1989). Labeled proteins were resolved using SDS-PAGE.
NADPH-diaphorase histochemistry. A previously described modified diaphorase method (Park et al., 1998) was used to reveal nNOS activity in fixed whole-mount preparations of the snail buccal ganglion (n = ∼100). Thirty cerebral ganglia were processed in exactly the same way to compare diaphorase staining in the CGCs with that in the B2 neurons.
RESULTS
Paradoxical expression of nNOS mRNA
Here, we report that a functional nNOS mRNA is expressed by two uniquely identified neurons in the Lymnaea CNS, the B2 motoneuron, and the CGC (see below). The expression of NOS protein by the B2 motoneuron was first reported by Moroz et al., (1994a,b), who showed that they are stained by the NOS-selective NADPH-diaphorase histochemical method (Matsumoto et al., 1993) and immunolabeled by an antibody to the neuronal isoform of NOS. The NADPH-diaphorase method responds to NOS enzyme activity in aldehyde-fixed cells and is an established indicator of the NOS enzyme in the mammalian nervous system (Dawson et al., 1991; Hope et al., 1991). We have verified that this technique also works reliably in invertebrate preparations (Elphick et al., 1995), and to confirm the findings of Moroz et al. (1994a,b), we have used the NADPH-diaphorase technique on whole-mount preparations on the entire Lymnaea CNS (Park et al., 1998). Below, we show that, in 100% of the preparations examined, the cell bodies of the paired B2 motoneurons in the buccal ganglion are strongly NADPH-diaphorase-positive, as expected. Paradoxically, the CGC is almost always NADPH-diaphorase-negative. This result is puzzling because it shows that the nNOS transcript can be present without the nNOS enzyme, suggesting translational control operates in the CGC to regulate nNOS protein expression. Results described below provide an unprecedented natural antisense mechanism involving a pseudogene that accounts for this paradoxical neuronal expression pattern.
An antisense-containing NOS pseudogene is transcribed in the CNS
We recently reported the cloning and expression of a full-length (5070 nt) mRNA encoding the first molluscan NOS (Korneev et al., 1998), the enzyme responsible for the calcium-regulated synthesis of the gaseous neurotransmitter NO (Bredt and Snyder 1992; Garthwaite and Boulton 1995). While screening a snail CNS cDNA library for other NOS-related transcripts, we isolated a smaller 2345 nt transcript, the sequence of which is shown in Figure 1 (GenBank accession number AF 165914). This small transcript shows >80% sequence identity over at least half its length with the larger nNOS transcript and would appear to be derived from another member of the same gene family. Although the transcript possesses features of a functional mRNA, such as the polyadenylation signal and a poly(A) tail, it cannot be translated into protein because of the presence of multiple stop codons in all three reading frames. This suggested to us that we had cloned a transcript from a NOS pseudogene, a conclusion confirmed by performing long-distance PCR on genomic DNA. The genomic sequence (data not shown) contains the exact sequence of the small transcript and includes introns, indicating that an unprocessed NOS pseudogene is actively transcribed in the snail CNS.
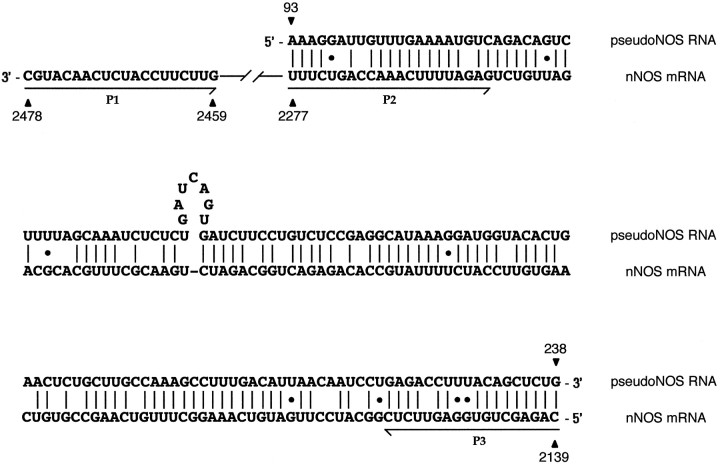
An unexpected feature of the pseudo-NOS transcript is the presence of a region ∼150 nt in length that is antisense to a region close to the middle of the functional nNOS mRNA (Fig.1A,B). This means that, in addition to being a pseudo-NOS RNA, the small transcript is also an example of atrans-encoded natural antisense RNA (Vanhe-Brossolleet and Vaquero, 1998). In Figure 2, we show an alignment of the antisense region in the pseudo-NOS transcript with the complimentary region of the functional nNOS mRNA. Note that the complementarity between the two RNA strands reaches 80% over a distance of ∼150 nt, and this should enable a stable duplex to form between the two transcripts in vivo.
Fig. 2.
Alignment of the antisense region (93–238 nt) of the pseudo-NOS transcript with its complementary counterpart in the nNOS mRNA. In this alignment, there is ∼80% complementarity. The non-Watson–Crick G–U base pairs that are common in RNA secondary structure are shown by dots. The positions of three primers used in the identification of RNA–RNA duplexes areunderlined and named. Primers in positions expected to be protected from ribonuclease A (within the proposed duplex) are P2 and P3. The primer located outside of the protected area is called P1. Further explanation of the ribonuclease A protection experiment is provided in Figure 5. Details of procedures are in Materials and Methods.
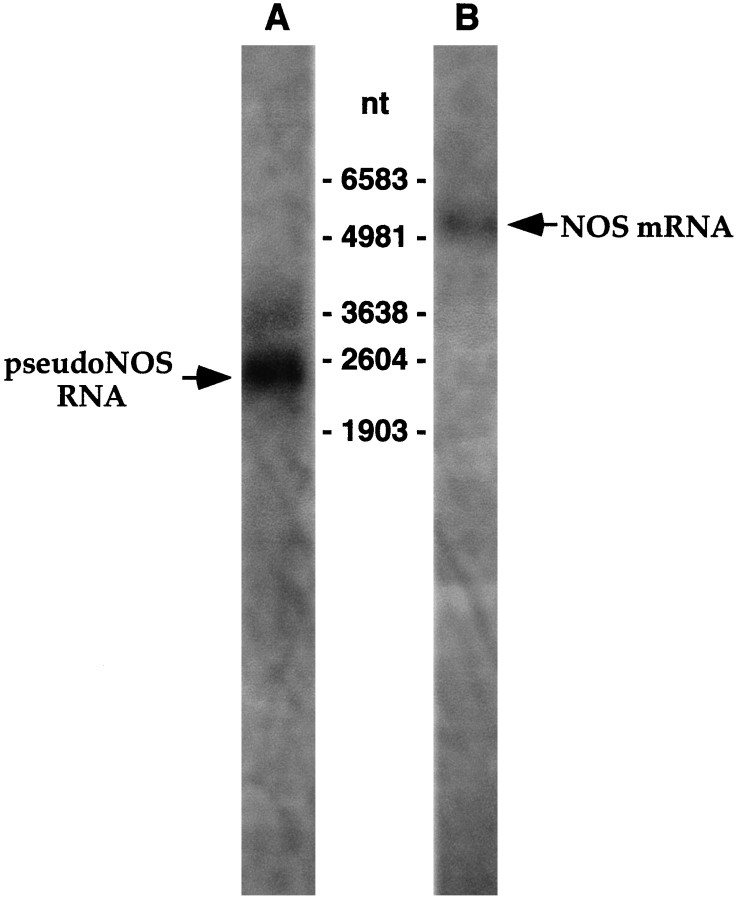
Natural antisense RNAs have been proposed to mediate the regulation of gene expression in eukaryotes (Wightman et al., 1993; Shayiq, 1997), and so it was of considerable interest to determine whether our pseudo-NOS RNA regulates nNOS expression. To address this question, we first showed that a transcript corresponding to the cloned pseudo-NOS cDNA is actually expressed in the CNS. This was achieved by Northern blot hybridization using a probe specific to the antisense region of the pseudo-NOS transcript. Results of this experiment are shown in Figure 3 in which the expression of transcripts containing the antisense region is compared with the expression of the larger 5070 nt functional nNOS mRNA. Importantly, the RNA revealed by the probe complementary to the antisense region of pseudo-NOS is of the expected size (2345 nt), indicating that the pseudo-NOS transcript we cloned is full-length and is indeed expressed in the CNS.
Fig. 3.
The pseudo-NOS and nNOS-encoding transcripts are expressed in the Lymnaea CNS. Northern blot analysis ofLymnaea CNS poly(A+) RNA using a probe specifically recognizing the antisense region of the pseudo-NOS transcript identifies a prominent band of the expected size (∼2500 nt) in lane A (arrow). A less prominent transcript of ∼3200 nt is also revealed in the experiment. This suggests that there are other RNA molecules in the CNS that are antisense to the nNOS-encoding transcript. In lane B, the result of the hybridization with a probe recognizing the 3′ untranslated end of nNOS mRNA is shown. As expected, a single transcript of ∼5000 nt is revealed (arrow).
Neuron-specific coexpression of nNOS mRNA and pseudo-NOS RNA
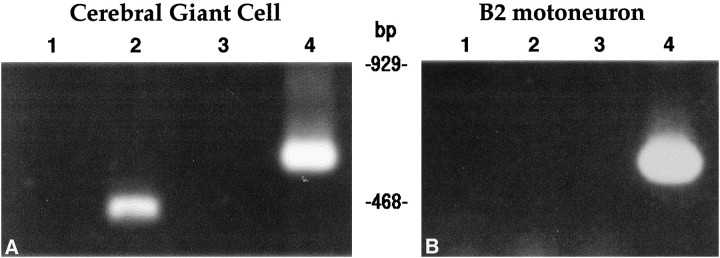
The extensive complementarity between the antisense region of the pseudo-NOS RNA and a corresponding region of the nNOS-encoding mRNA (Fig. 2) suggests strongly that a stable RNA–RNA duplex will form in neurons that contain both transcripts. But do neurons that coexpress pseudo-NOS and functional nNOS transcripts actually exist in the CNS of the snail? To answer this question, we have exploited the advantage of the existence in the snail CNS of two large identifiable nNOS-expressing neurons. These are the paired B2 motoneuron of the buccal ganglion (Park et al., 1998) and the paired CGCs of the cerebral ganglion (Korneev et al., 1998). In the present study, RNA was isolated from up to 20 individually dissected cell bodies of the CGC and B2 neurons, and reverse transcription (RT)-PCR experiments designed to identify pseudo-NOS and functional nNOS transcripts were performed (see Materials and Methods). Using this approach, both transcripts were detected in the CGC but not in the B2 neuron in which only the functional nNOS transcript is present (Fig.4). The identity of all PCR products was confirmed by cloning and sequencing.
Fig. 4.
A uniquely identified neuron (the CGC) coexpresses functional NOS mRNA and pseudo-NOS RNA. InA, the results of RT-PCR experiments on RNA purified from isolated identified CGCs are illustrated. Lane 4shows that a PCR product of the expected size (598 bp) is generated by primers specific to nNOS mRNA. Similarly, in lane 2, a PCR product of the expected size (431 bp) generated from the same RNA sample by primers specific to the pseudo-NOS RNA is detected.Lanes 1 and 3 show the results of PCR experiments designed to control for possible DNA contamination of the samples analyzed in lanes 2 and 4, respectively. In these experiments, reverse transcriptase was omitted, and as a consequence, no products were generated. InB, the results from isolated B2 motoneurons are presented. Lane 4 shows a PCR product of the expected size (598 bp) generated by the same nNOS-specific primers as used inA. Note that there is no PCR product in lane 2 in which the pseudo-NOS RNA-specific primers were used.Lanes 1 and 3 represent control experiments in which reverse transcriptase was omitted. The absence of any PCR products in these lanes proves that the RNA sample used in the RT-PCR experiments was free from DNA contamination. All RT-PCR products shown have been cloned and sequenced to confirm their identity.
From the data obtained from identified neurons, two major conclusions can be drawn: first, that nNOS mRNA and pseudo-NOS RNA are coexpressed in the CGC, and secondly, that the transcription of the corresponding genes can be independently regulated in a neuron-specific manner.
Stable RNA–RNA duplexes form in vivo
The neuron-specific coexpression of the nNOS-encoding and antisense NOS transcripts is strongly supportive of a role for the pseudo-NOS gene in the translational regulation of nNOS expression. However, this could only occur if the nNOS and pseudo-NOS transcripts form stable RNA–RNA duplex molecules. A method commonly used to detect such duplexes involves the treatment of purified cellular RNA with RNase A, an enzyme that specifically cleaves single-stranded RNA. Such treatment ought to leave our hypothesized RNA–RNA duplex intact. It is known, however, that RNase A can attack mismatched areas in double-stranded RNA, and because complementarity between the nNOS and pseudo-NOS transcripts does not reach 100%, RNase A might have some activity within the proposed duplex. If this activity were significant, all or most of the duplexed RNA molecules might be destroyed, making this method for detecting the duplex inappropriate. Therefore, before analyzing the results of experiments on RNase A-treated cytoplasmic RNA, we first performed a ribonuclease protection assay using synthetic nNOS mRNA and synthetic pseudo-NOS RNA (see Materials and Methods for details). Importantly, this in vitro control experiment showed that the RNase A activity was substantially suppressed within the RNA–RNA duplex (results not shown) under standard conditions for RNase A treatment (Shayiq, 1997). Therefore, we were confident that, by using the same conditions, it would be possible to detect the natural RNA–RNA hybrid in cytoplasmic RNA isolated from the snail CNS.
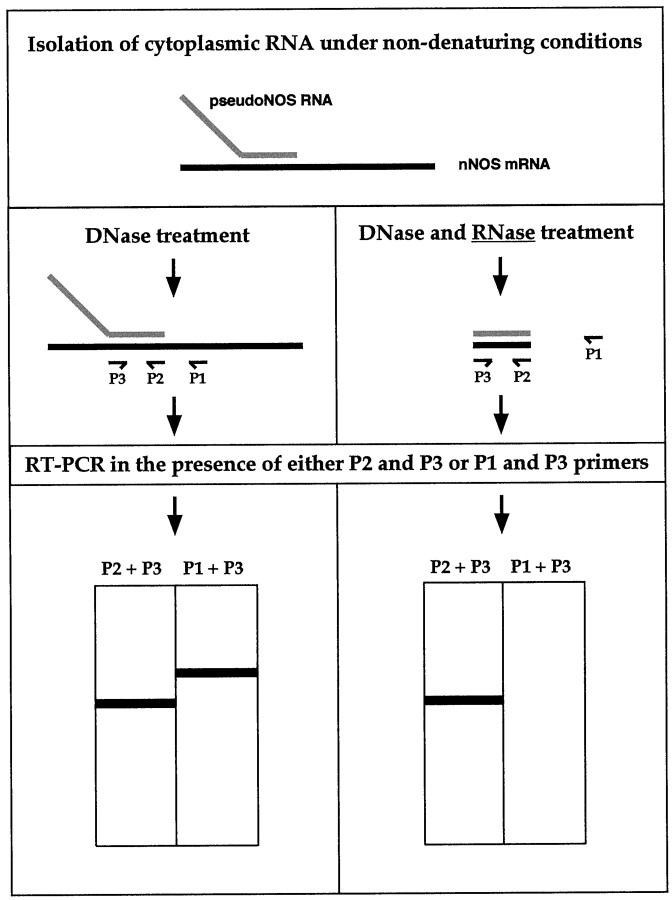
The logic of the experiment using cytoplasmic RNA is explained in Figure 5. Briefly, cytoplasmic RNA was purified under nondenaturing conditions to preserve possible RNA–RNA hybrids and then treated with RNase A. To identify the duplex, we first performed two reverse transcription reactions using two primers specific to nNOS mRNA. One primer was located within the duplex area (primer P2), and the other was located on single-stranded RNA outside the duplex area (primer P1). The cDNA generated by the P2 primer was then amplified by PCR in the presence of primer P2 and a second primer located within the duplex (primer P3). The cDNA generated by P1 was similarly amplified using P1 and P3 primers. If the duplex exists, a single product would be generated by the RNase A-treated sample (from P2 and P3), whereas nothing would be generated in the presence of the P1 and P3 primers (Fig. 5).
Fig. 5.
A schematic diagram showing the major steps of the ribonuclease protection procedure used to detect RNA–RNA duplexesin vivo. To preserve possible RNA–RNA hybrids, cytoplasmic RNA was purified from the CNS under nondenaturing conditions. To identify our hypothesized RNA–RNA duplex, the RNA has to be treated with RNase A, an enzyme that cleaves single-stranded but not double-stranded RNA molecules. After RNase A treatment, reverse transcription reactions in the presence of either P2 primer (located within the protected area) or P1 primer (located outside the protected area) are performed. After adding the P3 primer, a cDNA generated in the first reaction could be amplified using PCR and then will be revealed as a single band of the expected size by electrophoresis. In contrast, no cDNA could be produced in the second reverse transcription reaction, and subsequently, no PCR product is expected. In theleft column, the predicted results of the control experiments (no RNase A treatment) are summarized. Two RT-PCR products should be detected in the reverse transcription reaction: one generated by P2 and P3 and the other by P1 and P3.
The results of the experiment are shown in Figure6. A single product of the expected size (139 bp) is detected in the experiment in which the P2 and P3 primers were used (Fig. 6, lane 2). In contrast, no product was detected with the P1 and P3 primers (lane 4). Similar experiments were performed in parallel on purified RNA that was not treated with RNase A. As expected, a PCR product of 139 bp was generated by the P2 and P3 primers, and a 340 bp product was generated by the P1 and P3 primers (lanes 1, 3). The correct identity of the 139 and 340 bp products have been confirmed by cloning and sequencing. An exactly complementary experiment to that illustrated in Figure 5 was performed using primers designed to amplify the pseudo-NOS strand of the duplex. In this experiment, the antisense region of the pseudo-NOS transcript was protected from RNase A action.
Fig. 6.
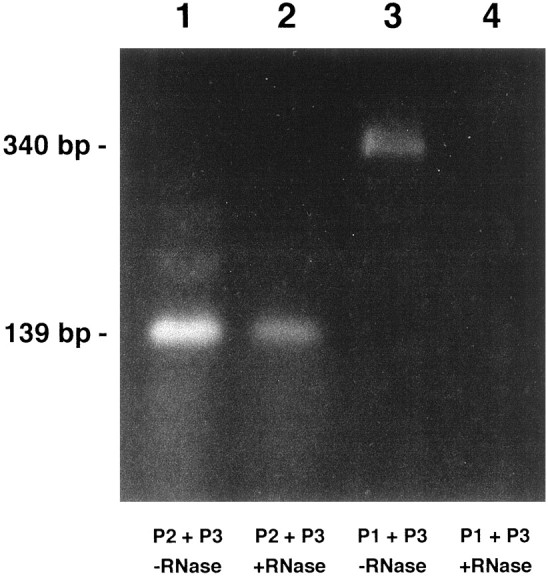
The predicted RNA–RNA duplex exists in the CNS. The experiment was performed according to the procedure described in Figure 5. Lanes 1 and 3 show the products of RT-PCR generated by RNA treated with DNase only. Lanes 2 and 4 show the products of RT-PCR generated on RNA treated with both DNase and RNase A. In lanes 1 and2, RNA was reverse transcribed with the P2 primer located within the predicted duplex and then amplified in the presence of the P2 and P3 primers (see Figs. 3 and 5 for location of the primers). Lanes 3 and 4 show the results generated when RNA was reverse transcribed with the P1 primer located outside the predicted duplex and then amplified in the presence of P1 and P3 primers. A product of the same predicted size (139 bp) is generated by the RNA sample that was treated with DNase only and with DNase plus RNase A (lanes 1 and 2). The band shown in lane 3 is the size predicted (340 bp) of a product generated by the P1 and P3 primers. Note that there is no product in lane 4. Importantly, these results correspond precisely to those predicted in Figure 5. All of the RT-PCR products shown have been cloned and sequenced to confirm their identity.
These experiments leave little doubt that stable RNA–RNA duplex molecules do form in vivo between the antisense pseudo-NOS transcript and the nNOS mRNA in Lymnaea CNS.
Antisense regulation of nNOS protein synthesisin vitro
To show that this RNA–RNA duplex is able to prevent the synthesis of nNOS protein, we have performed in vitro translation experiments. Using T7 RNA polymerase, we generated large quantities of the major part of the nNOS-encoding mRNA and the complete antisense region of the pseudo-NOS RNA. When translated alone, the synthetic nNOS cRNA provides a template for the synthesis of an nNOS protein of the correct size, and this can be seen in Figure7 (lane 1). When the nNOS cRNA is incubated with the antisense pseudo-NOS transcript, the synthesis of nNOS protein is completely suppressed (lane 2). Furthermore, when the antisense pseudo-NOS cRNA was replaced with sense pseudo-NOS cRNA, which cannot form a duplex with nNOS cRNA, there is no inhibition of translation (lane 3).
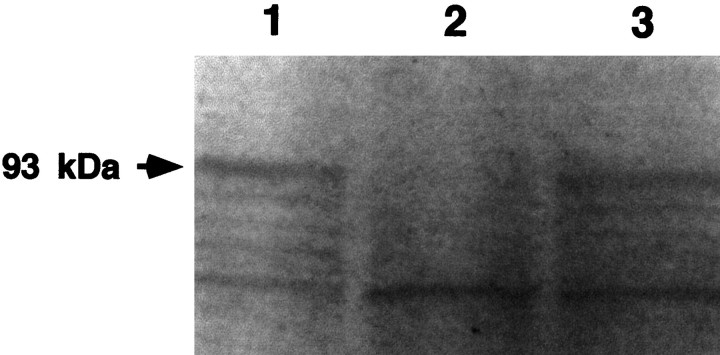
Fig. 7.
Synthesis of nNOS protein in vitrois suppressed by the antisense pseudo-NOS RNA. Lane 1represents the result of the translation of NOS cRNA and shows the main labeled product is a protein of the expected (93 kDa) size (arrow). Lane 2 illustrates the effect of incubating the 0.2 μg of NOS cRNA with 2 μg of antisense pseudo-NOS cRNA before translation. Note strong suppression of translation of the nNOS protein. As a control for any effects on translation that are not related to the formation of a duplex, we also preincubated the 0.2 μg of NOS cRNA with a 2 μg of a sense version of the pseudo-NOS cRNA (lane 3). No inhibition of NOS protein synthesis is observed. Similar results, although weaker, have been obtained, even when the ratio between nNOS cRNA and pseudo-NOS transcripts was 2.5:1. A second major protein of ∼50 kDa can be seen in eachlane. This protein is present in the cell-free wheat germ system and performs a useful function as an internal control. Note that it is not diminished in intensity in lane 2.
Importantly, the results of in vitro translation experiments show that the duplex formed between the pseudo-NOS transcript and nNOS mRNA is sufficient to block the synthesis of nNOS protein. They do not, however, show that an antisense mechanism suppresses the synthesis of nNOS protein in particular snail neurons in vivo.
In vivo antisense regulation of nNOS protein expression
It is the existence of two large, identified nNOS mRNA-containing neurons in the CNS (the paired B2 and the CGC) that has provided an opportunity to test our idea that a natural antisense RNA can prevent the expression of nNOS protein in vivo. Crucially, as described above, although both identified neurons express nNOS mRNA, only the CGC coexpresses the NOS pseudogene transcript. If our conclusions are valid, there should be significant suppression of nNOS enzyme activity correlated with the neuronal coexistence of the functional and pseudo-NOS transcripts in the CGC.
Enzyme activity associated with nNOS can be rapidly and reliably localized to cells using the NOS-specific NADPH-diaphorase histochemical method. In both nervous and peripheral tissue and in both vertebrates and invertebrates, NADPH-diaphorase and NOS activity are regarded as identical (Dawson et al., 1991; Matsumoto et al., 1993; Elphick et al., 1995). Our previous work on the buccal ganglion (Park et al., 1998) shows that the B2 neuron is strongly and consistently NADPH-diaphorase-positive (100% of B2 neurons observed in >100 preparations), indicating that all B2 neurons always contain an active nNOS protein (Fig.8A). Using exactly the same NADPH-diaphorase protocol, experiments were performed on cerebral ganglia preparations from 30 animals, each of which contains a pair of CGCs. Remarkably, in only one preparation did we find bilateral and strongly positive staining, comparable with staining in the B2 neuron. In four preparations, there was trace staining in one member of the pair, and for the remaining 54 neurons, no staining above background was seen. At best therefore, in the animals examined, only <10% (n = 60) of the CGCs show nNOS enzyme activity, although we know from the results of published in situhybridization experiments (Korneev et al., 1998) and from the single neuron RT-PCR experiments reported here that this neuron does contain the nNOS transcript (Figs. 4, 8B,C). An illustration of nonexpression and the exceptional sporadic expression of nNOS enzyme activity in the CGC is provided in Figure9, A and B. Importantly, these results confirm the existence of translational mechanism-suppressing nNOS expression in the CGC and demonstrate that, under some circumstances, it is possible for the neuron to synthesize nNOS protein.
Fig. 8.
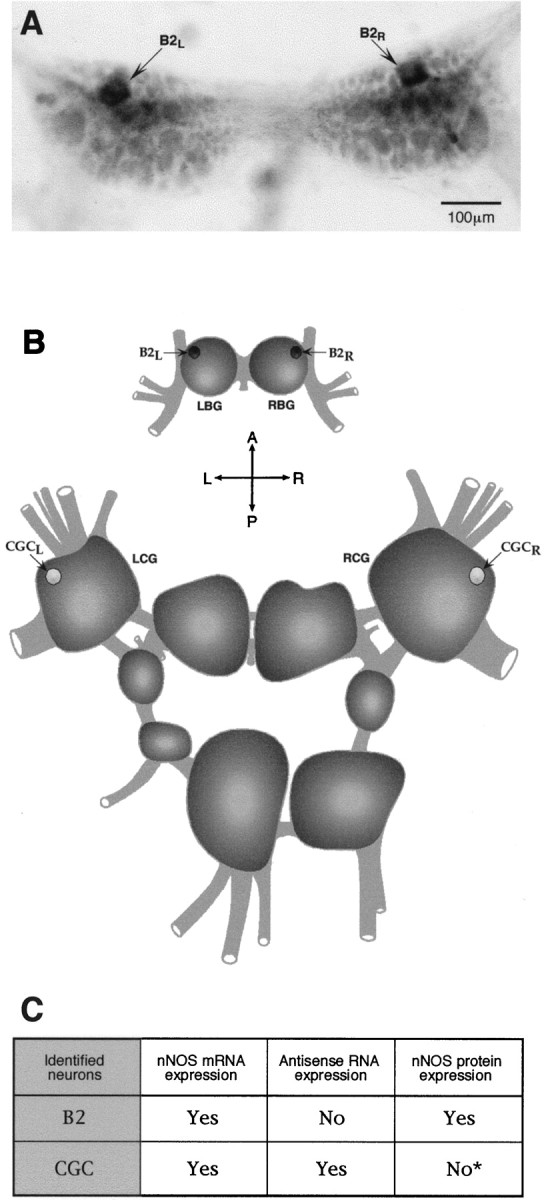
Antisense RNA-mediated regulation of nNOS protein expression in vivo. A, NADPH-diaphorase staining of the buccal ganglia. A pair of symmetrical NOS-containing diaphorase-positive B2 motoneurons is indicated byarrows. B, A diagram of theLymnaea CNS showing the positions of identified neurons referred to in this investigation. Dark cell bodies of B2 motoneurons reflect the fact that the neurons are strongly and consistently NADPH-diaphorase-positive, i.e., they always contain an active nNOS protein (A, C). In contrast, <10% of all CGCs examined showed nNOS enzyme activity. To emphasize this result, the cell bodies of CGCs are shown in light color. C, A summary of our experiments on the expression of nNOS mRNA, antisense RNA, and nNOS protein in the identified neurons B2 and CGC. The asterisk indicates that nNOS protein in the CGC is not entirely absent but is detected only in ∼10% of the cells observed (n = 60).
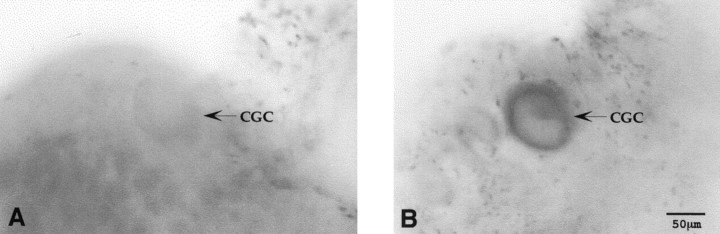
Fig. 9.
NADPH-diaphorase staining of the cerebral ganglia.A, The majority of CGCs shows no NADPH-diaphorase activity. B, One of the very few clearly NADPH-diaphorase-positive CGCs. Cell bodies of the CGCs are indicated by arrows.
To summarize, an identified neuron that contains the nNOS mRNA but not the pseudo-NOS RNA consistently expresses a functional NOS protein. In contrast, in a neuron in which both transcripts are colocalized, NOS enzyme activity is practically undetectable. These in vivoobservations support the view that the antisense pseudo-NOS transcript suppresses the translation of functional nNOS mRNA in neurons in which the two transcripts are colocalized. We conclude that the suppression of nNOS enzyme activity in the CGC is caused by the hybrid arrest of translation mediated by an endogenous antisense-containing RNA transcribed from a NOS pseudogene (see Discussion).
DISCUSSION
Our results have implications on two fronts. First, they show that, in a eukaryotic system, a natural antisense mechanism can regulate the synthesis of an important neuronal signaling molecule. Second, they suggest that novel regulatory functions for some transcribed pseudogenes can arise during the course of evolution.
Although our findings are conclusive with respect to the ability of a natural antisense-containing transcript to suppress nNOS protein synthesis, they do not speak to the mechanism of antisense interference. Two quite different mechanisms have been proposed to explain natural antisense-mediated regulation of translation: hybrid arrest of translation and digestion of double-stranded (ds) RNA with specific ribonucleases (Nellen and Lichteinstein, 1993). Although the second possibility cannot be fully excluded, there is a number of observations that cannot be explained if our RNA duplex is a target for dsRNA-activated RNases. For example, and most significantly, we have demonstrated the existence of the RNA duplex in vivoand that both transcripts involved in duplex formation are present in the CGCs. We therefore favor a mechanism involving antisense-mediated hybrid arrest of translation. The precise steps in the chain of events leading to the inhibition of nNOS protein synthesis, however, are not clear. They might include, for example: (1) steric alterations in the sense RNA structure that prevent translation; (2) trapping of specific RNA-binding proteins by the antisense transcript; and (3) inhibition of initiation of protein synthesis by activated dsRNA-dependent protein kinases, etc. (Vanhe-Brossolleet and Vaquero, 1998). Whatever its precise mechanism, this compelling example of natural antisense-mediated suppression suggests that the phenomenon of RNA duplex formation is an important mechanism of translational regulation in eukaryotes. Recently, this conclusion has found support in unexpected observations that the introduction of dsRNA molecules can effectively and specifically suppress gene expression inCaenorhabditis elegans and Drosophila (Fire et al., 1998; Kennerdell and Carthew, 1998).
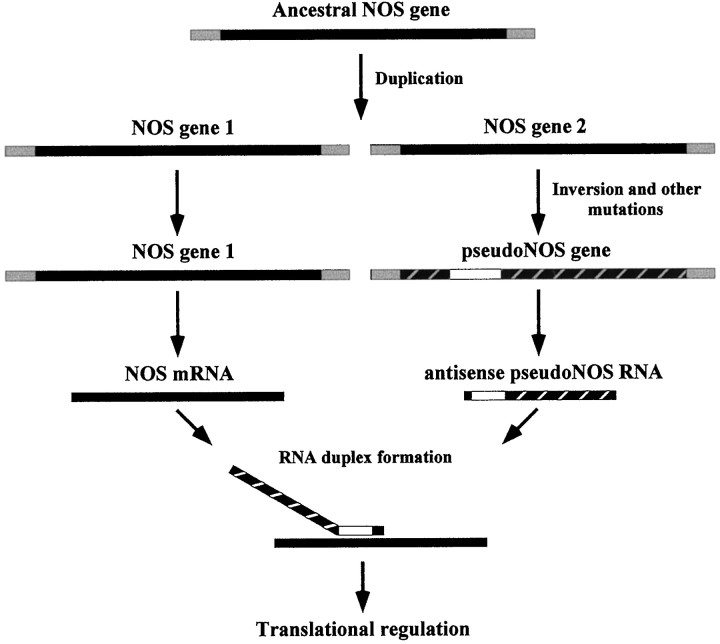
Our results show that nNOS protein synthesis is usually suppressed in the CGC by an antisense-mediated mechanism, but also that there is sporadic use of nNOS protein by this neuron (Fig. 9). According to our model (Fig. 10), these changes in nNOS expression are mediated by the differential transcription of the NOS pseudogene. Specifically, the active transcription of the pseudogene will lead to the suppression of nNOS protein synthesis, and on the other hand, the inhibition of pseudogene transcription will permit nNOS production. Importantly, a switch from the “off” to the “on” mode of nNOS expression would be achieved rapidly because the functional nNOS gene is already active in both modes and nNOS mRNA could be available immediately for translation once the suppressive effect of the NOS pseudogene is removed. We therefore propose that in the CGC antisense-mediated translational control, supplemented by transcriptional regulation of the NOS pseudogene, provides an effective molecular mechanism for achieving rapid changes in nNOS protein production in response to some internal or external signals.
Fig. 10.
A proposed model for the evolution of NOS pseudogene function.
Functionally, the important issue is to determine when and why the CGC in Lymnaea needs to express nNOS. At present, we can only speculate on the basis of what is known about the role of this neuron and its homologs in other molluscs. Crucially, we need to gain an understanding of the natural processes that cause sporadic expression of nNOS protein in the CGC of Lymnaea to occur. Comparison with other molluscan species might be helpful in this respect, and specifically, it is of some interest that the CGC homolog in the gastropod Pleurobranchaea consistently expresses nNOS enzyme (Moroz and Gillette, 1996). In Lymnaea, the CGCs are very well studied serotonergic neurons that have “gating” and modulatory functions in the neural circuit controlling feeding behavior (Kemenes et al., 1994). Their homologs in Aplysia [the metacerebral giant cells (MCCs)] and in Pleurobranchaea mediate the effects of arousal on feeding behavior (Gillette and Davis, 1977;Kupfermann and Weiss, 1982). Thus, it would appear that neurons of this type play important roles in aspects of behavioral plasticity related to feeding. Perhaps in Lymnaea, these or related functions might be reinforced by NO when the CGCs express the nNOS enzyme. In Aplysia, the homologous MCC, which does not stain using NADPH-diaphorase (Jacklet and Gruhn, 1994), generates a very slow EPSP in response to the release of NO after the stimulation of an identified presynaptic nitrergic neuron (Jacklet, 1995). This NO-induced slow depolarization of the MCC would appear to be mediated by cGMP (Koh and Jacklet, 1999). Thus, in Aplysia, the homolog of the CGC is sensitive to NO, and this might contribute to MCC-mediated arousal in the feeding system. If the CGC ofLymnaea is also activated by NO, NO might function as an autostimulatory neurotransmitter when the nNOS protein is expressed in this neuron.
Natural antisense-mediated regulation of gene expression is well known to exist in prokaryotic systems (Wagner and Simons, 1994) but is far less well recognized in eukaryotes (Vanhe-Brossolleet and Vaquero, 1998). Our experiments provide a clear example of a natural antisense mechanism operating in an eukaryotic system. The singular advantage of our system that has allowed us to demonstrate this is the ability to perform molecular experiments on single identified neurons. This has enabled us to show that a trans-encoded endogenous antisense RNA is involved in the translational regulation of the nNOS in a particular neuron. Of considerable additional interest is the finding that the endogenous antisense RNA molecule is derived from a pseudogene. Importantly, among the published examples of natural antisense RNAs in eukaryotes, none represent transcripts derived from pseudogenes. This new finding has fundamental implications for future investigations of antisense regulation. In particular, it suggests that a greater than expected number of antisense-containing transcripts may be encoded in the genomes of eukaryotes.
With respect to the evolution of regulatory functions of pseudogenes, we must now conclude that transcribed pseudogenes are not necessarily without function. Indeed, they would appear to be especially suited to roles involving the antisense regulation of the active genes to which they are related (Fig. 10). If this is true, there must be other examples of pseudogenes that are not translated but that may regulate the expression of proteins encoded by related functional genes. Although a comprehensive search of the available databases could provide an answer to this question, this is beyond the scope of the current paper. This is because an enormous amount of information would have to be analyzed without the benefit of a simple ready-to-use algorithm for detecting such sequences. Our future plans do include such an investigation, and as a prelude to this, we have recently analyzed the sequence ofjust 10 pseudogenes and their corresponding functional genes picked at random in the GenBank database. In this limited search, another example of a transcribed pseudogene (a cytochrome P-450-like pseudogene, accession numbers M12280 and M12287) (Zaphiropoulos et al., 1986) that has a region of significant antisense homology to the functional gene was detected. This suggests strongly that the regulatory role we attribute to the NOS pseudogene is unlikely to be an isolated example, and we may have uncovered the first member of an entirely new class of regulatory gene. We therefore believe that antisense-mediated regulation of gene expression is far more widespread in eukaryotic systems than currently recognized. In the brain, an organ in which the pattern of gene expression is highly complex and labile, such an expansion of the diversity of ways in which gene activity can be regulated is of particular significance.
Footnotes
This work is supported by the Biotechnology and Biological Sciences Research Council. We thank E. I. Korneeva, R. Philips, and M. Piper for their technical assistance and S. Saunders for her helpful comments on this manuscript.
Correspondence should be addressed to Michael O’Shea at the above address.
REFERENCES
- 1.Bredt DS, Snyder SH. Isolation of nitric oxide synthase, a calmodulin-requiring enzyme. Proc Natl Acad Sci USA. 1990;87:682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bredt DS, Snyder SH. Nitric oxide, a novel neuronal messenger. Neuron. 1992;8:3–11. doi: 10.1016/0896-6273(92)90104-l. [DOI] [PubMed] [Google Scholar]
- 3.Bredt DS, Hwang PM, Snyder SH. Localization of nitric oxide synthase indicating a neural role for nitric oxide. Nature. 1990;347:768–770. doi: 10.1038/347768a0. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Dawson TM, Snyder SH. Gases as biological messengers. Nitric oxide and carbon monoxide in the brain. J Neurosci. 1994;14:5147–5159. doi: 10.1523/JNEUROSCI.14-09-05147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson TM, Bredt DS, Fotuhi M, Hwang PM, Snyder SH. Nitric oxide synthase and neuronal NADPH-diaphorase are identical in brain and peripheral tissue. Proc Nat Acad Sci USA. 1991;88:7797–7801. doi: 10.1073/pnas.88.17.7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elphick MR, Rayne RC, Riveros-Moreno V, Moncada S, O’Shea M. Nitric oxide synthase in locust olfactory interneurons. J Exp Biol. 1995;198:821–829. doi: 10.1242/jeb.198.3.821. [DOI] [PubMed] [Google Scholar]
- 8.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 9.Garthwaite J, Boulton CL. Nitric oxide signaling in the central nervous system. Annu Rev Physiol. 1995;57:683–706. doi: 10.1146/annurev.ph.57.030195.003343. [DOI] [PubMed] [Google Scholar]
- 10.Garthwaite J, Charles SL, Chess-Williams R. Edothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain. Nature. 1988;336:385–388. doi: 10.1038/336385a0. [DOI] [PubMed] [Google Scholar]
- 11.Gillette R, Davis WJ. The role of the metacerebral giant neuron in the feeding behaviour of Pleurobranchaea. J Comp Physiol. 1977;116:129–159. [Google Scholar]
- 12.Hölscher C. Nitric oxide, the enigmatic neuronal messenger: its role in synaptic plasticity. Trends Neurosci. 1997;20:298–303. doi: 10.1016/s0166-2236(97)01065-5. [DOI] [PubMed] [Google Scholar]
- 13.Hope BT, Michael GJ, Knigge KM, Vincent SR. Neuronal NADPH-diaphorase is a nitric oxide synthase. Proc Natl Acad Sci USA. 1991;88:2811–2814. doi: 10.1073/pnas.88.7.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacklet JW. Nitric oxide is used as an orthograde cotransmitter at identified histaminergic synapses. J Neurophysiol. 1995;74:891–895. doi: 10.1152/jn.1995.74.2.891. [DOI] [PubMed] [Google Scholar]
- 15.Jacklet JW, Gruhn M. Nitric oxide as a putative transmitter in Aplysia: neural circuits and membrane effects. Neth J Zool. 1994;44:524–534. [Google Scholar]
- 16.Kemenes G, Hiripi L, Benjamin PR. Behavioural and biochemical changes in the feeding system of Lymnaea induced by the dopamine and serotonin neurotoxins 6-hydroxydopamine and 5,6-dihydroxytryptamine. Phil Trans R Soc Lond B Biol Sci. 1994;329:243–255. [Google Scholar]
- 17.Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 18.Koh H-Y, Jacklet JW. Nitric oxide stimulates cGMP production and mimics synaptic responses in metacerebral neurons of Aplysia. J Neurosci. 1999;19:3818–3826. doi: 10.1523/JNEUROSCI.19-10-03818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korneev SA, Piper MR, Picot J, Phillips R, Korneeva EI, O’Shea M. Molecular characterization of NOS in a mollusc: expression in a giant modulatory neuron. J Neurobiol. 1998;35:65–76. doi: 10.1002/(sici)1097-4695(199804)35:1<65::aid-neu6>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Kupfermann I, Weiss KR. Activity of an identified serotonergic neuron in free-moving Aplysia correlates with behavioral arousal. Brain Res. 1982;241:334–337. doi: 10.1016/0006-8993(82)91072-1. [DOI] [PubMed] [Google Scholar]
- 21.Lewin B. Genes IV. Oxford UP; New York: 1990. [Google Scholar]
- 22.Matsumoto T, Nakane M, Pollock JS, Kuk JE, Forstermann U. A correlation between soluble brain nitric oxide synthase and NADPH-diaphorase is seen only after exposure of the tissue to fixative. Neurosci Lett. 1993;155:61–64. doi: 10.1016/0304-3940(93)90673-9. [DOI] [PubMed] [Google Scholar]
- 23.Moroz LL, Gillette R. NADPH-diaphorase localization in the CNS and peripheral tissues of the predatory sea-slug Pleurobranchaea californica. J Comp Physiol [A] 1996;367:607–622. doi: 10.1002/(SICI)1096-9861(19960415)367:4<607::AID-CNE10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 24.Moroz LL, Bulloch AGM, Lukowiak K, Syed NI. Putative NOs-synthesizing neurons of Lymnaea in vivo and in vitro. Neth J Zool. 1994a;44:535–549. [Google Scholar]
- 25.Moroz LL, Winlow W, Turner RW, Bulloch AGM, Lukowiak K, Syed NI. Nitric oxide synthase-immunoreactive cells in the CNS and periphery of Lymnaea. NeuroReport. 1994b;5:1277–1280. doi: 10.1097/00001756-199406020-00031. [DOI] [PubMed] [Google Scholar]
- 26.Nellen W, Lichtenstein C. What makes an mRNA antisense-itive? Trends Biochem Sci. 1993;18:419–423. doi: 10.1016/0968-0004(93)90137-c. [DOI] [PubMed] [Google Scholar]
- 27.Park J-H, Straub VA, O’Shea M. Anterograde signaling by nitric oxide: characterization and in vivo reconstitution of an identified nitrergic synapse. J Neurosci. 1998;18:5463–5476. doi: 10.1523/JNEUROSCI.18-14-05463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz-Aellen M-F, Roulet E, Fischer-Lougheed J, O’Shea M. Synthesis of a heterodimer neurohormone precursor of locust adipokinetic hormone studied by in vitro translation and cDNA cloning. Neuron. 1989;2:1369–1373. doi: 10.1016/0896-6273(89)90075-5. [DOI] [PubMed] [Google Scholar]
- 29.Shayiq RM. Role of gene overlap in the regulation of mRNA translation for mitochondrial cytochrome P-450c27/25 in the rat. J Biol Chem. 1997;272:4050–4057. [PubMed] [Google Scholar]
- 30.Vanhe-Brossolleet C, Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- 31.Wagner EG, Simons RW. Antisense RNA control in bacteria, phages and plasmids. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 32.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 33.Zaphiropoulos PG, Folk WR, Coon MJ. Isolation and characterization of a novel cytochrome P-450-like pseudogene. Biochem Biophys Res Commun. 1986;134:499–505. doi: 10.1016/s0006-291x(86)80448-x. [DOI] [PubMed] [Google Scholar]