Abstract
Cell surface metallo-endopeptidases play important roles in cell communication by controlling the levels of bioactive peptides around peptide receptors. To understand the relative relevance of these enzymes in the CNS, we characterized a metallo-endopeptidase in the CNS of Aplysia californica, whose peptidergic pathways are well described at the molecular, cellular, and physiological levels. The membrane-bound activity cleaved Leu-enkephalin at the Gly3–Phe4 bond with an inhibitor profile similar to that of the mammalian neutral endopeptidase (NEP). This functional homology was supported by the molecular cloning of cDNAs from the CNS, which demonstrated that the Aplysiaand mammalian NEPs share all the same amino acids that are essential for the enzymatic activity. The protein is recognized both by specific anti-Aplysia NEP (apNEP) antibodies and by the [125I]-labeled NEP-specific inhibitor RB104, demonstrating that the apNEP gene codes for the RB104-binding protein. In situ hybridization experiments on sections of the ganglia of the CNS revealed that apNEP is expressed in neurons and that the mRNA is present both in the cell bodies and in neurites that travel along the neuropil and peripheral nerves. When incubated in the presence of a specific NEP inhibitor, many neurons of the buccal ganglion showed a greatly prolonged physiological response to stimulation, suggesting that NEP-like metallo-endopeptidases may play a critical role in the regulation of the feeding behavior inAplysia. One of the putative targets of apNEP in this behavior is the small cardioactive peptide, as suggested by RP-HPLC experiments. More generally, the presence of apNEP in the CNS and periphery may indicate that it could play a major role in the modulation of synaptic transmission in Aplysia and in the metabolism of neuropeptides close to their point of release.
Keywords: Aplysia, neutral endopeptidase, CNS, neuropeptide degradation, buccal ganglion, SCP
Specific behaviors and various physiological functions from yeast to mammals are controlled by a wide range of bioactive peptide hormones. The use of peptides as messengers usually involves the following steps: production and release of the peptide by a specific cell, interaction of the peptide with a receptor on the surface of the target cell, and degradation of the peptide to terminate its action. The first and last steps of this scheme require the participation of proteases/peptidases. It is now acknowledged that a small number of membrane peptidases, with a broad range of specificity, act together to put an end to the biological actions of neuropeptides (McKelvy and Blumberg, 1986; Maroux, 1987; Turner, 1986;Turner et al., 1987). Most of these neuropeptidases are zinc integral membrane proteins with their active site facing the exoplasmic side of the cell (Maroux, 1987). One of the best known of these peptidases is probably neutral endopeptidase-24.11 (NEP, enkephalinase, neprilysin, CALLA), which has been implicated in the physiological degradation of several bioactive peptides (for review, see Kenny, 1993;Roques et al., 1993).
To study the importance of neuropeptide-degrading enzymes in the CNS, we used the marine snail Aplysia californica. This animal has been used extensively to study a wide range of behaviors and physiological functions. The simplicity and accessibility of its neuronal components contributed to link cellular, biochemical, molecular, and physiological studies and to finely characterize peptidergic pathways (Miller et al., 1993a,b; Brezina et al., 1995;Byrne and Kandel, 1996).
So far, three peptidase activities have been characterized and linked to the extracellular metabolism of peptides in Aplysia. A leucine aminopeptidase activity (Squire et al., 1991), an aminopeptidase N activity (Bawab et al., 1992), and a neutral endopeptidase activity (Bawab et al., 1993). In mammals, NEP is a cell surface metallo-endopeptidase ubiquitously distributed in the CNS and the peripheral organs (Roques et al., 1993). Previous studies demonstrated that NEP plays a critical role in atrial natriuretic factor-mediated hypotension and diuresis (Gros et al., 1989, 1990;Seymour et al., 1995; Thompson and Morice, 1996), enkephalin-mediated analgesia (Roques et al., 1980), tachykinin-mediated modulation of synaptic transmission (Barnes et al., 1993; Saleh et al., 1996), endothelin-mediated vasoconstriction (Vijayaraghavan et al., 1990), and peptide-mediated inflammatory responses (for review, see Connelly et al., 1985; Martins et al., 1990; Shipp et al., 1990, 1991; for review, see Kenny, 1993; Roques et al., 1993).
In a previous study, we identified and characterized a neutral endopeptidase activity in the kidney membranes of A. californica (Bawab et al., 1993). As a means to better define the physiological role of apNEP in Aplysia, we have characterized a NEP-like activity in the CNS and cloned the corresponding cDNA. We have characterized apNEP by Western blotting and apNEP mRNA in the CNS by in situ hybridization. We have also demonstrated that inhibitors of the NEP-like activity potentiate the action of endogenous neuropeptides in the buccal ganglion, and in particular of small cardioactive peptide (SCP). All together these results support the importance of peptidases in the modulation of synaptic transmission and will further our investigation into the role of the extracellular regulation of neuropeptides in behavior.
MATERIALS AND METHODS
Peptides, chemicals, and solutions. Peptides Tyr-Gly-Gly and [Leu]enkephalin were purchased from IAF Biochem International (Montréal, Quebec, Canada), l-tyrosine was from Life Technologies-BRL (Burlington, Ontario, Canada), and amastatin, 1–10 phenanthroline, phosphoramidon, phenylmethyl-sulfonyl fluoride (PMSF), and 1-O-n-octyl-B-d-glucopyranoside (octylglucoside) were from Sigma (St. Louis, MO). Captopril was obtained from Squibb (Princeton, NJ). Thiorphan, (3-hydroxyamino-carbonyl-2-benzyl-1-oxopropyl)-glycine (HACBO-Gly) and 2[(3-iodo-hydroxy)phenylmethyl]-4-N-[3-(hydroxyamino-3-oxo-1-phenylmethyl)propyl]amino-4-oxobutanoic acid (RB104) were obtained from Bernard P. Roques (UniversitéRené Descartes, Paris, France). The labeled substrate (tyrosyl-3,5-3H)[Leu]enkephalin was obtained from New England Nuclear (Boston, MA). [125I]Na was purchased from Amersham (Ontario, Canada). Phosphoramidon (Sigma) was added directly to a static bath (2 ml volume) to obtain a final desired concentration. Artificial seawater (ASW) contained (in mm): NaCl 460, KCl 10, CaCl2 11, MgCl2 55, and HEPES buffer 10, pH 7.6.
Animals and preparations. A. californica(200–250 gm) were purchased from Marine Specimen Unlimited (Pacific Palisades, CA) or from the Aplysia Resource Facility (Miami, FL). They were maintained in a large 900 l tank at 15°C. All physiological experiments were performed at room temperature (22°C) on isolated buccal ganglia. Before dissection, the mollusks were anesthetized with an injection of an isotonic MgCl2solution (385 mm) corresponding to approximately one-third of their volume. Dissection of the buccal ganglion was performed in an extracellular medium made from equal volumes of isotonic MgCl2 and ASW. The ganglia were pinned to the bottom of a Sylgard-coated chamber (3 ml volume) filled with 2 ml of ASW. Both branches of the radula nerve were aspirated into a suction electrode for electrical stimulation. All preparations were rested under constant superfusion of ASW for at least 90 min before the start of an experiment.
Enzyme assays and metabolite analysis. A. californica plasma membranes were prepared as described (Bawab et al., 1992). For the enkephalin-degrading activity, 5–8 μg membrane proteins were preincubated for 15 min at 25°C in 100 μl of 50 mm 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.5, in the presence of amastatin, at a concentration of 10 μm, alone or combined with different peptidase inhibitors. The labeled substrate [3H][Leu]enkephalin [(tyrosyl-3,5-3H)leu-enkephalin] (30–40 Ci/mmol) was added, and the metabolites were separated from the substrate by RP-HPLC as described previously (Bawab et al., 1993). For the SCPB-degrading activity, 50 μg membrane proteins were preincubated for 15 min at 25°C in 100 μl of 50 mm MES, pH 6.5, in the presence of 10 μm amastatin and 1 μm captopril. The substrate SCPB (10 μg) was added and incubated for 1 hr at 25°C, and the metabolites were separated from the substrate by RP-HPLC on a μBondapak C-18 column (Waters). A linear gradient from 95% solvent A [0.1% trifluoroacetic acid (TFA) in water]/5% solvent B (80% acetonitrile/0.1% TFA) to 100% solvent B was developed for 50 min at a flow rate of 1 ml/min.
Molecular identification of [125I]RB104 binding proteins in Aplysia tissues. RB104 was iodinated by the chloramine T method and purified as described previously (Bawab et al., 1993). Membrane preparations fromAplysia CNS were solubilized for 1 hr at 4°C in Tris-buffered saline, pH 7.5, containing 1% (w/v) octylglucoside. The solubilized proteins were separated by electrophoresis, electroblotted to a nitrocellulose membrane, and labeled with [125I]RB104 as described previously (Bawab et al., 1993).
Molecular cloning of the apNEP cDNA. Filter replicates of a λJ1 genomic library were hybridized at low stringency with a 760 bpHindIII–ApaI fragment (nucleotides 1616–2376) isolated from the rabbit cDNA, in 6× SSC, 5× Denhardt’s solution, 20% formamide, 0.5% SDS, and 100 μg/ml denatured salmon sperm DNA, at 42°C for 16 hr. Filters were washed in 2× SSC, 0.1% SDS at 42°C for 1 hr. Restriction fragments of the genomic DNA were subcloned into pUC19 and sequenced. To clone the corresponding cDNA, a [32P]-labeled 68 bp genomic exon was used to screen random-primed λGT10 CNS and ovotestis cDNA libraries. Filters were hybridized at 42°C for 16 hr in 6× SSC, 5× Denhardt’s solution, 0.5% SDS, 50% formamide, and 100 μg/ml denatured salmon sperm DNA. After hybridization, filters were washed in 0.1× SSC, 0.1% SDS at 55°C for 1 hr and exposed to Kodak x-ray film at −80°C. Positive clones were identified, purified, and subcloned into pBluescript (Stratagene, La Jolla, CA). Double-stranded DNA was sequenced by the dideoxynucleotide method (Sanger et al., 1977) according to Sequenase protocols (United States Biochemical Corp.). The 5′ end of the cDNA was cloned by 5′-RACE (rapid amplification of 5′ cDNA extremities) using Aplysia CNS poly(A+) RNA as described by Chen (1996). The first-strand cDNA was synthesized with SuperScript reverse transcriptase (Life Technologies, Burlington, ON) using a specific primer CTTGACGATCCACTTTTTCCCC (nucleotides 639–660). An oligo (dA) anchor was added to the 3′ end of the first strand cDNA with terminal deoxynucleotidyltransferase. A short 12-cycle round of PCR was performed as described by Chen (1996) with the same specific 3′ primer and the 5′ anchor primer TGAGGTGGTTGCCACAGGAGG(T)20.The product of this PCR reaction was subjected to a second amplification using a nested, specific 3′ primer TCAAGGCTGCTGAGTCTTTGGG (nucleotides 601–622) and the 5′ anchor primer TGAGGTGGTTGCCACAGGAGG. The product was subcloned into the pCR II plasmid (Invitrogen, Carlsbad, CA) and sequenced.
cRNA probes. cRNA probes of 930 bp were obtained by in vitro transcription of the HindIII–EcoRI fragment of the apNEP cDNA, subcloned in pBluescript. Probes were labeled with digoxigenin-UTP (Boehringer Mannheim, Laval, Quebec, Canada) using T7 or T3 RNA polymerase (Pharmacia Biotechnology, Baie d’Urfé, Quebec, Canada) according to the manufacturer’s instructions. The size and amounts of labeled RNAs were evaluated by Northern blotting after separation on a formaldehyde-agarose gel. Probes were aliquoted and stored at −80°C until use.
In situ hybridization. In situ hybridization was performed essentially as described in Panoskaltsis-Mortari and Bucy (1995) on either frozen or paraffin tissue sections. Sections were hybridized with 3 ng of heat-denatured cRNA probe in 100 μl of 50% deionized formamide, 2× SSC, 500 μg/ml heat-denatured herring sperm DNA, 250 μg/ml yeast tRNA, 10% dextran sulfate, for 16 hr at 50°C. After hybridization, slides were successively washed in 2× SSC for 5 min at room temperature, treated with RNase A (40 mg/ml in 500 mm NaCl, 20 mm Tris-HCl, pH 7.5, 1 mm EDTA) at 37°C for 30 min, washed in 2× SSC, 50% formamide at 50°C for 15 min, and in 1× and 0.5× SSC at room temperature for 5 min each. Positive signals were detected using anti-digoxigenin antibodies (Boehringer Mannheim). Tissues were equilibrated for 1 min in antibody dilution buffer (100 mmTris-HCl, pH 7.5, 150 mm NaCl), blocked for 30 min in the same buffer containing 2% normal goat serum, and incubated at room temperature for at least 1 hr with sheep anti-digoxigenin antibodies diluted 1:500. Sections were then washed in the antibody dilution buffer for 5 min, transferred to the detection buffer (100 mm Tris-HCl, pH 9.5, 100 mm NaCl, 50 mm MgCl2) for 10 min, and incubated in 340 μg/ml nitroblue tetrazolium/175 μg/ml 5-bromo-4-chloro-3-indolyl-phosphate/4 toluidine salt (Boehringer Mannheim) in detection buffer. Staining was allowed to proceed overnight in the dark at 4°C. The coloring reaction was stopped in 10 mm Tris-HCl, pH 8.0, 1 mm EDTA. Sections were mounted in 33% glycerol, 1× PBS, and stored at 4°C.
Antibodies and immunoblotting. Antibodies directed against apNEP were produced by injecting rabbits with a pool of bacterially expressed C-terminal (amino acid 288–453) and N-terminal (amino acids 454–761) apNEP protein fragments fused to a 6-His tag (Qiagen, Mississauga, ON). Immunoblot analysis was performed using horseradish peroxidase-conjugated anti-rabbit IgG antibodies (Dako, Mississauga, ON) and the SuperSignal substrate (Pierce, Rockford, IL) as recommended by the manufacturer.
Electrophysiology. Intracellular microelectrodes were pulled from omega-dot borosilicate glass (WPI, Sarasota, FL) and filled with 2m KAc. Their resistances were between 10 and 20 MΩ. The experiments were performed in current-clamp mode, and the voltage signals were amplified using Axoclamp 2B amplifiers (Axon Instruments). Neurons in the buccal ganglion were identified on the basis of the classification suggested by Fiore and Meunier (1979): these were A neurons corresponding to cells B4 and B5 of Gardner’s classification (Gardner, 1971), and B neurons. A and B neurons and one or two other large silent neurons located near the B neurons were impaled in each experiment. The radula nerve was stimulated with a suction electrode with 3 msec pulses; at the beginning of the experiment, the stimulus intensity was adjusted to evoke several spikes in A neurons (usually 2–3 V). Then the radula nerve was stimulated with trains of 30–50 stimuli (20 Hz) to evoke several (two to three) waves of synaptic and electrotonic potentials in A and B neurons (see Fig. 9). The intertrain interval was 10 min. The evoked responses as well as the spontaneous background activity were continuously monitored during the experiment using a DASH iV (Astro-Med) chart recorder (25 mm/min chart speed).
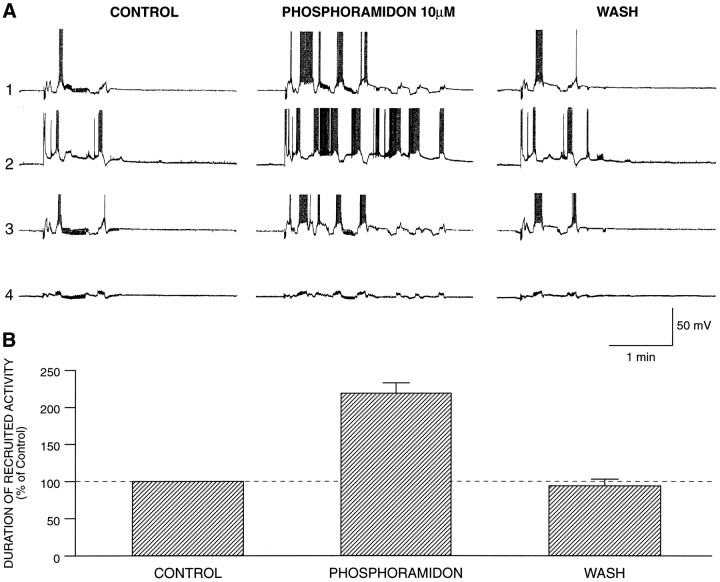
Fig. 9.
Phosphoramidon prolongs the responses of the buccal neurons to radula nerve stimulation. A, Simultaneous recordings from four neurons before, during, and after exposure to phosphoramidon (10 μm). In all cases the activity evoked is prolonged: trace 1, B neuron; trace2, A neuron; trace 3, B neuron; trace4, unidentified cell (see Results for details).B, Summary of five experiments (18 neurons) with phosphoramidon (10 or 100 μm). Prolongation of the responses evoked by radula nerve stimulation was observed in all the monitored neurons. The duration of the recruited activity was normalized to each respective control. The percentage average of every neuron in one experiment contributed to the average score of that experiment.
RESULTS
Evidence for a neutral endopeptidase-like activity in theAplysia CNS
To reveal the presence of a membrane-associated neutral endopeptidase activity in the Aplysia CNS, we incubated plasma membranes from pooled ganglia with [3H][Leu]enkephalin. Amastatin was added at a concentration of 10 μm to reduce as much as possible the strong aminopeptidase N activity present in this tissue (Bawab et al., 1992). The resulting metabolites were analyzed by RP-HPLC (Fig.1). As expected for a NEP-like activity, a peak that comigrated with the Tyr-Gly-Gly peptide is visible (Fig.1A). It corresponds to the degradation of 1.6 pmol of substrate per milligram of protein per minute. The tyrosine peak is probably generated by the residual aminopeptidase N activity (Bawab et al., 1992).
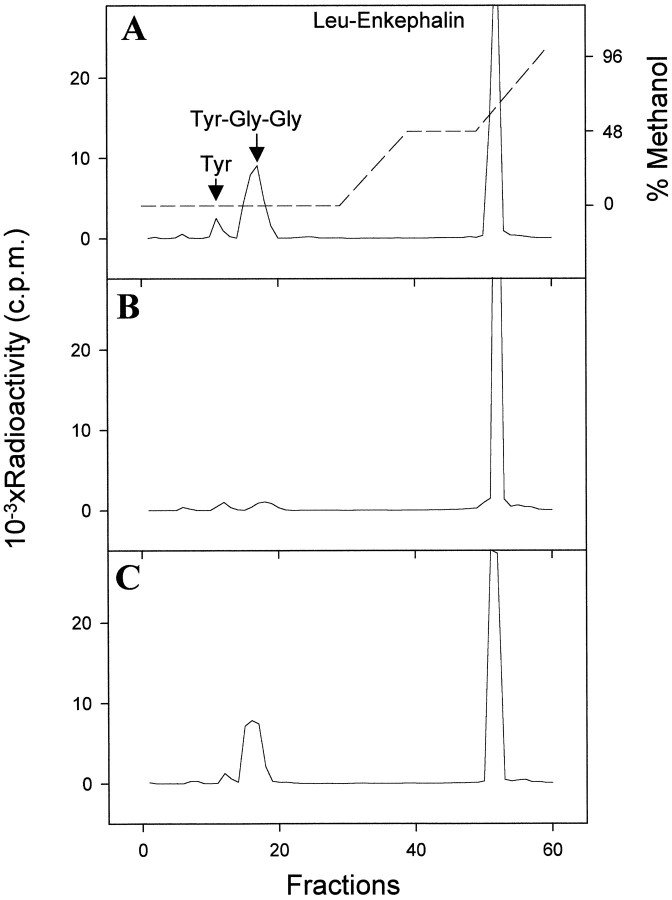
Fig. 1.
RP-HPLC analysis of degradation metabolites of [3H][Leu]enkephalin in theAplysia CNS. The substrate was incubated with CNS plasma membranes, in the absence (A) or presence of 1 μm RB104 (B) or 1 μm captopril (C).Arrows indicate the elution position of standard peptides. The dashed line represents the methanol gradient used in the HPLC.
To characterize the nature of the [Leu]enkephalin-degrading activity, we used various peptidase inhibitors. The cation chelating agent 1-10-phenanthroline completely inhibited the hydrolysis of [3H][Leu]enkephalin (Table1), suggesting that the activity is produced by a metallopeptidase. NEP inhibitors such as RB104 (Fig.1B), HACBO-Gly, thiorphan, and phosphoramidon (Table1) were shown to abolish the Tyr-Gly-Gly peak. In contrast, captopril, an inhibitor of the dipeptidylcarboxypeptidase (Fig. 1C), and PMSF, an inhibitor of serine proteases (Table 1), had no effect on the activity of our enzyme preparation. All of these results suggest that a metallopeptidase with an inhibitor profile similar to that of the NEPs found in Aplysia kidney and in mammals is present in the CNS of Aplysia.
Table 1.
Comparison of the action of peptidase inhibitors on apNEP activity present in Aplysia californica CNS and kidney membranes
| Inhibitors | Concentration (μm) | % Inhibition (head ganglia membranes) | % Inhibition (kidney membranes)1-a |
|---|---|---|---|
| RB104 | 1 | 87 | 91 |
| HACBO-Gly | 10 | 91 | 74 |
| Thiorphan | 10 | 41 | 33 |
| Phosphoramidon | 10 | 41 | 45 |
| 1-10 Phenantroline | 5 | 100 | 100 |
| Captopril | 10 | 11 | 0 |
| PMSF | 100 | 0 | 0 |
Plasma membranes from head ganglia or kidney [results from Bawab et al. (1993)] were incubated, before the addition of the [3H]Leu-enkephalin, in the absence or presence of various peptidase inhibitors. The enzymatic assays were performed as indicated in Materials and Methods. Results are expressed as a percentage of inhibition of substrate degradation. The percentage inhibition was calculated by comparing the radioactivity under the Tyr-Gly-Gly peak in the presence and absence of inhibitors.
Binding of the highly specific NEP inhibitor [125I]RB104 to NEP-like proteins in theAplysia CNS
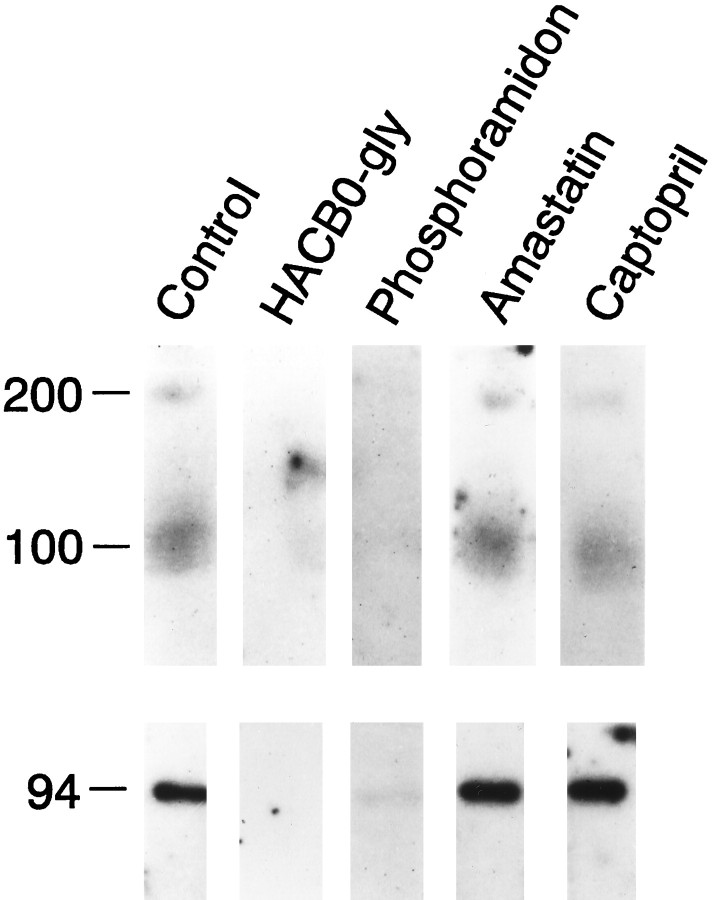
RB104 is a highly specific NEP inhibitor that was shown to detect as little as 2 ng of rat NEP on a Western blot (Fournié-Zaluski et al., 1992). We first tested the affinity of the enkephalin-degrading enzyme in CNS plasma membranes for [125I]RB104 and found that the KD is similar to that of the rat NEP and the Aplysia kidney enzyme (Table2) (Fournié-Zaluski et al., 1992;Bawab et al., 1993). [125I]RB104 was then used in inhibitor gel electrophoresis experiments. Solubilized CNS membrane proteins or purified rabbit NEP, which was used as a control, were separated by SDS-PAGE, transferred to nitrocellulose membranes, and incubated with [125I]RB104. As shown in Figure2, [125I]RB104 binds to proteins of 100 and 200 kDa in the Aplysia CNS membranes and to the 94 kDa rabbit protein; this binding was completely abolished by NEP inhibitors such as HACBO-Gly (10 μm) and phosphoramidon (10 μm). In contrast, the labeling was not affected by specific inhibitors of other peptidases such as captopril (10 μm) or amastatin (10 μm). These results demonstrate that NEP-like proteins are expressed in the CNS and that their molecular sizes are different from that of the 140 kDa NEP-like enzyme already observed in the Aplysia kidney membranes. However, their active site is likely to be structurally and functionally similar, because they all bind [125I]RB104 with high affinity. These results raise the question of whether the NEP-like proteins in the kidney and CNS are differentially glycosylated isoforms of the same protein or whether they are expressed from two closely related NEP-like genes.
Table 2.
Comparison of the NEP-like enzymes in the CNS and kidney ofAplysia californica
| Characteristics | CNS | Kidney2-a |
|---|---|---|
| Activity2-b | 1.6 pmol/mg protein per minute | 3.5 pmol/mg protein per minute |
| RB104-KD2-c | 0.16 × 10−10 | 0.26 × 10−10 |
| RB104-Bmax2-e | 12 fmol/mg protein | 20 fmol/mg protein |
| RB104-inhibitor gel | ||
| electrophoresis (M.W.) | 100 and 200 kDa | 140 kDa |
| Western blot (M.W.)2-d | 100 and 200 kDa | 140 kDa |
| Northern blot2-e | +f | ++ |
Membrane preperations were incubated with [3H][Leu]enkephalin, in the presence of amastatin at a concentration of 10 μm. The metabolites were separated by RP-HPLC, and the number of counts per minute under the Tyr-Gly-Gly peak were measured as described in Bawab et al. (1993).
Kidney plasma membranes were incubated in the presence of [125I]RB104 and different dilutions of cold I-RB104. The KD andBmax values were calculated from Scatchard analysis as reported previously (Bawab et al., 1993).
Western blot realized with specific apNEP antibodies.
Northern blot realized with an apNEP cDNA probe.
fIntensity of the hybridization signal.
Fig. 2.
Inhibitor gel electrophoresis with [125I]RB104 and different peptidase inhibitors. Solubilized Aplysia CNS membrane proteins (top panel) and purified rabbit NEP (bottom panel) were separated by SDS-PAGE and transferred onto nitrocellulose membranes. NEP-like proteins were labeled with 100 pm [125I]RB104 in the presence or absence of peptidase inhibitors: absence of inhibitor (Control); HACBO-gly at 10 μm; Phosphoramidon at 10 μm;Amastatin at 10 μm;Captopril at 10 μm.
Isolation of cDNA clones encoding an apNEP
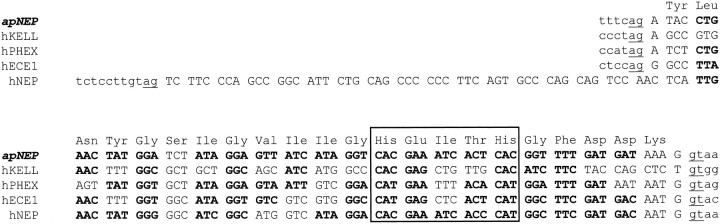
To answer this question, we cloned cDNAs coding for the NEP-like activity. We first screened an Aplysia genomic library at low stringency, using a 760 bp rabbit NEP cDNA fragment as a probe (Devault et al., 1987). One of the 13 clones (λNEPg1) found was further characterized, and a 400 bp fragment was subcloned and sequenced. A short segment of 68 bp, flanked by splicing consensus sequences, showed high sequence similarity to the rabbit NEP sequence (Fig. 3). Interestingly, the 3′ splicing site is identical to the one described for all of the NEP-like family members, and the 5′ splicing site is common to endothelin-converting enzyme (ECE), a human phosphate-regulating gene with homologies to endopeptidases on the X-chromosome (PHEX) and kell blood group protein (KELL) but not to NEP (Fig. 3). Considering the high level of conservation of exon/intron boundaries, these results not only suggest that apNEP is a member of the NEP family, but also indicate that the apNEP and mammalian NEP-like enzymes are likely to be derived from a common ancestor (see also below).
Fig. 3.
Comparison of the apNEP (λNEPg1 clone), hNEP, hECE1, hKELL, and hPHEX exons that code for the zinc-binding domain. Nucleotide sequences of exons and flanking introns are shown incapital and small letters, respectively. Splicing consensus sequences are underlined. The deduced amino acid sequence of the apNEP exon is shown above the nucleotide sequence. The codons for identical amino acids are inbold type, and the pentapeptide consensus sequences (His-Glu-Xaa-Xaa-His) that are part of the metalloprotease zinc-binding domain are boxed.
This 68 bp segment was PCR-amplified, subcloned, and used as a probe to screen Aplysia CNS and ovotestis cDNA libraries by plaque hybridization. Of eight positive recombinant phages, the inserts of the λ5.1, λNEPc, λNEPe, and λNEPf clones were sequenced. Their sequences indicated that they represented overlapping cDNAs derived from the same apNEP mRNA but that the 5′ region of the coding region was missing. Because we did not succeed in cloning the 5′ part of the cDNA by rescreening the libraries, we performed a 5′-RACE protocol using a set of nested specific internal primers and mRNA isolated from the CNS. This yielded one overlapping PCR product that covered the missing coding sequence and part of the 5′ UTR. The first ATG is found at position 164 and is followed by an open reading frame of 2361 nucleotides that codes for a putative apNEP protein of 787 amino acids (Fig. 4). This protein is ∼35% identical to human NEP.
Fig. 4.
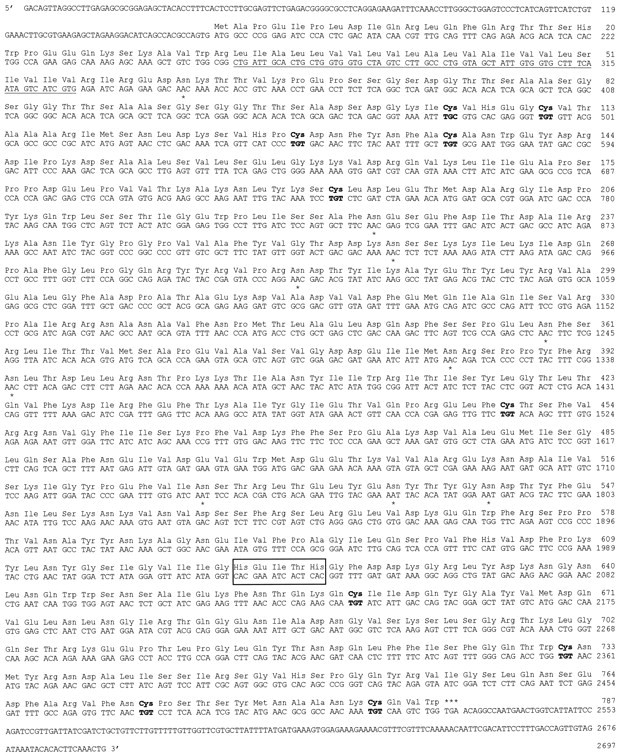
Nucleotide and deduced amino acid sequence of theAplysia neutral endopeptidase. Amino acids are numbered starting at the first ATG of the open reading frame. The putative transmembrane region is underlined. Potential sites ofN-glycosylation are indicated by anasterisk, and the cysteine residues arebold. The zinc-binding signature HEXXH isboxed. The nucleotide sequence has been submitted to the GenBank Data Bank with accession number AF104361.
Southern blot analysis of the apNEP gene
The cloning of one small exon of the apNEP gene suggests that it could be fragmented into many exons as observed for the mammalian homologs. To assess this point, a Southern blot ofA. californica genomic DNA was digested withBglII, EcoRI, HindIII,SacI, and XbaI and hybridized with a short probe. Considering the fact that no SacI or BglII site and only one XbaI restriction site exists in this probe, the multiple bands that hybridized in each lane indicate that this small cDNA region corresponds to at least three exons in the genomic DNA (Fig. 5A). Consistent with the cloning of a small exon (see above), this result suggests that the genomic organization of the apNEP gene may be similar to that of the members of the NEP-like family, which are all fragmented into several exons (D’Adamio et al., 1989).
Fig. 5.
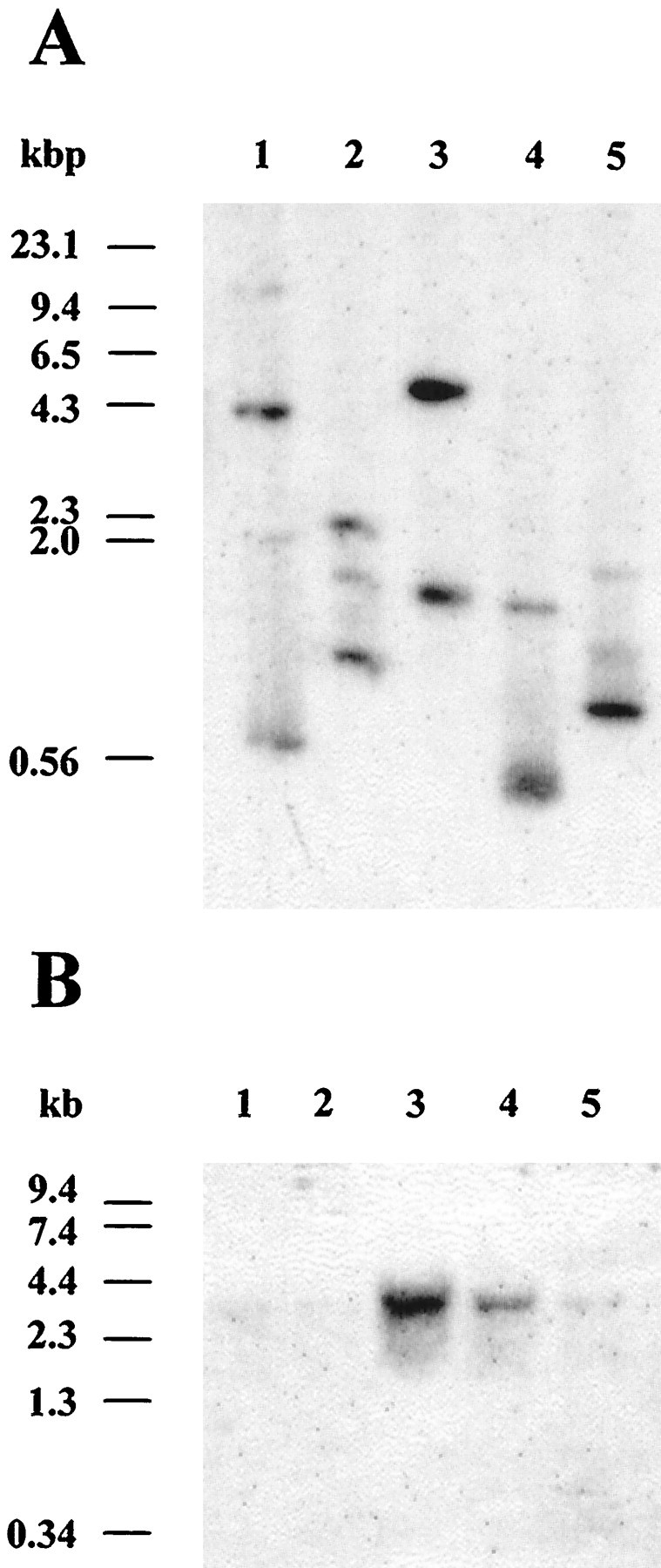
Southern and Northern blot analysis of theapNEP gene. A, Genomic DNA was isolated from ovotestis and digested with either BglII (lane 1), EcoRI (lane 2),HindIII (lane 3), SacI (lane 4), or XbaI (lane 5). Digested DNA (10 μg/lane) was run on a 0.8% agarose gel, transferred to a nitrocellulose membrane, and hybridized at high stringency with the [32P]-labeledHindIII–AccI apNEP fragment (nucleotides 1142–1458) as described previously (Wickham and DesGroseillers, 1991). DNA molecular weight markers are indicated in kilobase pairs (kbp) on the left. B, Northern blot analysis of the apNEP transcript. Total RNA was extracted from different tissues, and poly(A+) RNA (5 μg) isolated from gill (lane 1), heart (lane 2), ovotestis (lane 3), kidney (lane 4), and CNS (lane 5) was fractionated on a 1% formaldehyde/agarose gel, blotted to a nitrocellulose membrane, and hybridized at high stringency with the [32P]-(HindIII–AccI) apNEP fragment, as performed previously (Auclair et al., 1994). RNA molecular weight markers are indicated in kilobases (kb) on the left. To control the amounts of RNA in each lane, filters were stripped and rehybridized with an Aplysiaactin probe (data not shown).
Tissue expression of the apNEP mRNA, and cellular localization in the Aplysia CNS
Northern blots of poly(A+) RNA extracted from various tissues were probed with a 316 bp apNEP cDNA fragment and used to determine the size of the apNEP transcript and its specificity of expression (Fig. 5B). A single transcript of ∼3.8 kb was abundantly present in ovotestis and kidney and very little was expressed in the CNS, gill, and heart where the signal could only be detected after a long period of exposure. By comparison with the size of the cDNA, it is likely that additional 5′ and/or 3′ untranslated sequences are present in the transcript. The presence of apNEP in these tissues was confirmed by Western blot experiments (see below). These results confirm that apNEP is expressed in both the CNS and kidney as well as in many other organs.
To determine the type(s) of cells that express apNEP in theAplysia CNS, we performed preliminary in situhybridization experiments on paraffined sections of the ganglia. A positive signal can be observed with a cRNA probe in many neurons of all the ganglia (Fig.6A), demonstrating that neurons are the source of apNEP in the CNS. The signal is not restricted to the cell bodies and can also be observed in the neuropil and ganglion peripheral nerves in structures that look like neurites (Fig. 6C). The specificity of the signal was confirmed by the absence of any signal when the same experiments were performed on adjacent sections using a sense probe (Fig. 6B). At this point we did not try to identify individual neurons.
Fig. 6.
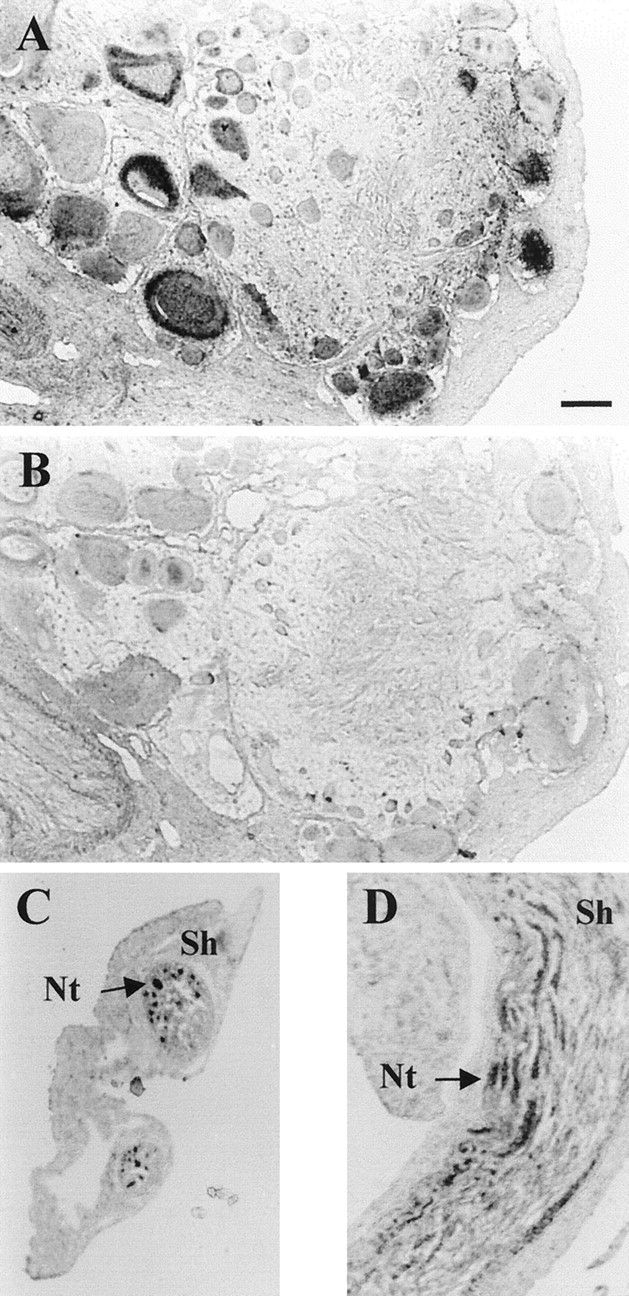
In situ hybridization of apNEP on paraffined sections of Aplysia ganglia. Sections of the abdominal ganglion (A, B) and of a buccal ganglion nerve (C, D) were hybridized with either an apNEP cRNA antisense (A, C, D) or sense (B) probe. Positive signal is seen in neurons (A) and neurites (Nt) extending into the nerve. No signal is detected in the sheath (Sh). The same results were obtained with sections from all the major ganglia. Scale bar, 100 μm.
Primary structure of apNEP
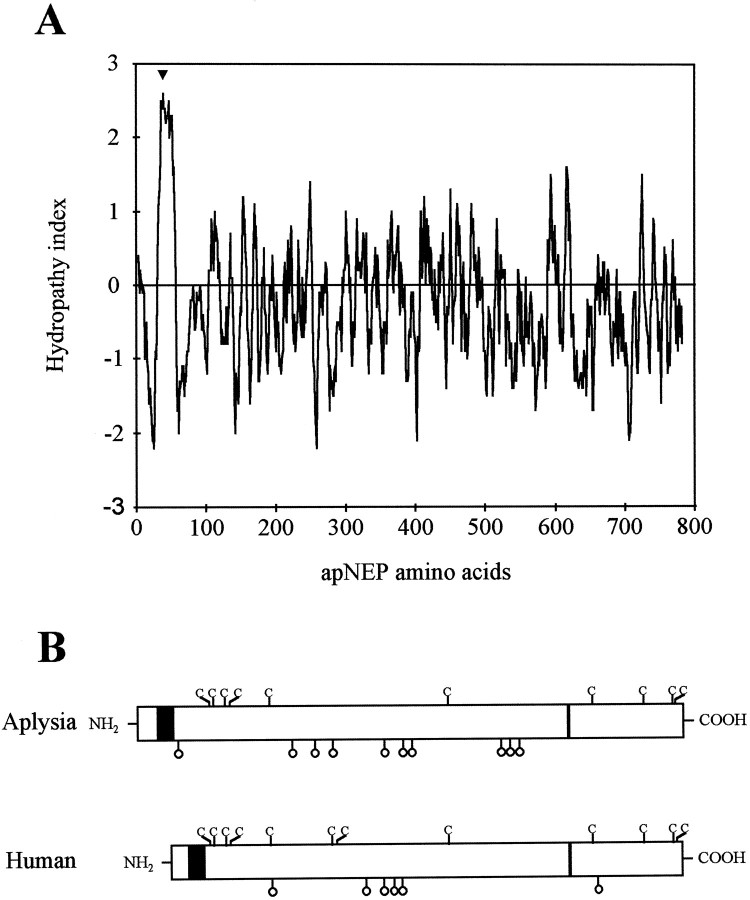
The apNEP cDNA sequence encodes a putative protein of 88 kDa, which shares important structural features with mammalian NEP. (1) As predicted by the Kyte and Doolittle (1982) hydrophobicity plot (Fig.7A), apNEP is a type II integral membrane protein with a short N-terminal cytoplasmic tail of 31 amino acids, a hydrophobic region of 23 residues, which represents a putative transmembrane helix, and a large extracellular C-terminal domain of 686 amino acids. (2) The extracellular portion of apNEP contains the highly conserved zinc-binding motif (residues 622–626) (Fig. 6B) and thus probably constitutes the catalytic domain. (3) apNEP contains 10 putative sites for N-glycosylation (Asn-Xxx-Ser/Thr), suggesting that apNEP is highly glycosylated. (4) The 10 cysteine residues found in the extracellular domain of apNEP coalign with those of the mammalian NEP (Fig. 7B). (5) Nearly all of the amino acids that are essential for the enzymatic activity of the mammalian NEP (for review, see Roques et al., 1993) are found in the same position on the cDNA encoding apNEP (Table3). All together, these results suggest that the apNEP cDNA codes for an Aplysia neutral endopeptidase homolog.
Fig. 7.
Molecular structure of apNEP. A, Hydropathy analysis of apNEP. The 787 amino acid-long apNEP sequence was scanned using the computer program of Kyte and Doolittle (1982).Numbers on the horizontal axis refer to the amino acid sequence. Negative values correspond to hydrophilic regions and positive values to hydrophobic regions. The arrowheadindicates the only potential membrane-spanning segment of apNEP.B, Schematic representation of the primary sequences of the human and Aplysia NEP proteins. The cysteine residues in the two proteins are indicated by the one-letter codeC. The black rectangle represents the transmembrane region, and the thin rectangle represents the HEXXH gluzincin domain. The position of the possibleN-glycosylation sites is indicated by open lollipops.
Table 3.
Comparison of the essential amino acids of thermolysin (TLN), mammalian neutral endopeptidase (mamNEP), and Aplysia californica neutral endopeptidase (apNEP)
| Action | TLN3-a | mamNEP | apNEP | References |
|---|---|---|---|---|
| Coordination of the zinc atom | His 142 | His 5833-b | His 622 | Colman et al., 1972; Hangauer et al., 1984; Devault et al., 1988; Le Moual et al., 1993 |
| His 146 | His 5873-b | His 626 | Colman et al., 1972; Hangauer et al., 1984; Devault et al., 1988; Le Moual et al., 1993 | |
| Glu 166 | Glu 6463-b | Glu 684 | Colman et al., 1972; Le Moual et al., 1991, 1993 | |
| Catalysis | Glu 143 | Glu 5843-b | Glu 623 | Colman et al., 1972; Weaver et al., 1977; Devault et al., 1988 |
| Substrate binding | Arg 1023-b | Ala 152 | Bateman et al., 1989; Beaumont et al., 1991; Kim et al., 1992 | |
| Arg 203 | Arg 7173-b | Arg 755 | Colman et al., 1972; Holmes and Matthews, 1982; Marie-Claire et al., 1997 | |
| Asn 112 | Asn 5423-b | Asn 581 | Roderick et al., 1989; Dion et al., 1995 | |
| Ala 113 | Ala 543 | Ala 582 | Weaver et al., 1977 | |
| Stabilization of the transition state | His 231 | His 7113-b | His 749 | Colman et al., 1972; Dion et al., 1993 |
| Asp 170 | Asp 6503-b | Asp 688 | Colman et al., 1972; Christianson and Alexander, 1990; Le Moual et al., 1994 |
Determined by crystallographic studies.
Determined by site-directed mutagenesis experiments.
The CNS 100 kDa and the kidney 140 kDa [125I]RB104-binding proteins are likely to be coded by the apNEP gene
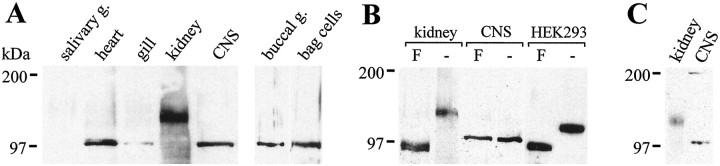
To determine whether the NEP-like molecules in the CNS and kidney membranes are both expressed from the apNEP gene, immunoblots of membrane extracts from the kidney and CNS were performed, using anti-apNEP antibodies. As expected from the inhibitor gel electrophoresis experiment, a band of 140 kDa was detected in the kidney plasma membranes, whereas a single band of 100 kDa was detected in the membranes of the CNS (Fig.8A). Under nonreducing electrophoresis conditions, an additional band of ∼200 kDa was detected in the membranes of the CNS (Fig. 8C), suggesting strongly that the 200 kDa protein is a dimer of the 100 kDa protein, as observed in mammals (Kenny and Maroux, 1982). These results demonstrate the presence of apNEP in both tissues and clearly link the RB104-binding proteins in the membranes of both the CNS and kidney to the product of the apNEP gene.
Fig. 8.
Immunoblot analysis of the expression and glycosylation of apNEP in different A. californicatissues. A, Twenty micrograms of solubilized membrane proteins (salivary gland, heart, gill, kidney, and CNS) and 30 μg of total protein extracts (buccal ganglion and bag cells) were separated on a 6% SDS-polyacrylamide gel under reducing conditions, blotted, and detected with an anti-apNEP antisera. B, Plasma membrane protein extracts isolated from Aplysia tissues (kidney, CNS) or from transiently transfected mammalian HEK293 cells (HEK293) were incubated in the absence (−) or presence (F) of PNGase F, before loading on the gel.C, SDS-PAGE under nonreducing conditions. Thearrow indicates the position of the 200 kDa band.
The discrepancy in the apparent molecular mass of the CNS and kidney NEP-like enzymes in Aplysia membranes may be the result of post-translational modifications, such as glycosylation. To examine this point, membrane extracts from these tissues were deglycosylated with PNGase F; the resulting proteins were separated by SDS-PAGE and detected by Western blotting. After PNGase F treatment, the molecular size of apNEP in the kidney was reduced to ∼88 kDa (Fig.8B), which is the predicted size from the cDNA sequence. This demonstrates that the 140 kDa protein is highly glycosylated and confirms that it is probably the product of theapNEP gene. On the other hand, the size of the 100 kDa protein in the CNS (Fig. 8B), heart, and gill was unchanged. To determine whether PNGase F can remove sugars from glycoproteins expressed in the CNS membranes of Aplysia, we probed the blot with antibodies directed against 5-HTap1, another highly glycosylated protein (Angers et al., 1998). This protein was not deglycosylated either (data not shown), indicating that several glycosylated proteins in the CNS are PNGase F resistant.
We cloned the apNEP cDNA from the CNS in pCDNA3/RSV, and the recombinant plasmid was introduced into mammalian HEK 293 cells, as reported previously (Angers et al., 1998). Plasma membranes were purified and the protein was detected by Western blotting after PNGase F treatment. As seen in Figure 8B, the results suggest that the enzyme found in Aplysia kidneys is likely to be coded by the same gene as the cDNA we isolated from the CNS because they are of the same size.
The application of a NEP-specific inhibitor potentiates the action of endogenous neuropeptides on the buccal ganglion and prevents thein vitro degradation of SCPB byAplysia CNS membranes
In situ hybridization and Western blotting experiments (Fig. 8A) showed that apNEP is present in the buccal ganglion of Aplysia. To determine whether apNEP could be responsible for the inactivation of neuropeptides in vivo, we studied a well understood behavior in invertebrates, which is feeding. In Aplysia, feeding consists of a number of different rhythmic motor patterns, including biting, swallowing, and rejection (Kupfermann, 1974; Weiss et al., 1986). Different reports have characterized the critical roles of several neuropeptides, including SCPB, FMRFamide, egg-laying hormone (ELH), buccalin, and myomodulin, as well as serotonin and acetylcholine, in the modulation of various aspects of the feeding behavior (Kreiner et al., 1987; Lloyd et al., 1987; Sossin et al., 1987; Lloyd, 1988; Miller et al., 1993a,b). Inhibition of apNEP by an apNEP-specific inhibitor should potentiate the action of secreted peptides that are normally substrates for this enzyme. Therefore, to recruit at least some of the peptidergic neurons in the ganglion, we decided to stimulate the radula nerve, because this nerve contains processes of SCP-containing neurons (Miller et al., 1994). Trains of stimuli to the radula nerve were delivered every 10 min (see Materials and Methods for more details); the evoked responses were recorded in A and B neurons and one or two other large cells located near the B cells. After three to four control responses, phosphoramidon (10–100 μm) was added to the bath, and three to four responses were monitored in the presence of the drug; 5–10 more responses were recorded after the inhibitor was washed out. The results of an experiment, in which 10 μmphosphoramidon was added, are shown in Figure9. In the control period during the stimulation itself, there was in general a burst of action potentials and a burst of PSPs with oscillations of membrane potentials in the monitored neurons (Fig. 9A). The later parts of the evoked responses were greatly prolonged in the presence of phosphoramidon. In the example in Figure 9A, one can notice that the delayed firing is increased in three of the neurons. These effects were reversible after washout. The summary of five experiments (18 neurons) is shown in Figure 9B. These results suggest that the action of several endogenous peptides in the buccal ganglia can be enhanced because of the decrease of their degradation by a NEP-like enzyme present in this ganglion.
Because exogenous applications of SCPB induce the same physiological responses on these neurons as those obtained after radula nerve stimulation (data not shown), we tested whether SCPBis a substrate for apNEP in vitro. Using RP-HPLC, we showed that SCPB is cleaved by Aplysia CNS membrane extracts and that this cleavage is inhibited by the NEP inhibitor phosphoramidon (10 μm). As seen in Figure10, the peak corresponding to the uncleaved SCP peptide is clearly preserved in the presence of phosphoramidon. In the absence of the inhibitor, this peak is strongly reduced, and other peaks appeared, probably corresponding to the metabolites resulting from the degradation of SCPB by a NEP-like enzyme present in the membrane protein extract.
Fig. 10.
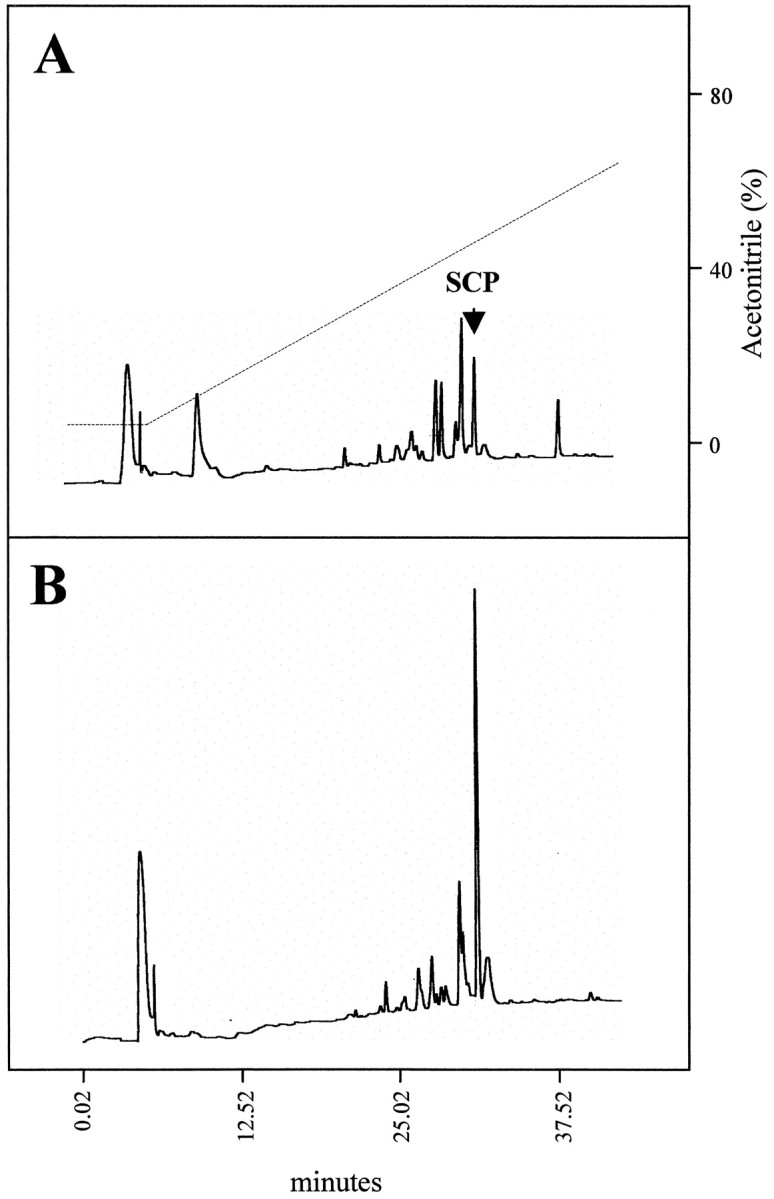
SCPB is degraded by anAplysia CNS NEP-like enzyme. SCPB was incubated with CNS plasma membranes, in the absence (A) or presence of 10 μmphosphoramidon (B). The arrowindicates the elution position of the uncleaved SCPB. Thedashed line represents the acetonitrile gradient used in the HPLC.
DISCUSSION
Endopeptidase activity in the CNS
Previous studies showed that an endopeptidase with catalytic properties similar to those of neutral endopeptidase-24.11 is present in the kidney of A. californica (Bawab et al., 1993). In this paper, we demonstrate that this activity also exists in the CNS of this mollusk. The HPLC profiles of [Leu]enkephalin degradation, the sensitivity of this activity to specific NEP inhibitors, and the binding of [125I]RB104 to the protein all strongly suggest that the CNS and kidney endopeptidases are similar. However, the CNS endopeptidase migrates as a 100 kDa protein band on a Western blot. Although consistent with the size of the mammalian (Kenny et al., 1987; Fournié-Zaluski et al., 1992) and the mollusk Mytilus edulis (Shipp et al., 1990) NEPs, the CNS protease is much smaller than the one found in the Aplysia kidney (Bawab et al., 1993). Our results demonstrate that not only is the pattern of glycosylation of apNEP different in the kidney and CNS, as observed in mammals (Roques et al., 1993), but the nature of the sugars that are added to the glycoproteins is likely to be different in these tissues. This could be attributable to the presence of a fucose residue on the first N-acetylglucosamine of the oligosaccharide chain in the CNS; this addition is known to inhibit the cleavage of sugar chains by PNGase F, and fucose residues have been reported in different glycoproteins isolated from the CNS of Aplysia (Thompson et al., 1976; Ambron et al., 1985; Goldberg and Ambron, 1986; Cleary and Schwartz, 1987). The meaning of this differential glycosylation is unknown because both proteins seem to exhibit similar [Leu]enkephalin-degradation activities, affinities for RB104 (Table2), and responses to different NEP inhibitors (Table 1).
More significantly, our results with [125I]RB104 and the anti-apNEP antibodies clearly link the RB104-binding protein in the plasma membranes of both the kidney and CNS to the product of theapNEP gene. We do not yet know whether the enkephalin-degrading activity in these membranes is generated by apNEP, although the binding of [125I]RB104 to a single protein in both the CNS and kidney is a strong indication for the expression of a single NEP-like gene in these tissues.
Structure/function of apNEP
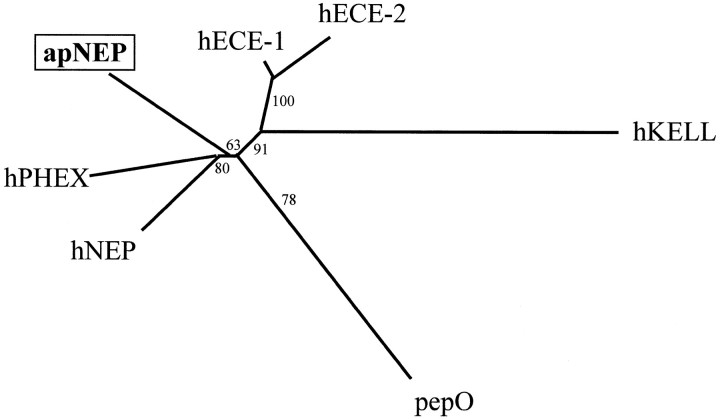
As described previously for the kidney endopeptidase, the activity of the CNS enzyme is low and does not allow us to fully characterize it. The molecular cloning of the apNEP cDNA represents a first step toward achieving this goal. The predicted molecular size, topological localization in the membranes, and peptidic sequence of the protein places apNEP in the large family of NEP-like enzymes (Turner and Tanzawa, 1997). Indeed, a phylogenetic analysis localizes apNEP at the branching point of mammalian NEP-like enzymes, suggesting thatapNEP may be considered as the ancestor of these genes (Fig.11). After the separation of vertebrates from invertebrates, it is likely that the ancestorNEP gene duplicated and diverged to generate peptidases involved in several physiological processes (NEP) (Roques et al., 1993), in bone and tooth mineralization processes (PHEX) (The HYPConsortium, 1995; Ruchon et al., 1998), in the control of blood pressure (ECEs) (Turner and Murphy, 1996; Webb et al., 1997), or in a still uncharacterized function in erythrocytes (KELL) (Lee et al., 1991). Consistent with this hypothesis, most of the residues that have been shown to be essential for the activity and/or conformation of human NEP are conserved at the same position in apNEP (Table 3). Such high conservation in the nature and position of all these residues is very significant when we consider that the two proteins originated from organisms whose ancestors diverged 600 million years ago, and it suggests that these residues were subjected to severe evolutionary constraints to keep the proper folding of its active site. In particular, the 10 cysteine residues in the ectodomain of apNEP and PHEX, which contribute to the stabilization of the active enzyme’s conformation (Tam et al., 1985), not only align perfectly in each protein but are also conserved in NEP, ECEs, and KELL. This again suggests that the structure of apNEP is close to that of the ancestor protein and that some of the mammalian NEP-like enzymes may have evolved by acquiring extra pairs of cysteine residues. Interestingly, the cluster of four cysteines (C-X4-C-X17-C-X7-C), which is located a few amino acids downstream from the transmembrane domain in all members of the NEP-like family, is separated from the transmembrane domain by a spacer of 50 amino acids in apNEP. This spacer contains many serine and threonine residues, suggesting that it may allow O-glycosylation of the protein and/or a better exposure of the active site at the cell surface. Alternatively, it may promote the cleavage of apNEP by a specific protease. Such a feature, which would either modulate the activity of apNEP at the membrane or liberate the protein into the extracellular fluid, has been described for human NEP (hNEP) (Almenoff et al., 1984; Johnson et al., 1985; Deschodt-Lanckman et al., 1989; Soleilhac et al., 1996). This may be particularly useful inAplysia, which has an open circulatory system with arteries leading directly to open tissue spaces (Kandel, 1979). The presence of soluble peptidases in the hemocel may be a more efficient way to degrade peptides, a possibility supported by the description of metallopeptidase activities in Aplysia hemolymph (Squire et al., 1991; Bawab et al., 1992; Owens et al., 1992; Rothman et al., 1992).
Fig. 11.
Phylogenetic analysis of the members of the NEP-like family. Sequences were aligned using the Clustal V program (Thompson et al., 1994). The phylogenetic tree was constructed using the Neighbor Joining method (Saitou and Nei, 1987) with a bootstrap analysis that calculates the probability of occurrence of the presented branching for 100 possible trees (Felsenstein, 1993).hNEP, Human neutral endopeptidase (accession numberM26605); hECE-1, human endothelin-converting enzyme 1 (accession number Z35307); hECE-2, human endothelin-converting enzyme 2 (accession number AB011179);apNEP, A. californica neutral endopeptidase (accession number AF104361); hPHEX, human phosphate-regulating gene with homologies to endopeptidases on the X-chromosome (accession number Y10196); hKELL, human kell blood group protein (accession number M64934);pepO, lactococcus lactis PepO gene (accession number L04938). Sequences were aligned, and only the peptide regions that could be aligned with the PepO sequence were retained for the analysis; this roughly corresponds to the extracellular parts of the human and mollusk enzymes.
Arg102 is the only functional residue that is not shared by apNEP and mammalian NEP. It is known to play a role in substrate binding and to interact with the free carboxy group of the P′2 residue of some substrates (e.g., enkephalins), allowing a dipeptidyl-carboxypeptidase-like activity (Beaumont et al., 1991). The absence of this arginine in the active site of apNEP could explain the weak enzymatic activity of apNEP toward enkephalins. In addition, we demonstrated previously that the enkephalin-degrading enzyme in kidney plasma membranes is a real endopeptidase because it degrades [Leu]enkephalinamide, a peptide that is protected from degradation by carboxypeptidases (Bawab et al., 1993).
Physiological role of apNEP
As observed in mammals (Roques et al., 1993), apNEP is found in many tissues, suggesting that it could be involved in the regulation of different peptidergic pathways. Indeed, neuropeptides are ubiquitously present in Aplysia, and many of them are potential substrates for apNEP. Localization of apNEP by in situhybridization and/or immunohistochemistry can provide important clues concerning its physiological roles and may guide the search for its physiologically relevant substrates. Colocalization of apNEP and specific peptides, and potentialization of the action of the peptides by specific enzyme inhibitors in vivo, are the two most important criteria to establish the physiological relevance of a peptidase in the regulation of a peptidergic pathway.
Our results suggest that apNEP-like peptidases in the buccal ganglion may be involved in the regulation of the feeding behavior. apNEP is expressed in this ganglion, and NEP-inhibitors potentiate the action of the peptides, most likely by controlling their rate of degradation. In this pathway, SCPs, myomodulin, and buccalin are potential substrates (Kreiner et al., 1987; Lloyd et al., 1987; Sossin et al., 1987; Miller et al., 1992). We have shown that one of these peptides, SCPB, is effectively degraded by a CNS NEP-like enzyme. Similarly, in the abdominal ganglion, α-bag cell peptide (α-BCP) (Owens et al., 1992), which is a neuropeptide that mediates the bag cell-induced inhibition of left upper quadrant cells (LUQ) and acts together with ELH to coordinate long- and short-lasting events in the egg-laying program (Rothman et al., 1985), was reported to be rapidly degraded by endogenous peptidases when applied to the abdominal ganglion in the absence of peptidase inhibitors (Rothman et al., 1985). Analysis of the metabolites revealed that among other peptidases, a NEP-like activity is involved in α-BCP degradation. The expression of apNEP by the LUQ cells (data not shown) and the presence of apNEP in the bag cell extracts (Fig. 7A) is consistent with the possibility that it could be involved in this α-BCP-degrading activity.
As observed in mammals (Barnes et al., 1988; Roques et al., 1993), theapNEP gene is expressed in neurons. This suggests that the protein may be present in proximity to peptide receptors where it can play a major role in the modulation of synaptic transmission by controlling the metabolism of neuropeptides close to their site of action. The presence of apNEP mRNA in neurites that come from the ganglia via peripheral nerves suggests that a finer regulation in the level of apNEP may be exerted by local translation of the transcript in neurites. Transport and local translation of mRNAs is now well documented (Wilhelm and Vale, 1993; Steward, 1997), although the significance of this phenomenon is not completely understood. There is building evidence that local translation of mRNA in neurites serves to locally modulate the action of the translated product in response to changing physiological conditions (Van Minnen, 1994; Martin et al., 1997).
The Aplysia nervous system uses a wide variety of neuropeptides to modulate its behavior and physiological functions, and several peptidases are responsible for the regulation of the actions of these peptides. A global understanding of the function of any neuropeptide requires knowledge of its synthesis, release, target tissues, and regulation. The present study provides insight into the nature and distribution of the Aplysia neuropeptidase apNEP and provides the necessary tools to further investigate the role that the extracellular regulation of neuropeptides plays in behavior.
Footnotes
This work was supported by grants from the Medical Research Council of Canada (MRC) and Fonds pour la Formation de Chercheurs et l’Aideá la Recherche (FCAR) to L.D.G. and V.F.C. We thank Jeanne Lavoie and Mireille Fyfe for excellent technical support, as well as Manon Moreau and Gaston Lambert for expert photographic work. We thank Dr. Philippe Crine for providing us with purified rabbit NEP, Dr. Richard H. Scheller for the generous gift of the Aplysia genomic library, and Dr. Bernard P. Roques for generously providing RB104, HACBO-Gly, and thiorphan. We also thank Dr. Hervé Le Moual for critical reading of this manuscript.
Correspondence should be addressed to Dr. Luc DesGroseillers, Département de Biochimie, Université de Montréal, C.P. 6128, Succursale Centre-Ville, Montréal, Québec, Canada H3C 3J7.
Dr. Yang’s present address: Department of Cancer Immunology and AIDS, Dana-Farber Institute, Harvard Medical School, Boston, MA 02115.
Dr. Storozhuk’s present address: Bogomoletz Institute of Physiology, Kiev, Ukraine.
REFERENCES
- 1.Almenoff J, Teirstein AS, Thornton JC, Orlowski M. Identification of a thermolysin-like metallo-endopeptidase in serum: activity in normal subjects and in patients with sarcoidosis. J Lab Clin Med. 1984;103:420–431. [PubMed] [Google Scholar]
- 2.Ambron RT, Schachen S, Rayport SG. Proteins rapidly transported to the synapses of a single identified neuron of Aplysia californica. J Neurosci. 1985;5:2866–2873. doi: 10.1523/JNEUROSCI.05-11-02866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angers A, Storozhuk MV, Duchaine T, Catellucci VF, DesGroseillers L. Cloning and functional expression of an Aplysia 5-HT receptor negatively coupled to adenylate cyclase. J Neurosci. 1998;18:5586–5593. doi: 10.1523/JNEUROSCI.18-15-05586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auclair D, Lang BF, Forest P, DesGroseillers L. Analysis of genes encoding highly conserved lysine-rich proteins in Aplysia californica and Saccharomyces cerevisiae. Eur J Biochem. 1994;220:997–1003. doi: 10.1111/j.1432-1033.1994.tb18704.x. [DOI] [PubMed] [Google Scholar]
- 5.Barnes K, Turner AJ, Kenny AJ. Electromicroscopic immunocytochemistry of pig brain shows that endopeptidase-24.11 is localized in neuronal membranes. Neurosci Lett. 1988;94:64–69. doi: 10.1016/0304-3940(88)90271-6. [DOI] [PubMed] [Google Scholar]
- 6.Barnes K, Turner AJ, Kenny AJ. An immunoelectron microscopic study of pig substantia nigra shows co-localization of endopeptidase-24.11 with substance P. Neuroscience. 1993;53:1073–1082. doi: 10.1016/0306-4522(93)90490-7. [DOI] [PubMed] [Google Scholar]
- 7.Bateman RC, Jr, Jackson D, Slaughter CA, Unnithan S, Chai YG, Moomaw C, Hersh LB. Identification of the active-site arginine in rat neutral endopeptidase 24.11 (enkephalinase) as arginine 102 and analysis of a glutamine 102 mutant. J Biol Chem. 1989;264:6151–6157. [PubMed] [Google Scholar]
- 8.Bawab W, Querido E, Crine P, DesGroseillers L. Identification and characterization of aminopeptidases from Aplysia californica. Biochem J. 1992;286:967–975. doi: 10.1042/bj2860967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bawab W, Aloyz RS, Crine P, Roques BP, DesGroseillers L. Identification and characterization of a neutral endopeptidase activity in Aplysia californica. Biochem J. 1993;296:459–465. doi: 10.1042/bj2960459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaumont A, Le Moual H, Boileau G, Crine P, Roques BP. Evidence that both arginine 102 and arginine 747 are involved in substrate binding to neutral endopeptidase (EC 3.4.24.11). J Biol Chem. 1991;266:214–220. [PubMed] [Google Scholar]
- 11.Brezina V, Bank B, Cropper EC, Rosen C, Vilim FS, Kupfermann I. Nine members of the myomodulin family of peptide cotransmitters at the B16-ARC neuromuscular junction of Aplysia. J Neurophysiol. 1995;74:54–72. doi: 10.1152/jn.1995.74.1.54. [DOI] [PubMed] [Google Scholar]
- 12.Byrne JH, Kandel ER. Presynaptic facilitation revisited: state and time dependence. J Neurosci. 1996;16:425–435. doi: 10.1523/JNEUROSCI.16-02-00425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z. Simple modifications to increase specificity of the 5′RACE procedure. Trends Genet. 1996;12:87–88. doi: 10.1016/0168-9525(96)81412-0. [DOI] [PubMed] [Google Scholar]
- 14.Christianson DW, Alexander RS. Another catalytic triad? Nature. 1990;346:225. doi: 10.1038/346225b0. [DOI] [PubMed] [Google Scholar]
- 15.Cleary LJ, Schwartz JH. Movement of newly synthesized membrane by fast transport along the axon of an identified Aplysia neuron. J Comp Neurol. 1987;263:92–105. doi: 10.1002/cne.902630108. [DOI] [PubMed] [Google Scholar]
- 16.Colman PM, Jansonius JN, Matthews BW. The structure of thermolysin: an electron density map at 2–3 A resolution. J Mol Biol. 1972;70:701–724. doi: 10.1016/0022-2836(72)90569-4. [DOI] [PubMed] [Google Scholar]
- 17.Connelly JC, Skidgel RA, Schulz WW, Johnson AR, Erdos EG. Neutral endopeptidase 24.11 in human neutrophils: cleavage of chemotactic peptide. Proc Natl Acad Sci USA. 1985;82:8737–8741. doi: 10.1073/pnas.82.24.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Adamio L, Shipp MA, Masteller EL, Reinherz EL. Organization of the gene encoding common acute lymphoblastic leukemia antigen (neutral endopeptidase 24.11): multiple miniexons and separate 5′ untranslated regions. Proc Natl Acad Sci USA. 1989;86:7103–7107. doi: 10.1073/pnas.86.18.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deschodt-Lanckman M, Michaux F, DePrez E, Abramovicz D, Vanherweghem JL, Goldman M. Increased serum levels of endopeptidase 24.11 (“enkephalinase”) in patients with end-stage renal failure. Life Sci. 1989;45:133–141. doi: 10.1016/0024-3205(89)90287-7. [DOI] [PubMed] [Google Scholar]
- 20.Devault A, Lazure C, Nault C, Le Moual H, Seidah NG, Chretien M, Kahn P, Powell J, Mallet J, Beaumont A, Roques BP, Crine P, Boileau G. Amino acid sequence of rabbit kidney neutral endopeptidase 24.11 (enkephalinase) deduced from a complementary DNA. EMBO J. 1987;6:1317–1322. doi: 10.1002/j.1460-2075.1987.tb02370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devault A, Sales V, Nault C, Beaumont A, Roques BP, Crine P, Boileau G. Exploration of the catalytic site of endopeptidase 24.11 by site-directed mutagenesis. Histidine residues 583 and 587 are essential for catalysis. FEBS Lett. 1988;231:54–58. doi: 10.1016/0014-5793(88)80701-4. [DOI] [PubMed] [Google Scholar]
- 22.Dion N, Le Moual H, Crine P, Boileau G. Kinetic evidence that His-711 of neutral endopeptidase 24.11 is involved in stabilization of the transition state. FEBS Lett. 1993;318:301–304. doi: 10.1016/0014-5793(93)80533-z. [DOI] [PubMed] [Google Scholar]
- 23.Dion N, Le Moual H, Fournié-Zaluski MC, Roques BP, Crine P, Boileau G. Evidence that Asn542 of neprilysin (EC 3.4.24.11) is involved in binding of the P2′ residue of substrates and inhibitors. Biochem J. 1995;311:623–627. doi: 10.1042/bj3110623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenstein J. PHYLIP (phylogeny inference package) 3.5.1c. University of Washington; Seattle, WA: 1993. [Google Scholar]
- 25.Fiore L, Meunier JM. Synaptic connections and functional organization in Aplysia buccal ganglia. J Neurobiol. 1979;10:13–29. doi: 10.1002/neu.480100103. [DOI] [PubMed] [Google Scholar]
- 26.Fournié-Zaluski MC, Soleilhac JM, Turcaud S, Laï-Kuen R, Crine P, Beaumont A, Roques BP. Development of [125I]RB104, a potent inhibitor of neutral endopeptidase 24.11, and its use in detecting nanogram quantities of the enzyme by “inhibitor gel electrophoresis.”. Proc Natl Acad Sci USA. 1992;89:6388–6392. doi: 10.1073/pnas.89.14.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner D. Bilateral symmetry and interneuronal organization in the buccal ganglia of Aplysia. Science. 1971;173:550–553. doi: 10.1126/science.173.3996.550. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg DJ, Ambron RT. Consequences of partial axotomy for production of neurotransmitter vesicles and routing of rapidly transported membrane glycoproteins in the axonal tree. J Neurosci. 1986;6:1712–1718. doi: 10.1523/JNEUROSCI.06-06-01712.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gros C, Souque A, Schwartz JC, Duchier J, Cournot A, Baumer P, Lecomte JM. Protection of atrial natriuretic factor against degradation: diuretic and natriuretic responses after in vivo inhibition of enkephalinase (EC 3.4.24.11). Proc Natl Acad Sci USA. 1989;86:7580–7584. doi: 10.1073/pnas.86.19.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gros C, Souque A, Schwartz JC. Inactivation of atrial natriuretic factor in mice in vivo: crucial role of enkephalinase (EC 3.4.24.11). Eur J Pharmacol. 1990;179:45–56. doi: 10.1016/0014-2999(90)90400-z. [DOI] [PubMed] [Google Scholar]
- 31.Hangauer DG, Monzigo AF, Matthews BW. An interactive computer graphics study of thermolysin-catalysed peptide cleavage and inhibition by N-carboxymethyl dipeptides. Biochemistry. 1984;23:5730–5741. doi: 10.1021/bi00319a011. [DOI] [PubMed] [Google Scholar]
- 32.Holmes MA, Matthews BW. Structure of thermolysin refined at 1.6 A resolution. J Mol Biol. 1982;160:623–639. doi: 10.1016/0022-2836(82)90319-9. [DOI] [PubMed] [Google Scholar]
- 33.Johnson AR, Coalson JJ, Ashton J, Larumbide M, Erdos EG. Neutral endopeptidase in serum samples of patients with adult respiratory distress syndrome. Comparison with angiotensin-converting enzyme. Am Rev Respir Dis. 1985;132:1262–1267. doi: 10.1164/arrd.1985.132.6.1262. [DOI] [PubMed] [Google Scholar]
- 34.Kandel ER. Behavioral biology of Aplysia. W. H. Freeman; San Francisco: 1979. [Google Scholar]
- 35.Kenny AJ. Endopeptidase-24.11: putative substrate and possible roles. Biochem Soc Trans. 1993;21:663–668. doi: 10.1042/bst0210663. [DOI] [PubMed] [Google Scholar]
- 36.Kenny AJ, Maroux S. Topology of microvillar membrane hydrolases of kidney and intestine. Physiol Rev. 1982;62:91–128. doi: 10.1152/physrev.1982.62.1.91. [DOI] [PubMed] [Google Scholar]
- 37.Kenny AJ, Stephenson SL, Turner AJ. Cell surface peptidases. In: Kenny AJ, Turner AJ, editors. Mammalian ectoenzymes. Elsevier Science Publishers; London: 1987. pp. 169–210. [Google Scholar]
- 38.Kim YA, Shriver B, Quay T, Hersh LB. Analysis of the importance of arginine 102 in neutral endopeptidase (enkephalinase) catalysis. J Biol Chem. 1992;267:12330–12335. [PubMed] [Google Scholar]
- 39.Kreiner T, Kirk MD, Scheller RH. Cellular and synaptic morphology of a feeding motor circuit in Aplysia. J Comp Neurol. 1987;264:311–325. doi: 10.1002/cne.902640304. [DOI] [PubMed] [Google Scholar]
- 40.Kupfermann I. Feeding behavior in Aplysia: a simple system for the study of motivation. Behav Biol. 1974;10:1–26. doi: 10.1016/s0091-6773(74)91644-7. [DOI] [PubMed] [Google Scholar]
- 41.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 42.Lee S, Zambas E, Marsh WL, Redman CM. Molecular cloning and primary structure of Kell blood group protein. Proc Natl Acad Sci USA. 1991;88:6353–6357. doi: 10.1073/pnas.88.14.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Moual H, Devault A, Roques BP, Crine P, Boileau G. Identification of glutamic acid 646 as a zinc-coordinating residue in endopeptidase-24.11. J Biol Chem. 1991;266:15670–15674. [PubMed] [Google Scholar]
- 44.Le Moual H, Roques BP, Crine P, Boileau G. Substitution of potential metal-coordinating amino acid residues in the zinc-binding site of endopeptidase 24.11. FEBS Lett. 1993;324:196–200. doi: 10.1016/0014-5793(93)81392-d. [DOI] [PubMed] [Google Scholar]
- 45.Le Moual H, Dion N, Roques BP, Crine P, Boileau G. Asp650 is crucial for catalytic activity of neutral endopeptidase 24.11. Eur J Biochem. 1994;221:475–480. doi: 10.1111/j.1432-1033.1994.tb18760.x. [DOI] [PubMed] [Google Scholar]
- 46.Lloyd PE. Fast axonal transport of modulatory neuropeptides from central ganglia to components of the feeding system in Aplysia. J Neurosci. 1988;8:3507–3514. doi: 10.1523/JNEUROSCI.08-09-03507.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lloyd PE, Frankfurt M, Stevens P, Kupfermann I, Weiss KR. Biochemical and immunocytological localization of the neuropeptides FMRFamide, SCPA, SCPB, to neurons involved in the regulation of feeding in Aplysia. J Neurosci. 1987;7:1123–1132. doi: 10.1523/JNEUROSCI.07-04-01123.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marie-Claire C, Ruffet E, Antonczak S, Beaumont A, O’Donohue M, Roques BP, Fournié-Zaluski MC. Evidence by site-directed mutagenesis that arginine 203 of thermolysin and arginine 717 of neprilysin (neutral endopeptidase) play equivalent critical roles in substrate hydrolysis and inhibitor binding. Biochemistry. 1997;36:13938–13945. doi: 10.1021/bi9712495. [DOI] [PubMed] [Google Scholar]
- 49.Maroux S. Structural and topological aspects. In: Kenny AJ, Turner AJ, editors. Mammalian ectoenzymes. Elsevier Science Publishers; London: 1987. pp. 15–45. [Google Scholar]
- 50.Martin KC, Casadio A, Huixiang Z, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse specific long-term facilitation of Aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 51.Martins MA, Shore SA, Gerard NP, Gerard C, Drazen JM. Peptidase modulation of the pulmonary effects of tachykinins in tracheal superfused guinea pig lungs. J Clin Invest. 1990;85:170–176. doi: 10.1172/JCI114408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKelvy JF, Blumberg S. Inactivation and metabolism of neuropeptides. Annu Rev Neurosci. 1986;9:415–434. doi: 10.1146/annurev.ne.09.030186.002215. [DOI] [PubMed] [Google Scholar]
- 53.Miller MW, Alevizos A, Cropper EC, Kupfermann I, Weiss KR. Distribution of buccalin-like immunoreactivity in the central nervous system and peripheral tissues of Aplysia californica. J Comp Neurol. 1992;320:182–195. doi: 10.1002/cne.903200204. [DOI] [PubMed] [Google Scholar]
- 54.Miller MW, Beuchausen S, Cropper EC, Eisenger K, Stamm S, Vilim FS, Vitek A, Zajc A, Kupfermann I, Brosius J, Weiss KR. The buccalin-related neuropeptides: isolation and characterization of an Aplysia cDNA clone encoding a family of peptides cotransmitters. J Neurosci. 1993a;13:3346–3357. doi: 10.1523/JNEUROSCI.13-08-03346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller MW, Beuchausen S, Vitek A, Stamm S, Kupfermann I, Brosius J, Weiss KR. The myomodulin-related neuropeptides: characterization of a gene encoding a family of peptides cotransmitters in Aplysia. J Neurosci. 1993b;13:3358–3367. doi: 10.1523/JNEUROSCI.13-08-03358.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller MW, Rosen SC, Schissel SL, Cropper EC, Kupfermann I, Weiss WR. A population of SCP-containing neurons in the buccal ganglion of Aplysia are radula mechanoafferents and receive excitation of central origin (1994). J Neurosci. 1994;14:7008–7023. doi: 10.1523/JNEUROSCI.14-11-07008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Owens DF, Menon JG, Rothman BS. Structure-activity relationship of neurotransmitter alpha-bag cell peptide on Aplysia LUQ neurons: implications regarding its inactivation in the extracellular space. J Neurobiol. 1992;23:656–670. doi: 10.1002/neu.480230605. [DOI] [PubMed] [Google Scholar]
- 58.Panoskaltsis-Mortari A, Bucy RP. In situ hybridization with digoxigenin-labeled RNA probes: facts and artifacts. Biotechniques. 1995;18:300–307. [PubMed] [Google Scholar]
- 59.Roderick SL, Fournié-Zaluski MC, Roques BP, Matthews BW. Thiorphan and retro-thiorphan display equivalent interactions when bound to crystalline thermolysin. Biochemistry. 1989;28:1493–1497. doi: 10.1021/bi00430a011. [DOI] [PubMed] [Google Scholar]
- 60.Roques BP, Fournié-Zaluski MC, Soroca E, Lecomte JM, Malfroy B, Llorens C, Schwartz JC. The enkephalinase inhibitor thiorphan shows antinociceptive activity in mice. Nature. 1980;288:286–288. doi: 10.1038/288286a0. [DOI] [PubMed] [Google Scholar]
- 61.Roques BP, Noble F, Daugé V, Fournié-Zaluski MC, Beaumont A. Neutral endopeptidase 24.11: structure, inhibition, and experimental and clinical pharmacology. Pharmacol Rev. 1993;45:87–146. [PubMed] [Google Scholar]
- 62.Rothman BS, Mayeri E, Scheller RH. The bag cell neurons of Aplysia as a possible peptidergic multitransmitter system: from genes to behavior. In: Zomzely-Neurath C, Walker WA, editors. Gene expression in brain. Wiley; New York: 1985. pp. 235–274. [Google Scholar]
- 63.Rothman BS, Dekruyff S, Talebian M, Menon JG, Squire CR, Yeh CH, Lee TD. Aplysia peptide neurotransmitters beta-bag cell peptide, Phe-Met-Arg-Phe-amide, and small cardioexcitatory peptide B are rapidly degraded by a leucine amino-like activity in hemolymph. J Biol Chem. 1992;267:25135–25140. [PubMed] [Google Scholar]
- 64.Ruchon AF, Marcinkiewicz M, Siegfried G, Tenenhouse HS, Desgroseillers L, Crine P, Boileau G. PEX mRNA is localized in developing mouse osteoblasts and odontoblasts. J Histochem Cytochem. 1998;48:459–468. doi: 10.1177/002215549804600405. [DOI] [PubMed] [Google Scholar]
- 65.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 66.Saleh TM, Kombian SB, Zidichouski JA, Pittman QJ. Peptidergic modulation of synaptic transmission in the parabrachial nucleus in vitro: importance of degradative enzymes in regulating synaptic efficacy. J Neurosci. 1996;16:6046–6055. doi: 10.1523/JNEUROSCI.16-19-06046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seymour AA, Abboa-Offei BE, Smith PL, Mathers PD, Asaad MM, Rogers WL. Potentiation of natriuretic peptides by neutral endopeptidase inhibitors. Clin Exp Pharmacol Physiol. 1995;22:63–69. doi: 10.1111/j.1440-1681.1995.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 69.Shipp MA, Stephano GB, D’Adamio L, Switzer SN, Howard FD, Sinisterra J, Scharrer B, Reinhertz EL. Downregulation of enkephalin-mediated inflammatory responses by CD10/neutral endopeptidase 24.11. Nature. 1990;347:394–396. doi: 10.1038/347394a0. [DOI] [PubMed] [Google Scholar]
- 70.Shipp MA, Stephano GB, Switzer SN, Griffin JD, Reinhertz EL. CD10 (CALLA)/neutral endopeptidase 24.11 modulates inflammatory peptide-induced changes in neutrophil morphology, migration, and adhesion proteins and is itself regulated by neutrophil activation. Blood. 1991;78:1834–1841. [PubMed] [Google Scholar]
- 71.Soleilhac JM, Lafuma C, Porcher JM, Auburtin G, Roques BP. Characterization of a soluble form of neutral endopeptidase-24.11 (EC 3.4.24.11) in human serum: enhancement of its activity in serum of underground miners exposed to coal dust particles. Eur J Clin Invest. 1996;26:1011–1017. doi: 10.1046/j.1365-2362.1996.2420580.x. [DOI] [PubMed] [Google Scholar]
- 72.Sossin WS, Kirk MD, Scheller RH. Peptidergic modulation of neuronal circuitry controlling feeding in Aplysia. J Neurosci. 1987;7:671–681. doi: 10.1523/JNEUROSCI.07-03-00671.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Squire CR, Talebian M, Menon JG, Dekruyff S, Lee TD, Shively JE, Rothman BS. Leucine aminopeptidase-like activity in Aplysia hemolymph rapidly degrades biologically active alpha-bag cell peptide fragments. J Biol Chem. 1991;266:22355–22363. [PubMed] [Google Scholar]
- 74.Steward O. mRNA localization in neurons: a multipurpose mechanism? Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- 75.Tam LT, Engelbrecht S, Talent JM, Gracy RW, Erdos EG. The importance of disulfide bridges in human endopeptidase (enkephalinase) after proteolytic cleavage. Biochem Biophys Res Commun. 1985;133:1187–1198. doi: 10.1016/0006-291x(85)91262-8. [DOI] [PubMed] [Google Scholar]
- 76.The HYP consortium. A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. Nat Genet. 1995;11:130–135. doi: 10.1038/ng1095-130. [DOI] [PubMed] [Google Scholar]
- 77.Thompson EB, Schwartz JH, Kandel ER. A radioautographic analysis in the light and electron microscope of identified Aplysia neurons and their processes after intrasomatic injection of L-(3H)fucose. Brain Res. 1976;112:251–281. doi: 10.1016/0006-8993(76)90283-3. [DOI] [PubMed] [Google Scholar]
- 78.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thompson JS, Morice AH. Neutral endopeptidase inhibitors and the pulmonary circulation. Gen Pharmacol. 1996;27:581–585. doi: 10.1016/0306-3623(95)02051-9. [DOI] [PubMed] [Google Scholar]
- 80.Turner AJ. Processing and metabolism of neuropeptides. Essays Biochem. 1986;22:69–119. [PubMed] [Google Scholar]
- 81.Turner AJ, Murphy LJ. Molecular pharmacology of endothelin converting enzymes. Biochem Pharmacol. 1996;51:91–102. doi: 10.1016/0006-2952(95)02036-5. [DOI] [PubMed] [Google Scholar]
- 82.Turner AJ, Tanzawa K. Mammalian membrane metallopeptidases: NEP, ECE, KELL, and PEX. FASEB J. 1997;11:355–364. doi: 10.1096/fasebj.11.5.9141502. [DOI] [PubMed] [Google Scholar]
- 83.Turner AJ, Hooper NM, Kenny AJ. Metabolism of neuropeptides. In: Kenny AJ, Turner AJ, editors. Mammalian ectoenzymes. Elsevier Science Publishers; London: 1987. pp. 211–248. [Google Scholar]
- 84.Van Minnen J. RNA in the axonal domain: a new dimension in neuronal functioning? Histochem J. 1994;26:377–391. doi: 10.1007/BF00160050. [DOI] [PubMed] [Google Scholar]
- 85.Vijayaraghavan J, Scicli AG, Carretero OA, Slaughter C, Moomaw C, Hersh LB. The hydrolysis of endothelins by neutral endopeptidase 24.11 (enkephalinase). J Biol Chem. 1990;265:14150–14155. [PubMed] [Google Scholar]
- 86.Weaver LH, Kester WR, Matthews BW. A crystallographic study of the complex of phosphoramidon with thermolysin. A model for the presumed catalytic transition state and for the binding of extended substances. J Mol Biol. 1977;114:119–132. doi: 10.1016/0022-2836(77)90286-8. [DOI] [PubMed] [Google Scholar]
- 87.Webb DJ, Monge JC, Rabelink TJ, Yanagisawa M. Endothelin: new discoveries and rapid progress in the clinic. Trends Pharmacol Sci. 1997;19:5–8. doi: 10.1016/s0165-6147(97)01144-9. [DOI] [PubMed] [Google Scholar]
- 88.Weiss KR, Chiel HJ, Koch U, Kupfermann I. Activity of an identified histaminergic neuron, and its possible role in arousal of feeding behavior in semi-intact Aplysia. J Neurosci. 1986;6:2403–2415. doi: 10.1523/JNEUROSCI.06-08-02403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wickham L, DesGroseillers L. A bradykinin-like neuropeptide precursor gene is expressed in neuron L5 of Aplysia californica. DNA Cell Biol. 1991;10:249–258. doi: 10.1089/dna.1991.10.249. [DOI] [PubMed] [Google Scholar]
- 90.Wilhelm JE, Vale RD. RNA on the move: the mRNA localization pathway. J Cell Biol. 1993;2:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]