Abstract
During cortical development, embryonic neurons migrate from germinal zones near the ventricle into the cortical plate, where they organize into layers. Mechanisms that direct neuronal migration may include molecules that act as chemoattractants. In rats, GABA, which localizes near the target destination for migrating cortical neurons, stimulates embryonic neuronal migration in vitro. In mice, glutamate is highly localized near the target destinations for migrating cortical neurons. Glutamate-induced migration of murine embryonic cortical cells was evaluated in cell dissociates and cortical slice cultures. In dissociates, the chemotropic effects of glutamate were 10-fold greater than the effects of GABA, demonstrating that for murine cortical cells, glutamate is a more potent chemoattractant than GABA. Thus, cortical chemoattractants appear to differ between species. Micromolar glutamate stimulated neuronal chemotaxis that was mimicked by μm NMDA but not by other ionotropic glutamate receptor agonists (AMPA, kainate, quisqualate). Responding cells were primarily derived from immature cortical regions [ventricular zone (vz)/subventricular zone (svz)]. Bromodeoxyuridine (BrdU) pulse labeling of cortical slices cultured in NMDA antagonists (μm MK801 or APV) revealed that antagonist exposure blocked the migration of BrdU-positive cells from the vz/svz into the cortical plate. PCR confirmed the presence of NMDA receptor expression in vz/svz cells, whereas electrophysiology and Ca2+ imaging demonstrated that vz/svz cells exhibited physiological responses to NMDA. These studies indicate that, in mice, glutamate may serve as a chemoattractant for neurons in the developing cortex, signaling cells to migrate into the cortical plate via NMDA receptor activation.
Keywords: mouse, chemotaxis, cortex, development, slice, chemoattractant
During CNS development, cortical neurons are generated in specialized germinal zones adjacent to the ventricle, termed the neuroepithelium. After terminal mitosis, neurons migrate from the neuroepithelium through the cell-sparse intermediate zone (iz) into the cortical plate (cp). After entering the cp, neuroblasts migrate through the established neuronal lamina and settle onto the outermost layer, forming an “inside-out” gradient of maturation, a process which is essential to cortical neuronal lamination (Jacobson, 1991).
As neurons travel from the germinal zones, many of them move along the fibers of radial glial cells (Rakic, 1972, 1990; Hatten, 1990), which extend from the mitotic zone to the pial surface. Some neurons also migrate along alternative pathways. Neurons in embryonic slice preparations have been observed to move orthogonally, along nonradial pathways that are perpendicular to the axis of glial processes (O’Rourke et al., 1992, 1995). This suggests that, during development, mechanisms other than direct contact with radial glial fibers may also influence nerve cell movement and thus contribute to the structural creation of the cortex.
Extracellular factors may provide directional cues to migrating neuroblasts. In the developing spinal cord, embryonic motoneurons transplanted into the dorsal half of the cord migrate back to the ventral horns, suggesting that locally mediated factors in the target region provide positional cues to the cells (Eisen, 1991). The positional cues may result from gradients of diffusible molecules, which act as chemoattractants to direct the cells toward their target destinations. Molecules present in the microenvironment of migrating nerve cells in vivo modulate the movement of the cellsin vitro. Among these, platelet-derived growth factor, brain-derived neurotrophic factor, and GABA promote movement of embryonic cells acutely dissociated from the developing rat CNS (Armstrong et al., 1990; Behar et al., 1994, 1996). Komuro and Rakic (1993) reported that granule cell migration in postnatal rat cerebellar slice preparations was inhibited by antagonists of ionotropic glutamate receptors, suggesting that glutamate provides motility signals to migratory neurons. Glutamate is also a likely candidate for a chemoattractant in the developing mouse cerebral cortex. The Cajal-Retzius (C-R) cells, which are generated first and are located in layer 1 of the cp, contain glutamate during the final week of gestation (del Rio et al., 1995), when neurons are actively migrating into this region (Jacobson, 1991). Thus, glutamate is found in an appropriate location to serve as a chemoattractant.
We used an in vitro chemotaxis assay (Falk et al., 1980) to examine whether glutamate influences embryonic cortical nerve cell movement. The assay was used to measure the effects of glutamate on the migration of acutely dissociated murine embryonic cortical cells. In addition, we examined the effects of glutamate on neuronal migration using organotypic slice preparations. Modulation of cortical neuroblast migration with glutamate receptor agonists and antagonists suggests that receptors for NMDA (NMDA-Rs) mediate the motility signals of glutamate in a subset of cortical neurons.
MATERIALS AND METHODS
Dissection and dissociation of the cells
Cortical cells from C57BL/6 pups at embryonic days 13–18 (E13–E18) were used in these experiments. Dams were killed with CO2, embryos were removed by cesarean section, and embryonic age was determined by measurement of crown-rump length (Schambra et al., 1992). Brains from littermates were removed and placed into Earle’s balanced salt solution (EBSS) (Life Technologies, Grand Island, NY). After removing the meninges, the cerebral cortices were dissected, minced, and dissociated into single cell suspensions using a papain digest described previously (Behar et al., 1994). Dissociated cells were resuspended in EBSS supplemented with 16 mm glucose at a concentration of 106 cells/ml for the migration studies. For quantitative analysis using immunocytochemistry, 2.5 × 105 cells were seeded onto 35 mm Nunc (Naperville, IL) culture dishes precoated with poly-d-lysine [>450,000 molecular weight (MW), 20 μg/ml] (Collaborative Research, Bedford, MA) and allowed to adhere onto the dishes for 1 hr at 37°C.
Preparation of ventricular zone and cp cell suspensions.Detailed description of the separation of the two brain regions was described previously (Behar et al., 1998). Briefly, brains from littermates were removed. A McIlwaine tissue chopper (Brinkman, Westbury, NY) was used to prepare 350 μm coronal slices of the telencephalon. Slices were transferred to a dish and were microdissected through the cortical intermediate zone, which divided the cortex into two tissue segments designated as the cp and the ventricular zone (vz). Tissue designated “cp” included the cp, subplate (sp), and the upper half of the iz. Tissue designated “vz” included the vz, svz, and the lower half of the iz. Tissue segments from each region were enzymatically dissociated into single cell suspensions with papain, and the cells were either immunostained or used in the microchemotaxis assay.
Quantitative analysis of TUJ1 and nestin staining in the vz and cp dissociates. Acutely adherent cells on culture plates (dissociated from the vz and cp preparations) were fixed 30 min in 4% paraformaldehyde (PF) and washed in PBS, pH 7.2. Cells were incubated at 4°C overnight in a mixture of rabbit anti-nestin antibody (a progenitor cell marker; Tohyama et al., 1992) and the monoclonal TUJ1 antibody (a neuronal marker; Lee et al., 1990). The cells were washed three times in PBS and then incubated 1 hr at 21°C in appropriate secondary antibodies [donkey anti-rabbit FITC and rat anti-mouse tetramethylrhodamine isothiocyanate (TRITC); 1:50; Jackson ImmunoResearch, West Grove, PA]. PBS supplemented with 0.05% saponin and 0.1% BSA was used for the antibody diluent. Immunolabeled cells were examined on a Zeiss (Thornwood, NY) Axiophot microscope equipped with epifluorescence and the appropriate filters for the coincident visualization of fluoresceine and rhodamine. The percentage of positively stained cells from both cp and vz regions was determined by dividing the number of fluorescently labeled cells in a field by the total number of cells in the same field (visualized under phase contrast using a water immersion 40× objective). Ten random fields were counted per dish (1000 cells per dish) for duplicate samples from seven separate experiments. Data were subjected to ANOVA to determine variation within groups.
Chemotaxis assay
Migration of the acutely dissociated cells was measured using a microchemotaxis assay described previously (Behar et al., 1994). Before assembly, the filter was coated with poly-d-lysine (>450,000 MW; 30 μg/ml; Collaborative Research) to promote adhesion.
Cells were incubated in the presence of attractants for 18 hr at 37°C with 10% CO2 and 90% air. After the incubation, the chambers were disassembled, and the cells on the upper surface of the filter that failed to migrate were scraped off. Migrated cells on the underside of the membrane were fixed, stained, and counted as described below.
Quantification of migration. For quantitative assays, migrated cells were fixed 30 min in 4% PF with 0.1% glutaraldehyde in 0.1 m phosphate buffer, pH 7.2, and stained 5 min in 0.1% cresyl violet. The membranes were mounted cell side up onto 2 × 3 inch glass slides, allowed to air dry, and covered with immersion oil. Cells were counted under bright field using oil immersion 16× or 40×Zeiss planapo objectives on a Zeiss photomicroscope.
Only cells that passed completely through the pores and migrated out onto the underside surface of the membrane were considered migrated (positive) cells. Spontaneous (random) migration of cells in control wells containing buffer only was also assessed in all assays. Each attractant condition was run in triplicate wells, and 5–10 fields of stained cells were counted for each well. The average number of migrated cells per square millimeter for each attractant condition was calculated, and the statistical significance was analyzed using ANOVA followed by Fisher’s PLSD test. Illustrations of data pertaining to the chemotaxis assay represent individual trials unless noted.
Chemotaxis (the directed migration of cells toward regions of higher concentrations of a chemical attractant) was distinguished from chemokinesis (stimulation of increased random cell motility) by placing the same concentration of attractant in both the upper and lower wells of the chamber, thereby eliminating a chemical gradient. The number of cells migrating under these conditions was considered a measure of chemokinesis.
Characterization of migrated cells by immunocytochemistry
In some assays, migrated cells on the underside of the filter were fixed, stained with cresyl violet, and then analyzed for expression of neurofilament (NF) using a monoclonal anti-NF tissue culture supernatant (1:8; gift of Dr. C. Gibbs, National Institute of Neurological Diseases and Stroke, National Institutes of Health, Bethesda, MD). The cells were washed three times and incubated in peroxidase-conjugated secondary antisera (goat anti-mouse IgM; 1:40; Jackson ImmunoResearch) for 1 hr at room temperature. Immunoreaction product was visualized using a diaminobenzidine (DAB) substrate. Cresyl violet staining was used to determine the number of total cells, whereas NF was used to indicate neurons only. Cells were counted under bright field using a 40× planapo objective.
Viability of cells in the presence of inhibitors. The starting population of cells, suspended in EBSS with 16 mmglucose, was seeded onto 35 mm Nunc culture dishes precoated with poly-d-lysine (>450,000 MW, 20 μg/ml; Collaborative Research) at 2.5 × 105 cells per dish. Cells were allowed to adhere onto the dishes for 1 hr at 37°C and then were fed with either control medium (EBSS with 16 mm glucose) or EBSS with glucose containing attractant (1 μm glutamate or NMDA) only or attractant plus inhibitor [10 μg/ml BAPTA-AM, 10 μm (+)-5-methyl-10,11-dihydro-5H-dibenzo [a,d] cyclohepten-5,10-imine maleate (MK801), or 10 μmR(−)-2-amino-5-phosphonovaleric acid (APV)]. Cells were incubated in the presence of the ligands for 18 hr at 37°C, and then the viability of the cells was determined using trypan blue exclusion. In control media (EBSS plus glucose), cell viability at 18 hr was 94.3 ± 0.4%. Viability of cells maintained in 1 μm glutamate was 96.2 ± 3.3%, whereas the viability of cells incubated in NMDA was 95.7 ± 1.2%. In the presence of each inhibitor, the viability equaled or surpassed that of the controls (BAPTA-AM, 98.5 ± 0.2%; MK801, 94.1 ± 2.2%; APV, 98.7 ± 0.9%).
Culture of embryonic cortical slices
E17 brains from littermates were removed and placed into cold (4°C) slicing medium (in mm: 120 NaCl, 5 KCl, 1.2 KH2PO4, 14 dextrose, 26 NaHCO3, and 1.24 MgSO47H2O plus 5 mg/ml phenol red), and 300 μm coronal slices of the telencephalon were prepared using a vibratome. Slices were transferred onto Millipore (Bedford, MA) filters and placed into six-well Costar (Cambridge, MA) plates containing ice-cold slicing medium. The slices were allowed to recover for 2 hr at 4°C and were then transferred on the filters to plates containing growth medium (Neuralbasal medium supplemented with B27 and glutamine; Life Technologies). At the beginning of the incubation period, the growth medium was supplemented with 50 μg/ml of bromodeoxyuridine (BrdU). After 18 hr, BrdU-containing medium was replaced with growth medium lacking BrdU. To control for anatomical gradients of cortical maturation, as well as possible differences in cortical development among embryos of the same litter, slices from one cortical hemisphere cultured under control conditions was compared with slices of the contralateral hemisphere cut from the same embryo and maintained in media containing antagonists at NMDA receptors (100 μm MK801 or APV). Antagonists were added at high micromolar levels to ensure that the ligands penetrated through the thick sections. Slices were cultured at 37°C in a mixture of 10% CO2 and 90% air, and medium was replaced every 3 d.
BrdU immunolabeling. Two or 6 d after removing the BrdU from the cultures, the slices were fixed for 1 hr in 4% PF and then permeabilized for 1 hr in 0.5% Triton X-100. After rinsing in PBS, the slices were first incubated for 10 min in cold (4°C) 0.1N HCl and then incubated for 25 min at 37°C in 1N HCl. The HCl was removed and the pH of the slices was neutralized by adding 0.1 mTris-HCl buffer, pH 8.0. The slices were then rinsed two times in PBS and incubated overnight at 4°C in the anti-BrdU antibody (diluted 1:33 in PBS) (Boehringer Mannheim, Indianapolis, IN). Immunolabeling for BrdU was visualized using the Elite Vectastain ABC kit (Vector Laboratories, Burlingame, CA) and DAB. After immunostaining, slices were briefly (30 sec) counterstained with 0.05% cresyl violet for visualization of cytoarchitectural features. Slices were examined on aZeiss Axiophot microscope, and the density of BrdU-labeled cells, cortical plate thickness, and total cp cells per unit area were measured using NIH Image analysis software. Control and experimental data were taken from corresponding anatomical positions. In every case, the measurement was an experimental value normalized to control and expressed as a percent. A total of 30 measurements from six coronal slices from three different littermates were collected and averaged for each trial. Illustrations present the averages (± SEM) from five trials. Image analysis data were analyzed using Student’s ttest.
Density of BrdU-labeled cells. Using the NIH Image software, the pixel density of BrdU-labeled nuclei was measured within five fixed areas of the cortical plate in the control. The average pixel density per unit area in the control slice was compared with similar measurements on pixel density of labeled nuclei in the same fixed area and position on the treated contralateral slice. Each capture was exposed to the same analysis subroutine and density slice thresholding.
Cortical plate thickness. Using the NIH Image software, the perpendicular linear distance (in pixels) was measured from the innermost edge of cortical layer VI to the outermost edge of cortical layer I. This distance was assessed in four places in a control slice and compared with values measured in matched positions on the contralateral treated slice.
Packing cell density in the cp. The average number of cells per unit area within the cortical plate of control slices was compared with the average number of cells per the same unit area in contralateral treated slices.
Semiquantitative PCR
A hot-start reverse transcription (RT)-PCR protocol was used to measure gene expression. Gene-specific primers were designed from GenBank sequences using the Oligo software (National Biosciences, Plymouth, MA). The sequences were as follows: NR1f, ATAGTGACAATCCACCAAGAGCC; NR1r, GTAGCTCGCCCATCATTCCGTT; NMDA2Af, CTCCAATATGTCCAACATGAACTCC; NMDA2Ar, GTCAACATCGCTACAGTCCTTGGG; NMDA2Bf, TCCGCCGAGAGTCCTCCGT; NMDA2Br, CTGCGTTGCCCTCGATGTT; NMDA2Df, GAACAGCAGACCCTCCGCC; NMDA2Dr, ACCCTTGCAGCATCTCTTCTCC. Total RNA was isolated from cortices of E17–E18 mice. RNAstat 60 (Tel-Test, Inc., Friendswood, TX) and its recommended protocol was used for the RNA isolation. To confirm purity of the product RNA, absorption ratios at 260 nm/280 nm were determined to be >1.8 for all samples. The samples were adjusted to 200 ng/μl for reverse transcription and PCR according to absorption at 260 nm. PCR [Perkin-Elmer (Norwalk, CT) GeneAmp RNA PCR kit; Applied Biosystems, Foster City, CA] involved preheating a mixture of Taq antibody (TaqStart; Clontech, Palo Alto, CA), primers, cDNA, and PCR components to 97°C for 90 sec before amplification. PCR cycle was 30 sec at 95°C (dissociation), 45 sec at 60°C (annealing), and 60 sec at 72°C (extension). Amplification was within the exponential range. Control RNA (transcribed from PAW 108 plasmid DNA; PAW 108 forward and reverse primers; Applied Biosystems) in all reverse transcription and PCR amplification reactions allowed for a ratiometric method for determining gene expression levels and permitted detection of inefficient PCR reactions.
To control for artifacts caused by DNA contamination, in some studies, RT-PCR was performed without reverse transcriptase. In the absence of reverse transcriptase, no bands were visualized. In addition, RNA samples in some studies were verified to be free of DNA contaminants as described previously (Somogyi et al., 1995).
Numbers were obtained by calculating the densitometry (NIH Image) of PCR products separated on polyacrylamide gradient gels. The ratios of sample bands to corresponding control bands was used as the measurement of gene expression. For every gene, ratiometric data for each group of triplicate reactions were averaged. PCR product identities were confirmed by restriction enzyme digestion. The NR2A cDNA was cleaved with AvrII enzyme, generating fragments of 435 and 160; NR2B cDNA was cleaved with PstI, generating fragments of 149 and 310; NR1 cDNA was cleaved with ApaLI, generating fragments of 230 and 89; and NR2D cDNA was cleaved with EarI, generating a fragment of 228.
Viability of cells in the cultured slices. The viability of cells in the slice cultures was monitored using propidium iodide (PI), which intercalates into double-stranded DNA and fluoresces a brilliant orange using the FITC filters. Cultured slices were transferred from the filters onto glass microscope slides and were completely covered with a drop of PI (50 μg/ml; Sigma, St. Louis, MO). After a 5 min incubation at room temperature, the sections were coverslipped and examined immediately on a Zeiss Axiophot microscope, equipped with epifluorescence and the appropriate filters for the visualization fluoresceine. To induce cell death in the slices, one set of sections was incubated at room temperature in ambient air for 4 hr and then was transferred to glass slides, overlayed with the propidium iodide, and examined on the microscope. Slices cultured for 3 d and acutely labeled with PI had very few cells with labeled nuclei; however, a few PI-positive (PI+) cells were observed scattered along the outermost edges of the slices. No difference in cell viability was noted between the antagonist-treated and the control slices. To ensure that the PI was able to penetrate through the slice, one set of slice cultures was allowed to stand in ambient air for 4 hr at room temperature and then was exposed to PI. The slices incubated at room temperature had an abundance of PI+ cells distributed throughout the cerebral wall.
Ca2+ recording of vz cells by digital videomicroscopy
Unless otherwise stated, cell recordings were performed in normal physiological medium (NPM) composed of the following reagents (in mm): 145 NaCl, 5 KC1, 1.8 CaCl2, 0.8 MgCl2, 10 glucose, and 10 HEPES, pH 7.3 (osmolarity, 290 mOsm). Ca2+-free recording media contained (in mm): 145 NaCl, 5 KCl, 0.8 MgCl2, 5 EGTA-NaOH, 10 glucose, and 10 HEPES, where Ca2+ was estimated to be <10 nm. NMDA and APV were obtained from Research Biochemicals (Natick, MA). After dissociation, vz cells were washed in NPM and plated at a density of 3 × 104 cells/cm2 on poly-d-lysine coated, photo-etched gridded coverslips (Bellco Glass Inc., Vineland, NJ) preglued to 35 mm tissue culture dishes (MatTek Corp., Ashland, MA). After being allowed to adhere for 1 hr at 37°C, the cells were loaded with 2 μm fura-2 AM (Molecular Probes, Eugene, OR) for 1 hr at 37°C. At the end of the incubation period, the cells were rinsed in NPM.
Fura-2-loaded cells were recorded using the Zeiss Attofluor Ratio-Vision work station (Atto Instruments, Rockville, MD) equipped with an Axiovert 135 inverted microscope (Zeiss, Thornwood, NY) and an ICCD camera (Atto Instruments). The fura-2 dye was sequentially excited at 500 msec intervals with a 100 W mercury arc lamp filtered at 334 ± 5 and 380 ± 5 nm and the respective emissions acquired through a 510 nm dichroic mirror and 520 nm long-pass filter set. All filters were obtained from Chroma Technology Corp. (Brattleboro, VT). To collect the fura-2 fluorescence data, either square- or polygonal-shaped regions of interest (ROI) were electronically drawn around each of up to 69 cells per recording field. The fluorescence intensities from each ROI were digitized with a Matrox Graphics (Dorval, Quebec, Canada) image processing board and plotted as line graphs using Attograph for Windows analysis software (Atto Instruments).
Fura-2 fluorescence emissions were converted into estimated [Ca2+]c concentrations using the following equation:
where KD is the fura-Ca2+ binding constant (225 nm),R is a ratio of fura-2 fluorescence at 334 and 380 nm,Rmin and Rmax are values of R in Ca2+-free and normal [Ca2+]o medium, respectively, using fura-2 Penta K+ salt (Molecular Probes) as a Ca2+ indicator, andF0/F∞ is the ratio of fura-2 fluorescence at 380 nm in Ca2+-free and 1.8 mm [Ca2+]o medium. The data were calibrated on-line using the Attofluor RatioVision acquisition software (Atto Instruments).
Cell responses to NMDA were examined by exposing the cells to 10 μm NMDA (Research Biochemicals) in the presence or absence of 100 μm APV. The source of Ca2+ contribution to [Ca2+]c was evaluated by exposing the cells to NMDA in Ca2+-free recording media. All media were delivered to the cells using gravity-driven perfusion at flow rates of ∼2 ml/min. All measurements were performed at 37°C.
Electrophysiology
The current responses to NMDA were examined in cells of the vz preparation using electrophysiology. All recordings were performed at room temperature (22–25°C) on a Nikon (Tokyo, Japan) inverted microscope. Before recording, dishes were removed from the incubator, and the culture medium was completely replaced with Tyrode’s solution containing (in mm): 145 NaCl, 5.4 KCl, 1.8 CaCl2, 0.8 MgCl2, 10 glucose, and 10 HEPES-NaOH, pH 7.4 (310 mOsm). Standard patch-clamp recordings (Hamill et al., 1981) were made with pipettes pulled in three stages from 1.5 mm outer diameter glass capillary tubes (World Precision Instruments, Sarasota, FL) with a computer-controlled pipette puller (BB-CH-PC; Mecanex SA, Nyon, Switzerland). These pipettes had a resistance of 3–5 MΩ when filled with internal solution composed of (in mm): 145 CsCl, 2 MgCl2, 0.1 CaCl2, 1.1 EGTA, 5 HEPES, 5 ATP (potassium salt), and 5 phosphocreatine, pH 7.2, (290 mOsm). Whole-cell currents were recorded with a L/M EPC-7 patch-clamp amplifier (Medical Systems Corp., Greenvale, NY) at a gain of 5 mV/pA. Series resistance was compensated for more than 70%. Current signals were digitized with a Digidata 1200 (Axon Instruments, Foster City, CA) and acquired with Axoscope 7.0 (Axon Instruments) on a Pentium-based personal computer. Recorded cells were continuously superfused with a perfusion system composed of a locally made perfusion controller and miniature electric solenoid valves (The Lee Co., Essex, CT) that allows fast switching (<200 msec complete solution exchange time) among different solutions (Liu et al., 1999). The perfusion rate (0.3–0.5 ml/min) was controlled by the air pressure applied to the solution reservoirs. Significance of responses to 1 μm NMDA were evaluated using Student’st test to compare 8300 points recorded at the holding current with 8300 points recorded during exposure to 1 μmNMDA.
Immunostaining embryonic cortical sections
Brains from E13 or E16 mice were removed, fixed for 48 hr in 4% PF with 0.1% glutaraldehyde, and then equilibrated in 30% sucrose. Frozen coronal sections (12 μm) were prepared using a cryostat. Serial sections were incubated for 48 hr at 4°C in rabbit anti-glutamate antibody (1:100; Signature Biologicals, Salt Lake City, UT), rinsed in three changes of PBS, and then incubated 1 hr at room temperature in TRITC-conjugated donkey anti-rabbit IgG (1:50; Jackson ImmunoResearch). Sections were examined using 10×, 16×, and 25× objectives on an Axiophot microscope (Zeiss) equipped with epifluorescence and the appropriate filters for the visualization of rhodamine.
RESULTS
Glutamate is present in the embryonic murine cortex during the stages of neurogenesis and neuronal migration
Cryostat sections of embryonic cortex were immunostained with anti-glutamate antibodies at two embryonic ages to identify the distribution of glutamatergic cells and fibers during the prenatal period. At E13, high levels of glutamate were detected in the fibers and cells of the primordial plexiform layer (Fig.1A). At this stage, the neuroepithelium did not label with the anti-glutamate antibodies. By E16, glutamate-immunoreactive elements were identified in the outer half of the cortical plate (Fig. 1B), whereas the cells of the subplate were not immunopositive. Within layer I, fibers also immunolabeled with the anti-glutamate antibodies. Thus, at the ages examined, glutamate-immunoreactive structures were detected near the target destinations for migrating cortical neurons.
Fig. 1.
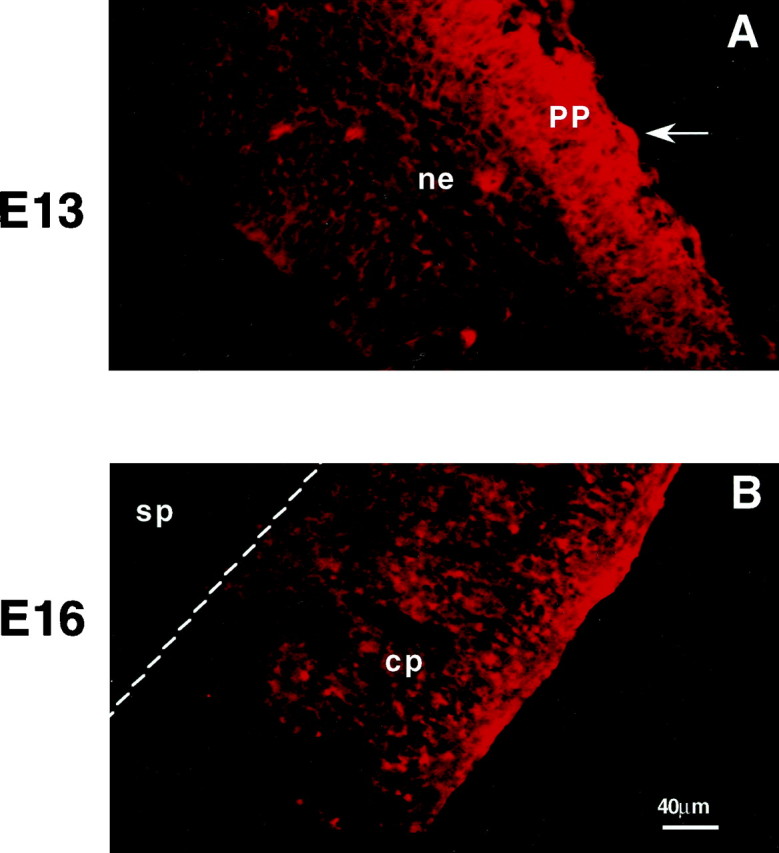
Glutamate immunoreactivity in the embryonic murine cortex. Photomicrographs of coronal sections of the embryonic cortex immunostained with anti-glutamate antisera.A, At E13, glutamate-immunoreactive cells (arrow) and processes are evident in the primordial plexiform layer (PP). B, By E16, fibers in the outer half of the cortical plate are highly immunoreactive for glutamate. ne, Neuroepithelium; cp, cortical plate; sp, subplate. Scale bar, 40 μm.
Glutamate stimulates migration of dissociated embryonic cortical cells in vitro
An in vitro microchemotaxis assay was used to analyze the effects of glutamate on the migration of cortical cells acutely dissociated from embryonic (E13–E18) C57BL mice. At E17, glutamate stimulated cell motility in a dose-dependent manner (Fig.2A). Glutamate concentrations ranging from 5 nm to 5 μmproduced levels of migration that were significantly higher than spontaneous random motility to buffer only (1–5 cells/mm2). Maximum migration to glutamate was seen at 500 nm. GABA also induced significant levels of migration compared with spontaneous values. However, fewer cells moved in response to GABA compared with glutamate, and the effective concentration range of GABA was more limited (Fig.2A). At optimal concentrations (500 nm), the number of cells migrating to glutamate was 10-fold greater than the number of cells responding to GABA. Approximately 5–20% of the starting population migrated to glutamate during the 18 hr assay, indicating that the amino acid stimulated motility in a subpopulation of cells. Significant migration to glutamate was observed in dissociates of each of the six embryonic ages analyzed; however, peak motility to glutamate occurred at E17–E18 (Fig.2B).
Fig. 2.
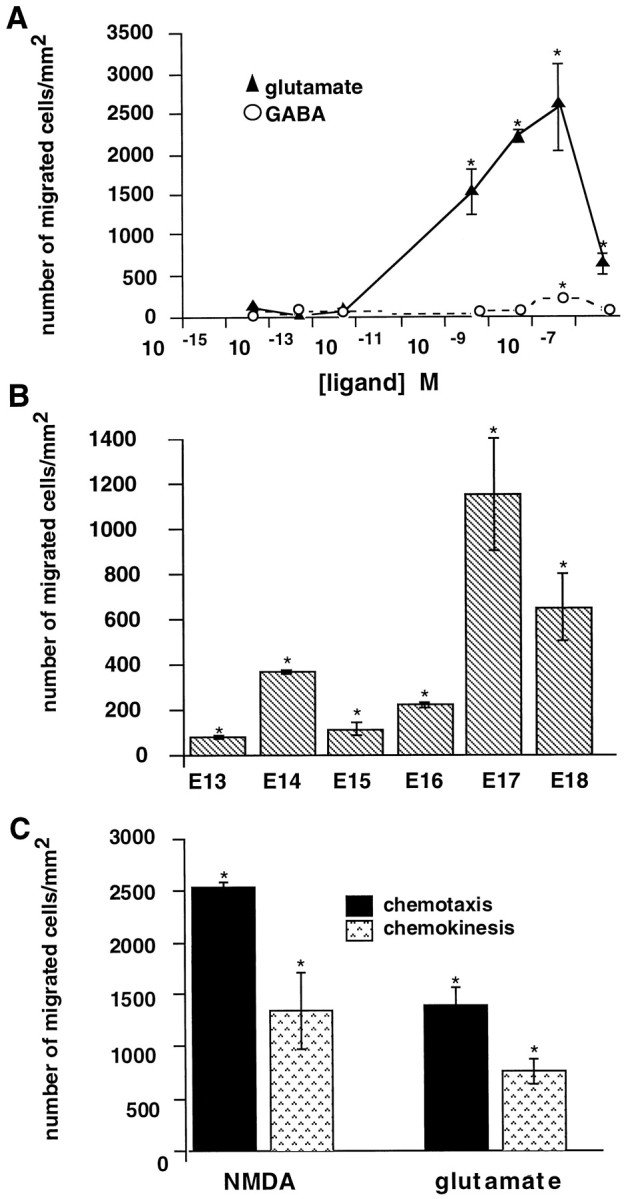
Glutamate is a more potent chemoattractant than GABA for dissociated murine embryonic cortical cells. A, Glutamate and GABA stimulate migration in a dose-dependent manner. At E17, distinctive ranges of glutamate (▴) or GABA (○) stimulate cells to migrate. Maximum numbers of migrating cells occur at 500 nm glutamate; however, significant migration (>50 cells/mm2) is observed at glutamate concentrations ranging between 5 nm and 5 μm. Nanomolar GABA also stimulates motility but at lower levels. At all effective concentrations, glutamate stimulates more migration than GABA.B, Response by age. Glutamate (1 μm) stimulates migration from E13 onward. Peak migratory responses to glutamate are observed at E17. C, Characterization of migration. Glutamate (1 μm) or NMDA stimulate both directed migration (chemotaxis) and random motility (chemokinesis). Approximately twice as many cells migrate in the presence of a chemical gradient (•) than in the absence of one (▧). Error bars indicate SEM. *p ≤ 0.01; ANOVA followed by Fisher’s PLSD test. Separate trials: A, Glutamate,n = 6; GABA, n = 4;B, E13, n = 3; E14,n = 3; E15, n = 3; E16,n = 3; E17, n = 6; E18,n = 3; C, n = 4.
We characterized the migratory response to glutamate in terms of the requirement for a chemical gradient across the membrane in the chemotaxis chamber. The number of cells undergoing gradient-dependent migration (chemotaxis) was significantly greater than the number exhibiting gradient-independent random motility (chemokinesis) (Fig.2C).
The chemotropic effects of glutamate are mimicked by NMDA and blocked by BAPTA-AM
To determine which type(s) of glutamate receptor mediates cell motility, agonists of different ionotropic glutamate receptors were assessed for their ability to stimulate migration. Only NMDA (1 μm) mimicked the effects of 1 μm glutamate, stimulating a similar number of cortical cells to migrate (Fig.3A). NMDA stimulated both chemotaxis and chemokinesis; however, nearly twice as many cells exhibited gradient-dependent migration compared with gradient-independent random motility (Fig. 2C). AMPA, kainate, and quisqualate failed to stimulate motility that was significantly different from spontaneous migration in controls. The conformational isomer, d-glutamate, also failed to induce migration. Thus, the signal for glutamate-induced migration appears to be mediated via NMDA-Rs. To confirm the involvement of NMDA-Rs, migration was assessed in the presence of 1 μm glutamate and serial dilutions of the NMDA-R antagonists MK801 or APV. In the presence of MK801 (100 nm to 100 μm), a noncompetitive receptor antagonist (Monaghan and Wenthold, 1997), glutamate-induced migration was reduced to 50% (Fig. 3B). APV, a competitive receptor antagonist (Monaghan and Wenthold, 1997), blocked glutamate-induced migration in a dose-dependent manner (Fig.3B), providing further evidence that glutamate-induced motility involves NMDA-Rs. High concentrations of APV (10–100 μm) inhibited glutamate-induced migration by nearly 67%; lower levels of APV failed to block significant levels of migration. Both chemotactic and chemokinetic responses to 1 μmglutamate or NMDA were attenuated in the presence of 10 μm either antagonist (data not shown). In the absence of [Mg2+]o, migration induced by lower levels of NMDA was potentiated (data not shown).
Fig. 3.
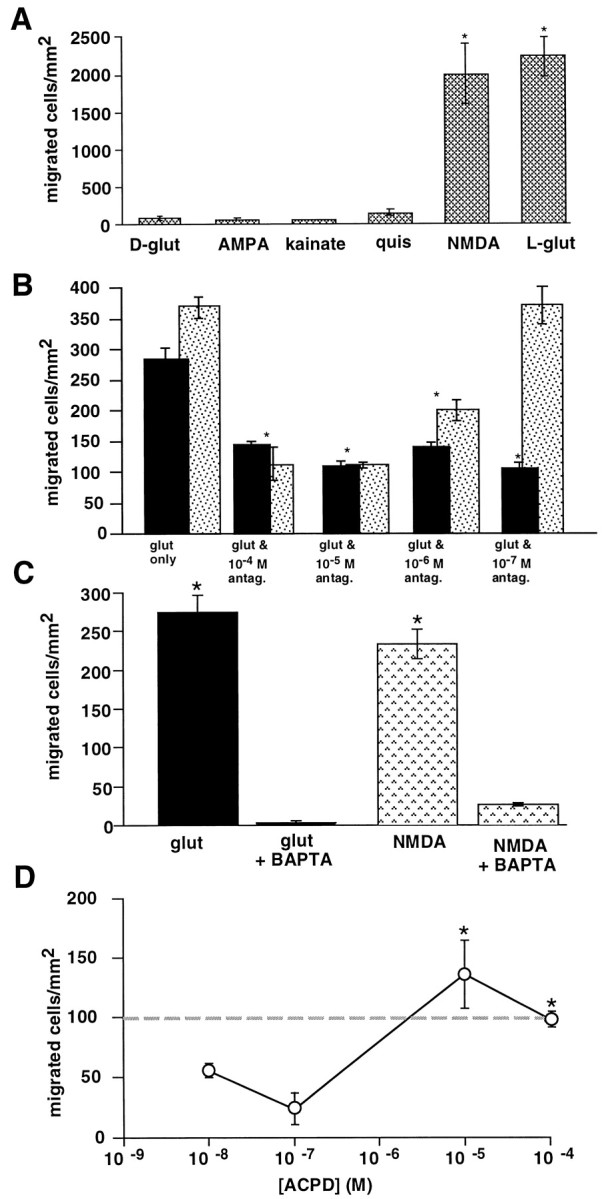
NMDA receptors mediate glutamate-induced cortical cell migration. A, Only NMDA mimics the effects of glutamate. E17 cells were migrated to 1 μml-glutamate, d-glutamate, kainate, quisqualate, NMDA, or AMPA. Only NMDA stimulates a similar level of motility asl-glutamate. B, MK801 or APV, antagonists at NMDA receptors, reduce the number of cells migrating to glutamate. Glutamate (1 μm) was mixed with serial dilutions of MK801 or APV (100 nm to 100 μm). At all concentrations of MK801, migration to glutamate was inhibited ∼50%. APV blocked glutamate-induced migration in a dose-dependent manner.C, Inhibition of migration by BAPTA-AM. Migration of E17 cells to 1 μm glutamate (•) or NMDA (▧) is entirely blocked in the presence of 10 μm BAPTA-AM.D, The metabotropic receptor agonist ACPD stimulates migration in a dose-dependent manner. Migratory responses to micromolar concentrations of ACPD (1–100 μm) are similar to migration induced by 1 μm glutamate (dashed line). Lower levels of ACPD fail to stimulate significant migration (>50 cells/mm2). *p≤ 0.01; ANOVA followed by Fisher’s PLSD test. Separate trials:A, n = 5; B,n = 3; C, n = 3.
We also assessed the effect of the Ca2+-chelator BAPTA-AM on NMDA- and glutamate-induced chemotaxis. BAPTA (10 μm) was added directly to the wells containing the cells during the incubation period. In the presence of BAPTA-AM, cell migration to glutamate and NMDA was completely blocked (Fig.3C), indicating that NMDA- or glutamate-induced elevations in cytosolic Ca2+([Ca2+]c) are required for chemotaxis to occur.
In these studies, NMDA was the only ionotropic glutamate receptor agonist that mimicked the chemotropic effects of glutamate. However, exposure to the NMDA-R antagonists resulted in only a partial block of glutamate-induced cortical cell migration. This suggests that a subpopulation of cells migrates to glutamate via activation of a different class of receptor, such as the metabotropic receptors. Therefore, we migrated E17 cortical cells to decreasing concentrations of trans-(±)-1-amino-1,3-cyclopentanedicarboxylic acid (ACPD), an agonist of metabotropic glutamate receptors (Monaghan and Wenthold, 1997). E17 cortical cells migrated to ACPD in a dose-dependent manner (Fig. 3D); micromolar ACPD stimulated a robust migratory response, whereas nanomolar concentrations failed to induce migration that was significantly higher than spontaneous motility to buffer only.
Viability studies demonstrated that the 18 hr exposure of the dissociated cells to the ligands used in the study did not alter cell survival (see Materials and Methods). Thus, the observed decreases in the number of migrating cells when BAPTA or the antagonists was present reflects inhibition of motility responses and not death of the cell.
Glutamate stimulates chemotaxis of vz neurons
To identify the anatomical origins of the migrating cells, E17 cortices were first microdissected into ventricular zone/subventricular zone (vz) and cortical plate/subplate (cp) regions. Immunocytochemistry was used to confirm that the microdissection effectively separated differentiating neurons of the cp (TUJ1+ only) (Lee et al., 1990) from the more immature neurons and precursors of the vz (TUJ1+/nestin+ and TUJ1−/nestin+, respectively). Immunostaining demonstrated that the two dissociates were comprised of cells at different stages of maturation. In the vz preparation, ∼45% of the cells expressed nestin only (Table 1), indicating that they were precursors or radial glia (Tohyama et al., 1992). The remaining vz cells (55.7 ± 2.4%) were TUJ1+ neurons. Many of the vz neurons also expressed nestin, indicating that they had recently undergone terminal mitosis. In contrast, the cp preparation contained primarily neurons (82.7 ± 5.0% TUJ1+) (Table 1), and the majority of TUJ1+ cp cells (72.4%) did not express nestin, demonstrating that they were more differentiated.
Table 1.
Characterization of the vz and cp dissociates by immunocytochemistry
| Region | % Total nestin+ | % Total TUJ1+ | % Nestin+ only | % TUJ1+ only | % Both |
|---|---|---|---|---|---|
| vz | 76.7 | 55.7 ± 2.4 | 45.0 ± 5.2 | 24.0 | 31.7 |
| cp | 27.6 | 82.7 ± 5.0 | 17.3 ± 4.1 | 72.4 | 10.3 |
After dissociation, the two populations were double labeled with anti-nestin and TUJ1 antibodies. Approximately half of vz cells express nestin only (45.0%), indicating that they are precursor cells. Neurons identified by TUJ1 staining comprise 55.7% of the vz population. The majority of cp cells were neurons (82.7%), and only 17.3% of the cp cells stained for nestin only. Values are means ± SEM;n = 7.
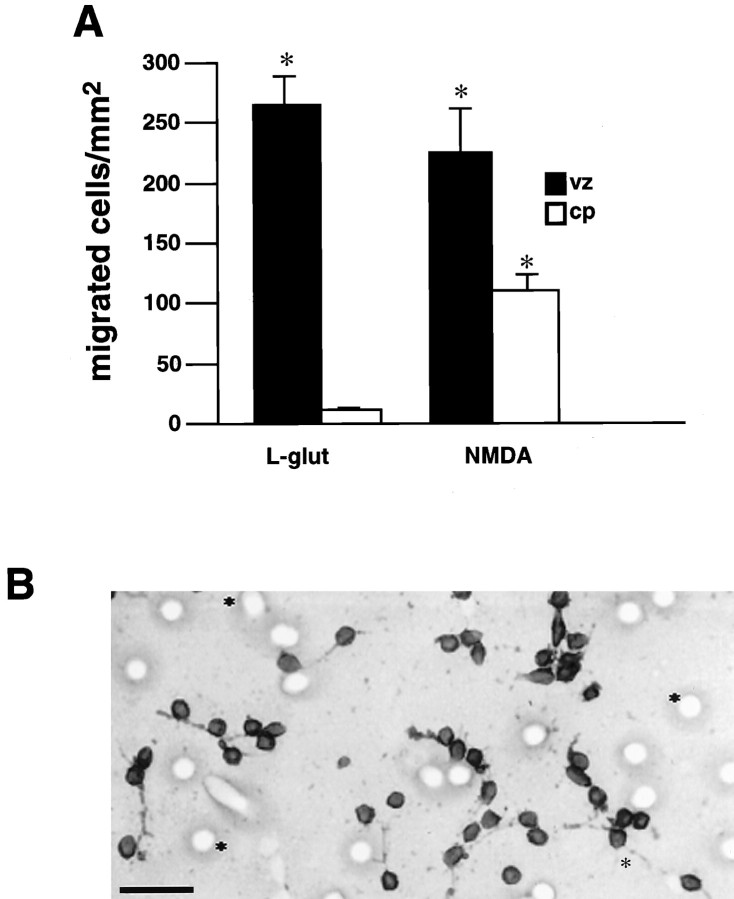
The migration of cellular dissociates from each region were examined in the microchemotaxis assay. In vitro, the dissociated vz cells demonstrated a greater migratory response to glutamate or NMDA than the cp cells (Fig.4A). Moreover, migration was limited to neuronal cells because immunostaining demonstrated that all of the migrated vz cells expressed NF protein (Fig. 4B).
Fig. 4.
Response by region. A, In vitro migratory responses of vz and cp cells. Vz cells (filled bars) exhibit greater migration to 1 μm glutamate or NMDA than cp cells (open bars) Significant numbers of cp cells do not respond to glutamate. Error bars indicate SEM. *p ≤ 0.01; ANOVA followed by Fisher’s PLSD test. B, Vz neurons migrate to glutamate. Vz cells were immunolabeled for neurofilament protein after migrating to glutamate in the chemotaxis assay. All vz cells that migrate to glutamate in vitro express neurofilament protein, indicating that they are neurons.Asterisks denote 8 μm pore in the membrane. Scale bar, 20 μm. Separate trials: A, B,n = 7.
Vz cells express NMDA-Rs
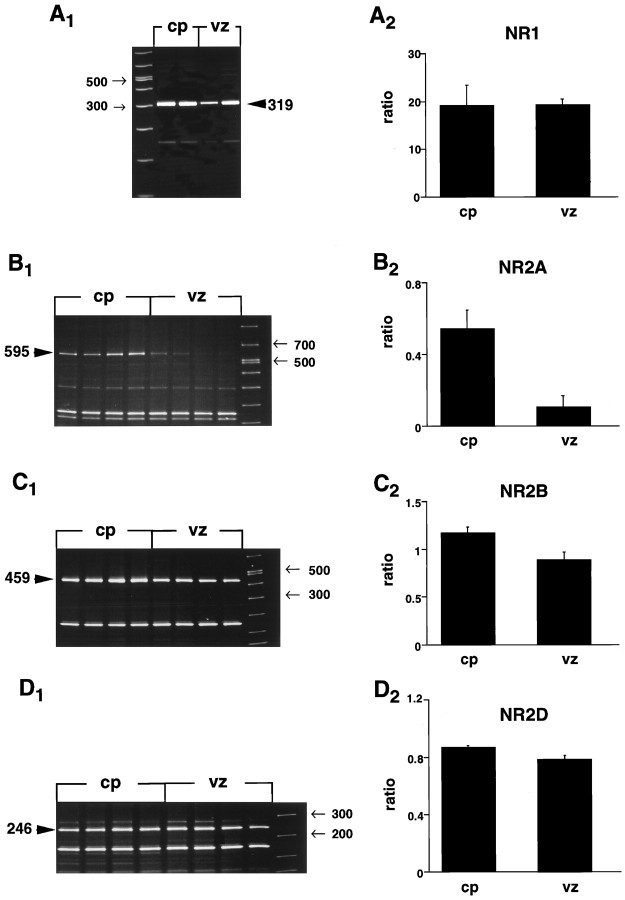
Vz and cp tissue segments were microdissected from E17–E18 cortices and were analyzed by semiquantitative PCR for expression of mRNA encoding NMDA-R subunit proteins. Transcripts detecting NR1 and three NR2 subunits (NR2A, NR2B, and NR2D) were evident in the cp cells (Fig.5A1–D1). In contrast, vz cells only expressed transcripts for NR1, NR2B, and NR2D (Fig. 5A1–D1). Densitometry demonstrated that the relative abundance of transcripts in cp cells was NR1 > NR2B > NR2D > NR2A (Fig.5A2–D2). In the vz cells, NR1 > NR2B > NR2D (Fig.5A2–D2). Both the cp and vz cells expressed similar levels of mRNA encoding NR1 (Fig.5A2). The results of the present PCR studies demonstrated that cells derived from both the immature (vz) and more differentiated (cp) regions of the cortex express mRNA encoding NMDA receptor subunit proteins. Western blotting confirmed the presence of NR1, NR2A, and NR2B proteins in embryonic cortical homogenates (data not shown).
Fig. 5.
Vz cells express mRNA encoding NMDA receptors subunits. Semiquantitative PCR was used to probe E17 vz and cp dissociates for mRNA encoding NR1 (A1,A2), NR2A (B1,B2), NR2B (C1,C2), and NR2D (D1,D2) subunits. Densitometry (A2, B2,C2, D2) revealed average relative abundance of transcripts in tissue homogenates from each region. For each region, n = 3.
NMDA-induced physiological responses of cells in the vz dissociate
Digital video microscopy was used to assess the effect of NMDA on [Ca2+]c levels in vz cells preloaded with the Ca2+ indicator dye, fura-2. NMDA evoked either rapidly rising (31 ± 12 sec to peak) or slowly rising (238 ± 56 sec to peak) elevations in [Ca2+]c in 15 and 9% of the recorded vz cells (n = 117), respectively (Fig.6). Typically, the more rapid response was associated with a larger increase in [Ca2+]c (830 ± 517 nm) when compared with the increase that occurred during the slower response (94 ± 47 nm). The former response was predominantly observed in larger (>10 μm in diameter), process-bearing cells. Both responses were antagonized by APV and were dependent on extracellular Ca2+[Ca2+]o. In some studies, cells were fixed and labeled with TUJ1 antibody after the fura-2 recording. All of the cells that responded to NMDA with increased [Ca2+]c immunolabeled with the TUJ1 antibody, indicating they were postmitotic neurons.
Fig. 6.
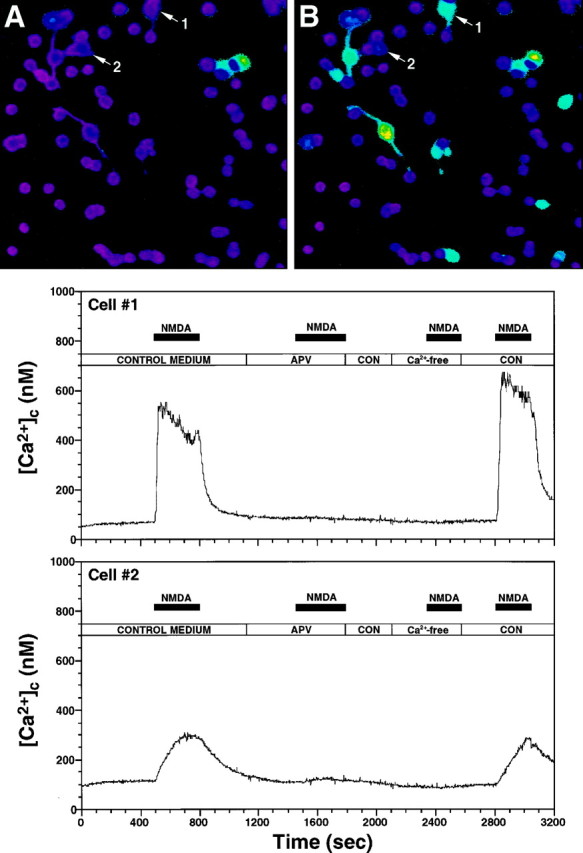
NMDA induces increases in [Ca2+]c in vz cells. After recording their resting [Ca2+]c in control (CON) medium (A), the cells were sequentially exposed to 10 μm NMDA (B), 100 μm APV plus 10 μm NMDA, followed by 10 μm NMDA in [Ca2+]o-free saline (see logabove the traces for the duration of each exposure). The data show rapid (Cell #1) and slow (Cell #2) [Ca2+]cresponses evoked by NMDA, typical of those recorded in the vz population. Both responses were reversibly blocked by APV and eliminated in [Ca2+]o-free saline.
Electrophysiology
NMDA-induced currents were recorded in cells of the vz preparation using conventional patch-clamp recording techniques. When membrane potential was clamped at −80 mV, low concentrations of NMDA (1 μm) induced a just-detectable inward current deflection (1.12 ± 0.01 pA), which was significantly different (p < 0.001) from the holding current (n = 2 of 7). Higher levels of NMDA induced larger currents with means of 6.62 ± 0.04 pA at 10 μm NMDA (n = 3 of 10) and 13.83 ± 0.08 pA at 50 μm NMDA (n = 5 of 10), which were superimposed with obvious fluctuations, presumably reflecting the underlying single-channel activity. The currents induced by 50 μm NMDA were almost completely blocked by equimolar concentrations of the NMDA antagonist MK801 (0.16 ± 0.03 pA) (Fig. 7).
Fig. 7.
NMDA-induced currents in vz cells.A, At 1 μm, NMDA induces a small (1.12 ± 0.01 pA; p < 0.001) inward current deflection that is significantly different from the holding current. At higher concentrations (B, C), NMDA induces larger currents with obvious superimposed open-channel noise. At the higher concentrations, the mean currents are 6.62 ± 0.04 pA at 10 μm (B) and 13.83 ± 0.08 pA at 50 μm (C). The current induced by 50 μm NMDA (C) is almost completely blocked by equimolar MK801 (0.16 ± 0.03).A and B were recorded from the same cell in the vz preparation; C was recorded from a different cell in the vz dissociate. Membrane potential was clamped at −80 mV.
Neuronal migration in cultured cortical slices is mediated via NMDA-Rs
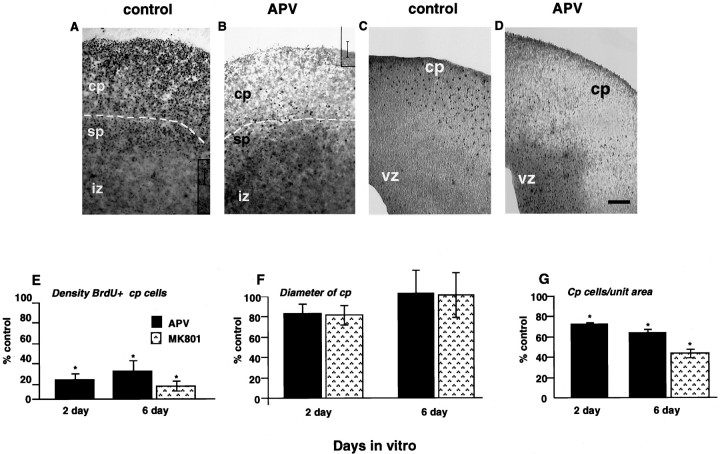
The studies on the dissociated cells revealed that direct application of glutamate stimulates chemotaxis of dissociated embryonic cortical neurons. To determine whether glutamate generated by cells within the cortex influences cell motility, we analyzed the effect of NMDA-R antagonists on cellular migration in organotypic cortical slice cultures. In theses studies, no exogenous glutamate was added to the slice cultures. In the developing cortex, the proliferating cells are located in the ventricular and subventricular zones (Jacobson, 1991). Because proliferating cells incorporate BrdU, we exposed the cultured slices to a pulse of BrdU and then used changes in the anatomical distribution of BrdU-labeled cells in the slices over time as an indication of postmitotic cell migration. Patterns of migration were assessed by administering an 18 hr pulse of BrdU to acutely prepared E17 slices. Culture medium was then replaced with growth media with or without NMDA-R antagonists, and slices were cultured for 2 or 6 additional days. Slices were fixed and immunostained for BrdU, and the distribution of labeled cells within the slices was examined. After 2 d of culture under these conditions, virtually all of the BrdU-labeled cells in control slices were localized to the cp and sp (Fig. 8A); the ventricular and intermediate zones in the control slices were virtually devoid of labeled cells (Fig. 8C). Because proliferation occurs within the ventricular regions, these results suggest that the cells that incorporated BrdU during the pulse migrated away from the germinal regions into the sp and cp. In contrast, contralateral slices maintained for 48 hr in the presence of APV (100 μm) had few labeled cells in the cp (Fig. 8B). The BrdU-positive cells in the treated slices were distributed throughout the ventricular and intermediate zones (Fig. 8D). Similar results were observed when slices were cultured for 6 d in the presence of APV or 100 μm MK801 (data not shown).
Fig. 8.
Glutamate-induced migration of cells in cultured cortical slices. A, Control slice maintained in vitro for 48 hr after the BrdU pulse. Almost all BrdU-labeled cells are in the cp and sp. B, The contralateral slice treated for 48 hr with 100 μm APV has few BrdU-labeled cells in the cp or sp. Scale bar, 80 μm. C, Low magnification of an untreated control slice shows few labeled cells in the vz or iz. D, The contralateral treated slice has an abundance of labeled cells in the vz and iz. Scale bar, 80 μm.E, Densitometry of cultured slices. Densitometry of contralateral slices treated with with either 100 μmAPV (•) or MK801 (▧). E, Density of BrdU-labeled nuclei in the cortical plate. At 2 and 6 d, there is a significant reduction in the number of BrdU-labeled cells in the cp of antagonist-treated slices compared with controls. F, Cortical plate thickness. At 2 d, antagonist treatment results in a minor decrease in cp thickness. By 6 d, the thickness of the cortical plate is not significantly diminished. G, Density of total cells in the cortical plate. At both 2 and 6 d, antagonist treatment results in significantly fewer cells per unit area when compared with controls. *p ≤ 0.05; Student’st test. Separate trials: n = 5.
Densitometry of the BrdU-labeled slices demonstrated that, in the presence of the NMDA-R antagonists, cellular migration into the cp was significantly attenuated (Fig. 8E). Slices maintained in APV for 48 hr showed an 80 ± 5.1% decrease in the number of BrdU-labeled cells in the cp. Slice cultures maintained for an extended period up to 6 d showed similar results. After 6 d of culture in APV, labeled cp cells decreased by 74 ± 10.3%. Similarly, slices maintained for 6 d in MK801 had an 85 ± 4.8% reduction of BrdU-labeled cells in the cp.
Effects of antagonists on cortical thickness in slice cultures
Because fewer BrdU-labeled cells migrated into the cp in antagonist-treated slices, the overall thickness of the cp might be expected to decrease. Therefore, we measured the thickness of the cp (from the innermost edge of layer VI through outermost edge of layer I) in treated and untreated slices. Antagonist treatment did not result in a significant decrease in cortical plate thickness. After 48 hr of culture, APV exposure resulted in only a 17 ± 10.3% decrease in cp thickness (Fig. 8F). Similarly, MK801-treated slices showed only a minor reduction of 18 ± 11.0% in the overall thickness of the cp when compared with controls. After 6 d of culture, treated and untreated cultures exhibited no significant difference in cp thickness (Fig. 8F).
Effects on cell density within the cp
These studies indicated that, although antagonist treatment resulted in a marked reduction in the number of BrdU-labeled cells in the cp, the overall cp thickness was not markedly reduced. To determine whether the antagonist treatment resulted in increased extracellular space between cp cells, we measured the relative “packing” density of total cells within the cp in control versus treated slices. Nontreated control slices contained a greater number of cells per unit area than treated slices (Fig. 8G). This was evident in slices treated with antagonists for 2 and 6 d. APV treatment for 2 d decreased the number of cells per unit area by 25 ± 1.3% compared with nontreated slices. After 6 d in culture, the number of cells per unit area in treated slices was also reduced (by 35 ± 1.8% in APV and 45 ± 2.4 in MK801) (Fig.8G). Thus, although the overall thickness of the cp was not markedly diminished, the packing density of cells in treated cultures was significantly less, indicating that exposure to NMDA-R antagonists resulted in fewer BrdU-labeled cells and an overall decrease in the number of total cells in the developing cp.
DISCUSSION
Our studies demonstrate that murine embryonic cortical neurons migrate in response to glutamate via mechanisms that involve NMDA receptors. NMDA was the only ionotropic agonist that mimicked the effects of glutamate on acutely dissociated cortical cells, stimulating a similar number of cells to migrate. Glutamate-induced migration of dissociated cells was inhibited by either MK801 or APV, antagonists at NMDA receptors (Monaghan and Wenthold, 1997). Thus, in a subpopulation of acutely dissociated murine embryonic cortical cells, NMDA receptors appear to mediate motility responses to glutamate. The cortical slice cultures supported the findings in the dissociated cells, providing further evidence that NMDA-Rs are involved in migratory responses to glutamate. Slices maintained in NMDA-R antagonists had an abundance of BrdU+ cells in the germinal zones but lacked BrdU-labeled cells in the cp region, suggesting that in the slices, NMDA-R antagonists inhibited postmitotic cell migration into the cp. Together, these results suggest that, in vivo, glutamate serves as a chemoattractant, stimulating the migration of murine embryonic cortical neurons toward their target destinations in the cp.
If chemoattractants provide positional cues to cells, then they should localize near the target destinations of migrating cells. In the embryonic mouse cortex, glutamate is highly expressed in the outer half of the cortical plate and in the C-R cells which lie within the primordial plexiform layer, near the target destination of migratory neurons (del Rio et al., 1995). In the developing rat cortex, GABA exhibits a similar distribution (Imamoto et al., 1994; Behar et al., 1996), and GABA has been shown to be a potent chemoattractant for rat embryonic cortical cells (Behar et al., 1996). In the present study, glutamate was a more effective chemoattractant for murine cortical cells than GABA. Thus, although the ligands differ, both rat and mouse appear to use motility-signaling molecules found in the developing cortical plate and C-R cells.
If chemoattractants play a relevant role in development, then migrating cells should express receptors for the attractants. During the final week of gestation, in situ hybridization studies have revealed that cells throughout the embryonic mouse cortex express mRNA transcripts encoding NMDA-R subunits (Feldmeyer and Cull, 1996;Monaghan and Wenthold, 1997). Here, RT-PCR revealed that both vz and cp dissociates express mRNA encoding NR1, an NMDA-R subunit protein that is an essential component of NMDA receptors (Monaghan and Wenthold, 1997). In addition, transcripts for NR2B and NR2D subunits were detected in dissociates from the two anatomical regions. Previous studies reported that embryonic cells in the cp of the developing cortex express functional NMDA-Rs (LoTurco et al., 1991); our findings demonstrate that cells derived from immature cortical regions also exhibit physiological responses to NMDA. Electrophysiology and Ca2+ imaging studies revealed that, after application of NMDA, cells in the vz preparation showed an increased current and exhibited increases in [Ca2+]c. In vitro, neurons dissociated from immature cortical regions exhibited migratory responses to NMDA. Together, these results demonstrate that neurons in the vz, svz, or lower intermediate zone express functional NMDA receptors. Thus, when cortical lamination occurs in vivo, both the attractant and its receptors are expressed in appropriate anatomical locations to form a physiologically relevant circuit.
In vivo, diffusible gradients of glutamate released by cells in the superficial regions of the cortex may act as chemoattractants for newly generated postmitotic neurons in the germinal zones, directing their migration into the cortical plate. Embryonic cortical sections immunostained for glutamate demonstrated that glutamate was most highly expressed in the cortical plate, the target region for migrating embryonic cortical neurons. In studies on the dissociated cells, glutamate primarily stimulated directed migration of neurons from immature regions: (1) more of the vz cells migrated to glutamate or NMDA than cp cells, (2) all of the responding vz cells were neuronal, and (3) the predominant mode of motility was directed migration, or chemotaxis. Peak migration to glutamate was observed at E17, suggesting that glutamate prominently influences the migration of neurons destined for cortical layers II and III (Angevine and Sidman, 1961; Schmidt and Lent 1987; Polleux et al., 1997). At earlier stages of development, migrating cells destined for the deep cortical lamina (V and VI) may lack the appropriate receptors and/or signaling mechanisms that mediate chemotactic responses to glutamate.
Glutamate-induced migration requires increases in [Ca2+]c. Migration to glutamate or NMDA was abolished in the presence of the Ca2+-chelating agent BAPTA-AM, which maintains [Ca2+]c at nanomolar levels. These findings are consistent with previous studies reporting that cellular locomotion requires increases in [Ca2+]c (Komuro and Rakic, 1992, 1996;Hinrichsen, 1993). In this regard, NMDA-Rs could directly influence cell motility by triggering increases in [Ca2+]c that modulate cytoskeletal dynamics required for cell movement. In the present study, application of NMDA resulted in a rapid flux of current in responding vz cells, whereas the Ca2+ imaging experiments revealed a slow rather than immediate rise in [Ca2+]cafter NMDA exposure. The slow rise in cytosolic Ca2+suggests that NMDA receptor activation leads to local increases in [Ca2+]c, which in turn trigger Ca2+ release from intracellular stores and/or stimulate further [Ca2+]o entry via other pathways.
NMDA-Rs appear to mediate motility signals in a cortical cell subpopulation. In other subpopulations, metabotropic receptors may mediate the motility signals of glutamate. NMDA-R antagonists only partially blocked migration to glutamate, suggesting that some cells migrate to glutamate via activation of a different type of receptor. ACPD, a metabotropic receptor agonist (Monaghan and Wenthold, 1997), also stimulated dissociated cells to migrate, indicating that glutamate receptors coupled to GTP binding proteins can induce nerve cell movement. In rats, GABA promotes motility of cortical cells via multiple classes of receptors, some of which couple to GTP binding proteins (Behar et al., 1998). A similar situation may exist in the developing murine cortex in which multiple classes of glutamate receptors stimulate migration in distinct subpopulations of embryonic neurons via different intracellular signaling mechanisms.
Cortical plate cells may express a non-NMDA-R that, when activated, arrests cell movement. Although NMDA stimulated migration in both vz cell and cp cell dissociates, the cp cells failed to migrate to glutamate. This demonstrates that the cp cells had the capacity to migrate in response to NMDA-R activation; however, they failed to move in the presence of the natural ligand. In rats, the GABA receptors that promote cp cell motility (GABAB- and GABAC-like) differ from those that arrest cp cell movement (GABAA) (Behar et al., 1998). A parallel mechanism may exist in the cp cells of the mouse; whereas NMDA-Rs promote motility, activation of a second class of glutamate receptor may attenuate migration. In vivo, migrating neurons may acquire a functional form of this second class of receptor soon after entering the cp. High concentrations of glutamate encountered near the target destinations could activate the receptor, providing a “stop” signal to migrating cells approaching their final positions.
Slice culture studies suggest that glutamate released from cortical cells stimulates embryonic neuronal migration toward the cp via NMDA-R activation
Blockade of NMDA-Rs prevents BrdU+ postmitotic cells from migrating into the cp. In untreated slice cultures, 2 d after the pulse, most BrdU+ cells were located in the cp. Only proliferating cells in ventricular regions incorporate BrdU; hence, the vz cells that took up the label during the pulse apparently migrated into the cp within 2 d. In contrast, antagonist-treated slices had few BrdU+ cells in the cp after 2 d; most labeled cells in these slices were observed in the vz, svz, and iz. The propidium iodide studies confirmed that the antagonist treatment of the slices did not affect cell viability (see Materials and Methods). Thus, the absence of BrdU+ cells in the cp of treated slices was not a result of cell death. Rather, the BrdU-labeled cells remained in the immature regions and apparently failed to migrate into the cp. Although the cp thickness was not diminished in antagonist-treated slices, the overall cell density was decreased. Hence, by the end of the culture period, slices maintained in NMDA-R antagonists had fewer total cp cells. These results demonstrate that inhibition of NMDA-Rs during corticogenesis leads to fewer cells in the cp, suggesting that in vivo, glutamate released from cortical cells directs the movement of postmitotic neurons into this region.
Our studies provide evidence that NMDA-Rs mediate motility signals in embryonic cortical neurons. These results appear to contradict earlier findings on transgenic mice that lack the NR1 subunit, which suggested that NMDA receptors are not involved cortical neuronal migration (Forrest et al., 1994; Messersmith et al., 1997). In an early study on mice lacking functional NR1 genes, gross observations of the cortex indicated that it had developed normally, although the authors did not conduct a careful microscopic examination of the cerebral cortex that would reveal subtle abnormalities in cortical lamination patterns (Forrest et al., 1994). Thus, the authors concluded that they could not exclude the possibility that NMDA receptors influence cortical nerve cell movement. In a subsequent report, Messersmith et al. (1997)suggested that NMDA receptors are not involved in cortical neuronal migration (Messersmith et al., 1997); however, their study examined the migration of cells destined for deep cortical lamina (layers V and VI). Here, we provide evidence that glutamate-induced migration primarily affects the cells destined for superficial cortical lamina (layers II and III). Furthermore, transgenic mice lacking NR1 subunits have reportedly developed compensatory mechanisms for fluxing Ca2+, which could directly influence cell motility (Messersmith et al., 1997). Finally, there may be a redundancy of mechanisms involved in glutamate-induced cortical cell migration. Our studies suggest that both ionotropic and metabotropic receptors are involved in cortical nerve cell movement. Compensatory mechanisms, coupled with a redundancy in glutamate receptor mechanisms that promote cortical cell movement, could account for the reportedly “normal” morphology of the cerebral cortex observed in transgenic mice that lack NR1 subunits.
The results of the present study provide evidence that rats and mice use different chemoattractants to promote the migration of cells destined for cortical layers II and III. In rats, GABA stimulates robust migration, whereas glutamate is a more potent attractant for murine cortical cells. In each species, the appropriate attractant is found highly expressed near the target locations for migrating cells; rat C-R and cp cells are GABAergic (Imamoto et al., 1994; Behar et al., 1996), whereas murine C-R and cp cells are glutamatergic (del Rio et al., 1995). It is unknown, however, whether the converse is true; rat cp cells may express high levels of glutamate, whereas murine cp cells may express GABA. Double-immunolabeling studies for GABA and glutamate would reveal whether the two molecules are detectable in the cortical plates of each species. Furthermore, GABA was a weak attractant for murine cells at E17; however, the molecule may stimulate robust chemotaxis at other developmental stages. Thus, a systematic study in rats and mice that compares the chemotropic effects of GABA and glutamate throughout the period of cortical histogenesis is warranted. These types of studies would resolve whether or not the two species use different signaling molecules to regulate cell chemotaxis in the developing cortex.
In summary, our results on the dissociated cells suggest that glutamate promotes chemotaxis of newly differentiated neurons within the developing embryonic mouse cortex. Pharmacological studies indicate that NMDA-Rs mediate the motility signals of glutamate in a subpopulation of cells. The studies on organotypic slice cultures confirmed the pharmacology demonstrated on the dissociated cells and provided evidence that, in vivo, glutamate released from cortical cells acts as a chemoattractant for embryonic neurons migrating toward the cp. Glutamate has been reported to stimulate neuronal movement via NMDA-Rs in the developing cerebellum (Komuro and Rakic, 1993), suggesting a widespread role for glutamate as a chemoattractant in the developing nervous system.
Footnotes
Correspondence should be addressed to T. N. Behar, Building 36, Room 2C02, 36 Convent Drive, Bethesda, MD 20892-4066.
REFERENCES
- 1.Angevine JB, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong RC, Harvath L, Dubois-Dalcq M. Type 1 astrocytes and oligodendrocyte-type 2 astrocyte glial progenitors migrate toward distinct molecules. J Neurosci Res. 1990;27:400–407. doi: 10.1002/jnr.490270319. [DOI] [PubMed] [Google Scholar]
- 3.Behar TN, Schaffner AE, Colton CA, Somogyi R, Olah Z, Lehel C, Barker JL. GABA-induced chemokinesis and NGF-induced chemotaxis of embryonic spinal cord neurons. J Neurosci. 1994;14:29–38. doi: 10.1523/JNEUROSCI.14-01-00029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behar TN, Li YX, Tran HT, Ma W, Dunlap V, Scott C, Barker JL. GABA stimulates chemotaxis and chemokinesis of embryonic cortical neurons via calcium-dependent mechanisms. J Neurosci. 1996;16:1808–1818. doi: 10.1523/JNEUROSCI.16-05-01808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behar TN, Schaffner AE, Scott CA, O’Connell C, Barker JL. Differential response of cortical plate and ventricular zone cells to GABA as a migration stimulus. J Neurosci. 1998;18:6378–6387. doi: 10.1523/JNEUROSCI.18-16-06378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.del Rio J, Martinez A, Fonseca M, Auladell C, Soriano E. Glutamate-like immunoreactivity and fate of Cajal-Retzius cells in the murine cortex as identified with calretinin antibody. Cereb Cortex. 1995;5:13–21. doi: 10.1093/cercor/5.1.13. [DOI] [PubMed] [Google Scholar]
- 7.Eisen JS. Determination of primary motoneuron identity in developing zebrafish embryos. Science. 1991;252:569–572. doi: 10.1126/science.1708527. [DOI] [PubMed] [Google Scholar]
- 8.Falk W, Goodwin RH, Leonard EJ. A 48 well micro-chemotaxis assembly for rapid and accurate measurement of leukocyte migration. J Immunol Methods. 1980;33:239–247. doi: 10.1016/0022-1759(80)90211-2. [DOI] [PubMed] [Google Scholar]
- 9.Feldmeyer D, Cull CS. Functional consequences of changes in NMDA receptor subunit expression during development. J Neurocytol. 1996;25:857–867. doi: 10.1007/BF02284847. [DOI] [PubMed] [Google Scholar]
- 10.Forrest D, Yuzaki M, Soares HD, Ng L, Luk DC, Sheng M, Stewart CL, Morgan JI, Connor JA, Curran T. Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron. 1994;13:325–338. doi: 10.1016/0896-6273(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 11.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 12.Hatten ME. Riding the glial monorail: a common mechanism for glial-guided migration in different regions of the developing brain. Trends Neurosci. 1990;13:179–184. doi: 10.1016/0166-2236(90)90044-b. [DOI] [PubMed] [Google Scholar]
- 13.Hinrichsen RD. Calcium and calmodulin in the control of cellular behavior and motility. Biochim Biophys Acta. 1993;1155:277–293. doi: 10.1016/0304-419x(93)90010-a. [DOI] [PubMed] [Google Scholar]
- 14.Imamoto K, Karasawa N, Isomura G, Nagatsu I. Cajal-Retzius neurons identified by GABA immunohistochemistry in layer I of the rat cerebral cortex. Neurosci Res. 1994;20:101–105. doi: 10.1016/0168-0102(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson M. Developmental neurobiology, Ed 3, pp 401–448. Plenum; New York: 1991. [Google Scholar]
- 16.Komuro H, Rakic P. Selective role of N-type calcium channels in neuronal migration. Science. 1992;257:806–809. doi: 10.1126/science.1323145. [DOI] [PubMed] [Google Scholar]
- 17.Komuro H, Rakic P. Modulation of neuronal migration by NMDA receptors. Science. 1993;260:95–97. doi: 10.1126/science.8096653. [DOI] [PubMed] [Google Scholar]
- 18.Komuro H, Rakic P. Intracellular Ca2+ fluctuations modulate the rate of neuronal migration. Neuron. 1996;17:275–285. doi: 10.1016/s0896-6273(00)80159-2. [DOI] [PubMed] [Google Scholar]
- 19.Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter A. The expression and posttranslational modification of a neuron-specific β-tubulin isotype during chick embryogenesis. Cell Motil Cytoskeleton. 1990;17:118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q-Y, Schaffner AE, Barker JL. Electrophysiological studies on receptors in vitro. In: Boulton AA, Baker GB, Bateson AN, editors. In vitro neurochemical techniques. Humana; Totowa, NJ: 1999. pp. 37–61. [Google Scholar]
- 21.LoTurco J, Blanton M, Kriegstein A. Initial expression and endogenous activation of NMDA channels in early neocortical development. J Neurosci. 1991;11:792–799. doi: 10.1523/JNEUROSCI.11-03-00792.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messersmith EK, Feller MB, Zhang H, Shatz CJ. Migration of neocortical neurons in the absence of functional NMDA receptors. Mol Cell Neurosci. 1997;5:347–357. doi: 10.1006/mcne.1997.0646. [DOI] [PubMed] [Google Scholar]
- 23.Monaghan DT, Wenthold RJ. The ionotropic glutamate receptors. Humana; Totowa, NJ: 1997. [Google Scholar]
- 24.O’Rourke NA, Dailey ME, Smith SJ, McConnell SK. Diverse migratory pathways in the developing cerebral cortex. Science. 1992;258:299–302. doi: 10.1126/science.1411527. [DOI] [PubMed] [Google Scholar]
- 25.O’Rourke NA, Sullivan DP, Kaznowski CE, Jacobs AA, McConnell SK. Tangential migration of neurons in the developing cerebral cortex. Development. 1995;121:2165–2176. doi: 10.1242/dev.121.7.2165. [DOI] [PubMed] [Google Scholar]
- 26.Polleux F, Dehay C, Kennedy H. The timetable of laminar neurogenesis contributes to the specification of cortical areas in mouse isocortex. J Comp Neurol. 1997;385:95–116. [PubMed] [Google Scholar]
- 27.Rakic P. Mode of cell migration of the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–84. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 28.Rakic P. Principles of neural cell migration. Experientia. 1990;46:882–891. doi: 10.1007/BF01939380. [DOI] [PubMed] [Google Scholar]
- 29.Schambra UB, Lauder JM, Silver J. Atlas of the prenatal mouse brain. Academic; San Diego: 1992. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt SL, Lent R. Effects of prenatal irradiation on the development of cerebral cortex and corpus callosumof the mouse. J Comp Neurol. 1987;264:193–204. doi: 10.1002/cne.902640205. [DOI] [PubMed] [Google Scholar]
- 31.Somogyi R, Wen X, Ma W, Barker JL. Developmental kinetics of GAD family mRNAs parallel neurogenesis in the rat spinal cord. J Neurosci. 1995;15:2575–2591. doi: 10.1523/JNEUROSCI.15-04-02575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tohyama T, Lee V, Rorke L, Marvin M, McKay R, Trojanowski J. Nestin expression in embryonic human neuroepithelium and in human neuroepithelial tumor cells. Lab Invest. 1992;66:303–313. [PubMed] [Google Scholar]