Abstract
Recent studies have suggested that protein phosphorylation of glutamate receptors may play an important role in synaptic transmission. Specifically, the phosphorylation of AMPA receptors has been implicated in cellular models of synaptic plasticity. The phosphorylation of the glutamate receptor 1 (GluR1) subunit of AMPA receptors by protein kinase A (PKA), protein kinase C (PKC), and Ca2+/calmodulin-dependent protein kinase II (CaMKII) has been characterized extensively. Phosphorylation of this subunit occurs exclusively on the intracellular C-terminal domain. However, the GluR1 subunit C terminus shows low homology to the other AMPA receptor subunits. In this paper we characterized the phosphorylation of AMPA receptor subunit GluR4, using site-specific mutagenesis and biochemical techniques. We found that GluR4 is phosphorylated on serine 842 within the C-terminal domain in vitro and in vivo. Serine 842 is phosphorylated by PKA, PKC, and CaMKII in vitro and is phosphorylated in transfected cells by PKA. Two-dimensional phosphopeptide analysis indicates that serine 842 is the major phosphorylation site on GluR4. In addition, we identified threonine 830 as a potential PKC phosphorylation site. These results suggest that GluR4, which is the most rapidly desensitizing AMPA receptor subunit, may be modulated by phosphorylation.
Keywords: glutamate, AMPA receptors, GluR4, phosphorylation, PKA, PKC
Rapid excitatory neurotransmission in the CNS is mediated mostly by ionotropic glutamate receptors, which are ligand-gated ion channels that play a role in synaptic plasticity (Bliss and Collingridge, 1993; Linden, 1994), neuronal cell death (Choi, 1988), and in some forms of neuronal degeneration (Zuo et al., 1997). Ionotropic glutamate receptors can be divided according to their molecular structure, sensitivity to agonists, and physiological properties into AMPA, kainate, or NMDA receptors. Molecular cloning studies identified receptor subunits for these receptor classes (Hollmann and Heinemann, 1994): glutamate receptors 1–4 (GluR1–4) are AMPA receptor subunits, GluR5–7 and KA1 and KA2 comprise kainate receptors, whereas NMDA receptors are formed by assembly of the NR1 subunit with NR2A–NR2D subunits. Initially, glutamate receptors were viewed as members of the family of ligand-gated ion channels, typified by the nicotinic acetylcholine receptors, and thus were assumed to be pentamers; each subunit was thought to have four transmembrane domains and extracellular N and C termini. However, it now has been demonstrated that the topology profile for glutamate receptors consists of three transmembrane domains, a channel-lining reentrant membrane loop, an extracellular N-terminal domain, and an intracellular C-terminal domain (Hollmann et al., 1994; Bennett and Dingledine, 1995;Wo and Oswald, 1995). Moreover, some recent evidence suggests that ionotropic glutamate receptors may share a tetrameric structure with the voltage-gated potassium channels (Mano and Teichberg, 1998;Rosenmund et al., 1998).
GluR4 has a limited expression in the brain, but it is highly expressed in the thalamus and in the cerebellum (Keinänen et al., 1990;Monyer et al., 1991). GluR4 is particularly prominent in cerebellar granule cells, where, along with GluR2, it probably accounts for most of the non-NMDA receptor channels (Monyer et al., 1991; Mosbacher et al., 1994). Swanson and coworkers have shown that the GluR4flip channels expressed in human embryonic kidney (HEK) 293 cells have agonist-dependent conductance properties (Swanson et al., 1997) resembling those of native high-conductance channels in cerebellar granule cells (Wyllie et al., 1993). Alternative splicing of the “flip-flop” region of AMPA receptor subunits regulates the desensitization properties of AMPA receptors and has the largest effect on GluR4 desensitization. Homomeric AMPA GluR4flopreceptors expressed in oocytes have desensitization time constants of ∼1 msec, whereas the GluR4flip homomeric receptors desensitize approximately four times more slowly (Mosbacher et al., 1994), suggesting that rapid desensitization of AMPA receptors can be regulated by the expression and alternative splicing of GluR4. Indeed, fast synaptic transmission and submillisecond desensitization have been detected in the cerebellum granule cells by postnatal days 11–17 (Silver et al., 1992), when GluR4flop is expressed (Monyer et al., 1991). GluR4flop also is expressed in auditory cells of the cochlear nucleus, where the fast kinetics of the AMPA receptor are physiologically important in transmitting the signals necessary for sound localization (Raman et al., 1994) and selective targeting of GluR4 to auditory nerve synapses has been shown (Rubio and Wenthold, 1997). Preferential expression of GluR4 may be a common feature of AMPA receptors mediating fast neurotransmission in the brainstem.
Another example of preferential expression of GluR4 is in the spinal cord motor neurons (Tomiyama et al., 1996), where rapid neurotransmission has been recorded (Smith et al., 1991) and where calcium-permeable AMPA receptors may be involved in excitotoxicity (Carriedo et al., 1996). In fact, both the reticular nucleus of the thalamus, which exclusively expresses GluR4 (Keinänen et al., 1990), and basal forebrain cholinergic neurons, which preferentially express GluR4 (Page and Everitt, 1995), are highly susceptible to AMPA-induced neurotoxicity (Page and Everitt, 1995), suggesting that GluR4 may be important in AMPA receptor-mediated excitotoxicity.
Biochemical and physiology studies have demonstrated that the function of AMPA receptors is modulated by protein phosphorylation in heterologous cells expressing recombinant receptor subunits, in primary neuronal cultures, and in hippocampal slices. GluR1 has been shown to be phosphorylated by PKA on Ser-845 in transfected HEK 293 cells (Roche et al., 1996), and treatment of hippocampal slices with forskolin increased the phosphorylation of GluR1 Ser-845 (Mammen et al., 1997). Furthermore, phosphorylation by PKA of Ser-845 on GluR1 potentiates its response to glutamate (Roche et al., 1996). These results are in agreement with earlier studies that described an increase in the amplitude of AMPA receptor responses on PKA activation, both in cultured neurons (Liman et al., 1989; Greengard et al., 1991; Wang et al., 1991) and in oocytes expressing GluR1/GluR3 subunits (Keller et al., 1992). GluR1 also is phosphorylated on Ser-831 by PKC and CaMKII both in transfected HEK 293 cells and in hippocampal slices (Roche et al., 1996; Barria et al., 1997a; Mammen et al., 1997). Recent studies have shown that GluR1 phosphorylation is modulated during cellular models of synaptic plasticity, including long-term potentiation and long-term depression (Barria et al., 1997b; Kameyama et al., 1998; Lee at al., 1998).
In the present study, we characterized the phosphorylation of GluR4in vitro and in situ, using site-specific mutagenesis. We demonstrate that GluR4 is phosphorylated in vitro on Ser-842 by PKA, PKC, and CaMKII, whereas Thr-830 is phosphorylated in vitro by PKC. In addition, Ser-842 is phosphorylated basally in HEK 293T cells in situ, and this phosphorylation is highly regulated by the PKA activator forskolin. These results indicate that GluR4 is phosphorylated directly and that phosphorylation of GluR4-containing receptors may regulate rapid excitatory synaptic transmission.
MATERIALS AND METHODS
Materials. GluR4 cDNA was a kind gift of Dr. Steve Heinemann (Salk Institute, San Diego, CA). PKC and the catalytic subunit of PKA were purified as previously described (Reimann and Behman, 1983; Woodgett and Hunter, 1987). Radioisotopes were purchased from DuPont New England Nuclear (Boston, MA), and cellulose thin layer chromatography (TLC) plates were from Kodak (Rochester, NY). Forskolin, phorbol 12-myristate 13-acetate (PMA), and CaMKII were obtained from Calbiochem (La Jolla, CA). Restriction enzymes were purchased from New England Biolabs (Beverly, MA), and polyvinylidene difluoride membrane was from Millipore (Bedford, MA).
Cell culture, transfection, and metabolic labeling. HEK 293T cells maintained at 37°C and 5% CO2 were transiently transfected with 10 μg of cDNA, using the calcium phosphate coprecipitation method, as previously described (Tingley et al., 1997;Mammen et al., 1999). Cells were used 48 hr after transfection. For metabolic labeling the cells were incubated with 1 mCi/ml [32P]orthophosphate in phosphate-free minimal essential medium (Life Technologies, Gaithersburg, MD) for 4 hr, and treated for 15 min with drugs, as indicated. Cells were scraped in 150 μl of 1% SDS in IP buffer containing 10 mm sodium phosphate, pH 7.0, 100 mm NaCl, and protease and phosphatase inhibitors; the cells were diluted by adding 750 μl of 1% Triton X-100 in ice-cold IP buffer and then sonicated. The residual insoluble fraction was removed by centrifugation at 14,000 ×g for 10 min at 4°C. The GluR4 protein or GluR4 mutant proteins were isolated by immunoprecipitation, using anti-GluR4 antibodies, as described previously for other AMPA receptor subunits (Roche et al., 1996). Samples were analyzed on a 7.5% polyacrylamide gel and visualized by autoradiography.
Fusion proteins. Fusion proteins containing the C-terminal region of GluR4 (amino acids 815–882) or truncated segments of the C-terminal region (polypeptides corresponding to amino acids 815–838 and to amino acids 815–852 of GluR4) were constructed by PCR amplification of GluR4 cDNA, by subcloning into restriction endonuclease sites (BamHI and EcoRI for the whole-length C terminal; BamHI and SalI for the truncated segments) in the pGEX4T-2 vector (Pharmacia, Piscataway, NJ), and by expression in BL21 Escherichia coli. Transformed bacteria were grown in 200 ml cultures, induced with isopropyl-1-thio-β-galactopyranoside (100 μm) for 2 hr, and then lysed in PBS containing 1% Triton X-100 and protease inhibitors. The cells were sonicated and shaken at 4°C for 30 min; the insoluble fraction was removed by centrifugation at 12,000 ×g for 10 min at 4°C. Fusion proteins were purified by glutathione–Sepharose affinity chromatography, according to the protocol of the manufacturer (Pharmacia), and were dialyzed overnight against TBS.
GluR4 fusion protein phosphorylation. Phosphorylation of 1 μg of purified fusion proteins was performed by incubation with 300 ng of purified kinases at 30°C for 30 min in a 100 μl total volume. Phosphorylation reaction buffer for PKC and CaMKII reactions contained 100 mm HEPES, pH 7.4, 20 mmMgCl2, 200 μmCaCl2, and 250 μm ATP. For PKC reactions, 50 μg/ml phosphatidylserine and 5 μg/ml diacylglycerol were added. Phosphorylation of fusion proteins by PKA was performed in 20 mm HEPES, pH 7.0, 10 mmMgCl2, and 250 μm ATP. Reactions included 5 μCi of [γ-32P] ATP and were stopped with SDS-PAGE sample buffer; the phosphorylated fusion proteins were resolved by SDS-PAGE and visualized by autoradiography. The phosphorylated proteins were excised from gels for phosphopeptide mapping.
Phosphopeptide mapping and phosphoamino acid analysis.Phosphopeptide mapping of phosphorylated proteins was performed essentially as previously described (Blackstone et al., 1994). Briefly, the polyacrylamide gel fragments containing the phosphorylated GluR4 or GluR4 fusion proteins were excised from the gel, digested with 0.3 mg/ml trypsin, and spotted onto TLC plates. The tryptic digests were separated by electrophoresis (400 V) in acetic acid/pyridine/H2O (19:1:89, v/v) in the first dimension and by ascending chromatography in pyridine/butanol/acetic acid/H2O (15:10:3:12, v/v) in the second dimension. The TLC plates were exposed in PhosphoImager cassettes and visualized with a Molecular Dynamics PhosphoImager (Sunnyvale, CA).
For phosphoamino acid analysis, peptides resulting from trypsin digestion were hydrolyzed in 6 m HCl and spotted onto TLC plates, along with phosphoserine, phosphothreonine, and phosphotyrosine standards. The phosphoamino acids were separated by electrophoresis (400 V) in pH 1.9 buffer (formic acid/acetic acid/H2O; 1:10:89, v/v) for 5 cm and then in pH 3.4 buffer (acetic acid/pyridine/H2O; 19:1:89, v/v) for 9 cm more. The phosphoamino acid standards were visualized with ninhydrin, and the TLC plate was exposed to film to visualize the 32P-labeled amino acids.
RESULTS
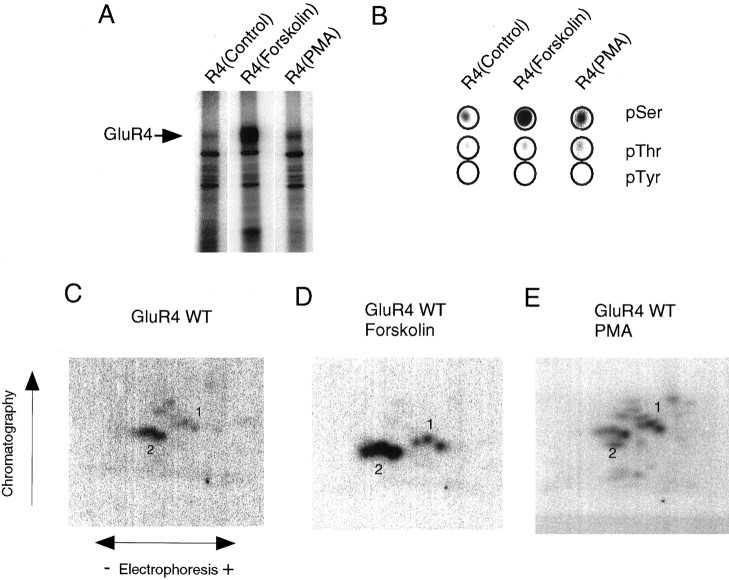
To examine the phosphorylation of the AMPA receptor GluR4 subunit, we transfected HEK 293T cells with GluR4 cDNA and labeled them with [32P] orthophosphate. GluR4 was immunoprecipitated via an anti-GluR4 antibody. GluR4 migrated on SDS-PAGE as a protein of ∼105 kDa that showed basal phosphorylation, which was increased on cell treatment with forskolin (Fig.1A). Phorbol esters had little effect on the phosphorylation of GluR4 (Fig.1A). GluR4 protein was excised from the gel, and two-dimensional phosphopeptide mapping and phosphoamino acid analysis were performed. Phosphorylation of recombinant GluR4 occurred predominantly on serine residues (Fig. 1B), but some phosphothreonine signal was detected. The increase in phosphorylation in response to forskolin was on serine residues. Tryptic phosphopeptide mapping of GluR4 revealed two main clusters of phosphopeptides (Fig.1C). Treatment of the 293T cells with forskolin or with phorbol esters did not change the phosphopeptide map patterns significantly (Fig. 1D,E). No novel major phosphopeptides appeared on cell stimulation with forskolin, although forskolin greatly increased GluR4 phosphorylation (Fig.1A), indicating that the site phosphorylated by PKA is phosphorylated basally in 293 cells.
Fig. 1.
Phosphorylation of GluR4 AMPA receptor subunit transiently expressed in HEK 293T cells. A, GluR4 was immunoprecipitated from labeled cells, resolved by SDS-PAGE, and visualized by autoradiography. B, After digestion with trypsin, GluR4 was hydrolyzed with 6N HCl, and the resulting amino acids were separated by electrophoresis. The circlesindicate the migration of ninhydrin-stained phosphoamino acid standards. GluR4 phosphoamino acids were visualized by autoradiography.C–E, Phosphorylated GluR4 was excised from the gel and digested with trypsin; the resulting phosphopeptides were spotted onto chromatography plates and resolved in two dimensions. HEK 293T cells expressing GluR4 were labeled with 1 mCi/ml [32P] orthophosphate and treated for 15 min with control solution (C), with 10 μm forskolin (D), or with 200 nm PMA (E).
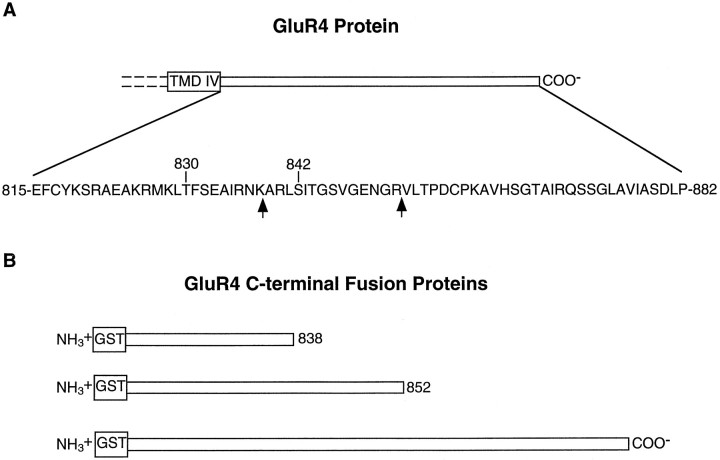
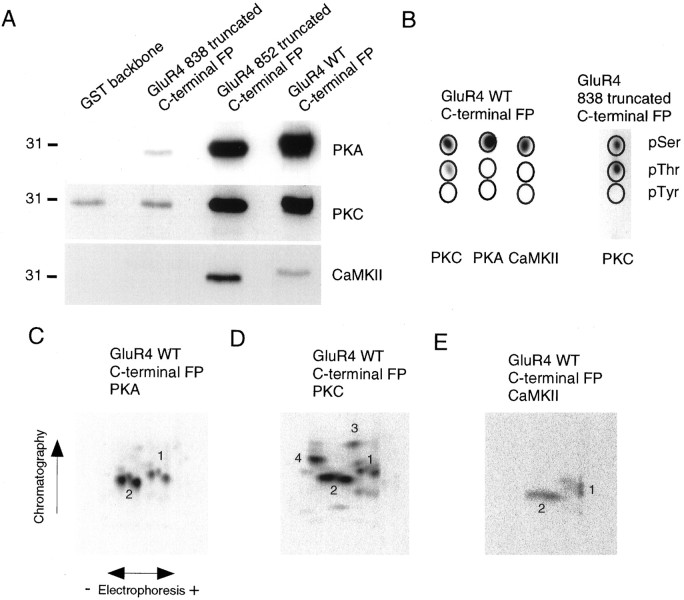
To identify kinases that directly phosphorylate GluR4 and to narrow down the regions likely to include the phosphorylation sites, we constructed glutathione S-transferase fusion proteins containing the C-terminal domain of GluR4 or truncated segments of the C terminus (Fig. 2). Purified bacterial fusion proteins were phosphorylated in vitro with purified kinases in the presence of [γ-32P] ATP (Fig.3). PKA phosphorylated the C terminal of GluR4 as well as the peptide corresponding to amino acids 815–852 of GluR4 (Fig. 3A). However, PKA phosphorylation was eliminated in the fusion protein containing GluR4 amino acids 815–838, suggesting that major phosphorylation sites for PKA are in the region between amino acids 838 and 852. Indeed, the tryptic phosphopeptide map of the PKA phosphorylated GluR4 C-terminal fusion protein (Fig. 3C) was identical to that of 852 truncated C-terminal fusion protein (data not shown). In both cases the phosphopeptide maps were identical to those generated by GluR4 isolated from transfected HEK 293T cells (see Fig. 1D). Very low phosphorylation of the 838 truncated C-terminal fusion protein was observed, and phosphopeptide mapping revealed only a minor phosphopeptide for this fusion protein (data not shown). These results demonstrate that the majority of C-terminal GluR4 phosphorylation by PKA occurs on residues between 838 and 852 of GluR4. PKA phosphorylated the fusion protein on serine residues only (Fig. 3B).
Fig. 2.
GluR4 C terminal. A, Schematic diagram of the C-terminal region of GluR4, including the fourth transmembrane domain (TMD IV). Theexpanded region shows the amino acid sequence of the C terminal, and the residues subjected to site-directed mutagenesis are indicated by their residue numbers. Thearrows indicate the ending of the truncated C-terminal bacterial fusion proteins. B, Schematic diagram of the fusion proteins containing segments of GluR4 C terminal.
Fig. 3.
Phosphorylation of GluR4 fusion proteins.A, Bacterial fusion proteins containing GluR4 C terminal or partial segments of GluR4 C terminal (amino acids 815–838 or 815–852) were incubated with purified kinases, as indicated, in the presence of [γ-32P] ATP, resolved by SDS-PAGE, and analyzed by autoradiography. B, Phosphoamino acid analysis of phosphorylated GluR4 fusion proteins. Phosphoamino acid analysis was performed as described in Figure 1B.C–E, The phosphorylated GluR4 C-terminal fusion proteins were excised from the gel and digested with trypsin; the phosphopeptides were resolved in two dimensions in TLC plates and visualized by autoradiography. In every case the phosphopeptide map for the fusion protein containing amino acids 815–852 of GluR4 protein was identical to that for the full-length C-terminal GluR4 fusion protein.
To characterize PKC phosphorylation of GluR4, we phosphorylated GluR4 C-terminal fusion proteins in vitro by purified PKC, and the resulting tryptic phosphopeptides were resolved (Fig. 3A,D). PKC phosphorylated the full-length GluR4 C-terminal fusion protein as well as the 852 truncated fusion protein (Fig. 3A). However, as with PKA, the majority of PKC phosphorylation was eliminated in the fusion protein containing amino acids 815–838, suggesting that the major phosphorylation sites for PKC are in the region between amino acids 838 and 852. The tryptic phosphopeptide map of the PKC phosphorylated GluR4 C-terminal fusion protein (Fig. 3D) was identical to that of the 852 truncated C-terminal fusion protein (data not shown). In contrast, the major phosphopeptides were absent in the 838 truncated C-terminal fusion protein. The PKC phosphorylated wild-type and 852 truncated C-terminal fusion proteins contained the same phosphopeptide clusters (phosphopeptides 1 and 2) that were observed for recombinant GluR4 isolated from HEK 293T cells; however, several additional phosphopeptides (phosphopeptides 3 and 4) were present in the fusion protein. These phosphopeptides were still phosphorylated by PKC in the 838 truncated C-terminal fusion protein. The majority of this phosphorylation occurred on phosphothreonine residues (Fig. 3B).
CaMKII phosphorylation of the fusion proteins produced phosphopeptides identical to those produced by PKA and PKC phosphorylation. Basically, the same two clusters of phosphopeptides (1, 2) were observed, both for the full-length C-terminal GluR4 fusion protein (Fig. 3E) and for the fusion protein including amino acids 815 to 852 (data not shown). CaMKII phosphorylated exclusively serine residues (Fig.3B). The fusion protein containing the shorter GluR4 C-terminal fragment was not phosphorylated by CaMKII (Fig.3A).
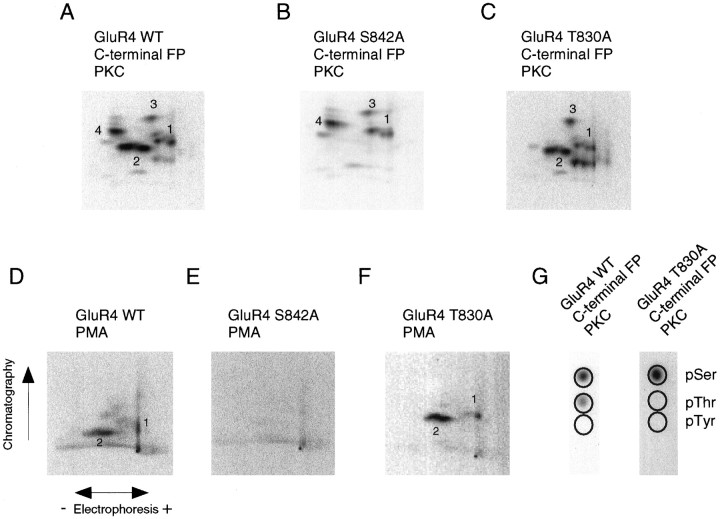
Taking into account that the major phosphorylation sites on the GluR4 C-terminal domain are in the region between amino acids 838 and 852, we used site-directed mutagenesis to identify which residues are phosphorylated. Conversion of serine residue 842 to alanine, in the GluR4 C-terminal fusion protein, resulted in tryptic phosphopeptide maps of PKA and PKC phosphorylated fusion proteins that lacked the major phosphopeptides (phosphopeptides 2) (Figs.4B, 5B). Similarly, phosphopeptides 2 were missing in the phosphopeptide map for the GluR4 S842A mutant C-terminal GST fusion protein phosphorylatedin vitro by purified CaMKII (data not shown). Moreover, mutation of serine 842 to alanine in full-length recombinant GluR4 eliminated the majority of the GluR4 phosphorylation seen in HEK 293T cells. Phosphopeptide maps of the mutant GluR4 lacked phosphopeptides 2 in unstimulated cells as well as in cells stimulated with forskolin (Fig. 4D) or with phorbol esters (Fig.5E). These results suggest that S842 is the major phosphorylation site in HEK cells.
Fig. 4.
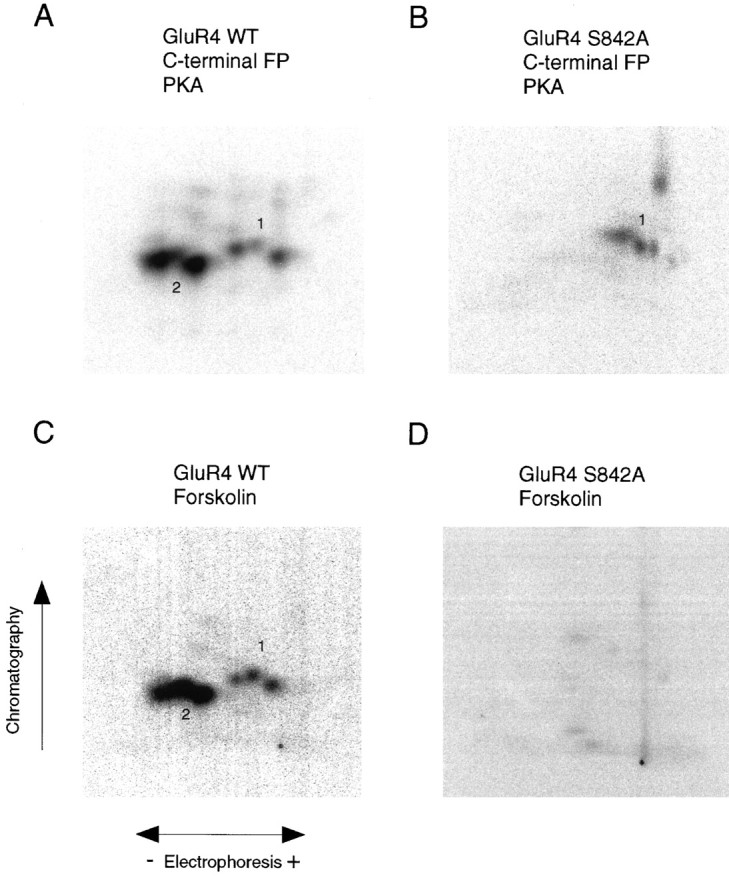
Identification of a PKA phosphorylation site on the GluR4 subunit. A, B, Purified fusion proteins containing the C terminal of GluR4 (A) or of GluR4 S842A mutant (B) were phosphorylatedin vitro with purified PKA. The phosphorylated fusion proteins were subjected to phosphopeptide mapping. C, D, HEK 293T cells expressing GluR4 or GluR4 S842A mutant were labeled with [32P] orthophosphate and stimulated with forskolin. Phosphorylated wild-type GluR4 (C) or the S842A GluR4 mutant (D) were immunoprecipitated, resolved in SDS-PAGE, and analyzed by tryptic phosphopeptide mapping.
Fig. 5.
Identification of PKC phosphorylation sites on the GluR4 subunit. A–C, PKC phosphorylation of wild-type GluR4 (A), S842A GluR4 (B), and T830A GluR4 (C) C-terminal GST fusion proteins. Fusion proteins were incubated with purified PKC in the presence of [γ-32P] ATP, resolved by SDS-PAGE, excised from the gel, and digested with trypsin for phosphopeptide mapping (A–C) or for phosphoamino acid analysis (G). For phosphoamino acid analysis the tryptic phosphopeptides were hydrolyzed and resolved by electrophoresis on a TLC plate, along with phosphoamino acid standards (migration is indicated by the circles). D–F, Wild-type GluR4 (D), the S842A GluR4 mutant (E), or the T830A GluR4 mutant (F) were expressed in HEK 293T cells, which were labeled with [32P] orthophosphate before stimulation with phorbol esters. Wild-type GluR4 and its mutants were isolated by immunoprecipitation and analyzed by phosphopeptide mapping.
Although S842 is the major site of phosphorylation, GluR4 expressed in HEK 293T cells also was phosphorylated on threonine residues (see Fig.1B). Interestingly, PKC phosphorylated the C-terminal GST fusion proteins in vitro on threonine residues. Threonine phosphorylation was observed on all C-terminal fusion proteins, including the fusion protein containing the shortest C-terminal fragment (amino acids 815–838), indicating that GluR4 C terminus is phosphorylated by PKC on threonine residues in the region between amino acids 815 and 838. Because Thr-830 is the only threonine residue in this region and because it is a consensus site (KXT; Pearson and Kemp, 1991) for PKC phosphorylation, we mutated Thr-830 to alanine in the GluR4 C-terminal fusion protein and in the full-length GluR4 subunit. In vitro phosphorylation by PKC of the T830A mutant GluR4 C-terminal fusion protein produced a phosphopeptide map lacking phosphopeptide 4 (Fig. 5C), and phosphothreonine was no longer detected (Fig. 5G). However, we have not been able to detect phosphopeptide 4 in situ for wild-type GluR4 (see Fig. 1C–E), and the map for the T830A GluR4 mutant expressed in HEK 293T cells (Fig. 5F) did not reveal any differences relative to the wild type.
DISCUSSION
Protein phosphorylation plays a critical role in the regulation of glutamate receptor function and is an important mechanism of synaptic plasticity (Roche et al., 1994; Moss and Smart, 1996). It has been demonstrated that AMPA receptor function is regulated by the phosphorylation of Ser-845 by PKA (Roche et al., 1996) and the phosphorylation of Ser-831 of GluR1 by both PKC (Roche et al., 1996) and CaMKII (Barria et al., 1997a; Mammen et al., 1997). However, the phosphorylation sites described for GluR1 are not conserved in other AMPA receptor subunits, and the phosphorylation of the GluR2–4 subunits has not been well characterized. Previous studies, using phosphorylation site-specific antibodies, suggested that Ser-696 in GluR2 and the corresponding site in other AMPA receptor subunits is phosphorylated on AMPA receptor activation with agonists (Nakazawa et al., 1995). However, according to the currently accepted model of the transmembrane topology of glutamate receptors, this region of GluR2 is extracellular and therefore inaccessible to intracellular kinases.
In this study we characterized the phosphorylation of the GluR4 subunitin vitro and in transfected cells, using site-specific mutagenesis and phosphopeptide mapping techniques. We demonstrated that PKA, PKC, and CaMKII directly phosphorylate GluR4 C-terminal fusion proteins in vitro and that Ser-842 is phosphorylated basally in GluR4 in transfected cells. Forskolin treatment of the transfected cells dramatically increased the phosphorylation of Ser-842, suggesting that PKA phosphorylates this site in situ. In addition, we showed that PKC phosphorylates Thr-830 in vitro, but we were not able to detect the phosphopeptide (phosphopeptide 4) containing Thr-830 in situ (see Fig. 5A). However, interestingly, the Thr-830 mutant GluR4 C-terminal fusion protein did not show any phosphothreonine signal when it was phosphorylated by PKC, indicating that Thr-830 is the major threonine that is phosphorylated by PKC in GluR4. Because PKA and CaMKII did not phosphorylate GluR4 fusion proteins on threonine residues, phosphorylation of Thr-830 may account for most of the phosphothreonine signal, both in GluR4 fusion proteins and in recombinant GluR4 expressed in HEK 293T cells.
GluR4 has high homology in the C-terminal with GluR2c, an alternatively spliced form of GluR2, which contains a longer C-terminal domain (Köhler et al., 1994), and both Ser-842 and Thr-830, described here as phosphorylation sites for GluR4, are conserved in GluR2c. However, only a minor fraction of the GluR2 transcripts encodes the longer C terminus, which could not be detected in the rat brain byin situ hybridization with a specific probe (Köhler et al., 1994).
Fast synaptic transmission and submillisecond desensitization have been detected in several parts of the brain, where they have important physiological function, for instance as specializations for temporal coding in the auditory neurons of the cochlear nucleus (Raman et al., 1994; Rubio and Wenthold, 1997). In all cases, rapid kinetics seems to be associated with heavy GluR4 expression (Monyer et al., 1991; Smith et al., 1991; Silver et al., 1992; Wyllie et al., 1993; Mosbacher et al., 1994; Raman et al., 1994; Tomiyama et al., 1996; Rubio and Wenthold, 1997; Swanson et al., 1997). We have described Ser-842 as a major phosphorylation site in GluR4 and identified Thr-830 as a potential phosphorylation site for PKC. These results suggest that, similar to other glutamate receptor subunits, the phosphorylation of GluR4 may be an important mechanism in regulating its functional properties and in modulating excitatory synaptic transmission.
Footnotes
A.L.C. was supported by a fellowship from the Portuguese Research Council (PRAXIS XXI).
Correspondence should be addressed to Dr. Richard L. Huganir, Howard Hughes Medical Institute, Johns Hopkins University, Department of Neuroscience, 904A PCTB, 725 North Wolfe Street, Baltimore, MD 21205.
REFERENCES
- 1.Barria A, Derkach V, Soderling T. Identification of the Ca2+/calmodulin-dependent protein kinase II regulatory phosphorylation site in the α-amino-3-hydroxy-5-methyl-4-isoxazole-propionate-type glutamate receptor. J Biol Chem. 1997a;272:32727–32730. doi: 10.1074/jbc.272.52.32727. [DOI] [PubMed] [Google Scholar]
- 2.Barria A, Muller D, Derkach V, Griffith LC, Soderling TR. Regulatory phosphorylation of AMPA-type glutamate receptors by CaM-KII during long-term potentiation. Science. 1997b;276:2042–2045. doi: 10.1126/science.276.5321.2042. [DOI] [PubMed] [Google Scholar]
- 3.Bennett JA, Dingledine R. Topology profile for a glutamate receptor: three transmembrane domains and a channel-lining reentrant membrane loop. Neuron. 1995;14:373–384. doi: 10.1016/0896-6273(95)90293-7. [DOI] [PubMed] [Google Scholar]
- 4.Blackstone C, Murphy TH, Moss SJ, Baraban JM, Huganir RL. Cyclic AMP and synaptic activity-dependent phosphorylation of AMPA-preferring glutamate receptors. J Neurosci. 1994;14:7585–7493. doi: 10.1523/JNEUROSCI.14-12-07585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bliss TVP, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 6.Carriedo SG, Yin HZ, Weiss JH. Motor neurons are selectively vulnerable to AMPA/kainate receptor-mediated injury in vitro. J Neurosci. 1996;16:4069–4079. doi: 10.1523/JNEUROSCI.16-13-04069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi DW. Glutamate neurotoxicity and diseases of the nervous system. Neuron. 1988;1:623–624. doi: 10.1016/0896-6273(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 8.Greengard P, Jen J, Nairn AC, Stevens CF. Enhancement of the glutamate response by cAMP-dependent protein kinase in hippocampal neurons. Science. 1991;253:1135–1138. doi: 10.1126/science.1716001. [DOI] [PubMed] [Google Scholar]
- 9.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 10.Hollmann M, Maron C, Heinemann S. N-glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron. 1994;13:1331–1343. doi: 10.1016/0896-6273(94)90419-7. [DOI] [PubMed] [Google Scholar]
- 11.Kameyama K, Lee H-K, Bear MF, Huganir RL. Involvement of a postsynaptic protein kinase A substrate in the expression of homosynaptic long-term depression. Neuron. 1998;21:1163–1175. doi: 10.1016/s0896-6273(00)80633-9. [DOI] [PubMed] [Google Scholar]
- 12.Keinänen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-sensitive glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- 13.Keller BU, Hollmann M, Heinemann S, Konnerth A. Calcium influx through subunits GluR1/GluR3 of kainate/AMPA receptor channels is regulated by cAMP-dependent protein kinase. EMBO J. 1992;11:891–896. doi: 10.1002/j.1460-2075.1992.tb05127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köhler M, Kornau H-C, Seeburg PH. The organization of the gene for the functionally dominant α-amino-3-hydroxy-5-methyl-isoxazole-4-propionic acid receptor subunit GluR-B. J Biol Chem. 1994;269:17367–17370. [PubMed] [Google Scholar]
- 15.Lee H-K, Kameyama K, Huganir RL, Bear MF. NMDA induces long-term synaptic depression and dephosphorylation of the GluR1 subunit of AMPA receptors in hippocampus. Neuron. 1998;21:1151–1162. doi: 10.1016/s0896-6273(00)80632-7. [DOI] [PubMed] [Google Scholar]
- 16.Liman ER, Knapp AG, Dowling JE. Enhancement of kainate-gated currents in retinal horizontal cells by cyclic AMP-dependent protein kinase. Brain Res. 1989;481:399–402. doi: 10.1016/0006-8993(89)90822-6. [DOI] [PubMed] [Google Scholar]
- 17.Linden DJ. Long-term synaptic depression in the mammalian brain. Neuron. 1994;12:457–472. doi: 10.1016/0896-6273(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 18.Mammen A, Kameyama K, Roche KW, Huganir RL. Phosphorylation of the α-amino-3-hydroxy-5-methyl isoxazole-4-propionic acid receptor GluR1 subunit by calcium-calmodulin-dependent kinase II. J Biol Chem. 1997;272:32528–32533. doi: 10.1074/jbc.272.51.32528. [DOI] [PubMed] [Google Scholar]
- 19.Mammen A, Kamboj S, Huganir RL. Protein phosphorylation of ligand-gated ion channels. Methods Enzymol. 1999;294:353–370. doi: 10.1016/s0076-6879(99)94022-3. [DOI] [PubMed] [Google Scholar]
- 20.Mano I, Teichberg VI. A tetrameric subunit stoichiometry for a glutamate receptor channel complex. NeuroReport. 1998;9:327–331. doi: 10.1097/00001756-199801260-00027. [DOI] [PubMed] [Google Scholar]
- 21.Monyer H, Seeburg PH, Wisden W. Glutamate-operated channels: developmentally early and mature forms arise by alternative splicing. Neuron. 1991;6:799–810. doi: 10.1016/0896-6273(91)90176-z. [DOI] [PubMed] [Google Scholar]
- 22.Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP. A molecular determinant for submillisecond desensitization in glutamate receptors. Nature. 1994;266:1059–1062. doi: 10.1126/science.7973663. [DOI] [PubMed] [Google Scholar]
- 23.Moss SJ, Smart TG. Modulation of amino acid-gated ion channels by protein phosphorylation. Int Rev Neurobiol. 1996;39:1–52. doi: 10.1016/s0074-7742(08)60662-5. [DOI] [PubMed] [Google Scholar]
- 24.Nakazawa K, Mikawa S, Hashikawa T, Ito M. Transient and persistent phosphorylation of AMPA-type glutamate receptor subunits in cerebellar Purkinje cells. Neuron. 1995;15:697–709. doi: 10.1016/0896-6273(95)90157-4. [DOI] [PubMed] [Google Scholar]
- 25.Page KJ, Everitt BJ. The distribution of neurons coexpressing immunoreactivity to AMPA-sensitive glutamate receptor subtypes (GluR1–4) and nerve growth factor receptor in the rat basal forebrain. Eur J Neurosci. 1995;7:1022–1033. doi: 10.1111/j.1460-9568.1995.tb01090.x. [DOI] [PubMed] [Google Scholar]
- 26.Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- 27.Raman IM, Zhang S, Trussel LO. Pathway-specific variants of AMPA receptors and their contribution to neuronal signaling. J Neurosci. 1994;14:4998–5010. doi: 10.1523/JNEUROSCI.14-08-04998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reimann ER, Behman RB. Catalytic subunit of cAMP-dependent protein kinase. Methods Enzymol. 1983;99:51–55. doi: 10.1016/0076-6879(83)99039-0. [DOI] [PubMed] [Google Scholar]
- 29.Roche KW, Tingley WG, Huganir RL. Glutamate receptor phosphorylation and synaptic plasticity. Curr Opin Neurobiol. 1994;4:383–388. doi: 10.1016/0959-4388(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 30.Roche KW, O’Brien RJ, Mammen AL, Bernhardt J, Huganir RL. Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron. 1996;16:1179–1188. doi: 10.1016/s0896-6273(00)80144-0. [DOI] [PubMed] [Google Scholar]
- 31.Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- 32.Rubio ME, Wenthold RJ. Glutamate receptors are selectively targeted to postsynaptic sites in neurons. Neuron. 1997;18:939–950. doi: 10.1016/s0896-6273(00)80333-5. [DOI] [PubMed] [Google Scholar]
- 33.Silver RA, Traynelis SF, Cull-Candy SG. Rapid-time-course miniature and evoked excitatory currents at cerebellar synapses. Nature. 1992;355:163–166. doi: 10.1038/355163a0. [DOI] [PubMed] [Google Scholar]
- 34.Smith DO, Franke C, Rosenheimer J, Zufall F, Hatt H. Glutamate-activated channels in adult rat spinal cord cells. J Neurophysiol. 1991;66:369–379. doi: 10.1152/jn.1991.66.2.369. [DOI] [PubMed] [Google Scholar]
- 35.Swanson GT, Kamboj SK, Cull-Candy SG. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J Neurosci. 1997;17:58–69. doi: 10.1523/JNEUROSCI.17-01-00058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tingley WG, Ehlers MD, Kameyama K, Doherty C, Ptak JB, Riley CT, Huganir RL. Characterization of protein kinase A and protein kinase C phosphorylation of the N-methyl-d-aspartate receptor NR1 subunit using phosphorylation site-specific antibodies. J Biol Chem. 1997;272:5157–5166. doi: 10.1074/jbc.272.8.5157. [DOI] [PubMed] [Google Scholar]
- 37.Tomiyama M, Rodriguez-Puertas R, Cortés R, Christnacher A, Sommer B, Pazos A, Palacios JM, Mengod G. Differential regional distribution of AMPA receptor subunit messenger RNAs in the human spinal cord as visualized by in situ hybridization. Neuroscience. 1996;75:901–915. doi: 10.1016/0306-4522(96)00321-1. [DOI] [PubMed] [Google Scholar]
- 38.Wang L-Y, Salter MW, MacDonald JF. Regulation of kainate receptors by cAMP-dependent protein kinase and phosphatases. Science. 1991;253:1132–1135. doi: 10.1126/science.1653455. [DOI] [PubMed] [Google Scholar]
- 39.Wo ZG, Oswald RE. A topological analysis of goldfish kainate receptors predicts three transmembrane segments. J Biol Chem. 1995;270:2000–2009. doi: 10.1074/jbc.270.5.2000. [DOI] [PubMed] [Google Scholar]
- 40.Woodgett JR, Hunter T. Isolation and characterization of two distinct forms of protein kinase C. J Biol Chem. 1987;262:4836–4843. [PubMed] [Google Scholar]
- 41.Wyllie DJA, Traynelis SF, Cull-Candy SG. Evidence for more than one type of non-NMDA receptor in outside-out patches from cerebellar granule cells of the rat. J Physiol (Lond) 1993;463:193–226. doi: 10.1113/jphysiol.1993.sp019591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo J, De Jager PL, Takahashi KA, Jiang W, Linden DJ, Heintz N. Neurodegeneration in Lurcher mice caused by mutation in D2 glutamate receptor gene. Nature. 1997;388:769–773. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]