Abstract
Desensitization of cannabinoid receptor signaling by a G-protein coupled receptor kinase (GRK) was examined using theXenopus oocyte expression system. Application of a CB1 agonist, WIN 55,212-2, evoked a concentration-dependent increase in K+ conductance (Kir3) in oocytes coexpressing rat CB1 with the G-protein-gated, inwardly rectifying K+ channels Kir3.1 and Kir3.4. Desensitization was slight during continuous agonist application in the absence of GRK and arrestin. However, coexpression of GRK3 and β-arrestin 2 (β-arr2) caused profound homologous CB1 receptor desensitization, supporting the hypothesis that GRK3 and β-arr2 effectively produce CB1 receptor desensitization. To identify the regions of the CB1 receptor responsible for GRK3- and β-arr2-mediated desensitization, we constructed several CB1 receptor mutants. Truncation of the C-terminal tail of CB1 receptor at residue 418 (Δ418) almost completely abolished desensitization but did not affect agonist activation of Kir3. In contrast, truncation at residues 439 and 460 did not significantly affect GRK3- and β-arr2-dependent desensitization. A deletion mutant (Δ418–439) did not desensitize, indicating that residues within this region are important for GRK3- and β-arr2-mediated desensitization. Phosphorylation in this region was likely involved in desensitization, because mutation of either of two putative phosphorylation sites (S426A or S430A) significantly attenuated desensitization. CB1 receptors rapidly internalize after activation by agonist. Phosphorylation of S426 or S430 was not necessary for internalization, because the S426A/S430A CB1 mutant internalized when stably expressed in AtT20 cells. These studies establish that CB1 desensitization can be regulated by a GRK and that different receptor domains are involved in GRK- and β-arrestin-dependent desensitization and CB1 internalization.
Keywords: cannabinoid, desensitization, inwardly rectifying potassium channel, G-protein-coupled receptor, β-arrestin, G-protein coupled receptor kinase, phosphorylation
Cannabinoids produce their characteristic behavioral effects as a consequence of binding to a G-protein-coupled receptor, the CB1 cannabinoid receptor (Matsuda et al., 1990; Matsuda, 1997). The abundance of these receptors and the discovery of several endogenous ligands (Devane et al., 1992; Stella et al., 1997) suggest that an endogenous cannabinoid neuromodulatory system serves an important physiological role (DiMarzo et al., 1994). Cellular consequences of CB1 receptor activation include inhibition of adenylyl cyclase, activation of mitogen-activated protein kinase, and modulation of ion channels (Pertwee, 1993). The CB1 receptor activates at least two classes of potassium channels, the voltage-dependent potassium A current and inwardly rectifying potassium channels (GIRK or Kir3 channels) (Matsuda, 1997). Potassium channels often control the resting membrane potential of neurons and play a major role in determining excitability. Previous studies from our laboratories demonstrated that CB1 receptor activated Kir3 or GIRK channels in Xenopus oocytes (Henry and Chavkin, 1995) and the corticotroph-like cell line AtT20 (Mackie et al., 1995; Garcia et al., 1998), suggesting that activation of these channels may be a key effector mechanism for cannabinoid action.
Tolerance develops rapidly during the chronic administration of cannabinoids. Receptor desensitization or uncoupling has been consistently implicated as one of the molecular events underlying the onset of tolerance in many systems (Appleyard et al., 1997; Kovoor et al., 1997; Smith et al., 1988). Desensitization of G-protein-coupled receptors (GPCRs) is often associated with phosphorylation of the receptor by G-protein-coupled receptor kinases (GRKs), followed by binding of β-arrestin (β-arr) and a reduction in affinity for G-proteins (Zhang et al., 1997; Krupnick and Benovic, 1998). This sequence of events effectively attenuates signaling by the GPCR and its ligand. Recent studies have demonstrated that GRK and β-arr are required for δ opioid receptor desensitization (Kovoor et al., 1997). β-Arr also may serve other roles in the regulation of GPCR signaling. For example, β-arr has been proposed to function as an adaptor between phosphorylated β2-adrenergic receptors and clathrin-coated endocytic pits, thus directing phosphorylated receptor to this endocytotic pathway (Lin et al., 1997). CB1 receptor exhibits agonist-induced receptor internalization (Hsieh et al., 1999). However, the mechanisms underlying these phenomena for CB1 receptors have not been elucidated. In particular, it is unknown whether β-arr is involved in CB1 internalization. In the present study we used theXenopus oocyte expression system and AtT20 cells stably expressing CB1 receptors to study CB1 receptor coupling to G-protein-gated, inwardly rectifying K+ channels. CB1-expressing AtT20 cells were also used to study the relationship between desensitization and internalization of the CB1 receptor. We determined that GRK3 and β-arr2 were able to mediate agonist-dependent CB1 receptor desensitization, and we found that distinct domains of the CB1 receptor were involved in desensitization and internalization.
MATERIALS AND METHODS
CB1 receptor mutagenesis. Amplicons for CB1 receptor truncation mutants Δ418, Δ439, and Δ460 were produced by PCR as follows. One nanogram of rat CB1 template (Mackie et al., 1995), 60 pmol of dNTPs (Life Technologies, Grand Island, NY), 4.0 U ofPfu polymerase (Stratagene, La Jolla, CA), and 25 pmol of each primer (Table 1; Life Technologies) were combined in a final reaction volume of 100 μl. PCR was performed in a DNA thermal cycler (model 480; Perkin-Elmer, Foster City, CA) for 28 cycles with annealing temperatures appropriate for the oligonucleotide sequences. The CB1 sense primer (Table 1) was used for all mutants in combination with the appropriate antisense primer (Table1).
Table 1.
Primers used to construct CB1 cannabinoid receptor mutants
| Primer | Sequence (5′–3′) |
|---|---|
| CB1 sense | CGGGATCCATGAAGTCGATCCTAGATGGCTTG |
| CB1 antisense | CGGAATTCACAAAAGCAGCTCACAGAGC |
| Δ418 antisense | CGGAATTCTTATTCGCACGAAGGGAACATGCTTCG |
| Δ439 antisense | CGGAATTCTTAGTTGGCGTGCTTGTGCAG |
| Δ460 antisense | CGGAATTCTTACTTCGCGATCTTAACCGTGCTCTTGAT |
| T419A antisense | TAGAGGCTGTGCGGCTTCGCA |
| T419A sense | TGCGAAGGCGCCGCACAGCCTCTA |
| S426A antisense | GTGCTTGTGCAGGCAGTCTGAGTCCCC |
| S426A sense | GGGGACTCAGACTGCCTGCACAAGCAC |
| S430A antisense | GCAGTCTGCGTCCCCCATGCTGTT |
| S430A sense | AACAGCATGGGGGACGCAGACTGC |
| S426A/S430A antisense | GTCTGCGTCCCCCATGGCGTTGTCTAGAGGCTGTGCGGCGCC |
| S426A/S430A sense | GGCGCCGCACAGCCTCTAGACAACGCCATGGGGGACGCAGACGAC |
| ΔG418-N438 antisense | TTCGCACGAAGGGAACATGC |
| ΔG418-N438 sense | ATGTTCCCTTCGTGCGAAAACACAGCCAGCATGCACAGG |
Sense refers to the forward primer, and antisense to the reverse primer for each reaction.
PCR products were ethanol-precipitated, pelleted, washed in 70% ethanol, vacuum-dried, resuspended in 10 mm Tris and 0.1 mm EDTA, pH 8.0 (T1/10E) buffer, and gel-purified on 1% Seaplaque agarose (FMC Bioproducts, Rockland, ME). Amplicons detected by ethidium bromide staining were excised and purified with 0.22 μm Micropure separators (Amicon, Beverly, MA). Eluted DNA was phenol-chloroform-extracted and ethanol-precipitated.
CB1 receptor mutants T419A, S426A, S430A, S426A/S430A and ΔG418-N438 were produced by overlap extension of the PCR (Ho et al., 1989) using wild-type rat CB1 as template. For each mutant, two rounds of PCR were performed. In the first round, two separate reactions were performed. In the first the primers were CB1 sense and the mutant antisense (Table1). In the second the primers were the mutant sense and CB1 antisense (Table 1). CB1 receptor mutant S426A/S430A was used as starting template in overlap–extension of the PCR to produce CB1 receptor mutant T419A/S426A/S430A.
Amplicon templates produced with Pfu polymerase in the first round of PCR were joined in overlap–extension of the PCR with 2.5 U ofTaq DNA polymerase (Qiagen, Valencia, CA) using CB1 sense and antisense primers (Table 1). Purified PCR products and vector pcDNA3 (Invitrogen, Carlsbad, CA) were digested with BamHI and EcoRI restriction endonucleases (New England Biolabs, Beverly, MA), phenol-chloroform-extracted, ethanol-precipitated, pelleted, washed in 70% ethanol, vacuum-dried, and resuspended in T1/10E. The vector was dephosphorylated with shrimp alkaline phosphatase (Boehringer Mannheim, Indianapolis, IN) and gel-purified as above. Inserts and vector were ligated with T4 DNA ligase (New England Biolabs) overnight at 16°C. Orientation of the inserts allowed the T7 promoter to direct RNA transcripts (see below). Competent XL-1 BlueEscherichia coli (Stratagene) was transformed and plated on LB-amp-tet agar plates. Plasmid DNA was isolated from broth cultures started from amp-tet-resistant bacterial colonies (Qiagen), and the plasmid DNA was screened by BamHI and EcoRI restriction endonuclease (New England Biolabs) digestion. Mutations were confirmed by automated DNA sequencing (Applied Biosystems, Perkin-Elmer).
cDNA clones and cRNA synthesis. cDNA for Kir3.1 channel was obtained from Dr. Henry Lester (California Institute of Technology, Pasadena, CA) (GenBank accession number U01071). Kir3.4 [clone provided by Dr. John Adelman (Vollum Institute, Portland, OR), GenBank accession number X83584] and β-arr2 cDNA [clone provided by Dr. Robert Lefkowitz (Duke University, Durham, NC), GenBank accession number M91590] was first amplified by the utilization of Amplitaq DNA Polymerase (Perkin-Elmer Cetus, Norwalk, CT) in a standard PCR using oligonucleotides designed to add a T7 promoter region and a 45 base poly A tail. The rat GRK3 cDNA was provided by Dr. Shaun Coughlin (University of California at San Francisco, San Francisco, CA) (Vu et al., 1991). Plasmid templates were linearized before cRNA synthesis, and mMESSAGE MACHINE kit (Ambion) was used to generate capped cRNA.
Oocyte culture and injection. Oocytes were prepared as described (Kovoor et al., 1995) and were incubated in ND96 (96 mm NaCl, 2 mm KCl, 1 mmMgCl2, 1 mm CaCl2, and 5 mm HEPES, pH 7.5) solution supplemented with sodium pyruvate (2.5 mm) and gentamycin (50 μg/ml). cRNA was injected into oocytes (50 nl/oocyte) with a Drummond microinjector. cRNAs injected into each oocyte were as follows: CB1 receptor and its mutants, 2.5–4 ng; GRK3, 0.5 ng; β-arr2, 5 ng; GIRK Kir3.1 and Kir3.4, 0.02 ng each; and δ opioid receptor, 0.4 ng. All recordings were performed 2–4 d after injection.
Oocyte electrophysiology. Oocytes were clamped at −80 mV with two electrodes filled with 3 m KCl having resistances of 0.5–1.5 mΩ using a Geneclamp 500 amplifier and pCLAMP 6 software (Axon Instruments, Foster City, CA). All data were digitally recorded (Digidata, Axon Instruments, and Intel 386PC) and filtered at 500 HZ. Membrane current traces were also recorded using a chart recorder. To measure inwardly K+ currents flowing through the Kir3 channels, K+ concentration in the oocyte saline buffer was increased from 2 to 16 mm. The concentration of NaCl was decreased to maintain iso-osmolality.
AtT20 cell electrophysiology. Kir currents were recorded from AtT20 cells stably expressing wild-type or Δ418 truncated CB1 receptors as previously described (Mackie et al., 1995;Garcia et al., 1998). In the whole-cell configuration of the patch clamp (Hamill et al., 1981), cells were held at −45 mV and hyperpolarized to −100 mV for 50 msec every 5 sec. The extracellular potassium concentration was 40 mm, and the Kircurrent was defined as the component of the current blocked by 1 mm Ba2+. The average current during the hyperpolarization was determined and plotted versus time.
Internalization. The CB1 receptor mutant S426A/S430A was stably expressed in AtT20 cells (Mackie et al., 1995). Cells were stimulated and CB1 receptors were detected with a CB1-specific antibody in fixed cells with confocal microscopy as previously described (Hsieh et al., 1999).
Statistical analysis. The Student’s t test was used for comparison of independent means. Statistical significance was defined as p < 0.05. Data from the dose–response experiments were fitted to a simple Emax model using the nonlinear regression analysis package NFIT to determine agonist EC50and 95% confidence intervals (95% CI).
Chemicals. [d-Pen2,5]enkephalin was from Peninsula Laboratories (San Carlos, CA). WIN 55,212-2 was from Research Biochemicals International (Natick, MA). SR 141716A was from the National Institute on Drug Abuse drug supply program (Research Triangle Park, NC).
RESULTS
To characterize CB1 receptor coupling to G-protein-gated inwardly rectifying K+ channels, cRNAs for CB1 receptor Kir3.1 and Kir3.4 channel subunits were injected into Xenopus oocytes. As shown in Figure1A, basal inward currents were observed when the KCl concentration increased from 2 to 16 mm in normal oocyte saline buffer. Superfusion of WIN 55,212-2 (WIN, 1 μm) further increased the inward current. The current activated by WIN showed inward rectification (Fig.1B). The WIN-evoked response was concentration-dependent, with an EC50 of 55 nm(45–64 nm, 95% CI; Fig. 1C). Activation of the inwardly rectifying current by WIN was completely reversed by application of the competitive antagonist SR 141716A (1 μm) (Fig. 1A). In the oocytes expressing only CB1 receptor and Kir3.1 and Kir3.4, modest desensitization (25 ± 3.3%;n = 9) was observed after prolonged (8 min) perfusion of 1 μm WIN (Fig. 1A). In this study, the amount of desensitization was defined as the percent decrease in WIN activation of the Kir3 current after 8 min of WIN application.
Fig. 1.

Coupling of the CB1 receptor to the G-protein-activated inward rectifier potassium channels Kir3.1 and Kir3.4. A, Representative trace showing the change in current during a typical experiment. A large inward current was apparent as the K+ concentration was increased from 2 to 16 mm in normal oocyte saline buffer. WIN (1 μm) in the buffer (16 mm K+) further increased the current. To detect any change in basal current after the agonist treatment (8 min), SR 141716A (1 μm), a CB1 receptor antagonist, was applied to displace WIN and reverse CB1-mediated activation of the current. The interpolated decrease in baseline current was plotted, as shown by the dashed line. The amount of desensitization was calculated as the percent change in response to WIN after 8 min. Current traces presented in subsequent figures show only the agonist-activated current adjusted for the change in baseline. B, WIN activation of inwardly rectifying K+ channels. TheI–V relationship was generated by steps from −140 to 40 mV after subtracting the current at the same potential in 16 mm K+ buffer. C, Concentration–response curve of WIN. Cumulatively higher concentrations of WIN were applied to the bath followed by perfusion with SR 141716A (1 μm). The agonist response at each concentration was normalized as a percentage of the maximal WIN response. Each point represents the mean response measured in 10–14 different oocytes.
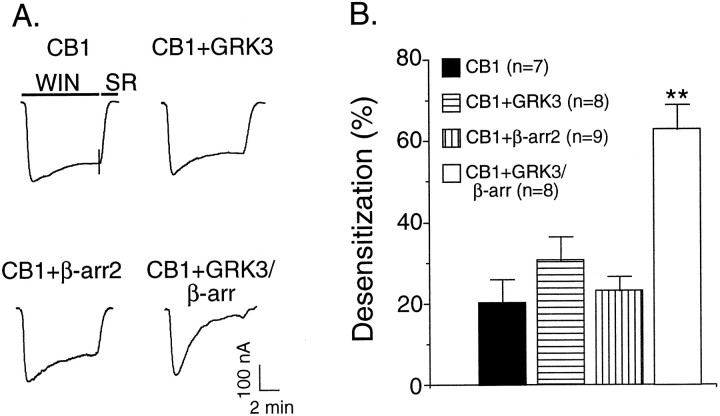
To examine whether GRK and β-arr enhanced CB1 receptor desensitization, GRK3 and β-arr2 were coexpressed with CB1 receptor and Kir3.1 and Kir3.4 channels. As shown above, treatment with WIN for 8 min only produced a very small desensitization in the absence of GRK3 and β-arr 2 (25 ± 3.3%). Expression of CB1 and Kir3.1 and Kir3.4 along with either GRK3 or β-arr2 alone did not significantly increase the desensitization rate (Fig.2A). However, coexpression both GRK3 and β-arr2 caused profound agonist-dependent desensitization (63 ± 6.1%; n = 8) (Fig.2A). These results suggested that GRK3 and β-arr2 were sufficient to induce CB1 receptor desensitization. To determine whether GRK3 and β-arr2 target the CB1 receptor, we next tested whether the GRK3- and β-arr2-mediated desensitization was homologous or heterologous. In the oocytes that were also injected with cRNA for the δ opioid receptor, brief application of 1 μm DPDPE (a δ opioid agonist) elicited a large inward current, 336 ± 39 nA (n = 10; Fig.3A). During perfusion with WIN, prominent desensitization of the CB1-mediated response was apparent (Fig. 3A,B). However, the amplitude of the current activated by a subsequent application of DPDPE was identical to the DPDPE-induced current in non-WIN-55,212-2-treated oocytes (Fig.3A,B). These data indicate that GRK3 and β-arr2 mediate homologous CB1 receptor desensitization, suggesting that they target CB1 receptors rather than a common downstream effector, such as the channel, G-protein, or accessory proteins.
Fig. 2.
GRK3 and β-arr2 were required for CB1 receptor desensitization. A, Representative traces showing that the coexpression of GRK3 and β-arr2 significantly increased desensitization of Kir current activated by WIN. Top left, Oocytes injected with mRNA for CB1 and Kir3 show modest desensitization during an 8 min exposure to 1 mm WIN 55,212-2. Top right, Addition of GRK3 mRNA to the injection mix does not enhance desensitization.Bottom left, Addition of β-arr2 mRNA also does not enhance desensitization. Bottom right, Addition of both GRK3 and β-arr2 mRNA to the injection mix significantly enhances desensitization. The short vertical lines through thefirst trace indicate the time of buffer switch from WIN to SR 141716A perfusion. Response was adjusted by baseline subtraction.B, Summary of data. Data are mean ± SEM; **p < 0.05 compared with oocytes not coexpressing GRK3 and β-arr2.
Fig. 3.
GRK3 and β-arr2 mediated homologous CB1 receptor desensitization. A, Representative traces show that current activated by WIN desensitized during prolonged treatment, whereas the DPDPE-mediated response was unaffected. OR, Opioid receptor; SR, SR 141716A. B, Summary of data. Open bars, Current at initial agonist application; filled bars, represent the current elicited by the indicated agonist after WIN application. DPDPE-elicited current was not affected by an intervening application of WIN, whereas the WIN-elicited current strongly desensitizes. Data are presented as mean ± SEM; **p < 0.05 (WIN-elicited current at the beginning vs end of WIN application).
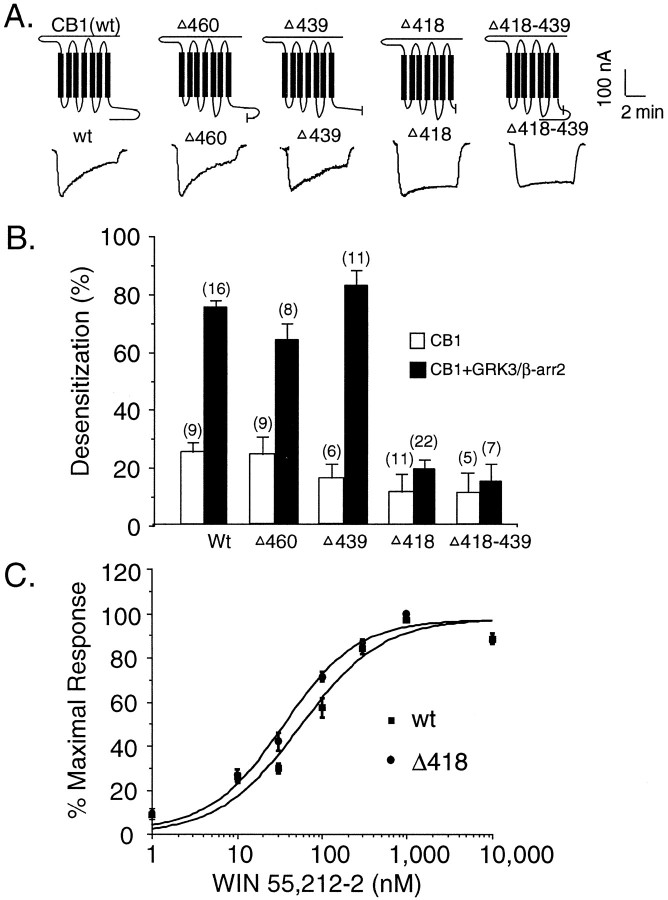
It is believed that phosphorylation of serine and threonine residues in either the third cytoplasmic loop or the C-terminal tail by GRK is the mechanism of GRK- and arrestin-mediated desensitization of many GPCRs (Freedman and Lefkowitz, 1996). To identify regions of the CB1 receptor required for desensitization, we first constructed several receptor mutants by successively shortening the CB1 receptor C-terminal tail (Fig. 4A). Truncation at residues 439 and 460 did not significantly affect WIN activation of Kir3 current activation or the GRK3- and β-arr2-mediated desensitization observed (Fig. 4A,B). In a manner similar to wild-type CB1 receptors, Kir3 activation by CB1 mutants Δ439 and Δ460 significantly desensitized (82 ± 5%;n = 11; and 63 ± 6%; n = 8, respectively). However, truncation at residue 418 caused a dramatic attenuation of desensitization (19 ± 3%; n = 22; Fig. 4A,B). To determine whether the lack of desensitization in the mutant Δ418 was attributable to gross alteration of receptor properties, concentration–response curves for the Δ418 mutant and wild-type receptor were generated (Fig.4C). The EC50 for WIN was similar for both wild-type CB1 and Δ418 mutant (55 and 36 nm, respectively). This result indicates that Δ418 mutant activation of Kir3 currents was similar to activation by wild-type CB1 receptor. Because only slight desensitization was observed with the Δ418 mutant, the 20 amino acid residues between 418 and 439 are likely to be critical for GRK- and β-arr2-mediated CB1 receptor desensitization. To test this hypothesis, we constructed a deletion mutant (Δ418–439) with these 20 amino acids removed. Indeed, this deletion mutant behaved similarly to Δ418, showing little GRK3- and β-arr2-mediated desensitization (15 ± 6%; n = 7; Fig. 4A,B).
Fig. 4.
The region between G418 and N419 in the CB1 receptor was critical for GRK3- and β-arr2-mediated CB1 receptor desensitization. A, Schematic of the rat CB1 receptor.Short vertical lines indicate the sites of truncation or deletion. Representative traces show that truncation mutant Δ418 and deletion mutant (Δ418–439) eliminated GRK3- and β-arr2-mediated desensitization. B, Summary of data. Data were collected from at least three separate experiments with three different oocyte donors. C, Concentration response curves of WIN in wild-type CB1 receptor and mutant (ΔG418). Cumulatively increasing concentrations of WIN were applied to the bath followed by perfusion with SR 141716A. Oocytes were injected with cRNA for Kir3.1 and Kir3.4 and CB1 receptors. Each pointrepresents the mean response measured in seven oocytes.wt, Wild type.
The oocyte system offers the convenience of allowing rapid screening of receptor mutants. However, we also wanted to determine whether Kir current activation desensitized in an excitable cell constitutively expressing Kir channels, without exogenous β-arr2 or GRK. In addition we wished to determine whether the domains of the receptor critical for desensitization were also involved in agonist-induced internalization (below). Because we measure internalization in AtT20 cells stably transfected with CB1 receptors, it was important to determine whether activation of Kir by cannabinoids in these cells desensitized in a manner similar to that inXenopus oocytes. For both these reasons we stably expressed CB1 and the CB1 truncation mutant Δ418 in AtT20 cells. We then determined the degree of desensitization of the current during 1–2 min of agonist exposure. Figure 5Ashows that for an individual cell expressing wild-type CB1, the current activated by 200 nm WIN 55,212-2 rapidly desensitizes. In contrast, the current activated in cells expressing the CB1 truncation Δ418 shows little desensitization (Fig. 5B). Figure 5,C and D, shows the current amplitude before and during WIN 55,212-2 application as a percent of the maximum current for several cells. The current activated by WIN 55,212-2 in cells expressing wild-type CB1 declines with a τ of ∼30 sec (temperature, 22–23°), whereas the current activated in cells expressing the Δ418 truncation shows no detectable decrease during this time. These results suggest that similar regions in CB1 are involved in desensitization of Kir current in AtT20 cells and oocytes.
Fig. 5.
The C terminus of CB1 is also required for desensitization of GIRK activation in AtT20 cells. A, In AtT20 cells stably expressing rat CB1 receptors, 200 nm WIN 55,212-2 activates an inward current that rapidly desensitizes.B, In AtT20 cells stably expressing the CB1 truncation mutant ΔG418, 200 nm WIN 55,212-2 activates an inward current, but this current shows little desensitization.C, Aggregate data showing the rapid desensitization of inward current activated by WIN 55,212-2 in cells expressing rat CB1 (n = 5). D, Aggregate data showing no desensitization of the inward current activated by WIN 55,212-2 in cells expressing the CB1 truncation ΔG418 (n = 10).
Was desensitization attributable to the phosphorylation of residues between residues 418 and 439 in CB1 receptor? There are three possible phosphorylation sites (T419, S426, and S430) in this region (Fig.6A). To test whether phosphorylation of any of these three sites was necessary for desensitization, we constructed five mutants in which combinations of the sites were mutated to alanine as diagrammed (Fig.6A) Mutation of T419 to alanine did not prevent desensitization (Fig. 6B), suggesting phosphorylation at this site by GRK3 is not critical. In contrast, the single mutation of either S426A or S430A significantly attenuated the desensitization (S426A, 23 ± 4%; S430A, 26 ± 3.5). A similar low degree of desensitization was observed with the double mutant S426A/S430A (25 ± 3%; Fig. 6). Our data indicate that phosphorylation of both S426 and S430 of the CB1 receptor by GRK3 may be responsible for its homologous desensitization by GRK3 and β-arr2.
Fig. 6.
Phosphorylation of serine 426 and serine 430 in the C terminus of CB1 likely underlies GRK3- and β-arr2-mediated CB1 receptor desensitization. A, Schematic representation of rat CB1 receptor. The amino acid sequence between G418 and N439 is shown using single-letter amino acid abbreviations. Five mutants were constructed based on sequentially substituting serine and threonine residues with alanine in this region. B, Summary data showing that single mutation of S426 or S430 but not T419 effectively blocks GRK- and β-arr-mediated desensitization. Data were collected from at least three separate experiments with three different oocyte donors. Data are means ± SEM.
We next determined the role of S426/S430 in agonist-induced internalization of the CB1 receptor. When expressed in AtT20 cells, CB1 receptors readily internalize in response to agonist (Hsieh et al., 1999). In cells expressing wild-type CB1 receptors, the receptor is primarily found at the cell surface (Fig.7A). When these cells are stimulated with 100 nm WIN 55,212-2 for 30 min, CB1 receptors internalize (Fig. 7B). Previously, we found that residues 460–464 of the receptor were required for internalization (Hsieh et al., 1999). We now wanted to determine whether phosphorylation of S426 or S430 was also required. That is, were the residues phosphorylated by GRK3, and presumably involved in β-arrestin 2 binding, necessary for agonist-induced internalization of the CB1 receptor? In AtT20 cells stably expressing the CB1 S426A/S430A mutant, the receptor was found at the plasma membrane (Fig.7C). However when these cells were stimulated with 100 nm WIN for 30 min, substantial internalization of the receptor was evident (Fig. 7D). Thus in AtT20 cells, phosphorylation of the CB1 receptor at S426 and S430 is not required for its internalization. These results suggest that different receptor domains are required for GRK/β-arr-dependent desensitization and receptor internalization.
Fig. 7.
S426 and S430 are not required for CB1 receptor internalization. A, In unstimulated AtT20 cells stably expressing wild-type CB1 receptor, the receptor is primarily found on the cell surface (arrows). B, After 30 min stimulation with 100 nm WIN 55,212-2, CB1 receptors are predominantly intracellular (arrowheads).C, In unstimulated AtT20 cells stably expressing the mutant CB1 receptor S426A/S430A, the receptor is primarily found on the cell surface (arrows). After 30 min stimulation with 100 nm WIN 55,212-2, CB1 receptors are internalized, as in cells expressing the wild-type receptor (arrowheads).
DISCUSSION
The principal finding of this study is that GRK3 and β-arrestin can cause profound desensitization of CB1 cannabinoid receptor-mediated activation of Kir3 channels. Using Xenopusoocytes and a mutagenic strategy, we defined a region (residues 418–439) of the CB1 receptor critical for GRK3- and β-arr2-mediated desensitization. It is likely that GRK3 phosphorylation of the CB1 receptor in this region underlies desensitization, because mutating either of two serines (S426 or S430) in this region eliminates desensitization. Furthermore, phosphorylation of this region of the receptor does not seem to be involved in agonist-induced internalization in AtT20 cells, because internalization proceeds normally when both S426 and S430 are mutated to alanine. These results also suggest that a β-arrestin interaction with phosphorylated S426 and S430 is not required for CB1 internalization. The results of this study, combined with those of our earlier work, clearly demonstrate that distinct domains of the CB1 receptor are involved in CB1 receptor internalization and desensitization.
Sustained administration of cannabinoids leads to rapid development of tolerance in both animals and humans (Abood and Martin, 1992; Martin et al., 1994). Tolerance does not involve changes in pharmacokinetics (Dewey, 1986). It also does not correlate with changes in receptor density (Abood and Martin, 1992; Pertwee, 1997). Chronic administration of Δ-(9)-tetrahydrocannabinol leads to an uncoupling of CB1 receptors from G-proteins, as measured by a decrease in WIN 55,212-2-stimulated GTPγS binding (Sim et al., 1996). Our results suggest a possible mechanism for this uncoupling, namely the phosphorylation of the receptor by a G-protein-coupled receptor kinase.
At least two families of protein kinases, GRKs and second messenger-dependent kinases, are involved in phosphorylation of GPCRs (Freedman and Lefkowitz, 1996). G-protein-coupled receptor kinases specifically phosphorylate agonist-activated receptors, facilitating the binding of an inhibitory protein (arrestin) to the phosphorylated receptor, thereby uncoupling the receptor from its G-protein(s) and inducing receptor-specific desensitization (homologous desensitization). Desensitization of several GPCRs [β2-adrenergic receptors (AR), α1-AR, α2-AR, and m2-muscarinic, thrombin, and opioid receptors, among others] involves GRK-mediated phosphorylation (Inglese et al., 1993; Freedman and Lefkowitz, 1996; Kovoor et al., 1997). When determined, the sites of phosphorylation are localized to serines and threonines in the carboxyl-terminal tail or the third of intracellular loop (Bouvier et al., 1988; Dohlman et al., 1987; Liggett et al., 1992). Whether a serine or threonine is phosphorylated by a GRK appears to depend on the overall topological structure of the activated receptor rather than on a defined linear recognition sequence (Chen et al., 1993). Second messenger-dependent kinases (e.g., PKA and PKC) phosphorylate a variety of proteins. This phosphorylation may mediate a generalized cellular hyporesponsiveness, thus sometimes causing heterologous desensitization. (Freedman and Lefkowitz, 1996).
CB1 and opioid receptors share common features in their signal transduction and pharmacology (Martin et al., 1994; Pertwee, 1997). Opioid receptor desensitization requires GRKs and arrestin. Opioid receptor desensitization was blocked by the expression of a dominant negative GRK and enhanced by overexpression of GRKs (Raynor et al., 1994; Pei et al., 1995). In Xenopus oocytes, homologous agonist-induced δ opioid receptor desensitization of GIRK activation requires coexpression of GRK3 and β-arrestin 2. Phosphorylation of serine and threonine residues in the receptor cytoplasmic tail by GRK3 underlies the desensitization (Kovoor et al., 1997). The present study found, similarly to the δ opioid receptor, that desensitization of the CB1 receptor is also GRK3- and β-arrestin 2-dependent. This is the first evidence that GRKs and arrestin may produce desensitization of a CB1 receptor-mediated response. Furthermore, our results indicate that phosphorylation of serines (S426 and/or S430) in the cytoplasmic tail of the CB1 receptor may be the molecular mechanism of homologous CB1 receptor desensitization.
Together, the results of this and our earlier study show that for the CB1 cannabinoid receptor the processes of desensitization and internalization can be clearly dissociated. Previously we found that residues 460–464 of the CB1 receptor were required for internalization (Hsieh et al., 1999). The results from the present study demonstrate that these residues are not needed for desensitization of Kir current activation by the CB1 receptor inXenopus oocytes (Fig. 4). Furthermore, because the nondesensitizing S426A/S430A CB1 mutant internalizes normally, it is likely that the residues phosphorylated by a GRK and involved in β-arrestin binding are not required for internalization. Additional support that the region involved in desensitization is unimportant for internalization comes from the observation that a nondesensitizing Δ418–439 CB1 deletion mutant internalizes similarly to the wild-type CB1 receptor (H. Kim, A. Sorom, and K. Mackie, unpublished observation). Internalization of the CB1 cannabinoid receptor occurs via clathrin-coated pits (Hsieh et al. 1999) in a dynamin-dependent manner (Kim and Mackie, unpublished observation). Thus, if β-arrestin is important for CB1 internalization, it must be interacting with residues other than those that we have found in the current study to be critical determinants for desensitization.
In summary, our results suggest that phosphorylation of the CB1 receptor by a G-protein receptor kinase followed by binding of β-arrestin may underlie the tolerance that develops during prolonged administration of cannabis or cannabinoids. Receptor internalization does not seem to be involved in rapid desensitization, because either process can proceed independently of the other.
Footnotes
This work was supported by National Institutes of Health Research Grants DA08934, DA00286, DA11322, NS01588, and DA04123 (to C.C.) and the W. M. Keck Foundation.
Correspondence should be addressed to Ken Mackie, Department of Anesthesiology, Box 356540, University of Washington, Seattle, WA 98195-6540.
REFERENCES
- 1.Abood ME, Martin BR. Neurobiology of marijuana abuse. Trends Pharmacol Sci. 1992;13:201–206. doi: 10.1016/0165-6147(92)90064-d. [DOI] [PubMed] [Google Scholar]
- 2.Appleyard SM, Patterson TA, Jin WZ, Chavkin C. Agonist-induced phosphorylation of the kappa-opioid receptor. J Neurochem. 1997;69:2405–2412. doi: 10.1046/j.1471-4159.1997.69062405.x. [DOI] [PubMed] [Google Scholar]
- 3.Bouvier M, Hausdorff WP, De BA, O’Dowd BF, Kobilka BK, Caron MG, Lefkowitz RJ. Removal of phosphorylation sites from the beta 2-adrenergic receptor delays onset of agonist-promoted desensitization. Nature. 1988;333:370–373. doi: 10.1038/333370a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen CY, Dion SB, Kim CM, Benovic JL. Beta-adrenergic receptor kinase. Agonist-dependent receptor binding promotes kinase activation. J Biol Chem. 1993;268:7825–7831. [PubMed] [Google Scholar]
- 5.Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 6.Dewey WL. Cannabinoid pharmacology. Pharmacol Rev. 1986;38:151–178. [PubMed] [Google Scholar]
- 7.DiMarzo V, Fontana A, Cadas H, Schinelli S, Cimino G, Schwartz JC, Piomelli D. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature. 1994;372:686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- 8.Dohlman HG, Bouvier M, Benovic JL, Caron MG, Lefkowitz RJ. The multiple membrane spanning topography of the beta 2-adrenergic receptor. Localization of the sites of binding, glycosylation, and regulatory phosphorylation by limited proteolysis. J Biol Chem. 1987;262:14282–14288. [PubMed] [Google Scholar]
- 9.Freedman NJ, Lefkowitz RJ. Desensitization of G protein-coupled receptors. Recent Prog Horm Res. 1996;51:319–351. [PubMed] [Google Scholar]
- 10.Garcia DE, Brown S, Hille B, Mackie K. Protein kinase C disrupts cannabinoid actions by phosphorylation of the CB1 cannabinoid receptor. J Neurosci. 1998;18:2834–2841. doi: 10.1523/JNEUROSCI.18-08-02834.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth F. Improved patch-clamp techniques for high resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 12.Henry DJ, Chavkin C. Activation of inwardly rectifying potassium channels (GIRK1) by co-expressed rat brain cannabinoid receptors in Xenopus oocytes. Neurosci Lett. 1995;186:91–94. doi: 10.1016/0304-3940(95)11289-9. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh C, Brown S, Derleth C, Mackie K (1999) Internalization and recycling of the CB1 cannabinoid receptor. J Neurochem, in press. [DOI] [PubMed]
- 14.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 15.Inglese J, Freedman NJ, Koch WJ, Lefkowitz RJ. Structure and mechanism of the G protein-coupled receptor kinases. J Biol Chem. 1993;268:23735–23738. [PubMed] [Google Scholar]
- 16.Kovoor A, Henry DJ, Chavkin C. Agonist-induced desensitization of the mu opioid receptor-coupled potassium channel (GIRK1). J Biol Chem. 1995;270:589–595. doi: 10.1074/jbc.270.2.589. [DOI] [PubMed] [Google Scholar]
- 17.Kovoor A, Nappey V, Kieffer BL, Chavkin C. Mu and delta opioid receptors are differentially desensitized by the coexpression of beta-adrenergic receptor kinase 2 and beta-arrestin 2 in Xenopus oocytes. J Biol Chem. 1997;272:27605–27611. doi: 10.1074/jbc.272.44.27605. [DOI] [PubMed] [Google Scholar]
- 18.Krupnick JG, Benovic JL. The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annu Rev Pharmacol Toxicol. 1998;38:289–319. doi: 10.1146/annurev.pharmtox.38.1.289. [DOI] [PubMed] [Google Scholar]
- 19.Liggett SB, Ostrowski J, Chesnut LC, Kurose H, Raymond JR, Caron MG, Lefkowitz RJ. Sites in the third intracellular loop of the alpha 2A-adrenergic receptor confer short term agonist-promoted desensitization. Evidence for a receptor kinase-mediated mechanism. J Biol Chem. 1992;267:4740–4746. [PubMed] [Google Scholar]
- 20.Lin FT, Krueger KM, Kendall HE, Daaka Y, Fredericks ZL, Pitcher JA, Lefkowitz RJ. Clathrin-mediated endocytosis of the beta-adrenergic receptor is regulated by phosphorylation/dephosphorylation of beta-arrestin1. J Biol Chem. 1997;272:31051–31057. doi: 10.1074/jbc.272.49.31051. [DOI] [PubMed] [Google Scholar]
- 21.Mackie K, Lai Y, Westenbroek R, Mitchell R. Cannabinoids activate an inwardly rectifying potassium conductance and inhibit Q-type calcium currents in AtT20 cells transfected with rat brain cannabinoid receptor. J Neurosci. 1995;15:6552–6561. doi: 10.1523/JNEUROSCI.15-10-06552.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin BR, Welch SP, Abood M. Progress toward understanding the cannabinoid receptor and its second messenger systems. Adv Pharmacol. 1994;25:341–397. doi: 10.1016/s1054-3589(08)60437-8. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda LA. Molecular aspects of cannabinoid receptors. Crit Rev Neurobiol. 1997;11:143–166. doi: 10.1615/critrevneurobiol.v11.i2-3.30. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- 25.Pei G, Kieffer BL, Lefkowitz RJ, Freedman NJ. Agonist-dependent phosphorylation of the mouse delta-opioid receptor: involvement of G protein-coupled receptor kinases but not protein kinase C. Mol Pharmacol. 1995;48:173–177. [PubMed] [Google Scholar]
- 26.Pertwee R. The evidence for the existence of cannabinoid receptors. Gen Pharmacol. 1993;24:811–824. doi: 10.1016/0306-3623(93)90154-p. [DOI] [PubMed] [Google Scholar]
- 27.Pertwee RG. Pharmacology of cannabinoid CB1 and CB2 receptors. Pharmacol Ther. 1997;74:129–180. doi: 10.1016/s0163-7258(97)82001-3. [DOI] [PubMed] [Google Scholar]
- 28.Raynor K, Kong H, Hines J, Kong G, Benovic J, Yasuda K, Bell GI, Reisine T. Molecular mechanisms of agonist-induced desensitization of the cloned mouse kappa opioid receptor. J Pharmacol Exp Ther. 1994;270:1381–1386. [PubMed] [Google Scholar]
- 29.Sim LJ, Hampson RE, Deadwyler SA, Childers SR. Effects of chronic treatment with Delta(9)-tetrahydrocannabinol on cannabinoid-stimulated [S-35]GTP gamma S autoradiography in rat brain. J Neurosci. 1996;16:8057–8066. doi: 10.1523/JNEUROSCI.16-24-08057.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith AP, Law P-Y, Loh HH. Role of opioid receptors in narcotic tolerance/dependence, Chap 13 (Pasternak GW, ed), pp 441–476. Humana; Clifton, NJ: 1988. [Google Scholar]
- 31.Stella N, Schweitzer P, Piomelli D. A second endogenous cannabinoid that modulates long-term potentiation. Nature. 1997;388:773–778. doi: 10.1038/42015. [DOI] [PubMed] [Google Scholar]
- 32.Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Ferguson SS, Barak LS, Aber MJ, Giros B, Lefkowitz RJ, Caron MG. Molecular mechanisms of G protein-coupled receptor signaling: role of G protein-coupled receptor kinases and arrestins in receptor desensitization and resensitization. Receptors Channels. 1997;5:193–199. [PubMed] [Google Scholar]