Abstract
Photic entrainment of insect circadian rhythms can occur through either extraretinal (brain) or retinal photoreceptors, which mediate sensitivity to blue light or longer wavelengths, respectively. Although visual transduction processes are well understood in the insect retina, almost nothing is known about the extraretinal blue light photoreceptor of insects. We now have identified and characterized a candidate blue light photoreceptor gene in Drosophila(DCry) that is homologous to the cryptochrome (Cry) genes of mammals and plants. TheDCry gene is located in region 91F of the third chromosome, an interval that does not contain other genes required for circadian rhythmicity. The protein encoded by DCry is ∼50% identical to the CRY1 and CRY2 proteins recently discovered in mammalian species. As expected for an extraretinal photoreceptor mediating circadian entrainment, DCry mRNA is expressed within the adult brain and can be detected within body tissues. Indeed, tissue in situ hybridization demonstrates prominent expression in cells of the lateral brain, which are close to or coincident with the Drosophila clock neurons. Interestingly, DCry mRNA abundance oscillates in a circadian manner in Drosophila head RNA extracts, and the temporal phasing of the rhythm is similar to that documented for the mouse Cry1 mRNA, which is expressed in clock tissues. Finally, we show that changes in DCry gene dosage are associated predictably with alterations of the blue light resetting response for the circadian rhythm of adult locomotor activity.
Keywords: circadian, cryptochrome, photoreceptor, blue light, Drosophila, extraretinal
Molecular genetic studies in the moldNeurospora, the fruit fly Drosophila, and the mouse have shown that phylogenetically conserved biochemical mechanisms underly the generation of biological rhythms (Dunlap, 1996; Darlington et al., 1998; Gekakis et al., 1998; Hall, 1998; Young, 1998). The same analysis has culminated in a detailed model describing the circadian timing device. The timing mechanism now can be described in terms of an autoregulatory feedback loop in which circadian changes in the abundance of clock proteins negatively regulate clock gene transcription. Similarly, the clock resetting mechanism can be understood at the molecular level: resetting stimuli such as light or temperature lead to rapid alterations in the abundance of a clock component, effectively shifting the clock to a new time of day (Crosthwaite et al., 1995; Hunter-Ensor et al., 1996; Lee et al., 1996;Myers et al., 1996; Zeng et al., 1996; Liu et al., 1998; Sidote et al., 1998). However, the photopigments that function in the circadian system and the phototransduction pathways that serve to reset the circadian clocks of animal species have not yet been subjected to a detailed molecular analysis.
Studies in the fruit fly Drosophila and other holometabolous insect species indicate that an extraretinal blue light photoreceptor mediates light input to the circadian clock regulating behavioral rhythmicity (Zimmerman and Ives, 1971; Truman, 1972; Helfrich, 1986;Blaschke et al., 1996; Suri et al., 1998; Yang et al., 1998). In silkmoths, for example, brain transplantation studies demonstrated that both the circadian clock and a photoreceptor for entrainment were located in the CNS (Truman, 1972). Similarly, retinally blind and eyeless mutants of Drosophila retain the capacity to entrain to environmental light/dark cycles (Helfrich, 1986), indicating that an extraretinal photoreceptor mediates circadian resetting. Indeed, recent studies show that degradation of the timeless (TIM) clock protein, which is correlated with light-induced circadian resetting, occurs in visual transduction mutants ofDrosophila (Yang et al., 1998). Finally, spectral resetting curves for the Drosophila activity rhythm show that the circadian clock is sensitive to both blue/green light and longer wavelengths (Blaschke et al., 1996; Suri et al., 1998). Sensitivity to the two portions of the spectrum is thought to be mediated by the retinally based opsin system and an extraretinal blue/green photoreceptor.
Although the retinal phototransduction process is well understood in insects (Zuker, 1996), little is known about the molecular basis of extraretinal (blue light) photoreception in these species. In contrast, blue light photoreception has been well characterized in plants (Ahmad and Cashmore, 1996) and depends on photopigments known as cryptochromes (CRY1 and CRY2), which are homologous to DNA repair (photoreactivating) enzymes known as photolyases but which themselves completely lack DNA repair activity (Ahmad and Cashmore, 1993; Malhotra et al., 1995; Todo et al., 1996). Photoreception through CRY1 and CRY2 are required for many different blue light responses, including plant phototropism, the inhibition of hypocotyl elongation, and the early photomorphogenesis of seedlings (Ahmad and Cashmore, 1993; Ahmad et al., 1998b). In addition, CRY1 also has been implicated in signaling pathways necessary for the circadian regulation of plant catalase expression (Zhong et al., 1997), and CRY2 functions in the photic regulation of flowering time (Guo et al., 1998). The phytochromes (PhyA and PhyB), another well studied class of plant photopigments that are sensitive to red/far red light, participate in some of these photically stimulated responses (Zhong et al., 1997; Ahmad et al., 1998a; Guo et al., 1998), and it recently has been suggested that CRY1 physically interacts with PhyA in vivo (Ahmad et al., 1998a).
Cry-gene homologs encoding related cryptochromes recently have been identified and characterized in mammalian species (Hsu et al., 1996). In the mouse, both Cry1 and Cry2 are expressed in the ganglion cell and inner nuclear layer of the retina, and it has been hypothesized that the encoded cryptochromes mediate light input to the circadian clock in the suprachiasmatic nuclei (SCN;Miyamoto and Sancar, 1998). Interestingly, Cry1 also is expressed in the SCN, and the mRNA oscillates in abundance during the circadian cycle (Miyamoto and Sancar, 1998), although the role ofCry1 in the SCN is not understood. Nonetheless, evidence from both plant and animal species implicates the cryptochromes in circadian photoreception. Therefore, we have characterized a cryptochrome homolog in Drosophila to use a molecular genetic approach to evaluate its role in circadian photoreception.
MATERIALS AND METHODS
Culture conditions and behavioral analysis. All fly stocks were obtained from the Indiana University DrosophilaStock Center (Bloomington, IN). Df(3R)Dl-BX12 removes region 91F1-2 to 92D3-6, whereas Df(3R)Cha7 is an overlapping deletion removing region 90F1-4 to 91F5. To generate siblings carrying one or two copies of the DCry+ gene, we crossed Df(3R)Dl-BX12/TM6B(DCry+) flies tow1118 individuals, which carry two normal copies of the gene. Cultures were reared on Drosophilacornmeal medium according to standard lab procedures (Newby and Jackson, 1993). Adult activity rhythms were monitored and light-induced phase shifts were calculated as previously described (Newby and Jackson, 1993; Levine et al., 1994). Blue light pulses of ∼200 lux were delivered by using a Kodak 47B wratten filter with a peak transmittance at 430 nm. This filter has <0.1% transmittance at wavelengths above 500 nm.
RNA and DNA analyses. The DrosophilaCanton-Special (C-S) strain was used for all Northern, Southern, and RNase protection studies. All three procedures were performed exactly as previously described (Newby and Jackson, 1993, 1996). RNA was prepared from different developmental stages and tissues according to standard procedures (see Newby and Jackson, 1993). Samples of adult heads and bodies were prepared by freeze-fracturing adult flies (previously frozen at −80°C) and separating the different body parts by sieving on ice in the cold (4°C).
RNase protection analysis was used to examine the expression ofDCry RNA in different tissues and to characterize daily changes in abundance. To characterize oscillations in DCryRNA abundance, we entrained flies to a cycle of 12 hr of light and 12 hr of dark (LD 12:12) for 4–5 d before collection. As probes, we usedin vitro transcribed [32P]-labeled RNAs representing a 264 base pair (bp) fragment from the 5′ end of the HL03779 cDNA clone and a 112 bp fragment representing the rp49 RNA. RNA probes representing DCry and rp49 were annealed overnight at 42°C to 10 μg of total RNA, and RNA/RNA duplexes then were digested at 37°C for 1 hr with a cocktail of RNase A and T1 according to the manufacturer’s instructions (Ambion, Austin, TX). The sizes of protected fragments were determined by denaturing polyacrylamide electrophoresis, using MspI-digested pBR322 DNAs as size standards. Gels were subjected to film autoradiography, and signals were quantitated exactly as previously described (McNeil et al., 1998) by densitometric scanning of films. DCry RNA abundance was quantified relative to rp49 abundance, which does not change during the diurnal cycle.
Probes representing the HL03779 or GM03047 (Drosophila 6-4 photolyase) cDNAs were [32P]-labeled by random hexamer priming for use in Northern analysis. A labeled rp49probe (see above) was used as a control for gel loading. Probes were hybridized to RNA blots containing 5 μg poly(A+) RNA or 20 μg of total RNA per lane. Hybridizations contained ∼0.25–1 × 106 cpm probe/ml and were performed in ExpressHyb solution (Clontech, Palo Alto, CA) for 2 hr or overnight at 45°C (DNA probes) or 65°C (rp49 probe). Then the blots were washed three times (15 min each) at room temperature (∼23°C) and twice (30 min each) at 50°C before film autoradiography at −80°C to detect hybridization signals.
We performed in situ hybridization of sense and antisenseDCry probes to paraffin-embedded adult tissues, using standard methods (Lehmann and Tautz, 1994). Adults were collected at the middle of the subjective day, which corresponds to the high point of DCry RNA abundance (see Results). A 300 bp digoxigenin-labeled RNA probe representing bases 1524–1823 of the HL03779 cDNA was generated for these studies.
Chromosome in situ hybridizations were performed as described by Engels et al. (1986). A cDNA fragment representing the entire HL03779 clone was labeled with Biotin-dATP by nick translation and was used in chromosome localization. Chromosomes were stained lightly with Giemsa (0.4%; Sigma, St. Louis, MO) after the detection reaction. The polytene chromosome maps of Sorsa (1988) were used to localize the position of the DCry gene.
Sequence comparisons. Comparisons of protein sequences were performed with the software of the Genetics Computer Group (Wisconsin Package Version 9.0, Genetics Computer Group, Madison, WI). Direct comparisons among DCry and hCRY1 and hCRY2 were performed by the GAP program with a gap weight of 12 and length weight of 4. The multiple sequence alignment was constructed by using PILEUP with a gap creation penalty of 12 and a gap extension penalty of 4. Sequences were displayed by using PRETTY, and the shading of amino acid residues was accomplished with Microsoft Word. Distance matrices were generated by either DISTANCES or OLDDISTANCES, using the Kimura method of corrected multiple substitutions. A similarity plot for full-length sequences was generated with OLDDISTANCES and a threshold of 2; identity was calculated with a threshold of 4. The phylogram was produced with GROWTREE to generate a UPGMA tree, using a DISTANCES matrix for full-length proteins. Resampling of the data set with the bootstrapping technique yielded trees that were essentially the same as that shown in Figure 1.
Fig. 1.
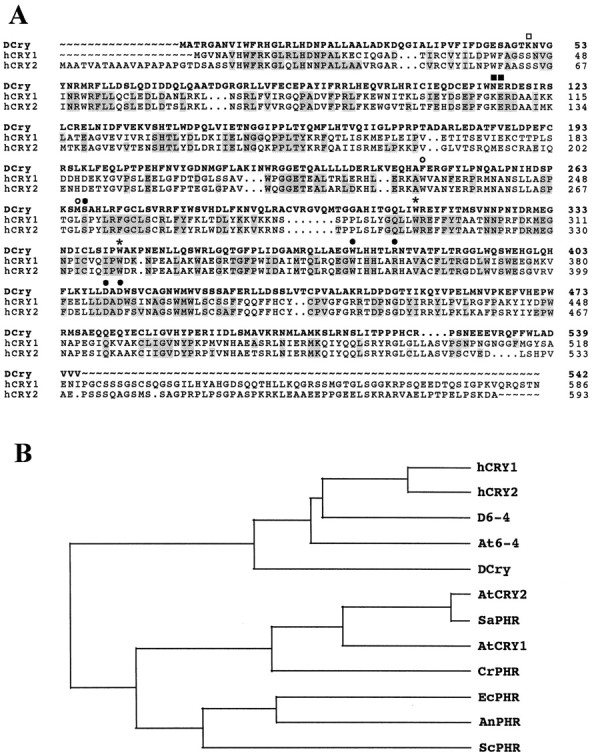
Sequence of a Drosophilacryptochrome-like protein and comparison to cryptochrome and photolyase sequences from several other organisms. A, Comparison of DCry to the human cryptochromes hCRY1 and hCRY2. Gray highlighting indicates residues in cryptochrome and photolyase sequences that are identical to the new Drosophilacryptochrome-like sequence. Circles,squares, and stars indicate similarities to the Escherichia coli PHR protein. Openand filled squares indicate positions that are similar and identical, respectively, to residues known to be important for folate binding.Open and filled circles indicate positions with similarity and identity, respectively, to residues important for FAD binding. Stars indicate tryptophan (W) residues that are conserved in all blue light photoreceptors and photolyases. The first Wis known to be important for CPD binding to E. coli PHR; the second is thought to be important for electron transfer involving FAD. B, Phylogram indicating relatedness of mammalian, insect, plant, and microbial cryptochromes and 6-4 photolyases.hCRY1, Human cryptochrome 1 (accession D83702);hCRY2, human cryptochrome 2 (accession AB014558);D6-4, Drosophila 6-4 photolyase (accession S74530); At6-4, Arabidopsis6-4 photolyase (accession AB003687); AtCRY1,Arabidopsis cryptochrome 1 (accession S66909);AtCRY2, Arabidopsis cryptochrome 2 (accession U43397); SaPHR, Sinapsis albacryptochrome (accession P40115); CrPHR,Chlamydomonas reinhardtii cryptochrome (accessionS57795); EcPHR, E. coli PHR photolyase (accession P00914); AnPHR, Anacystis nidulans PHR (accession P05327); ScPHR,Saccharomyces cerevisiae PHR (accession P05066).
RESULTS
Identification and sequence of a Drosophilacryptochrome-like gene
On the basis of the reported sequences of mouse and human cryptochromes, we searched the Berkeley Drosophila Genome Project (BDGP) expressed sequence tag (EST) database for similar genes in Drosophila. A number of cDNA clones representing two different Drosophila genes were identified in the initial search. One of these genes is the Drosophila 6-4 photolyase, which previously was reported to be similar to blue light photoreceptors (Todo et al., 1996). This gene is expressed at high levels only in female ovaries, consistent with the known photolyase activity of its encoded product and its role in the photoreactivation of damaged DNA (Todo et al., 1996). The second Drosophilagene, however, was identified from an EST representing a novel cDNA clone from a head cDNA library. That gene is represented in the BDGP database by two cDNA clones. One of those clones (HL03779), which appeared to contain the entire translational open reading frame, was obtained from Genome Systems and sequenced in its entirety. Not counting a polyA tail of 20 adenylate residues (As), the sequence is 1823 bp in length and contains a consensus polyadenylation signal at bp 1801 and an open reading frame between bp 121 and 1746 that predicts a cryptochrome-like protein of 542 amino acids (Fig.1A). This protein is ∼50% similar and 40% identical to either hCRY1 or hCRY2 in direct sequence comparisons (using the Genetics Computer Group GAP program; see Materials and Methods). Therefore, we provisionally designated the fly protein DCry for Drosophila cryptochrome and the relevant gene DCry.
Cryptochromes are members of a large protein family that includes blue light photoreceptors, 6-4 photolyases (DNA photoreactivating enzymes), and microbial Class I CPD (cyclobutane pyrimidine dimers) photolyases (Ahmad and Cashmore, 1993; Malhotra et al., 1995; Todo et al., 1996). Figure 1A shows the predicted sequence of DCry and a comparison of the fly protein to the human CRY1 and CRY2 proteins. As indicated in the Distances Matrix of Table1 (see Materials and Methods), DCry is most similar to the human cryptochromes, with similarities of 46 and 45% to hCRY1 and hCRY2, respectively. DCry is more similar to mammalian cryptochromes than plant cryptochromes or Class I CPD photolyases, the latter exhibiting similarities ranging from 23 to 30% (Table 1). The generation of a phylogram for 12 different cryptochromes and photolyases (see Materials and Methods) underscores the relatedness of DCry to the hCRY1 and hCRY2 proteins (Fig. 1B), although DCry also is related similarly to both theDrosophila and Arabidopsis 6-4 photolyases. There is little or no homology among DCry and higher eukaryotic Class II CPD photolyases (data not shown in Table 1 or Fig. 1). Therefore,DCry can be considered a new member of the protein subfamily that includes mammalian cryptochromes and 6-4 photolyases. The aggregate of our comparisons is consistent with the idea that DCry functions as a blue light photoreceptor rather than a photolyase. This conclusion is supported by the functional analysis described below, which indicates that the DCry protein mediates blue light resetting of the Drosophila circadian clock.
Table 1.
Distance matrix for photolyase and Cry proteins*
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.0000 | 0.7730 | 0.5556 | 0.5438 | 0.4594 | 0.2969 | 0.3074 | 0.2867 | 0.2833 | 0.2966 | 0.2996 | 0.2283 |
| 2 | 1.0000 | 0.5759 | 0.5307 | 0.4539 | 0.2816 | 0.3094 | 0.2901 | 0.2749 | 0.3093 | 0.3017 | 0.2283 | |
| 3 | 1.0000 | 0.5400 | 0.4315 | 0.2926 | 0.3094 | 0.2944 | 0.2833 | 0.3136 | 0.3037 | 0.2333 | ||
| 4 | 1.0000 | 0.4227 | 0.3091 | 0.3174 | 0.3054 | 0.2756 | 0.3157 | 0.3037 | 0.2365 | |||
| 5 | 1.0000 | 0.2897 | 0.3014 | 0.2860 | 0.3063 | 0.3008 | 0.3079 | 0.2325 | ||||
| 6 | 1.0000 | 0.9261 | 0.5899 | 0.4690 | 0.3496 | 0.3554 | 0.2761 | |||||
| 7 | 1.0000 | 0.6387 | 0.5250 | 0.3432 | 0.3388 | 0.2954 | ||||||
| 8 | 1.0000 | 0.4493 | 0.3623 | 0.3368 | 0.2602 | |||||||
| 9 | 1.0000 | 0.3729 | 0.3492 | 0.2655 | ||||||||
| 10 | 1.0000 | 0.4682 | 0.3983 | |||||||||
| 11 | 1.0000 | 0.3595 | ||||||||||
| 12 | 1.0000 |
*Threshold of comparison; 2; denominator; length of shorter sequence without gaps; key for column and row indices, 1, hCRY1; 2, hCRY2; 3, D6-4; 4, At6-4; 5, DCry; 6, AtCRY2; 7, SaPHR; 8, AtCRY1; 9, CrPHR; 10, EcPHR; 11, AnPHR; 12, ScPHR. The numbers represent the percentage identity for the various comparisons.
Similar to other cryptochromes, DCry has conserved domains that include residues known to be important for the noncovalent binding of the cofactors pterin (folate) and flavin adenine dinucleotide (FAD; see Fig. 1 legend). In plant and mammalian cryptochromes, these cofactors determine light absorption spectra (Lin et al., 1995; Malhotra et al., 1995; Hsu et al., 1996). Also like other cryptochromes, the DCry protein has a nonconserved C terminus (of 41 residues) that is completely unrelated to photolyases, cryptochromes, or any other sequence in the protein databases. It has been suggested that the novel C termini of cryptochromes might be important for interactions with effector molecules (Hsu et al., 1996), and it has been demonstrated that the C terminus of the human CRY2 protein can physically interact with and inhibit the phosphatase activity of the tetratricopeptide repeat (TPR)-containing protein PP5 (Zhao and Sancar, 1997). The DCry protein does not contain an N-terminal extension similar to those of microbial and plant photolyases that have been implicated in mitochondrial and nuclear targeting (Yasui et al., 1992).
A single Drosophila Cry gene maps to chromosome region 91F
Because there are two Cry genes present in the genomes of certain plants and mammals, we wondered whether additional cryptochrome genes existed in Drosophila. Southern hybridizations at reduced stringency, however, revealed genomic fragments corresponding to the new Drosophilacryptochrome-like gene or to the Drosophila 6-4 photolyase gene, but no additional hybridizing fragments were observed (data not shown). Therefore, there is no evidence for additional genes, although we cannot exclude the possibility of more highly divergent fly cryptochrome-like genes. However, both in humans andArabidopsis, the two Cry genes are much more similar to each other than they are to their counterparts in the other species (see Table 1). This also suggests that the Cry gene pairs of these different species evolved from independent duplication events after the divergence of plants and animals.
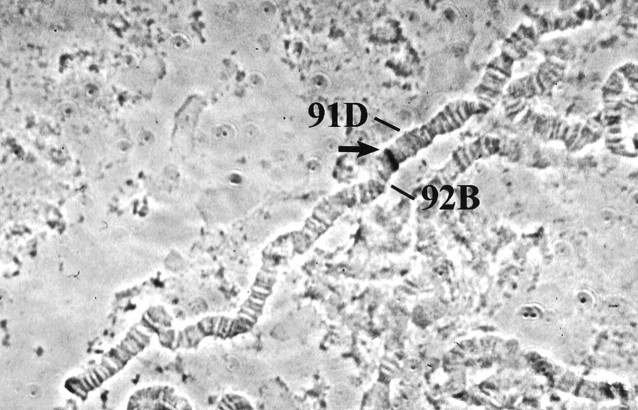
There are a number of Drosophila rhythm genes that have not been characterized molecularly (see Jackson, 1993). Because we wondered whether any of them corresponded to DCry, we determined the cytogenetic location of the Drosophila gene by hybridization of cDNA sequences to larval salivary gland polytene chromosomes (see Materials and Methods). A single site of hybridization was observed within polytene region 91F of the third chromosome distal to band 91F5 (Fig. 2). No circadian rhythm genes besidesDCry have been mapped to this cytogenetic interval.
Fig. 2.
Cytogenetic localization of theDCry gene. The arrow points to region 91F of the third chromosome. The numbers indicate adjacent chromosomal intervals.
The DCry mRNA is expressed in the adult brain and other nonretinal tissues
The HL03779 cDNA clone was isolated from a fly cDNA library generated from brain and sensory organ RNA (BerkeleyDrosophila Genome Project/HHMI EST Project; unpublished data), and we have confirmed that the DCry mRNA is expressed in head tissues. In head RNA preparations a doublet of ∼2 kb is detected in either total RNA or poly(A+) RNA fractions (Fig. 3A; data not shown). As the HL03779 cDNA is 1823 bp in length and most mRNAs contain 100–200 As at their 3′ end, we assume that this clone represents most or all of the DCry mRNA sequence.
Fig. 3.
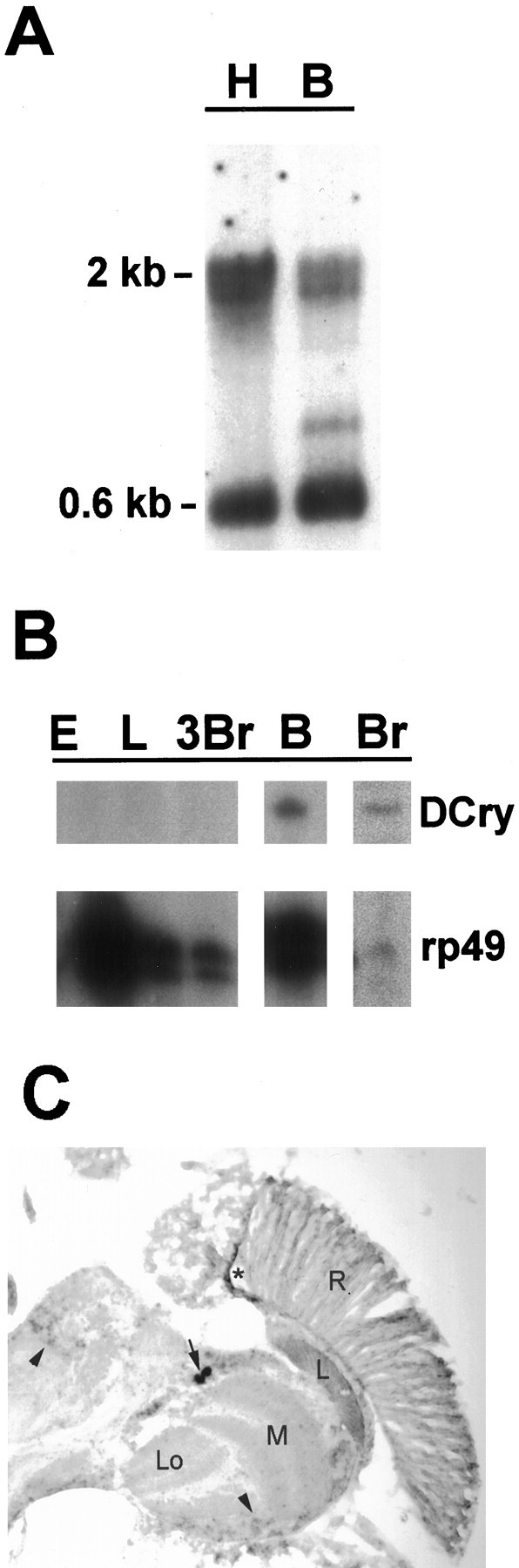
Tissue and developmental expression ofDCry mRNA. A, Northern analysis showing expression in head and body tissues. H denotes 5 μg of head poly(A+) RNA; B denotes 5 μg of body poly(A+) RNA. Size markers indicate theDCry mRNA doublet (2 kb) andrp49 mRNA (0.6 kb). B, RNase protection analysis of DCry expression in different tissues and developmental stages. Protected bands representing DCry and control rp49 mRNAs are indicated. Br, Adult brain; E, 0–24 hr embryo; L, third instar larva; 3Br, third instar larval brain; B, body. The body preparation used for RNase protection analysis was examined by light microscopy to be certain that no contaminating heads were present. C,In situ hybridization to a horizontal section of an adult head. The arrow indicates the position ofDCry-expressing cells in the lateral CNS. Thearrowheads show specific signal in other portions of the brain. The star indicates nonspecific staining, which also was seen with the sense probe. A similar spatial pattern of expression was observed in two independent experiments.R, Retina; L, optic lamina;M, optic medulla; Lo, lobula.
In contrast to DCry expression, a probe representing theDrosophila 6-4 photolyase gene (see Materials and Methods) did not detect a signal when hybridized to a Northern containing the same head RNA preparations (data not shown). This result is not surprising, because our database searches found 6-4 photolyase-homologous cDNAs only in an EST collection representing aDrosophila ovary cDNA library. As previously reported (Todo et al., 1996), the fly 6-4 photolyase gene is expressed at significant levels only in ovary tissues.
We used RNase protection methods to examine various developmental stages and tissues for DCry expression. To determine whether the DCry RNA was expressed in the brain, we prepared RNA samples from hand-dissected adult brains, which were completely devoid of eyes and ocelli. As judged by the control rp49 signal (Fig.3B, lane Br), DCry mRNA can be detected in a modest amount of total RNA (lane Br), indicating that the message is enriched in the brain. This result also demonstrates that the message is expressed in an extraretinal manner and suggests that it encodes the cryptochrome mediating circadian photoreception. Consistent with expression in the brain, DCry mRNA was detected readily in head tissues of eyes absent (eya) mutants, which entirely lack compound eyes (data not shown). Interestingly,DCry mRNA can be detected in body tissues (lane Bin Fig. 3A,B), which previously have been shown to contain photoreceptive clocks (Plautz et al., 1997), although the relative abundance of the mRNA is apparently lower in the body. Finally,DCry message could not be detected in moderate-to-large amounts of total RNA from 0–24 hr embryos (Fig. 3B, lane E), whole third instar larvae (lane L), or third instar larval brains (lane 3Br), suggesting that a different photoreceptor might mediate circadian resetting at these developmental stages.
In situ hybridization techniques were used to examine the spatial localization of DCry mRNA within the adult nervous system. As shown in Figure 3C, a low level of expression could be detected throughout the cell body layer of the CNS (seearrowheads in Fig. 3C). A much stronger signal, however, was observed in large cells of the lateral CNS (Fig. 3C, arrow), which are close to or coincident with the ventral group ofDrosophila clock neurons (see Discussion). Specific expression also was detected in adult non-neural tissues, including the gut (data not shown). Importantly, sections hybridized with aDCry sense probe did not show any signal within brain or gut cells (data not shown). A small amount of reaction product was observed within the retina (R) with both the sense and antisense DCryprobes; thus, we conclude there is no specific signal forDCry mRNA within retinal tissues.
The DCry mRNA oscillates in abundance during the circadian cycle
As the mouse Cry1 mRNA had been reported to oscillate in abundance during the diurnal cycle, we determined whether the same might be true of the DCry mRNA. As shown in Figure4, the DCry message is more abundant in head RNA samples during the day than at night, relative torp49 abundance, which does not change during the diurnal cycle (Hardin et al., 1990). In two independent experiments,DCry mRNA was 6- and 11-fold more abundant at peak during the day than it was at the trough of the rhythm during the night (Fig.4B). Indeed, the amplitude of the DCryrhythm was greater than that observed for the mouse Cry1mRNA, which oscillates in abundance in the suprachiasmatic nuclei (SCN;Miyamoto and Sancar, 1998). Additional experiments showed thatDCry mRNA did not show immediate increases in abundance in response to the lights-on signal (data not shown), indicating thatDCry gene expression is not light-inducible. Similar to mouse Cry1 mRNA, the rhythm in DCry abundance persisted in constant conditions (Fig. 4C), demonstrating that it is under circadian regulation.
Fig. 4.
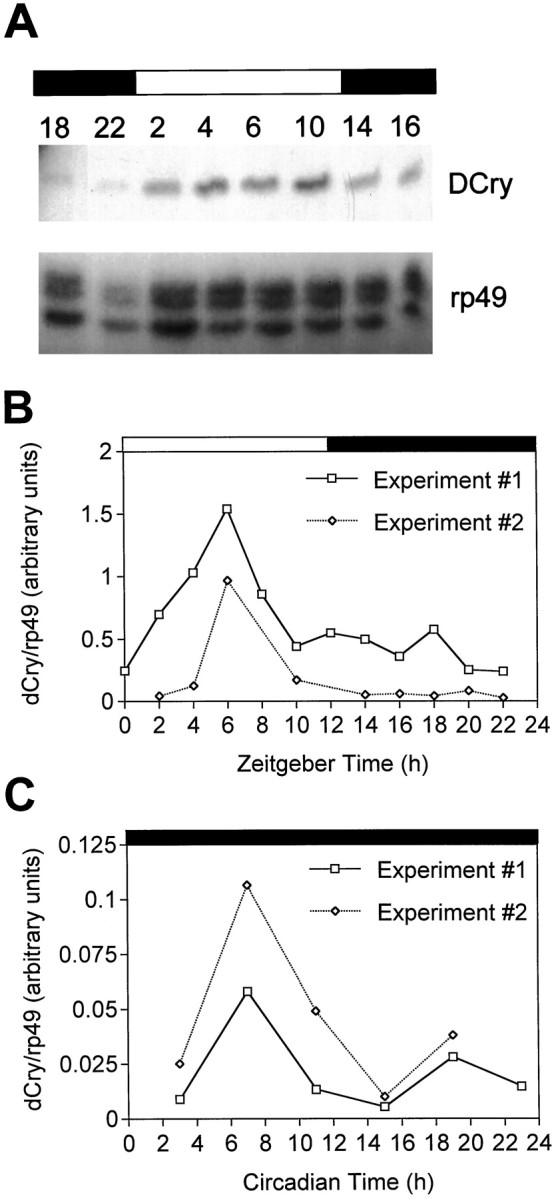
The DCry mRNA oscillates in abundance in head tissues during the diurnal cycle. A, RNase protection gel showing the relative abundances ofDCry (above) and rp49(below) RNAs at different times of the day.Numbers above the gel indicate zeitgeber time (ZT, in hours). B, Quantitation of DCry RNA abundance relative to rp49 abundance in two independent LD experiments. The open and filled rectangles above A and B show the light and dark portions of the LD 12/12 cycle to which adults were entrained.Numbers refer to zeitgeber time (ZT). Similar results were obtained in two other experiments. C, Quantitation of DCry RNA abundance in constant darkness (DD).Numbers refer to circadian time (CT). Data are shown for two independent experiments. In two other DD experiments not shown here, similar circadian changes in RNA abundance were documented, although peak RNA abundance was several hours earlier than that shown in C. In those experiments RNA abundance changed by approximately four- to fivefold during the circadian cycle.
Changes in DCry gene dosage affect the blue light resetting response
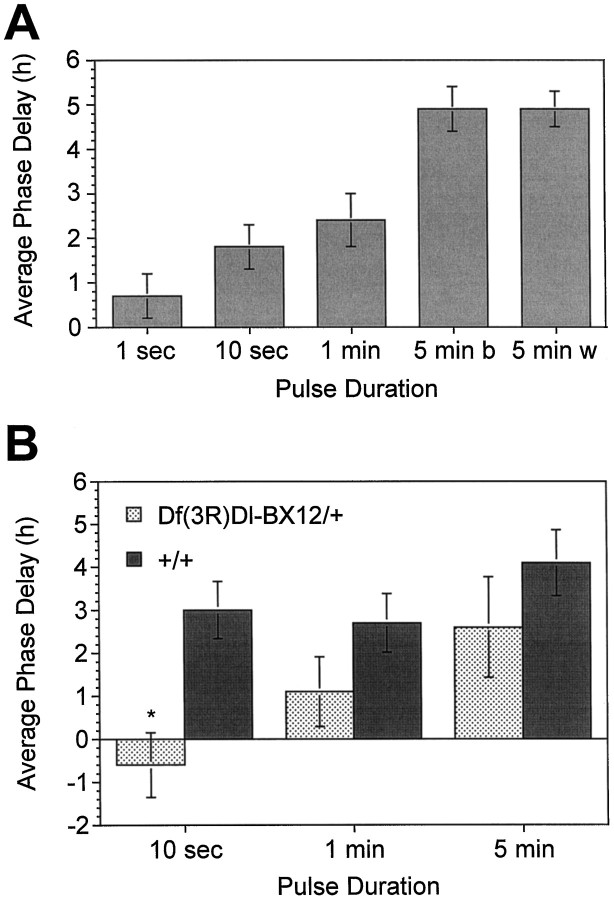
We conducted behavioral genetic experiments to test the notion that the Drosophila cryptochrome mediates blue light resetting of the circadian clock. As a prelude to these experiments, we first examined blue light resetting in normal flies. As shown in Figure5A, normal individuals exhibited phase shifts of increasing magnitude in response to ∼200 lux blue light pulses of increasing duration. Flies receiving 5 min of blue light (5 min b) or 5 min of 2500 lux white light (5 min w) exhibited phase delays of identical magnitude, suggesting that this duration of blue light constituted a saturating light pulse. Importantly, these data indicate that 10 sec and 1 min pulses of blue light cause submaximal phase shifts, and thus such resetting pulses might be appropriate for detecting behavioral alterations that result from changes in DCry gene dosage.
Fig. 5.
Blue light resetting responses in normal flies and individuals deficient for DCry gene product.A, Average phase delays (hr ± SEM) ofw1118 flies (DCry+/DCry+) in response to blue light pulses of ∼200 lux ranging from 1 sec to 5 min. Flies receiving pulses for 5 min saw either blue light (5 min b) or full spectrum white light of ∼2500 lux (5 min w). Sample sizes ranged from 7 to 13 flies per group.B, Phase delays of Df(3R)Dl-BX12/+ and +/+ siblings at three different pulse durations. For the 10 sec and 1 and 5 min pulses, respectively, we examined 11, 10, and 6 flies forDf(3R)Dl-BX12/+ and 14, 15, and 10 flies for the +/+ siblings. *p < 0.001 as compared with +/+ siblings. Both Df(3R)Dl-BX12/+ and +/+ siblings had white eye color, similar to the w1118flies of A. The average phase delay values shown inA and B were corrected by subtracting the small phase delay that results after transfer from LD to DD (Levine et al., 1994). The negative phase delay value forDf(3R)Dl-BX12/+ flies indicates that the delay that followed the LD to DD transition was actually slightly larger than the delay induced by the blue light pulse; effectively, these flies showed no phase delay in response to the 10 sec pulse. Even without the usual correction, however, the difference between deletion-bearing flies and siblings was statistically significant.
To determine whether changing DCry dosage affected blue light resetting, we characterized the resetting responses of flies carrying one or two doses of the gene. We used two different third chromosome deletions in these experiments: Df(3R)Dl-BX12 andDf(3R)Cha7. Based on our localization of the DCrygene (in distal 91F), the Dl-BX12 deletion was predicted to remove the gene, whereas Cha7 was expected to delete a region adjacent to the gene (see Materials and Methods). Control flies heterozygous for the Cha7 deletion exhibited phase delays at all three pulse durations that were within the wild-type range and not significantly different from those of normal flies (data not shown), which confirmed that this deletion did not remove the DCrygene. For Df(3R)Dl-BX12, resetting responses were characterized in sibling flies of similar genetic background (see Materials and Methods), which were either heterozygous for the deletion (Df/+) or homozygous for the normalDCry+ allele (+/+). As shown in Figure5B, flies heterozygous for Df(3R)Dl-BX12 had significantly smaller phase delays than normal siblings in response to a 10 sec pulse of blue light (p < 0.001). Phase delays for such flies were progressively larger in response to longer pulses (Fig. 5B), but not significantly different from those of siblings, presumably because the system was at or near saturation at the longer pulse durations. These data indicate that flies deficient for DCry product have decrements in blue light resetting.
DISCUSSION
Identification of a Drosophila Cry homolog
Cryptochrome (Cry) proteins lacking photolyase activity have been identified in mammals and plants (Ahmad and Cashmore, 1993; Malhotra et al., 1995), and these proteins have been demonstrated to mediate blue light photoreception in Arabidopsis. We have identified and characterized a new Drosophila gene (DCry) encoding a cryptochrome protein (DCry) with significant similarity to the cryptochromes previously described in plant and animal species. Although DCry also has similarity to 6-4 photolyase proteins, it is most similar to the mammalian cryptochromes (hCRY1 and hCRY2) that have been implicated in circadian photoreception (Miyamoto and Sancar, 1998).
Given that the DCry gene is expressed inDrosophila brain and body tissues, we postulate that the encoded protein serves as the extraretinal blue light photoreceptor mediating the entrainment of circadian behavioral rhythms. Previous studies have indicated that a circadian photoreceptor is localized in the insect brain (Truman, 1972; Helfrich, 1986), and recent work has demonstrated the existence of photoreceptive circadian clocks in a variety of Drosophila tissues (Plautz et al., 1997). Consistent with a role for cryptochrome in light resetting of the circadian clock, Drosophila strains that are deficient forDCry product show correlated changes in the resetting response to blue light. In addition, work reported by Stanewsky et al. (1998a) indicates that a mutation in the DCry gene we have characterized leads to altered light resetting of circadian rhythms. The aggregate of these results suggests that the DCry photoreceptor mediates extraretinal light input to the Drosophilacircadian clock. Because brain-localized photoreceptors mediate clock resetting in other holometabolous insect species (Truman, 1972), it is likely that cryptochromes similar to DCry function in circadian phototransduction in such species.
How might Cry-mediated phototransduction reset the circadian clock?
Although DCry mRNA is expressed within the CNS, the identities of the cell types relevant for extraretinal photoreception have not been positively determined. However, relative to other parts of the nervous system, DCry mRNA is expressed at high levels in large cells of the lateral CNS. Although we have not yet performed double-labeling experiments with clock cell antibodies, on the basis of the size and position of the hybridizing cells we postulate that they correspond to the ventral group of the so-called “lateral clock neurons.” These cells are known to be critical for circadian function (Frisch et al., 1994; Helfrich-Förster, 1998) and contain the clock proteins Period and Timeless (Young, 1998) as well as a neuropeptide known as pigment-dispersing hormone (PDH;Helfrich-Förster, 1995). We hypothesize that the clock neurons themselves are directly light-sensitive by virtue of containing the DCry cryptochrome. Perhaps less likely, DCry protein might be localized within dedicated circadian photoreceptor cells that are close to the clock neurons and innervate them.
The signaling pathway that transduces light information from Cry proteins to the clock mechanism has not been elucidated in any species. In Drosophila such a signaling mechanism ultimately must result in the degradation of Timeless (TIM) protein, which serves as the sensor for light input to the clock (Hunter-Ensor et al., 1996; Lee et al., 1996; Myers et al., 1996; Zeng et al., 1996). Thus, Cry-mediated phototransduction must involve the activation of an appropriate protease, perhaps indirectly via a kinase or phosphatase intermediary. In this regard, it is of interest that Zhao and Sancar (1997) have shown that human CRY proteins can interact with members of the TPR family of proteins, including serine/threonine phosphatase 5 (PP5). Moreover, the activity of PP5 can be inhibited by interaction with hCRY proteins, although the observed inhibition is not light-dependent (Zhao and Sancar, 1997). It also is known that there is a blue light-mediated and CRY-dependent autophosphorylation of anArabidopsis protein kinase known as NPH1 (Ahmad et al., 1998b). Therefore, it is reasonable to suppose that a light-induced alteration of a specific phosphatase and/or protein kinase might be required for the activation of the protease that mediatesDrosophila TIM degradation.
Circadian oscillations in DCry mRNA abundance
DCry mRNA abundance changes in a circadian manner, with peaks of abundance occurring during the photoperiod of a light/dark cycle. Although the daily phasing of the DCry cycle is similar in Drosophila and the mouse (Miyamoto and Sancar, 1998), the significance of the RNA oscillation is not understood in either species. Indeed, one would expect the abundance of a photoreceptor protein either to be constant during the circadian cycle or to be higher during the night, at the time when the clock is maximally sensitive to resetting stimuli. This could be the case for DCry protein if there is a temporal lag in translation of theDCry message. Alternatively, the DCry rhythm might be required for another aspect of circadian clock function in addition to photoreception.
Although Cry rhythmicity might be relevant to circadian photoreception or another circadian function, it is also possible that the mammalian and Drosophila Cry rhythms reflect the ancestral photolyase activity of the proteins. For photolyases, a higher DNA repair activity might be advantageous during the photoperiod. In this regard, it is known that the expression of certain photolyase genes can be induced by light (Batschauer, 1993; Ahmad et al., 1997), although it is unclear whether the expression of these genes is under clock control. Irrespective of the physiological significance of the Cry mRNA rhythm, the remarkable similarity in the phasing of rhythmicity in different species suggests a conservation of function and/or gene regulatory mechanisms. A more detailed molecular genetic analysis of the DCry RNA rhythm may yield insights about its function.
Note. While this paper was in review, other labs reported the genetic and molecular characterization of theDrosophila DCry gene (Emery et al., 1998; Stanewsky et al., 1998b). Their work reports the behavioral analysis of aDCry mutant and provides additional molecular evidence that DCry protein functions as a circadian photoreceptor.
Footnotes
This work was supported by grants from the National Institutes of Health and National Science Foundation. We thank Yin Xu and Dale Hunter for performing tissue in situ hybridizations, Mike Byrne and the Tufts Biochemistry Facility for help with DNA sequencing, Rob Willson of the Tufts Imaging Facility for densitometric scanning of films, and Jeff Hall for communicating unpublished results. We also thank the Flybase consortium, the Berkeley DrosophilaGenome Project, and the Indiana University DrosophilaStock Center for access to database information and fly stocks. The DNA sequence of cDNA clone HL03779 and the predicted protein sequence have been deposited in GenBank.
All authors contributed equally to this work; with the exception of the corresponding author (F.R.J.), they are listed alphabetically.
Correspondence should be addressed to Dr. F. Rob Jackson, Department of Neuroscience, Tufts University School of Medicine, 136 Harrison Avenue, Boston, MA 02111.
REFERENCES
- 1.Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad M, Cashmore AR. Seeing blue: the discovery of cryptochrome. Plant Mol Biol. 1996;30:851–861. doi: 10.1007/BF00020798. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad M, Jarillo JA, Klimczak LJ, Landry LG, Peng T, Last RL, Cashmore AR. An enzyme similar to animal type II photolyases mediates photoreactivation in Arabidopsis. Plant Cell. 1997;9:199–207. doi: 10.1105/tpc.9.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998a;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature. 1998b;392:720–723. doi: 10.1038/33701. [DOI] [PubMed] [Google Scholar]
- 6.Batschauer A. A plant gene for photolyase: an enzyme catalyzing the repair of UV-light-induced DNA damage. Plant J. 1993;4:705–709. doi: 10.1046/j.1365-313x.1993.04040705.x. [DOI] [PubMed] [Google Scholar]
- 7.Blaschke I, Lang P, Hofbauer A, Engelmann W, Helfrich-Förster C. Preliminary action spectra suggest that the clock cells of Drosophila are synchronized to the external LD cycle by the compound eyes plus extraretinal photoreceptors. In: Elsner N, Schnitzler H-U, editors. Brain and evolution. Thieme Verlag; Stuttgart: 1996. p. 30. [Google Scholar]
- 8.Crosthwaite SK, Loros JJ, Dunlap JC. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- 9.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 10.Dunlap JC. Genetic and molecular analysis of circadian rhythms. Annu Rev Genet. 1996;30:579–601. doi: 10.1146/annurev.genet.30.1.579. [DOI] [PubMed] [Google Scholar]
- 11.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 12.Engels WR, Preston CR, Thompson P, Eggleston WB. In situ hybridization to Drosophila salivary chromosomes with biotinylated DNA probes and alkaline phosphatase. Focus. 1986;8:6–8. [Google Scholar]
- 13.Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 14.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 15.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 16.Hall JC. Genetics of biological rhythms in Drosophila. Adv Genet. 1998;38:135–184. doi: 10.1016/s0065-2660(08)60143-1. [DOI] [PubMed] [Google Scholar]
- 17.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 18.Helfrich C. Role of the optic lobes in the regulation of the locomotor activity rhythm of Drosophila melanogaster: behavioral analysis of neural mutants. J Neurogenet. 1986;3:321–343. doi: 10.3109/01677068609106857. [DOI] [PubMed] [Google Scholar]
- 19.Helfrich-Förster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci USA. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helfrich-Förster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol [A] 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- 21.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang R-P, Todo T, Wei Y-F, Sancar A. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35:13871–13877. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- 22.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 23.Jackson FR. Circadian rhythm mutants of Drosophila. In: Young MW, editor. Molecular genetics of biological rhythms, cellular clocks series. Dekker; New York: 1993. pp. 91–121. [Google Scholar]
- 24.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER–TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann R, Tautz D. In situ hybridization to RNA. In: Goldstein LSB, Fyrberg EA, editors. Methods in cell biology, Vol 44. Academic; New York: 1994. pp. 575–598. [DOI] [PubMed] [Google Scholar]
- 26.Levine JD, Casey CI, Kalderon DD, Jackson FR. Altered circadian pacemaker functions and cyclic AMP rhythms in the Drosophila learning mutant dunce. Neuron. 1994;13:967–974. doi: 10.1016/0896-6273(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 27.Lin C, Ahmad M, Gordon D, Cashmore AR. Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-A, and green light. Proc Natl Acad Sci USA. 1995;92:8423–8427. doi: 10.1073/pnas.92.18.8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. Academic; New York: 1992. [Google Scholar]
- 29.Liu Y, Merrow M, Loros JJ, Dunlap JC. How temperature changes reset a circadian oscillator. Science. 1998;28:825–829. doi: 10.1126/science.281.5378.825. [DOI] [PubMed] [Google Scholar]
- 30.Malhotra K, Kim S-T, Batschauer A, Dawut L, Sancar A. Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapsis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry. 1995;34:6892–6899. doi: 10.1021/bi00020a037. [DOI] [PubMed] [Google Scholar]
- 31.McNeil GP, Zhang XL, Genova G, Jackson FR. A molecular rhythm mediating circadian clock output in Drosophila. Neuron. 1998;20:297–303. doi: 10.1016/s0896-6273(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 32.Miyamoto Y, Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci USA. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers MP, Wagner-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 34.Newby LM, Jackson FR. A new biological rhythm mutant of Drosophila melanogaster that identifies a gene with an essential embryonic function. Genetics. 1993;135:1077–1090. doi: 10.1093/genetics/135.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newby LM, Jackson FR. Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J Neurobiol. 1996;31:117–128. doi: 10.1002/(SICI)1097-4695(199609)31:1<117::AID-NEU10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 36.Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 37.Sidote D, Majercak J, Parikh V, Edery I. Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol Cell Biol. 1998;18:2004–2013. doi: 10.1128/mcb.18.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorsa V. Chromosome maps of Drosophila, Vol II. CRC; Boca Raton, FL: 1988. [Google Scholar]
- 39.Stanewsky R, Kaneko M, Emery P, Rosbash M, Kay S, Hall JC (1998a) A new rhythm mutant found by real-time reporting of period gene expression is defective in fly cryptochrome [abstract]. J Neurogenet, in press.
- 40.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998b;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 41.Suri V, Qian Z, Hall JC, Rosbash M. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron. 1998;21:225–234. doi: 10.1016/s0896-6273(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 42.Todo T, Ryo H, Yamamoto K, Toh H, Inui T, Ayaki H, Nomura T, Ikenaga M. Similarity among the Drosophila (6-4) photolyase, a human photolyase homolog, and the DNA photolyase blue-light photoreceptor family. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- 43.Truman JW. Physiology of insect rhythms. II. The silkmoth brain as the location of the biological clock controlling eclosion. J Comp Physiol. 1972;81:99–114. [Google Scholar]
- 44.Yang Z, Emerson M, Su HS, Sehgal A. Response of the timeless protein to light correlates with behavioral entrainment and suggests a nonvisual pathway for circadian photoreception. Neuron. 1998;21:215–223. doi: 10.1016/s0896-6273(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 45.Yasui A, Yajima H, Kobayashi T, Eker APM, Oikawa A. Mitochondrial DNA repair by photolyase. Mutat Res. 1992;273:231–236. doi: 10.1016/0921-8777(92)90084-g. [DOI] [PubMed] [Google Scholar]
- 46.Young MW. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu Rev Biochem. 1998;67:135–152. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- 47.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 48.Zhao S, Sancar A. Human blue-light photoreceptor hCRY2 specifically interacts with protein serine/threonine phosphatase 5 and modulates its activity. Photochem Photobiol. 1997;66:727–731. doi: 10.1111/j.1751-1097.1997.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhong HH, Resnick AS, Straume M, McClung CR. Effects of synergistic signaling by phytochrome A and cryptochrome 1 on circadian clock-regulated catalase expression. Plant Cell. 1997;9:947–955. doi: 10.1105/tpc.9.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmerman WF, Ives D. Some photophysical aspects of circadian rhythmicity in Drosophila. In: Menaker M, editor. Biochronometry. National Academy of Sciences; Washington, DC: 1971. pp. 381–391. [Google Scholar]
- 51.Zuker CS. The biology of vision in Drosophila. Proc Natl Acad Sci USA. 1996;93:571–576. doi: 10.1073/pnas.93.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]