Abstract
Conformational properties of several similar FMRFamide-like neuropeptides from mollusks were investigated by nuclear magnetic resonance (NMR) spectroscopy. It was found that amino acid substitutions in the N-terminal variable regions of the peptides had dramatic effects on the populations of reverse turns in solution. The populations of turns, as measured by two independent NMR parameters, were found to be highly correlated (r2 = 0.93 and 0.82) with IC50 values using receptor membrane preparations fromHelix aspersa (Payza, 1987; Payza et al., 1989). These results suggest that the amount of turn in the free peptide can influence the receptor binding affinities of that peptide. On the basis of these observations, a model was developed in which only a single species from a conformational ensemble of an unbound peptide will bind to a particular receptor. Thus, the conformational ensemble reduces the effective concentration of a particular peptide with respect to a particular receptor.
Keywords: FMRFamide-like peptides, NMR, structure–function relations, conformational averaging, dynamics, three-dimensional structure, reverse turn
Neuropeptides are a major source of neurochemical diversity, with functions as wide ranging, for example, as the modulation of the action of morphine and feeding behavior in mammals (Yang et al., 1985; Sakurai et al., 1998), the regulation of cardiac stimulation and pacemakers in mollusks (Payza, 1987; Simon et al., 1992), the stimulation or inhibition of oviduct contractions in locust (Wang et al., 1995a,b), and the modification of lobster pattern-generating circuits (Dickinson et al., 1990). Currently the largest, most widely distributed, and most diverse family of neuropeptides are the FMRFamide-like peptides (FLPs). FLPs are thought to be present in all animals (Greenberg and Price, 1992) and have amino acid sequence similarities to the “parent” peptide Phe-Met-Arg-Phe-NH2 (FMRFa), which was first discovered in the clam (Price and Greenberg, 1977).
FLPs, like other neuropeptides, are encoded by precursor proteins that are processed into mature peptides in the secretory pathway (Sossin et al., 1989). Some FLP precursor proteins produce multiple copies of the same peptide. The most dramatic example of multiple copies is fromAplysia, with 28 copies of FMRFamide and one copy of FLRFamide (Taussig and Scheller, 1986). Other precursor proteins encode several different FLPs, which tend to be longer than four amino acids and have common C-terminal amino acid sequences and variable N-terminal extensions. Large numbers of different FLPs are often present in a given animal. In nematodes, for example, Ascaris suum has at least 20 different FLPs (Cowden et al., 1989; Cowden and Stretton, 1993, 1995; Edison et al., 1997), and Caenorhabditis eleganshas 20 FLP genes (Rosoff et al., 1992; Wilson et al., 1994; Bargmann, 1998; Nelson et al., 1998; Chris Li, personal communication). If all of the C. elegans genes were expressed and fully processed, they would produce at least 56 different FLP peptides (Bargmann, 1998; Nelson et al., 1998).
The functional role of multiple diverse peptides is currently ambiguous. A deletion in C. elegans of a FLP precursor protein encoding for eight closely related peptides produces five distinct phenotypes (Nelson et al., 1998), but it is unknown whether individual peptides or a “bouquet” (Greenberg and Price, 1992) is responsible for the specific phenotypes. Substitutions of a single amino acid in an FLP from locust produces opposite G-protein-mediated responses (Wang et al., 1995a,b). Replacement of an aspartic acid by tyrosine lowers the receptor binding affinity of a molluscan FLP (Payza, 1987; Payza et al., 1989). In contrast, two similar FLPs have essentially identical effects on the crab stomatogastric ganglion (Weimann et al., 1993), and a group of six peptides from the same precursor protein in Ascaris (Edison et al., 1997) produces similar effects on Ascaris muscle (Davis and Stretton, 1996).
Here we report the conformational properties of individual peptides from a group of related FLPs from mollusks using nuclear magnetic resonance (NMR) spectroscopy and correlate the conformations with previously published receptor binding affinities (Payza, 1987; Payza et al., 1989). The organization of this paper is as follows. The technical NMR details are primarily limited to Materials and Methods. The Results section has four parts: NMR chemical shifts, NMR distance measurements, NMR pH dependence, and conformational models. Each NMR section has an introduction to the parameter with a description of its significance to the present study. The conformational model section presents a three-dimensional reverse turn structure that is most consistent with the NMR data and the known conformational properties of the amino acids comprising the peptides. The Discussion section begins with a summary of published results from receptor binding studies of the same set of peptides (Payza, 1987; Payza et al., 1989). We compare experimental conditions between the NMR and receptor binding experiments and show that the NMR conformations are directly correlated with binding affinities. The strong correlation leads to a model in which the active conformation of a peptide is only a fraction (up to 100%) of the ensemble of conformations in solution. We end by discussing the biological significance of the conformational ensemble model and propose a set of experiments to further test its validity.
MATERIALS AND METHODS
Peptide synthesis. Peptides were synthesized by Alfred Chung (University of Florida Interdisciplinary Center for Biotechnology Research Protein Chemistry Core) at the 0.25 mm scale. After the synthesis of the conserved PFLRF-NH2 C terminus, the reaction was split for the individual synthesis of the different N-terminal extensions. The peptides were made with a 432A Peptide Synthesizer (Applied Biosystems, Foster City, CA) using standard 4-(fluorenylmethyloxy)carbonyl chemistry and were cleaved from the solid support with trifluoroacetic acid in the presence of appropriate scavengers. The scavengers were extracted with t-butyl methyl ether, and the peptides were diluted with acetic acid and freeze-dried. When necessary, peptides were purified by HPLC. All peptides used for NMR were at least 95% pure, as estimated by analytical HPLC and mass spectrometry.
NMR spectroscopy. NMR data were collected at 600 MHz using a Varian Unity 600 (Palo Alto, CA) at the University of Florida Center for Structural Biology. Approximately 1–5 mmsamples were prepared with 90% H2O, 10% D2O, and 0.3 mm 3-trimethylsilyl (2,2,3,3-2H4) propionic acid for an internal chemical shift standard (0.0 ppm). Unless otherwise indicated, the pH (uncorrected for D2O) of each sample was adjusted to between 4.5 and 5.5. The temperature of the samples during data collection was 4°C unless indicated otherwise. For each peptide, we compared one-dimensional 1H spectra between samples under the following conditions: high concentration peptide (1–5 mm) and low salt, high concentration peptide and 150 mm KCl, and a 10-fold dilution of the high concentration samples. None of the chemical shifts for any of the samples deviated by more than ∼0.02 ppm, showing the absence of any aggregation or salt effects.
Data were obtained with the transmitter centered on the H2O peak, which was reduced using presaturation or by excitation sculpting (Callihan et al., 1996). Proton spectral widths were between 10 and 13 ppm. One-dimensional data were generally collected with 16,384 points; two-dimensional data were collected with 2048 or 4096 complex and 512 complex points in the acquisition and indirect dimensions, respectively.
One-dimensional 1H pH titrations were from pH 2 to 5.6 at roughly 0.2 pH increments; the pH was adjusted by adding small volumes of concentrated KOH or HCl to the samples. Two-dimensional total correlation spectroscopy (Braunschweiler and Ernst, 1983) data were collected using an MLEV-17 mixing sequence applied for 60 msec with an 8.8 kHz radio frequency (rf) field strength. Rotating Overhauser effect spectroscopy (ROESY) (Bothner-By et al., 1984) data were collected using continuous wave irradiation during the mixing time with an rf field strength of 2.2–3 kHz and mixing times ranging from 150 to 300 msec. Quadrature detection in the indirect dimensions was achieved by the method of States and coworkers (1982).
Data were processed using the computer software NMRPipe (Delaglio et al., 1995) by first eliminating the residual water peak using spectral deconvolution followed by appodization with squared cosine functions, zero-filling to twice the original data size, Fourier transformation, and baseline correction. Data were sequentially assigned using standard methods (Wüthrich, 1986) using NMRview (Johnson and Blevins, 1994). Peak positions and intensities were determined using the automatic peak-picking routine in NMRview (Johnson and Blevins, 1994). Intensities were also checked by extracting one-dimensional slices through the peaks of interest.
RESULTS
NMR data were collected for the naturally occurring and modified peptides shown in Table 1 (Linacre et al., 1990; Price et al., 1990; Saunders et al., 1991; Lutz et al., 1992; Kellett et al., 1994). The series of peptides were chosen because of detailed binding studies that revealed large differences in receptor binding affinities as a function of amino acid sequence (Payza, 1987;Payza et al., 1989). We used standard NMR methods to assign each resonance (peak) in the NMR spectrum to a particular atom in the peptide (Wüthrich, 1986). From the peak assignments, we obtained structural information from the positions of the peaks (chemical shifts), the pH dependence of the peaks, and interactions between atoms closer than 5 Å in space.
Table 1.
Peptides from this study and their measured NMR parameters
| Sequencea | INN/IαNF4–L51-b | Δδ pH F4 (ppm)1-c |
|---|---|---|
| PFLRFad | 0.0e | NAe |
| DPFLRFad | 0.20f | 0.128f |
| SDPFLRFa | 0.48 (4°C) | 0.33 |
| 0.51 (15°C) | ND | |
| 1.58 (26°C) | ND | |
| GDPFLRFa | 0.45 (4°C) | 0.31 |
| 0.72 (15°C) | ND | |
| 1.54 (26°C) | ND | |
| NDPFLRFa | 0.24 | 0.33 |
| pQDPFLRFa | 0.28 | 0.35 |
| GYPFLRFad | 0.0 | NA |
| pQYPFLRFad | 0.0 | NA |
NA, Not applicable (no titrating group); ND, not done.
One-letter amino acid code with “pQ” = pyroglutamic acid and with “a” = NH2.
Ratio of intensities of amide-to-amide (INN) and α-to-amide (IαN) ROESY cross peaks between F4 and L5.
Magnitude of the shift (Δ) in chemical shift (δ) of the amide proton of F4 as a function of pH. The values represent the shift at highest pH minus the shift at lowest pH.
dNot naturally occurring.
Data are for F2 and L3.
fData are for F3 and L4.
Short linear peptides can be very flexible and often are completely averaged in solution. However, many examples can be cited in which short peptides have high populations of turns or helices (Dyson et al., 1988a,b; Mayo et al., 1991; Miick et al., 1992; Yao et al., 1994;Millhauser et al., 1996; Yeagle et al., 1997). It must be stressed thatall of the NMR data shown below represent populations of averaged structures (Jardetzky, 1980; Bradley et al., 1990;Dz̆akula et al., 1992). However, the population averaging varies in the series of peptides in this study from approximately zero to 80% turn.
Chemical shifts
Chemical shifts are extremely sensitive probes of molecular conformation. Despite recent advances in empirical (Wishart and Sykes, 1994) and theoretical (Oldfield, 1995) understanding of chemical shifts in proteins, conformational averaging and interactions with solvents currently prevent a quantitative use of chemical shifts in small peptides. Each atom in each amino acid has a characteristic and known random coil chemical shift value when the amino acid is completely random and without structure (Wishart and Sykes, 1994). Useful qualitative information can be gleaned from groups of similar peptides by comparing their chemical shifts with each other and with random coil values (Wishart and Sykes, 1994).
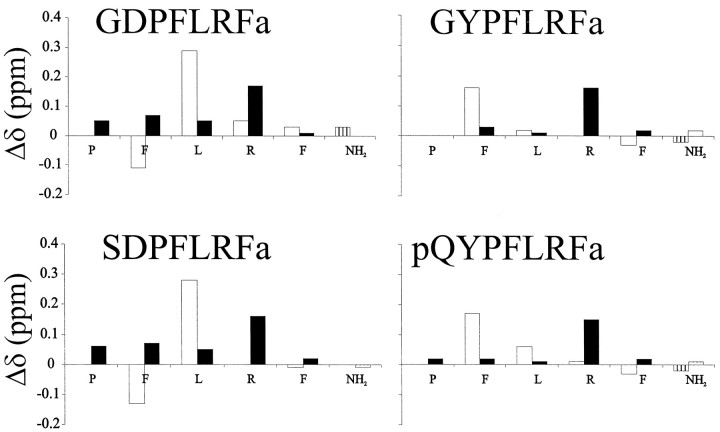
Figure 1 shows differences between random coil values of the amide (HN) and α (Hα) proton chemical shifts of the conserved PFLRF-NH2 region of representative peptides from Table 1. The histograms of Figure 1 provide a “fingerprint” of the overall conformational ensemble of each peptide. The major feature of Figure 1is that the peptides can be classified into DPFLRF-NH2 (DP) and YPFLRF-NH2 (YP) subgroups. It must be reemphasized that the shifts presented in Figure 1 are common to all peptides. Therefore, chemical shift data show that long-range conformational changes occur in these peptides as a result of the amino acid N terminal to the proline. A second feature of Figure 1 is that the deviations from random coil chemical shift values, in general, are larger for the DP than for the YP subgroup, suggesting that the DP peptides have higher populations of “nonrandom” conformations.
Fig. 1.
Deviations from random coil values (Wishart and Sykes, 1994) of HN (open bars) and Hα (solid bars) chemical shifts. The C-terminal amide values were subtracted from the average of all the values recorded in this study. Vertical axes are in parts per million, and the horizontal axes represent the conserved PNFLRF-NH2region of each peptide. The peptide pQYPFLRFa had a significant amount of overlap and conformational heterogeneity, preventing us from making complete assignments of all the side-chains, but the backbone assignments shown in Figure 1 are complete. Data were collected at approximately pH 5.5 and 4°C.
Internuclear distances
Chemical shifts are extremely sensitive to molecular conformation and thus provide a good probe for comparing similar molecules. However, details about particular conformations such as turns can best be obtained by direct measurement of internuclear distances. Two-dimensional ROESY experiments yield cross peaks between protons closer than 5 Å in space; the closer the protons, the stronger the peak.
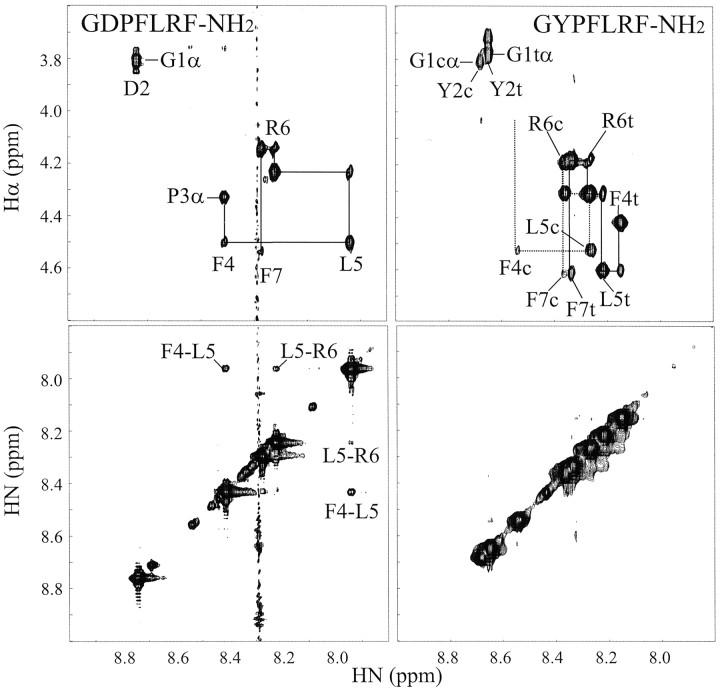
Figure 2 shows the α-to-amide and amide-to-amide regions of ROESY spectra from GDPFLRF-NH2and GYPFLRF-NH2, which are representative of other DP and YP peptides. One of the most notable features in Figure 2 is the large difference in the amide region (bottom panels) between the two peptides. The ROESY spectrum of GDPFLRF-NH2 has strong cross peaks between the amide protons of F4 and L5 and weaker peaks between L5 and R6. In contrast, GYPFLRF-NH2 has no measurable amide-to-amide cross peaks except between F7 and the NH2 group (not included in Fig. 2), a feature present in all of the dozens of amidated peptides that we have studied (our unpublished observations). The pattern of ROESY amide-to-amide cross peaks in GDPFLRF-NH2 is consistent with a reverse turn (Dyson et al., 1988a); the lack of similar ROESY cross peaks in GYPFLRF-NH2 indicates that it is extended or randomly oriented.
Fig. 2.
α-to-amide (top) and amide-to-amide (bottom) regions of ROESY spectra of GDPFLRF-NH2 (left) and GYPFLRF-NH2 (right). The sequential assignments are shown in the top panels, with the amino acid labels referring to the amide proton positions. The cross peaks in the amide-to-amide region representing the more compact structure in GDPFLRF-NH2 are labeled. The stripe at 8.3 ppm in the GDPFLRF-NH2 spectrum was an instrumental artifact. The temperature for both spectra was 4°C, the pH was ∼5.5, and mixing times were 250 msec.
The α-to-amide region (Fig. 2, top panels) shows two main features. First, GYPFLRF-NH2 has significant heterogeneity associated with the two forms (cis and trans) of the Y–P peptide bond. GDPFLRF-NH2, on the other hand, is predominantly trans, with <10% of the population in the cis form. All peptides examined in this study hadtrans X-Pro peptide bonds as the major species, as evidenced by ROESY cross peaks between Pro-Hδ and X-Hα or X-Hβ (Dyson et al., 1988a,b). We estimate that ∼90% of the DP and 70% of the YP subgroups have trans X-Pro peptide bonds. Second, the chemical shift dispersion, a good indicator of structure, is greater in GDPFLRF-NH2 than in GYPFLRF-NH2.
Interactions between amide protons of adjacent amino acids are particularly sensitive to secondary structure: amide-to-amide distances are large in extended structures but short in turns or helical structures. In contrast, the distance between an α proton of one amino acid and the amide proton of the next amino acid is characteristically short for extended structures but long in turns or helical structures (Wüthrich, 1986; Dyson et al., 1988a,b). Thus, the ratio of the intensity of ROESY amide-to-amide and α-to-amide cross peaks (INN/IαN) can provide a measure of the percentage of compact structure (Bradley et al., 1990). Table 1 listsINN/IαN values for interactions between F4 and L5 for all of the peptides in this study. TheINN/IαN values range from 0.0 for all of the YP peptides to >1.5 for the DP peptides. These numbers will be more thoroughly examined in Discussion.
All of the data presented to this point have been collected at 4°C to match experimental binding conditions (see Discussion). For two of the peptides (GDPFLRF-NH2 and SDPFLRF-NH2), we collected ROESY data at higher temperatures. To our surprise, the INN/IαNvalues became larger as the temperature increased, suggesting that the turn population increases at room temperature.
pH dependence
The pH dependence of the chemical shifts of titratable groups in peptides can provide important data about hydrogen bonding (Bundi and Wüthrich, 1979). Within the pH range investigated in this study (1.9–5.5), the only titratable group in the peptides of Table 1 is aspartic acid (D). Generally, the peptide C terminus is titratable, but each peptide in this study has a C-terminal amide because all of the FLPs are amidated in vivo. Amide, α, and side-chain protons within aspartic or glutamic acids undergo intrinsic shifts that, in general, contain little conformational information. Protons outside of titratable amino acids often show little or no dependence on pH. However, protons that interact closely through hydrogen bonding with carboxylic acid side-chains will exhibit chemical shift changes as a function of pH and thus provide direct evidence of long-range interactions.
Figure 3 shows the pH dependence of the amide protons of GDPFLRF-NH2, pQDPFLRF-NH2, and DPFLRF-NH2. The other XDPFLRF-NH2 (X = S, N) peptides were similar to GDPFLRF-NH2 and are not shown. Each peptide shows the characteristic upfield shift of the aspartic acid (D) HN (Bundi and Wüthrich, 1979) and very large downfield shifts of the amide protons of the phenylalanine after the proline. These data clearly demonstrate strong hydrogen bonding interactions between the carboxylate side-chain of aspartic acid and the amide proton of phenylalanine-4 (F4) in GDPFLRF-NH2. The smaller magnitude of the shifts in DPFLRF-NH2 suggests more moderate interactions or a lower population of the hydrogen-bonded conformation. The magnitude of the shifts of F4 (F3 in DPFLRF-NH2) over the entire pH range for each peptide are given in Table 1.
Fig. 3.
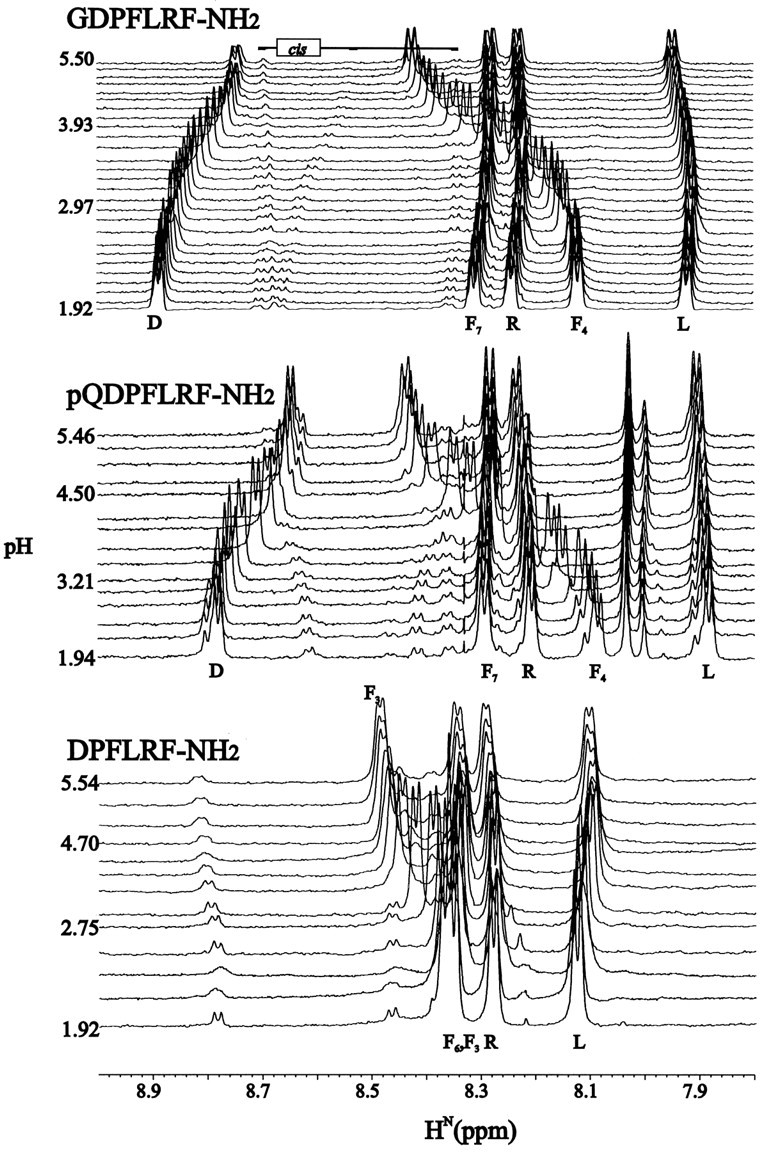
Amide regions of one-dimensional pH titrations for GDPFLRF-NH2 (top), pQDPFLRF-NH2(middle), and DPFLRF-NH2(bottom). The total magnitudes (highest pH minus lowest pH) of the changes in the phenylalanine chemical shift for each peptide studied are shown in Table 1. Series of one-dimensional 1H spectra are drawn from lowest to highest pH (bottom totop), with some of the values of pH indicated. Minor peaks in the spectra correspond to cis D–P peptide bonds. All data were collected at 4°C.
On the basis of data from pH titrations of bovine pancreatic trypsin inhibitor, Bundi and Wüthrich (1979) estimated that a 100% population of an amide to carboxylate hydrogen bond will give rise to a 0.4 ppm downfield shift. Assuming linearity, they claimed that a 0.04 ppm downfield amide shift corresponds to a 10% hydrogen-bonded population. Those estimates translate into ∼80% of GDPFLRF-NH2 and SDPFLRF-NH2 and 30% of DPFLRF-NH2 in hydrogen-bonded conformations.
Conformational models
The NMR data are all consistent with significant populations of reverse turns in the DP subgroup and predominantly extended or “random” conformations in the YP subgroup. From the NMR data alone, we are unable to specify a particular type of turn. However, on the basis of the following facts, we are confident that the peptides in the DP subgroup form type I reverse turns. First, aspartic acid and proline are very commonly found in the first and second positions, respectively, of type I turns (Wilmot and Thornton, 1988). Second, crystal structures of proteins with DP in the first two positions of a type I turn often reveal the aspartic acid side-chain hydrogen bonded to the amide proton of amino acid after the proline (Wilmot and Thornton, 1988). This hydrogen bonding is identical to what we observe through the pH titration data, with large downfield shifts of the amide protons of the phenylalanine after the proline.
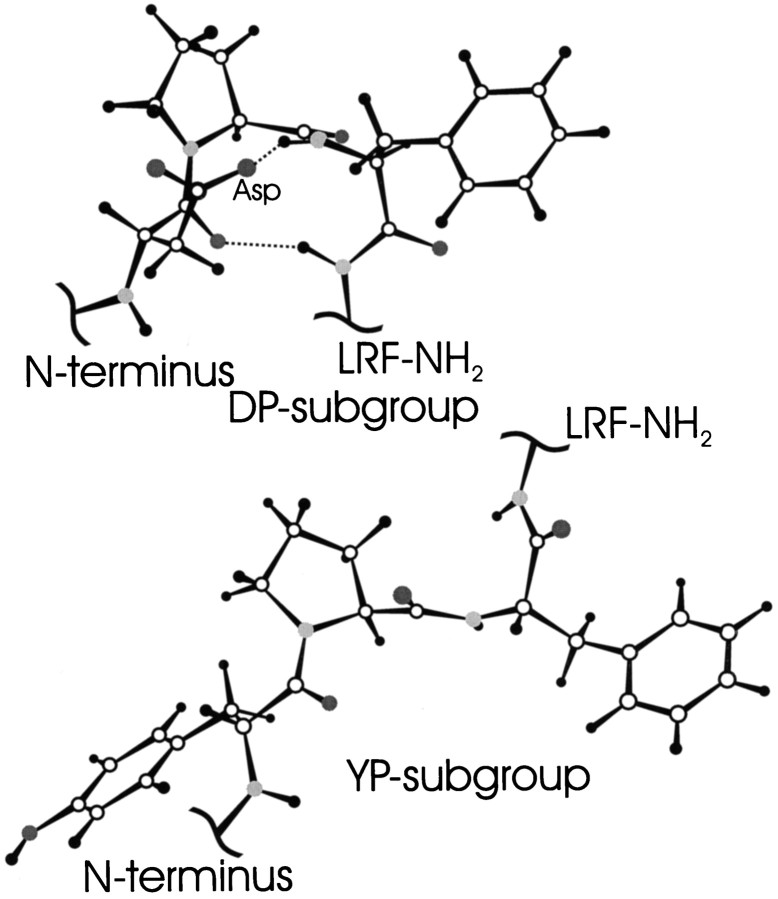
Models that are consistent with the NMR data for both the DP and YP peptides are shown in Figure 4. The most populated conformation in the DP peptides is a turn, and the YP peptides do not have any well defined structure. In addition, we see no well defined structure in the entire “LRF-NH2” C-terminal region in any of the peptides. The conserved C terminus is directly involved in receptor binding and must be able to fit into an active site, and our data suggest that this fit requires a flexible group.
Fig. 4.
Molecular models of DP (top) and YP (bottom) subgroups. Both models include the amino acids XPF (X = D or Y) and the amide group of L. The top is a type I reverse turn with a geometry that is consistent with the NMR data, including the close proximity of the D side-chain to the F amide proton and the F and L amide protons. The YP subgroup was drawn in one of many possible extended structures, representing a complete lack of close contacts seen in the DP peptides. The YP model and all amino acids not represented here from both groups are completely averaged. Models were made using Insight II (Biosym).
DISCUSSION
Receptor binding studies
The peptides for this study were chosen because of previously published detailed physiological and ligand displacement studies of the same peptides on membrane preparations from the brains of the snailHelix aspersa (Payza, 1987; Payza et al., 1989). The receptor binding results most relevant to this study are shown in Table2.
Table 2.
Summary of receptor binding studies in Helixbraina
| Peptide2-b | IC50(μm)2-c |
|---|---|
| FMRFa | 0.53 |
| FLRFa | 1.3 |
| PFLRFa | 0.86 |
| DPFLRFa | 3.4 |
| NDPFLRFa | 9 |
| SDPFLRFa | 15 |
| pQDPFLRFa | 10 |
| pQYPFLRFa | 0.22 |
| daYPFLRFa | 0.20 |
This table is composed from data in Table 2 of Payza et al. (1989).
Abbreviations include “a” = NH2, “pQ” = pyroglutamic acid, “da” = des-amino, “nL” = norleucine.
Concentration needed to displace 50%125I-da YFnLRFa from a Helix brain membrane preparation.
Table 2 shows that the peptides with the sequence XDPFLRFa (X = S, N, pQ) need between 3 and 20 times the concentration to displace a reporter molecule than FLRFa, PFLRFa, or DPFLRFa. These effects are not simply a result of a peptide that is too long, as illustrated by the high receptor affinity of pQYPFLRFa (almost 10 times greater than FLRFa). Nor can these results be easily understood in terms of unfavorable interactions with the receptor because, for example, the pQ at the N terminus of DPFLRFa lowers the receptor affinity by a factor of three, whereas pQ at the N terminus of daYPFLRFa has no effect on binding.
Comparison of NMR and receptor binding experimental conditions
The receptor binding experiments were performed with assay buffer of 80 mmpiperazine-n,n′-bis(2-hydroxypropanesulfonic acid), pH 7.9, 1% BSA (Payza, 1987; Payza et al., 1989). To minimize the degradation of peptides, the receptor binding assays were performed on ice at 0°C.
NMR measurements have several experimental constraints. First, for solution studies in aqueous samples, the temperature must be above freezing. We used 4°C for most of the NMR results presented in this report. Second, amide protons exchange too rapidly for observation around pH 8. The pH titrations shown above clearly demonstrate that at around pH 5.5, the aspartic acid side-chains are fully deprotonated; no further change in these or any other side-chains will take place as the pH is increased. The N-terminal amino group is the only part of the peptides that would still be affected by differences in pH between the binding and NMR studies. Third, NMR measurements are notoriously insensitive and require close to 1 mm concentrations of sample, leading to the possibility of aggregation. We tested for aggregation in each sample by making 10-fold dilutions and recording one-dimensional 1H spectra. In the dilution studies, no peak moved more than 0.02 ppm, demonstrating that aggregation is negligible at the concentrations used for NMR analysis. Finally, NMR is sensitive to salt concentrations, so all of the data presented above are in the minimal salt needed to adjust the pH. To rule out effects of salt, we added 150 mm KCl to each sample and measured the one-dimensional 1H spectra. As with the dilution studies, we saw no change in chemical shift >0.02 ppm between high- and low-salt conditions, demonstrating that KCl has little or no influence on these samples.
Thus, we are confident that the NMR experimental conditions are at least relevant, if not closely matched, to the receptor binding assays. The only factor that we suspect may be different between the binding and NMR data is the degree of protonation of the N-terminal amino groups between pH 5.5 and 7.9.
Comparison of NMR and receptor binding results
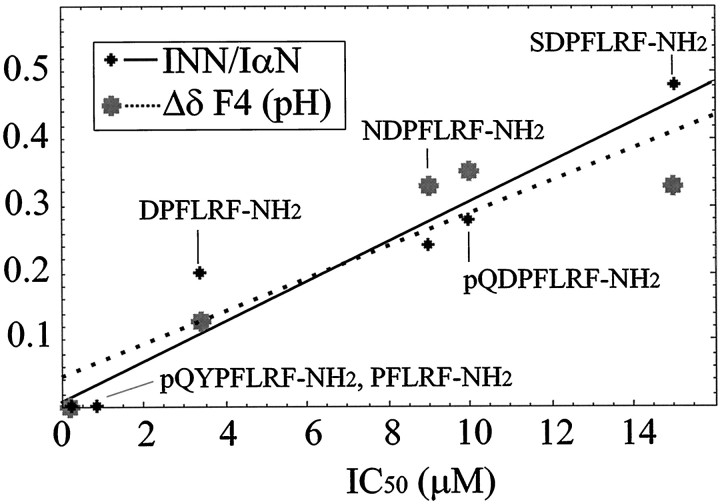
Figure 5 is a plot of the measured NMR parameters that are sensitive to turn population (INN/IαN and pH shifts from Table 1) versus the IC50 values measured for the same peptides (Payza, 1987; Payza et al., 1989). Both NMR parameters increase with increasing values of IC50. The simplest interpretation of the NMR data is a rapid two-state equilibrium between a reverse turn and fully extended structure. For the DP peptides, the equilibrium is shifted in varying amounts toward the turn. For the YP peptides and PFLRF-NH2, the equilibrium is nearly 100% in favor of the extended structure.
Fig. 5.
Plots ofINN/IαN(small black dots and solid line) and pH titration (large gray dots and dotted line) data versus IC50 measurements. The vertical axis is parts per million for the pH titration and unitless forINN/IαN. The IC50 data represent the concentration of peptide needed to displace 50% of 125I-daYFnLRF-NH2 from aHelix brain membrane preparation (Payza, 1987; Payza et al., 1989). TheINN/IαNand pH titration data are from Table 1 and are described in the text. Data were fit to linear equations, IC50 =a (NMR) + b(INN/IαNdata: a = 33.33, b = −0.27,r2 = 0.93; pH data:a = 41.67, b = −1.79,r2 = 0.82). A data point for the pH dependence of pQYPFLRF-NH2 at 0 ppm was added, because there is no titratable group and thus no pH dependence.
A simple two-state model will lead to a linear relationship between the percentage of turn and the magnitude of bothINN/IαN and the pH shifts. Thus, we used a linear relationship to fit the NMR parameters to the IC50 data. The correlation coefficients (r2) for the fits were quite good at 0.93 and 0.82 for theINN/IαN and pH data, respectively. Deviations from linearity could result from either more than two predominant conformations in the unbound state or from additional (or different) factors involved in the binding data, such as direct interactions (favorable or unfavorable) between the ligands and receptor. The effects of multiple conformations are currently under investigation in modeling studies. We present a number of IC50 predictions below to further test our model.
Biological roles of conformational ensembles
The strong linear structure–function relationships in Figure 5suggest that the observed IC50 data can best be explained by the conformation of the unbound peptides. Namely, peptides with high populations of turn displace radio-labeled ligands at higher concentrations, and the turns are preventing the peptides from interacting with the receptors in the binding assay. These relationships suggest a simple modification to elementary ligand–receptor equations. In the simplest case of binding, a ligand (L) binds to a receptor (R) to form a ligand–receptor complex (LR) with an equilibrium dissociation constant Keq:
The fraction of bound receptors (PB) for this simple equilibrium is:
The effect of our model is to reduce the active form of the ligand to LA, which in the current study represents peptides in extended or random conformations:
The inactive form of the ligand, LI(here representing molecules in a turn conformation), is not able to bind directly to the receptor, thus reducing the fraction of bound receptors:
It must be stressed that active or inactive forms of a ligand are defined with respect to a particular receptor.
The model and data described above apply to a single peptide ligand and a single receptor that binds the active ligand (LA) conformation. However, receptor heterogeneity can play a large role in our model. As described above, the role of conformational ensembles of a single ligand interacting with a single receptor will lead to changes in the bound population of that receptor. If there is a second receptor that preferentially binds to the inactive form (defined with respect to the first receptor) of the same ligand, then more complicated signaling can occur. Depending on the degree of coupling between the two receptors, the resultant signals from the two different conformations of the peptide could be divergent or convergent, leading to increased “physiological flexibility” (Brezina et al., 1996).
Payza and coworkers found that the HelixXDPFLRF-NH2 peptides have a complicated physiology. At concentrations <0.03 μm they are ∼30–50 times more cardiostimulatory than FMRF-NH2, but at concentrations >0.03 μm they are cardioinhibitory (Payza, 1987). From these observations, Payza hypothesized that the XDPFLRF-NH2 peptides must interact with different or additional receptors. We showed above that the XDPFLRF-NH2peptides have up to ∼80% reverse turn in solution. Therefore,if we assume that the XDPFLRF-NH2 peptides bind with the same affinity to both the stimulatory and inhibitory receptors, then our model would predict that stimulatory receptor preferentially binds the turn conformation (dominant conformation) and the inhibitory receptor preferentially binds the extended conformation (minor conformation). Clearly, more data are needed to prove this relationship.
There are currently two cloned FLP receptors: a peptide-gated sodium channel (Green et al., 1994; Linguelglia et al., 1995) and a G-protein-coupled receptor (Tensen et al., 1998). In addition to the two sequences, several other FLP receptors have been biochemically characterized (Payza, 1987; Payza et al., 1989; Chin et al., 1994; Wang et al., 1995a,b). All available evidence, therefore, points to the presence of multiple FLP receptors (Tensen et al., 1998). We hypothesize that the effect of conformational ensembles in differential binding to multiple receptors will be widespread.
Most of the focus in ligand–receptor interactions has naturally involved the ligand–receptor complex, and these direct contacts with receptors are obviously important. However, our data clearly show the importance of the unbound conformational state in ligand–receptor interactions. From our model we can make a number of predictions for future binding assays under the same conditions used by Payza and coworkers (1987, 1989). (1) On the basis of our NMR measurements (Table1), GDPFLRF-NH2 and GYPFLRF-NH2 will have IC50 values of 13 μm and <1 μm, respectively. (2) YDPFLRF-NH2 will have IC50 values near the other DP peptides (10–15 μm). (3) SDZPFLRF-NH2 (Z any amino acid except D, N, or P) will have IC50 values near the YP or tetra-peptides (<1 μm). (4) SNPFLRF-NH2 will have IC50 values near SDPFLRF-NH2 (15 μm), because asparagine (N) has hydrogen bonding properties and type I turn propensities (Wilmot and Thornton, 1988) similar to those of aspartic acid (D).
We have demonstrated that populations of reverse turns in a series of neuropeptides are inversely correlated with their receptor-binding affinities. This finding suggests that one of the roles for diverse N-terminal amino acid sequences in FLPs is to provide different unbound conformational ensembles and thus lead to different receptor binding affinities.
Footnotes
These studies were supported by a grant from the Florida Affiliate of the American Heart Association (A.S.E.), the University of Florida Howard Hughes Medical Institute Pilot Studies program, and the University of Florida Brain Institute. Dan Plant and James Rocca, University of Florida Center for Structural Biology, provided important technical support. We thank Matt Carrigan and Professor Glen Cottrell for helpful and stimulating discussions and Professors Ben Dunn and Gerry Shaw for critically reading this manuscript. We give special thanks to the reviewer whose helpful comments led to a greatly improved manuscript.
Correspondence should be addressed to Dr. Arthur S. Edison, Department of Biochemistry and Molecular Biology, Center for Structural Biology, University of Florida Brain Institute, and National High Magnetic Field Laboratory, University of Florida, Gainesville, FL 32610-0245.
REFERENCES
- 1.Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- 2.Bothner-By AA, Stephens RL, Lee J, Warren CD, Jeanloz RW. Structure determination of a tetrasaccharide: transient nuclear Overhauser effects in the rotating frame. J Am Chem Soc. 1984;106:811–813. [Google Scholar]
- 3.Bradley EK, Thomason JF, Cohen FE, Kosen PA, Kuntz ID. Studies of synthetic helical peptides using circular dichroism and nuclear magnetic resonance. J Mol Biol. 1990;215:607–622. doi: 10.1016/S0022-2836(05)80172-X. [DOI] [PubMed] [Google Scholar]
- 4.Braunschweiler L, Ernst RR. Coherence transfer by isotropic mixing: application to protein correlation spectroscopy. J Magn Reson. 1983;53:521–528. [Google Scholar]
- 5.Brezina V, Orekhova IV, Weiss KR. Functional uncoupling of linked neurotransmitter effects by combinatorial convergence. Science. 1996;273:806–810. doi: 10.1126/science.273.5276.806. [DOI] [PubMed] [Google Scholar]
- 6.Bundi A, Wüthrich K. Use of amide 1H-NMR titration shifts for studies of polypeptide conformation. Biopolymers. 1979;18:299–311. [Google Scholar]
- 7.Callihan D, West J, Kumar S, Schweitzer BI, Logan TM. Simple, distortion-free homonuclear spectra of peptides and nucleic acids in water using excitation sculpting. J Magn Reson B. 1996;112:82–85. doi: 10.1006/jmrb.1996.0114. [DOI] [PubMed] [Google Scholar]
- 8.Chin GJ, Payza K, Price DA, Greenberg MJ, Doble KE. Characterization and solubilization of the FMRFamide receptor of squid. Biol Bull. 1994;187:185–189. doi: 10.2307/1542241. [DOI] [PubMed] [Google Scholar]
- 9.Cowden C, Stretton AOW. AF2, an Ascaris neuropeptide: isolation, sequence, and bioactivity. Peptides. 1993;14:423–430. doi: 10.1016/0196-9781(93)90127-3. [DOI] [PubMed] [Google Scholar]
- 10.Cowden C, Stretton AOW. Eight novel FMRFamide-like neuropeptides isolated from the nematode Ascaris suum. Peptides. 1995;16:491–500. doi: 10.1016/0196-9781(94)00211-n. [DOI] [PubMed] [Google Scholar]
- 11.Cowden C, Stretton AOW, Davis RE. AF1, a sequenced bioactive neuropeptide isolated from the nematode Ascaris suum. Neuron. 1989;2:1465–1473. doi: 10.1016/0896-6273(89)90192-x. [DOI] [PubMed] [Google Scholar]
- 12.Creighton TE. Proteins: structures and molecular properties. W. H. Freeman; New York: 1993. [Google Scholar]
- 13.Davis RE, Stretton AOW. The motornervous system of Ascaris: electrophysiology and anatomy of the neurons and their control by neuromodulators. Parasitology. 1996;113:S97–S117. doi: 10.1017/s0031182000077921. [DOI] [PubMed] [Google Scholar]
- 14.Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 15.Dickinson PS, Mecsas C, Marder E. Neuropeptide fusion of two motor-pattern generator circuits. Nature. 1990;344:155–158. doi: 10.1038/344155a0. [DOI] [PubMed] [Google Scholar]
- 16.Dyson HJ, Rance M, Houghten RA, Lerner RA, Wright PE. Folding of immunogenic peptide fragments of proteins in water solution. I. Sequence requirements for the formation of a reverse turn. J Mol Biol. 1988a;201:161–200. doi: 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]
- 17.Dyson HJ, Rance M, Houghten RA, Wright PE, Lerner RA. Folding of immunogenic peptide fragments of proteins in water solution: II. The nascent helix. J Mol Biol. 1988b;201:201–217. doi: 10.1016/0022-2836(88)90447-0. [DOI] [PubMed] [Google Scholar]
- 18.Dz̆akula Z̆, Westler WM, Edison AS, Markley JL. The CUPID method for calculating the continuous probability distribution of rotamers from NMR data. J Am Chem Soc. 1992;114:6195–6199. [Google Scholar]
- 19.Edison AS, Messinger LA, Stretton AOW. afp-1: A gene encoding multiple transcripts of a new class of FMRFamide-like neuropeptides in the nematode Ascaris suum. Peptides. 1997;18:929–935. doi: 10.1016/s0196-9781(97)00047-8. [DOI] [PubMed] [Google Scholar]
- 20.Green KA, Falconer SWP, Cottrell GA. The neuropeptide FMRFamide directly gates two ion channels in an identified Helix neurone. Pflügers Arch. 1994;428:232–240. doi: 10.1007/BF00724502. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg MJ, Price DA. Relationships among the FMRFamide-like peptides. In: Joosse J, Buijs RM, Tilders FJH, editors. Progress in brain research. Elsevier; Amsterdam: 1992. pp. 25–37. [DOI] [PubMed] [Google Scholar]
- 22.Jardetzky O. On the nature of molecular conformations inferred from high-resolution NMR. Biochim Biophys Acta. 1980;621:227–232. doi: 10.1016/0005-2795(80)90174-9. [DOI] [PubMed] [Google Scholar]
- 23.Johnson BA, Blevins RA. NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 24.Kellett E, Saunders SE, Li KW, Staddon JW, Benjamin PR, Burke JF. Genomic organization of the FMRFamide gene in Lymnaea: multiple exons encoding novel neuropeptides. J Neurosci. 1994;14:6564–6570. doi: 10.1523/JNEUROSCI.14-11-06564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linacre A, Kellett E, Saunders S, Bright K, Benjamin PR, Burke JF. Cardioactive neuropeptide FMRFamide and novel related peptides are encoded in multiple copies by a single gene in the snail Lymnaea stagnalis. J Neurosci. 1990;10:412–419. doi: 10.1523/JNEUROSCI.10-02-00412.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linguelglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995;378:730–733. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 27.Lutz EM, MacDonald M, Hettle S, Price DA, Cottrell GA, Sommerville J. Structure of cDNA clones and genomic DNA encoding FMRFamide-related peptides (FaRPs) in Helix. Mol Cell Neurosci. 1992;3:373–382. doi: 10.1016/1044-7431(92)90049-8. [DOI] [PubMed] [Google Scholar]
- 28.Mayo KH, Parra-Diaz D, McCarthy JB, Chelberg M. Cell adhesion promoting peptide GVKGDKGNPGWPGAP from the collagen type IV triple helix: cis/trans proline-induced multiple 1H NMR conformations and evidence for a KG/PG multiple turn repeat motif in the all-trans proline state. Biochemistry. 1991;30:8251–8267. doi: 10.1021/bi00247a022. [DOI] [PubMed] [Google Scholar]
- 29.Miick SM, Martinez GV, Fiori WR, Todd AP, Millhauser GL. Short alanine-based peptides may form 310-helices and not α-helices in aqueous solution. Nature. 1992;359:653–655. doi: 10.1038/359653a0. [DOI] [PubMed] [Google Scholar]
- 30.Millhauser GL, Stenland CJ, Bolin KA, van de Ven FJM. Local helix content in an alanine-rich peptide as determined by the complete set of 3JHNα coupling constants. J Biomol NMR. 1996;7:331–334. doi: 10.1007/BF00200434. [DOI] [PubMed] [Google Scholar]
- 31.Nelson LS, Rosoff ML, Li C. Disruption of a neuropeptide gene, flp-1, causes multiple behavioral defects in Caenorhabditis elegans. Science. 1998;281:1686–1690. doi: 10.1126/science.281.5383.1686. [DOI] [PubMed] [Google Scholar]
- 32.Oldfield E. Chemical shifts and three dimensional protein structures. J Biomol NMR. 1995;5:217–225. doi: 10.1007/BF00211749. [DOI] [PubMed] [Google Scholar]
- 33.Payza K. FMRFamide receptors in Helix aspersa. Peptides. 1987;8:1065–1074. doi: 10.1016/0196-9781(87)90138-0. [DOI] [PubMed] [Google Scholar]
- 34.Payza K, Greenberg MJ, Price DA. Further characterization of Helix FMRFamide receptors: kinetics, tissue distribution, and interactions with the endogenous heptapeptides. Peptides. 1989;10:657–661. doi: 10.1016/0196-9781(89)90156-3. [DOI] [PubMed] [Google Scholar]
- 35.Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science. 1977;197:670–671. doi: 10.1126/science.877582. [DOI] [PubMed] [Google Scholar]
- 36.Price DA, Lesser W, Lee TD, Doble KE, Greenberg MJ. Seven FMRFamide-related and two SCP-related cardioactive peptides from Helix. J Exp Biol. 1990;154:421–437. doi: 10.1242/jeb.154.1.421. [DOI] [PubMed] [Google Scholar]
- 37.Rosoff ML, Bürglin TR, Li C. Alternatively spliced transcripts of the flp-1 gene encode distinct FMRFamide-like peptides in Caenorhabditis elegans. J Neurosci. 1992;12:2356–2361. doi: 10.1523/JNEUROSCI.12-06-02356.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S, Arch FRS, Buckingham RE, Haynes AC, Carr SA, Annan RS, McNulty DE, Liu W-S, Terrett JA, Elshourbagy NA, Bergsma DJ, Yanagisawa M. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 39.Saunders SE, Bright K, Kellett E, Benjamin PR, Burke JF. Neuropeptides GDPFLRFamide and SDPFLRFamide are encoded by an exon 3′ to FMRFamide in the snail Lymnaea stagnalis. J Neurosci. 1991;11:740–745. doi: 10.1523/JNEUROSCI.11-03-00740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon TW, Opdyke CA, Calabrese RL. Modulatory effects of FMRF-NH2 on outward currents and oscillatory activity in heart interneurons of the medicinal leech. J Neurosci. 1992;12:525–537. doi: 10.1523/JNEUROSCI.12-02-00525.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sossin WS, Fisher JM, Scheller RH. Cellular and molecular biology of neuropeptide processing and packaging. Neuron. 1989;2:1407–1417. doi: 10.1016/0896-6273(89)90186-4. [DOI] [PubMed] [Google Scholar]
- 42.States DJ, Haberkorn RA, Ruben DJ. A two dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J Magn Reson. 1982;48:286–292. [Google Scholar]
- 43.Taussig R, Scheller RH. The Aplysia FMRFamide gene encodes sequences related to mammalian brain peptides. DNA. 1986;5:453–461. doi: 10.1089/dna.1.1986.5.453. [DOI] [PubMed] [Google Scholar]
- 44.Tensen CP, Cox KJA, Smit AB, van der Schors RC, Meyerhof W, Richter D, Planta RJ, Hermann PM, van Minnen J, Geraerts WPM, Knol JC, Burke JF, Vreugdenhil E, van Heerikhuizen H. The Lymnaea cardioexcitatory peptide (LyCEP) receptor: a G-protein-coupled receptor for a novel member of the RFamide neuropeptide family. J Neurosci. 1998;18:9812–9821. doi: 10.1523/JNEUROSCI.18-23-09812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Orchard I, Lange AB, Chen X, Starratt AN. A single receptor transduces both inhibitory and stimulatory signals of FMRFamide-related peptides. Peptides. 1995a;16:1181–1186. doi: 10.1016/0196-9781(95)00093-y. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Lange AB, Orchard I. Coupling of a single receptor to two different G proteins in the signal transduction of FMRFamide-related peptides. Biochem Biophys Res Commun. 1995b;212:531–538. doi: 10.1006/bbrc.1995.2002. [DOI] [PubMed] [Google Scholar]
- 47.Weimann JM, Marder E, Evans B, Calabrese RL. The effects of SDRNFLRFamide and TNRNFLRFamide on the motor patterns of the stomatogastric ganglion of the crab Cancer borealis. J Exp Biol. 1993;181:1–26. doi: 10.1242/jeb.181.1.1. [DOI] [PubMed] [Google Scholar]
- 48.Wilmot CM, Thornton JM. Analysis and prediction of the different types of β-turn in proteins. J Mol Biol. 1988;203:221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- 49.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 50.Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 51.Wüthrich K. NMR of proteins and nucleic acids. Wiley; New York: 1986. [Google Scholar]
- 52.Yang H-Y, Fratta W, Majane EA, Costa E. Isolation, sequencing, synthesis, and pharmacological characterization of two brain neuropeptides that modulate the action of morphine. Proc Natl Acad Sci USA. 1985;82:7757–7761. doi: 10.1073/pnas.82.22.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao J, Dyson HJ, Wright PE. Three-dimensional structure of a type VI turn in a linear peptide in water solution: evidence for stacking of aromatic rings as a major stabilizing factor. J Mol Biol. 1994;243:754–766. doi: 10.1016/0022-2836(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 54.Yeagle PL, Alderfer JL, Salloum AC, Ali L, Albert AD. The first and second cytoplasmic loops of the G-protein receptor, rhodopsin, independently form β-turns. Biochemistry. 1997;36:3864–3869. doi: 10.1021/bi962403a. [DOI] [PubMed] [Google Scholar]