Abstract
The NMDA and AMPA classes of ionotropic glutamate receptors are concentrated at postsynaptic sites in excitatory synapses. NMDA receptors interact via their NR2 subunits with PSD-95/SAP90 family proteins, whereas AMPA receptors bind via their GluR2/3 subunits to glutamate receptor-interacting protein (GRIP), AMPA receptor-binding protein (ABP), and protein interacting with C kinase 1 (PICK1). We report here a novel cDNA (termed ABP-L/GRIP2) that is virtually identical to ABP except for additional GRIP-like sequences at the N-terminal and C-terminal ends. Like GRIP (which we now term GRIP1), ABP-L/GRIP2 contains a seventh PDZ domain at its C terminus. Using antibodies that recognize both these proteins, we examined the subcellular localization of GRIP1 and ABP-L/GRIP2 (collectively termed GRIP) and their biochemical association with AMPA receptors. Immunogold electron microscopy revealed the presence of GRIP at excitatory synapses and also at nonsynaptic membranes and within intracellular compartments. The association of native GRIP and AMPA receptors was confirmed biochemically by coimmunoprecipitation from rat brain extracts. A majority of detergent-extractable GluR2/3 was complexed with GRIP in the brain. However, only approximately half of GRIP was associated with AMPA receptors. Unexpectedly, immunocytochemistry of cultured hippocampal neurons and rat brain at the light microscopic level showed enrichment of GRIP in GABAergic neurons and in GABAergic nerve terminals. Thus GRIP is associated with inhibitory as well as excitatory synapses. Collectively, these findings support a role for GRIP in the synaptic anchoring of AMPA receptors but also suggest that GRIP has additional functions unrelated to the binding of AMPA receptors.
Keywords: AMPA receptor, GRIP, excitatory synapse, PDZ domain, immunoprecipitation, postsynaptic density
At excitatory synapses, ionotropic glutamate receptors of the NMDA class are highly concentrated in the postsynaptic density (PSD). The NR2 subunits of the NMDA receptors bind through their C-terminal cytoplasmic tails to the PDZ domains of PSD-95/SAP90 and related proteins, which are major components of the PSD (Kornau et al., 1995; Müller et al., 1996; Niethammer et al., 1996). The NR1 subunit of the NMDA receptor interacts with a distinct set of intracellular proteins, including α-actinin (Wyszynski et al., 1997), neurofilament-L (Ehlers et al., 1998), and yotiao (Lin et al., 1998). The interaction between NMDA receptors and PSD-95 family proteins may be important for the localization of NMDA receptors at postsynaptic sites. In Drosophila, the PSD-95 homologDiscs-large is essential for the synaptic clustering of its PDZ binding partners Shaker and Fasciclin II (Tejedor et al., 1997;Thomas et al., 1997; Zito et al., 1997). In addition, because of its multidomain structure, PSD-95 can also bind to several signaling and cytoskeletal proteins, including neuronal nitric oxide synthase (Brenman et al., 1996), synGAP (Chen et al., 1998; Kim et al., 1998), and CRIPT (Niethammer et al., 1998), thereby potentially bringing these proteins together in a complex (for review, see Craven and Bredt, 1998). By acting as scaffold proteins, PSD-95 and related molecules can organize a specific cytoskeletal-signaling complex that is physically linked to the NMDA receptor.
Ionotropic glutamate receptors of the AMPA class (composed of GluR1–4 subunits) are also concentrated at postsynaptic sites. Although they largely colocalize with NMDA receptors in excitatory synapses, AMPA receptors do not interact with PSD-95 family proteins. Instead, the AMPA receptor subunits GluR2/3 bind specifically to other PDZ proteins, termed glutamate receptor-interacting protein (GRIP) (Dong et al., 1997), AMPA receptor-binding protein (ABP) (Srivastava et al., 1998), and protein interacting with C kinase 1 (PICK1) (Xia et al., 1999). GRIP contains seven PDZ domains and no other recognizable motif, in contrast to PSD-95, which has three PDZ domains plus an Src homology 3 domain and a guanylate kinase-like domain. ABP resembles GRIP in primary sequence; it differs from GRIP most notably in lacking the C-terminal seventh PDZ domain. The C-terminal sequence (-ESVKI) shared by AMPA receptor GluR2/3 subunits is reported to bind selectively to the forth and fifth PDZ domains (PDZ4/5) of GRIP (Dong et al., 1997) and the third, fifth, and sixth PDZ domains of ABP (Srivastava et al., 1998). However, neither Dong et al. (1997) nor Srivastava et al. (1998)confirmed a biochemical association of GRIP and AMPA receptors in brain. We have recently found that GRIP is only modestly enriched in the PSD fraction when compared with PSD-95 (Wyszynski et al., 1998). These observations beg the question of how substantially AMPA receptors interact with GRIP and ABP in vivo.
We report here the cloning of a novel GRIP-like protein (termed ABP-L/GRIP2) that is virtually identical to ABP but that contains additional N-terminal and C-terminal sequences, including a seventh PDZ domain. ABP and ABP-L/GRIP2 appear to be splice variants of the same gene containing six or seven PDZ domains, respectively; thus the domain organizations, as well as the primary sequence, of the ABP and GRIP genes are highly conserved. We will henceforth use the term GRIP1 for the original GRIP polypeptide described by Dong et al. (1997). Using antibodies that recognize both GRIP1 and ABP-L/GRIP2 (these related proteins collectively termed GRIP), we show a wider subcellular distribution of these proteins in neurons than previously appreciated, including enrichment in GABAergic nerve terminals. Using a semiquantitative coimmunoprecipitation assay, we show that a majority of the detergent-extractable GluR2/3 is complexed with GRIP in adult rat brain tissue; however, a considerable fraction of GRIP is not associated with GluR2/3. Taken together, these results show for the first time the biochemical association of AMPA receptors and GRIPin vivo but suggest additional functions for GRIP other than that of anchoring AMPA receptors at synapses.
MATERIALS AND METHODS
Cloning of GRIP1 and ABP-L/GRIP2. Yeast two-hybrid screening was performed as described previously using the L40 yeast strain harboring HIS3 and β-galactosidase as reporter genes (Niethammer et al., 1996). Approximately 2 × 106 clones were screened of a rat brain cDNA library constructed in the GAL4 activation domain vector pGAD10 (Clontech, Palo Alto, CA). The two-hybrid bait consisted of a C-terminal peptide, -GRISYDL, fused to LexA (this peptide was an artifactual sequence generated aberrantly during PCR construction of another bait). The two-hybrid screen yielded multiple isolates of two distinct clones (clones 5 and 14). Clone 5 was a cDNA fragment of GRIP1, encoding amino acids 363–1112 (containing PDZ4 through the C terminus). Clone 14 encoded amino acids 556–1002 of ABP-L/GRIP2 (beginning within PDZ5 and extending through the C terminus). Additional GRIP1 and ABP-L/GRIP2 cDNA sequences were obtained by hybridization screening of λZAP II rat cortical and hippocampal cDNA libraries (Stratagene, La Jolla, CA) using as probes the GRIP1 and ABP-L/GRIP2 cDNA fragments described above. The 5′ ends of GRIP1 and ABP-L/GRIP2 were obtained by 5′ rapid amplification of cDNA ends using a Marathon-Ready rat brain cDNA library (Clontech). DNA sequences were obtained by automated sequencing. The nucleotide sequence of ABP-L/GRIP2 has been deposited in GenBank under accession number AF112182
Northern blots. A rat poly-A mRNA multi-tissue Northern blot (Clontech) was probed with 32P-labeled ABP-L/GRIP2 cDNA fragments (corresponding to amino acids 556–1002) under high-stringency conditions using ExpressHyb (Clontech) and exposed at −80°C on XAR-5 film (Eastman Kodak, Rochester, NY) for 50 hr.
Antibodies. GRIP antibodies (termed “1756” and “C8399”) were raised by immunizing two different rabbits with a hexahistidine (H6)-tagged fusion protein incorporating GRIP1 amino acids 664–1112 (includes PDZ6 and extends to the C terminus of the protein); 1756 antibodies were then affinity-purified on a Sulfolink column (Pierce, Rockford, IL) coupled to a soluble H6-tagged fusion protein of GRIP1 containing PDZ4–PDZ6 (residues 463–761) and have been described (Wyszynski et al., 1998). C8399-I antibodies were affinity-purified on a Sulfolink column coupled to a soluble glutathione S-transferase fusion protein incorporating GRIP1 amino acids 664–1112. The rabbit anti-PSD-95 antibody (CSK) has been previously described (Kim and Sheng, 1996). The following antibodies were obtained from commercial sources: polyclonal anti-GluR2/3 and monoclonal anti-GluR2-N antibodies (Chemicon, Temecula, CA); anti-NR1 monoclonal antibody 54.1 (PharMingen, San Diego, CA); anti-glutamic acid decarboxylase (GAD) monoclonal antibodies (Boehringer Mannheim, Indianapolis, IN); anti-GAD-6 monoclonal antibodies (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA); and nonimmune purified rabbit IgG and anti-synaptophysin monoclonal antibodies (Sigma, St. Louis, MO). The rabbit and guinea pig anti-GluR1 antibodies were gifts from Richard L. Huganir (Johns Hopkins University School of Medicine, Baltimore, MD).
Immunohistochemistry on brain and cultured neurons. Brain immunohistochemistry was performed on Vibratome-cut 50 μm floating brain sections from Sprague Dawley rats (∼6 weeks of age) perfused transcardiacally with 4% paraformaldehyde and treated with proteinase K as described (Lin et al., 1998). Primary anti-GAD monoclonal antibodies were used at 1 μg/ml. Hippocampal neuron cultures were prepared from hippocampi dissected from embryonic day 18 rat embryos by the methods of Banker and Cowan (1977) and Goslin and Banker (1991). Neurons were fixed at 3 weeks in culture with 4% paraformaldehyde and 4% sucrose in phosphate buffer, permeabilized with 0.25% Triton X-100, and immunolabeled using mouse anti-GluR1, mouse anti-GluR2-N, mouse anti-synaptophysin, or mouse anti-GAD-6 antibodies. All double labeling was performed with rabbit anti-GRIP antibody 1756 antibodies (1 μg/ml). Staining was visualized using Cy3- or FITC-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove PA).
Immunoelectron microscopy. For electron microscopy, five Sprague Dawley rats were perfusion-fixed with a mixture of 2–4% paraformaldehyde and 2% glutaraldehyde; brains were post-fixed for 2–4 hr. Fifty-micrometer-thick transverse sections were cut on aVibratome and processed for osmium-free embedment (for details, seePhend et al., 1995). Thin sections were cut and collected on 300-mesh uncoated nickel grids and stained for GRIP using anti-GRIP1 antibodies C8399-I and 1756. After treatment with 4% paraphenylenediamine in Tris-buffered saline containing detergent (TBS/T; 0.1 mTris, pH 7.6, with 0.005% Tergitol NP-10) for 1 hr, grids were rinsed and incubated overnight at 37°C in primary antibody (1:100), rinsed in TBS/T, pH 7.6, transferred to TBS/T, pH 8.2, and incubated for 1 hr in the secondary antibody [goat anti-rabbit IgG conjugated to 10 nm (Amersham, Arlington Heights, IL) or 18 nm (E-Y Labs, San Mateo, CA) gold particles]. Grids were then rinsed and counterstained with uranyl acetate and Sato’s lead. In control experiments, in the absence of primary rabbit serum, virtually no gold particles were detected; after exposure to nonimmune rabbit serum, sparse gold particles were seen that showed no obvious pattern. Grids were examined at 80 kV on a JEOL (Tokyo, Japan) 200CX electron microscope. For quantitation, micrographs at 29,000–40,000× original magnification were digitized on a flat-bed scanner. Locations of gold particles were measured using NIH Image software running on a Macintosh platform. For each grid studied, the first 10 synapses with clearly visible synaptic structures, labeled with at least one gold particle within 100 nm on either side of the postsynaptic membrane, were considered. To define “axodendritic” position, we measured the distance between the center of each gold particle and the outer leaflet of the postsynaptic membrane. To define “lateral” synaptic position, we measured the distance from each end of the active zone to a line drawn perpendicular to the synapse running through the center of the particle.
Immunoprecipitation. Coimmunoprecipitation of GRIP, GluR2/3, and GluR1 complexes from rat cortex was done on sodium deoxycholate (DOC)-extracted rat brain membranes as described (Luo et al., 1997). In brief, DOC extracts were prepared by resuspending rat brain cortex P2 fraction in a suitable volume (1 ml/gm of tissue) of ice-cold Tris-EDTA buffer (10 mm Tris-HCl and 5 mm EDTA, pH 7.4) and sonicated briefly. One-tenth volume of ice-cold DOC buffer (10% sodium deoxycholate and 500 mm Tris-HCl, pH 9.0) was added to the sonicate, followed by incubation at 36°C for 30 min. After the addition of 0.1 volume of Triton X-100 buffer (1% Triton X-100 and 500 mm Tris-HCl, pH 9.0), the samples were dialyzed against ice-cold binding buffer (50 mm Tris-HCl, pH 7.4, and 0.1% Triton X-100). Unextractable material was pelleted at 37,000 ×g for 40 min, and the supernatant was used for immunoprecipitation.
For immunoprecipitation, GRIP (C8399-I), GluR2/3, GluR2-N, or rabbit IgG antibodies were prebound to protein A-Sepharose beads by incubating the antibodies with protein A-Sepharose beads (Sigma) at a ratio of 20 μg of antibodies/40 μl of protein A-Sepharose beads in 200 mm Na-borate buffer, pH 8.0, at 4°C for 12 hr. The protein A-Sepharose/antibody-bound beads were then washed with 200 mm Na-borate buffer, pH 9.0, added to 100 μg of DOC-extracted protein (see above), and allowed to incubate at 4°C for 2–3 hr. The immunoprecipitation reaction was then centrifuged at high speed in a microcentrifuge; the supernatant was saved, and the immunoprecipitate pellet was washed three times with binding buffer.
RESULTS
Cloning of ABP-L/GRIP2, a splice variant of ABP containing seven PDZ domains
By yeast two-hybrid screening (see Materials and Methods), we obtained not only multiple isolates of a cDNA fragment (clone 5) from the original GRIP gene (Dong et al., 1997; GRIP1) but also a cDNA fragment (clone 14) that we subsequently found to be virtually identical to ABP within the region of overlap (Srivastava et al., 1998). We obtained the full-length coding sequence of this ABP-related gene, which showed interesting differences from the full-length ABP sequence published by Srivastava et al. (1998). Compared with ABP, our open reading frame contained an N-terminal extension of 52 amino acids, an internal deletion of 41 amino acids between PDZ3 and PDZ4, and a C-terminal extension that contains an additional seventh PDZ domain (PDZ7; Fig. 1). Otherwise, the two proteins are essentially identical, suggesting that the extensions and deletions noted in our clone arise from differential splicing. We called the novel protein encoded by our cDNA “ABP-L/GRIP2,” because it is a longer splice variant of ABP, and because it is clearly a close relative of GRIP1 (Dong et al., 1997), both in terms of primary sequence and domain organization. Because ABP is essentially contained within ABP-L/GRIP2, we presume that ABP and ABP-L/GRIP2 are splice variants of the same gene. In this paper, we use the name GRIP1 for the original GRIP described by Dong et al. (1997) and ABP-L/GRIP2 for the long splice variant of ABP. The generic term GRIP will be used to refer to GRIP1 and ABP-L/GRIP2 proteins as a set.
Fig. 1.
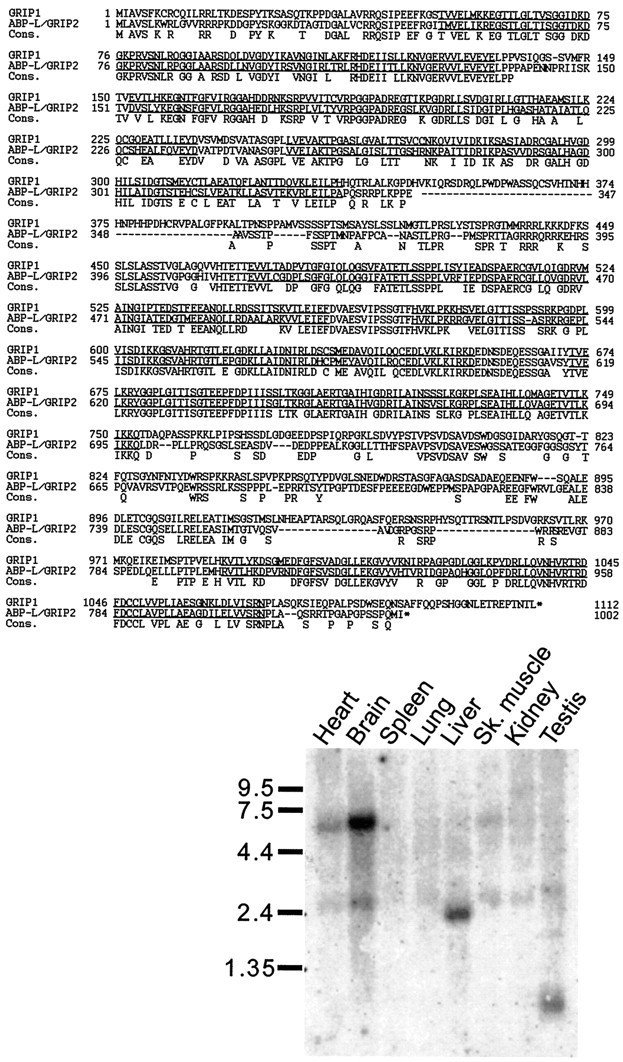
Top, Amino acid sequence alignment of GRIP1 and ABP-L/GRIP2. The third line(Cons.) shows the consensus amino acids that are common between GRIP1 and ABP-L/GRIP2. The seven PDZ domains areunderlined. Bottom, Rat poly-A mRNA Northern blot showing the tissue distribution of ABP-L/GRIP2 mRNAs. Positions of RNA molecular size markers are shown in kilobases.
Over its entire length, ABP-L/GRIP2 is 56% identical and 68% similar to GRIP1 at the amino acid level (Fig. 1). The greatest similarity between GRIP1 and ABP-L/GRIP2 occurs over the respective PDZ domains 1–7, with highest identity for PDZ6 (93%) and lowest identity for PDZ3 (62%). There is also significant homology between GRIP1 and ABP-L/GRIP2 in the intervening sequences between each PDZ domain. During the cloning and sequencing of overlapping cDNAs, presumptive alternative splice forms of both GRIP1 and ABP-L/GRIP2 were noted that contained small insertions or deletions at certain sites in both cDNA sequences (data not shown). The particular splice variant of ABP-L/GRIP2 protein shown in Figure 1 is 1002 amino acids long and has a predicted Mr of 107,733.
By Northern analysis, ABP-L/GRIP2 mRNA is expressed predominantly in the brain, in the form of a major ∼6 kb transcript and a minor ∼2.8 kb transcript (Fig. 1, inset). Similar-size mRNAs were present in the heart, but less abundantly. ABP-L/GRIP2 mRNAs of smaller size were detected in liver (∼2.4 kb) and in testis (∼1 kb). Thus ABP-L/GRIP2 mRNA shows a tissue expression pattern distinct from GRIP1, which is predominantly expressed in brain and testis and expressed at a low level in kidney (Wyszynski et al., 1998).
Ultrastructural localization of GRIP protein in rat brain
Two different polyclonal rabbit antisera were raised against fusion proteins containing PDZ6 through the C terminus of GRIP1 and affinity-purified (Wyszynski et al., 1998). The affinity-purified antibodies (termed 1756 and C8399-I; see Materials and Methods) specifically and effectively recognize both GRIP1 and ABP-L/GRIP2, as assessed by immunoblotting of recombinant bacterial fusion proteins and rat brain extracts (Wyszynski et al., 1998; data not shown). The cross-reactivity of 1756 and C8399 antibodies is not surprising, because the PDZ6 and PDZ7 domains are highly conserved between GRIP1 and ABP-L/GRIP2 (Fig. 1). Although unable to distinguish between GRIP1 and ABP-L/GRIP2, these antibodies are nevertheless specific for GRIP and hence useful for probing the set of GRIP1 and ABP/GRIP2 proteins in the brain. Using these “pan-GRIP” antibodies, we have recently described the immunohistochemical localization of GRIP in various regions of rat brain (Wyszynski et al., 1998). At the light microscopy (LM) level, GRIP is widely distributed within brain neurons, showing a punctate staining pattern on top of a diffuse intracellular labeling (Wyszynski et al., 1998).
To obtain higher resolution, we studied the subcellular localization of GRIP by postembedding immunogold electron microscopy (EM) of adult rat neocortex (Fig. 2). A similar labeling pattern was seen using two GRIP antibodies prepared from different rabbits and purified in different ways (C8399-I and 1756), supporting the specificity of immunogold labeling. In all blocks, the highest density of gold particles coding for GRIP was associated with asymmetric synapses (Fig. 2A,D,E). Labeled symmetric synapses were rare; of 114 labeled synapses studied in three rats, only 2 were symmetric. Labeling (at a lower level density) could also be seen over nonsynaptic plasma membranes (Fig.2B,C). Considerable intracellular labeling was present within spines and within dendritic shafts (Fig.2C,F); in dendritic shafts particles were typically associated with microtubules (Fig. 2F). In some cases, GRIP immunoreactivity was seen in axons, axon terminals, and somatic endoplasmic reticulum (data not shown). Thus based on immuno-EM, GRIP is present at excitatory synapses but additionally associated with nonsynaptic membranes and intracellular compartments.
Fig. 2.
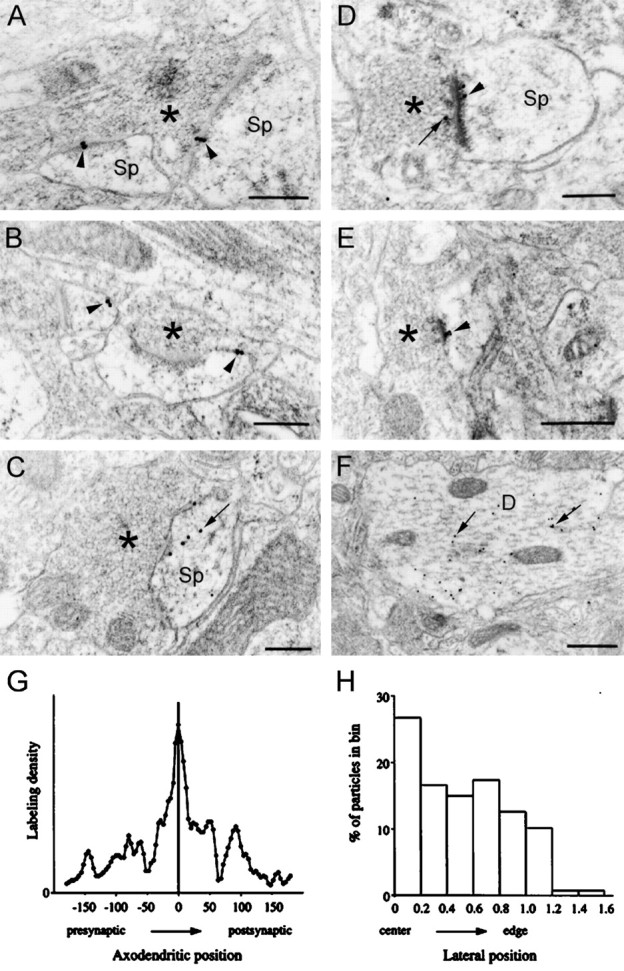
EM photomicrographs of immunogold labeling (18 nm gold particles) for GRIP in layers II/III of rat somatic sensory cortex. A, Gold particles (arrowheads) label two asymmetric synapses made by the same terminal onto spines.B, GRIP immunogold labeling (arrowheads) at nonsynaptic sites of the plasma membrane of two spines.C, Gold particles labeling the postsynaptic membrane of a perforated synapse with additional intracellular labeling within the dendritic spine (arrow). D, Gold particle labeling encroaches into the presynaptic cytoplasm (arrow); the postsynaptic labeling (arrowhead) coincides with the “inner” margin of the PSD. E, Immunogold labeling (arrowhead) at an asymmetric synapse extends into the postsynaptic cytoplasm.F, Large dendritic profile immunopositive for GRIP;arrows point to labeling adjacent to microtubules.Sp, Spine; asterisks mark presynaptic terminals. Scale bars: A–D, 0.25 μm; E, F, 0.5 μm. G, H, Quantitative analysis of the distribution of GRIP immunogold particles at the synapse. G, Distribution of gold particles in the axodendritic axis. One hundred sixty particles lying within 175 nm of the postsynaptic membrane were measured in 70 random electron micrographic fields, selected to reveal clear synaptic membranes; data were pooled from three animals. Abscissa, Distance from the center of each gold particle to the postsynaptic membrane (in nanometers); ordinate, labeling density (arbitrary units); zero density corresponds to the x-axis.H, Lateral distribution of gold particles along the synapse (only those within 100 nm of the postsynaptic membrane were considered). Lateral position is normalized; center of the active zone corresponds to 0; and edge corresponds to 1.0.
The synaptic labeling for GRIP was concentrated over the postsynaptic membrane and the postsynaptic density and was generally sparser over the presynaptic profile. Approximately one-third (87 of 266) of asymmetric synapses were labeled for GRIP. This is probably an underestimate, considering the limited sensitivity of the method and our use of single, rather than serial, sections to assess each synapse. To quantify the staining pattern at synapses, we measured the distributions of gold particles associated with synapses in 70 randomly selected fields from three animals. Along the axodendritic axis, particle density was highest at the postsynaptic membrane with some spread (up to 150 nm) toward the presynaptic and postsynaptic compartments (Fig. 2G). The labeling density appeared higher at the center of the active zone, declining toward the edge of the PSD (Fig. 2H).
Localization of GRIP in cultured hippocampal neurons
We also assessed the cellular and subcellular localization of GRIP by immunocytochemistry of hippocampal neurons in low-density culture (Fig. 3A–D). A similar labeling pattern was seen using anti-GRIP antibodies 1756 and C8399-I. GRIP showed fine punctate and diffuse staining in cell bodies and dendrites (Fig. 3A–D). In addition, GRIP appeared to be concentrated in sparse puncta of larger diameter (0.5–2 μm) along the somata and dendrites of many neurons (Fig.3B,C). Double-label immunocytochemistry with GRIP 1756 and synaptophysin antibodies revealed GRIP localization to a subset of synaptic sites (Fig.3A). A large fraction of the GRIP puncta, however, appeared nonsynaptic.
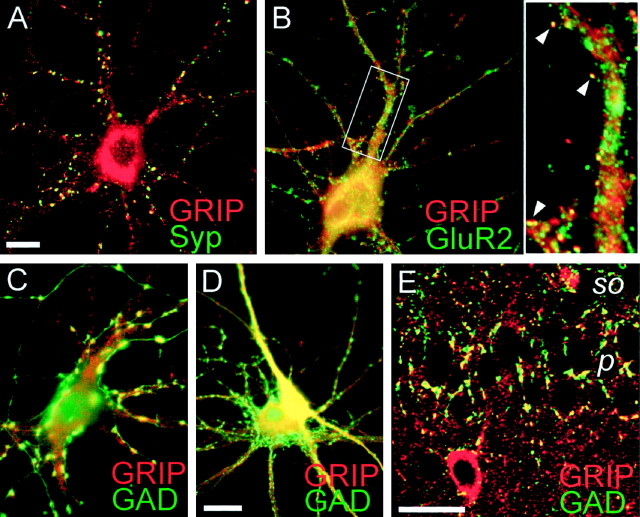
Fig. 3.
Immunocytochemical localization of GRIP in cultured hippocampal neurons and in rat brain.A–E, Double-label immunocytochemistry of hippocampal neurons (3 weeks in low-density culture) with antibodies to GRIP (1756, A–D) and synaptophysin (Syp, A), GluR2 (B), or glutamic acid decarboxylase (GAD, C, D). GRIP is visualized by Cy3 secary antibody (red), and synaptophysin, GluR2, and GAD are visualized by FITC secondary antibody (green). Colocalization is shown inyellow. A subset of GRIP-immunoreactive puncta showed colocalization with synaptophysin (A) and GluR2 (B). Inset, Enlargement of theboxed region in B, showing examples of GRIP/GluR2 colocalization in dendritic spine-like structures (arrowheads). There is also extensive overlap of GRIP and GluR2 staining in the cell body (B). The larger GRIP puncta colocalized with a subset of GAD-labeled inhibitory terminals (C). GABAergic neurons also had higher levels of diffuse somatodendritic GRIP immunoreactivity than did pyramidal neurons (D). E, Confocal image showing partial colocalization of GRIP and GAD in area CA1 of hippocampus. Note colocalized immunoreactive puncta on pyramidal cell bodies and adjacent neuropil. p, Stratum pyramidale;so, stratum oriens. Scale bars: A,D, 10 μm (for A–D); E, 40 μm.
Double labeling with 1756 and anti-GluR2 antibodies showed extensive overlap of GRIP and AMPA receptor staining in cell bodies and major dendritic shafts but only limited colocalization of GRIP and GluR2 in dendritic puncta [∼10% of GluR2 puncta colocalized with GRIP along dendrites (Fig. 3B, arrowheads)]. On the other hand, double labeling with 1756 antibodies and an antibody against GAD, to reveal inhibitory presynaptic sites, showed extensive colocalization of GRIP and GAD in the larger puncta, although colocalization was not complete (Fig. 3C). Interestingly, diffuse cytoplasmic GRIP immunoreactivity was also much higher in GABAergic neuronal somata and their processes than in pyramidal neurons (Fig. 3D). Association of GRIP with inhibitory neurons and synapses has also been observed with independent GRIP antibodies (R. L. Huganir, personal communication). Overall, the pattern of immunostaining in embryonic hippocampal cultures indicated that GRIP is highly expressed in GABAergic neurons and enriched in inhibitory nerve terminals. The nature of the nonsynaptic GRIP staining is unclear, but it is consistent with the GRIP labeling of nonsynaptic membranes and intracellular compartments seen by EM (Fig. 2). Earlier biochemical fractionation had also suggested a widespread subcellular distribution of GRIP (Wyszynski et al., 1998).
Immunocolocalization of GRIP and GAD was confirmed in vivoby double labeling of rat brain (for detailed information on the pattern of cellular labeling, see Burette et al., 1999). Punctate GRIP staining in the neuropil of rat brain is greatly enhanced by light protease treatment of the sections. In hippocampus a subset of these GRIP immunoreactive puncta colocalized with GAD-positive puncta on cell bodies and proximal dendrites of pyramidal neurons (Fig.3E), consistent with the localization of GRIP in inhibitory synapses. Unfortunately, the protease digestion was incompatible with synaptophysin staining; thus we were unable to perform double labeling to test whether the GRIP puncta in general were synaptic in brain sections (data not shown).
Association of GRIP and AMPA receptors in vivo
Although our immuno-EM data revealed the presence of GRIP at excitatory synapses in rat brain, it does not prove that GRIP is biochemically associated with AMPA receptors at these sites. To test whether GRIP and AMPA receptors exist in a protein complex, we optimized a coimmunoprecipitation assay that allowed us to immunoprecipitate GRIP or AMPA receptor subunits virtually quantitatively from detergent extracts prepared from rat brain membranes (Fig. 4A). Because our antibodies recognize both GRIP1 and ABP/GRIP2, we can assess AMPA receptor association with the entire known family of GRIP proteins.
Fig. 4.
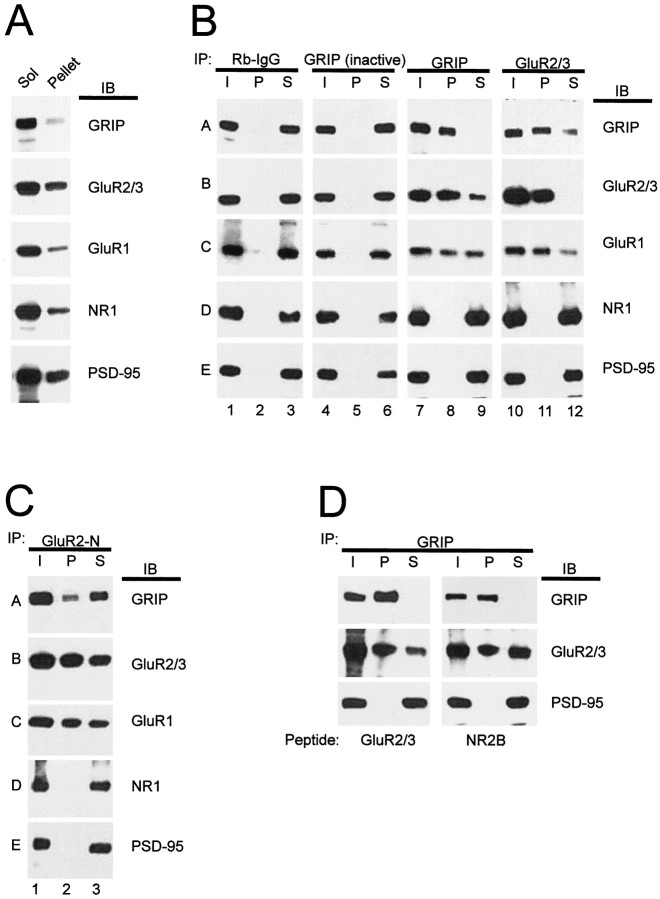
Coimmunoprecipitation of native complexes containing GRIP and AMPA receptor subunits from rat brain.A, Detergent solubility of GRIP and AMPA receptor subunits. Crude synaptosomal membrane fraction from rat cortex was extracted with sodium deoxycholate (DOC); equal percentages of the resulting extract (Sol) and pellet fractions were immunoblotted for GRIP, GluR2/3, GluR1, NR1, and PSD-95. B, Membrane proteins from adult rat cortex were detergent-solubilized as described in Materials and Methods. Parallel samples of brain extract were immunoprecipitated (IP) with nonspecific rabbit IgGs or affinity-purified antibodies to GRIP or GluR2/3, as indicated. In lanes 4–6, the GRIP antibodies were heat-inactivated by boiling before use. Each immunoprecipitation experiment is shown in three lanes:I, input to the immunoprecipitation reaction;P, immunoprecipitated pellet; and S, supernatant remaining after the immunoprecipitation. Equal fractions of each sample (∼10%) were loaded, thus allowing direct estimate of the fraction of each protein immunoprecipitated (the amount of protein inI = P + S). TheI, P, and S fractions of the various immunoprecipitation reactions were immunoblotted (IB) for GRIP, GluR2/3, GluR1, NR1, and PSD-95, as indicated to the right of each row.Rows are also labeled (A–E) for reader orientation (see Results). C, D, Same as forB, except that GluR2-N antibodies were used for immunoprecipitation in C, and solubilization and immunoprecipitation was performed in the presence of either GluR2/3 or NR2B C-terminal competitor peptide (0.5 mm final concentration) in D.
Our extraction conditions using deoxycholate detergent solubilized the large majority of GRIP and AMPA receptors (typically >80%; Fig.4A). Using GRIP antibodies for immunoprecipitation, close to all of the extracted GRIP could be pelleted with the protein A-Sepharose beads, leaving little or undetectable amounts of GRIP in the supernatant (Fig. 4B, compare row A, lanes 8, 9). This GRIP immunoprecipitate contained most of the GluR2/3 in the extract, leaving behind a minor fraction of GluR2/3 in the supernatant (Fig.4B, row B, lanes 8, 9). In multiple experiments, the fraction of GluR2/3 that was coimmunoprecipitated with GRIP ranged from ∼40 to ∼80%. Furthermore, approximately half of the extracted GluR1 (an AMPA receptor subunit that does not bind directly to GRIP) was coimmunoprecipitated with GRIP (Fig. 4B, row C, lanes 8, 9). Control experiments showed that the NMDA receptor subunit NR1 and PSD-95 were not coimmunoprecipitated with GRIP antibodies (Fig. 4B, rows D, E, lane 8). Because PSD-95 can be efficiently coimmunoprecipitated with NR1 under the same conditions (data not shown), this result indicates that GRIP-GluR2/3-containing complexes are distinct from complexes containing NR1 and PSD-95.
In the converse reaction, GluR2/3 antibodies were able to quantitatively immunoprecipitate GluR2/3 from the extract and to coimmunoprecipitate approximately half of the extracted GRIP protein (Fig. 4B, rows A, B, lanes 11, 12; data not shown). In multiple experiments, the fraction of GRIP that was coimmunoprecipitated with GluR2/3 ranged from ∼30 to ∼50%. The same immunoprecipitates contained a majority of the GluR1 (Fig.4B, row C, lanes 11, 12). Again, the GRIP-GluR2/3 coimmunoprecipitation is specific, because no detectable NR1 and PSD-95 was pelleted with GluR2/3 (Fig. 4B,rows D, E, lane 11). In further negative controls, equal amounts of nonspecific rabbit immunoglobulins or heat-inactivated GRIP antibodies were unable to precipitate GRIP, GluR2/3, GluR1, NR1, or PSD-95 (Fig. 4B, rows A–E, lanes 2, 5). Similar results were reproduced in several experiments.
We also used an independent antibody (GluR2-N) directed against the extracellular N-terminal region of GluR2 to perform the immunoprecipitation (Fig. 4C). GluR2-N immunoprecipitated approximately half of the GluR2/3 and GluR1 immunoreactivity from the brain extract, consistent with the association of these subunits in heteromeric AMPA receptors (Fig. 4C, rows B, C, lanes 2, 3). A significant fraction of GRIP was specifically coimmunoprecipitated with GluR2-N antibodies (Fig. 4C,row A, lanes 2, 3). However, no detectable NR1 and PSD-95 was pelleted with GluR2-N (Fig. 4C, rows D, E, lane 2). Thus using multiple antibodies we could show substantial association of AMPA receptors and GRIP in adult rat brain.
If the immunoprecipitation of GRIP-AMPA receptor subunit complexes from brain detergent extracts was attributable to artifactual association of these proteins occurring after solubilization, a synthetic peptide comprising the last 10 residues of the GluR2/3 C terminus should compete for PDZ domain binding and hence should reduce or abolish the coimmunoprecipitation of GRIP and GluR2/3 by competing for binding to the GRIP PDZ domain. However, even in the presence of a 0.5 mm concentration of GluR2/3 C-terminal peptide, GluR2/3 showed equally substantial coimmunoprecipitation with GRIP (Fig.4D). These results indicate that AMPA receptor subunits are largely associated with GRIP in native complexes in rat brain, but that a significant fraction of GRIP is not associated with AMPA receptors.
GRIP and GluR2/3 were also coimmunoprecipitated from juvenile rat brain (postnatal day 7) with similar efficiency to adult, indicating that GRIP and AMPA receptors are highly associated in immature brain. However, in neuronal culture (3 weeks in culture), we found poor association (<10%) of GRIP and AMPA receptor subunits by coimmunoprecipitation (data not shown). This poor coimmunoprecipitation in cultured neurons correlates with the weak colocalization of GRIP and GluR2 seen by immunocytochemistry (Fig. 3).
DISCUSSION
ABP and ABP-L/GRIP2 are splice variants of the same gene
ABP, a GluR2/3-binding protein with six PDZ domains, has been recently reported (Srivastava et al., 1998). Based on sequence comparisons with the ABP-L/GRIP2 protein described here, ABP and ABP-L/GRIP2 are almost certainly alternative splice variants of the same gene. ABP is a splice form that has been truncated at the C-terminal region, thereby losing the seventh PDZ domain. The reported size of ABP on SDS-PAGE matches the minor 97 kDa band that we detect consistently with our pan-GRIP antibodies (Wyszynski et al., 1998); furthermore, Srivastava et al. (1998) also reported higher molecular weight bands on their ABP Western blots that probably correspond, at least in part, to “full-length” ABP-L/GRIP2 polypeptides. It will be of interest to identify the binding partners of PDZ7 in GRIP, because its absence in ABP implies that the function of PDZ7 can be uncoupled from that of the other six PDZ domains. The significance of the alternative splicing of ABP/ABP-L/GRIP2 remains to be determined.
A major fraction of GluR2/3 is associated with GRIP
The binding of GluR2/3 to GRIP has been previously studied onlyin vitro and in heterologous expression systems (Dong et al., 1997; Srivastava et al., 1998). We show here that AMPA receptor subunits are associated with GRIP in vivo. Indeed, a major fraction of GluR2/3 appears to be complexed with GRIP in the brain under optimal conditions for immunoprecipitation. Approximately half of the extractable GluR1 is also associated with GRIP, presumably via heteromultimerization with GluR2/3, because GluR1 cannot bind directly to the PDZ domains of GRIP. That a lesser fraction of GluR1 than of GluR2/3 is associated with GRIP in rat brain cortex may be accounted for in part by the existence of AMPA receptors that contain GluR1 but not GluR2/3 (Martin et al., 1993; Kharazia et al., 1996; Wenthold et al., 1996; Kondo et al., 1997). When it is not in a heteromeric configuration with GluR2/3, GluR1 may be associated with scaffold proteins other than GRIP. It is intriguing, given that much of GluR1 appears not to be complexed with GRIP, that a recent report showed GluR1 to be coimmunoprecipitated with SAP97, a PSD-95-related protein (Leonard et al., 1998).
Although most of the extractable GluR2/3 is complexed with GRIP, a substantial proportion of GRIP appears not to be associated with GluR2/3. The simplest interpretation of this observation is that AMPA receptors interact with only a subset of GRIP in the brain. The remaining fraction of GRIP may be involved in the assembly of protein complexes that do not include AMPA receptor subunits.
Several findings support the idea that GRIP may play additional roles unrelated to the anchoring of AMPA receptors at synapses. First, GRIP is only modestly enriched in synaptic or PSD fractions when compared with PSD-95, the prototypical postsynaptic scaffold protein (Wyszynski et al., 1998). Second, significant amounts of GRIP are present in subcellular fractions that are depleted of synaptic membranes, such as the cytosolic fraction S3 and the synaptic vesicle-enriched fraction LP2 (Wyszynski et al., 1998). Third, although immunogold EM shows a concentration of GRIP at asymmetric synapses in adult brain neurons, it also reveals GRIP immunoreactivity at nonsynaptic membranes and in intracellular compartments of dendritic spines and shafts. Fourth, immunocytochemistry of cultured embryonic neurons from hippocampus reveals that a prominent signal for GRIP is in GABAergic neurons and their nerve terminals, and a large fraction of GRIP immunoreactivity is nonsynaptic. Finally, GRIP expression is high very early during cortical development and does not correlate temporally with AMPA receptor expression or synaptogenesis (Wyszynski et al., 1998). Taken together, these data argue that a large proportion of GRIP is not associated with excitatory synapses, where AMPA receptors are presumed to be concentrated. Perhaps this nonsynaptic fraction of GRIP is represented by the subset of GRIP that does not coimmunoprecipitate with GluR2/3. Based on the precedent of PSD-95, it is quite likely that there are multiple binding partners for each individual PDZ domain of GRIP. Thus at synaptic sites, GRIP may be complexed with AMPA receptors, whereas at nonsynaptic sites, GRIP may be complexed with a different set of interacting proteins. It is also possible that the GRIP–AMPA receptor interaction occurs only after both proteins reach the synapse, in which case the nonsynaptic GRIP may be in an unbound (AMPA receptor-free) form in transit to its final location.
The above discussion is prejudiced toward the idea that AMPA receptors are highly concentrated in excitatory postsynaptic membranes, and that the site of GluR2/3-GRIP interaction is primarily at synaptic sites. However, to reach their synaptic destination, AMPA receptor proteins themselves have to be sorted via specific vesicle trafficking pathways emanating from the cell body. Furthermore, recent evidence points to a regulated trafficking of AMPA receptors at postsynaptic sites and the existence of intracellular and subsynaptic pools of AMPA receptor subunits (Molnár et al., 1993; Baude et al., 1995; Nusser et al., 1998; Petralia et al., 1999). These considerations imply the existence of AMPA receptor proteins residing in intracellular membranous compartments on the way to, and in the vicinity of, the synapse. Consistent with this, we note that the staining pattern of AMPA receptors in neurons is typically more diffuse than that of NMDA receptors, suggesting a more widespread subcellular distribution for AMPA receptors (Rao et al., 1998). If GluR2/3 subunits are as highly associated with GRIP as the coimmunoprecipitations would suggest, then it is possible that some of the nonsynaptic GRIP that we observe is complexed with intracellular pools of AMPA receptors. Thus GRIP could play a role in AMPA receptor trafficking in addition to its function in synaptic anchoring of AMPA receptors. In this regard, it will be important to determine at what stage of the secretory pathway the GRIP-GluR2/3 interaction takes place.
Localization of GRIP in GABAergic neurons and terminals
A surprising finding was that in hippocampal neuron cultures GRIP immunoreactivity colocalized more prominently with GAD, a marker of GABAergic neuronal somata and terminals, than with AMPA receptors. The association of GRIP with GABAergic neurons in culture is consistent with immunohistochemistry of rat brain sections, which showed the most striking expression of GRIP protein in interneurons of hippocampus and cortex (Wyszynski et al., 1998). The immunostaining pattern in cultured neurons indicates the enrichment of GRIP in GABAergic somata and dendrites and nerve terminals. A partial colocalization of GRIP- and GAD-immunoreactive puncta was also seen in brain by confocal microscopy, supporting the idea that GRIP is concentrated in GABAergic presynaptic terminals in vivo. The particularly abundant expression of GRIP in GABAergic neurons is of interest. GABAergic interneurons contain AMPA receptor clusters that are more insoluble in Triton X-100 than AMPA receptors in pyramidal cells (Allison et al., 1998). Moreover, relative to pyramidal neurons, GABAergic neurons show a particularly high content and low variability of AMPA receptors at excitatory postsynaptic sites (Nusser et al., 1998). Whether these postsynaptic characteristics are related to the high levels of GRIP in these neurons remains to be determined.
Synthesis of LM and EM findings: multiple pools of GRIP
In contrast to LM immunocytochemistry of hippocampal cultures, we did not observe a concentration of GRIP signal over nerve terminals at symmetric (presumed inhibitory) synapses by immuno-EM of neocortex. Instead, the highest density of GRIP labeling occurred over asymmetric synapses, as might be expected for an AMPA receptor-binding protein. We are confident of the specificity of each staining pattern because they were consistently obtained with multiple independent GRIP antibodies in appropriately controlled experiments. The apparently different results in hippocampal culture and adult rat neocortex may reflect different developmental stages of these preparations and/or the differential strengths and weakness of LM versus postembedding EM immunolocalization. The postembedding immunogold EM technique is especially sensitive to the density of the antigen and may miss antigens that are relatively soluble and easily lost during the processing of the thin sections (such as may occur for GRIP in GABAergic nerve terminals). The accessibility or “visibility” of the antigen may also vary with the different methods. Proteins in the PSD may be less accessible to antibodies and often are more difficult to stain by LM immunohistochemistry than the same proteins in cytoplasmic compartments (e.g., see Watanabe et al., 1998). In support of this, we found that punctate GRIP staining in the brain at the LM level is dramatically revealed by protease treatment of brain sections. On the other hand, the thin sectioning of postembedding immuno-EM can directly reveal antigens in hard-to-access places like the PSD.
Alternatively, it is possible that real differences exist between the subcellular distribution of GRIP in hippocampal culture and in the intact brain. In particular, we note that in contrast to the brain, GRIP and AMPA receptor subunits are poorly associated in neuron cultures, as assessed by coimmunoprecipitation. This correlates with the limited colocalization of GRIP and AMPA receptor subunits at synaptic puncta in cultured neurons. In this sense, cultured hippocampal neurons may represent an imperfect model for GRIP–AMPA receptor interactions.
Because of the apparent discrepancies discussed above, it is important to synthesize the results from multiple immunohistochemical approaches and to combine them with more quantitative biochemical fractionation studies. By doing this, we conclude that multiple pools of GRIP exist in neurons spread among several subcellular locations. GRIP is not only present at the excitatory postsynaptic membrane; it is also at inhibitory synapses and in intracellular compartments close to and distant from the synapse. Moreover, only approximately half of GRIP is associated with AMPA receptors. The future challenge will be to define the molecular roles played by these various different subsets of GRIP and to relate these functions to the trafficking and function of AMPA receptors.
Footnotes
This work was supported by National Institutes of Health Grants NS35050 (to M.S.), CA66268 (to M.W.), NS35527 (to R.W.), and NS34361 (to D.G.S.) and a grant from the Korea Research Foundation (to E.K.). M.S. is Assistant Investigator of the Howard Hughes Medical Institute. We thank Carlo Sala, Fu-chia Yang, and Sheila Rudolph-Correia for experimental help. We are especially grateful to Anuradha Rao and Ann Marie Craig for double-label immunocytochemistry of hippocampal neurons with antibodies to GRIP and glutamic acid decarboxylase.
Correspondence should be addressed to Dr. Morgan Sheng, Howard Hughes Medical Institute, Wellman 423, Massachusetts General Hospital, 50 Blossom Street, Boston, MA 02114.
REFERENCES
- 1.Allison DW, Gelfand VI, Spector I, Craig AM. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banker GA, Cowan WM. Rat hippocampal neurons in dispersed cell culture. Brain Res. 1977;126:397–425. doi: 10.1016/0006-8993(77)90594-7. [DOI] [PubMed] [Google Scholar]
- 3.Baude A, Nusser Z, Molnár E, McIlhinney AJ, Somogyi P. High-resolution immunogold localizaiton of AMPA-type glutamate receptor subunits at synaptic and non-synaptic sites in rat hippocampus. Neuroscience. 1995;69:1031–1055. doi: 10.1016/0306-4522(95)00350-r. [DOI] [PubMed] [Google Scholar]
- 4.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 5.Burette A, Wyszynski M, Valtschanoff JG, Sheng M, Weinberg RJ (1999) Characterization of GRIP-immunopositive neurons in rat brain. J Comp Neurol, in press. [PubMed]
- 6.Chen JJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- 7.Craven SE, Bredt DS. PDZ proteins organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 8.Dong H, O’Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 9.Ehlers MD, Fung ET, O’Brien RJ, Huganir RL. Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J Neurosci. 1998;18:720–730. doi: 10.1523/JNEUROSCI.18-02-00720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goslin K, Banker G, editors. Culturing nerve cells. MIT; Cambridge, MA: 1991. [Google Scholar]
- 11.Kharazia VN, Wenthold RJ, Weinberg RJ. GluR1-immunopositive interneurons in rat neocortex. J Comp Neurol. 1996;368:399–412. doi: 10.1002/(SICI)1096-9861(19960506)368:3<399::AID-CNE6>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Kim E, Sheng M. Differential K+ channel clustering activity of PSD-95 and SAP97, two related membrane-associated putative guanylate kinases. Neuropharmacology. 1996;35:993–1000. doi: 10.1016/0028-3908(96)00093-7. [DOI] [PubMed] [Google Scholar]
- 13.Kim JH, Liao D, Lau LF, Huganir RL. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 14.Kondo M, Sumino R, Okado H. Combinations of AMPA receptor subunit expression in individual cortical neurons correlate with expression of specific calcium-binding proteins. J Neurosci. 1997;17:1570–1581. doi: 10.1523/JNEUROSCI.17-05-01570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornau H-C, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 16.Leonard AS, Davare MA, Horne MC, Garner CC, Hell JW. SAP97 is associated with the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem. 1998;273:19518–19524. doi: 10.1074/jbc.273.31.19518. [DOI] [PubMed] [Google Scholar]
- 17.Lin JW, Wyszynski M, Madhavan R, Sealock R, Kim JU, Sheng M. Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J Neurosci. 1998;18:2017–2027. doi: 10.1523/JNEUROSCI.18-06-02017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BB. The majority of N-methyl-d-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B). Mol Pharmacol. 1997;51:79–86. doi: 10.1124/mol.51.1.79. [DOI] [PubMed] [Google Scholar]
- 19.Martin LJ, Blackstone CD, Levey AI, Huganir RL, Price DL. AMPA glutamate receptor subunits are differentially distributed in rat brain. Neuroscience. 1993;53:327–358. doi: 10.1016/0306-4522(93)90199-p. [DOI] [PubMed] [Google Scholar]
- 20.Molnár E, Baude A, Richmond SA, Patel PB, Somogyi P, McIlhinney RAJ. Biochemical and immunocytochemical characterization of antipeptide antibodies to a cloned GluR1 glutamate receptor subunit: cellular and subcellular distribution in the rat forebrain. Neuroscience. 1993;53:307–326. doi: 10.1016/0306-4522(93)90198-o. [DOI] [PubMed] [Google Scholar]
- 21.Müller BM, Kistner U, Kindler S, Chung WJ, Kuhlendahl S, Lau L-F, Veh RW, Huganir RL, Gundelfinger ED, Garner CC. SAP102, a novel postsynaptic protein that interacts with the cytoplasmic tail of the NMDA receptor subunit NR2B. Neuron. 1996;17:255–265. doi: 10.1016/s0896-6273(00)80157-9. [DOI] [PubMed] [Google Scholar]
- 22.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niethammer M, Valtschanoff JG, Kapoor TM, Allison DW, Weinberg TM, Craig AM, Sheng M. CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron. 1998;20:693–707. doi: 10.1016/s0896-6273(00)81009-0. [DOI] [PubMed] [Google Scholar]
- 24.Nusser Z, Lujan R, Laube G, Roberts JD, Molnar E, Somogyi P. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron. 1998;21:545–559. doi: 10.1016/s0896-6273(00)80565-6. [DOI] [PubMed] [Google Scholar]
- 25.Petralia RS, Esteban JA, Wang Y-X, Partridge JG, Zhao H-M, Wenthold RJ, Malinow R. Selective acquisition of AMPA receptors over postnatal development suggests a molecular basis for silent synapses. Nat Neurosci. 1999;2:31–36. doi: 10.1038/4532. [DOI] [PubMed] [Google Scholar]
- 26.Phend KD, Rustioni A, Weinberg RJ. An osmium-free method of Epon embedment that preserves both ultrastructure and antigenicity for postembedding immunocytochemistry. J Histochem Cytochem. 1995;43:469–479. doi: 10.1177/43.3.7532656. [DOI] [PubMed] [Google Scholar]
- 27.Rao A, Kim E, Sheng M, Craig AM. Heterogeneity in the molecular composition of excitatory postsynaptic sites during development of hippocampal neurons in culture. J Neurosci. 1998;18:1217–1229. doi: 10.1523/JNEUROSCI.18-04-01217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB. Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 29.Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. Essential role for dlg in synaptic clustering of shaker K+ channels in vivo. J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas U, Kim E, Kuhlendahl S, Ho Koh Y, Gundelfinger ED, Sheng M, Garner CC, Budnik V. Synaptic clustering of the cell adhesion molecule fasciclin II by discs-large and its role in the regulation of presynaptic structure. Neuron. 1997;19:787–799. doi: 10.1016/s0896-6273(00)80961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe M, Fukaya M, Sakimura K, Manabe T, Mishina M, Inoue Y. Selective scarcity of NMDA receptor channel subunits in the stratum lucidum (mossy fibre-recipient layer) of the mouse hippocampal CA3 subfield. Eur J Neurosci. 1998;10:478–487. doi: 10.1046/j.1460-9568.1998.00063.x. [DOI] [PubMed] [Google Scholar]
- 32.Wenthold RJ, Petralia RS, Blahos J, II, Niedzielski AS. Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J Neurosci. 1996;16:1982–1989. doi: 10.1523/JNEUROSCI.16-06-01982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 34.Wyszynski M, Kim E, Yang F-C, Sheng M. Biochemical and immunocytochemical characterization of GRIP, a putative AMPA receptor anchoring protein, in rat brain. Neuropharmacology. 1998;37:1335–1344. doi: 10.1016/s0028-3908(98)00120-8. [DOI] [PubMed] [Google Scholar]
- 35.Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 36.Zito K, Fetter RD, Goodman CS, Isacoff EY. Synaptic clustering of fasciclin II and shaker: essential targeting sequences and role of dlg. Neuron. 1997;19:1007–1016. doi: 10.1016/s0896-6273(00)80393-1. [DOI] [PubMed] [Google Scholar]