Abstract
The molecular mechanisms underlying the targeting and localization of glutamate receptors at postsynaptic sites is poorly understood. Recently, we have identified a PDZ domain-containing protein, glutamate receptor-interacting protein 1 (GRIP1), which specifically binds to the C termini of AMPA receptor subunits and may be involved in the synaptic targeting of these receptors. Here, we report the cloning of GRIP2, a homolog of GRIP1, and the characterization of the GRIP1 and GRIP2 proteins in the rat CNS. GRIP1 and GRIP2 are ∼130 kDa proteins that are highly enriched in brain. GRIP1 and GRIP2 are widely expressed in brain, with the highest levels found in the cerebral cortex, hippocampus, and olfactory bulb. Biochemical studies show that GRIP1 and GRIP2 are enriched in synaptic plasma membrane and postsynaptic density fractions. GRIP1 is expressed early in embryonic development before the expression of AMPA receptors and peaks in expression at postnatal day 8–10. In contrast, GRIP2 is expressed relatively late in development and parallels the expression of AMPA receptors. Immunohistochemistry using the GRIP1 antibodies demonstrated that GRIP1 is expressed in neurons in a somatodendritic staining pattern. At the ultrastructural level, DAB and immunogold electromicroscopy studies showed that GRIP1 was enriched in dendritic spines near the postsynaptic density and was expressed in dendritic shafts and in peri-Golgi regions in the neuronal soma. GRIP1 appeared to be clustered at both glutamatergic and GABAergic synapses. These results suggest that GRIP1 and GRIP2 are AMPA receptor binding proteins potentially involved in the targeting of AMPA receptors to synapses. GRIP1 also may play functional roles at both excitatory and inhibitory synapses, as well as in early neuronal development.
Keywords: GRIP1, GRIP2, AMPA receptor, neuronal synapses, GAD, postsynaptic density, vesicle, development
Glutamate receptors mediate the majority of excitatory synaptic transmission in the CNS and are involved in neuronal development, excitotoxicity, and synaptic plasticity (Seeburg, 1993; Hollmann and Heinemann, 1994). Glutamate receptors (GluR) can be subdivided based on their pharmacology into three major classes: AMPA, kainate, and NMDA receptors. AMPA receptors mediate rapid excitatory synaptic transmission and are heteromeric complexes composed of four homologous subunits (GluR1–GluR4) that differentially combine to form a variety of AMPA receptor subtypes (Seeburg, 1993; Hollmann and Heinemann, 1994). These subunits are thought to have a large extracellular N-terminal domain, three transmembrane domains, and an intracellular C-terminal domain (Seeburg, 1993; Hollmann and Heinemann, 1994).
Synaptic localization and clustering of ion channels and neurotransmitter receptors are necessary for normal synaptic transmission (Gomperts, 1996; Sheng, 1996; Colledge and Froehner, 1998). Recent studies have indicated that the regulation of the synaptic targeting of glutamate receptors, such as AMPA receptors, may play important roles in synaptic plasticity (Sheng and Kim, 1996;Kennedy, 1997; O’Brien et al., 1998). However, the molecular mechanisms that underlie the targeting and localization of AMPA receptors at postsynaptic membranes have not yet been elucidated.
Recently, proteins that contain the protein–protein interaction motifs called postsynaptic density-95/Discs large/zona occludens-1 (PDZ) domains have been suggested to play a role in the process of receptor targeting and localization (Ehlers et al., 1996; Sheng, 1996; Sheng and Kim, 1996; Kornau et al., 1997; Sheng and Wyszynski, 1997; O’Brien et al., 1998). PDZ domains specifically bind the C termini of a variety of membrane proteins, including neurotransmitter receptors. We have previously identified a PDZ domain-containing protein, glutamate receptor-interacting protein (GRIP1), that specifically interacts with the C termini of the GluR2 and GluR3 subunits of AMPA receptors (Dong et al., 1997). GRIP1 has seven PDZ domains and may serve as an adapter protein that links AMPA receptors to other cellular proteins that may play a critical role in clustering AMPA receptors at excitatory synapses.
Here, we report the cloning and characterization of GRIP2, a gene highly homologous to GRIP1, and the further characterization of GRIP1. GRIP1- and GRIP2-specific antibodies were used to compare the expression of the two proteins and their interaction at biochemical level. We also further characterized the distribution and expression of GRIP1 using immunohistochemical approaches. These results demonstrate that GRIP1 and GRIP2 are neuronal proteins that are expressed in various brain regions and are enriched in postsynaptic density (PSD) but are differentially regulated during development. GRIP1 is also found associated with vesicular structures in dendritic spines and shafts and within the soma near the Golgi. Surprisingly, we found that GRIP1 was found at both glutamatergic and GABAergic synapses, indicating that it may regulate both excitatory and inhibitory synaptic function. These results suggest that GRIP1 and GRIP2 are neuronal-specific synaptic proteins that may be involved in the targeting of AMPA receptors and other receptors at synapses.
MATERIALS AND METHODS
Yeast cotransformation assay for protein interaction.Yeast assays were performed as described previously (Dong et al., 1997), using the Y190 yeast strain harboring HIS3 and β-galactosidase as reporter genes. Various combinations of GRIP1 PDZ domains were generated by PCR with specific primers and subcloned in-frame into pPC97 or pPC86 yeast expression vectors to obtain Gal4 DNA binding domain fusion proteins and transcriptional activation domain fusion proteins. The yeast constructs used for GRIP1 were as follows: pPC97-PDZ456 [amino acids (aa) 412–795 containing PDZ domains 4–6, with an additional 60 N-terminal aa and 35 C-terminal aa]; pPC97-PDZ6 (aa 669–760); pPC97-PDZ123 (aa 30–331); pPC86-PDZ456 (the same as described above for pPC97); pPC86-PDZ4 (aa 470–557); pPC86-PDZ5 (aa 572–663); pPC86-PDZ6 (the same as described above for pPC97); pPC86-PDZ456 (aa 470–760); pPC86-PDZ45 (aa 470–663); pPC86-PDZN4 (aa 412–557); pPC86-PDZ45 (aa 412–557); pPC86-PDZ456s (aa 412–760); pPC86-PDZ6c (aa 669–795); pPC86-PDZ56c (aa 572–795); and pPC86-L2 (aa 798–904).
Cloning of GRIP2. Mouse Expressed Sequence Tag (EST) database searches with the GRIP1 sequence found many homologous clones. Two cDNA clones were ordered from Genome Systems (St. Louis, MO) and used as hybridization probes to screen a rat hippocampal λ phage cDNA library to obtain the full-length GRIP2 cDNA sequence.
Generation of antibodies. GRIP1- and GRIP2-specific antibodies were generated using a 20 amino acid peptide corresponding to the C-terminal 19 amino acids of GRIP1 and a 19 amino acid peptide unique to GRIP2 (YTPQVAVRSVTPQEWRSSR) cross-linked to thyroglobulin, which was then used to generate polyclonal antisera in rabbits. Antibodies (anti-GRIP1, anti-GRIP2) were affinity purified using BSA-conjugated antigen peptides cross-linked to Affi-Gel 10 resin (Bio-Rad, Hercules, CA). The GluR1 antibody has been described previously (Blackstone et al., 1992), and the monoclonal anti-GluR2 antibody was obtained from Chemicon (Temecula, CA). Mouse monoclonal anti-glutamic acid decarboxylase (GAD) antibodies were obtained from Boehringer Mannheim (Indianapolis, IN). Mouse monoclonal and rabbit polyclonal primary antibodies were visualized with anti-mouse or anti-rabbit IgG antibodies linked to rhodamine (red) or FITC (green) (Jackson ImmunoResearch, West Grove, PA).
Immunoblotting. Brain and other tissues were homogenized in 25 mm Tris-HCl, pH 7.4, 100 mm NaCl, with 5 mm EDTA, 5 mm EGTA, 0.1 mmphenylmethylsulfonyl fluoride (PMSF), 20 U/ml aprotinin, and 10 μg/ml each of leupeptin, anti-papain, pepstatin, and chymostatin. Homogenates of tissue preparations were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. The blots were imaged with enhanced chemiluminescence reagents (RenaissanceR ECL kit; NEN Life Science Products, Boston, MA).
Subcellular fractionations and PSD preparations. Subcellular fractionation and preparation of synaptic plasma membranes (SPMs) were performed essentially as described previously (Blackstone et al., 1992; Huttner et al., 1983). In brief, the lysed brain membrane fractions were separated on a discontinuous sucrose gradient containing 0.8, 1.0, and 1.2 m sucrose and centrifuged at 65,000 × g for 2 hr in a Beckman Instruments (Fullerton, CA) SW-28 rotor. Light membrane fractions were recovered from the 0.8 m sucrose layer. Other membranes were recovered in the layer between 0.8 and 1m sucrose. SPMs were recovered in the layer between 1.0 and 1.2 m sucrose. Mitochondria were recovered from the bottom of the sucrose gradient.
Synaptosome and PSD fractions were prepared from rat brains as described previously (Carlin et al., 1980; Cho et al., 1992) with the following modifications. The synaptosome fraction isolated by discontinuous sucrose gradient centrifugation was solubilized in ice-cold 0.5% Triton X-100 for 15 min and centrifuged at 32,000 × g for 20 min to obtain the PSD I pellet. This pellet was either resuspended and solubilized in 0.5% Triton X-100 and then centrifuged at 201,800 × g for 1 hr to obtain the PSD II pellet, or it was resuspended and solubilized in ice-cold 3% sarcosyl (Sigma, St. Louis, MO) for 10 min and then centrifuged at 201,800 × g for 1 hr to obtain the PSD III pellet. All pellets were resuspended in 40 mm Tris-HCl, pH 8.0. Protein concentrations were determined by BCA assay (Pierce, Rockford, IL). Samples were then analyzed by SDS-PAGE and immunoblotted as described above.
Transfection of HEK293T cells and coimmunoprecipitation in vitro. cDNAs subcloned into the vectors pBKCMV (full-length GRIP1), pRK5 (GluR2), or pRK5 with an N-terminal myc- or HA-tag (partial GRIP1) were transfected into HEK293T cells using calcium phosphate coprecipitation as described previously (Dong et al., 1997). The various GRIP1 constructs used were as follows: myc- PDZ456 (aa 435–969); myc-PDZ45 (aa 412–557); myc-PDZ56 (aa 572–795); and myc- PDZ6 (aa 669–795). After transfection for 36–48 hr, 293T cells were solubilized in buffer T (25 mm Tris-HCP with 100 mm NaCl, 5 mm EDTA, 5 mm EGTA, 20 U/ml Trasylol, and 0.1 mm PMSF) with 1% Triton X-100. Solubilized lysates (400 μl) were centrifuged at 15,000 ×g in microcentrifuge for 15 min at 4°C, and the supernatants were immunoprecipitated with either 10 μg of anti-GRIP1 antibody or 1 μg of monoclonal anti-myc ascites conjugated with 100 μl of a 1:1 slurry of protein A-Sepharose beads in buffer T. Immunoprecipitates were washed once in 1% Triton X-100 buffer T, twice in 1% Triton X-100 buffer T with 0.5 m NaCl, and three times with buffer T. Samples were then eluted with SDS sample buffer, separated by SDS-PAGE, and subjected to immunoblot analysis.
Coimmunoprecipitation in vivo. GRIP1 and GluR2 complexes from rat cortex were immunoprecipitated from sodium deoxycholate (DOC)-extracted rat brain membranes prepared as described previously (Luo et al., 1997). In brief, DOC extracts were prepared by resuspending rat brain cortex crude synaptosomal fraction (P2) in a suitable volume (3–5 mg/ml) of ice-cold TE buffer (10 mm Tris-HCl and 5 mm EDTA, pH 7.4) and pipette up and down briefly. One-tenth volume of cold DOC buffer (10% DOC and 500 mm Tris-HCl, pH 9.0) was added, followed by incubation at 36°C for 30 min. After the addition of one-tenth volume of Triton X-100 buffer (1% Triton X-100 and 500 mm Tris-HCl, pH 9.0), the samples were dialyzed against the binding buffer (50 mm Tris-HCl, pH 7.4, and 0.1% Triton X-100) overnight. The dialyzed sample was pelletted at 37,000 × g for 40 min, and the supernatant was used for immunoprecipitation. Anti-GRIP1 antibodies were conjugated to protein A-Sepharose beads by incubating 20 μg of antibody per 40 μl of protein A-Sepharose beads in 200 mm Na-borate buffer, pH 8.0 and rotated for 1–2 hr at 4°C. The protein A-Sepharose–antibody-bound beads were washed with 200 mm Na-borate buffer, pH 9.0, added to 200 μg of DOC-extracted proteins, and incubated at 4°C for 2–3 hr. Monoclonal anti-GluR2 antibodies (or unrelated monoclonal anti-myc and anti-NR1 antibodies) were conjugated to protein A-Sepharose beads by incubating 6 μg of antibody with 100 μl of protein A-Sepharose beads in binding buffer and rotated for 1–2 hr at 4°C. The protein A–antibody-bound beads were washed with binding buffer twice, added to 200 μg of DOC-extracted proteins, and rotated at 4°C for 2–3 hr. The immunoprecipitation reaction was then centrifuged at high-speed briefly in a microcentrifuge, and the supernatant was saved. The immunoprecipitate was washed three times with binding buffer, eluted with sample buffer, analyzed by SDS-PAGE, and immunoblotted using antibodies against GluR2 or GRIP1.
Immunocytochemistry for light microscopy. Male Sprague Dawley rats (200–350 gm) were anesthetized with pentobarbital (50 mg/kg) and perfused transcardially with 100 ml of cold 0.1m 0.9% PBS, followed by cold 4% paraformaldehyde prepared in 0.1 m phosphate buffer, pH 7.3. Brains were removed and post-fixed for 2 hr at 4°C and cryoprotected in 30% sucrose–PBS overnight at 4°C. Coronal and sagittal sections (40 μm) of rat brain were made with a sliding microtome and transferred to cold 0.02% sodium azide–PBS. Floating 40 μm brain sections were washed with PBS three times, incubated in 0.6% H2O2 (9 ml of 1× PBS, 1 ml of ethanol, and 200 μl of 30% H2O2) for 15 min at room temperature, washed with PBS three times, blocked, and permeabilized in blocking buffer [10% normal goat serum (NGS), 2% skim milk, and 0.2% Triton X-100 in PBS] for 1 hr at room temperature. For some preparations, freeze–thaw permeabilization was performed. The brain sections in a support dish were immersed in liquid nitrogen for 30 sec to 1 min, thawed in PBS for 10 min at room temperature, and then blocked in 10% NGS–2% skim milk–PBS for 1 hr at room temperature. Sections were incubated with rabbit anti-GRIP1 antibodies at a concentration of 1 μg/ml in 2% NGS–2% skim milk–PBS overnight at 4°C. Control sections were incubated with anti-GRIP1 antibody preabsorbed with an excess of antigen GRIP1 peptide. Subsequently, sections were rinsed in PBS three times, incubated with 1:300 biotinylated goat anti-rabbit antibody in PBS for 1 hr at room temperature, washed three times in PBS, and incubated with avidin–biotin–peroxidase complex for 1 hr at room temperature (Vectastain ABC kit; Vector Laboratories, Burlingame, CA). Sections were washed twice in PBS, twice in 0.05 mm Tris, pH 7.6, incubated with the substrate 3,3-diaminobenzidine tetrahydrochloride (DAB) (1 ml of 10 mg/ml, plus 17 ml of 0.05 mm Tris-HCl and 60 μl of 3% H2O2) for 5–15 min and washed three times in 0.05 mm Tris buffer. Sections were then mounted onto gelatin-treated slides, dried overnight, and dehydrated serially through one change each of 50% ethanol, 70% ethanol, 95% ethanol, two changes of 100% ethanol, and two changes of xylene. Slides were then coverslipped using the mounting solution DPX (Fluka BioChemika, Ronkonkoma, NY). DAB-stained brain sections were viewed in a Zeiss (Oberkochen, Germany) Axioskop microscope. Images were taken with a digital camera (Princeton Instruments, Trenton, NJ) and analyzed with the MetaMorph Imaging System (Universal Imaging, West Chester, PA).
Immunoperoxidase preembedding EM. Adult rats were preperfused with 50 ml of heparin (1000 U/ml) in PBS and then perfused with 70 ml of 3.75% acroline with 2% paraformaldehyde and 400 ml 2% paraformaldehyde (three rats), or straight 4% paraformaldehyde (two rats) in 0.1 m phosphate buffer, pH 7.3. Brains were removed and post-fixed in the same fixative for 1 hr, and 100 μm sections were cut using a vibratome. Sections were cyroprotected in 30% sucrose–PBS for 1 hr and transferred into nylon mesh baskets, which were quickly immersed into liquid nitrogen for 10–15 sec, and then thawed in PBS at room temperature. Sections were then treated with 1% sodium borohydride in PBS for 30 min. Immunoperoxidase staining was processed the same as for light microscopy analysis, except that Triton X-100 was omitted. After the DAB reaction, selected sections were fixed in 1% osmium tetroxide in PBS for 1 hr at room temperature and washed in PBS for 10 min with three changes. Sections were dehydrated in a graded series of ethanol, subjected to overnight infiltration in Dureapam Acm. Epoxy Resin (Electron Microscopy Sciences, Fort Washington, PA), flat embedding, and polymerization at 55°C for 48 hr. Embedded tissues were mounted on resin stubs and sectioned at 90 nm with an Ultracut E ultramicrotome. Sections were viewed and photographed with a Zeiss 600 EM10A transmission electron microscope at 60 kV.
Postembedding immunogold. For postembedding immunogold electron microscopy was performed as described previously (Petralia et al., 1997; Rubio and Wenthold, 1997; Wang et al., 1998). Male Sprague Dawley rats were anesthetized and perfused with 0.12m PBS, followed by 4% paraformaldehyde plus 0.5% glutaraldehyde in 0.12 × mPBS. Brains were post-fixed for 2 hr and washed three times for 1 hr. Washing and sectioning were performed in 0.1 mPBS with 4% glucose. Sections were prepared with a Microslicer at 200–300 μm, cryoprotected using a series of 10, 20, and 30% glycerol in 0.1 m PB, and then plunge-frozen in liquid propane at −184°C in a Leica (Nussloch, Germany) EM CPC (National Institutes of Health, Bethesda, MD). Frozen tissues were immersed in 1.5% uranyl acetate in methanol at −90°C in a Leica AFS freeze-substitution unit [in flow-through Reichert (Deerfield, IL) capsules], infiltrated with Lowicryl HM 20 resin at −45°C, and polymerized with UV light (−45°C to 0°C). Thin sections were cut with an ultramicrotome and placed on grids coated with Coat-quick G coating pen (Electron Microscopy Sciences). Grids were attached to Hiraoka support plates (Electron Microscopy Sciences) and incubated in 0.1% sodium borohydride and 50 mm glycine in Tris-buffered saline–0.1% Triton X-100 (TBST) for 10 min. After incubation in 10% NGS–TBST for 10 min, grids were incubated with anti-GRIP1 antibody (0.5 μg/μl at 1:5 to 1:25) in 1% NGS–TBST for 2 hr. Grids were then washed in TBST, blocked in 1% NGS–TBST, and incubated with 1:20 10 nm of immunogold conjugated goat anti-rabbit IgG (Amersham, Arlington Heights, IL) in 1% NGS–TBST plus 0.5% polyethylene glycol (20,000 molecular weight) for 1 hr. After further washes, sections were dried and stained with 1% uranyl acetate and 0.3% lead citrate.
Cell culture and immunocytochemistry. Hippocampal neuronal cultures were prepared from embryonic day 18 (E18) rats and maintained in serum-free medium above a glial monolayer as described previously (Banker and Cowan, 1977; Goslin and Banker, 1990). Cultured neurons (2–3 weeks old) on coverslips were fixed in freshly made, precooled 4% paraformaldehyde with 4% sucrose in PBS at 4°C for 15–20 min, and then permeabilized with 0.2% Triton X-100 in PBS at 4°C for 5 min. Coverslips were blocked in 10% normal donkey serum in PBS at room temperature for 1 hr. Immunostaining with anti-GRIP1 antibody, anti-GluR2, or anti-GAD antibodies was performed as described previously (Craig et al., 1994). Coverslips were mounted onto slides by Perma Fluor Aqueour (Lipshaw Immunon, Pittsburgh, PA) containing 0.25% 1,4-diazabicyclo [2.2.2]-octane (Aldrich, Milwaukee, WI). Coverslips were dried in the dark and viewed using a Zeiss Axioskop microscope. Images were obtained with a digital camera (Princeton Instruments) and analyzed with the MetaMorph Imaging System (Universal Imaging).
RESULTS
Cloning of GRIP2 and generation of GRIP1- and GRIP2-specific antibodies
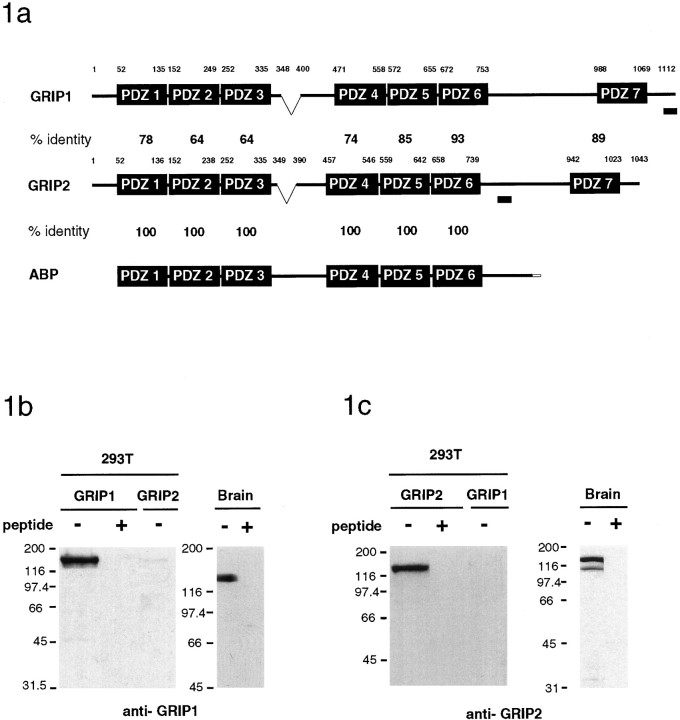
AMPA type glutamate receptors play an important role in fast excitatory synaptic transmission and are highly enriched at synapses. Recently, we identified a PDZ domain containing protein GRIP1 (Dong et al., 1997) that can interact with the C terminus of the GluR2 and GluR3 subunits of the AMPA receptor and may serve as an adaptor protein involved in the targeting of AMPA receptors to synapses. Through EST database searches, we found several cDNAs that had a high sequence homology with GRIP1. Using these cDNAs as probes, we screened a rat hippocampal cDNA library and isolated full-length cDNAs encoding GRIP2. Recently, a GluR2/3 binding protein homologous to GRIP1, AMPA receptor-binding protein (ABP), has been described (Srivastava et al., 1998). ABP is apparently a short splice variant of GRIP2 that lacks the N terminus and the seventh PDZ domain of GRIP2 (Fig.1a). In addition, a full-length GRIP2 cDNA was recently reported by Bruckner et al. (1999). Similar to GRIP1, GRIP2 contains seven PDZ domains that are very homologous to GRIP1 within the PDZ domains (64–93% identity) but has little sequence similarity in the linker regions between the PDZ domains (Fig. 1a).
Fig. 1.
GRIP1 and GRIP2 structure and characterization of GRIP1- and GRIP2-specific antibodies.a, Domain structure and sequence alignment of members of the GRIP protein family. GRIP2 is highly homologous to GRIP1, especially in the PDZ domains. ABP is a shorter splice variant of GRIP2, lacking the N-terminal sequence and the seventh PDZ domain. Regions of GRIP1 and GRIP2 used to generate GRIP-specific antibodies are indicated by the shaded bar. The region indicated between PDZ3 and PDZ4 of GRIP1 and GRIP2 is alternatively spliced. b, Brain lysates or HEK293T cells transfected with GRIP1 or GRIP2 were analyzed using the GRIP1-specific antibodies. c, Brain lysates or HEK293T cells transfected with GRIP1 or GRIP2 were analyzed using the GRIP2-specific antibodies. b, c, Preabsorption of both antibodies with the respective immunogenic peptides blocked immunorecognition.
GRIP1- and GRIP2-specific antibodies were generated against unique GRIP1 and GRIP2 peptides. The GRIP1 and GRIP2 antibodies specifically detected GRIP1 and GRIP2 as 135 and 130 kDa proteins, respectively, expressed in transfected 293T cells (Fig. 1b,c). The GRIP1 protein was expressed predominantly as a 135 kDa doublet in brain. The major forms of GRIP2 in brain had apparent molecular weights of 130 and 106 kDa, corresponding to GRIP2 and ABP, respectively. Preabsorption of the GRIP1 and GRIP2 antibodies with their respective immunogenic peptides completely blocked immunorecognition.
Differential distribution and regulated expression of GRIP1 and GRIP2
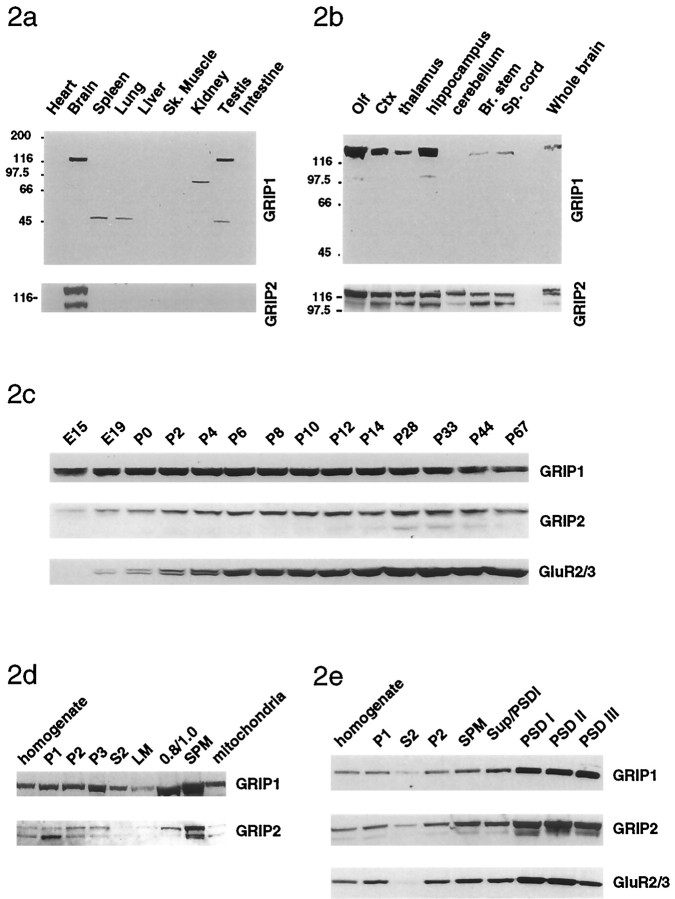
To specifically examine the tissue distribution of GRIP1 and GRIP2, various rat tissues were analyzed by Western blot techniques using the GRIP1- and GRIP2-specific antibodies. The GRIP1 protein was expressed in brain and testis but was not detected in heart, spleen, lung, liver, skeletal muscle, kidney, and intestine, whereas the major forms of GRIP2 were selectively expressed in brain (Fig.2a). GRIP1 and GRIP2 had similar distributions in rat brain regions and were enriched in the olfactory bulb, cortex, and hippocampus and were also expressed at lower levels in thalamus, cerebellum, and spinal cord (Fig.2b). GRIP1 expression in rat brain was detected in early embryonic stages (as early as E10; data not shown), gradually increased throughout early development, peaked at approximately postnatal day 6–8, then slightly decreased and remained relatively stable in the adult (Fig. 2c). Interestingly, in contrast, GRIP2 expression was relatively low early in development and increased postnatally, reaching a peak at postnatal day 14, similar to what is observed for GluR2 (Fig. 2c).
Fig. 2.
Distribution and subcellular fractionation of GRIP1 and GRIP2 protein in brain. a, Tissue distribution of GRIP1 and GRIP2. GRIP1 is a 135 kDa doublet in brain and testis, whereas GRIP2 expressed as 130 and 106 kDa proteins in brain. b, Distribution of GRIP1 and GRIP2 in various regions of the brain. GRIP1 and GRIP2 were enriched in olfactory bulb, cortex, and hippocampus. c, Developmental profile of GRIP1 and GRIP2 expression in brain. GRIP1 expression occurs early in development and increases during development, peaking at approximately postnatal day 6–8, and then decreasing slightly in the adult. In contrast, GRIP2 is expressed postnatally and more closely parallels the expression of GluR2. d, GRIP1 and GRIP2 are found in several subcellular fractions, including the nuclear (P1), cytosol (S2), crude synaptosomal fraction (P2), lysated synaptosomal membrane (P3), light membrane (LM), and other membrane (0.8/1.0) but is most highly enriched in SPM. e, GRIP1 and GRIP2 are enriched in PSD fractionations. GRIP1 and GRIP2 are found in soluble fraction (S2) and the crude synaptosomal fraction (P2) but were enriched in SPM andPSD I (1 Triton X-100 extraction), PSD II(2 Triton X-100 extractions), and PSD III (1 Triton X-100 and 3% sarcosyl extraction).
Subcellular fractionation of rat brain was used to examine the distribution of GRIP1 and GRIP2 using the GRIP-specific antibodies (Huttner et al., 1983; Blackstone et al., 1992). GRIP1 and GRIP2 were widely expressed in various fractions but were most highly enriched in SPM fractions (Fig. 2d). Treatment of synaptic plasma membrane fractions with various detergents to generate PSD fractions demonstrated that GRIP1 and GRIP2 were enriched in the PSD (Fig.2e). Although the majority of GRIP1 and GRIP2 were associated with membrane fractions, a significant portion of GRIP1 and GRIP2 were also found in cytosolic fractions. Whether membrane association is via protein–protein interactions with membrane proteins or by other mechanisms, such as palmitoylation (Topinka and Bredt, 1998), is not yet clear. To assess the detergent solubility of membrane-associated GRIPs, the P2 fraction of rat brain cortex (Luo et al., 1997) was treated with 1% Triton X-100, 1.0% deoxycholate, or 1% SDS. Both GRIP1 and GRIP2 were partially solubilized by 1% Triton X-100 and totally solubilized in 1.0% deoxycholate and 1% SDS (data not shown).
GRIP1 and GRIP2 can form homomultimers and heteromultimers
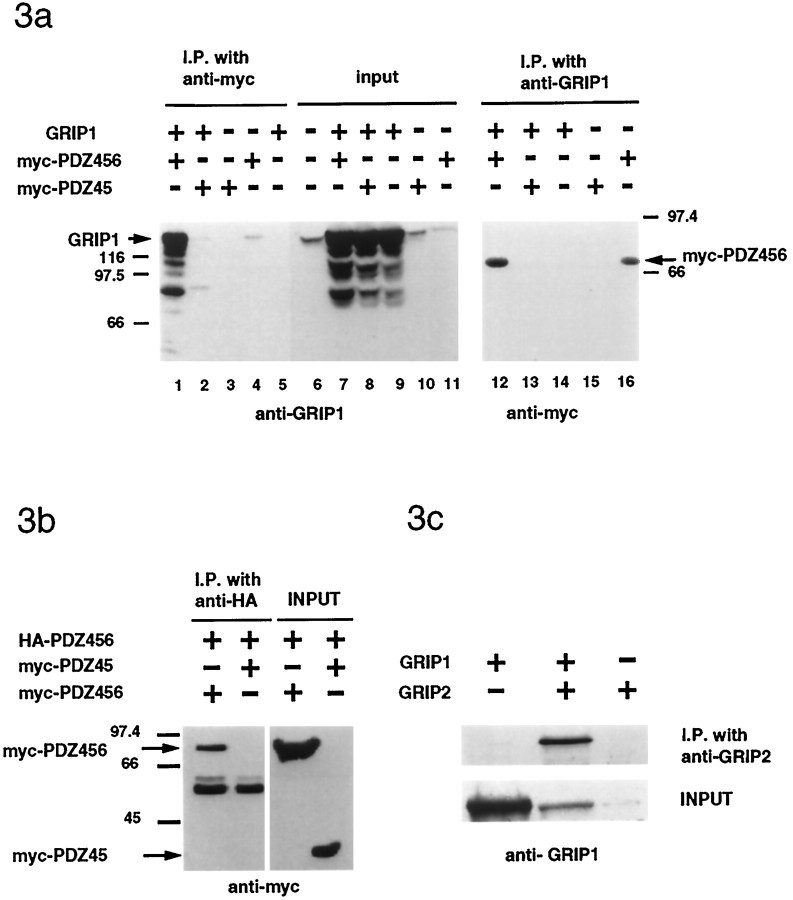
Previous studies have shown that PDZ–PDZ domain interactions can occur between proteins (Brenman et al., 1996). Recently, ABP has been shown to interact with GRIP1 (Srivastava et al., 1998). To examine whether GRIP1 and GRIP2 can form multimers and to determine the domains that mediate these interactions, we performed in vitrocoimmunoprecipitation experiments in 293T cells. Full-length GRIP1 could be coimmunoprecipitated with a myc-tagged protein containing PDZ domains 4, 5, and 6 of GRIP1 (Fig.3a, lanes 1,12). In contrast, GRIP1 did not coimmunoprecipitate with shorter constructs, such as myc-PDZ45 (Fig. 3a, lanes 2, 13). Interestingly, 293T cells have a low endogenous level of GRIP1, which could be coimmunoprecipitated with myc-PDZ456 when it was transfected alone (Fig. 3a, lanes 4,16).
Fig. 3.
Homomultimerization and heteromultimerization of GRIP1 and GRIP2. a, In cotransfected HEK293T cells, full-length GRIP1 protein could be coimmunoprecipitated with myc-PDZ456 (lane 1) but not with a myc-PDZ45 construct lacking PDZ6 (lane 2). Myc-PDZ456 (lane 12), but not myc-PDZ45 (lane 13), could also be coimmunoprecipitated by anti-GRIP1 antibody.b, Myc-PDZ456, but not myc PDZ45, could be coimmunoprecipitated by HA-PDZ456. c, In transfected 293T cells, full-length GRIP1 coimmunoprecipitated with GRIP2.
To test whether the PDZ domain 456 self-interacts, we used two different tagged PDZ456 constructs. As shown in Figure 3b, HA-PDZ456 can pull down myc-PDZ456 but not myc-PDZ45, indicating that the GRIP1 self-interaction was mediated via the self-interaction of the PDZ456 region (Fig. 3b). Similar results were observed in yeast two-hybrid assays, which showed that yeast constructs containing GRIP1 PDZ456 could interact with itself but not with PDZ456 constructs with shorter sequences or with PDZ123 or PDZ7 (data not shown). To examine whether GRIP1 and GRIP2 could form heteromers, we cotransfected GRIP1 and GRIP2 in HEK293T cells and analyzed GRIP2 immunoprecipitates for the presence of GRIP1. GRIP1 was specifically coimmunoprecipitated with GRIP2 using the GRIP2 antibody (Fig. 3c). These results indicate that GRIP1 may be able to form homomultimers or heteromultimers with GRIP2, allowing the formation of very large macromolecular complexes.
In vivo and in vitro interaction of full-length GRIP1 and AMPA receptors
In our previous study, we were unable to coimmunoprecipitate GRIP with AMPA receptors from brain because of difficulties in solubilizing GRIP1 from synaptic plasma membranes using nondenaturing detergents (Dong et al., 1997). However, using a modified protocol fromLuo et al. (1997), we have now been able to solubilize significant amounts of GRIP1 using 1% deoxycholate at pH 9.0. GRIP1 could be specifically coimmunoprecipitated with GluR2 from deoxycholate-solubilized brain membrane preparations (P2) using a monoclonal GluR2 antibody (Fig.4a). Conversely, GluR2 could also be coimmunoprecipitated with GRIP1, and this coimmunoprecipitation could be blocked by preabsorption of the antibody with the antigenic peptide (data not shown).
Fig. 4.
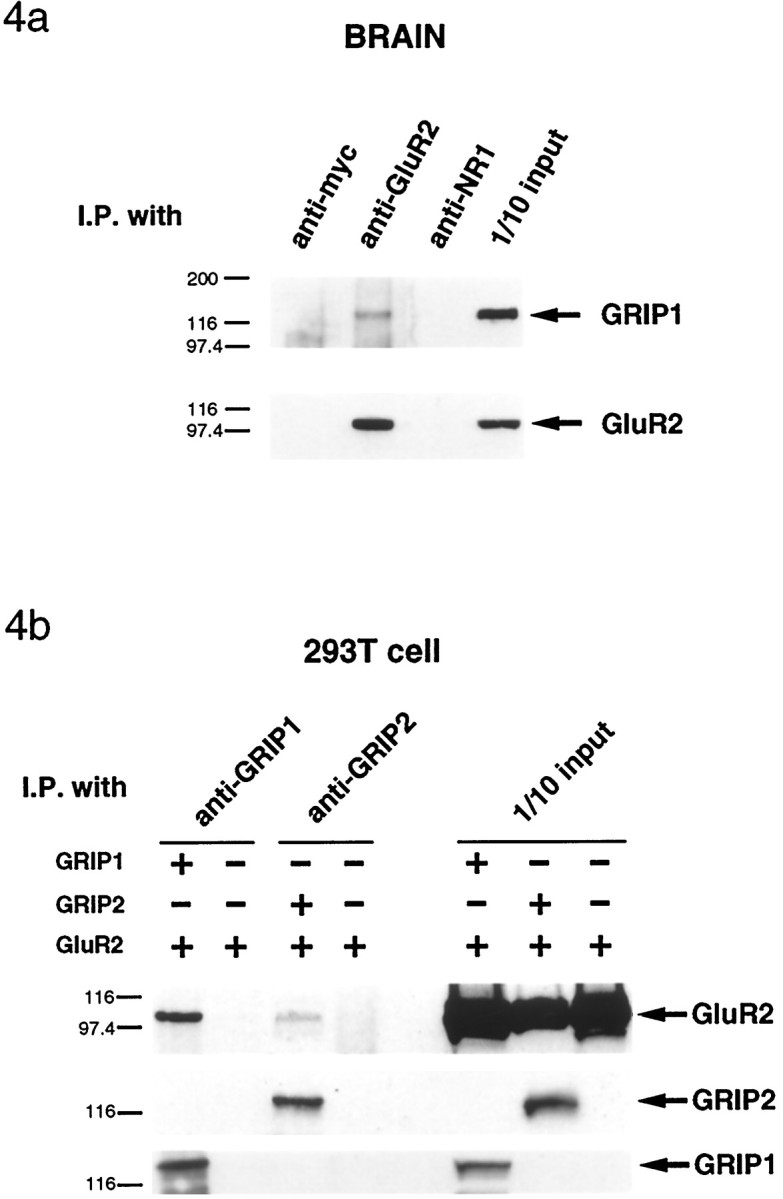
Coimmunoprecipitation of GRIP1 with AMPA receptorsin vitro and in vivo. a, When rat cortex membrane proteins were solubilized with 1% deoxycholate at pH 9, GRIP1 protein could be coimmunoprecipitated by monoclonal anti-GluR2 antibody. Coimmunoprecipitation was negative when nonrelated monoclonal antibodies (anti-myc and NR1) were used for coimmunoprecipitation. b, In transfected HEK293T cells, the GluR2 subunit could be coimmunoprecipitated together with GRIP1 and GRIP2 proteins but not when GluR2 was transfected alone.
Although we have been unable to coimmunoprecipitate GRIP2 with GluR2 from rat brain, experiments using transfected HEK293T cells demonstrated that full-length recombinant GRIP2 can associate with GluR2 in situ. GRIP2 antibodies specifically coimmunoprecipitated GluR2 with GRIP2 from 1% Triton X-100-solubilized lysates from HEK293T cells cotransfected with GluR2 and GRIP2 (Fig.4b). Similar results were seen with GRIP1. When GluR2 was transfected alone, low levels of GluR2 were coimmunoprecipitated with the anti-GRIP1 antibody as a result of the endogenous low levels of GRIP1 expressed in these cells (data not shown). These results demonstrate that GRIP1 and GRIP2 can associate with AMPA receptorsin situ in transfected cells and that GRIP1 is associated with AMPA receptors in vivo in brain.
Immunocytochemical localization of GRIP1 in rat brain
The distribution of GRIP1 in rat brain was examined at higher resolution with immunocytochemical techniques using light and electron microscopy. Immunohistochemical staining of rat brain sections using the GRIP1-specific C-terminal peptide antibody (Fig.5a–c) demonstrated that GRIP1 was expressed in a somatodendritic pattern in neurons throughout the rat brain. Prominent staining was observed in the cerebral cortex and hippocampus and other regions of the brain, consistent with the Western analysis of rat brain regions. This staining was completely eliminated when the antibody was preincubated with immunogen (Fig.5d–f; data not shown). In the cerebral cortex, immunolabeling with the GRIP1 antibody was highest in the pyramidal neurons of layer II–III and in layers IV and V (Fig. 5a). Moderate staining was evident in numerous nonpyramidal cells of layer IV and VI. In addition, in the deepest part of layer VI, just adjacent to the white matter, horizontal cells were densely stained with GRIP1 (data not shown). Neuropils were also stained throughout the cortical layers. Pyramidal neurons were prominently stained in their cell soma and apical dendrites, as well as some proximal basal dendrites.
Fig. 5.
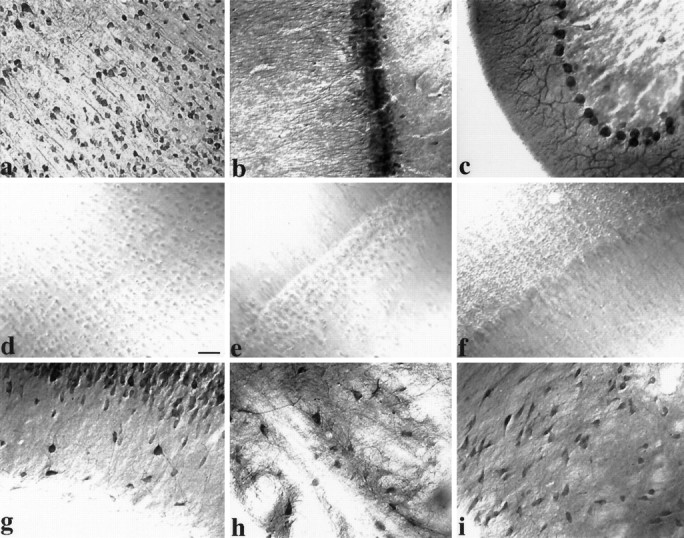
Immunohistochemical localization of GRIP1 in rat brain. Coronal rat brain sections were immunostained with GRIP1 antibody and visualized by DAB. GRIP1 immunoreactivity was observed in cerebral cortex (a), hippocampal formation (b) and cerebellum (c), with no staining in corresponding controls (d–f; antibody preincubated with immunogenic peptide). a, Immunostaining of cell soma and apical and basal dendrites of pyramidal neurons in the cerebral cortex. b, Labeling of hippocampus CA1 pyramidal cells in a somatodendritic manner. Note scattered interneuron immunoreactivity and neuropil staining in stratum oriens and stratum radiatum. c, Staining of Purkinje neurons extending from cell soma throughout major dendritic arborizations. g, Strong staining of scattered inhibitory interneurons in the stratum oriens of the CA3 area. GRIP1 immunoreactivity was also observed in many other areas, such as striatum (h) and substantia nigra (i). Scale bar, 40 μm.
All regions of the hippocampus contained immunoreactive neurons with a somatodendritic staining pattern for GRIP1 (Fig. 5b). The cell bodies of pyramidal cells in the CA1, CA2, and CA3 regions were prominently stained, and labeled apical dendrites were observed to extend into branches in the stratum radiatum. Interneurons scattered within the stratum oriens, as well as occasionally in the stratum radiatum, were heavily stained for GRIP1 (Fig. 5g). The granule cell layer of the dentate gyrus was also labeled for GRIP1. A large number of scattered polymorph neurons in the hilus were labeled as well (data not shown). In the cerebellum, GRIP1 immunoreactivity was observed prominently in cell bodies and throughout the dendritic arborizations of Purkinje cells in all folia, whereas the granule cell layer was not stained (Fig. 5c). Some neurons of the cerebellar deep nucleus were also labeled (data not shown).
Consistent with regional distribution of GRIP1 detected by Western blots, all the olfactory regions stained densely with GRIP1 antibodies, with diffuse neuropil staining in both external plexiform layer and glomerular layer (data not shown). GRIP1 immunoreactivity was also observed in many multipolar neurons in other brain regions in a somatodendritic pattern, including the striatum (Fig. 5h), substantia nigra (Fig. 5i), septum, amygdala, midbrain, brainstem, thalamus, hypothalamus, and mammaliary bodies (data not shown). These results demonstrate that GRIP1 is expressed in neurons, in a somatodendritic pattern throughout the brain similar to the distribution pattern of GluR2/3/4c (Petralia and Wenthold, 1992; Martin et al., 1993).
Ultrastructral localization of GRIP1 in rat cortex and hippocampus: immunoperoxidase
To investigate the localization of GRIP1 at the ultrastructural level, we first examined the distribution of GRIP1 using preembedding immunoperoxidase methods. Using a low concentration (2.5 ng/μl) of the GRIP1-specific antibody, we examined GRIP1 immunoreactivity in the cerebral cortex (Fig.6d–f) and the hippocampal CA1 region (Fig. 6a–c) from three adult rats. Positive labeling for GRIP1 was observed in the perikaryon, dendrites, and dendritic spines of neurons (Fig. 6, arrows). Preabsorption of the GRIP1 antibody with the immunogen eliminated immunoperoxidase labeling (data not shown). GRIP1 staining was often found with vesicular profiles of variable sizes and shapes (Fig. 6,arrows). In the perikaryon, staining was associated with peri-Golgi complexes and the ER, as well as scattered in the cytoplasm, and was more frequently found on one side of the cell soma (basal side) (Fig. 6d). Dendritic shafts (Fig.6c,f, arrows) and large dendrite spines (Fig. 6b, arrows) were more strongly stained for GRIP1 near small vesicular structures. Interestingly, the labeled vesicles were associated with the postsynaptic density in spines (Fig. 6e) or were found clustered directly underneath the plasma membrane of a dendrite in direct contact with an axonal terminal (Fig. 6f).
Fig. 6.
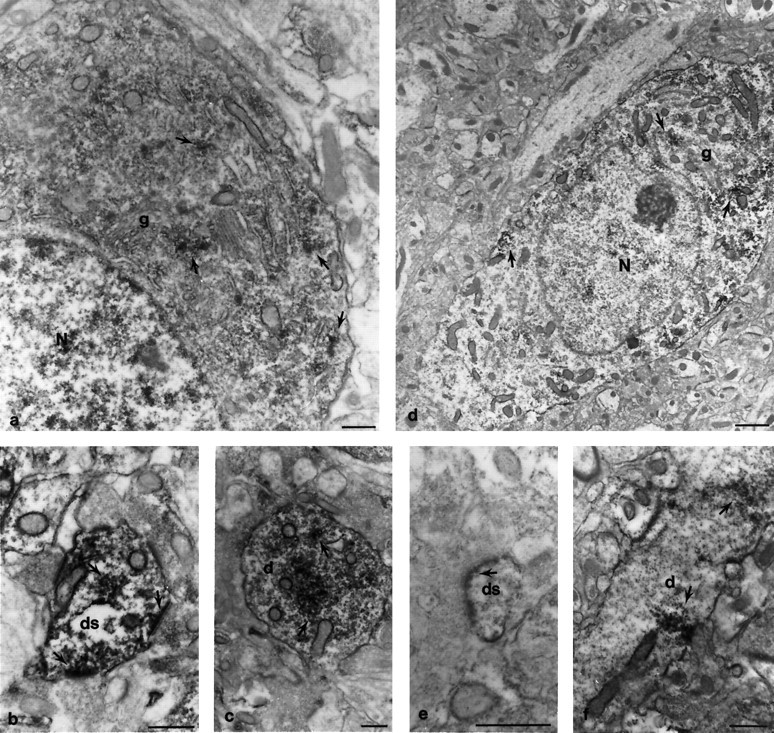
Immunoperoxidase electron microscopic localization of GRIP1 protein in cortex and hippocampus. Coronal rat brain sections were immunostained with the GRIP1-specific antibody and visualized by DAB. The hippocampal CA1 region (a–c) and the cortex (d–f) were analyzed by electron microscopy. The loosely packed, irregular vesicular profiles in the perikaryon and dendritic spines often formed dense patches that were masked with peroxidase product. In the perikaryon of pyramidal cells, the clustered vesicles were often near peri-Golgi complexes, rough endoplasmic reticulum, or scattered in the cytosol (a,d, arrows) and were more condensed in the basal side of neurons (d). Dendritic shafts and large spines contained vesicles with substantial staining (c, f, arrows). Inf, a line of stained vesicles was localized directly underneath the postsynaptic membrane plasma membrane. In large dendritic spines (b, arrow), vesicles were found either clustered in the cytosal or individually merged with stained postsynaptic densities. Some presynaptic nerve terminals were also stained (the terminal in b, right arrow). The electron density of the postsynaptic membrane was significantly enhanced because of immunoperoxidase precipitation. This phenomenon was found in either large dendritic spines (b, arrow) or spine heads (e, arrow). N, Nucleus;g, Golgi complex; d, dendritic shaft;ds, dendritic spine. Scale bar, 0.5 μm.
The postsynaptic membranes of asymmetric synapses (Fig.6b,e, arrows) appeared to be specifically labeled in comparison to unstained spine heads (Fig.6b, bottom). The immunostaining was not just restricted to areas within the postsynaptic membrane but also extended to adjacent areas in the plasma membrane (Fig.6b,e). The immunoreactivity was also occasionally observed in the postsynaptic density (Fig. 6e) within a regular array of vesicular profiles. GRIP1 staining was also occasionally found on the outer membrane of mitochondria and in nuclei (Fig. 6).
Electron microscopy: immunogold
On the basis of the patterns of GRIP1 distribution using immunoperoxidase detection, immunogold methods were used to confirm these results and to obtain higher resolution. The overall immunogold labeling was similar to that seen with immunoperoxidase. In the perikaryon, clusters of gold particles were found near the peri-Golgi complex and associated with vesicular-like structures (Fig.7a, arrows). Immunogold labeling was also observed over the rough ER or scattered in the cytoplasm. Most frequently, the gold clusters appeared in dendrites (Fig. 7b, arrows) and were typically associated with vesicles, sometimes clearly outlining a vesicular profile. These gold-labeled vesicles were often very close to microtubule filaments. Immunogold label was also found in postsynaptic spines, frequently near the PSD region (Fig. 7c–e, arrows), and was occasionally found within the PSD (Fig. 7c,arrows). Although decreasing the concentration of antibody from 100 to 25 ng/μl lowered the average number of gold particles per cluster, the pattern of staining remained the same.
Fig. 7.
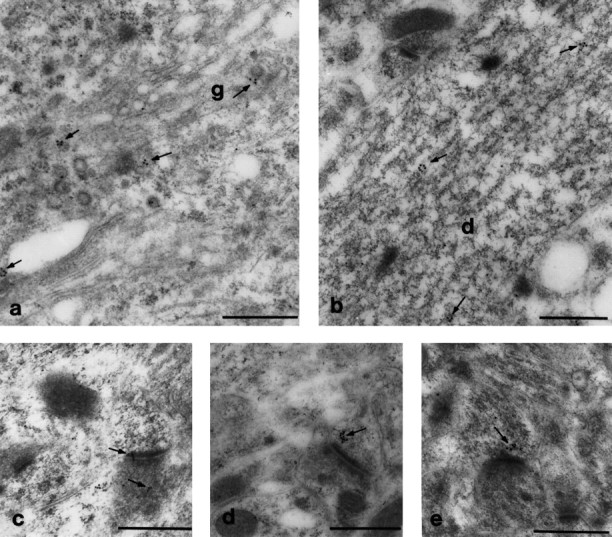
Immunogold electron microscopy localization of GRIP1 protein in CNS. Consistent with results in Figure 6, immunogold particles were found in small clusters, which were frequently associated with vesicular structures and localized near the peri-Golgi and endoplasmic reticulum in the perikaryon (a) and dendritic shafts (b). In b, gold particles outlined a vesicle near microtubule filaments. Gold particles were occasionally found in the PSD (c) but more frequently localized near the PSD (d,e, arrows). Gold labeling was also observed infrequently in terminals (c, bottom arrow). g, Golgi apparatus; d, dendritic shaft. Scale bar, 0.5 μm.
Immunogold labeling was also found in a few presynaptic terminals (Fig.7c, bottom arrow). In 41 labeled synaptic profiles in the cerebral cortex and hippocampus, 26% were labeled in presynaptic elements, whereas 74% were labeled in postsynaptic elements. Gold particles were also detected at mitochondria and nuclei at levels slightly above background staining for these organelles. In control samples using 2% normal serum or an irrelevant antibody (anti-rapsyn antibody), single gold particles were occasionally observed (data not shown).
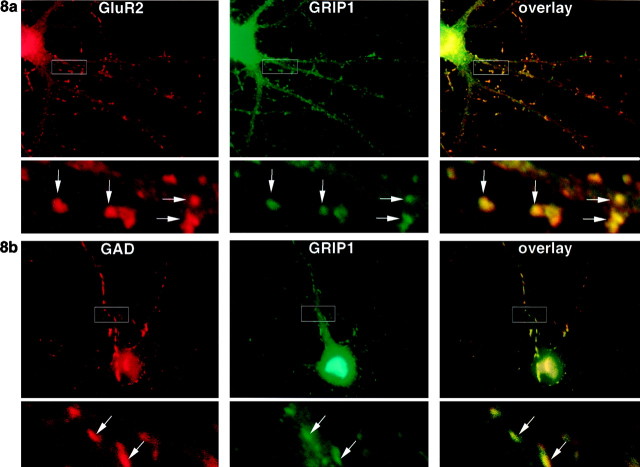
Colocalization of GRIP1 with glutamatergic and GABAergic synapses in cultured hippocampal neurons
To examine whether GRIP1 is localized to excitatory or inhibitory synapses, cultured hippocampal neurons were either double-labeled with monoclonal anti-GluR2 and GRIP1 antibodies (Fig.8a) or double-labeled with monoclonal anti-GAD and GRIP1 antibodies (Fig. 8b). GRIP1 (green) was distributed in small puncta along dendrites (Fig. 8a,b, middle) and directly colocalized in many cases with AMPA receptors (red) at excitatory synapses on dendritic spines of pyramidal neurons (Fig.8a). Surprisingly, many of the GRIP1 puncta also colocalized with GAD (red), a marker for GABAergic synapses on the dendritic shafts of both GABAergic neurons (data not shown) and pyramidal neurons (Fig. 8b).
Fig. 8.
GRIP1 colocalizes with AMPA receptors at excitatory synapses and with GAD at inhibitory synapses. Hippocampal neurons in culture (3 weeks) were double-labeled with anti-GRIP1 (green) and monoclonal anti-GluR2 antibodies (a, red), or anti-GRIP1 and monoclonal anti-GAD antibodies (b, red). GRIP1 staining (green) was punctate and was found throughout the neuronal cell body and neurites and colocalized with AMPA at dendritic spines (a, arrows). GRIP1 also colocalized with GAD at inhibitory synapses on dendritic shafts (b, arrow). The bottom panels show an enlarged portion of the area within thewhite rectangles in the top panels.
DISCUSSION
In an effort to reveal the molecular machinery underlying AMPA receptor targeting and localization, we have previously used the yeast two-hybrid approach to identify GRIP1, a novel multiple PDZ domain-containing protein that interacts with AMPA receptor subunits GluR2 and GluR3 (Dong et al., 1997). We now report the identification and characterization of GRIP2 and the further characterization of GRIP1. The structure of GRIP2 is very similar to that of GRIP1 and contains seven PDZ domains, which are highly homologous to the corresponding PDZ domains in GRIP1. The sequence of GRIP2 is identical to a recently described AMPA receptor binding protein ABP, except that ABP appears to be a short splice variant of GRIP2 that is missing the seventh PDZ domain and part of the N-terminal sequence. The cloning of GRIP2 was also recently reported by Bruckner et al. (1999).
We have found that the expression of the GRIP1 and GRIP2 proteins is relatively brain-specific. Both were expressed in most brain regions, were enriched in synaptic membrane fractions, and cofractionated with AMPA receptors in PSD preparations. GRIP1 immunoreactivity was distributed in various neuronal types and was found in the dendritic spines and near the PSD. Moreover, GRIP1 could be coimmunoprecipitated together with GluR2 subunits, both in transfected 293T cells and from brain lysates, indicating that GRIP1 interacts with AMPA receptorsin vivo. These results are consistent with our original hypothesis that GRIP1 is involved in the regulation of AMPA receptor function and clustering at excitatory synapses. However, GRIP1 was also found associated with membrane vesicles in the cell soma near the peri-Golgi region and in dendritic shafts. These vesicular structures may be derived from smooth endoplasmic reticulum and/or the trans-Golgi network (Spacek and Harris, 1997), suggesting that GRIP1 could play a role in the sorting and transport (and/or recycling) of membrane proteins from cytoplasm to dendrite to synapse. This is interesting in light of recent studies demonstrating that the regulation of receptor surface expression may play critical roles in synaptic plasticity at both excitatory and inhibitory synapses (Liao et al., 1995; Maletic-Savatic et al., 1998; Nusser et al., 1998).
Intriguingly, we found that GRIP1 is not only found at excitatory synapses but is also highly concentrated at inhibitory synapses. The specific association and enrichment of GRIP1 in both excitatory and inhibitory synapses suggests that GRIP1 may play a more general role in the sorting, transportation, and organization of proteins at many types of synapses. Moreover, GRIP1 is expressed in testis and is also expressed early in development before the expression of GluR2/3/4C. These results indicate that GRIP1 is likely to play a pleiotropic role as an adaptor protein in several contexts. In contrast, GRIP2 expression, similar to ABP expression (Srivastava et al., 1998), parallels GluR2 expression and may be more specifically involved in synapse formation.
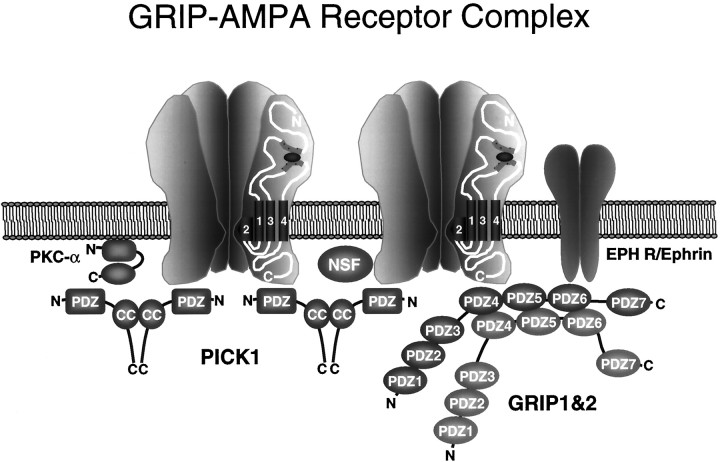
GRIP1 and GRIP2 contain seven PDZ domains with different ligand specificities (Dong et al., 1997), indicating that GRIPs may simultaneously bind multiple ligands to form a large adaptor complex. GRIP has recently been shown to interact with ephrine (EPH) receptors and ephrins (Torres et al., 1998) and with novel neuronal proteins we have termed GRASPs for GRIP-associated proteins (B. Ye, H. Dong, and R. Huganir, unpublished results). In addition, we have demonstrated that GRIP can form homomeric and heteromeric multimers through the interaction of PDZ domains 456. AMPA receptors are heteromeric complexes that may contain several copies of GluR2 and GluR3, providing multiple binding sites for GRIPs. The C termini of GluR2/3 has also recently been shown to bind to the PDZ domain of the PKC-interacting protein PICK1 (Xia et al., 1999), as well as to the membrane fusion protein N-ethylmaleimide-sensitive factor (NSF) (Nishimune et al., 1998; Osten et al., 1998; Song et al., 1998).
These results indicate that GRIP1, GRIP2, and AMPA receptors can form very large complexes containing a constellation of proteins, which may include EPH receptors, ephrins, PICK1, PKC, and NSF (Fig.9). The regulation of this complex may be very dynamic. GRIP1, GRIP2, and PICK1 all bind to the last few amino acids of the C termini of GluR2/3 and should compete with each other for binding. It is interesting to speculate that GRIP1/2 and/or PICK1 binding to GluR2/3 may be differentially regulated to selectively modulating the interaction of GluR2/3 with GRIPs and PICK1. For example, GRIP may bind GluR2/3 and assist in the sorting and export of AMPA receptors from the cell soma to the dendrite, where PICK1 may bind and stabilize GluR2/3 at the synapse. Future studies characterizing the regulation of the interaction of GRIP with these ligands will help to clarify functional role of GRIPs in the CNS.
Fig. 9.
GRIP protein complex. GRIP1 and GRIP2 may be associated with a large protein network containing AMPA receptors, PICK1, EPH receptors, ephrins, PKC, and NSF. These complexes may play an important role in the regulation of the function and/or the subcellular targeting of AMPA receptors. In addition, this large complex may regulate novel downstream signal transduction pathways from the AMPA receptor.
Footnotes
This work was supported by the Howard Hughes Medical Institute and the National Institutes of Health (National Institute of Neurological Diseases and Stroke). We thank X. Zhang for her technical assistance in preparing hippocampal neuronal cultures and C. A. Doherty for her help in antibody purification. We also thank J.-H. Kim for her assistance in preparing the figures and D. Bury for her assistance in preparing this manuscript.
Correspondence should be addressed to Dr. Richard L. Huganir, Howard Hughes Medical Institute, Department of Neuroscience, The Johns Hopkins University School of Medicine, 725 N. Wolfe Street, PCTB 904, Baltimore, MD 21205.
REFERENCES
- 1.Banker GA, Cowan WM. Rat hippocampal neurons in dispersed cell culture. Brain Res. 1977;126:397–342. doi: 10.1016/0006-8993(77)90594-7. [DOI] [PubMed] [Google Scholar]
- 2.Blackstone CD, Moss SJ, Martin LJ, Levey AI, Price DL, Huganir RL. Biochemical characterization and localization of a non-N-methyl-d-aspartate glutamate receptor in rat brain. J Neurochem. 1992;58:1118–1126. doi: 10.1111/j.1471-4159.1992.tb09370.x. [DOI] [PubMed] [Google Scholar]
- 3.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 4.Bruckner K, Pablo Labrador J, Scheiffele P, Herb A, Seeburg PH, Klein R. EphrinB ligands recruit GRIP family PDZ adaptor proteins into raft membrane microdomains. Neuron. 1999;22:511–524. doi: 10.1016/s0896-6273(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 5.Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 7.Colledge M, Froehner SC. Signals mediating ion channel clustering at the neuromuscular junction. Curr Opin Neurobiol. 1998;8:357–363. doi: 10.1016/s0959-4388(98)80061-5. [DOI] [PubMed] [Google Scholar]
- 8.Craig AM, Blackstone CD, Huganir RL, Banker G. Selective clustering of glutamate and gamma-aminobutyric acid receptors opposite terminals releasing the corresponding neurotransmitters. Proc Natl Acad Sci USA. 1994;91:12373–12377. doi: 10.1073/pnas.91.26.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong H, O’Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 10.Ehlers MD, Mammen AL, Lau LF, Huganir RL. Synaptic targeting of glutamate receptors. Curr Opin Cell Biol. 1996;8:484–489. doi: 10.1016/s0955-0674(96)80024-x. [DOI] [PubMed] [Google Scholar]
- 11.Gomperts SN. Clustering membrane proteins: it’s all coming together with the PSD-95/SAP90 protein family. Cell. 1996;84:659–662. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 12.Goslin K, Banker G. Rapid changes in the distribution of GAP-43 correlate with the expression of neuronal polarity during normal development and under experimental conditions. J Cell Biol. 1990;110:1319–1331. doi: 10.1083/jcb.110.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollmann M, Heinemann S. Cloned glutamate receptors. Ann Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 14.Huttner WB, Schiebler W, Greengard P, De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy MB. The postsynaptic density at glutamatergic synapses. Trends Neurosci. 1997;20:264–268. doi: 10.1016/s0166-2236(96)01033-8. [DOI] [PubMed] [Google Scholar]
- 16.Kornau HC, Seeburg PH, Kennedy MB. Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 17.Liao D, Hessler NA, Malinow R. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature. 1995;375:400–404. doi: 10.1038/375400a0. [DOI] [PubMed] [Google Scholar]
- 18.Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BW. The majority of N-methyl-d-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B). Mol Pharmacol. 1997;51:79–86. doi: 10.1124/mol.51.1.79. [DOI] [PubMed] [Google Scholar]
- 19.Maletic-Savatic M, Koothan T, Malinow R. Calcium-evoked dendritic exocytosis in cultured hippocampal neurons. II. Mediation by calcium/calmodulin-dependent protein kinase II. J Neurosci. 1998;18:6814–6821. doi: 10.1523/JNEUROSCI.18-17-06814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin LJ, Blackstone CD, Levey AI, Huganir RL, Price DL. AMPA glutamate receptor subunits are differentially distributed in rat brain. Neuroscience. 1993;53:327–358. doi: 10.1016/0306-4522(93)90199-p. [DOI] [PubMed] [Google Scholar]
- 21.Nishimune A, Isaac JT, Molnar E, Noel J, Nash SR, Tagaya M, Collingridge GL, Nakanishi S, Henley JM. NSF binding to GluR2 regulates synaptic transmission. Neuron. 1998;21:87–97. doi: 10.1016/s0896-6273(00)80517-6. [DOI] [PubMed] [Google Scholar]
- 22.Nusser Z, Lujan R, Laube G, Roberts JD, Molnar E, Somogyi P. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron. 1998;21:545–559. doi: 10.1016/s0896-6273(00)80565-6. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien RJ, Lau LF, Huganir RL. Molecular mechanisms of glutamate receptor clustering at excitatory synapses. Curr Opin Neurobiol. 1998;8:364–369. doi: 10.1016/s0959-4388(98)80062-7. [DOI] [PubMed] [Google Scholar]
- 24.Osten P, Srivastava S, Inman GJ, Vilim FS, Khatri L, Lee LM, States BA, Einheber S, Milner TA, Hanson PI, Ziff EB. The AMPA receptor GluR2 C terminus can mediate a reversible, ATP-dependent interaction with NSF and α- and β-SNAPs. Neuron. 1998;21:99–110. doi: 10.1016/s0896-6273(00)80518-8. [DOI] [PubMed] [Google Scholar]
- 25.Petralia RS, Wenthold RJ. Light and electron immunocytochemical localization of AMPA-selective glutamate receptors in the rat brain. J Comp Neurol. 1992;318:329–354. doi: 10.1002/cne.903180309. [DOI] [PubMed] [Google Scholar]
- 26.Petralia RS, Wang YX, Mayat E, Wenthold RJ. Glutamate receptor subunit 2-selective antibody shows a differential distribution of calcium-impermeable AMPA receptors among populations of neurons. J Comp Neurol. 1997;385:456–476. doi: 10.1002/(sici)1096-9861(19970901)385:3<456::aid-cne9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Rubio M, Wenthold RJ. Glutamate receptors are selectively targeted to postsynaptic sites in neurons. Neuron. 1997;18:939–950. doi: 10.1016/s0896-6273(00)80333-5. [DOI] [PubMed] [Google Scholar]
- 28.Seeburg PH. The TINS/TiPS lecture. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci. 1993;16:359–365. doi: 10.1016/0166-2236(93)90093-2. [DOI] [PubMed] [Google Scholar]
- 29.Sheng M. PDZs and receptor/channel clustering: rounding up the latest suspects. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 30.Sheng M, Kim E. Ion channel associated proteins. Curr Opin Neurobiol. 1996;6:602–608. doi: 10.1016/s0959-4388(96)80091-2. [DOI] [PubMed] [Google Scholar]
- 31.Sheng M, Wyszynski M. Ion channel targeting in neurons. Bioessays. 1997;19:847–853. doi: 10.1002/bies.950191004. [DOI] [PubMed] [Google Scholar]
- 32.Song I, Kamboj S, Xia J, Dong H, Liao D, Huganir RL. Interaction of the N-ethylmaleimide sensitive factor with AMPA receptors. Neuron. 1998;21:393–400. doi: 10.1016/s0896-6273(00)80548-6. [DOI] [PubMed] [Google Scholar]
- 33.Spacek J, Harris KM. Three-dimensional organization of smooth endoplasmic reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. J Neurosci. 1997;17:190–203. doi: 10.1523/JNEUROSCI.17-01-00190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB. Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 35.Topinka JR, Bredt DS. N-terminal palmitoylation of PSD-95 regulates association with cell membranes and interaction with K+ channel Kv1.4. Neuron. 1998;20:125–134. doi: 10.1016/s0896-6273(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 36.Torres R, Firestein BL, Dong H, Staudinger J, Olson EN, Huganir RL, Bredt DS, Gale NW, Yancopoulos GD. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Wenthold RJ, Ottersen OP, Petralia RS. Endbulb synapses in the anteroventral cochlear nucleus express a specific subset of AMPA-type glutamate receptor subunits. J Neurosci. 1998;18:1148–1160. doi: 10.1523/JNEUROSCI.18-03-01148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]