Abstract
Accurate proteolytic processing of neuropeptide and peptide hormone precursors by members of the kexin/furin family of proteases is key to determining both the identities and activities of signaling peptides. Here we identify amontillado (amon), theDrosophila melanogaster homolog of the mammalian neuropeptide processing protease PC2, and show that in contrast to vertebrate PC2, amontillado expression undergoes extensive regulation in the nervous system during development.In situ hybridization reveals that expression ofamontillado is restricted to the final stages of embryogenesis when it is found in anterior sensory structures and in only 168 cells in the brain and ventral nerve cord. After larvae hatch from their egg shells, the sensory structures and most cells in the CNS turn off or substantially reduce amontillado expression, suggesting that amontillado plays a specific role late in embryogenesis. Larvae lacking the chromosomal region containingamontillado show no gross anatomical defects and respond to touch. However, such larvae show a greatly reduced frequency of a hatching behavior of wild-type Drosophila in which larvae swing their heads, scraping through the eggshell with their mouth hooks. Ubiquitous expression of amontillado can restore near wild-type levels of this behavior, whereas expression ofamontillado with an alanine substitution for the catalytic histidine cannot. These results suggest thatamontillado expression is regulated as part of a programmed modulation of neural signaling that controls hatching behavior by producing specific neuropeptides in particular neurons at an appropriate developmental time.
Keywords: Drosophila melanogaster, hatching behavior, neuropeptide, PC2, protease, development, nervous system
Scission after basic amino acid residues is required for the production of most neuropeptides; the active peptide signals are typically synthesized as parts of larger precursors that contain multiple copies of one neuropeptide or a collection of several different neuropeptides (Andrews et al., 1989). PC2 and PC1/3, two mammalian members of a family of Kex2-like proteases, are expressed only in neural and neuroendocrine cells (Smeekens et al., 1991) and are sorted along with neuropeptide precursors and prohormones to nascent secretory granules where proteolytic processing reactions begun in the trans-Golgi network (TGN) are completed (Itoh et al., 1996). Various co-expression, antisense, and biochemical experiments have demonstrated that these two proteases cleave neuropeptide precursors correctly after dibasic sitesin vivo and in vitro (Zhou and Lindberg, 1993;Friedman et al., 1994; Hoflehner et al., 1995; Johanning et al., 1996;Paquet et al., 1996; Rovere et al., 1996). Recently, PC2 knock-out mice have been shown to be hypoglycemic, slow-growing animals that are defective in processing proglucagon, prosomatostatin-14, and proinsulin (Furuta et al., 1997, 1998). A human patient deficient in PC1/3, with extreme childhood obesity and low insulin levels, has also been identified (Jackson et al., 1997). Thus, definitive proof has been obtained that these enzymes are responsible for processing critical peptide hormone precursors and at least one neuropeptide precursor.
Such proteolysis is not just required for production of active peptides, but also can regulate signal identity within the endocrine system. For example, the precursor proopiomelanocortin is processed to generate different sets of peptides in different parts of the pituitary (Mains and Eipper, 1979; Eipper and Mains, 1980). Exactly which peptides are produced is determined by whether PC1/3 or PC2, or both of these proteases, is expressed and therefore cleaves the precursor in a particular cell (Mains and Eipper, 1979; Eipper and Mains, 1980; Zhou et al., 1993; Rouille et al., 1994, 1995). Regulation of the expression of these proteases is a key control point for determining which signal is produced by a cell, and thus what response is produced on stimulation.
We sought to establish a simple system in which to study the regulatory roles of neuropeptide processing proteases, with the aim of being able to more easily identify relationships between particular processing events and particular functions of the nervous system than has yet been feasible in mammals. The genetically manipulable model organism Drosophila melanogaster was an attractive choice because it has a relatively simple nervous system yet can perform complicated behaviors such as flight and mating. The precursors for several neuropeptides such as FMRFamide (Nambu et al., 1988;Schneider and Taghert, 1988), Drosulfakinin (Nichols et al., 1988), Amnesiac (Feany and Quinn, 1995), and Eclosion hormone (Horodyski et al., 1993) have been identified in Drosophila and contain processing sites consisting of basic residues. Distinct peptides are produced from the Drosulfakinin and FMRFamide precursors in different cells (Nichols et al., 1995a,b; Nichols and Lim, 1996), pointing to the likelihood of neuropeptide production being regulated by Kex2-like proteases in Drosophila.
MATERIALS AND METHODS
Degenerate PCR reactions. Reaction mixtures (25 μl) contained 1 μg of total Drosophila genomic DNA, 8 μm each primer [(5′ to 3′) GGI/C GTI/C GGI/C GTN GCN TAT/C AA and CCC CAA/G CTN GCA/G CTA/G TAA/G/T AT], 0.25 UTaq DNA polymerase (Perkin-Elmer, Norwalk, CT), and 0.2 mm dNTP in 10 mm Tris HCl, pH 8.3, 50 mm KCl, 1.5 mmMgCl2, 0.01% gelatin. Thirty cycles were conducted of 1 min at 94°C, 3 min at 55°C, and 2 min at 72°C. After separation on 3% NuSieve, 1% SeaPlaque (FMC Bioproducts, Rockland, ME) low melting agarose gels, the PCR products were excised, purified by phenol extraction, blunt-ended using Klenow (NEB, Beverly, MA), cloned into BlueScript SK+(Stratagene, La Jolla, CA), and sequenced using the flanking T3 and T7 primers and standard dideoxy sequencing methods (Sambrook et al., 1989).
Cloning and sequencing of the cDNA. Filters corresponding to 400,000 plaques of an adult head Drosophila melanogaster phage library in lambda EXLX(+) (Palazzolo et al., 1989) were screened with DMH #5 fragment labeled with (32P)dCTP using random primers. Fourteen positive phages were identified and plaque-purified. Plasmid subclones of the phage insert were produced by the action of the crerecombinase on lox sites within the phage on infection of bacterial strain BM25.5. Deletions were constructed from the longest cDNA, #5–8, using Exo III nuclease and sequenced on both strands by the Stanford PAN facility using an ABI automated sequencer (Applied Biosystems, Foster City, CA).
RNA and quantitative DNA blots. Four-hour collections of eggs laid on feeding plates were obtained from large population cages of Canton S Drosophila melanogaster. These were either aged on standard Drosophila grape juice plates at 25°C for embryonic time points or seeded into medium-sized plastic containers with food and aged for larval and pupal stages (Ashburner, 1989). Larval stages were collected from the food at the midpoints of the appropriate times. Pupal stages were scraped from the walls. Collected samples were flash-frozen in liquid nitrogen and RNA was prepared, blotted, and hybridized as described (Sambrook et al., 1989). RNA blots were hybridized with radiolabeled DMH #5 fragment. Df(3R)Tl9QRX, Df(3R)ro80b, and Df(3R)roXB3 were obtained from K. Anderson (Memorial Sloan Kettering, New York, NY), Canton S from M. Krasnow (Stanford University, Stanford, CA). Southern blots of genomic DNA from Deletion Df(3R)Tl9QRX, Deletion Df(3R)ro80b, Df(3R)roXB3 and Canton S flies (Sambrook et al., 1989) were hybridized with radiolabeled fragments comprising the entire amontillado cDNA sequence and control fragments from branchless or trimmed cDNAs. Hybridization signals were quantified using a Molecular Dynamics (Sunnyvale, CA) Phosphorimager.
In situ hybridizations. Digoxigenin-labeled sense and antisense RNA probes were made using Boehringer Mannheim’s (Indianapolis, IN) RNA Labeling Kit by transcribing from the T3 and T7 promoters of plasmid pAP6, which contains a 2 kb piece of theamontillado cDNA from the ApaI to PstI in BlueScript (Stratagene). In situ hybridization was performed on stage 15–17 embryos as described previously (Kopczynski et al., 1996), with the modification inspired by an antibody staining protocol (Patel, 1994); after the first fixation 100 ul of embryos in 500 ul of methanol were sonicated at setting 1 on a Kontes (Vineland, NJ) small-tip sonicator for 3 sec. The embryos were then fixed a second time according to the Kopczynski (Kopczynski et al., 1996) protocol. Ten stage 17 embryos were analyzed in detail. To obtain first and second instar larvae for dissections, 2 hr egg cap collections from bottles of Canton S Drosophila were aged at 25°C for 34 hr and 46 hr, respectively. Nine first instar larvae were dissected in 1× PBS to free the gut and brain, still attached to the anterior in some larvae, from the body. Nine second instar larvae were pinned and cut open. These were then fixed for 45 min in 4% paraformaldehyde in PBS; six were incubated with the antisense RNA probe, and three were incubated with the sense RNA probe and then treated as in the standardin situ protocol. After development, second instar brains were freed from the body. Embryos and larval brains were mounted in 70% glycerol under an elevated coverslip (Patel, 1994) to allow rotation and analysis from all angles.
In situ salivary gland squashes. Dissected polytene salivary gland chromosomes from wandering third instar larvae were squashed and hybridized with a Biotin-16-dUTP (ENZO Biochemicals, Garden City, NY)-labeled EcoRI-EcoRV fragment from the amontillado cDNA. After incubation with a Vectastain avidin–biotinylated peroxidase complex (Vector Laboratories, Burlingame, CA), the location of the probe was visualized by a diaminobenzidine hydrogen peroxide reaction. Chromosomes were counterstained with Giemsa.
Deletion cross and observations of hatching. Virgin females were collected from Deletion Df(3R)Tl9QRX/TM3Sb 35uz-2 lacZ and were mated in bottles containing fresh yeast to males carrying Deletion Df(3R)ro80b/TM3Sb 35uz-2 lacZ for 3 d. Four-hour collections on grape juice agar plates were aged 14 hr at 25°C and then fixed and stained with antibodies against β-galactosidase, myosin, and horse radish peroxidase (HRP) (see below). Df Tl9QRX/Dfro80b embryos were identified by the absence of lacZ expression in the Ultrabithorax pattern. Four separate collections as above were aged 32 hr at 25°C and examined to determine the number of unhatched larvae; the total number of eggs in each ranged from 148 to 209. For videotaping of hatching, eggs were collected as above or from wild-type Canton S, aged 12–16 hr, and videotaped for 6 hr. Individual larvae (5–12) were lined up on an egg cap with a wick to a surrounding shallow water bath to preserve humidity. Eight Canton S larvae and at least 40 larvae from each of the crosses were analyzed. Videotaping was performed using a Panasonic VCR connected to a DAGE-MTI (Michigan City, IN) CCD-100 camera mounted on aZeiss dissecting microscope, with illumination from a Nikon (Garden City, NJ) MK-II fiber optic light source. Videotapes were viewed on a Sony SSM-930 black and white monitor, and the occurrence and duration of any movements were noted. To examine the behavior of larvae carrying two copies of the TM3Sb balancer chromosome, Canton S females were mated to Ki/TM3Sb males. From their progeny, virgin +/TM3Sb females and males were selected; collections and videotaping were performed as above. As expected, 27% of the laid eggs failed to hatch, namely the TM3Sb/TM3Sb larvae; these die because of homozygosity of the Sb mutation. Larvae from such a cross were videotaped as above.
Rescue of hatching. Transgenic rescue lines were created using a construct containing anEcoRI-EcoRV fragment of theamontillado cDNA subcloned intoEcoRI-HpaI cut pCaSper-hs to allow ubiquitous exogenous amontillado expression under the control of the heat shock promoter; a control line that would produce a nonfunctional protease was created by replacing the KpnI-SphI fragment in pCaSperhs:amon with one containing a mutation of the active site histidine to an alanine, accomplished via PCR with a mutagenic primer. Sequencing of the final plasmid on a PE Applied Biosystems (Foster City, CA) ABI Prism 310 Genetic Analyzer confirmed the construction; the His to Ala change was present at position 237, as well as an Asp to Glu change at position 232. Inserts of both of these constructs into line w1118 were mapped genetically according to standard protocols. All subsequent crosses to create the final strains used to test rescue were performed in parallel to assure identical genetic backgrounds. yw;D/TM3Sb Kr:GFP flies were generously provided before publication by D. Casso, F.-A. Ramirez-Weber, and T. B. Kornberg (University of California San Francisco). To test rescue, virgin female yw; P(w+)hs:amon; Df(3R)ro80b/TM3Sb Kr:GFP were crossed to yw; P(w+)hs:amon; Df(3R)Tl9QRX/TM3Sb Kr:GFP males as above. As a negative control, virgin femaleyw; P(w+)hs:amonH237A; Df(3R)ro80b/TM3Sb Kr:GFP were crossed to yw; P(w+)hs:amonH237A; Df(3R)Tl9QRX/TM3Sb Kr:GFP males. As a further control yw; Df(3R)ro80b/TM3Sb Kr:GFP flies were crossed to yw; Df(3R)Tl9QRX/TM3Sb Kr:GFP. Four-hour plate collections from the experimental and control crosses were aged 17 hr and heat-shocked by being placed in a 37°C room for 0.5 hr. Three hours later, larvae were examined using a Leica MZ12 fluorescent dissecting microscope under a GFP filter set. Those larvae that did not fluoresce in two spots within the mouth hooks were lined up on plates as described above and videotaped, starting at 4 hr after heat shock. After 4 hr of videotaping, the larvae were removed from their egg shells with bleach and double-checked under the microscope for a lack of fluorescence. The videotapes were replayed as above, and the number of swings observed for each nonfluorescing larva in a 3 hr period was counted, using 25 larvae for the lines expressing the H237A mutated amontillado control, 22 larvae in the control lacking any rescue construct, and 31 larvae expressing the wild-type amontillado. A 2 hr collection of wild-type Canton S embryos was aged 18 hr and then videotaped for 6 hr. The behavior of 20 of these Canton S embryos was analyzed for the 3 hr before their hatching. To test the consistency of counting, a randomly chosen videotape of six animals was rescored. There was a 1% difference in the mean number of swings determined in the two counts. Statistical analysis was conducted using a Student’s t test and a Bootstrap T Test (written by Arman Magbouleh of the Department of Linguistics), which produced nearly identical results.
Naming the gene. We named the geneamontillado, from the title of the Edgar Allen Poe story about a man who is walled-in while still alive (Poe, 1973).
Northern blot on transgenic hs:amon and hs:amonH237A. Two sets of 10 adult male flies were collected from the following three genotypes: Canton S,yw; P(w+)hs:amon; Df(3R)ro80b/TM3Sb Kr:GFP, yw; P(w+)hs:amonH237A; and Df(3R)ro80b/TM3Sb Kr:GFP. One set was heat-shocked by being placed in a 37°C water bath for 0.5 hr; the other vial was left at room temperature. After heat shock, the flies were left at room temperature to recover for 1 hr and then were flash-frozen in liquid nitrogen. RNA was prepared using TRIzol Reagent (Life Technolories, Rockville, MD). A Northern blot was prepared using 10 ug of RNA from each heat-shocked and unheat-shocked genotype and was hybridized with a (32P)dCTP-labeledEcoRI-EcoRV fragment of theamontillado cDNA (Sambrook et al., 1989).
Antibody stains. Embryos were fixed and stained as described (Patel, 1994). Rabbit antiserum against β-galactosidase (Cappell-ICN Pharmaceuticals, Aurora, OH) was used at 1:1500. Goat-anti HRP (Cappell) was used at 1:300. Biotinylated secondary antibodies were used at 1:300, followed by Vectastain avidin–horseradish peroxidase (Vector Laboratories, Burlingame, CA) histochemistry. Myosin antibody was a gift from Dr. Dan Kiehart (Duke University).
RESULTS
Identifying Kex2 homologs in Drosophila
To identify Kex2 homologs present in Drosophila melanogaster in a manner unbiased by differences in expression level, degenerate PCR was performed on genomic DNA. The primers that were used corresponded to two regions of almost absolute identity present in an alignment of yeast Kex2 with human furin and PC2. Three products of 220, 365, and 750 base pairs were seen in PCR reactions (data not shown). Sequence analysis demonstrated that each product contained a variably sized intron flanked by consensus splice sites. Two of the predicted translation products show considerable identity in the region between the primers to known mammalian Kex2 homologs (Fig.1A). DMH #5 displays 71% identity to human PC2. DMH #11 corresponds to theDrosophila furin homolog called Dfur2 (Roebroek et al., 1992). The predicted splice sites in both products are at the position known to be spliced in the human furin gene. The DMH #15 sequence was the most divergent, showing similarity to yeast Kex2. The DMH #15 PCR product hybridized on a developmental Northern blot to a 3.5 kb transcript expressed only during the pupal stages (data not shown) and may correspond to a novel member of the Kex2 family. Because of the role of mammalian PC2s in differential processing of neuropeptides and hormones (Benjannet et al., 1991; Smeekens et al., 1991; Thomas et al., 1991), we focused on this putative PC2 homolog.
Fig. 1.
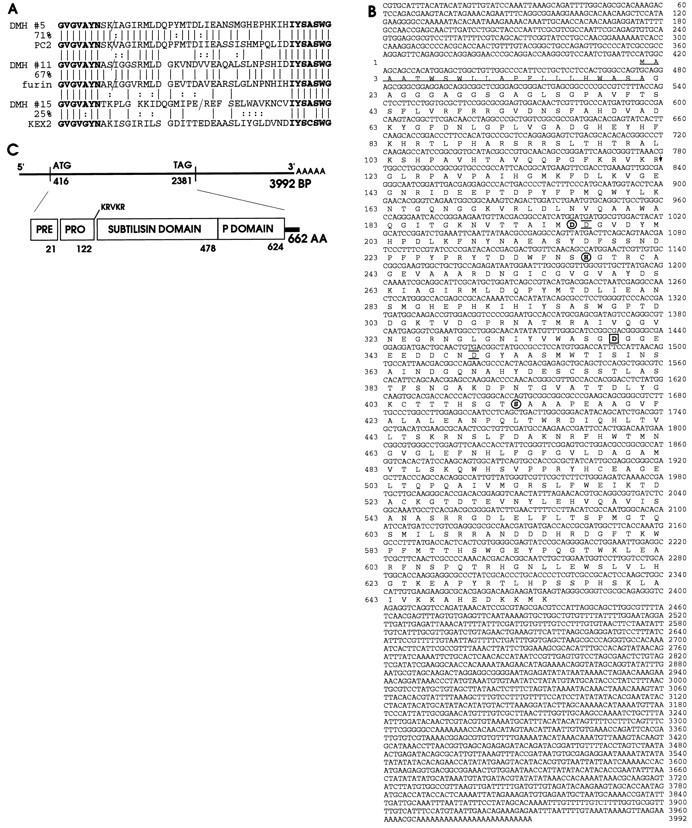
A, Drosophila melanogaster PCR products with homology to the Kex2 protease family. PCR reactions were conducted on genomic DNA fromDrosophila melanogaster. A translation of the DNA sequence of the three PCR products obtained, DMH #5,DMH #11, and DMH #15, is shown; below each is the Kex2 family member to which it has the greatest amino acid identity: PC2, furin, and Kex2, respectively. Theslashes indicate conceptual splice sites in theDrosophila products at consensus splice sequences and the known splice site in human furin and PC2. The amino acid sequences corresponding to the primers used are printed in bold; the percentage of identical amino acids found in the region between the primers for each Drosophila product and its closest homolog is also indicated. B, The sequence of theDrosophila PC2 cDNA and protein. DMH #5 was used to screen an adult Drosophila head library.A shows the complete nucleotide sequence of the longestamontillado cDNA obtained along with the amino acid sequence of its longest open reading frame. Catalytic triad residues are circled. The aspartate residue unique to PC2s at the position of the subtilisin oxyanion hole asparagine, which is predicted to stabilize the substrate oxyanion during catalysis, isboxed. The residues predicted by molecular modeling to underlie hPC2 specificity for dibasic residues areunderlined and overlined. The predicted signal peptide is underlined; the predicted autoproteolytic cleavage site is indicated by an arrow. These sequence data have been submitted to the DDBJ/EMBLJ/GenBank databases under accession number AF033117. C, A schematic summary indicates the positions of the nucleotides at the beginning and end of the open reading frame, the length of the cDNA and predicted protein, the domain structure of the predicted protein, and the amino acid at which each domain ends.
Drosophila contains a Kex2 homolog closely related to the human neuropeptide processing protease PC2
Because mammalian PC2 is highly expressed in the brain, we used the DMH #5 PCR product to screen an adult Drosophila head cDNA library (Palazzolo et al., 1989). Fourteen hybridizing plaques were identified from a total of 400,000 that were screened; all contained related cDNAs as assessed by restriction mapping with four different enzymes. Figure 1B,C shows the complete sequence and conceptual translation of the longest cDNA. This 4 kb transcript ends with a polyA tail that is preceded by a consensus polyadenylation signal and corresponds in length to the band seen on a Northern blot (Fig. 2). The open reading frame (ORF) begins with the first methionine 418 bp into the cDNA and is preceded by stops in all three reading frames. Sequences surrounding this methionine match only two of the nine nucleotides expected before a translational initiation codon from the Kozak rules yet maintain the optimal G following the ATG (Kozak, 1984), thereby falling into the 2–5% of Drosophila start codons containing a pyrimidine at −3 and a purine at +4 (Cavener, 1987). The protein predicted by the ORF shares the structure of the Kex2 family of proprotein-processing proteases: it contains a predicted signal sequence, a putative pro-domain, a region of homology to the subtilisin protease family containing the active site residues, and a region of homology unique to the Kex2 family called the P-domain (Gluschankof and Fuller, 1994) (Fig. 1C). It also exhibits features shared by and unique to all the PC2 homologs identified in other organisms. Most notable among these is the presence of an aspartic acid at the catalytically important position termed the oxyanion hole [Asn155 in subtilisin (Bryan et al., 1986)]. This position is occupied by an aspartic acid in all PC2s in place of the asparagine found in all other subtilisin and Kex2 family proteases. The ORF encoded by this cDNA bears much higher identity to PC2 than to any other Kex2 family member. Within the catalytic domain, this ORF is 75–81% identical to all PC2s but only 51–55% identical to all other mammalian Kex2-like proteases and 50–51% identical to the Drosophila melanogaster furins (% identity in subtilisin domain of amontillado and various family members: hPC2, 75; xPC2, 75; aPC2, 76; cePC2, 81; hfur, 55; hPC1, 53; hPACE4, 52; rPC7, 52; mPC4, 51; mPC6, 51; Dfur2, 51; Dfur1, 50; % identity between different domains of amontillado and hPC2: PrePro, 30; subtilisin, 75; P-domain, 54; C-tail, 31; overall, 66). The P-domain of this Drosophila protein also shows a degree of similarity to the mammalian PC2s (54% identity). Finally, like all known PC2 homologs, this predicted protein has a short C-terminal extension beyond the P-domain and no transmembrane domain. We have named this gene amontillado (abbreviatedamon; see Materials and Methods).
Fig. 2.
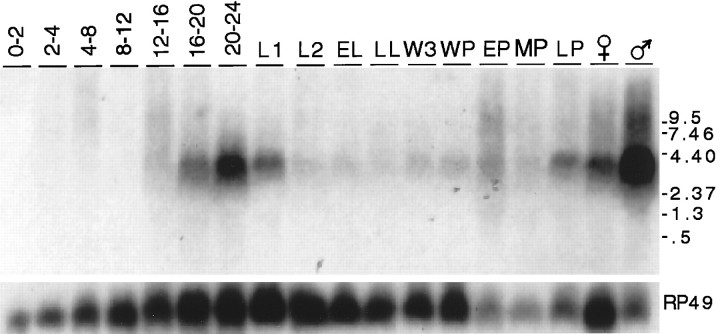
Temporal expression pattern ofamontillado RNA during development. The PCR product corresponding to amontillado was hybridized to a developmental Northern blot prepared with 10 μg of total RNA per lane. The lanes correspond to seven different time periods encompassing the first 24 hr of embryogenesis, first and second instar larvae (L1 and L2), early and late third instar larvae (EL and LL), wandering third instar larvae (W3), white prepupae (WP), early, mid, and late pupae (EP, MP, andLP) and female and male adult flies. The Northern blot was subsequently hybridized to RP49 as a loading control, which is known to drop in levels in adult male flies (Al-Atia et al., 1985). The higher expression in adult males as compared with females was not observed in a second independent collection from flies raised in small bottles rather than in larger cages.
Expression of amontillado peaks at the end of embryogenesis in a subset of the nervous system and gut and peaks again in adults
To determine the temporal expression pattern of the gene, a blot of total RNA from each developmental stage of the Drosophilalife cycle was probed with the amontillado cDNA. A single 4 kb transcript was first observed at ∼12–16 hr (stage 15, 16) and peaked at the end of embryogenesis (late stage 17) (Fig. 2). The transcript level dropped in the first larval stage (L1) and by the second larval stage (L2) reached a very low level of expression observed throughout the remainder of larval and most of pupal life. Expression increased in late pupae and peaked once again in adults. This same temporal profile was confirmed in a Northern blot in which RNA was isolated from an independent series of collections (data not shown).
We were intrigued by this late embryonic peak of amontilladoexpression, because it coincides with the developmental and physiological transition of embryo to larva. To determine the tissue distribution of amontillado RNA at this time, we performedin situ hybridizations with RNA probes on whole-mount embryos whose cuticle had been removed by sonication. In good correlation with the results from the Northern blot, expression was first detected ∼11.5 hr after egg laying during stage 15 of embryogenesis (Fig. 3A) in a segmentally repeated, bilaterally symmetrical pattern of cell clusters in the nerve cord. Cells within the brain and the subesophageal ganglion, near the pharynx, and in the lateral bipolar dendritic (lbd) sensory neuron of every segment, along with additional cells in each segment of the nerve cord, began to express amontilladoduring stage 16 (Fig. 3B).
Fig. 3.
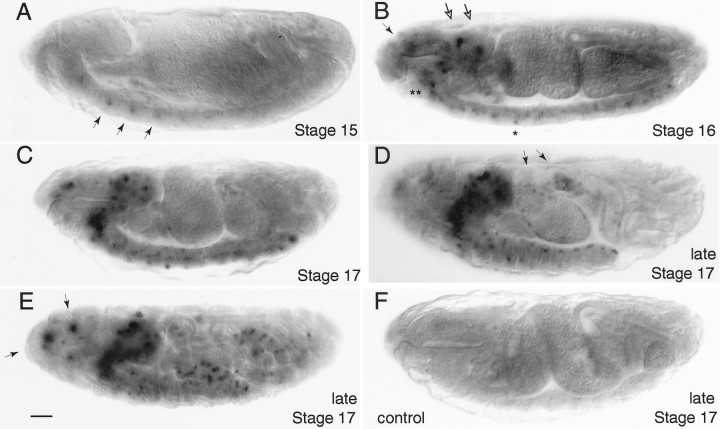
In situ hybridization ofamontillado to embryos. Antisense RNA probes corresponding to half of the amontillado cDNA were hybridized to Drosophila embryos. A, Signal was first observed at stage 15 in a segmentally repeated pattern in the nervous system (arrows). B, During stage 16, signal appears in the brain (hollow arrows) and near the pharynx (filled arrow), in the subesophageal ganglion (∗∗), and at unique positions in the nerve cord (one example shown with ∗). C, During early stage 17 the number of cells expressing in the brain and the intensity of all expressing cells increases. By late stage 17 (shown in Dand E, E being a later time point and different focal plane), staining in the nerve cord can be roughly distinguished at a dorsal, middle, and ventral level (D), and staining begins in the gut (arrow). Sensory structures at the anterior of the embryo also show staining (E, arrows).F, No hybridization was observed at any of these stages with a sense control probe (stage 17 shown). Scale bar, 50 μm.
By stage 17 the full complement of embryonic amontilladoexpression became apparent. In this stage, expression was seen in the CNS in bilaterally symmetrical clusters of one to five cells in the brain and nerve cord, representing 2% of the total cells in the nerve cord (Goodman and Doe, 1993). During stage 17, amontilladomRNA appeared in the endocrine ring gland and in epitracheal cells in every segment; one cell was located near the junction of the dorsal trunk and the transverse connective of the tracheal system (data not shown). In the peripheral nervous system, anterior sensory structures (Fig. 3E) and the stomatogastric frontal ganglion also displayed amontillado mRNA by late stage 17 (data not shown), and expression remained in the lbd neuron; two sensory cells in each segment, one on each side of the embryo at a ventrolateral position that we estimate corresponds to the campaniform sensilla, also expressed amontillado. During late stage 17, when the ventral nerve cord had retracted to 75% of egg length, a few regions of the gut began to show a punctate pattern of cells producingamontillado (Fig. 3D,E). amontilladoexpression was thus highly restricted and found only in small subsets of the nervous and endocrine systems and of the gut.
The expression of amontillado mRNA disappears from anterior sensory structures and decreases sharply in the brain after hatching
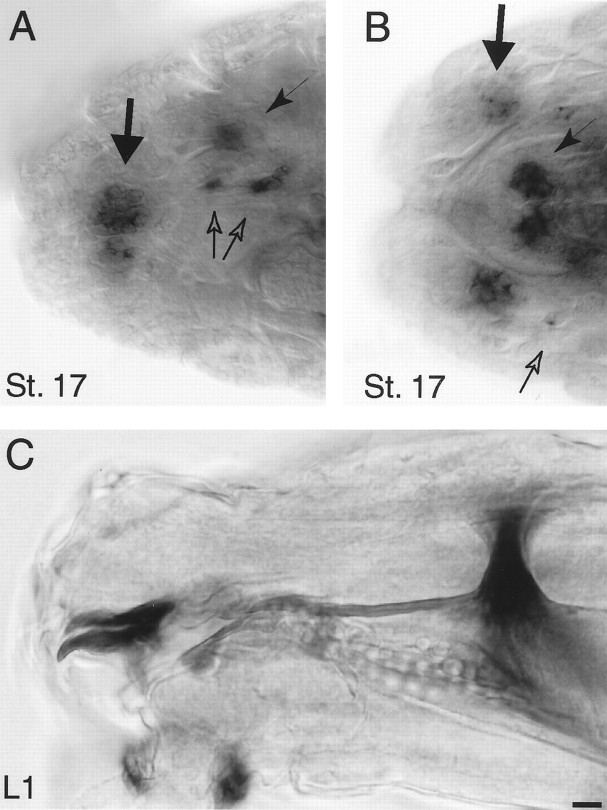
The late embryonic peak in amontillado mRNA levels observed in the RNA blot suggested that amontillado might be involved in certain functions needed only at the time of hatching. To determine whether this regulation of amontillado might reflect a particular role within the nervous system, we performedin situ hybridization on dissected mid-first and mid-second instar larval nervous systems. As described above,amontillado was found in sensory structures at the anterior of the developing larva during stage 17. Figure4A,B shows a close-up of the expression of amontillado during stage 17 in 12 cells in the larval touch-response organ, called the antennal-maxillary complex, as well as approximately seven cells in or around the epiphysis, a sensory organ associated with the pharynx. Two other cells near the esophagus also expressed amontillado. Once larvae reached mid-first instar, no expression of amontillado could be detected in these regions by in situ hybridization (Fig.4C).
Fig. 4.
amontillado RNA expression in anterior sensory structures during stage 17. A, A lateral view of the anterior of whole-mount stage 17 embryo hybridized to antisense amontillado RNA probe. B, A dorsal view of such a stage 17 embryo. C, A lateral view of a dissected first instar larvae similarly hybridized. Note that the expression in all structures seen in stage 17 is completely absent in L1: in the antennal maxillary complex (thick arrow), the epiphysis (thin arrow), and two other cells lying near the esophagus (hollow arrows). Expression was seen in the attached brain in the first instars examined as a positive control for the staining reactions. Scale bar, 10 μm.
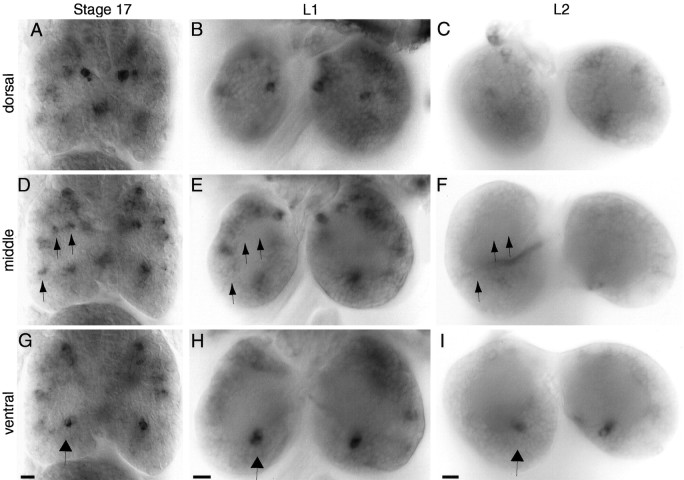
amontillado was expressed in ∼70 cells in the brain at late embryonic stage 17 and in 26 bilaterally symmetrical clusters at several focal planes throughout all parts of the brain (Fig.5A,D,G; summarized in Fig. 7). Only four cells in the middle protocerebrum of the brain maintained the expression of amontillado at a high level throughout all larval stages (Fig.5G,H,I,arrows). All of the other cells in the brain decreased or turned off amontillado expression by the middle of the first larval instar, only 15 hr later (Fig.5B,E,H; summarized in Fig. 7). At least 44 cells in the brain, 62% of the original expressors, turned off amontillado RNA expression completely. An average of 20 cells in the brain, ∼30% of the original number, showed medium to low level amontilladoexpression at this time. The location of these 20 cells was variable and not always bilaterally symmetrical. Some appeared to be in places unrelated to the earlier expression, implying a dynamic process of regulation in which some new cells were turning onamontillado at low levels and others were turning it off. Simultaneously, a very low level of amontillado expression was seen in larger clusters in the superior protocerebrum and midprotocerebrum that continued throughout larval development.amontillado was turned off in additional cells during the second larval instar. By mid-second instar, aside from the four continuously, strongly expressing mid-protocerebral cells, an average of only 10 cells in the brain showed even a low level ofamontillado expression (Fig.5C,F,I). The precise location of these low-level expressing cells varied from brain to brain.
Fig. 5.
In situ hybridizations ofamontillado to stage 17, first and second instar larval brains. amontillado antisense RNA probes were hybridized to whole-mount stage 17 embryos (A, D,G) and to dissected first (B,E, H) and second (C, F, I) larval instar brains. Dorsal (A–C), midbrain (D–F), and ventral (G–I) focal planes are shown for each of the three different stages from a brain selected for having the greatest number of hybridizing cells of the brains examined from that stage. Note that most of the cells exhibit decreased or no amontillado expression by first instar, and almost all expression is absent by the second larval instar. In D, E, and F,arrows mark some of the positions of cells that undergo such regulation. Arrows in G throughI indicate the two midprotocerebral cells present in each brain hemisphere with consistently strong expression during these three stages. Scale bars, 10 μm.
Fig. 7.
Schematic of results of amontillado in situ hybridization to the CNS of stage 17, L1, and L2. The results summarize amontilladoin situhybridization to the CNSs from at least six different larvae in stage 17, L1 and L2. The outlines of the nervous systems are traced from photographs. Scale bar, 25 μm. Each markindicates that expression was observed in a cell at that location in at least one-half of the examined animals. In stage 17, differentcolors and shapes are used to indicate the different focal planes from dorsal to ventral as shown by thekey. Where the identity of cells expressing at later stages could be reasonably assigned based on their similar location, the colored shape was maintained throughout subsequent stages. Lighter colors indicate a lower level of expression. During first and second instar larval stages the location of cells expressing in the brain was quite variable and not always bilaterally symmetric.
The expression of amontillado RNA in the ventral nerve cord also decreases after hatching
In the ventral nerve cord, 98 cells expressedamontillado at stage 17; these were assigned to five groups based on their positions. (1) At or near the ventral surface of the nerve cord lay two pairs of cells in segments T2-A4 (Fig.6C, arrows; Fig.7, blue squares). (2) In the middle level of the nerve cord there was a cluster of three cells at the lateral edge of each of the thoracic nerve cord segments, in a similar position to the FMRFamide-expressing cells designated T1–3v (Schneider et al., 1991) (Fig. 6B,filled arrows; Fig. 7, green circles). (3) In the same focal plane in segments A7–8 there were clusters of two to three cells along the midline and in segments A8 bilaterally flanking the midline (Fig. 6B, hollow arrows; Fig. 7,green circles). (4) At the dorsal surface of the nerve cord a chain of one small cell per segment stretched from T1 to A7 (Fig.6A, hollow arrows; Fig. 7, red triangles). In A1-A7 the chain increased in density to three cells per segment, with the two additional cells in a different focal plane. (5) Two strongly expressing cells surrounded by zero to three others were positioned at the top of the nerve cord (Fig.6A, top arrow; Fig. 7, red triangles).
Fig. 6.
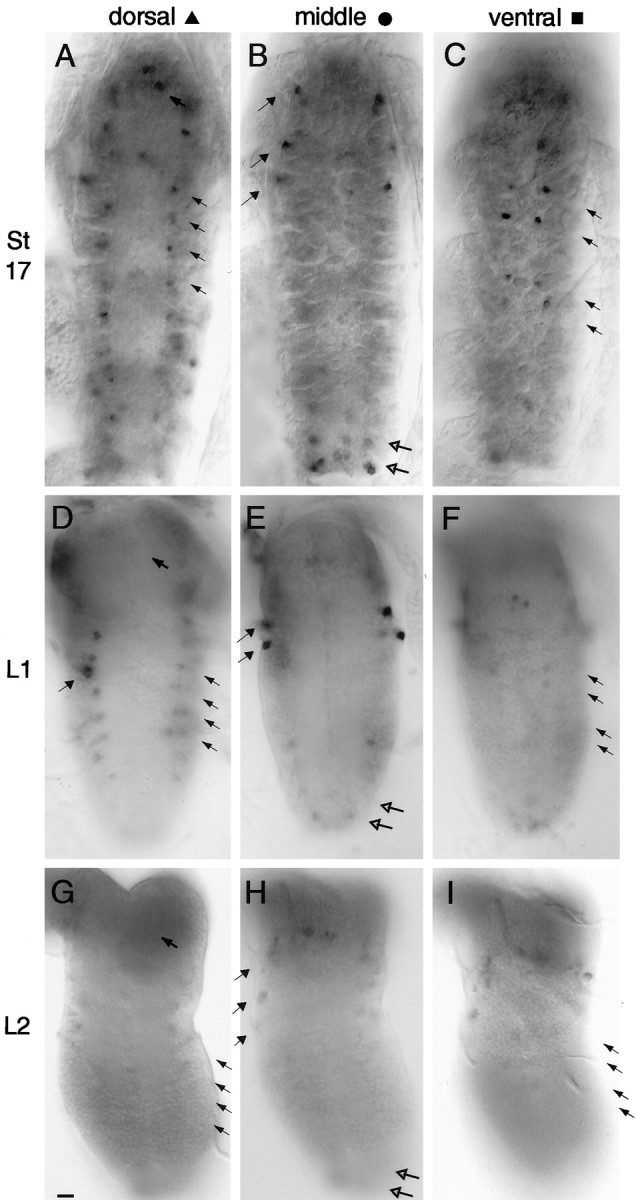
In situ hybridizations ofamontillado to stage 17, first and second instar larval ventral nerve cords. amontillado antisense RNA probes were hybridized to whole-mount stage 17 embryos (A–C) and to dissected L1 (D–F) and L2 (G–I) ventral nerve cords. Dorsal (A, D,G), middle (B, E,H), and ventral (C,F, I) focal planes are shown for each of the three stages. Compare A withD and G, and compare Cwith F and I where arrowsindicate some of the cells that show strong amontilladoexpression in stage 17 but severely reduced or abolished expression by L1. In B, E, and H, thesolid arrows indicate the cells in which expression remains strong in L1 but drops in L2. (The third group of cells indicated with a solid arrow in B is in a more dorsal focal plane in the L1 larva, so is seen in Drather than E.) Hollow arrows indicate the cells in which expression has already dropped by L1. The two dark anterior circles in E, F,H, and I are the dorsohaemal appendages and do not represent hybridizing cells. Scale bar, 10 μm.
By the middle of the first larval instar only 12 cells at the lateral edge of the thoracic segments of the nerve cord still maintained a level of amontillado RNA comparable to that in stage 17 (Fig. 6B,E); in the remaining cellsamontillado had been dramatically downregulated. The cells at the ventral surface had all but eliminated amontilladoexpression (Fig. 6C,F). The long chain of dorsally positioned cells had strongly reduced the level of their expression. The clusters of cells at the anterior of the nerve cord and in A7-A8 had either eliminated or substantially downregulated the level of amontillado RNA (Fig.6B,E; summarized in Fig. 7). By the middle of the second larval instar only faint amontilladoexpression remained in a few cells in the nerve cord: the 12 cells in the thoracic segments and the two cells at the anterior of the nerve cord (Figs. 6G–I, 7).
Animals lacking the region containing amontilladoappear normal but fail to hatch
We localized the amontillado gene to chromosomal bands 97D1–2 by in situ hybridization of the cDNA to salivary gland chromosomes (Fig.8A). To confirm this map position we examined the hybridization of amontillado to deficiencies in this region (Fig. 8D). Hybridization was absent on chromosomes carrying either deletion DfTl9QRX, which extends from 97B to a break-point in Toll at 97D1–2, or Dfro80b, which extends from 97D1 to 97D15 (Fig. 8B,C).amontillado must therefore map in the overlap of these two deficiencies in the 97D1–2 region (Fig. 8D). Quantitative analysis of DNA blots confirmed that DNA that hybridizes to the entire amontillado cDNA was eliminated by both deletions (data not shown).
Fig. 8.
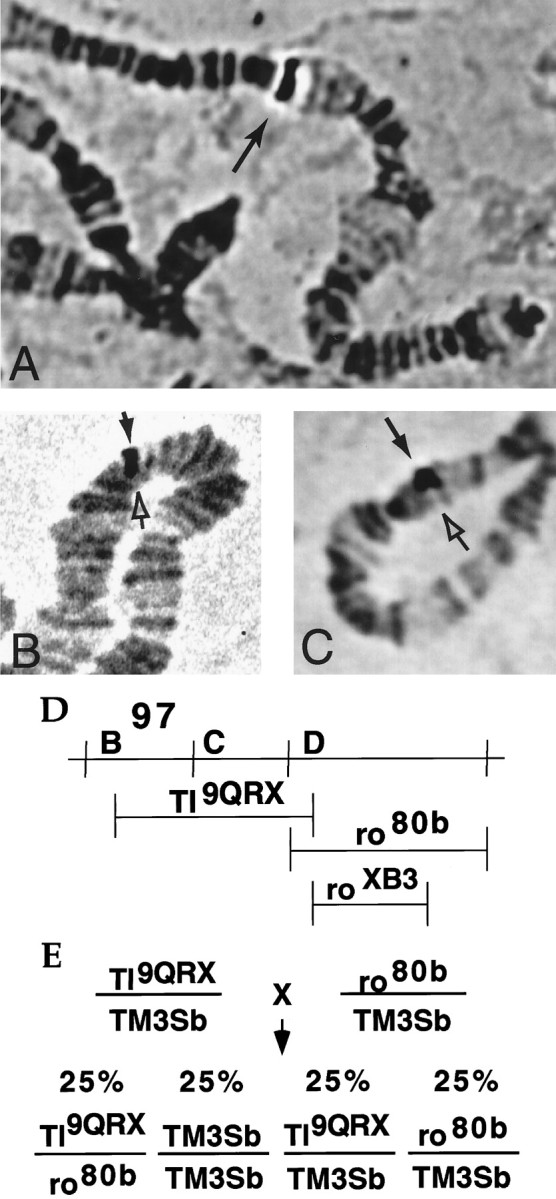
Elimination of the chromosomal region containingamontillado results in a hatching defect. Polytene salivary gland chromosomes obtained from wandering third instar larvae were squashed and hybridized with an amontillado probe. Glands were taken from wild-type Oregon R larvae (A) or from larvae carrying Deletion Df(3R)Tl9QRX(B) or Deletion Df(3R)ro80b (C) over a balancer chromosome. Hybridization was seen on the right arm of the third chromosome in region 97D (filled arrow). Hybridization was absent in the half of the polytene corresponding to the deletion chromosome (hollow arrow), indicating that both deficiencies remove the amontillado gene and their overlap at 97D1–2 contains the amontilladotranscription unit. D, The region from the right arm of the third chromosome from band 97B to 97E is depicted schematically.E, The extent of the genomic region eliminated by Deletion Df(3R) Tl9QRXand Deletion Df(3R) ro80b is indicated by horizontal lines. A cross of two lines that each carry a separate deficiency over a balancer is diagrammed. The expected Mendelian frequencies of the genotypes of the progeny are shown.
As a first step in determining the phenotypic effects of the absence ofamontillado we examined a plate collection of a cross of DfTl9QRX/TM3Sb to Dfro80b/TM3Sb. Because these two overlapping deficiencies both lack the 97D1–2 region, embryos heterozygous for both deficiencies (DfTl9QRX/Dfro80b) will completely lackamontillado. Because TM3Sb homozygotes die before hatching, 50% of the eggs laid from such a collection would hatch (Fig.8E) if the absence of the amontilladoregion caused embryonic lethality, whereas 75% would hatch if it did not cause embryonic lethality. In four separate collections, a total of 55 ± 1.5% (SE) of the eggs failed to hatch and 45 ± 1.5% did hatch. As a control we used different deficiency, DfroXB3 (97D1–2 to 97D9), which breaks in Toll and removes much of the region deleted byro80b yet does not removeamontillado as assessed by quantitative Southern blots (data not shown). Df Tl9QRX/DfroXB3 embryos did hatch. In a cross between DfroXB3/TM3Sb and Df(3R) Tl9QRX/TM3Sbonly 26% of the eggs, presumably the TM3Sbhomozygotes, failed to hatch. We conclude that the absence of the 97D1–2 region containing amontillado caused the embryos to die at some time during embryogenesis.
To determine more precisely when this defect occurred and what tissues might be affected, we fixed and microscopically examined DfTl9QRX/Dfro80b embryos. Embryos that lacked the amontillado region developed apparently normally to stage 17, the final stage of embryogenesis. Staining with an anti-myosin or an anti-HRP antibody revealed no obvious morphological defects, either in the gut, nervous system, or musculature (data not shown). Because embryonic development through the latest stage appeared to occur normally yet the larvae failed to hatch, we hypothesized that the absence of amontillado caused a defect in hatching behavior.
The hatching behavior of wild-type Drosophila
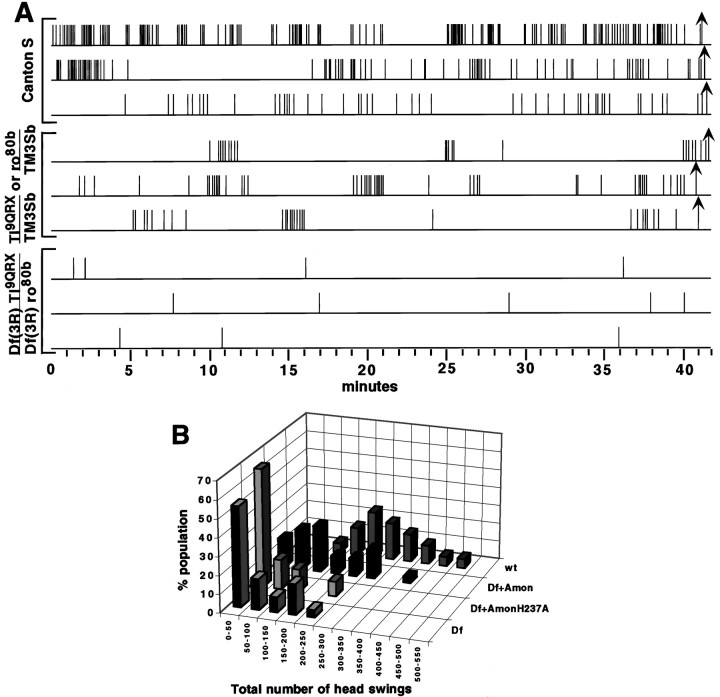
We were unable to find a description of the hatching behavior ofDrosophila melanogaster larvae in the literature; at a later time we discovered a report generally but not quantitatively describing the movements of dechorionated larvae (Kaliss, 1939). We therefore videotaped the normal hatching process in eight wild-type Canton S larvae starting at 16 hr after egg laying to facilitate observation and quantitation of the process. For 4.5–5 hr before hatching at room temperature, wild-type larvae scraped their mouth hooks against the vitelline membrane and chorion, in a lateral semi-circular arc ranging from 20 to 120°; the majority of arcs encompassed 45–90°. These arcs mainly traced along the so-called “hatching region,” an apposition of two discontinuous halves of endochorion where the integrity of the eggshell is maintained before hatching only by a fragile inner endochorion and a layer of exochorion (Margaritis, 1985). Depending on the individual larva, from 1 to 20% of swings observed consisted of arcs scraping vertically. Sequences of all head motions were very frequently episodic, with bursts of reiterated head swinging consisting of one swing every 1–30 sec, lasting from 40 sec to 7 min. These bursts were followed by periods with no concerted motions that ranged from 31 sec to 17 min (Fig.9A). Single head swings were also observed. Once a larva had created enough of a hole to get its head out, continual strong waves of peristalsis in the body wall musculature propelled the larva forward and out of the egg shell, usually between 22 and 24 hr after egg laying (AEL).
Fig. 9.
Evidence that amontillado is required for a hatching behavior: larval head swinging.A, The absence of the amontillado region results in a significant reduction in the frequency of head swinging. Stage 17 embryos were videotaped, and the tapes were replayed for analysis. Each head swing is indicated by a vertical line. Hatching is indicated by an arrow. Thehorizontal axis shows increasing time in minutes. The behavioral profiles of three separate individuals from each class are shown, chosen to represent the range of profiles observed. Thetop three graphs are from wild-type Canton S larvae. Themiddle and bottom three graphs are embryos from the cross of Df Tl9QRXand Df ro80b. The middle set corresponds to embryos that hatched, namely DfTl9QRX/TM3Sb or Dfro80b/TM3Sb. The bottom set corresponds to motile nonhatching larvae carrying one copy of each of the two overlapping deletions Df(3R)Tl9QRX/Df(3R)ro80b.B, Expression of wild-type, but not of mutant,amontillado rescues the head-swinging defect. Shown is the distribution of the frequency of swinging behavior in wild-type and mutant embryos with and without amontillado. The number of swings observed within a 3 hr period was counted for 22 heat-shocked Df(3R)Tl9QRX/Df(3R)ro80blarvae, 31 heat-shocked Df(3R)Tl9QRX/Df(3R)ro80blarvae carrying the hs:amon transgene, and 25 heat-shocked Df(3R)Tl9QRX/Df(3R)ro80blarvae carrying the hs:amonH237Atransgene. Twenty wild-type Canton S larvae were analyzed for 3 hr before hatching. The percentage of the population of each strain whose frequency of swinging fell into each of the bins indicated on thex-axis was plotted for each genotype. The mean (±SE) for each of the four different classes, from front to back, was 75 (± 16), 58 (± 13), 147 (± 17), and 275 (± 21).
Mutants lacking the amontillado region fail to perform normal hatching behavior
To determine what effect the absence of the amontilladochromosomal region had on this behavior, we videotaped larvae from a cross of DfTl9QRX/TM3Sb35uz-2lacZ and Dfro80b/TM3Sb35uz-2lacZ within their eggshells during the period of 18–24 hr AEL. We then aged the plates to 36 hr AEL and determined which of the videotaped larvae eventually hatched. The offspring of this cross fell into three classes: one-half hatched, one-fourth did not hatch and showed no movement, and one-fourth did not hatch but did show a small amount of head-swinging behavior. We videotaped a control cross in which the only unhatched embryos carried two copies of the TM3Sb balancer and found that this genotype resulted in completely immobile larvae (see Materials and Methods). We therefore concluded that the nonmotile, nonhatching one-fourth of the embryos from our experimental cross corresponded to the TM3Sb/TM3Sb genotype. Hatching larvae, the DfTl9QRX/TM3Sb and Dfro80b/TM3Sbgenotypes, always showed robust head-swinging before hatching, although the reiterated cycles of swinging were shorter and the pauses between them were longer than in Canton S (Fig. 9A). This may have been caused by strain variations or a dosage effect from the absence of one copy of the 97D1–2 region. By inference we assigned the nonhatching larvae that showed some head swinging to the genotype DfTl9QRX/Dfro80b. We confirmed this conclusion in later experiments using a GFP-labeled balancer (kindly provided before publication by D. Casso, F.-A. Ramirez-Weber, and T. B. Kornberg), which allowed us to directly identify the Df Tl9QRX/Dfro80b larvae (see below). Many of the swings observed by this class of larvae traced arcs of <20°, although an occasional 120° swing was seen. Significantly, reiterated cycles of head swinging were almost never observed, and the frequency of head swinging was reduced by 97%. These DfTl9QRX/Dfro80b larvae we had observed that lack the genomic region containing amontillado could perform the normal motor program of a head swing but did so much less frequently. On mechanical removal from the eggshell at 36 hr AEL, these larvae appeared normal in their morphology and responded to a touch on the side by twitching their bodies and retracting their heads, just as wild-type larvae did, although some mutants were slower to respond. Although the larvae lacking the amontillado chromosomal region displayed much reduced hatching behavior, many aspects of their CNS must be functional, because they were capable of performing coordinated motion and retained the ability to sense and react to an external stimulus.
Expression of a wild-type form of amontillado can rescue the defect in hatching behavior
The Df Tl9QRX/Dfro80b deficiency heterozygote larvae lack the 97D1–2 chromosomal region that has been shown in a saturation ethylmethane sulfonate screen to contain three lethal complementation groups (A.R. Kidd, D. Tolla, and M. Bender, personal communication). To determine whether the greatly reduced hatching behavior seen in these larvae was caused by the absence of theamontillado gene, we tested whether reintroducingamontillado would rescue the defect. We created transgenic lines that after induction by heat shock ubiquitously express either the wild-type Amontillado (hs:amon) or a mutant Amontillado (hs:amonH237A) whose predicted active site histidine had been changed to an alanine. Such a substitution in the related serine protease subtilisin causes a 106 decrease in catalytic activity (Carter and Wells, 1988). We also used a third chromosome TM3Sb Kr:GFP balancer (D. Casso, F.-A. Ramirez-Weber, and T. B. Kornberg, unpublished observations) to allow us to directly identify by the absence of fluorescence the living larvae lacking the balancer that thus carried one copy of each deletion. We compared the hatching behavior of heat-shocked larvae lacking the amontilladoregion that expressed no transgene, expressed the catalytic mutant form of Amontillado (hs:amonH237A), or expressed wild-type Amontillado (hs:amon). Deficiency heterozygote embryos lacking the 97D1–2 region showed a distribution of behavior in which the great majority of the larvae swung their heads at a very low frequency, 0–50 swings during a 3 hr period, although a small percentage of the larvae performed up to 219 swings during the same period (Fig. 9B). Expression of the mutant protein, AmontilladoH237A, which should have no proteolytic activity, did not rescue the behavioral defects of larvae lacking the 97D1–2 region (Fig. 9B). The behavioral profile observed was visually and statistically indistinguishable from that seen in the absence of expressing theamonH237A transgene (t test, p = 0.38 for a difference in the mean frequency of head swings). However, ubiquitous expression of wild-type amontillado in deficiency heterozygote larvae resulted in a statistically significant increase in the frequency of swinging (t test, p = 0.0026) (Fig.9B). The percentage of larvae swinging at the lowest level dropped from 55 to 16% (Fig. 9B), and some larvae exhibited up to 374 head swings during the observation period. Ubiquitous expression of transgenic amontillado under heat-shock promoter control did not completely rescue all larvae lacking 97D1–2 to wild-type levels of behavior. Canton S embryos showed a distribution of swinging that ranged from 114 to 485 swings with a mean of 275 ± 21 (SE) (Fig. 9B).
There are two obvious explanations for the difference in the ability of the mutated and wild-type forms of the transgenicamontillado to rescue the hatching defect seen in absence of the amontillado region. Amontillado function could absolutely require the predicted active site histidine, or theamonH237A transgene could have inserted into a region of the chromosome that prevented its normal expression after heat shock. To distinguish between these possibilities, we carried out a Northern blot on the hs:amonand the hs:amonH237A transgenic lines; both showed equal levels of induction of the transgenic copy ofamontillado mRNA on heat shock (data not shown). We therefore conclude that expression of wild-type amontilladocan rescue the head-swinging behavioral defect caused by the removal of the 97D1–2 chromosomal region containing amontillado.
DISCUSSION
amontillado is predicted to encode a PC2-like protease
We have identified a Drosophila melanogaster gene,amontillado, that is predicted to encode a member of the Kex2 family of proteases most similar to the neuropeptide processing protease PC2. Amontillado shares 75% sequence identity with human PC2 in the catalytic domain, including all three catalytic triad residues, the diagnostic oxyanion hole aspartate, and the two residues predicted by molecular modeling to underlie PC2’s specificity for dibasic sites (Lipkind et al., 1995). Amontillado has the same domain structure as all known PC2s and maintains a 66% identity to human PC2 over the whole protein. In addition, the expression of amontillado,like that of all other previously identified PC2s, is restricted to neural and neuroendocrine cells. Finally, we have shown that the function of Amontillado depends on the presence of one of three predicted catalytic residues common to serine proteases (Carter and Wells, 1988). Therefore, although the enzymatic activity of theamontillado gene product remains to be detected directly, we conclude that Amontillado is most likely an ortholog of mammalian PC2, functioning in proteolytic processing of neuropeptides and prohormones.
amontillado is dynamically expressed in a small set of neurons
In contrast to vertebrate PC2s, which are expressed broadly throughout the brain at relatively constant levels (Schafer et al., 1993, Zheng et al., 1994, Pu et al., 1995), amontillado is expressed in only ∼2% of the embryonic nervous system, and its expression level changes strikingly during the Drosophilalife cycle. From the peak of expression in 168 cells in the brain and ventral nerve cord during the final stage of embryogenesis,amontillado mRNA levels fall precipitously after hatching, disappearing or sharply diminishing in >85% of previously expressing neurons. Our data also define multiple populations of neurons that display distinct temporal patterns of amontilladoexpression: those that turn off after stage 17, those that turn off after L1, and those that express from stage 17 through L2. Each of these groups can be subdivided further into classes in which expression either decreases during these times or does not. The expression profile of amontillado contrasts with that of the previously identified Drosophila Kex2 family member Dfur1, which is found in a much larger fraction of the embryonic brain and nerve cord and is not regulated extensively (Roebroek et al., 1993). To our knowledge, we provide the first description of such dramatic developmental regulation within the nervous system of the expression of any Kex2 family member, raising the possibility that extensive temporal changes in neuropeptide production could be occurring.
Hatching behavior and a proposed role for neuropeptides
The largest population of amontillado-expressing cells in the nervous system is the one in which amontillado mRNA expression appears during the latest stages of embryogenesis and vanishes by the first larval instar. Drosophila development requires 16 hr at 25°C for the unicellular egg to undergo the cell divisions, movements, and fate determinations that produce the final structure of a stage 16 embryo. During the last 7 hr of embryogenesis, the embryo undergoes the transition to a free-living creature. Movements of the gut and digestion of the yolk start during this time (Skaer, 1993). Motor neurons fire, causing peristaltic contractions in the body wall musculature (Broadie and Bate, 1993). Yet these neural activities continue throughout larval life. The one process that begins and ends during the latest stage of embryogenesis is hatching, and in particular the behavioral pattern we describe in this work. The larva swings its head reiteratively through a semicircular arc, using its mouth hooks to tear apart the chorion in front of it, freeing it from entombment within the egg shell.
Insects also free themselves from the external relic of a previous developmental stage during the molts from one larval stage to the next and when adults escape from their pupal cases. Motor programs distinct from those observed in hatching are used in each of these cases, but each shares the characteristic of consisting of a reiterated set of stereotyped movements: a rhythmic dimpling of the larval body wall of the silkmoth Manduca sexta during pre-ecdysis and peristaltic contractions during ecdysis (Copenhaver and Truman, 1986;Miles and Weeks, 1991), and head expansion, head thrusting, and abdominal contractions by adult Drosophila during eclosion (McNabb et al., 1997). In both of these organisms, molting and eclosion behavior are coordinated and controlled by the release of a hierarchy of neuropeptides (Ewer et al., 1994; Zitnan et al., 1996; Ewer et al., 1997; Gammie and Truman, 1997). Given these observations and the fact that neuropeptides modulate many other behavioral motor programs such as eating (de Bono and Bargmann, 1998; Sakurai et al., 1998), pain response (Konig et al., 1996; Zimmer et al., 1998), and egg laying inAplysia (Bernheim and Mayeri, 1995), we propose that hatching behavior is regulated by neuropeptides.
amontillado is required for hatching behavior
Our data show that when the region at 97D1–2 that containsamontillado has been eliminated, the frequency of this hatching behavior is reduced by >97% compared with the wild-type, although these larvae can perform head swings and respond to touch. Our evidence strongly suggests that this defect is attributable to the loss of amontillado. The absence of this 97D1–2 region removesToll, the only other characterized gene in this region; however, embryos lacking zygotic Toll activity hatch normally (Gerttula et al., 1988). A saturation screen over the 97D1–2 region identified two other lethal complementation groups (A. R. Kidd, D. Tolla, and M. Bender, personal communication), implying that only three essential genes are removed in these experiments. Most importantly, ubiquitous expression of wild-type but not a catalytic mutant form of Amontillado increased the frequency of this motor behavior to wild-type levels in many larvae, strongly supporting the conclusion that the decrease in swinging behavior is caused by the absence of a proteolytically active Amontillado.
The inability of hs:amon to restore this behavior to wild-type levels in all larvae could be attributable to the artificial and ubiquitous nature of heat shock-dependent expression of the gene or to a partial requirement for another gene (or genes) within the deficiency for hatching behavior. We are currently attempting to isolate mutations that only affect amontillado, but have screened 8250 random transposon insertions without finding one located in the 5′ end of the amontillado gene (data not shown).
Control of the timing of hatching behavior
The location and timing of amontillado expression suggest that amontillado may be part of a program that is not only required for the hatching behavior to occur but may also be crucial for determing when the behavior occurs. Hatching behavior appears to be produced in response to sensing an anterior barrier, because it ceases abruptly just when the larval head breaks through the eggshell. As a free first-instar larva, it responds differently to an anterior barrier, by backing up and turning. We observeamontillado mRNA in sensory structures at the anterior of the larva only during the time just preceding and during hatching. Possibly these anterior structures sense the continued presence of the encircling egg shell, and because of the action of Amontillado can release neuropeptides that trigger mouth-hook scraping behavior only during this developmental stage. We suggest that the actual behavior is controlled by neuropeptides produced by amontillado during its peak of expression in a circuit of cells within the subesophageal ganglion, brain, and ventral nerve cord.
There are no known Drosophila neuropeptide precursors whose temporal profile of expression in the brain or nerve cord peaks during stage 17. Thus, there are no obvious candidates for Amontillado-dependent peptides that could trigger hatching behavior. The Drosophila neuropeptides and neuropeptide precursors known to be expressed during stage 17, FMRFamide, Drosulfakinin, and Dromyosuppressin, are found in a handful of cells and display constant or increased production during later stages (McCormick and Nichols, 1993; Nichols et al., 1995a,b; Nichols and Lim, 1996). However, it is possible that the hypothesized neuropeptide precursor substrate of Amontillado in late embryogenesis does not vary in its level, but rather that Amontillado appearance and disappearance in some neural cells, for example the subesophageal ganglion cells where the FMRFamide precursor is found, results in a temporally restricted pattern of differential processing to release a specific set of peptide products that function during hatching. In the adult rat, the presence of PC2 in the neurointermediate lobe of the anterior pituitary appears to be responsible for additional processing of the proopiomelanocortin precursor-derived peptides adrenocorticotrophic hormone and β-lipotrophic hormone to α-melanocyte stimulating hormone and β-endorphin, respectively. The absence of PC2 in the anterior lobe results in the release of a different profile of peptides from the anterior and the intermediate pituitary lobes (Mains and Eipper, 1979; Benjannet et al., 1991; Seidah et al., 1991; Thomas et al., 1991). Differential spatial processing of a neuropeptide precursor has also been demonstrated in two cases in theDrosophila CNS (Nichols et al., 1995a,b; Nichols and Lim, 1996). Amontillado could play a role in a novel kind of differential processing, namely a genetically programmed temporal variation to produce distinct neuropeptides at a crucial developmental time. Such a mechanism may provide a way for the same stimulus to elicit different behavioral outputs at different developmental times without necessitating alterations in neural connectivity.
Footnotes
This research was supported by National Institutes of Health Grant GM39697 to R.S.F. D.S. was supported in part by National Institutes of Health training Grant 2T32GM07599. We thank M. A. Krasnow and members of his laboratory, particularly J. Jarecki, for technical guidance, encouragement, and stimulating scientific discussions. We thank A. Maghbouleh and the Stanford Statistics Department Consulting Service for help with statistical analysis. We thank G. Beitel, S. Dietrich, K. Guillemin, D. Micklem, Y. Nakajima, and members of the Fuller and Krasnow laboratories for comments on this manuscript. We thank M. Palazzolo for the use of theDrosophila head cDNA library, D. Kiehart for the use of a Drosophila myosin antibody, and D. Casso, F.-A. Ramirez-Weber, and T. B. Kornberg for use of the D/TM3SbKrGFP flies. We thank A. R. Kidd, D. Tolla, and M. Bender and D. Casso, F.-A. Ramirez-Weber and T. B. Kornberg for communication of results before publication.
Correspondence should be addressed to Dr. Robert S. Fuller at his present address: Department of Biological Chemistry, 5413 Medical Science I, 1301 Catherine Road, University of Michigan Medical School, Ann Arbor, MI, 48109-0606.
Dr. Siekhaus’s present address: Department of Molecular and Cell Biology, Barker Hall, University of California, Berkeley, CA 94720.
REFERENCES
- 1.Al-Atia GR, Fruscoloni P, Jacobs-Lorena M. Translational regulation of mRNAs for ribosomal proteins during early Drosophila development. Biochemistry. 1985;24:5798–5803. doi: 10.1021/bi00342a017. [DOI] [PubMed] [Google Scholar]
- 2.Andrews PC, Brayton KA, Dixon JE. Post-translational proteolytic processing of precursors to regulatory peptides. Experientia [Suppl] 1989;56:192–209. doi: 10.1007/BF01945356. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner M. Drosophila: a laboratory handbook. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1989. [Google Scholar]
- 4.Benjannet S, Rondeau N, Day R, Chretien M, Seidah NG. PC1 and PC2 are proprotein convertases capable of cleaving proopiomelanocortin at distinct pairs of basic residues. Proc Natl Acad Sci USA. 1991;88:3564–3568. doi: 10.1073/pnas.88.9.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernheim SM, Mayeri E. Complex behavior induced by egg-laying hormone in Aplysia. J Comp Physiol [A] 1995;176:131–136. doi: 10.1007/BF00197759. [DOI] [PubMed] [Google Scholar]
- 6.Broadie KS, Bate M. Development of the embryonic neuromuscular synapse of Drosophila melanogaster. J Neurosci. 1993;13:144–166. doi: 10.1523/JNEUROSCI.13-01-00144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryan P, Pantoliano MW, Quill SG, Hsiao HY, Poulos T. Site-directed mutagenesis and the role of the oxyanion hole in subtilisin. Proc Natl Acad Sci USA. 1986;83:3743–3745. doi: 10.1073/pnas.83.11.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter P, Wells JA. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 9.Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copenhaver PF, Truman JW. Identification of the cerebral neurosecretory cells that contain eclosion hormone in the moth Manduca sexta. J Neurosci. 1986;6:1738–1747. doi: 10.1523/JNEUROSCI.06-06-01738.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 12.Eipper BA, Mains RE. Structure and biosynthesis of pro-adrenocorticotropin/endorphin and related peptides. Endocr Rev. 1980;1:1–27. doi: 10.1210/edrv-1-1-1. [DOI] [PubMed] [Google Scholar]
- 13.Ewer J, De Vente VJ, Truman JW. Neuropeptide induction of cyclic GMP increases in the insect CNS: resolution at the level of single identifiable neurons. J Neurosci. 1994;14:7704–7712. doi: 10.1523/JNEUROSCI.14-12-07704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewer J, Gammie SC, Truman JW (1997) Control of insect ecdysis by a positive-feedback endocrine system: roles of eclosion hormone and ecdysis triggering hormone. J Exp Biol 869–881. [DOI] [PubMed]
- 15.Feany MB, Quinn WG. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science. 1995;268:869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- 16.Friedman TC, Loh YP, Birch NP. In vitro processing of proopiomelanocortin by recombinant PC1 (SPC3). Endocrinology. 1994;135:854–862. doi: 10.1210/endo.135.3.8070378. [DOI] [PubMed] [Google Scholar]
- 17.Furuta M, Yano H, Zhou A, Rouille Y, Holst JJ, Carroll R, Ravazzola M, Orci L, Furuta H, Steiner DF. Defective prohormone processing and altered pancreatic islet morphology in mice lacking active SPC2. Proc Natl Acad Sci USA. 1997;94:6646–6651. doi: 10.1073/pnas.94.13.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuta M, Carroll R, Martin S, Swift HH, Ravazzola M, Orci L, Steiner DF. Incomplete processing of proinsulin to insulin accompanied by elevation of Des-31,32 proinsulin intermediates in islets of mice lacking active PC2. J Biol Chem. 1998;273:3431–3437. doi: 10.1074/jbc.273.6.3431. [DOI] [PubMed] [Google Scholar]
- 19.Gammie SC, Truman JW. Neuropeptide hierarchies and the activation of sequential motor behaviors in the hawkmoth, Manduca sexta. J Neurosci. 1997;17:4389–4397. doi: 10.1523/JNEUROSCI.17-11-04389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerttula S, Jin YS, Anderson KV. Zygotic expression and activity of the Drosophila Toll gene, a gene required maternally for embryonic dorsal-ventral pattern formation. Genetics. 1988;119:123–133. doi: 10.1093/genetics/119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gluschankof P, Fuller RS. A C-terminal domain conserved in precursor processing proteases is required for intramolecular N-terminal maturation of pro-Kex2 protease. Embo J. 1994;13:2280–2288. doi: 10.1002/j.1460-2075.1994.tb06510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman CS, Doe CQ. Embryonic development of the Drosophila central nervous system. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1993. pp. 1131–1206. [Google Scholar]
- 23.Hoflehner J, Eder U, Laslop A, Seidah NG, Fischer CR, Winkler H. Processing of secretogranin II by prohormone convertases: importance of PC1 in generation of secretoneurin. FEBS Lett. 1995;360:294–298. doi: 10.1016/0014-5793(95)00127-u. [DOI] [PubMed] [Google Scholar]
- 24.Horodyski FM, Ewer J, Riddiford LM, Truman JW. Isolation, characterization and expression of the eclosion hormone gene of Drosophila melanogaster. Eur J Biochem. 1993;215:221–228. doi: 10.1111/j.1432-1033.1993.tb18026.x. [DOI] [PubMed] [Google Scholar]
- 25.Itoh Y, Tanaka S, Takekoshi S, Itoh J, Osamura RY. Prohormone convertases (PC1/3 and PC2) in rat and human pancreas and islet cell tumors: subcellular immunohistochemical analysis. Pathol Int. 1996;46:726–737. doi: 10.1111/j.1440-1827.1996.tb03541.x. [DOI] [PubMed] [Google Scholar]
- 26.Jackson RS, Creemers JW, Ohagi S, Raffin SM, Sanders L, Montague CT, Hutton JC, O’Rahilly S. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat Genet. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 27.Johanning K, Mathis JP, Lindberg I. Role of PC2 in proenkephalin processing: antisense and overexpression studies. J Neurochem. 1996;66:898–907. doi: 10.1046/j.1471-4159.1996.66030898.x. [DOI] [PubMed] [Google Scholar]
- 28.Kaliss N. The effect on development of a lethal deficiency in Drosophila melanogaster: with a description of the normal embryo at the time of hatching. Genetics. 1939;24:244–270. doi: 10.1093/genetics/24.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konig M, Zimmer AM, Steiner H, Holmes PV, Crawley JN, Brownstein MJ, Zimmer A. Pain responses, anxiety and aggression in mice deficient in pre-proenkephalin. Nature. 1996;383:535–538. doi: 10.1038/383535a0. [DOI] [PubMed] [Google Scholar]
- 30.Kopczynski CC, Davis GW, Goodman CS. A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science. 1996;271:1867–1870. doi: 10.1126/science.271.5257.1867. [DOI] [PubMed] [Google Scholar]
- 31.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipkind G, Gong Q, Steiner DF. Molecular modeling of the substrate specificity of prohormone convertases SPC2 and SPC3. J Biol Chem. 1995;270:13277–13284. doi: 10.1074/jbc.270.22.13277. [DOI] [PubMed] [Google Scholar]
- 33.Mains RE, Eipper BA. Synthesis and secretion of corticotropins, melanotropins, and endorphins by rat intermediate pituitary cells. J Biol Chem. 1979;254:7885–7894. [PubMed] [Google Scholar]
- 34.Margaritis LH. Structure and physiology of the eggshell. In: Kerkut GA, Gilbert LI, editors. Comprehensive insect physiology, biochemistry and pharmacology. Pergamon; Oxford: 1985. pp. 221–224. [Google Scholar]
- 35.McCormick J, Nichols R. Spatial and temporal expression identify dromyosuppressin as a brain-gut peptide in Drosophila melanogaster. J Comp Neurol. 1993;338:278–288. doi: 10.1002/cne.903380210. [DOI] [PubMed] [Google Scholar]
- 36.McNabb SL, Baker JD, Agapite J, Steller H, Riddiford LM, Truman JW. Disruption of a behavioral sequence by targeted death of peptidergic neurons in Drosophila. Neuron. 1997;19:813–823. doi: 10.1016/s0896-6273(00)80963-0. [DOI] [PubMed] [Google Scholar]
- 37.Miles CI, Weeks JC. Developmental attenuation of the pre-ecdysis motor pattern in the tobacco hornworm, Manduca sexta. J Comp Physiol [A] 1991;168:179–190. doi: 10.1007/BF00218410. [DOI] [PubMed] [Google Scholar]
- 38.Nambu JR, Murphy EC, Andrews PC, Feistner GJ, Scheller RH. Isolation and characterization of a Drosophila neuropeptide gene. Neuron. 1988;1:55–61. doi: 10.1016/0896-6273(88)90209-7. [DOI] [PubMed] [Google Scholar]
- 39.Nichols R, Lim IA. Spatial and temporal immunocytochemical analysis of drosulfakinin (Dsk) gene products in the Drosophila melanogaster central nervous system. Cell Tissue Res. 1996;283:107–116. doi: 10.1007/s004410050518. [DOI] [PubMed] [Google Scholar]
- 40.Nichols R, Schneuwly SA, Dixon JE. Identification and characterization of a Drosophila homologue to the vertebrate neuropeptide cholecystokinin. J Biol Chem. 1988;263:12167–12170. [PubMed] [Google Scholar]
- 41.Nichols R, McCormick J, Lim I, Caserta L. Cellular expression of the Drosophila melanogaster FMRFamide neuropeptide gene product DPKQDFMRFamide. Evidence for differential processing of the FMRFamide polypeptide precursor. J Mol Neurosci. 1995a;6:1–10. doi: 10.1007/BF02736754. [DOI] [PubMed] [Google Scholar]
- 42.Nichols R, McCormick JB, Lim IA, Starkman JS. Spatial and temporal analysis of the Drosophila FMRFamide neuropeptide gene product SDNFMRFamide: evidence for a restricted expression pattern. Neuropeptides. 1995b;29:205–213. doi: 10.1016/0143-4179(95)90062-4. [DOI] [PubMed] [Google Scholar]
- 43.Palazzolo MJ, Hyde DR, VijayRaghavan K, Mecklenburg K, Benzer S, Meyerowitz E. Use of a new strategy to isolate and characterize 436 Drosophila cDNA clones corresponding to RNAs detected in adult heads but not in early embryos. Neuron. 1989;3:527–539. doi: 10.1016/0896-6273(89)90211-0. [DOI] [PubMed] [Google Scholar]
- 44.Paquet L, Massie B, Mains RE. Proneuropeptide Y processing in large dense-core vesicles: manipulation of prohormone convertase expression in sympathetic neurons using adenoviruses. J Neurosci. 1996;16:964–973. doi: 10.1523/JNEUROSCI.16-03-00964.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: practical uses in cell and molecular biology. Academic; San Diego: 1994. pp. 446–485. [DOI] [PubMed] [Google Scholar]
- 46.Poe EA. The cask of amontillado. In: Stern PVD, editor. The portable poe. Penguin Books; Harmondsworth, England: 1973. pp. 309–317. [Google Scholar]
- 47.Pu LP, Hayes WP, Mill JF, Ghose S, Friedman TC, Loh YP. Frog prohormone convertase PC2 mRNA has a mammalian-like expression pattern in the central nervous system and is colocalized with a subset of thyrotropin-releasing hormone-expressing neurons. J Comp Neurol. 1995;354:71–86. doi: 10.1002/cne.903540107. [DOI] [PubMed] [Google Scholar]
- 48.Roebroek AJ, Creemers JW, Pauli IG, Kurzik DU, Rentrop M, Gateff EA, Leunissen JA, Van de Ven W. Cloning and functional expression of Dfurin2, a subtilisin-like proprotein processing enzyme of Drosophila melanogaster with multiple repeats of a cysteine motif. J Biol Chem. 1992;267:17208–17215. [PubMed] [Google Scholar]
- 49.Roebroek AJ, Creemers JW, Pauli IG, Bogaert T, Van de Ven W. Generation of structural and functional diversity in furin-like proteins in Drosophila melanogaster by alternative splicing of the Dfur1 gene. EMBO J. 1993;12:1853–1870. doi: 10.1002/j.1460-2075.1993.tb05834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouille Y, Westermark G, Martin SK, Steiner DF. Proglucagon is processed to glucagon by prohormone convertase PC2 in alpha TC1–6 cells. Proc Natl Acad Sci USA. 1994;91:3242–3246. doi: 10.1073/pnas.91.8.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rouille Y, Martin S, Steiner DF. Differential processing of proglucagon by the subtilisin-like prohormone convertases PC2 and PC3 to generate either glucagon or glucagon-like peptide. J Biol Chem. 1995;270:26488–26496. doi: 10.1074/jbc.270.44.26488. [DOI] [PubMed] [Google Scholar]
- 52.Rovere C, Barbero P, Kitabgi P. Evidence that PC2 is the endogenous pro-neurotensin convertase in rMTC 6–23 cells and that PC1- and PC2-transfected PC12 cells differentially process pro-neurotensin. J Biol Chem. 1996;271:11368–11375. doi: 10.1074/jbc.271.19.11368. [DOI] [PubMed] [Google Scholar]
- 53.Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S, Arch JR, Buckingham RE, Haynes AC, Carr SA, Annan RS, McNulty DE, Liu WS, Terrett JA, Elshourbagy NA, Bergsma DJ, Yanagisawa M. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1989. [Google Scholar]
- 55.Schafer MK, Day R, Cullinan WE, Chretien M, Seidah NG, Watson SJ. Gene expression of prohormone and proprotein convertases in the rat CNS: a comparative in situ hybridization analysis. J Neurosci. 1993;13:1258–1279. doi: 10.1523/JNEUROSCI.13-03-01258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider LE, Taghert PH. Isolation and characterization of a Drosophila gene that encodes multiple neuropeptides related to Phe-Met-Arg-Phe-NH2 (FMRFamide). Proc Natl Acad Sci USA. 1988;85:1993–1997. doi: 10.1073/pnas.85.6.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider LE, O’Brien MA, Taghert PH. In situ hybridization of the FMRFamide neuropeptide gene in Drosophila I. Restricted expression in embryonic and larval stages. J Comp Neurol. 1991;304:608–622. doi: 10.1002/cne.903040408. [DOI] [PubMed] [Google Scholar]
- 58.Seidah NG, Marcinkiewicz M, Benjannet S, Gaspar L, Beaubien G, Mattei MG, Lazure C, Mbikay M, Chretien M. Cloning and primary sequence of a mouse candidate prohormone convertase PC1 homologous to PC2, Furin, and Kex2: distinct chromosomal localization and messenger RNA distribution in brain and pituitary compared to PC2. Mol Endocrinol. 1991;5:111–122. doi: 10.1210/mend-5-1-111. [DOI] [PubMed] [Google Scholar]
- 59.Skaer H. The alimentary canal. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1993. pp. 941–1012. [Google Scholar]
- 60.Smeekens SP, Avruch AS, LaMendola J, Chan SJ, Steiner DF. Identification of a cDNA encoding a second putative prohormone related to PC2 in AtT20 cells and islets of Langerhans. Proc Natl Acad Sci USA. 1991;88:340–344. doi: 10.1073/pnas.88.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas L, Leduc R, Thorne BA, Smeekens SP, Steiner DF, Thomas G. Kex2-like endoproteases PC2 and PC3 accurately cleave a model prohormone in mammalian cells: evidence for a common core of neuroendocrine processing enzymes. Proc Natl Acad Sci USA. 1991;88:5297–5301. doi: 10.1073/pnas.88.12.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng M, Streck RD, Scott RE, Seidah NG, Pintar JE. The developmental expression in rat of proteases furin, PC1, PC2, and carboxypeptidase E: implications for early maturation of proteolytic processing capacity. J Neurosci. 1994;14:4656–4673. doi: 10.1523/JNEUROSCI.14-08-04656.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y, Lindberg I. Purification and characterization of the prohormone convertase PC1(PC3). J Biol Chem. 1993;268:5615–5623. [PubMed] [Google Scholar]
- 64.Zhou A, Bloomquist BT, Mains RE. The prohormone convertases PC1 and PC2 mediate distinct endoproteolytic cleavages in a strict temporal order during proopiomelanocortin biosynthetic processing. J Biol Chem. 1993;268:1763–1769. [PubMed] [Google Scholar]
- 65.Zimmer A, Zimmer AM, Baffi J, Usdin T, Reynolds K, Konig M, Palkovits M, Mezey E. Hypoalgesia in mice with a targeted deletion of the tachykinin 1 gene. Proc Natl Acad Sci USA. 1998;95:2630–2635. doi: 10.1073/pnas.95.5.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zitnan D, Kingan TG, Hermesman JL, Adams ME. Identification of ecdysis-triggering hormone from an epitracheal endocrine system. Science. 1996;271:88–91. doi: 10.1126/science.271.5245.88. [DOI] [PubMed] [Google Scholar]