Abstract
Neuropeptides are a ubiquitous class of signaling molecules. In our attempt to understand the generation of feeding behavior inAplysia, we have sought to identify and fully characterize the neuropeptides operating in this system. Preliminary evidence indicated that Mytilus inhibitory peptide (MIP)-like peptides are present and operating in the circuitry that generates feeding in Aplysia. MIPs were originally isolated from the bivalve mollusc Mytilus edulis, and related peptides have been identified in other invertebrate species, but no precursor has been identified. In this study, we describe the isolation and characterization of novel AplysiaMIP-related peptides (AMRPs) and their precursor. Several AMRPs appear to have some structural and functional features similar to vertebrate opioid peptides. We use matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to confirm that all 14 AMRPs predicted by the precursor are processed in isolated neurons. Northern analysis, whole-mount in situ hybridization, and immunohistochemistry are used to map the abundant expression of these peptides in the CNS and peripheral tissues such as the digestive tract, vasculature, and the reproductive organs. Physiological studies demonstrate that the rank order of the inhibitory actions of these peptides is different for three target muscles. These results underscore the importance of using a multidisciplinary approach to identifying and characterizing the actions of neuropeptides in an effort to gain understanding of their role in systems of interest. The widespread distribution of the AMRPs indicates that they may be operating in many different systems of Aplysia.
Keywords: Mytilus inhibitory peptide, neuropeptide, mollusc, Aplysia, cDNA cloning, immunohistochemistry, in situ hybridization, MALDI-TOF MS
Neuropeptides are a ubiquitous and diverse class of signaling molecules. Molluscs have been good models for studying the mechanisms of neurotransmission, which involves a number of neuropeptides as chemical messengers (Brezina et al., 1996;Vilim et al., 1996a,b). The opisthobranch gastropod, Aplysia, is one of the best-studied molluscs, in which various peptides have been identified, and the peptidergic neurotransmission has been shown to play crucial roles in the physiologically important processes such as feeding, egg laying, and cardioregulation (Scheller et al., 1983;Campanelli and Scheller, 1987; Brezina et al., 1995). Nevertheless, it seems likely that additional important peptide transmitters remain to be identified, even in this well-investigated animal. One such set of unidentified peptides in Aplysia would be a family ofMytilus inhibitory peptides (MIPs). These neuropeptides were originally isolated from the bivalve mollusc Mytilus edulis(Hirata et al., 1988; Fujisawa et al., 1991, 1993), and similar peptides have been identified in other molluscan species (Ikeda et al., 1992a,b; Ohtani et al., 1995; Li et al., 1996). In general, these peptides have inhibitory actions on target muscles and hyperpolarize central neurons (Yongsiri et al., 1989; Kiss, 1990; Kiss and Osipenko, 1997). Although these peptides are likely to play important roles in the function of many systems in a variety of molluscs, their precursor protein has yet to be identified in any species.
Neuropepstides are often processed from precursor proteins that code for other bioactive peptides as well. Often these peptides are considered to be functionally redundant, but typically have different potencies on single targets (Brezina et al., 1995; Hewes et al., 1998;Perry et al., 1998). Furthermore, peptides derived from a single precursor can affect multiple targets differentially, as in the case of pro-opiomelanocortin (POMC) or allostatins in insects (Bendena et al., 1997). Thus, it becomes important to characterize the precursor protein to peptides of interest in the effort to understand their role in systems of interest.
In preliminary studies, we found that in Aplysia numerous neurons in the cerebral and buccal ganglia and many axons in the cerebrobuccal connective showed MIP-like immunoreactivity, suggesting that MIP-like peptides may be involved in the central pattern generator for the feeding system of this animal. In addition, an MIP-related peptide, GAPRFVamide, was isolated from the extract of the digestive tract of Aplysia by bioassay using the crop-gizzard preparation (O. Matsushima, unpublished observations). These results led us to postulate that MIP-related peptides play important roles in the feeding, and possibly other systems, ofAplysia.
In the present study, we describe the identification of theAplysia MIP-related peptides (AMRPs) and their precursor using biochemical and molecular techniques. We use matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) to demonstrate the processing of the AMRPs from their precursor protein in isolated neurons. Expression of the AMRPs in both CNS and peripheral tissues is also investigated by Northern blot, in situ hybridization, and immunohistochemistry. The physiological actions of these peptides are investigated on several AMRP-innervated target muscles.
MATERIALS AND METHODS
Animals
Two species of the opisthobranch mollusc Aplysia were used, Aplysia kurodai and Aplysia californica.Animals were kept in tanks filled with artificial seawater (ASW) continuously aerated at 14–15°C. A. kurodai (50–300 gm) were caught in Hiroshima Bay, Hamada City (the coast of the Sea of Japan) and Onahama City (the coast of the Pacific Ocean) in Japan. These specimens were used for peptide purification within 8 hr of collection or within 10 d for physiological experiments. Specimens of A. californica (10–500 gm) were obtained fromAplysia Research Facility (Miami, FL), Pacific Biomarine (Venice, CA), and Marinus Inc. (Long Beach, CA). Several larger animals (up to 1000 gm) were collected off the Monterey Peninsula between January and July 1998. Animals were used between 3 and 14 d of receipt. Large animals (200–500 gm) were used for RNA extraction and MALDI MS, whereas both large and small (10–500 gm) animals were used for immunocytochemistry and in situ hybridization.
Peptide purification
The cerebral, pedal, and pleural ganglia were isolated from 290 specimens of A. kurodai and immediately frozen in liquid nitrogen. The ganglia were homogenized by Polytron homogenizer in five volumes of 80% acetone on ice. The homogenate was centrifuged at 15,000 × g at 4°C for 20 min, and the supernatant was collected. The obtained supernatant was evaporated to remove acetone and passed through C18 cartridges (Mega Bond Elut C18; Varian, Harbor, CA). The cartridges were washed first with 10% methanol containing 0.1% trifluoroacetic acid (TFA) and then eluted with 60% methanol–0.1% TFA. The eluate was concentrated to a small volume and loaded onto a reversed-phase HPLC column (Capcell Pak C18; Shiseido, Tokyo, Japan) and eluted with a linear gradient of acetonitrile (0–60% in 60 min) containing 0.1% TFA at a flow rate of 1.0 ml/min. Fractions of 1 ml each were collected, and an aliquot of each fraction was applied to a competitive ELISA using the anti-MIP antibody raised against GSPMFVamide (WM1) as described previously (Fujisawa, 1996). Immunopositive fractions were separately applied onto a cation-exchange column (TSKgel SP-5PW; Tosoh, Tokyo, Japan) equilibrated with 10 mm sodium phosphate buffer, pH 6.7, and eluted with a linear gradient of NaCl (0–0.6m for 60 min). The MIP-like immunoreactivity of each fraction was measured by the ELISA, and positive fractions were purified by alternating reversed-phase HPLC and ELISA to produce single UV-absorbance peaks.
Structural analysis
Amino acid sequences of the purified peptides were determined by automatic peptide sequencer (PSQ-1; Shimadzu, Kyoto, Japan). Fast atom bombardment mass spectrometry (FAB-MS) was performed for all the peptides (SX-102A; JEOL, Tokyo, Japan) to determine C-terminal amidation, because a peptide with free C terminus and that of an amidated form have different molecular masses. To confirm the structures, peptides with the predicted sequences were chemically synthesized on the automated peptide synthesizer (PSSM-8; Shimadzu), and co-chromatographed with the purified peptides on a reversed-phase HPLC column.
Cloning
Standard molecular techniques (Sambrook et al., 1989) were used except where noted. Aplysia californica ganglion cDNA library was a gift of Dr. Gregg Nagle. The library, a directional Uni-Zap Lambda phage library (Stratagene, La Jolla, CA), was used both as a template for PCR and for conventional hybridization screening. Seminested degenerate rapid amplification of cDNA ends was performed using two vector primers and antisense degenerate primers designed to GAPRFVG (CCI ACR AAI CKI GGI GCI CC). PCR was performed in two stages on a Robocycler Gradient 40 thermal cycler (Stratagene) using taq DNA polymerase and dNTPs from Perkin-Elmer (Norwalk, CT). Both stages were cycled 25 times for 30 sec at 95°C, 1 min at the annealing temperature, and 2 min at 72°C. Three separate annealing temperatures (50, 54, and 58°C) were run in parallel, and a set without the degenerate primer was used as a control. The reactions were hot started and not allowed to cool to <72°C between the stages. In the first stage, 10 μl reactions containing 0.1 μm vector primer (ACC ATG ATT ACG CCA AG), 0.1 μm degenerate primer, 100 μm dNTPs, and 0.1 μl of library were hot-started with 0.1 U of taq in 0.5 μl of reaction buffer. In the second stage, 50 μl of prewarmed (72°C) reaction mix containing 1 μm nested vector primer (AAT TAA CCC TCA CTA AAG), 1 μm degenerate primer, and 100 μm dNTPs was added to each tube then hot-started again with 1 U of taq.
The results of the PCR were assessed using agarose gel electrophoresis, and the highest temperature reactions showing significantly more product than the matched degenerate primerless control were polyethylene glycol-precipitated and TA-cloned (Invitrogen, Carlsbad, CA). Insert-bearing clones were identified using colony PCR, then cycle-sequenced with dye termination (Perkin-Elmer/Applied Biosystems). Inserts from promising degenerate clones were isolated and labeled using 32P-dCTP and random primers (New England Biolabs, Beverly, MA). These probes were then used to screen a library to identify full-length clones. At least two independent clones were sequenced for all regions using a combination of restriction, deletion, and primer walking. Sequence alignments were generated using Geneworks version 2.1, and consensus contigs were assembled manually.
Mass spectrometry
Cellular sample preparation. Ganglia with intact connectives and commissures were removed after an injection of 390 mm MgCl2 equal to 50% of each animal's body weight. In some cases, a moderate protease treatment (e.g., 1% protease Type IX for 30–60 min at 34°C) was used to soften the connective tissues before cell dissection. Extracellular salts were removed by a previously described approach (Garden et al., 1996). Briefly, the pleural ganglia were isolated and pinned down, and the physiological saline was replaced with an aqueous MALDI matrix solution, 10 mg/ml of 2,5-dihydroxybenzoic acid (DHB; ICN Pharmaceuticals, Costa Mesa, CA). Specific cells were identified and isolated based on the immunostaining results. Tungsten needles were used to isolate individual or group of cells onto a MALDI sample plate containing 0.5 μl of matrix solution. After drying at ambient temperature, samples were either frozen for future analysis or analyzed immediately.
Microbore-liquid chromatography of cellular homogenates.Fifty three abdominal ganglia were collected on dry ice and subsequently stored at −80°C. Peptide extracts were made in 500 μl of acidified acetone (40:6:1 acetone:water:HCl) according to previous methods (Floyd et al., 1999). Samples were homogenized in a microhomogenizer (Jencons Scientific Ltd.), sonicated for 5 min (model 2200; Branson, Danbury, CT), and centrifuged for 10 min at 13,000 rpm in a microcentrifuge (Baxter, McGaw Park, IL). The supernatant was removed, freeze-dried (Labconco; Fisher Scientific, Itasca, IL), and resuspended in 500 μl of 2% acetonitrile in 0.1% aqueous TFA. Two hundred and fifty microliters of the extract was injected in a reversed-phase microbore liquid chromatography (LC) instrument (Magic 2002; Michrom BioResources, Auburn, CA) consisting of a Reliasil C-18 column (0.5 × 150 mm) with 300 Å packing. The flow rate was 150 μl/min at ambient temperature. The column was equilibrated with solvent A (98% H2O, 0.1% TFA, and 1.9% acetonitrile), and a gradient was developed from 0–80% of solvent B (90% acetonitrile, 9.9% H2O, and 0.1% TFA) for 30 min and then from 80–98% of solvent B for 10 min. Samples were collected by a fraction collector (FC 203B; Gilson, Middletown, WI), and each fraction was screened by MALDI-TOF MS; 0.25 μl of each HPLC fraction was deposited onto a MALDI sample plate followed by the same volume of α-cyano-4-hydroxycinnamic acid (Sigma, St. Louis, MO) matrix solution.
MALDI-TOF MS. Mass spectra were obtained using a Voyager DE-STR mass spectrometer equipped with delayed ion extraction (PE Biosystems, Framingham, MA). A pulsed nitrogen laser (337 nm) was used as the desorption/ionization source, and positive-ion mass spectra were acquired using both linear and reflectron mode. Each representative mass spectrum shown is average of 128–256 laser pulses. Mass calibration was performed internally using identified peptides FMRFamide (m/z 599.33) and myomodulin C (MM-C;m/z 861.45) as calibrants. Laser power and delay time were optimized for each type of samples (i.e., single cells and LC fractions). Mass spectral peaks were assigned based on combination of observed masses and the knowledge of prohormone sequences.
Post-source decay. Equal volumes of an HPLC fraction and matrix solution were either premixed in the vial or mixed on the MALDI sample plate. The matrix used was 10 mg/ml α-cyano-4-hydroxy-cinnamic acid (dissolved in 6:3:1 acetonitrile:water:3% TFA) (Aldrich, Milwaukee, WI). Post-source decay (PSD) analysis was performed with a Voyager DE-STR mass spectrometer in reflectron mode. The total acceleration voltage was 20 kV with a 75 nsec delay time. Spectra were obtained by accumulating data from 100–256 laser shots. To obtain complete PSD spectra, a series of reflectron TOF spectral segments were acquired, each optimized to focus fragment ions within differentm/z ranges. Segments of each were stitched together to generate a composite PSD spectrum.
Northern analysis
The buccal, cerebral, pleural, pedal, and abdominal ganglia were separately dissected and pooled from five animals (A. californica) anesthetized with 50% volume of isotonic MgCl2. RNA was isolated by the acid-phenol method of Chomcyznski and Sacchi (1987). RNA was fractionated on a 3[N-morpholino]propanesulfonic acid (MOPS)/formaldehyde 1.5% agarose gel and downward transferred (turboblotter; Schleicher & Schuell, Keene, NH) overnight with 20× standard saline phosphate EDTA (SSPE) onto positively charged nylon (Biodyne B; Life Technologies, Gaithersburg, MD). The RNA was UV-crosslinked (Stratalinker; Stratagene), then washed with diethyl pyrocarbonate-treated water and stained with methylene blue (0.2% methylene blue/0.3m sodium acetate, pH 5.5). The blot was scanned to document the loading and transfer of the RNA. The positions of the lanes and the bands in the RNA marker lane (Novagen, Madison, WI) were noted on the membrane with a number 2 pencil. After complete destaining in 1% SDS–0.1× SSPE, the blots were prehybridized (50% formamide, 7% SDS, 250 mm sodium phosphate, pH 7.2, 10 mm EDTA, and 10% dextran sulfate) for 1 hr at 50°C in a rotary oven (Hybaid, Franklin, MA). The blot was then hybridized with random primer-labeled (New England Biolabs) probe overnight at 50°C. Washes were performed 2 × 15 min at room temperature (RT) with 2× SSPE–0.1% SDS, then at 50°C for 1 hr with 0.1× SSPE–0.1% SDS. Blots were wrapped in saran and exposed to film. Autoradiograms were aligned with the blots, and the positions of the markers were noted. They were then scanned and assembled into final figures using Photoshop 3.0.
New antibodies
The antigen was prepared by coupling GSPRFFamide (AnaSpec Inc., San Jose, CA) to BSA (Sigma, catalog #A0281) using either 1-ethyl-3-(dimethylaminopropyl)carbodiimide (EDC) (Sigma, catalog #E7750) or paraformaldehyde–glutaraldehyde (PfG) (EM Sciences, Fort Washington PA). The coupling was performed in a 0.5 ml volume of 50 mm NaH2PO4, pH 7.2, containing 5 mg of BSA, 1 mg of peptide, and either 10 mg of EDC or 1% paraformaldehyde and 0.1% glutaraldehyde. The mixture was allowed to react overnight at 4°C, and then the coupled antigen was purified from the reaction using a Microcon-30 (spinning at 13,800 × g for 30 min at 4°C to concentrate). After washing the retentate four times with 0.4 ml of 50 mmNaH2PO4, pH 7.2, it was resuspended in 0.5 ml of the same buffer and transferred to a new tube.
Two male Sprague Dawley rats (Teconic; 250–300 gm) were immunized by intraperitoneal injection with 12.5 μl (∼125 μg) of antigen in an emulsion of 0.5 ml of PBS and 0.5 ml of Freund's complete adjuvant. At 21 and 42 d after initial injection, the rats were boosted by intraperitoneal injection with 6.25 μl (∼62.5 μg) of antigen in an emulsion of 0.5 ml of PBS and 0.5 ml of Freund's incomplete adjuvant. The animals were killed by decapitation at 49 d after initial injection; the blood was harvested and processed for serum. Sera were aliquoted, frozen, and lyophilized, or stored at 4°C with EDTA (25 mm final) and thimerosal (0.1% final) added as stabilizers.
Of the two antibodies we made, EDC-coupled antigen gave more specific immunostaining than PfG-coupled antigen, because PfG stained almost twice as many neurons than EDC. Furthermore, EDC-immunopositive neurons were the most consistent with the in situhybridization-positive neurons and the Northern analysis of the distribution of the AMRP precursor mRNA in the different ganglia. Therefore, all subsequent immunostaining analysis we report here was done with the antibody made to EDC-coupled GSPRFFamide–BSA antigen. Immunostaining with this antibody was abolished by preincubation with 10−4m GSPRFFamide (data not shown).
In situ hybridization
The whole-mount in situ hybridization protocol used in this study is a modification of a method established for tunicate embryos (Makabe et al., 1992). Animals (A. californica) were anesthetized with 50% volume injection of isotonic MgCl2, and the desired ganglia and tissues were dissected and removed. The tissues were pinned out in the desired orientation in 50% isotonic MgCl2–50% ASW. All subsequent reagents and solutions used in the in situhybridization were made with diethyl pyrocarbonate-treated MilliQ water, and care was taken to avoid contamination with RNases. Tissues were fixed in 4% paraformaldehyde, 0.5 m NaCl, and 0.1 m MOPS, pH 7.5, for 3 hr at RT or overnight at 4°C, then washed for 3 × 10 min at RT in PBT (0.8% NaCl, 0.02% KCl, 0.3% Na2HPO4–12 H2O, 0.02% KH2PO4, and 0.1% Tween 20, pH 7.4). The tissue was digested with 50 μg/ml of proteinase K in PBT for 30 min at 37°C, then washed again with PBT for 3 × 10 min at RT. The tissue was post-fixed with 4% paraformaldehyde in PBT for 1 hr at RT, then washed once more for 3 × 10 min at RT with PBT. The tissue was prehybridized for 1 hr at 42°C in hyb-buffer (5× SSC, 1% blocking reagent, 50 μg/ml salmon sperm DNA, 0.1% sarkosyl, and 0.02% SDS) and then hybridized overnight at 42°C in hyb-buffer containing 1 μg/ml of the labeled oligo. Oligos were labeled by tailing with digoxigenin (DIG)–dUTP/dATP according to the manufacturer's instructions (Boehringer Mannheim, Indianapolis, IN). Unbound probe was washed out with 2× SSC and 0.01% SDS for 3 × 1 hr at 42°C then with PBT for 2 × 10 min at RT. The tissue was blocked with 1% blocking reagent (Boehringer, catalog #1096176) in 0.15 m NaCl and 0.1 mmaleic acid, pH 7.5, for 3 hr at RT and then incubated in 1:200 dilution of anti-DIG antibody labeled with alkaline phosphatase (Boehringer, catalog #1093274) in blocking solution for 24 hr at 4°C. Unbound antibody was washed out with PBT for 5 × 1 hr at RT, then washed with detection buffer (0.1 m Tris, 0.1m NaCl, 5 mmMgCl2, and 10 mmlevamisole) for 2 × 30 min at RT. The signal was developed for 30 min at RT with detection buffer containing 350 μg/ml nitroblue tetrazolium, 175 μg/ml 5-bromo-4-chloro-3-indolyl phosphate, and 0.1% Tween 20, and the reaction was then stopped by washing the tissue with PBT containing 1 mm EDTA (PBTE). The tissues were post-fixed with 4% paraformaldehyde in PBT for overnight at 4°C. After washing with PBT, they were stored protected from light in 50% glycerol and PBTE at 4°C. Selected preparations were photographed on a Nikon microscope, and the negatives were scanned and compiled into figures with Photoshop 3.0.
Immunocytochemistry
Immunocytochemistry was performed on A. californicaas previously described (Vilim et al., 1996a). Tissues were fixed in freshly prepared fixative (4% paraformaldehyde, 0.2% picric acid, 25% sucrose, and 0.1 mNaH2PO4, pH 7.6) for either 3 hr at RT or overnight at 4°C. After washes with PBS to remove the fixative, the ganglia from large animals were desheathed to expose the neurons. Ganglia from small animals (10–15 gm) were processed without desheathing. All subsequent incubations were done at RT with rocking. Tissue was permeabilized and blocked by overnight incubation in blocking buffer (BB; 10% normal donkey serum, 2% Triton X-100, 1% BSA, 154 mm NaCl, 10 mmNa2HPO4, 50 mm EDTA, and 0.01% thimerosal, pH 7.4). Primary antibody was diluted 1:250 in BB and incubated with the tissue for 4–7 d. The tissue was then washed twice a day for 2–3 d with washing buffer (WB; 2% Triton X-100, 1% BSA, 154 mmNaCl, 10 mmNa2HPO4, 50 mm EDTA, and 0.01% thimerosal, pH 7.4). After the washes, the tissue was incubated with 1:500 dilution of secondary antibody (lissamine–rhodamine donkey anti-rat; Jackson ImmunoResearch, West Grove, PA) for 2–3 d. The tissue was then washed twice with WB for 1 d and 4 times with storage buffer (SB; 1% BSA, 154 mm NaCl, 10 mmNa2HPO4, 50 mm EDTA, and 0.01% thimerosal, pH 7.4) for 1 d. The tissues were then stored at 4°C or viewed and photographed on a Nikon microscope equipped with epifluorescence (Morrell Instrument Co., Melville, NY). Negatives were scanned and compiled into figures using Photoshop 3.0.
Recording of muscle contractions
Animals (A. kurodai) were anesthetized by injecting isotonic MgCl2 solution before dissection. The esophagus, the dorsal longitudinal muscle of the inner body wall, and the penis retractor muscle (1–1.5 cm in length) were excised from the animal, and then both ends of them were tied with cotton thread. One end was fixed to a chamber filled with ASW (2 ml), and the other was fixed to a force transducer. In the case of the esophagus, the chamber was continuously aerated, and 20 μl of peptide solutions at 100× concentration was injected into the chamber. Tension changes of the spontaneous contractions of the esophagus were recorded on a thermal pen recorder. In the case of the body wall muscle and the penis retractor muscle, the chamber was not aerated, and electrical stimulation was applied to evoke tetanic contractions at 10 min intervals. Peptide solutions dissolved in ASW at final concentration were applied to the preparations by exchanging the whole chamber content.
RESULTS
Isolation of five AMRPs from the CNS of Aplysia
By means of the competitive ELISA combined with HPLC purification, five peptides were isolated from the acetone extract of the cerebral, pedal, and pleural ganglia of Aplysia kurodai. Figure1 shows an example of purification steps for one of the five peptides, GAPRFVamide. Results of amino acid sequence analysis and FAB-MS measurement indicated that the five peptides are all hexapeptides with the amidated C terminus (Table1). Co-chromatography experiments using the synthetic and the purified peptides confirmed the predicted structures (data not shown). Therefore, the structures of the isolated peptides were determined as follows: GAPRFVamide, GAPRFIamide, GPPRFIamide, GSPHFIamide, and GSPRFFamide. It is notable that GAPRFVamide was identified both in the gut and the CNS extracts. The five peptides thus isolated from the CNS of Aplysia were apparently homologous to the known MIP-related peptides; they possessed the conserved structure–PXFV/Iamide, characteristic to the MIP family, except for GSPRFFamide. However, the substitution of Phe6 is a conservative change, which in turn means that GSPRFFamide can be regarded as an MIP-related peptide. Therefore, we designated the five peptides AMRPs.
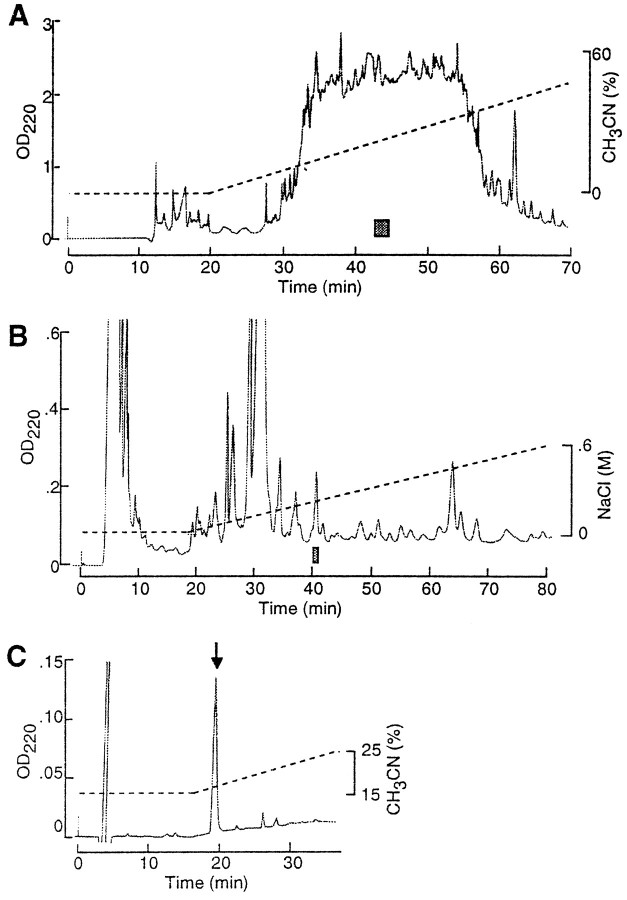
Fig. 1.
Purification of GAPRFVamide from the CNS ofAplysia using immunoassay. A, First separation step of reversed-phase HPLC. The column (Capcell Pak C18; 10 × 250 mm) was developed by 0–60% acetonitrile and 0.1% TFA. A shaded bar indicates the immunoreactive fraction from which GAPRFVamide was purified. B, Second step of cation-exchange HPLC. The column (TSKgel SP-5PW; 7.5 × 75 mm) was eluted with a gradient of 0–0.6 m NaCl and 10 mm sodium phosphate buffer, pH 6.7) A shaded bar indicates the immunoreactive fraction that was subjected to the next HPLC step. C, Final step of reversed-phase HPLC. The column (TSKgel ODS-80TM; 4.6 × 150 mm) was eluted by 15–25% acetonitrile and 0.1% TFA. Arrow indicates a peak of the purified peptide.
Table 1.
Amino acid sequence analysis and FAB-MS for the purified peptides
| Amino acid sequence analysis (pmol)1-a | Measured MW (M+H)+ | Theoretical MW (M+H)+ | Predicted structure |
|---|---|---|---|
| G(499)-A(569)-P(479)-R(274)-F(212)-V(89) | 645.3 | 645.4 | GAPRFVamide |
| G(305)-A(353)-P(273)-R(80)-F(125)-I(31) | 659.3 | 659.4 | GAPRFIamide |
| G(226)-P(217)-P(192)-R(97)-F(68)-I(19) | 685.3 | 685.4 | GPPRFlamide |
| G(434)-S(96)-P(352)-R(121)-F(278)-F(149) | 709.3 | 709.3 | GSPRFFamide |
| G(117)-S(23)-P(63)-H(34)-F(14)-I(+)1-b | 656.2 | 656.3 | GSPHFIamide |
Values in parentheses are amount of PTH amino acids detected in each cycle.
+ means <1 pmol.
Cloning of the Aplysia AMRP precursor mRNA
Because GAPRFVamide was isolated both on the basis of bioactivity and immunoreactivity, we decided to focus on this peptide to clone the precursor. The seminested degenerate PCR yielded several clones which, upstream of the degenerate primer to GAPRFVamide, coded for other peptides in the MIP family. This unequivocally demonstrated to us that we had the correct mRNA species. Sequencing of the cDNA clones isolated by library screening generated a 3262 bp consensus sequence (GenBank accession number AF160191). Northern analysis indicated that the mRNA is ∼3.3 kb (see Fig. 5), which suggests that the sequence is very near to full length and is expressed in the ganglia ofAplysia.
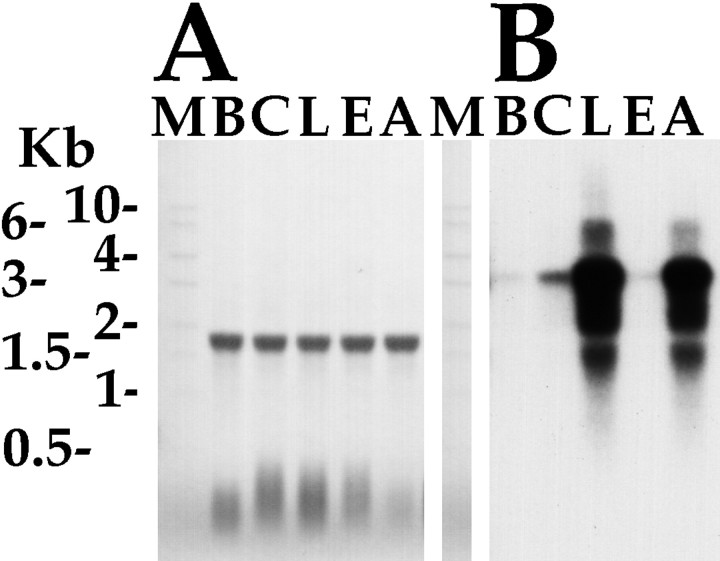
Fig. 5.
Northern analysis of AMRP mRNA. A,Methylene blue-stained Northern blot of total RNA isolated from the five different ganglia of Aplysia californica showing equal loading in all lanes. Aplysia ribosomal RNA runs as a single 18 S band. B, Hybridization of the same blot with AMRP coding sequence probe. Kb, Kilobase;M, RNA size marker; B, buccal ganglion;C, cerebral ganglion; L, pleural ganglion; E, pedal ganglion; A, abdominal ganglion.
The predicted mRNA contained a 2205 bp open reading frame that coded for a 735 amino acid precursor shown in Figure2. The precursor had a predicted hydrophobic signal peptide and a predicted cleavage site between Ser20 and Phe21 (Nielsen et al., 1997), indicating that the protein is targeted to the secretory pathway. There was an unusual glutamine-rich region on the precursor between the signal peptide and the first AMRP. The precursor coded for a total of 26 copies of 14 different predicted amidated peptides as indicated by C-terminal glycines, monobasic, dibasic, and tribasic residues (Eipper et al., 1992; Sediah and Chreiten, 1997). The distribution of these peptides on the precursor is shown in Figure3. At least 21 connecting or linker peptides were also predicted by the precursor, most of which are acidic in nature. In many precursors, these acidic connecting peptides are degraded and have been postulated to compensate for the basic nature of the processing sites. However, some connecting peptides have been demonstrated to survive processing and targeting, and to have bioactivity (Fan et al., 1997). The MALDI-TOF MS data described below indicate that at least some of these connecting peptides are present in the soma of AMRP-containing neurons (see below).
Fig. 2.
Predicted amino acid sequence of the AMRP precursor. Amino acids are numbered at right, and predicted amidated peptides are underlined. Monobasic, dibasic, and tribasic cleavage sites are shown in bold,and an asterisk denotes predicted signal sequence cleavage site (SS-FS).
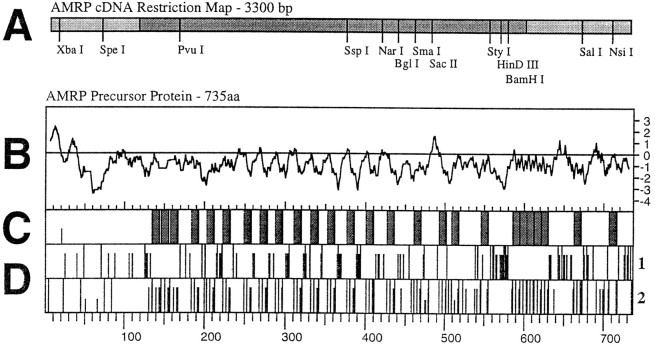
Fig. 3.
Organization of the AMRP precursor.A, Scale drawing of a partial restriction map of the AMRP precursor mRNA. Open reading frame is shown in a darker shade of gray. B–D are scale drawings comparing different aspects of the AMRP precursor protein.B, Kyte–Doolittle hydropathy plot of the AMRP precursor protein. The initial hydrophobic upward deflection denotes the signal peptide. C, The distribution of the predicted amidated peptides shown as gray bars. Connecting peptides are shown as the intervening white regions.Half-height line denotes position of predicted signal peptide cleavage. D, Distribution in the AMRP precursor of acidic (D1) and basic (D2) residues shown as vertical lines. For acidic residues (D1), full height lines represent glutamate residues, and two-thirds-height linesrepresent aspartate residues. For basic residues (D2), full height lines represent arginine residues, two-thirds-heightlines represent lysine residues, andone-third-height lines represent histidine residues. Note acidic nature of the connecting peptides and basic residues flanking the predicted amidated peptides. For comparison, scale is identical in parts B–D with amino acids numbered atbottom.
It is interesting to note that 17 of 26 copies of AMRPs predicted by the precursor are FFamides. This motif is similar to the recently discovered putative μ opiate receptor ligand neuropeptide endomorphin 2 (YPFFamide; Zadina et al., 1997). Furthermore, there are two AMRPs, SDPFFMamide and GAPRFLamide, of which the two C-terminal amino acids are identical to Met (YGGFM) and Leu (YGGFL) enkephalin, respectively (Harrison et al., 1998).
Processing of the AMRP precursor
Five of the amidated peptides predicted by the AMRP precursor have been biochemically isolated and sequenced, thus confirming that they are in fact made. The other nine peptides predicted by the precursor needed confirmation that they are in fact processed out from the precursor as predicted. This is especially true because some of the C-terminal single arginine processing sites do not fall cleanly into the known consensus sites (Sediah and Chreiten, 1997) K/R-Xn-K/R-cut where n = 0, 2, 4, 6, and 8 (i.e., GAAPKFFamide,n = 3; AAPRFFamide, n = 9, 10, 11). For this purpose, we used MALDI-TOF MS, which has been shown to be an excellent method for identification of peptide products of gene expression and post-translational processing (Jimenez et al., 1994;Garden et al., 1998; Li et al., 1998; Worster et al., 1998).
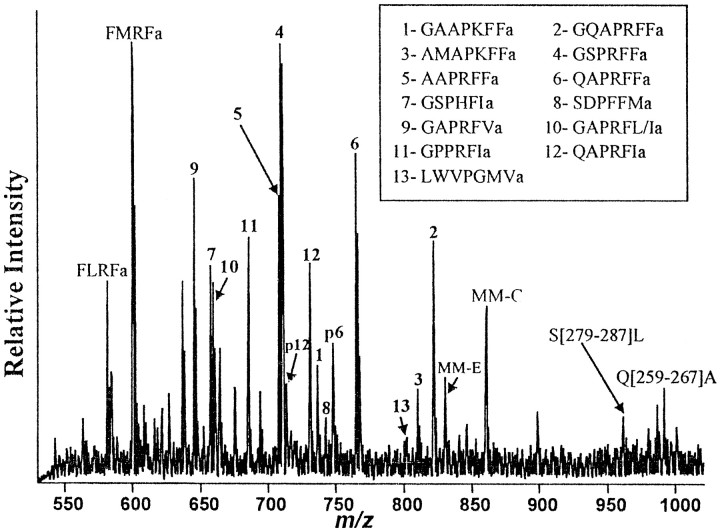
Figure 4 shows a representative mass spectrum obtained from a small group of cells (∼5–8 cells) in the right pleural ganglion close to the cerebropleural connective, which had been indicated to contain AMRPs by in situ hybridization and immunostaining (see below). FMRFamide and MM-C detected in the spectrum (Fig.4) were used as internal calibrants to provide improved mass accuracy for AMRPs. As clearly shown in Figure 4, molecular ion peaks corresponding to each of the 14 putative amidated peptides were observed, confirming the synthesis of all the 14 predicted peptides on the AMRP precursor. Furthermore, two peptides (QAPRFIamide and QAPRFFamide) contain an N-terminal Gln, and hence can form pyroglutamate (pGlu) forms. Both native and pGlu forms of these peptides were detected. The peptides GAPRFIamide and GAPRFLamide have the same molecular mass peak (as Ile and Leu are isomers), which was observed. In addition, the amidated peptides with n = 3 and n = 9, 10, 11 cleavage sites (see above) were found to be processed from the precursor. Table2 shows the mass accuracy of measurements in both cellular samples and HPLC fractions. The average error of measurements for both types of samples are 20 and 14 ppm, respectively, excellent compared to previous cell work (Jimenez et al., 1994; Garden et al., 1998; Li et al., 1998).
Fig. 4.
MALDI-TOF MS in single neurons of the pleural ganglion. A mass spectrum in the 550–1000 Da range from isolated pleural neurons. Assigned peaks are labeled with corresponding peptides. Peptides derived from three precursors, FMRFamide, myomodulin, and AMRP are detected. The inset box shows the numbering scheme used for the AMRP peptides. Pindicates a pGlu-modified form of the two AMRPs containing N-terminal Gln. Connecting peptide S[279–287]L is SDDNVALDL, and Q[259–267]A is QDDDIMIAA.
Table 2.
Detection of amidated peptides encoded by AMRP gene
| Peptide | [M+H]+Calcd | Cellular samples (R. pleural cells) | LC fractions (abdominal ganglion homogenate) | Confirmed with PSD | Isolated and sequenced | ||||
|---|---|---|---|---|---|---|---|---|---|
| [M+H]+Obsvd | % error | ppm | [M+H]+Obsvd | % error | ppm | ||||
| GAPRFVa | 645.38 | 645.367 | 0.002 | 20 | 645.400 | 0.0025 | 25 | Yes | Yes |
| GSPHFIa | 656.33 | 656.346 | 0.0024 | 24 | 656.334 | 0.00061 | 6.1 | Yes | |
| GAPRFI/La | 659.40 | 659.37 | 0.0045 | 45 | 659.384 | 0.0024 | 24 | Yes | |
| GPPRFIa | 685.39 | 685.408 | 0.0026 | 26 | 685.409 | 0.0028 | 28 | Yes | Yes |
| AAPRFFa | 707.40 | 707.398 | 0.0003 | 2.8 | 707.393 | 0.00099 | 9.9 | ||
| GSPRFFa | 709.38 | 709.374 | 0.0008 | 8.4 | 709.377 | 0.00042 | 4.2 | Yes | Yes |
| pQAPRFIa | 713.41 | 713.40 | 0.0014 | 14 | |||||
| QAPRFIa | 730.44 | 730.422 | 0.0025 | 25 | Partial | ||||
| GAAPKFFa | 736.40 | 736.377 | 0.0031 | 31 | |||||
| SDPFFMa | 742.32 | 742.33 | 0.0013 | 13 | |||||
| pQAPRFFa | 747.39 | 747.37 | 0.0027 | 27 | |||||
| QAPRFFa | 764.42 | 764.415 | 0.0006 | 6.5 | 764.41 | 0.0013 | 13 | Partial | |
| LWVPGMVa | 800.45 | 800.43 | 0.0025 | 25 | |||||
| AMAPKFFa | 810.43 | 810.417 | 0.0016 | 16 | |||||
| GQAPRFFa | 821.44 | 821.428 | 0.0015 | 15 | 821.435 | 0.00061 | 6.1 | ||
In addition to the amidated AMRP peptides, a number of presumed linker peptides are predicted from the prohormone, with most occurring at higher molecular masses. In several cases in gastropod molluscs, such peptides are rapidly degraded, and some of these intermediates can be detected with MALDI MS (de With et al., 1997; Garden et al., 1998). Several of these linker peptides predicted by the prohormone were also detected in the spectra (data not shown). Table3 lists several of the putative linker peptides cleaved between dibasic sites that we detected in these cell samples. It remains to be determined whether these peptides were in the process of being degraded or if they are retained and possess bioactivity.
Table 3.
Detection of linker peptides encoded by AMRP gene
| Linker peptides | [M+H]+Calcd | [M+H]+Obsvd | % error | ppm |
|---|---|---|---|---|
| S[279–287]L | 961.45 | 961.48 | 0.0031 | 31 |
| SDDNVALDL | ||||
| Q[259–267]A | 991.44 | 991.49 | 0.005 | 50 |
| QDDDIMIAA | ||||
| S[412–426]T | 1461.67 | 1461.62 | 0.0034 | 34 |
| SDGLDDGGNIIDVAT | ||||
| F[214–229]F | 1939.91 | 1939.90 | 0.0005 | 5.2 |
| D[632–661]V | 3390.67 | 3390.33 | 0.01 | 99 (Linear mode) |
| G[674–705]E | 3687.14 | 3687.27 | 0.0035 | 35 |
To confirm our assignments, primary structural information of a peptide detected with MALDI-MS can be obtained using PSD analysis (Kaufmann et al., 1993; Spengler, 1997). Because PSD analysis requires both more concentrated samples and a greater amount of samples than those generally provided by a cell or cells, multiple ganglia were pooled, homogenized, and separated using HPLC. PSD analysis was performed on several LC fractions to obtain full or partial sequence information from the most intense putative AMRP peptide peaks. We were able to use PSD (data not shown) to confirm that GAPRFVa, GPPRFIa, and GSPRFFa are authentic AMRPs, as predicted by the gene, and partially confirm (i.e., a partial sequence obtained) for QAPRFIa and QAPRFFa. Taken together, the simultaneous detection of all predicted amidated peptides and several putative linker peptides encoded by the AMRP gene in a single group of cells and sequencing five of them using PSD both confirmed the gene sequence and verified its expression in theAplysia CNS. Because not all the neurons expressing the AMRPs in the CNS have been analyzed, the possibility that not all of these neurons express all 14 AMRPs cannot be ruled out.
Distribution of AMRP-containing neurons and their processes in Aplysia
Cross-correlation of the results from Northern analysis, in situ hybridization, and immunocytochemistry was used to map AMRP-containing neurons and their processes. For selected identifiable neurons, MALDI-TOF MS was also used to confirm the presence of AMRPs. Each of these methods has its own strengths and limitations, which when combined, can increase the validity of mapping.
Northern analysis was performed to identify the length and overall distribution of the mRNA that codes for the AMRP precursor. The Northern blot was hybridized with a random primer32P-labeled a probe using a template corresponding to the peptide-coding region (∼1600–2300) of the sequence. Figure 5 shows that the mRNA coding for the AMRPs is a single band ∼3.3 kb in length that corresponds well with the size of longest clones isolated from the library. The mRNA has highest levels in the pleural ganglion, with somewhat lesser amount in the abdominal ganglion. The cerebral, buccal, and pedal have relatively low amounts of the AMRP precursor mRNA with approximately cerebral > buccal > pedal. The methylene blue staining of the ribosomal RNA shows equal loading in all the lanes. The relative amounts of mRNA in the different ganglia serve as a benchmark to assess the validity of the in situ hybridization and immunohistochemistry results.
In situ hybridization was performed to get a more precise distribution of the mRNA that codes for the AMRP precursor. Several controls were used to assess the validity of the in situhybridization results. First, hybridization with a control labeled oligo resulted in the absence of staining. Second, using two different antisense oligos directed against different parts of the AMRP mRNA sequence gave identical staining patterns (GAP 3a, TGC TGA CTC ACC AGA CGA CTT; GAP-ISH, CCA AAA AAT CTG GGY GAA CCT C; data not shown). One of these oligos (GAP-ISH) was designed against repetitive sequences in the AMRP mRNA, thus hybridizing multiply, and consequently giving stronger signals. The GAP-ISH oligo was subsequently used in all the analyses. Third, the distribution of in situhybridization-positive neurons can be correlated to the distribution of immunopositive neurons (see below). The distribution of the mRNA as shown by in situ hybridization is consistent with the results of the Northern analysis. The highest density of in situ hybridization-positive neurons was observed in the pleural and abdominal ganglia, with lower densities observed in the cerebral, buccal, and pedal ganglia. We also observed in situhybridization-positive neurons in the crop, filtering chamber, and in the stomatogastric ring.
Immunocytochemistry was used to map the AMRP-synthesizing neurons and their processes. Several controls were used to assess the validity of the AMRP immunostaining. First, the abolition of immunostaining by preadsorption of the antibody with peptide was confirmed. Second, the correlation of the amount of immunostaining in the different ganglia with the amount of mRNA as detected by Northern analysis was verified. Third, the correlation of the immunopositive neurons with in situ hybridization-positive neurons was confirmed. Initial experiments using the antibody to GSPMFVamide (the original MIP) immunostained more neurons than predicted from Northern andin situ results (e.g., many immunopositive neurons in the pedal ganglion), suggesting that additional cDNAs code for immunologically similar peptides. This result also necessitated that a more specific antibody be made, and GSPRFFamide was chosen as the antigen because it exists in 11 copies on the precursor. Of the two antibodies made, the EDC-coupled antigen gave results that correlated the best with Northern and in situ results and thus was used for all subsequent experiments. In addition, preadsorption of this antibody with 10−4m GSPRFFamide completely abolished immunostaining (data not shown).
The ganglia from both large (100–300 gm) and small (10–15 gm) animals were used for in situ hybridization and immunostaining with the AMRP antibody. The ganglia from small (10–15 gm) animals are shown in Figures 7-9, except where noted. There was some variability in the number and size of neurons staining in different animals, even in the same weight range. What we present are the typical results from both large and small animals. A diagram summarizing the distribution of AMRP-positive neurons in each ganglion represents the correlated results of in situ hybridization and immunocytochemistry. Locations of the nerves in the drawings are intended as landmarks, the relative positions of the neurons and nerves vary somewhat from animal to animal and depending on how the ganglia are pinned. To avoid redundancy, neurons that were observed to be both immunopositive andin situ hybridization-positive are referred to as AMRP-positive below. Because in situ hybridization cannot be used to define processes of neurons, cross-correlation is not possible. It is likely that immunostaining of processes reflects the presence of bona fide AMRPs because of the excellent cross-correlation of in situ hybridization and immunostaining of neuronal cell bodies.
Fig. 7.
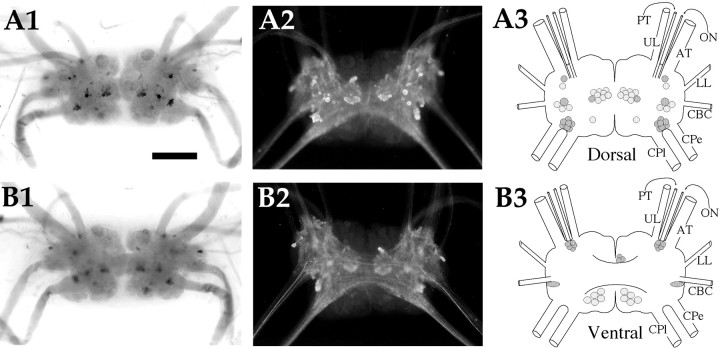
AMRP neurons in the cerebral ganglion.A1,In situ hybridization of dorsal surface. A2, Immunocytochemistry of dorsal surface.A3, Drawing of the AMRP neurons on the dorsal cerebral ganglion. B1,In situ hybridization of ventral surface. B2, Immunocytochemistry of ventral surface. B3, Drawing of the AMRP neurons on the ventral cerebral ganglion. UL, Upper labial nerve;PT, posterior tentacular nerve; ON, optic nerve; AT, anterior tentacular nerve; LL,lower labial nerve; CBC, cerebrobuccal connective;CPe, cerebropedal connective; CPl,cerebropleural connective. Scale bar: A1, 500 μm (same in all panels).
Fig. 8.
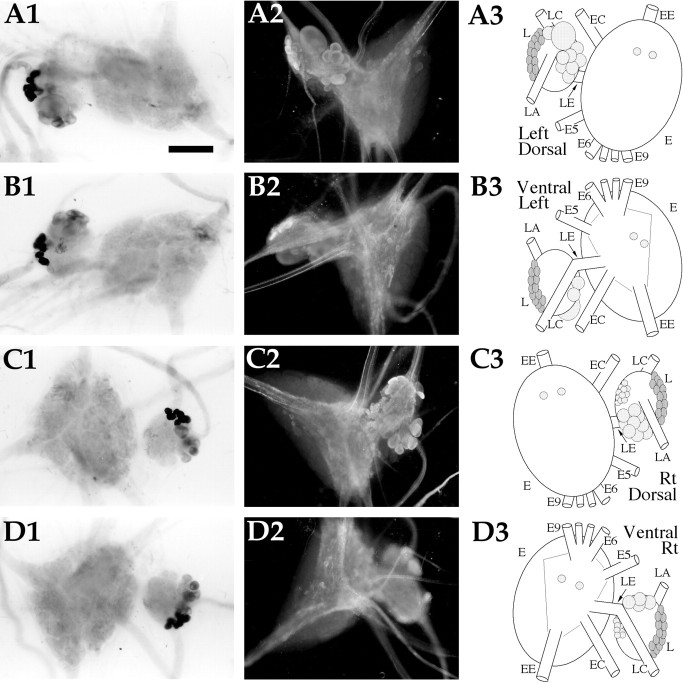
AMRP neurons in the pleural and pedal ganglia.A1,In situ hybridization of left ganglion pair dorsal surface. A2,Immunocytochemistry of the left ganglion pair dorsal surface.A3, Drawing of the AMRP neurons on the dorsal surface of the left ganglion pair. B1,In situhybridization of left ganglion pair ventral surface. B2,Immunocytochemistry of the left ganglion pair ventral surface.B3, Drawing of the AMRP neurons on the ventral surface of the left ganglion pair. C1,In situhybridization of right ganglion pair dorsal surface. C2,Immunocytochemistry of the right ganglion pair dorsal surface.C3, Drawing of the AMRP neurons on the dorsal surface of the right ganglion pair. D1,In situhybridization of right ganglion pair ventral surface.D2, Immunocytochemistry of the right ganglion pair ventral surface. D3, Drawing of the AMRP neurons on the ventral surface of the right ganglion pair. L, Pleural ganglion; E, pedal ganglion; LE,pleuropedal connective; EE, pedal commissure;EC, cerebropedal connective; LC,cerebropleural connective; LA, pleuroabdominal connective; E5, posterior tegumentary nerve (P5);E6, anterior parapodial nerve (P6); E9,posterior pedal nerve (P9). Not all nerves are drawn for simplicity. Scale bar: A1, 500 μm (same in all panels).
Fig. 9.
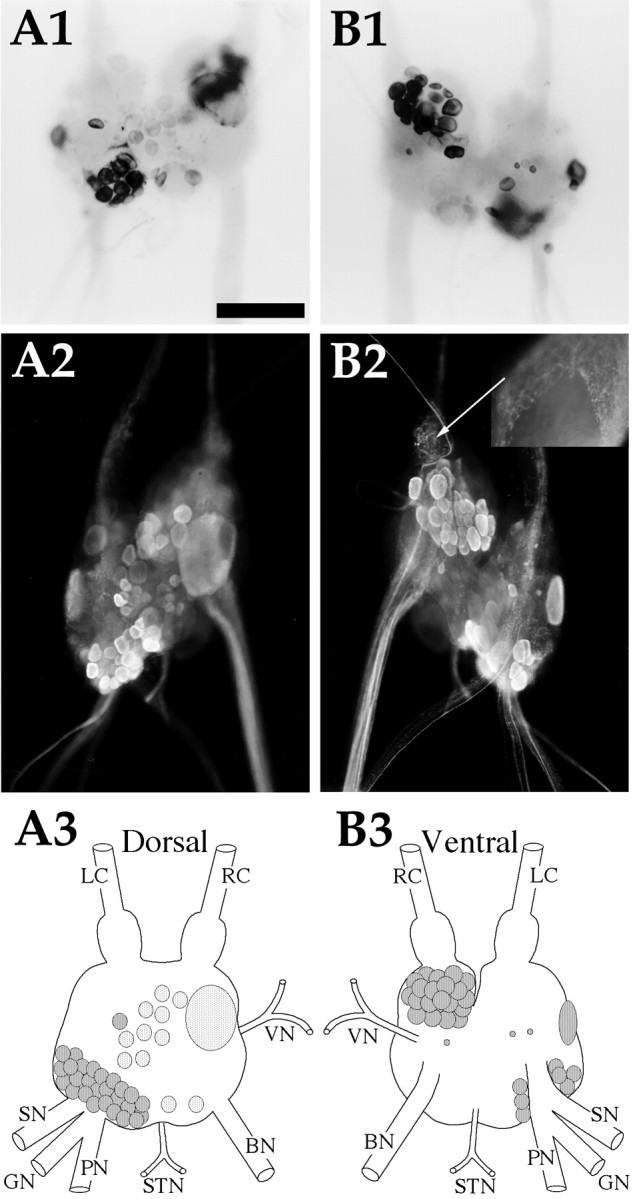
AMRP neurons in the abdominal ganglion.A1,In situ hybridization of dorsal surface. A2, Immunocytochemistry of dorsal surface.A3, Drawing of the AMRP neurons on the dorsal abdominal ganglion. B1,In situ hybridization of ventral surface. B2, Immunocytochemistry of ventral surface. Inset is the same region asarrow, partially desheathed bag cells from an adult animal showing dense immunopositive innervation of the sheath overlying the bag cells, but not the bag cells themselves. B3,Drawing of the AMRP neurons on the ventral abdominal ganglion.LC, Left pleuroabdominal connective; RC,right pleuroabdominal connective; VN, vulvar nerve;BN, branchial nerve; STN, spermathecal nerve; PN, pericardial nerve; GN, genital nerve; SN, siphon nerve. Scale bar: A1,500 μm (same in all panels, includinginset).
Buccal ganglion (Fig. 6)
Fig. 6.
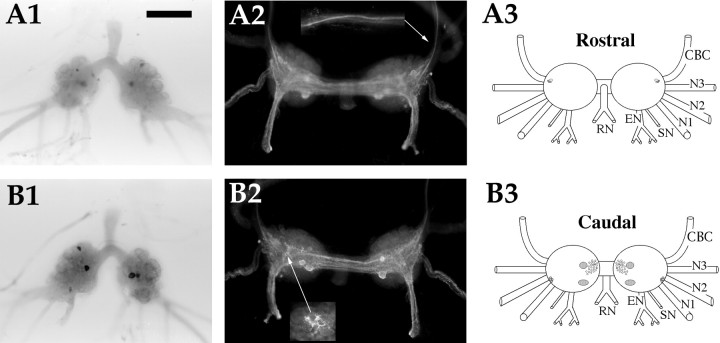
AMRP neurons in the buccal ganglion.A1,In situ hybridization of rostral surface. A2, Immunocytochemistry of rostral surface.Inset is same region as arrow from an adult animal showing immunoreactive axons in the CBC.A3, Drawing of the AMRP neurons on the rostral buccal ganglion. B1,In situ hybridization of caudal surface. B2, Immunocytochemistry of caudal surface. Inset is the same region asarrow in an adult animal showing dense innervation of the B1–B2 cluster. B3, Drawing of the AMRP neurons on the caudobuccal ganglion. CBC, Cerebrobuccal connective;N1, nerve 1 (B4); N2, nerve 2 (B5);N3, nerve 3 (B6); SN, salivary nerve (B3); EN, esophageal nerve (B2); RN,radula nerve (B1). Scale bar: A1, 500 μm (same in all panels, including insets).
Two intensely AMRP-positive neurons were observed in each hemiganglion. The neurons were typically present on the dorsocaudal aspect in the region of the sensory neurons. Dense immunostaining was observed in the neuropil of the buccal ganglion, particularly in the region of the B1 B2 cluster (Fig. 6B2, inset). These immunostained processes seemed to envelop the proximal axons of the neurons in this cluster. In the buccal nerves, a large number of immunostained axons were observed in the esophageal nerve. Several immunostained axons were also observed in the cerebrobuccal connective (Fig. 6A2, inset). Typically, only a few immunostained axons were observed in nerve 1, and none were observed in nerve 2, nerve 3, and radular nerve.
Cerebral ganglion (Fig. 7)
On the dorsal surface (Fig. 7A1–3), there were two clusters (20–30 neurons each) of 20–50 μm AMRP-positive neurons in each hemiganglion. One of these was in the F cluster [nomenclature after Jahan-Parwar and Fredman (1976)], the other was in the A cluster near the base of the cerebropedal connective. The A cluster AMRP-positive neurons ventralized in adult animals, although they were still located on top of the cerebropleural connective. A third cluster of very small (10–20 μm) AMRP-positive neurons was located in the M cluster between the upper labial and anterior tentacular nerves. In addition, there were two larger (50–100 μm) intensely staining AMRP-positive neurons in each dorsal hemiganglion, one in the D cluster, the other between the D and E clusters. On the ventral surface (Fig. 7B1–3), a strongly staining AMRP-positive neuron was observed in the E cluster at the base of the cerebrobuccal connective. Dense immunostained processes were seen throughout the neuropil of the cerebral ganglion, and immunostained axons were observed in all the cerebral nerves. A particularly high density of immunostained axons was observed in the cerebropleural connective, and a particularly large, intensely staining axon was observed in the cerebropedal connective. This fiber could be traced into the cerebral neuropil and was observed to cross to the opposite hemiganglion via the cerebral commissure.
Pleural ganglion (Fig. 8)
The highest concentration of AMRP-positive neurons were in the pleural ganglia, with right and left pleural ganglia showing different staining patterns. In the left pleural ganglion, the giant cell (LP1) and several large neurons (150–200 μm) in a cluster around the pleuroabdominal connective were AMRP-positive. This cluster extended along the anterior part of the ganglion on both the dorsal and ventral surfaces. In the right pleural ganglion, a similar cluster of AMRP-positive neurons (100–200 μm) was observed in the anterior part of the ganglion between the pleuroabdominal connective and the pleurocerebral connective. This cluster also extended from the dorsal to the ventral aspect of the anterior part of the ganglion, but contained more smaller AMRP-positive neurons (100 μm). These neurons were examined by MALDI-TOF analysis to independently confirm that they contain the AMRPs. The dorsal surface of right pleural ganglion contained a large number (∼50) of smaller (50–80 μm) AMRP-positive neurons near the pleuropedal connective. Immunostained processes were observed in the neuropil of both ganglia, and immunostained axons were observed in all the pleural nerves.
Pedal ganglion (Fig. 8)
The pedal ganglion had the lowest concentration of AMRP-positive neurons. Typically only one or two AMRP-positive neurons were observed on the ventral surface near the pleuropedal connective. Dense immunostained processes in the pedal neuropil and immunostained axons could be observed in most of the pedal nerves.
Abdominal ganglion (Fig. 9)
The abdominal ganglion also showed a high concentration of AMRP-positive neurons. On the dorsal surface (Fig. 9A1–3), the giant neuron R2 and several smaller neurons in its immediate vicinity were AMRP-positive. In the left hemiganglion, a cluster of AMRP-positive neurons is located at the base of the genital pericardial and siphon nerves. This cluster extended around the medial aspect of the left ganglion dorsally to almost encircle the genital pericardial nerve. Most of these neurons were of medium size, ∼100 μm, with a single larger (∼300 μm) neuron at the ventrolateral aspect of this cluster. There was also a strongly staining AMRP-positive cluster of 100–200 μm neurons in the right ventral hemiganglion located near the pleuroabdominal connective (Fig. 9B1–3). MALDI-TOF MS of neurons from these regions confirms the presence of all the AMRPs (data not shown). Numerous immunostained axons were observed in all the nerves exiting the abdominal ganglion. The sheath overlying the bag cells showed a dense immunostained innervation, but no immunostained processes were observed on the bag cell bodies themselves (Fig.9B2, inset). This innervation suggests that AMRPs may affect the bag cells and/or their processes.
Body wall (Fig.10A)
Fig. 10.
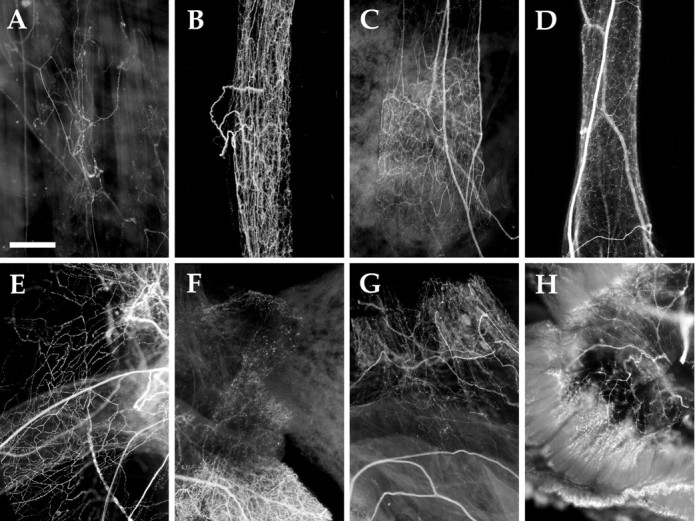
AMRP immunocytochemistry in cardiovascular system and other peripheral tissues. A, Body wall musculature (200 gm animal). B, Anterior aorta (10 gm animal).C, Abdominal aorta (10 gm animal). D,Gastrointestinal artery (10 gm animal). E, Pericardium (10 gm animal). F, Heart valves with heart atright, crista aortae at left, and abdominal aorta at bottom (10 gm animal).G, Crista aortae at bottom and pericardium (10 gm animal). H, Gill (10 gm animal). Scale bar: A, 500 μm (same in all panels).
Sparse immunopositive innervation was observed in the body wall, but none was observed in the foot. The immunopositive innervation was not uniformly distributed throughout the body wall, but was concentrated in restricted areas. Immunopositive axons were also observed in many of the nerves innervating the body wall.
Buccal mass
The buccal mass is a collection of muscles that control the movements of the radula, a cartilaginous structure that, in turn, controls the movement of food into the animal. We found no immunopositive innervation in either the entire buccal mass from a 10 gm animal or selected muscles from larger animals (data not shown). This suggests that none of the motor or sensory neurons that innervate the buccal musculature express these neuropeptides. The buccal ganglion innervates the buccal mass, and immunopositive axons were observed only in nerves that innervate the region of the esophagus which, as described below, is strongly immunopositively innervated.
Circulatory and respiratory systems (Fig. 10B–G)
Aplysia has an open circulatory system containing a single atrium and ventricle (Kandel, 1979; Skelton et al., 1992). Three major arteries, the anterior, the gastrointestinal, and the abdominal arteries, direct blood flow to different parts of the animal. In the cardiovascular system, no immunoreactivity was observed in the heart. In contrast, dense immunoreactive innervation was evident in pericardium as well as all the major arteries. The innervation was observed in the proximal arterial system, and was not evident in distal, smaller branches of the arterial system. Because most of the neurons responsible for cardiovascular control reside in the abdominal ganglion, it is reasonable to assume that the AMRP-immunopositive neurons innervating the major arteries reside in the abdominal ganglion. Immunopositive innervation was also detected in the gill (Fig. 10H) and kidney (data not shown) of the animal, both of which are heavily vascularized. The dense proximal immunopositive innervation of the proximal arterial system suggests that the AMRPs may be involved in vascular tone and redirection of blood flow (Skelton et al., 1992).
Reproductive system (Fig.11A–H)
Fig. 11.
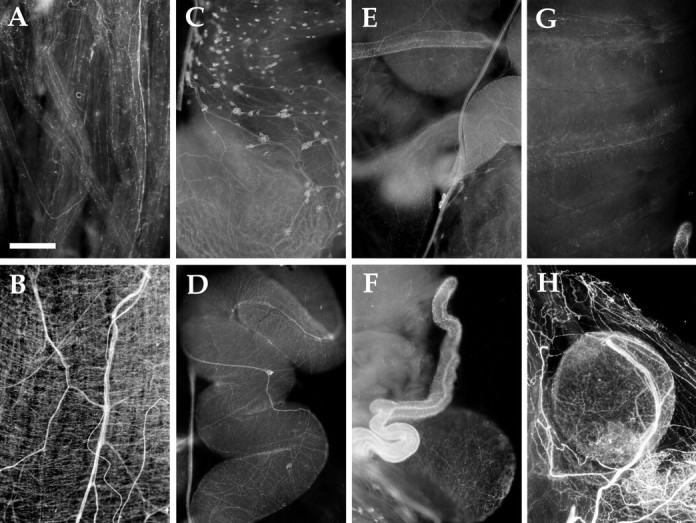
AMRP immunostaining in the reproductive system.A, Penis retractor muscle. B, Penis sheath. C, Large hermaphroditic duct. D,Small hermaphroditic duct. E, Junction of small hermaphroditic duct (top right), large hermaphroditic duct (bottom right), seminal receptacle (bottom left), and accessory genital mass (top left).F, As in E but viewed from the other side with seminal receptacle more in focus on bottom right.G, Accessory genital mass. H, Spermatheca (gametolytic gland) surrounded by pericardium. All panels from sexually mature 100 gm animal. Scale bar: A, 500 μm (same in all panels).
Aplysia is a hermaphrodite containing both male and female reproductive organs (Blankenship et al., 1977; Kandel, 1979;Painter et al., 1985). In the male reproductive system, the penis is housed inside the animal, but is everted during copulation. The position of the penis is controlled by two extrinsic penis retractor muscles, extrinsic penis extensor muscles, and longitudinal muscles in the penis sheath. Immunopositive innervation was detected in all these muscles, but was particularly dense in the penis sheath (Fig.11A,B). Occasionally, small (10–20 μm) immunopositive neurons could be seen along the nerve running adjacent to the penis sheath.
The female portion of the reproductive system of the animal is also densely immunopositively innervated (Fig. 11C–H). In addition, numerous immunopositive neuronal soma were observed. Approximately 50–100 immunopositive neurons were observed in the large hermaphroditic duct (Fig. 11C), and 10–20 immunopositive neurons were observed in the small hermaphroditic duct (Fig.11D) (near the entry point to the accessory genital mass). The small hermaphroditic duct was densely innervated with immunopositive fibers, especially near the entry point to the accessory genital mass. These fibers were radial in appearance, forming rib-like structures. Immunopositive innervation could also be seen in the accessory genital mass (Fig. 11G), spermatheca (also called the gametolytic gland; Fig. 11H), seminal receptacle (Fig. 11F), and large hermaphroditic duct. The far-ranging innervation of AMRP-immunopositive neurons throughout the reproductive system suggests that these peptides have important roles in its function. Additional effects of these peptides on reproduction may be exerted on the bag cells and/or their processes in the abdominal ganglion.
Digestive system (Fig. 12)
Fig. 12.
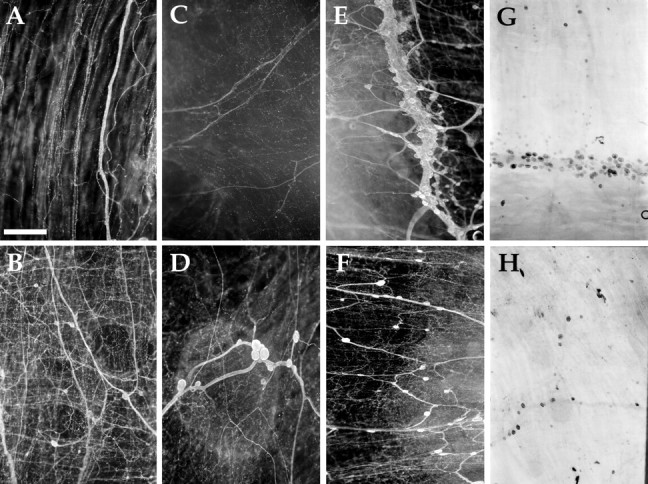
AMRP immunostaining and in situhybridization in the digestive system. A, Esophagus immunostaining (10 gm animal). B, Crop immunostaining (10 gm animal). C, Triturating stomach immunostaining (10 gm animal). D, Filtering chamber immunostaining (200 gm animal). E, Stomatogastric ring immunostaining (10 gm animal). F, Filtering chamber immunostaining (10 gm animal). G, Stomatogastric ring in situ(10 gm animal). H, Filtering chamber in situ (10 gm animal). Scale bar: A, 500 μm (same in all panels).
Of all the peripheral tissues examined immunohistochemically, the most striking pattern was observed in the anterior digestive tract. The digestive tract of Aplysia consists of the esophagus, the crop, the triturating stomach (anterior gizzard), the filtering chamber (posterior gizzard), intestine (true stomach), and the rectum (Kandel, 1979; Lloyd et al., 1988). Immunostained enteric neurons were observed in a ring-like structure, which we are calling the stomatogastric ring, at the junction of the crop and triturating stomach (Fig.12E). Small cardioactive peptide (SCP)-immunostaining neurons were previously reported in this structure (Lloyd et al., 1988). Additional immunopositive neurons were observed throughout the filtering chamber and posterior crop (Fig.12B,D,F). Few immunopositive enteric neurons were detected in the triturating stomach, and no immunopositive neurons were observed in the esophagus (Fig. 12A). In situ hybridization-positive neurons were detected with the same distribution in these structures, confirming the specificity of the immunostaining (Fig. 12G,H). A dense network of immunopositive fibers was observed throughout the anterior digestive tract, including the esophagus (Fig. 12A), crop (Fig.12B), triturating stomach (Fig. 12C), and filtering chamber (Fig. 12D,F). In contrast, the posterior part of the digestive tract, which is embedded in the hepatopancreas (including the true stomach, intestine, and cecum) is virtually devoid of immunostained neurons and processes (data not shown). The extensive innervation of the anterior part of digestive tract by AMRP-immunopositive neurons suggests that these peptides are exerting important effects in its function.
Physiological actions of AMRPs on Aplysia muscles
To obtain more physiological evidence that the peptides act as signaling molecules, we examined effects of the peptides on mechanical responses of some of the immunopositive tissues, including the esophagus, penis retractor muscle, and body wall muscle in A. kurodai.
Esophagus
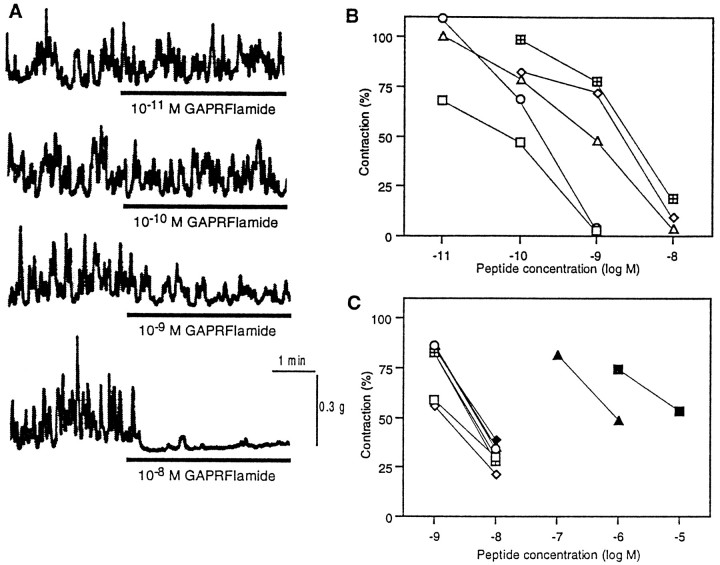
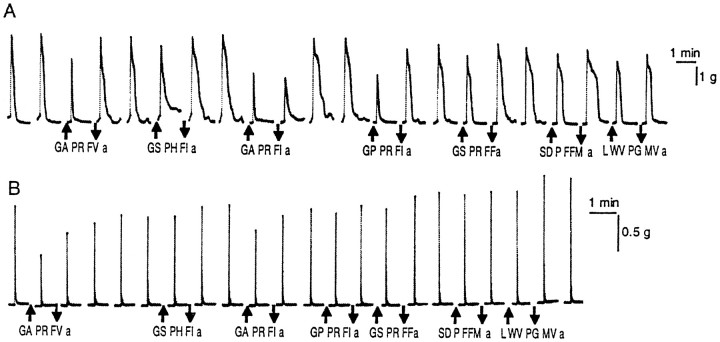
The isolated esophagus showed complex pattern of contractions for more than several hours. Application of AMRPs caused significant suppression of the spontaneous contraction of the esophagus in a dose-dependent, reversible manner (Fig.13). Figure 13A shows a representative suppression by GAPRFIamide. It is notable that the effects of the five isoforms that were biochemically isolated from the CNS were qualitatively similar, but the potency was slightly different from each other (Fig. 13B). The most potent isoform was GAPRFVamide, and the least potent one was GSPRFFamide; the potency difference between the two peptides was about one order of magnitude. Subsequently, another nine peptides that were predicted from the cDNA sequence were chemically synthesized, and their effects were examined. Seven of the nine showed inhibitory effects almost comparable to the above five purified peptides (Fig. 13C). However, LWVPGMVamide and SDPFFMamide were over two orders of magnitude less potent than the above seven peptides (Fig. 13C).
Fig. 13.
Effect of AMRPs on contractions of the esophagus of Aplysia. A, Dose-dependent inhibition of the esophagus motility by GAPRFIamide. B, Dose–response relationship for the five purified AMRPs. ■, GAPRFVamide; ○, GAPRFIamide; ▵, GPPRFIamide; ⋄, GSPHFIamide; ⊞, GSPRFFamide.C, Dose–response relationship for the nine predicted AMRPs. ■, GAAPKFFamide; ⋄, GQAPRFIamide; ○, AMAPKFFamide; ▵, AAPRFFamide; ⊞, QAPRFFamide, ▪, SDPFFMamide; ♦, GAPRFLamide; ●, QAPRFIamide; ▴, LWVPGMVamide. Note that LWGPGMamide and SDPFFMamide were much less potent than the others.
Penis retractor muscle
Figure 14A shows the inhibitory effect of the AMRPs on electrically induced tetanic contraction of the penis retractor muscle at 10−8m. The order of potency was different from that in the case of the esophagus; GAPRFIamide and GPPRFIamide were more potent than the others. SDPFFMamide and LWVPGMVamide, which were much less effective than the other AMRPs on the esophagus, showed significant inhibition comparable to one of the authentic isoforms, GSPHFIamide and GSPRFFamide.
Fig. 14.
Effect of AMRPs on contractions of other muscles in Aplysia. A, Effect on electrically induced contraction of the penis retractor muscle. Repetitive electrical pulses (20 V, 1 msec, 40 Hz, 40 pulses) were applied to the muscle at 10 min intervals. Peptides (10−8m) were applied for 9 min. B, Effect on electrically induced contraction of the dorsal longitudinal muscle of the body wall. Electrical pulses (20 V, 2 msec, 100 Hz, 20 pulses) were applied at 10 min intervals. Peptides (10−7m were applied for 9 min.
Body wall muscle
Contraction of the body wall muscle was also suppressed by the AMRPs (Fig. 14B). GAPRFVamide was again most effective among the isoforms tested. However, the order of potency of each peptide was different from those in the cases of the other muscles, and GSPHFIamide and LWVPGMVamide were ineffective at 10−7m.
DISCUSSION
In this paper, we describe the isolation of five peptides from the opisthobranch mollusc Aplysia kurodai. They were homologous to the MIP-family peptides and thus named AMRPs. Based on the sequence of these peptides, we cloned a cDNA encoding the AMRP precursor protein from a closely related species, Aplysia californica. This is the first report on the MIP-related peptides in the opisthobranch gastropods and the first demonstration of the precursor structure for the MIP-related peptides. The precursor contained 26 copies of 14 different amidated AMRPs, including the five purified peptides. The expression of these AMRPs was confirmed by MALDI-TOF MS. In addition, two of the AMRP peptides are N-terminally modified to pyroglutamate. Amidation is often indicative of bioactive peptides. In fact, the C-terminal amidation is required for bioactivity of a number of different peptides (Eipper et al., 1992). Mass spectra from the pleural neurons also indicated the presence of some of the connecting peptides predicted by the precursor.
In addition to sharing structural features with MIPs, the AMRPs share some structural and functional features with the vertebrate opioid peptides. In particular, 17 of 26 AMRPs end in PXFFamide (where X is a basic residue), which is similar to the recently discovered mammalian putative μ opiate receptor ligand neuropeptide endomorphin 2 (YPFFamide; Zadina et al., 1997). Furthermore, two other AMRPs, SDPFFMamide and GAPRFLamide share the same two C-terminal amino acids as Met (YGGFM) and Leu (YGGFL) enkephalin, respectively (Harrison et al., 1998). There have been several reports of Leu and Met enkephalin-like peptides inAplysia based on immunocytology, ligand binding, and physiological responses, but no amino acid sequences for these peptides in Aplysia have been reported (Lukowiak et al., 1982; Leung et al., 1986; Takayanagi and Takeda, 1988; Carpenter et al., 1995). The AMRPs may be some of the endogenous ligands inAplysia that account for these observations. For example, one of the AMRPS, SDPFFMamide, may be an endogenous ligand for the high-affinity binding site ofd-Ala2-Met-enkephalinamide (YdAGFMamide) that has been described (Carpenter et al., 1995). The AMRPs also share some functional features with opioid peptides in mammals. These include an inhibitory action on contractions of the digestive tract and presence in the reproductive system. In addition, the AMRPs are expressed at high levels in the pleural and abdominal ganglia, which contain the neurons controlling respiration and cardiovascular regulation, well known targets of opiate actions in mammals (Olson et al., 1998). Because the precursor to endomorphin-2 has not yet been published, it remains to be determined whether the similarities between AMRPs and these opioid peptides is functional or merely coincidental.
The bioactivity of MIPs in other species has, in general, been found to be inhibitory (Yongsiri et al., 1989; Kiss, 1990; Li et al., 1996; Kiss and Osipenko, 1997). We found that the AMRPs were also inhibitory on three Aplysia target muscles. Two important points emerge from these studies. First, the different peptides on the AMRP precursor have different dose–response relationships on one target. Differences in potency of different peptides arising from the same precursor has been documented in other systems (Cropper et al., 1994; Vilim et al., 1994; Brezina et al., 1995; van Golen et al., 1996; Hewes et al., 1998;Perry et al., 1998). Curiously, some of the structurally related peptides derived from a single precursor appear to lack bioactivity altogether when examined on single targets. Second, the rank order of the potency of different AMRPs is different among distinct targets. Different rank orders of potencies of the peptide products of a single precursor on different targets has been shown for allostatins in insects (Bendena et al., 1997) and for POMC in mammals. However, these peptides are released into the blood, thus are more endocrine in function, and their targets are more diverse. Here we show that for similar targets, the modulatory peptide products derived from a single precursor show differing rank orders of potencies.
Our results suggest that the different peptides produced from a single precursor are not simply functionally redundant. For example, peptides that apparently lack bioactivity on one target could possess bioactivity on other targets. If different receptor subtypes account for different rank orders of potencies, then the combination of receptors that the released peptide milieu encounters could determine its actions. Conversely, the combination of receptors expressed by a target could determine the response characteristics to the same peptide milieu released on it. In the case of different targets of the same neuron, differences in receptor expression could increase the diversity of response to the same range of peptide release. Furthermore, differential expression of peptidases in various tissues could also affect the rank order of peptide potencies. This underscores the importance of identifying the complement of bioactive peptides that are released from neurons of interest, because the actions of single peptides may yield erroneous interpretations of their function. Clearly, complementary multidisciplinary approaches are needed to obtain a more accurate understanding of the physiological function of modulatory peptide transmitters.
The peptides contained on the AMRP precursor may have an important role in the function of many systems, including the digestive tract. In fact, MIP-related peptides inhibit contractions of the digestive tract in other molluscan species (Li et al., 1996). Several lines of evidence conclusively demonstrate that these peptides are present in the gut ofAplysia. First, one of the peptides contained on the AMRP precursor, GAPRFVamide, was biochemically isolated from extracts of the gut based on its inhibitory actions on contractions. Second, neurons in the gut have been shown to contain mRNA coding for AMRPs using in situ hybridization. Third, immunocytochemistry indicates dense immunopositive innervation and many AMRP immunopositive neurons in the gut. Finally, AMRPs profoundly inhibit contractions of the gut.
In addition to innervating the gut, AMRP-containing processes also innervate a group of buccal neurons, the B1–B2 cluster that regulates gut motility (Lloyd et al., 1988). Because multiple AMRP-immunopositive axons are present in the esophageal nerve, which connects the gut with the buccal ganglion, the AMRP containing innervation of the B1–B2 cluster may originate in AMRP neurons of the gut. Thus, the AMRPs may be acting both locally in the gut and centrally in the buccal ganglion to regulate the contractions of the digestive tract. Because many neurons that have axons in the cerebrobuccal connective have been shown to be a part of the circuitry responsible for generating feeding inAplysia, the presence of AMRP-immunopositive axons in these connectives also suggests a function of AMRPs in the feeding behavior of Aplysia. One of the AMRPs, GAPRFVamide, has also been detected in the buccal nerves and esophagus of Lymnaea (Li et al., 1996), suggesting that MIP-related peptides may have similar functions in other gastropods.
In addition to functioning in the digestive system, the AMRPs may also have important functions in other systems of Aplysia. The presence of dense AMRP-immunopositive innervation in both the male and female reproductive systems suggests that these peptides have important roles in their function. The inhibitory actions of AMRPs on contractions of the penis retractor muscle support this notion. Although variable, AMRP-containing innervation is apparently present throughout the reproductive system. The source of this innervation appears, at least in part, to be of local origin because immunopositive neurons are present in these structures. However, it is likely that at least some of the immunopositive innervation arises from as yet unidentified AMRP neurons in the CNS. The presence of dense AMRP-immunopositive innervation in the sheath covering the bag cells suggests that these peptides may exert effects on the central neuroendocrine control of the reproductive system and/or be released into the hemolymph and have endocrine functions.
Another system in which AMRPs are likely to exert important effects is in cardiovascular control. Although no innervation was detected in the heart, dense immunopositive innervation was detected in the major arteries, pericardium, and the highly vascularized kidney and gill. The vascular innervation is likely to originate in the abdominal ganglion because most of the neurons involved in cardiovascular regulation are located in this ganglion (Skelton et al., 1992), and the AMRPs are abundantly expressed in the abdominal ganglion. Preliminary results (data not shown) indicate that AMRPs relax the vascular muscle and suggest that these peptides could redirect blood flow. Thus, AMRPs may serve a similar function in Aplysia as endothelin does in vertebrate vasculature. The abundant expression of AMRPs in the abdominal and pleural ganglia, as well as the dense immunopositive innervation in the gill suggests that these peptides may play a role in the plasticity associated with the gill withdrawal reflex (Kandel, 1979; Lukowiak et al., 1982).
We have demonstrated the presence of the AMRPs and the mRNA that codes for their precursor protein in the CNS and in peripheral tissues ofAplysia. Evidence for their presence in the innervation of multiple peripheral targets suggests that these peptides may function in many systems, including feeding, reproduction, circulation, respiration, excretion, and locomotion. Future work is likely to show that these peptides are important modulators in many of these systems.
Footnotes
This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan to Y. Fujisawa, Y. Furukawa, and O. Matsushima. Y. Furukawa was also supported by the Suntory Institute for Bioorganic Research and a traveling grant from the Brain Science Foundation. The support of the National Institute of Neurological Diseases and Stroke through Grant NS31609 to J.V.S. and the National Institute of Mental Health through Grants MH50235 and K05MH01427 to K.R.W. is gratefully acknowledged. We are grateful to Drs. M. Kurokawa (Tokyo Metropolitan University, Tokyo, Japan) and T. Nagahama (Kobe University, Kobe, Japan) for supplying the animals and to Dr. Gregg Nagle for the generous gift of the AplysiacDNA library. Aplysia californica were partially provided by the National Resource for Aplysia at the University of Miami under National Institutes of Health National Center for Research Resources Grant RR10294.
Correspondence should be addressed to Ferdinand S. Vilim, Box 1218, Department of Physiology and Biophysics, Mount Sinai School of Medicine, New York, NY 10029. E-mail: vilim@inka.mssm.edu.
REFERENCES
- 1.Bendena WG, Garside CS, Yu CG, Tobe SS. Allatostatins: diversity in structure and function of an insect neuropeptide family. Ann NY Acad Sci. 1997;814:53–66. doi: 10.1111/j.1749-6632.1997.tb46144.x. [DOI] [PubMed] [Google Scholar]
- 2.Blankenship JE, Rock MK, Hill J. Physiological properties of the penis retractor muscle of Aplysia. J Neurobiol. 1977;8:549–568. doi: 10.1002/neu.480080605. [DOI] [PubMed] [Google Scholar]
- 3.Brezina V, Bank B, Cropper EC, Rosen S, Vilim FS, Kupfermann I, Weiss KR. Nine members of the myomodulin family of peptide cotransmitters at the B16-ARC neuromuscular junction of Aplysia. J Neurophysiol. 1995;74:54–72. doi: 10.1152/jn.1995.74.1.54. [DOI] [PubMed] [Google Scholar]
- 4.Brezina V, Orekhova IV, Weiss KR. Functional uncoupling of linked neurotransmitter effects by combinatorial convergence. Science. 1996;273:806–810. doi: 10.1126/science.273.5276.806. [DOI] [PubMed] [Google Scholar]
- 5.Campanelli JT, Scheller RH. Histidine-rich basic peptide: a cardioactive neuropeptide from Aplysia neurons R3–14. J Neurophysiol. 1987;57:1201–1209. doi: 10.1152/jn.1987.57.4.1201. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter DO, Kemenes G, Elekes K, Leung M, Stefano G, Rozsa KS, Salanki J. Opioid peptides in the nervous system of Aplysia: a combined biochemical, immunocytochemical, and electrophysiological study. Cell Mol Neurobiol. 1995;15:239–256. doi: 10.1007/BF02073331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomcyznski P, Sacchi N. Single-step method of RNA isolation by guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Cropper EC, Brezina V, Vilim FS, Harish O, Price D, Rosen S, Kupfermann I, Weiss KR. FRF peptides in the ARC neuromuscular system of Aplysia: purification and physiological actions. J Neurophysiol. 1994;72:2181–2195. doi: 10.1152/jn.1994.72.5.2181. [DOI] [PubMed] [Google Scholar]
- 9.de With ND, Li KW, Jimenez CR, Vnok N, Dreisewerd K, Hillenkamp F, Karas M, Geraerts WPM. Intracellular degradation of C-peptides in molluscan neurons producing insulin-related hormones. Peptides. 1997;18:765–770. doi: 10.1016/s0196-9781(97)00020-x. [DOI] [PubMed] [Google Scholar]
- 10.Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: peptide alpha-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 11.Fan X, Croll RP, Wu B, Fang L, Shen Q, Painter SD, Nagle GT. Molecular cloning of a cDNA encoding the neuropeptides APGWamide and cerebral peptide 1: location of APGWamide-like immunoreactivity in the central nervous system and male reproductive organs of Aplysia. J Comp Neurol. 1997;387:53–62. doi: 10.1002/(sici)1096-9861(19971013)387:1<53::aid-cne5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Floyd PD, Li L, Moroz TP, Sweedler JV. Characterization of peptides from Aplysia using microbore liquid chromatography with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry guided purification. J Chromatogr A. 1999;830:105–113. doi: 10.1016/s0021-9673(98)00880-2. [DOI] [PubMed] [Google Scholar]
- 13.Fujisawa Y. Immunohistochemical localization and Ca2+-dependent release of Mytilus inhibitory peptides in the ABRM of Mytilus edulis. Zool Sci. 1996;13:795–801. [Google Scholar]
- 14.Fujisawa Y, Kubota I, Ikeda T, Minakata H, Muneoka Y. A variety of Mytilus inhibitory peptides in the ABRM of Mytilus edulis: isolation and characterization. Comp Biochem Physiol. 1991;100C:525–531. [PubMed] [Google Scholar]
- 15.Fujisawa Y, Takahashi T, Ikeda T, Muneoka Y, Kubota I, Minakata H, Nomoto K, Nose T, Miki W. Further identification of bioactive peptides in the anterior byssus retractor muscle of Mytilus: two contractile and three inhibitory peptides. Comp Biochem Physiol. 1993;106C:261–267. doi: 10.1016/0742-8413(93)90281-o. [DOI] [PubMed] [Google Scholar]
- 16.Garden RW, Moroz LL, Moroz TP, Shippy SA, Sweedler JV. Excess salt removal with matrix rinsing: direct profiling of neurons from marine invertebrates using MALDI-TOF mass spectrometry. J Mass Spectrom. 1996;31:1126–1130. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Garden RW, Shippy SA, Li L, Moroz TP, Sweedler JV. Proteolytic processing of the Aplysia egg-laying hormone prohormone. Proc Natl Acad Sci USA. 1998;95:3972–3977. doi: 10.1073/pnas.95.7.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison LM, Kastin AJ, Zadina JE. Opiate tolerance and dependence: receptors, G-proteins, and antiopiates. Peptides. 1998;19:1603–1630. doi: 10.1016/s0196-9781(98)00126-0. [DOI] [PubMed] [Google Scholar]
- 19.Hewes RS, Snowdeal EC, III, Saitoe M, Taghert PH. Functional redundancy of FMRFamide-related peptides at the Drosophila larval neuromuscular junction. J Neurosci. 1998;18:7138–7151. doi: 10.1523/JNEUROSCI.18-18-07138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirata T, Kubota I, Iwasawa N, Takabatake I, Ikeda T, Muneoka Y. Structures and actions of Mytilus inhibitory peptides. Biochem Biophys Res Commun. 1988;152:1376–1382. doi: 10.1016/s0006-291x(88)80437-6. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda T, Y-Kamatani Y, Minakata H, Kenny PTM, Nomoto K, Muneoka Y. Mytilus-inhibitory peptide analogues isolated from the ganglia of a pulmonate mollusc, Achatina fulica. Comp Biochem Physiol. 1992a;101C:245–249. [PubMed] [Google Scholar]
- 22.Ikeda T, Minakata H, Fujita T, Muneoka Y, Kiss T, Hiripi L, Nomoto K. Neuropeptides isolated from Helix pomatia Part 1. Peptides related to MIP, buccalin, myomodulin-CARP and SCP. In: Yanaihara N, editor. Peptide chemistry 1992. ESCOM; Osaka: 1992b. pp. 576–578. [Google Scholar]
- 23.Jahan-Parwar B, Fredman SM. Cerebral ganglion of Aplysia: cellular organization and origin of nerves. Comp Biochem Physiol. 1976;54A:347–357. doi: 10.1016/s0300-9629(76)80124-7. [DOI] [PubMed] [Google Scholar]
- 24.Jimenez CR, van Veelen PA, Li KW, Wildering WC, Geraerts WPM, Tjaden UR, van der Greef J. Neuropeptide expression and processing as revealed by direct matrix-assisted laser desorption ionization mass spectrometry of single neurons. J Neurochem. 1994;62:404–407. doi: 10.1046/j.1471-4159.1994.62010404.x. [DOI] [PubMed] [Google Scholar]
- 25.Kandel ER. Behavioral biology of Aplysia. W. H. Freeman; San Francisco: 1979. [Google Scholar]
- 26.Kaufmann R, Spengler B, Lutzenkirchen F. Mass spectrometric sequencing of linear peptides by product-ion analysis in a reflectron time-of-flight mass spectrometer using matrix-assisted laser desorption ionization. Rapid Commun Mass Spectrom. 1993;7:902–910. doi: 10.1002/rcm.1290071010. [DOI] [PubMed] [Google Scholar]
- 27.Kiss T. Effect of Mytilus inhibitory peptide on identified molluscan neurons. Comp Biochem Physiol. 1990;95C:207–212. [Google Scholar]
- 28.Kiss T, Osipenko ON. Effects of molluscan neuropeptide RAPYFVamide on identified Helix pomatia L. neurons. Gen Pharmacol. 1997;29:97–102. doi: 10.1016/s0306-3623(96)00531-9. [DOI] [PubMed] [Google Scholar]
- 29.Leung MK, Rozsa KS, Hall A, Kuruvilla S, Stefano GB, Carpenter DO. Enkephalin-like substance in Aplysia nervous tissue and actions of leu-enkephalin on single neurons. Life Sci. 1986;38:1529–1534. doi: 10.1016/0024-3205(86)90567-9. [DOI] [PubMed] [Google Scholar]
- 30.Li KW, Van Minnen J, Van Veelen PA, Van der Greef J, Geraertss WPM. Structure, localization and action of a novel inhibitory neuropeptide involved in the feeding of Lymnaea. Mol Brain Res. 1996;37:267–272. doi: 10.1016/0169-328x(95)00333-n. [DOI] [PubMed] [Google Scholar]
- 31.Li L, Moroz TP, Garden RW, Floyd PD, Weiss KR, Sweedler JV. Mass spectrometric survey of interganglionically transported peptides in Aplysia. Peptides. 1998;19:1425–1433. doi: 10.1016/s0196-9781(98)00094-1. [DOI] [PubMed] [Google Scholar]
- 32.Lloyd PE, Kupfermann I, Weiss KR. Central peptidergic neurons regulate gut motility in Aplysia. J Neurophysiol. 1988;59:1613–1625. doi: 10.1152/jn.1988.59.5.1613. [DOI] [PubMed] [Google Scholar]
- 33.Lukowiak K, Thornhill JA, Edstrom J. Methionine enkephalin increases CNS suppressive control exerted over gill reflex behaviours and associated neural activity in Aplysia california. Regul Pept. 1982;3:303–312. doi: 10.1016/0167-0115(82)90135-5. [DOI] [PubMed] [Google Scholar]
- 34.Makabe KW, Fujiwara S, Nishida H, Satoh N. Failure of muscle myosin heavy-chain gene expression in quarter ascidian embryos developed from the secondary muscle lineage cells. Zool Sci. 1992;9:569–573. [Google Scholar]
- 35.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Ohtani M, Muneoka Y, Takahashi T, Teranishi H. Bioactive peptides isolated from the clam Meretrix lusoria. Acta Biol Hung. 1995;46:2–4. [PubMed] [Google Scholar]
- 37.Olson GA, Olson RD, Vaccarino AL, Kastin AJ. Endogenous opiates: 1997. Peptides. 1998;19:1791–843. doi: 10.1016/s0196-9781(98)00137-5. [DOI] [PubMed] [Google Scholar]
- 38.Painter SD, Kalman VK, Nagle GT, Zuckerman RA, Blankenship JE. The anatomy and functional morphology of the large hermaphroditic duct of three species of Aplysia, with special reference to the atrial gland. J Morphol. 1985;186:167–194. doi: 10.1002/jmor.1051860204. [DOI] [PubMed] [Google Scholar]
- 39.Perry SJ, Straub VA, Kemenes G, Santama N, Worster BM, Burke JF, Benjamin PR. Neural modulation of gut motility by myomodulin peptides and acetylcholine in the snail Lymnaea. J Neurophysiol. 1998;79:2460–2474. doi: 10.1152/jn.1998.79.5.2460. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 41.Scheller RH, Jackson JF, McAllistar LB, Rothman BS, Mayer E, Axel R. A single gene encodes multiple neuropeptides mediating a stereotyped behavior. Cell. 1983;32:7–22. doi: 10.1016/0092-8674(83)90492-0. [DOI] [PubMed] [Google Scholar]
- 42.Sediah NG, Chreiten M. Eukaryotic protein processing: endoproteolysis of precursor proteins. Curr Opin Biotechnol. 1997;8:602–607. doi: 10.1016/s0958-1669(97)80036-5. [DOI] [PubMed] [Google Scholar]
- 43.Skelton M, Alevizos A, Koester J. Control of the cardiovascular system of Aplysia by identified neurons. Experientia. 1992;48:809–817. doi: 10.1007/BF02118413. [DOI] [PubMed] [Google Scholar]
- 44.Spengler B. Post-source decay analysis in matrix-assisted laser desorption/ionization mass spectrometry of biomolecules. J Mass Spectrom. 1997;32:1019–1036. [Google Scholar]
- 45.Takayanagi H, Takeda N. Coexistence of FMRFamide, met-enkephalin and serotonin in molluscan neurons. Comp Biochem Physiol A. 1988;91:613–620. doi: 10.1016/0300-9629(88)90643-3. [DOI] [PubMed] [Google Scholar]
- 46.van Golen FA, Li KW, Chen S, Jimenez CR, Geraerts WP. Various isoforms of myomodulin identified from the male copulatory organ of Lymnaea show overlapping yet distinct modulatory effects on the penis muscle. J Neurochem. 1996;66:321–329. doi: 10.1046/j.1471-4159.1996.66010321.x. [DOI] [PubMed] [Google Scholar]
- 47.Vilim FS, Cropper EC, Rosen SC, Tenenbaum R, Kupfermann I, Weiss KR. Structure, cellular localization and modulatory action of buccalin b–a novel bioactive peptide from Aplysia californica. Peptides. 1994;15:959–969. doi: 10.1016/0196-9781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 48.Vilim FS, Price DA, Lesser W, Kupfermann I, Weiss KR. Costorage and corelease of modulatory peptide cotransmitters with partially antagonistic actions on the accessory radula closer muscle of Aplysia californica. J Neurosci. 1996a;16:8092–8104. doi: 10.1523/JNEUROSCI.16-24-08092.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vilim FS, Cropper EC, Price DA, Kupfermann I, Weiss KR. Release of peptide cotransmitters in Aplysia: regulation and functional implications. J Neurosci. 1996b;16:8105–8114. doi: 10.1523/JNEUROSCI.16-24-08105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worster BM, Yeoman MS, Benjamin PR. Matrix-assisted laser desorption/ionization time of flight mass spectrometric analysis of the pattern of peptide expression in single neurons resulting from alternative mRNA splicing of the FMRFamide gene. Eur J Neurosci. 1998;10:3498–3507. doi: 10.1046/j.1460-9568.1998.00361.x. [DOI] [PubMed] [Google Scholar]
- 51.Yongsiri A, Takeuchi H, Kubota I, Muneoka Y. Effects of Mytilus inhibitory peptides on a giant neurone of Achatina fulica Ferussac. Eur J Pharmacol. 1989;171:159–165. doi: 10.1016/0014-2999(89)90104-0. [DOI] [PubMed] [Google Scholar]
- 52.Zadina JE, Hackler L, Ge LJ, Kastin AJ. A potent and selective endogenous agonist for the μ-opiate receptor. Nature. 1997;386:499–502. doi: 10.1038/386499a0. [DOI] [PubMed] [Google Scholar]