Abstract
The regulatory factor Sox10 is expressed in neural crest derivatives during development as well as in the adult CNS and peripheral nervous system. Mutations of the human Sox10 gene have been identified in patients with Waardenburg-Hirschsprung syndrome that is characterized by defects in neural crest development. Previous studies suggested that Sox10 might function as an important transcriptional regulator of neural crest development. No natural target genes of Sox10 have yet been identified. Although human Sox10 activates a synthetic promoter consisting of a TATA box and multiple Sox consensus sequences, no transcriptional activity of the rat Sox10 homolog has been detected. Here we report that the neuronal nicotinic acetylcholine receptor β4 and α3 subunit gene promoters are transactivated by rat Sox10 in a cell type-specific manner. The α3 and β4 subunits, in combination with the α5 subunit, make up the predominant nicotinic receptor subtype expressed in the peripheral nervous system. Transfections using Sox10 mutants indicate that the C-terminal region is dispensable for its ability to activate the β4 and α3 promoters. Rat Sox10 was originally identified as an accessory protein of the POU domain protein Tst-1/Oct6/SCIP in glial cells. Tst-1/Oct6/SCIP was shown previously to activate the α3 promoter. We now demonstrate that it can transactivate the β4 promoter as well. However, we were unable to detect any synergistic effects of Sox10 and Tst-1/Oct6/SCIP on β4 or α3 promoter activity. Finally, we present data suggesting that recombinant Sox10 protein can directly interact with a previously characterized regulatory region of the β4 gene.
Keywords: Sox10, gene expression, nACh receptor, ligand-gated ion channel, transcriptional regulation, POU
Eleven members of the gene family encoding neuronal nicotinic acetylcholine (nACh) receptor subunits have been identified and include α2–α9 and β2–β4 (Boyd, 1997). These subunits form heteromeric and homomeric receptors with distinct pharmacological and physiological profiles (Schoepfer et al., 1990;Elgoyhen et al., 1994; McGehee and Role, 1995; Role and Berg 1996;Gerzanich et al., 1997). The functional diversity exhibited by the neuronal nACh receptor family results in large part from the differential expression of the subunit genes and the subsequent incorporation of the subunits into mature receptors. The molecular mechanisms leading to formation of the various nACh receptor subtypes remain to be completely elucidated. However, it is clear that regulation at the level of transcription plays an important role (Boyd, 1997).
Our previous work focused on transcriptional regulation of three genomically clustered receptor genes, those encoding the β4, α3, and α5 subunits. These three subunits make up the predominant receptor subtype expressed in the peripheral nervous system (PNS) (Conroy et al., 1993; Conroy and Berg, 1995). We and others have identified several cis-acting elements (Boyd, 1994; Yang et al., 1994, 1997; Hu et al., 1995; Bigger et al., 1996; Fornasari et al., 1997; McDonough and Deneris, 1997) and trans-acting proteins (Yang et al., 1995; Fyodorov and Deneris, 1996; Milton et al., 1996; Bigger et al., 1997; Du et al., 1997, 1998; Fyodorov et al., 1998; Campos-Caro et al., 1999) that are important for the transcriptional regulation of the clustered genes. However, the mechanisms underlying the neuron-specific expression of the receptor genes remain elusive.
Recently, a novel member of the Sox protein family, Sox10, was identified (Kuhlbrodt et al., 1998a). Sox proteins have been implicated as important transcriptional regulators in a variety of developmental processes (Gubbay et al., 1990; Denny et al., 1992; Wright et al., 1993; Kamachi et al., 1995). Sox10 has been proposed to function as an important regulator during neural crest development (Bondurand et al., 1998). Consistent with this hypothesis is the demonstration that a Sox10 gene mutation in the Dominant megacolon(Dom) mouse gives rise to aganglionosis of the colon and pigmentation defects as well as to substantial losses of neurons and glia in the PNS and a complete loss of the enteric nervous system (Herbarth et al., 1998; Southard-Smith et al., 1998). Additionally, SOX10 mutations have been detected in patients with Waardenburg-Hirschsprung syndrome, which resembles the Domphenotype (Pingault et al., 1998). Despite this critical role in development, no natural target genes of Sox10 have yet been identified.
Interestingly, Sox10 is highly expressed in neuronal structures in which the clustered β4, α3, and α5 nACh receptor genes are also highly expressed. Furthermore, the α3 gene has been shown to be regulated by the POU domain protein Tst-1/Oct6/SCIP (Yang et al., 1994), a factor for which Sox10 has been shown to be an accessory protein (Kuhlbrodt et al., 1998a). Therefore, we investigated the possibility that Sox10 may participate in the transcriptional regulation of the β4 and α3 subunit genes.
MATERIALS AND METHODS
Plasmids. The wild-type rat β4/luciferase expression plasmid pX1B4FH, containing a 226 bpFokI/HindIII fragment spanning nucleotides −89 to +137, relative to the β4 transcription initiation site, was described previously (Hu et al., 1994). A 2.1 kbHindIII/SacI fragment of the rat α3 genomic clone λRG518B (Boulter et al., 1990) [generously provided by Jim Boulter (University of California, Los Angeles, Los Angeles, CA)], containing nucleotides −2036 to +64, relative to the α3 transcription start site, was subcloned into the promoterless luciferase expression vector pXP1 (Nordeen, 1988), yielding a wild-type α3/luciferase expression construct, pX1A3HS. All of the Sox10 expression constructs were kindly provided by Dr. Michael Wegner (Hamburg University, Hamburg, Germany) and are described in detail elsewhere (Kuhlbrodt et al., 1998a,b). The expression plasmid pCGS-SCIP, containing the SCIP-coding sequence inserted downstream of the cytomegalovirus (CMV) promoter (Monuki et al., 1993), was generously provided by Dr. Greg Lemke (The Salk Institute, San Diego, CA).
Site-directed mutagenesis. Site-directed mutagenesis of the wild-type β4/luciferase construct pX1B4FH was performed as described previously (Bigger et al., 1996) except that a different 5′ primer (5′-GAC GGA TCC CTC TCA GAC CCT CCC CTC CCC TGT GGC ACC AGC GCA TCC CAA-3′) was used.
Preparation of recombinant proteins. The T7-tag–Sox10 fusion protein construct pET28c/SX107.1.1.ΔN was also a kind gift of Dr. Michael Wegner (Kuhlbrodt et al., 1998a). This fusion protein contains amino acids 89–466 of the rat Sox10 protein fused in-frame with a T7-tag sequence. Expression and purification of T7-tag fusion protein were performed using a T7-tag monoclonal antibody purification kit (Novagen, Madison, WI).
Cell culture and transfections. HeLa, Neuro2A, and Rat2 cells were obtained from the American Type Culture Collection (Rockville, MD). Neuro2A cells (Klebe et al., 1970) were grown in minimal essential medium (Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS; Sigma, St. Louis, MO). HeLa (Gey et al., 1952) and SN17 cells (Hammond et al., 1990) were maintained in DMEM supplemented with 10% FBS. Sol8 cells (Daubas et al., 1988) were cultured in DMEM containing 20% FBS and 0.5% chicken embryo extract (Life Technologies). NIH3T3 cells (Jainchill et al., 1969) were grown in DMEM containing 10% calf serum (Sigma). Rat2 cells (Topp, 1981) were grown in DMEM containing 5% FBS. Pheochromocytoma 12 (PC12) cells (Greene and Tischler, 1976) were cultured and differentiated with nerve growth factor (Upstate Biotechnology, Lake Placid, NY) as described previously (Hu et al., 1994).
Neuro2A, SN17, Sol8, NIH3T3, and HeLa cells were transfected at 60% confluency in 60 mm dishes using a calcium phosphate method and a commercially available kit (5 Prime–3 Prime, Boulder, CO). Cells were transfected with 5 μg of test DNA (pX1B4FH or pX1A3HS), 5 μg of effector DNA [the empty pCMV5 vector (Invitrogen, Carlsbad, CA), pCMVSox10 constructs, or pCGS-SCIP], and 5 μg of a β-galactosidase expression vector, pCH110 (Pharmacia, Piscataway, NJ ). In some cases, no effector DNA was included in the transfections. To test dose dependency, we transfected Neuro2A cells with either pX1B4FH or pX1A3HS and 1, 2, 4, or 8 μg of wild-type pCMVSox10 expression plasmid. To insure that the calcium phosphate/DNA precipitates had equal amounts of DNA, we added appropriate quantities of pBluescript II SK DNA (Stratagene, La Jolla, CA) to each sample. Rat2 cells were transfected in 60 mm dishes at a density of 105 cells/ml using 30 μl of Lipofectamine (2 mg/ml; Life Technologies) and the same quantities of DNAs as described above. Forty-eight hours after transfection, cells were harvested and assayed for luciferase activity using a commercially available kit (Promega, Madison, WI) and an Autolumat LB953 luminometer (EG&G Berthold, Gaithersburg, MD). All transfections were done a minimum of two times with two different preparations of plasmid DNAs. To correct for differences in transfection efficiencies between dishes, we normalized the luciferase activity in each sample to the β-galactosidase activity in the same sample, which was measured using a commercially available kit (Galacto-Light; Tropix, Bedford, MA).
Western blotting. Preparation of cell lysates and Western blotting were performed as described previously (Bigger et al., 1997). Anti-Sox10 antibody was a generous gift from Dr. Michael Wegner (Kuhlbrodt et al., 1998a) and was used at a 1:3000 dilution in Blotto.
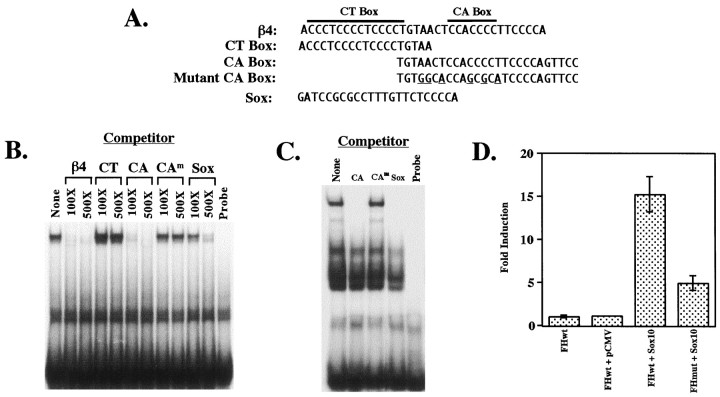
Electrophoretic mobility shift assays. Electrophoretic mobility shift assays were performed using either a32P-labeled double-stranded β4 oligonucleotide (see Fig. 5A, sequence) and 1 μg of recombinant T7-tag–Sox10 protein or a32P-labeled double-stranded CA box oligonucleotide (see Fig. 5A, sequence) and 3.5 μg of rat brain nuclear extract [prepared as described previously (Hu et al., 1995)]. Reaction mixtures containing protein, binding buffer (10 mm HEPES, pH 8.0, 50 mmNaCl, 5 mm MgCl2, 2 mm DTT, 0.1 mm EDTA, and 5% glycerol), and either 0.5 μg (for T7-tag–Sox10) or 2 μg (for nuclear extract) of poly(dI–dC) were preincubated for 5 min at room temperature (RT; for T7-tag–Sox10) or on ice (for nuclear extract) before the addition of 5 fmol of the end-labeled probe. After addition of the probe, binding reactions were further incubated for 15 min at RT. For competition experiments, unlabeled double-stranded β4 oligonucleotides or Sox oligonucleotide, containing a consensus binding site for Sox proteins (see Fig. 5A, sequence) (Kuhlbrodt et al., 1998a), were preincubated with the protein for 5 min before the addition of labeled oligonucleotides. Reaction mixtures were then electrophoresed through 6% native polyacrylamide gels. Radioactivity was detected by autoradiography of the dried gels.
Fig. 5.
Direct interactions between Sox10 and the β4 promoter region are functionally relevant. A, Sequences of the oligonucleotides used in electrophoretic mobility shift assays are shown. Mutations in the CA box oligonucleotide areunderlined. The sequence of an oligonucleotide corresponding to a consensus binding site for Sox proteins (Pevny and Lovell-Badge, 1997) is also shown. B, Electrophoretic mobility shift assays were performed using 5 fmol of a radiolabeled oligonucleotide (β4 in A) corresponding to nucleotides −82 to −48 of the β4 gene and 1 μg of T7-tag–Sox10 protein. The left lane shows DNA–protein complex formation in the absence of competitors. Competition experiments were performed using 100- and 500-fold molar excesses of competitor oligonucleotides (either unlabeled wild-type or mutated β4 oligonucleotides or the consensus Sox-binding site oligonucleotide). The right lane contains unbound β4 probe in the absence of Sox10 protein. C, Electrophoretic mobility shift assays were performed using 5 fmol of a radiolabeled oligonucleotide (CA Box in A) and 3.5 μg of rat brain nuclear extract. The left lane shows DNA–protein complex formation in the absence of competitors. Competition experiments were performed using 100-fold molar excess of competitor oligonucleotides (either unlabeled wild-type or mutated CA box oligonucleotides or the consensus Sox-binding site oligonucleotide). The right lane contains unbound CA box probe in the absence of protein. D, The mutations shown in A (Mutant CA Box) were introduced into the β4/luciferase construct pX1B4FH to create pX1B4FHmut. Wild-type pX1B4FH (FHwt) and pX1B4FHmut (FHmut) were cotransfected into Neuro2A cells with pCMVSox10. As controls, wild-type pX1B4FH was also transfected either alone (FHwt) or with pCMV5 (pCMV). Luciferase values were normalized to β-galactosidase expression as driven by the SV40 promoter. Fold induction was calculated relative to the normalized luciferase activity obtained by transfecting wild-type pX1B4FH alone. Error bars represent SDs. CAm, Mutant CA box.
RESULTS
Cell type-specific activity of the β4 promoter region
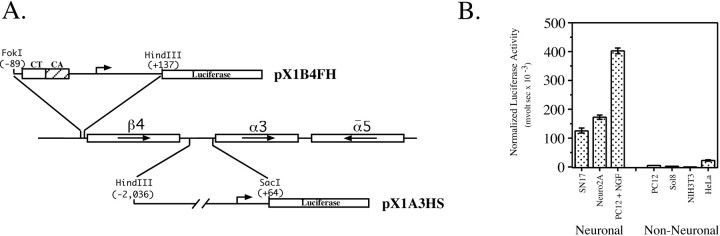
We described previously the isolation and partial characterization of the 5′-flanking region of the rat β4 subunit gene (Hu et al., 1994) and identified two transcriptional regulatory elements that are critical for β4 promoter activity (Hu et al., 1995; Bigger et al., 1996). The regulatory elements, a CT box and a CA box, are located within a 226 bp FokI/HindIII fragment that possesses strong transcriptional activity (Fig.1A) (Hu et al., 1995;Bigger et al., 1996). To determine whether the transcriptional activity is cell type dependent, we performed a series of transient transfection experiments using a variety of neuronal and non-neuronal cell lines. When neuronal cell lines were transfected with the β4/luciferase construct pX1B4FH (Fig. 1A), significant luciferase activity was detected (Fig. 1B). In contrast, no significant activity was seen in extracts of transfected non-neuronal cell lines (Fig. 1B). These data suggest that the transcriptional activity of pX1B4FH is cell type specific. The α3 promoter region has been extensively characterized by Deneris and colleagues (Yang et al., 1994, 1995, 1997; McDonough and Deneris, 1997).
Fig. 1.
A, β4 and α3 promoter/luciferase constructs. The genomic organization of the clustered nACh receptor subunit genes is presented. Thestraightarrows indicate directions of transcription. The 5′-flanking regions of the β4 and α3 subunit genes were subcloned upstream of the coding sequence for luciferase to generate pX1B4FH and pX1A3HS, respectively. Numberscorrespond to the positions of the 5′-most and 3′-most nucleotides of the fragments relative to the major transcription initiation sites (bentarrows). B, Cell type-specific activity of the β4 promoter. Neuronal cell lines [SN17, Neuro2A, and nerve growth factor (NGF)-treated PC12] and non-neuronal cell lines (untreated PC12, Sol8 muscle, NIH3T3 fibroblast, and HeLa cervical carcinoma) were transiently transfected with pX1B4FH as described in Materials and Methods. Luciferase values were normalized to correct for differences in transfection efficiencies (see Materials and Methods). Error bars represent SDs.
Sox10 transactivates the neuronal nACh receptor β4 and α3 subunit gene promoters in a cell type-specific manner
Because the temporal and spatial patterns of expression of Sox10 partially overlap those of the β4 and α3 subunit genes, we set out to investigate whether Sox10 can regulate the promoter activities of these genes. To determine whether Sox10 can transactivate the β4 and α3 gene promoters, transfection experiments were performed in various cell lines with the β4 or α3 promoter/luciferase reporter constructs alone or in combination with an expression construct in which the rat Sox10 gene is under the control of the CMV promoter (Fig.2). To confirm that Sox10 expression was involved in transactivation, we also transfected the reporter constructs with the pCMV vector alone (devoid of the Sox10-coding sequence). The cell lines included two rodent neuroblastoma lines, SN17 and Neuro2A, two fibroblast cell lines, NIH3T3 and Rat2, and one mouse muscle cell line, Sol8.
Fig. 2.
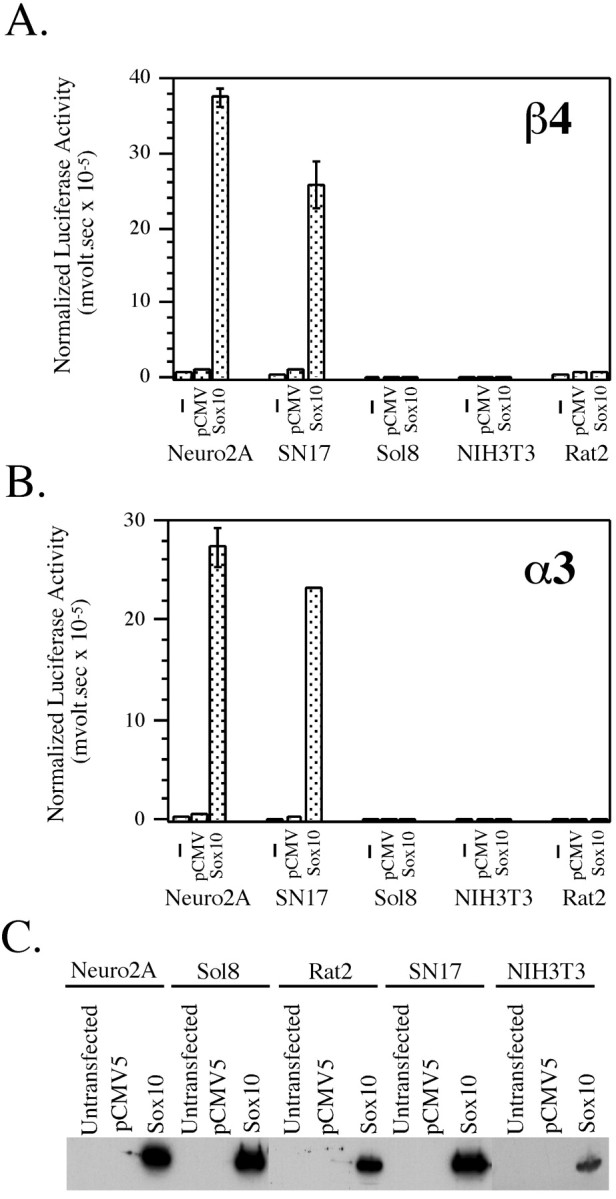
Sox10 transactivates the β4 and α3 gene promoters in a cell type-specific manner. A, B, The β4/luciferase construct pX1B4FH (A) and the α3/luciferase construct pX1A3HS (B) were transiently transfected into neuronal (Neuro2A and SN17) and non-neuronal (Sol8 muscle, NIH3T3, and Rat2 fibroblast) cell lines either alone (−) or with pCMV5 (pCMV) or pCMVSox10 (Sox10). Luciferase values were normalized to β-galactosidase expression as driven by the SV40 promoter. Error bars represent SDs. C, Qualitative Western blot analysis of transfected cells is shown. Cell extracts were prepared from untransfected cells or cells transfected with pCMV5 or pCMVSox10. For a given cell line, equivalent amounts of protein for each condition were analyzed by SDS-PAGE. After SDS-PAGE, Western blot analysis was done using anti-Sox10 antibody to demonstrate Sox10 expression in pCMVSox10-transfected cells.
Cotransfection of the β4 (Fig. 2A) and α3 (Fig.2B) reporter constructs with an “empty” pCMV vector had no effect on reporter gene expression in any of the cell types tested. However, when the β4 and α3 reporters were cotransfected with Sox10, a dramatic transactivation of reporter gene activity was observed in Neuro2A and SN17 cells (∼20- to 30-fold induction over background) but not in the muscle or fibroblast cell lines (Fig. 2A,B). To confirm that Sox10 protein was synthesized in the transfected cells, qualitative Western blot analysis was performed. As shown in Figure 2C, extracts of all five pCMVSox10-transfected cell lines had readily detectable levels of Sox10 protein, indicating that the lack of transactivation in the non-neuronal cell lines was most likely not caused by the lack of Sox10 protein. Furthermore, transactivation of the β4 and α3 promoter/luciferase constructs by Sox10 occurs in a dose-dependent manner in both Neuro2A (Fig. 3) and SN17 (data not shown) cells. Taken together, these results indicate that Sox10 can activate both the β4 and α3 subunit gene promoters and does so in a neuron-specific manner.
Fig. 3.
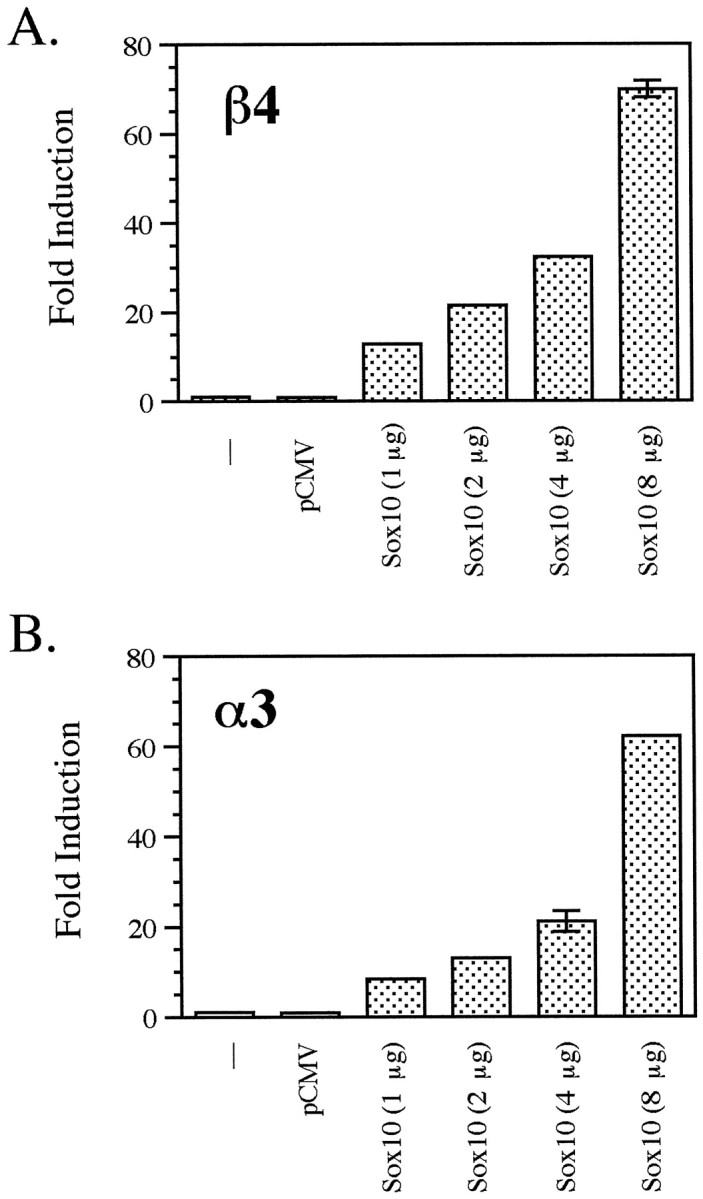
Dose-dependent activation of the β4 and α3 promoters by Sox10. The β4/luciferase construct pX1B4FH (A) and the α3/luciferase construct pX1A3HS (B) were cotransfected with pCMVSox10 (1, 2, 4, and 8 μg) in Neuro2A cells. Luciferase values were normalized to β-galactosidase expression as driven by the SV40 promoter. Fold induction was calculated relative to the normalized luciferase activity obtained by transfecting pX1B4FH or pX1A3HS alone. Error bars represent SDs.
Domains of Sox10 required for its transactivation activity
In the experiment described above rat Sox10 protein was used. It has been reported previously that rat Sox10 protein does not possess a transactivation activity of its own when tested in glial cells and using a synthetic promoter containing a TATA box and multiple Sox consensus binding sites (Kuhlbrodt et al., 1998a). In contrast, results of two reports on human SOX10 indicate the presence of a potent transactivation domain at the C terminal of the human protein (Kuhlbrodt et al., 1998b; Pusch et al., 1998). Interestingly, the data presented in Figure 2 demonstrate that in the context of neuronal cell lines, rat Sox10 does in fact possess transactivation activity. Therefore, to map the domains of rat Sox10 that are required for its ability to transactivate the β4 and α3 promoters in neuronal cells, transfection experiments were performed using a number of Sox10 mutants in both the Neuro2A (Fig. 4) and in SN17 (data not shown) cell lines. Mutant Sox10ΔN lacks the N-terminal 89 amino acids, whereas Sox10-HMG contains only the HMG domain of the protein (Fig. 4A). Four of the SOX10 mutations, WS029, MIC, 059, and 095 (Fig. 4A), were originally identified in patients with Waardenburg-Hirschsprung syndrome, a disease characterized by deafness, pigmentation defects, and aganglionic megacolon (Pingault et al., 1998). Mutant WS029 is a nonsense mutation that converts tyrosine 83 to a stop codon; MIC is also a nonsense mutation leading to a truncated protein of only 188 amino acids; mutant 059 lacks the last 106 amino acids as a consequence of a deletion of two nucleotides at position 1076 resulting in a frameshift (the frameshift creates a sequence of 40 unrelated amino acids at the C terminal of the protein); and mutant 095 carries an insertion of six nucleotides between positions 482 and 483 leading to the addition of a leucine and an arginine into the HMG box (the open reading frame remains intact). Each of these four mutations observed in patients with Waardenburg-Hirschsprung syndrome was subsequently introduced into the rat Sox10 cDNA (Kuhlbrodt et al., 1998b). The transactivation data indicate that only wild-type Sox10 and mutant 059 are capable of significantly transactivating the β4 and α3 promoters (Fig. 4). The fact that mutant 095 did not transactivate either the β4 or α3 promoter provides evidence that an intact HMG box is required for Sox10 function. However, sequences in addition to the HMG box are necessary for transactivation because Sox10-HMG, which contains just the HMG domain, also failed to activate either promoter. Similar to Sox10-HMG, Sox10ΔN failed in this assay, suggesting that the extreme N-terminal domain of rat Sox10 is required for its transcriptional activity. Furthermore, mutant MIC did not transactivate either nACh receptor gene promoter, indicating that the central and the C-terminal portions of the protein are required for Sox10 activity. However, the 106 amino acids located at the extreme C terminal do not appear to be critical for Sox10 function because mutant 059 activated both promoters to levels similar to those of wild-type Sox10. Although we cannot formally exclude the slight possibility that mutants Sox10ΔN, MIC, WS029, Sox10-HMG, or 095 failed to transactivate the β4 promoter in Neuro2A cells because they were either not expressed to a sufficient extent or not properly translocated to the nucleus, the data presented above strongly suggest that the N-terminal 89 amino acids, the HMG domain, and amino acids 189–360 are necessary for the transactivation function of Sox10.
Fig. 4.
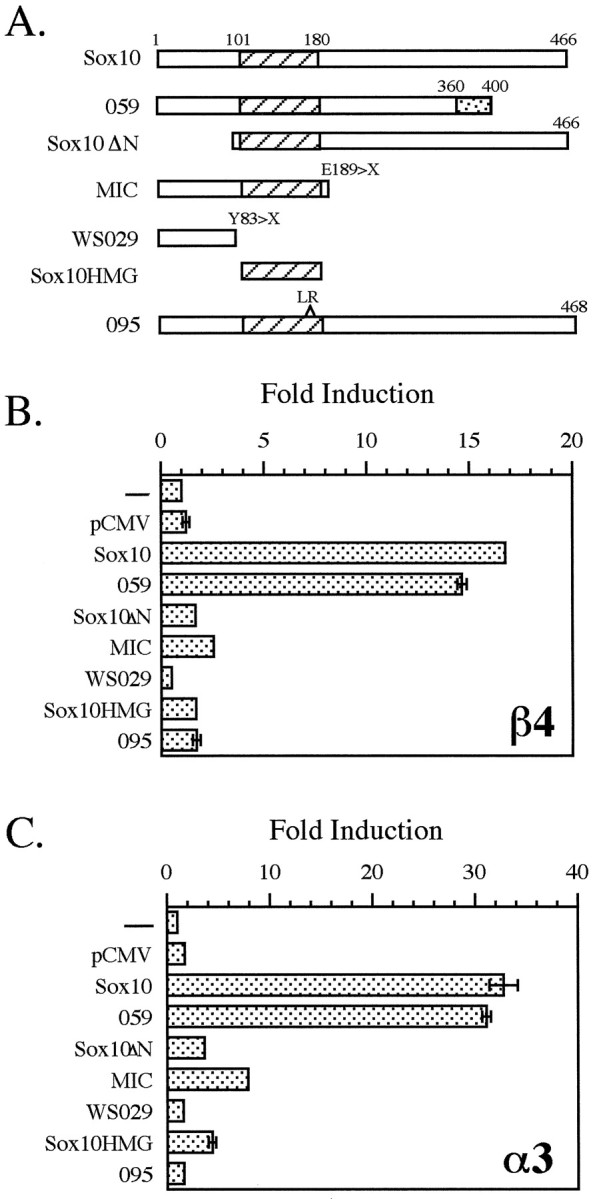
Protein domains of Sox10 required for its transcriptional activity. A, Schematic representations of the Sox10 expression constructs used for transfection analysis (see Results for details) are shown. The hatchedbox represents the HMG domain in each construct. Thedottedbox in mutant 059 represents 40 unrelated amino acids created by a frameshift (see Results). B, C, Neuro2A cells were transfected with the β4/luciferase construct pX1B4FH (B) or the α3/luciferase construct pX1A3HS (C) either alone (−) or with pCMV5, wild-type Sox10, or one of the Sox10 mutant constructs as indicated. Fold induction was calculated as described in Figure3.
Sox10 directly interacts with the β4 promoter region
As described above, the inability of mutant 095 to transactivate the β4 and α3 promoters indicates that an intact HMG domain is required for Sox10 function. The fact that the HMG box mediates protein–DNA interactions (Kuhlbrodt et al., 1998a), coupled with the observation that mutant 095 is unable to bind DNA (Kuhlbrodt et al., 1998b), suggests that Sox10 must bind to DNA to effect transactivation. No consensus binding site for Sox10 has yet been identified for any gene. However, to demonstrate the ability of Sox10 to bind to DNA,Kuhlbrodt et al. (1998a) used a synthetic oligonucleotide with a sequence that is recognized by several members of the Sox family (van de Wetering et al., 1993) in electrophoretic mobility shift assays. Although there are two classic Sox consensus binding sites, C(T/A)TTTG(T/A)(T/A) (Pevny and Lovell-Badge, 1997; Wegner, 1999), present in the promoter of α3 at positions −1275 and −1284 relative to the major transcription start site (Yang et al., 1994), this consensus sequence is not present in the 226 bpFokI/HindIII β4 promoter fragment used in the transfection experiments described above. Therefore, to demonstrate that Sox10 interacts directly with the β4 promoter, electrophoretic mobility shift assays were done using a double-stranded oligonucleotide encompassing the CT and CA boxes of the β4 promoter region as the probe (Fig. 5A). As discussed previously, these two regulatory elements are critical for β4 promoter activity. Incubation of recombinant T7-tag–Sox10 fusion protein with radioactively labeled β4 oligonucleotide led to the formation of a single specific Sox10/DNA complex (Fig. 5B). An unlabeled oligonucleotide corresponding to the consensus Sox-binding site also competed for binding to Sox10, but the competition was not complete (i.e., even at 500-fold molar excess of the Sox oligonucleotide, there was still Sox10/β4 complex formation; Fig.5B). This may reflect differences in the affinity of Sox10 for the two sites. In an attempt to localize the Sox10-binding site further, competition experiments were done using oligonucleotides containing either the CT box or the CA box (Fig. 5A). The CT box failed to compete for binding to Sox10, whereas the CA box competed as well as the β4 oligonucleotide (Fig. 5B). When a CA box oligonucleotide mutated in six positions (Fig. 5A) was used as a competitor, no competition was seen (Fig. 5B). These data suggest that a region of the β4 promoter that overlaps the CA box is involved in direct interactions with Sox10. As an initial attempt to determine whether Sox10/β4 interactions may occur in a more physiological context, electrophoretic mobility shift assays were performed using radioactively labeled CA box as a probe and nuclear extract prepared from adult rat brain as a protein source. As shown in Figure 5C, incubation of the CA box probe with brain nuclear extract resulted in the formation of several DNA–protein complexes. Formation of these complexes was competed by unlabeled CA box and by unlabeled consensus Sox-binding site oligonucleotides but not by the mutated CA box oligonucleotide (Fig. 5C). These data are consistent with the electrophoretic mobility shift results obtained using purified Sox10 (Fig. 5B) and provide further evidence of direct interactions between the β4 promoter and Sox10.
To test the functional significance of these interactions, the same point mutations were introduced into a β4 promoter/luciferase construct and subsequently used in transfection experiments. As shown in Figure 5D, Sox10 transactivation of the mutated promoter was significantly less than that of the wild-type promoter. Taken together, these data strongly suggest that Sox10 directly interacts with the β4 promoter in a functionally relevant manner.
Sox10 and Tst-1/Oct6/SCIP do not transactivate the nACh receptor β4 or α3 subunit gene promoters synergistically in Neuro2A or SN17 cells
Sox proteins have been proposed to function as cell type-specific accessory proteins for POU domain factors (Yuan et al., 1995). Indeed, the initial characterization of Sox10 showed that it can function synergistically with the POU domain protein Tst-1/Oct6/SCIP to activate transcription in glial cells (Kuhlbrodt et al., 1998a). This is particularly relevant to nACh receptor gene expression because it has been demonstrated that Tst-1/Oct6/SCIP is capable of activating the α3 promoter in vitro (Yang et al., 1994). Interestingly, activation of the α3 promoter by Tst-1/Oct6/SCIP appears to be indirect, because it occurs independently of the Tst-1/Oct6/SCIP-binding sites (Fyodorov and Deneris, 1996). We therefore tested, first, the ability of Tst-1/Oct6/SCIP to activate the β4 promoter and, second, the possible synergy of Sox10 and Tst-1/Oct6/SCIP in transactivating the β4 and α3 promoters. As shown in Figure 6A, Tst-1/Oct6/SCIP activated the β4 promoter ∼35-fold in SN17 cells and 20-fold in Neuro2A cells. Little or no transactivation was detected in Sol8 muscle cells or in the fibroblast cell line NIH3T3. Thus, both the β4 and α3 promoters can be activated by Tst-1/Oct6/SCIP. Moreover, the transactivation activity of Tst-1/Oct6/SCIP on the β4 promoter is cell type specific. As also shown in Figure 6, cotransfection of SN17 cells with Tst-1/Oct6/SCIP and Sox10 did not lead to an additive or a synergistic effect on β4 or α3 promoter activities, with the activation being comparable with that seen with Sox10 alone. Similar results were observed in Neuro2A cells as well (data not shown). This raises the possibility, then, that the synergistic interaction of Tst-1/Oct6/SCIP and Sox10 seen by Kuhlbrodt et al. (1998a) may be glial cell specific and that it does not occur in neuronal cells. Another possibility is that activation by Sox10 was at a saturated level in the above experiments, and therefore, no synergy would occur under such conditions. However, dose–response experiments performed using lower amounts of both Sox10 and Tst-1/Oct6/SCIP also failed to uncover any synergistic effects (data not shown).
Fig. 6.
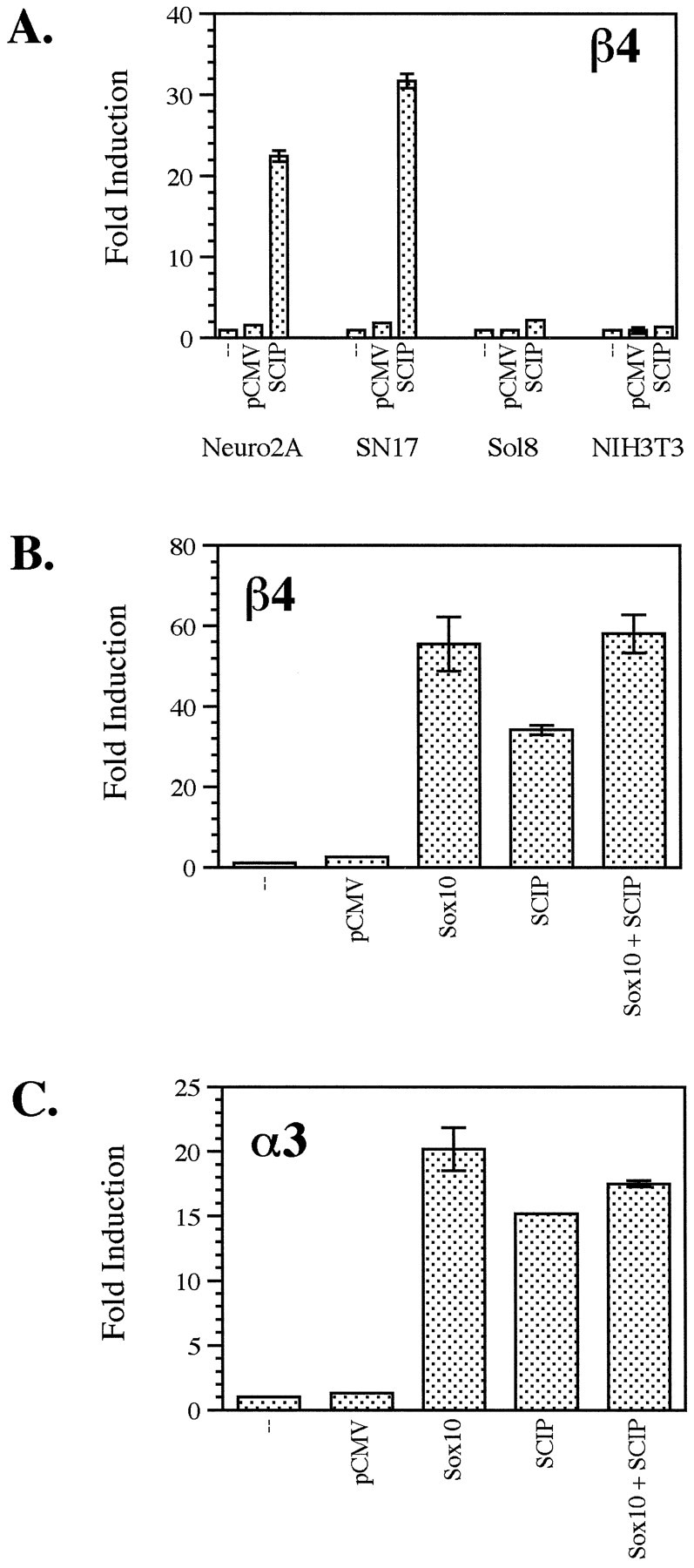
A, Tst-1/Oct6/SCIP transactivates the β4 gene promoter in a cell type-specific manner. The β4/luciferase construct pX1B4FH was transfected into neuronal cell lines (Neuro2A and SN17) and non-neuronal cell lines (Sol8 muscle and NIH3T3 fibroblast) either alone (−) or with pCMV5 (pCMV) or pCGS-SCIP (SCIP). Fold induction was calculated as described in Figure 3. B, C, Tst-1/Oct6/SCIP and Sox10 do not transactivate the β4 or α3 gene promoters additively or synergistically. SN17 cells were transfected with the β4/luciferase construct pX1B4FH (B) or the α3/luciferase construct pX1A3HS (C) either alone (−), with pCMV5 (pCMV), or with expression constructs for Sox10 and Tst-1/Oct6/SCIP (SCIP) individually and together. Fold induction was calculated as described above.
DISCUSSION
Sox10 belongs to the Sox family of transcription factors, members of which are characterized by the presence of a DNA-binding domain, an HMG box that is highly similar to the HMG domain of the mammalian sex determination factor Sry (Pevny and Lovell-Badge, 1997; Wegner, 1999). Sox proteins play important roles during a number of developmental processes such as sex determination, chondrogenesis, neurogenesis, and lens formation (Wegner, 1999). Sox10 has been proposed to be a regulator of neural crest development (Herbarth et al., 1998;Southard-Smith et al., 1998). Dom mice, which carry a Sox10 frameshift mutation, are characterized by several neural crest defects. Heterozygotes have pigmentation abnormalities and suffer from aganglionic megacolon; homozygotes have significant losses of neurons and glia in the PNS, and in addition, their entire enteric nervous system is lacking (Herbarth et al., 1998; Southard-Smith et al., 1998). Patients with Waardenburg-Hirschsprung syndrome carry mutations in one of the alleles of SOX10 and display phenotypic abnormalities similar to those found in Dom mice (Pingault et al., 1998). Despite the well-characterized phenotypes resulting from mutations in Sox10, no natural target genes for this factor have been documented.
The α3 and β4 subunits, together with α5, form the predominant nACh receptor subtype expressed in the PNS. Because these three genes are tightly clustered genomically, it is possible that they are subject to coordinate regulation. In agreement with this idea, Sp1 has been shown to transactivate these three genes (Yang et al., 1995; Bigger et al., 1996, 1997; Campos-Caro et al., 1999). We demonstrated previously that in addition to Sp1, the β4 promoter can be transactivated by another member of the Sp family, Sp3 (Bigger et al., 1997). Moreover, it appears that the transactivation potentials of Sp1 and Sp3 are differentially regulated by another protein capable of interacting with the β4 promoter, hnRNP K (Du et al., 1998). Deneris and colleagues identified a PC12 cell-specific enhancer, β43′, in the β4/α3 intergenic region that is capable of activating transcription from the α3 and β4 subunit gene promoters (McDonough and Deneris, 1997). A novel ETS-domain protein, Pet-1, can transactivate this enhancer in a cell type-specific manner (Fyodorov et al., 1998). However, the β43′ enhancer is not sufficient to direct reporter gene expression in the PNS, where α3 and β4 are expressed, as judged by transgenic analysis (McDonough and Deneris, 1997). Deneris and colleagues have also demonstrated that the promoter of α3 can be transactivated by a POU domain protein, Tst-1/Oct6/SCIP (Yang et al., 1994). In addition to regulation by Tst-1/Oct6/SCIP, α3 can be regulated by another class of POU domain proteins, Brn-3. Although Brn-3a activates the α3 promoter, Brn-3b and Brn-3c repress it (Milton et al., 1996). The β4 promoter appears to be unaffected by Brn-3 proteins (Milton et al., 1996). Transactivation of the α3 promoter by Tst-1/Oct6/SCIP was shown to be independent of the Tst-1/Oct6/SCIP DNA-binding sites, leading to the hypothesis that the observed effect is a consequence of protein–protein interactions (Yang et al., 1994). Interestingly, it appears that POU domain proteins can synergize and in some cases directly interact with Sox proteins (Yuan et al., 1995; Ambrosetti et al., 1997; Kuhlbrodt et al., 1998a,b). Tst-1/Oct6/SCIP can synergistically activate transcription with Sox10 (Kuhlbrodt et al., 1998a), making Sox10 a potential candidate for the regulation of nACh receptor gene expression. Furthermore, the expression patterns of Sox10, α3, and β4 spatially and temporally overlap in the developing nervous system (Zoli et al., 1995; Kuhlbrodt et al., 1998a). These observations prompted us to investigate whether Sox10 can regulate the α3 and β4 promoters.
The results presented here indicate that rat Sox10 can, by itself, significantly transactivate the α3 and β4 promoters in neuronal cell lines but has no effect on the activity of these promoters in non-neuronal cells. Our data are in contrast to the results ofKuhlbrodt et al. (1998a) who were unable to detect any autonomous transactivation activity of rat Sox10 in glial cells either on an artificial promoter containing Sox-binding sites or in a heterologous system in which Sox10 was fused to the GAL4 DNA-binding domain and subsequently cotransfected with a reporter containing GAL4 DNA-binding sites (Kuhlbrodt et al., 1998a). Instead, Sox10 was able to function as a transactivator only in synergy with other transcription factors such as Tst-1/Oct6/SCIP or Pax3 (Kuhlbrodt et al., 1998a). It is possible, then, that the transcriptional activity of rat Sox10 observed in neuronal cells results from it synergizing with endogenous transcription factors present in those cells. On the other hand, it is possible that the transactivation domain of rat Sox10 is masked in glial cells but is accessible in other cell types, such as neurons. It is important to note that human SOX10 does possess a strong transactivation domain in its C terminal as assayed in the GAL4 system in COS cells (Pusch et al., 1998). The C-terminal domains of rat and human Sox10 are virtually identical in this region, with only one amino acid difference occurring at residue 415 (Pusch et al., 1998).
To determine which regions of Sox10 are important for its ability to transactivate the α3 and β4 promoters in neuronal cells, we used a number of mutant Sox10 constructs. Our results indicate that the C-terminal 106 residues of rat Sox10 are dispensable for its activity in neuronal cells, suggesting that the C-terminal of rat Sox10 does not contain a transactivation domain in the context of neuronal cell lines. The basis for the functional differences between the C-terminal domains of human and rodent Sox10 remains to be elucidated. An intact HMG domain is required for Sox10's transactivation activity; however, this domain alone is not sufficient because a mutant Sox10 consisting of just the HMG domain proved to be inactive. In addition to the HMG domain, the N terminal of Sox10 is absolutely necessary, but again not sufficient, for its activity, and the central part of the protein up to amino acid 360 is required as well. Taken together, these data suggest that to activate the α3 and β4 promoters, Sox10 must bind DNA. Preferred binding sites for Sox10 have not been reported; however, Sox10 has been demonstrated to bind to a Sox protein consensus site (Kuhlbrodt et al., 1998a). Sequence analysis of the promoter regions of α3 and β4 revealed that the α3 promoter contains two consensus Sox-binding sites, whereas such elements were not found in the β4 promoter. However, we demonstrated that Sox10 can specifically interact with an oligonucleotide containing the CT and CA elements of the β4 promoter. These elements have been shown to be critical for β4 promoter activity (Hu et al., 1995; Bigger et al., 1996). Interestingly, it appears that the affinity of Sox10 for this region of the β4 promoter is higher than its affinity for the Sox consensus site.
As mentioned above, Sox10 can act synergistically with Tst-1/Oct6/SCIP (Kuhlbrodt et al., 1998a), a factor that can transactivate the α3 promoter (Yang et al., 1994). We extended these observations to show that Tst-1/Oct6/SCIP is able to transactivate the β4 promoter in a neuron-specific manner as well. Because the region of the β4 promoter used does not contain binding sites for POU domain proteins, we suggest that, analogous to α3 activation (Yang et al., 1994), Tst-1/Oct6/SCIP may be able to regulate the β4 promoter via interactions with other transcription factors. A similar mechanism has been proposed for the repression of the myelin P0 promoter by Tst-1/Oct6/SCIP (Monuki et al., 1990). We investigated the possibility that Tst-1/Oct6/SCIP can transactivate nACh receptor gene expression via functional interactions with Sox10 but were unable to detect any synergy between the two factors. There are several possibilities for this phenomenon. It has been demonstrated previously that synergistic interactions between Sox and POU domain proteins, such as those between Sox2 and Oct3, Sox10 and Tst-1/Oct6/SCIP, and Sox11 (and similarly Sox4) and Brn-1 and Brn-2, require binding of the respective factors to adjacent sites on DNA (Ambrosetti et al., 1997; Kuhlbrodt et al., 1998a,b). Therefore, because the α3 and β4 promoters do not contain binding elements for POU domain proteins, Tst-1/Oct6/SCIP and Sox10 may be unable to interact functionally. However, the HMG2 protein can synergistically interact with the Oct factors in the absence of DNA-binding sites for the former (Zwilling et al., 1995). This observation raises the possibility that the functional interactions between POU and HMG domain proteins, such as the Sox family members, might occur in a cell type-specific manner.
In summary, we have identified the nACh receptor β4 and α3 subunit genes as natural target genes of rat Sox10. Furthermore, we have shown that Sox10 transactivates the β4 and α3 gene promoters in a neuron-specific manner. In addition, we have shown that, similar to the α3 promoter, the POU domain protein Tst-1/Oct6/SCIP can transactivate the β4 gene promoter in a neuron-specific manner. However, no synergistic effect of Sox10 and Tst-1/Oct6/SCIP was observed in the neuronal cell lines tested. On the basis of these results, we hypothesize that Sox10 is a crucial regulatory factor involved in the neuron-specific expression of the β4 and α3 subunit genes.
Footnotes
This work was supported by grants to P.D.G. from The National Institutes of Health, The Council for Tobacco Research USA, and The Smokeless Tobacco Research Council, Inc. I.N.M. was supported by a POWRE grant from The National Science Foundation. We thank Jim Boulter, Patrick Burrola, Kirsten Kuhlbrodt, Greg Lemke, and Michael Wegner for generously providing reagents and Katie Reidel and Shyun Li for help during the early phases of this work. P.D.G. thanks B. P. and D. R. for inspiration.
Q.L. and I.N.M. contributed equally to this work.
Correspondence should be addressed to Dr. Paul D. Gardner, Department of Molecular Medicine, University of Texas Health Science Center, 15355 Lambda Drive, San Antonio, TX 78245-3207. E-mail:gardner@uthscsa.edu.
Dr. Hu's present address: Department of Neurobiology, Stanford University School of Medicine, Stanford, CA 94305-5125.
REFERENCES
- 1.Ambrosetti D-C, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigger CB, Casanova EA, Gardner PD. Transcriptional regulation of neuronal nicotinic acetylcholine receptor genes: functional interactions between Sp1 and the rat β4 subunit gene promoter. J Biol Chem. 1996;271:32842–32848. doi: 10.1074/jbc.271.51.32842. [DOI] [PubMed] [Google Scholar]
- 3.Bigger CB, Melnikova IN, Gardner PD. Sp1 and Sp3 regulate expression of the neuronal nicotinic acetylcholine receptor β4 subunit gene. J Biol Chem. 1997;272:25976–25982. doi: 10.1074/jbc.272.41.25976. [DOI] [PubMed] [Google Scholar]
- 4.Bondurand N, Kobetz A, Pingault V, Lemort N, Encha-Razavi F, Couly G, Goerich DE, Wegner M, Abitbol M, Goossens M. Expression of the SOX10 gene during human development. FEBS Lett. 1998;432:168–172. doi: 10.1016/s0014-5793(98)00843-6. [DOI] [PubMed] [Google Scholar]
- 5.Boulter J, O'Shea-Greenfield A, Duvoisin RM, Connolly JG, Wada E, Jensen A, Gardner PD, Ballivet M, Deneris ES, McKinnon D, Heinemann S, Patrick J. α3, α5, β4: three members of the rat neuronal nicotinic acetylcholine receptor-related gene family form a gene cluster. J Biol Chem. 1990;265:4472–4482. [PubMed] [Google Scholar]
- 6.Boyd RT. Sequencing and promoter analysis of the genomic region between the rat neuronal nicotinic acetylcholine receptor β4 and α3 genes. J Neurobiol. 1994;25:960–973. doi: 10.1002/neu.480250806. [DOI] [PubMed] [Google Scholar]
- 7.Boyd RT. The molecular biology of neuronal nicotinic acetylcholine receptors. Crit Rev Toxicol. 1997;27:299–318. doi: 10.3109/10408449709089897. [DOI] [PubMed] [Google Scholar]
- 8.Campos-Caro A, Carrasco-Serrano C, Valor LM, Viniegra S, Ballesta JJ, Criado M. Multiple functional Sp1 domains in the minimal promoter region of the neuronal nicotinic receptor α5 subunit gene. J Biol Chem. 1999;274:4693–4701. doi: 10.1074/jbc.274.8.4693. [DOI] [PubMed] [Google Scholar]
- 9.Conroy WG, Berg DK. Neurons can maintain multiple classes of nicotinic acetylcholine receptors distinguished by different subunit combinations. J Biol Chem. 1995;270:4424–4431. doi: 10.1074/jbc.270.9.4424. [DOI] [PubMed] [Google Scholar]
- 10.Conroy WG, Vernallis AB, Berg DK. The α5 gene product assembles with multiple acetylcholine receptor subunits to form distinctive receptor subtypes in brain. Neuron. 1993;9:679–691. doi: 10.1016/0896-6273(92)90031-8. [DOI] [PubMed] [Google Scholar]
- 11.Daubas P, Klarsfeld A, Garner I, Pinset C, Cox R, Buckingham M. Functional activity of the two promoters of the myosin alkali light chain gene in primary muscle cell cultures: comparison with other muscle gene promoters and other culture systems. Nucleic Acids Res. 1988;16:1251–1270. doi: 10.1093/nar/16.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny P, Swift S, Brand N, Dabhade N, Barton P, Ashworth A. A conserved family of genes related to the testis determining gene, SRY. Nucleic Acids Res. 1992;20:2887. doi: 10.1093/nar/20.11.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Q, Tomkinson AE, Gardner PD. Transcriptional regulation of neuronal nicotinic acetylcholine receptor genes: a possible role for the DNA-binding protein Purα. J Biol Chem. 1997;272:14990–14995. doi: 10.1074/jbc.272.23.14990. [DOI] [PubMed] [Google Scholar]
- 14.Du Q, Melnikova IN, Gardner PD. Differential effects of hnRNP K on Sp1- and Sp3-mediated transcriptional activation of a neuronal nicotinic acetylcholine receptor promoter. J Biol Chem. 1998;273:19877–19883. doi: 10.1074/jbc.273.31.19877. [DOI] [PubMed] [Google Scholar]
- 15.Elgoyhen AB, Johnson DS, Boulter J, Vetter DE, Heinemann S. α9: an acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell. 1994;79:705–715. doi: 10.1016/0092-8674(94)90555-x. [DOI] [PubMed] [Google Scholar]
- 16.Fornasari D, Battaglioli E, Flora A, Terzano S, Clementi F. Structural and functional characterization of the human α3 nicotinic subunit gene promoter. Mol Pharmacol. 1997;51:250–261. doi: 10.1124/mol.51.2.250. [DOI] [PubMed] [Google Scholar]
- 17.Fyodorov D, Deneris E. The POU domain of SCIP/Tst-1/Oct-6 is sufficient for activation of an acetylcholine receptor promoter. Mol Cell Biol. 1996;16:5004–5014. doi: 10.1128/mcb.16.9.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fyodorov D, Nelson T, Deneris E. Pet-1, a novel ETS domain factor that can activate neuronal nAChR gene transcription. J Neurobiol. 1998;34:151–163. [PubMed] [Google Scholar]
- 19.Gerzanich V, Kuryatov A, Anand R, Lindstrom J. “Orphan” α6 nicotinic AChR subunit can form a functional heteromeric acetylcholine receptor. Mol Pharmacol. 1997;51:320–327. [PubMed] [Google Scholar]
- 20.Gey GO, Coffman WD, Kubicek MT. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952;12:264–265. [Google Scholar]
- 21.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Münsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 23.Hammond DN, Lee HJ, Tonsgard JH, Wainer BH. Development and Characterization of clonal cell lines derived from septal cholinergic neurons. Brain Res. 1990;512:190–200. doi: 10.1016/0006-8993(90)90626-M. [DOI] [PubMed] [Google Scholar]
- 24.Herbarth B, Pingault V, Bondurand N, Kuhlbrodt K, Hermans-Borgmeyer I, Puliti A, Lemort N, Goossens M, Wegner M. Mutation of the Sry-related Sox10 gene in Dominant megacolon, a mouse model for human Hirschsprung disease. Proc Natl Acad Sci USA. 1998;95:5161–5165. doi: 10.1073/pnas.95.9.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu M, Whiting Theobald NL, Gardner PD. Nerve growth factor increases the transcriptional activity of the neuronal nicotinic acetylcholine receptor β4 subunit promoter in transfected PC12 cells. J Neurochem. 1994;62:392–395. doi: 10.1046/j.1471-4159.1994.62010392.x. [DOI] [PubMed] [Google Scholar]
- 26.Hu M, Bigger CB, Gardner PD. A novel regulatory element of a nicotinic acetylcholine receptor gene interacts with a DNA-binding activity enriched in rat brain. J Biol Chem. 1995;270:4497–4502. doi: 10.1074/jbc.270.9.4497. [DOI] [PubMed] [Google Scholar]
- 27.Jainchill JL, Aaronson SA, Todaro GJ. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol. 1969;4:549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamachi Y, Sockanathan S, Liu Q, Breitman M, Lovell-Badge R, Kondoh H. Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J. 1995;14:3510–3519. doi: 10.1002/j.1460-2075.1995.tb07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klebe RJ, Chen T, Ruddle FH. Controlled production of proliferating somatic cell hybrids. J Cell Biol. 1970;45:74–82. doi: 10.1083/jcb.45.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998a;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhlbrodt K, Schmidt C, Sock E, Pingault V, Bondurand N, Goossens M, Wegner M. Functional analysis of Sox10 mutations found in human Waardenburg-Hirschsprung patients. J Biol Chem. 1998b;273:23033–23038. doi: 10.1074/jbc.273.36.23033. [DOI] [PubMed] [Google Scholar]
- 32.McDonough J, Deneris E. β43′: an enhancer displaying neural-restricted activity is located in the 3′-untranslated exon of the rat nicotinic acetylcholine receptor β4 gene. J Neurosci. 1997;17:2273–2283. doi: 10.1523/JNEUROSCI.17-07-02273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGehee DS, Role LW. Physiological diversity of nicotinic acetylcholine receptors expressed by vertebrate neurons. Annu Rev Physiol. 1995;57:521–546. doi: 10.1146/annurev.ph.57.030195.002513. [DOI] [PubMed] [Google Scholar]
- 34.Milton NGN, Bessis A, Changeux J-P, Latchman DS. Differential regulation of neuronal nicotinic acetylcholine receptor subunit gene promoters by Brn-3 POU family transcription factors. Biochem J. 1996;317:419–423. doi: 10.1042/bj3170419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monuki E, Kuhn R, Weinmaster G, Trapp B, Lemke G. Expression and activity of the POU transcription factor SCIP. Science. 1990;249:1300–1303. doi: 10.1126/science.1975954. [DOI] [PubMed] [Google Scholar]
- 36.Monuki ES, Kuhn R, Lemke G. Cell-specific action and mutable structure of a transcription factor effector domain. Proc Natl Acad Sci USA. 1993;90:9978–9982. doi: 10.1073/pnas.90.21.9978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nordeen S. Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques. 1988;6:454–457. [PubMed] [Google Scholar]
- 38.Pevny LH, Lovell-Badge R. Sox genes find their feet. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 39.Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Prθhu M-O, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Smith JC, Read AP, Wegner M, Goossens M. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 40.Pusch C, Hustert E, Pfeifer D, Südbeck P, Kist R, Roe B, Wang Z, Balling R, Blin N, Scherer G. The SOX10/Sox10 gene from human and mouse: sequence, expression, and transactivation by the encoded HMG domain transcription factor. Hum Genet. 1998;103:115–123. doi: 10.1007/s004390050793. [DOI] [PubMed] [Google Scholar]
- 41.Role LW, Berg DK. Nicotinic receptors in the development and modulation of CNS synapses. Neuron. 1996;16:1077–1085. doi: 10.1016/s0896-6273(00)80134-8. [DOI] [PubMed] [Google Scholar]
- 42.Schoepfer R, Conroy WG, Whiting P, Gore M, Lindstrom J. Brain α-bungarotoxin binding protein cDNAs and MAbs reveal subtypes of this branch of the ligand-gated ion channel gene superfamily. Neuron. 1990;5:35–48. doi: 10.1016/0896-6273(90)90031-a. [DOI] [PubMed] [Google Scholar]
- 43.Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 44.Topp WC. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981;113:408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- 45.van de Wetering M, Oosterwegel M, van Norren K, Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright EM, Snopek B, Koopman P. Seven new members of the Sox gene family expressed during development. Nucleic Acids Res. 1993;21:744. doi: 10.1093/nar/21.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, McDonough J, Fyodorov D, Morris M, Wang F, Deneris ES. Characterization of an acetylcholine receptor α3 gene promoter and its activation by the POU domain factor SCIP/Tst-1. J Biol Chem. 1994;269:10252–10264. [PubMed] [Google Scholar]
- 49.Yang X, Fyodorov D, Deneris ES. Transcriptional analysis of acetylcholine receptor α3 gene promoter motifs that bind Sp1 and AP2. J Biol Chem. 1995;270:8514–8520. doi: 10.1074/jbc.270.15.8514. [DOI] [PubMed] [Google Scholar]
- 50.Yang X, Yang F, Fyodorov D, Wang F, McDonough J, Herrup K, Deneris E. Elements between the protein-coding regions of the adjacent β4-α3 acetylcholine receptor genes direct neuron-specific expression in the central nervous system. J Neurobiol. 1997;28:311–324. [PubMed] [Google Scholar]
- 51.Yuan HB, Corbi N, Basilico C, Daily L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 52.Zoli M, Le Novere N, Hill JA, Changeux J-P. Developmental regulation of nicotinic ACh receptor subunit mRNAs in the rat central and peripheral nervous systems. J Neurosci. 1995;15:1912–1939. doi: 10.1523/JNEUROSCI.15-03-01912.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zwilling S, K÷nig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]