Abstract
In the CNS kainate subtype glutamate receptors (GluRs) are likely to be heteromeric assemblies containing multiple gene products. However, although recombinant kainate receptors from the GluR5–GluR7 gene family have been studied extensively in their homomeric forms, there have been no tests to determine whether these subunits can coassemble with each other. We used the GluR5 selective agonists (RS)-2-amino-3-(3-hydroxy-5-tertbutylisoxazol-4-yl)propanoic acid (ATPA) and (S)-5-iodowillardiine (I-will) to test for the coassembly of GluR5 with GluR6 and GluR7 by measuring changes in rectification that occur for heteromeric receptors containing both edited and unedited Q/R site subunits. Birectifying ATPA and I-will responses resulting from polyamine block for homomeric GluR5(Q) became outwardly rectifying when GluR6(R) was coexpressed with GluR5(Q), although GluR6 was not activated by ATPA or I-will, indicating the formation of heteromeric receptors. Similar approaches showed the coassembly of GluR7 with GluR6 and GluR5. Heteromeric kainate receptors containing both GluR5 and GluR6 subunits exhibited novel functional properties, including reduced desensitization and faster recovery from desensitization than those recorded for homomeric GluR5. Coexpression of GluR6 with GluR5 also enhanced the magnitude of responses to GluR5 selective agonists. In contrast, the coassembly of GluR7 with GluR6 markedly decreased the amplitude of agonist responses. Our results indicate that, similar to AMPA receptors, the kainate receptor subunits GluR5–GluR7 exhibit promiscuous coassembly. The formation of heteromeric kainate receptors may help to explain why the functional properties of native kainate receptors differ from those that have been reported for recombinant kainate receptors.
Keywords: kainate receptors, polyamines, glutamate receptors, ATPA, iodowillardiine, coassembly, heteromeric glutamate receptors
Kainate receptors are encoded by two gene families: glutamate receptors (GluRs) GluR5–GluR7, which form functional homomeric receptors with distinct physiology and pharmacology (Hollmann and Heinemann, 1994; Schiffer et al., 1997;Dingledine et al., 1999), and KA1 and KA2, which form functional ion channels only after coassembly with GluR5–GluR7 (Werner et al., 1991;Herb et al., 1992). The discovery of kainate receptor synaptic currents in diverse areas of the CNS (Castillo et al., 1997; Vignes and Collingridge, 1997; Frerking et al., 1998; Li and Rogawski, 1998;DeVries and Schwartz, 1999; Li et al., 1999), possible modulatory actions of kainate receptors on neurotransmitter release (Clarke et al., 1997; Rodriguez-Moreno et al., 1997; Vignes et al., 1998), and studies on GluR6 knock-out mice (Mulle et al., 1998; Bureau et al., 1999) have stimulated recent interest in kainate receptor biology. The overlapping expression of mRNAs encoding kainate receptor subunits (Wisden and Seeburg, 1993; Bahn et al., 1994; Bischoff et al., 1997) makes it extremely likely that, similar to AMPA receptors, there exist diverse subtypes of heteromeric kainate receptor assemblies. The mechanisms regulating the assembly of glutamate receptor (GluR) subunits are poorly understood but must include signals that allow coassembly within and between some gene families while restricting coassembly with other gene families. Thus, AMPA receptor subunits, which show 68–73% amino acid sequence homology, show promiscuous coassembly with each other, but not with kainate receptor subunits (Partin et al., 1993; Brose et al., 1994; Puchalski et al., 1994). In contrast, although GluR5–GluR7 show similar low homology with both AMPA receptor subunits (39–41%) and KA1 and KA2 (43–44%), they coassemble only with the latter (Partin et al., 1993; Brose et al., 1994; Puchalski et al., 1994; Wenthold et al., 1994).
It seems likely that, similar to AMPA receptors, the coassembly of GluR5, GluR6, and GluR7 could generate heteromeric receptors with novel functional properties; however, experimental tests of this have not been reported. In addition to providing the fundamental knowledge required for a full understanding of the biology of the diverse subtypes of GluRs, information on the coassembly of kainate receptors would be especially useful at the present time, given some surprising results for kainate receptor-mediated mossy fiber synaptic responses in CA3 pyramidal neurons (Castillo et al., 1997; Vignes and Collingridge, 1997). Mossy fiber kainate receptor EPSCs are absent in GluR6−/− mice, suggesting a role for the GluR6 subunit (Mulle et al., 1998) but also are antagonized by decahydroisoquinolines that selectively block the activation of homomeric GluR5, but not GluR6 (Clarke et al., 1997; Vignes et al., 1997). It seems likely that the mossy fiber EPSC could be mediated by heteromeric kainate receptors formed by the coassembly of GluR5 and GluR6, perhaps with additional subunits. To test for the coassembly of kainate receptors from the GluR5–GluR7 gene family, we took advantage of the recent discovery that two agonists, ATPA, the tertbutyl analog of AMPA (Lauridsen et al., 1985), and (S)-5-iodowillardiine (I-will), selectively activate GluR5, but not GluR6 or GluR7 (Clarke et al., 1997; Swanson et al., 1998). By combining GluR5 subunit selective activation with the attenuation of polyamine block by the edited (R) forms of GluRs, we were able to demonstrate the coassembly of GluR5 with both GluR6 and GluR7. Then analogous approaches were used to demonstrate that GluR6 and GluR7 can coassemble also. Although further experiments will be required to determine the subunit composition of native kainate receptors, and in particular whether there exist heteromeric forms assembled from more than two subunits (for example, both GluR5 and GluR6 combined with KA1, KA2, or GluR7), the results of our experiments are the first step toward this difficult goal.
MATERIALS AND METHODS
Mutagenesis cell culture and expression of recombinant receptors. The GluR5 cDNA used in our experiments was the GluR5-2a splice variant described by Sommer et al. (1992); DNA sequencing confirmed that the construct used in our experiments had only 16 amino acids in the C terminus that follows the last membrane-spanning domain and no insert in N-terminal domain. The GluR6 construct we used (I567V and Y571C) was fully edited in the first membrane domain (Köhler et al., 1993). The GluR7 construct that was used was the GluR7a splice variant (Schiffer et al., 1997). To create GluR7(R), we performed site-directed mutagenesis by amplification in vitro of pBSGluR7a(Q), using complimentary mutagenic oligonucleotides (Life Technologies, Rockville, MD) andPfu polymerase (Stratagene, La Jolla, CA). Selection of mutants was achieved by the subsequent digestion of parental plasmid with DpnI. A 563 bp BglII fragment that included the Q to R mutation was sequenced completely and subcloned into a GluR7a eukaryotic expression vector construct in which a cytomegalovirus promoter controls the transcription of cDNAs (Keinänen et al., 1990). HEK 293 cells (CRL 1573, American Type Culture Collection, Manassas, VA) were maintained at 70–80% confluence in Life Technologies’ minimal essential medium with Earle’s salts, 2 mm glutamine, and 10% fetal bovine serum. Then 24 hr after being plated at low density (2 × 104 cells per ml) onto 35 mm Petri dishes, the cells were transfected via the calcium phosphate technique (Chen and Okayama, 1987). Kainate receptor cDNAs, 2.4 μg per 35 mm dish, were cotransfected with 0.6 μg of the cDNA for green fluorescent protein (S65T mutation) to aid in the identification of transfected cells. For the coexpression of different receptor subunit combinations, cDNAs were transfected at the desired ratios with the total cDNA maintained at 3 μg per dish. The cells were washed with PBS 12–18 hr after transfection and used for electrophysiological recordings after another 24–48 hr.
Recording conditions. Electrophysiological recordings were performed by using whole-cell patch clamp. The external solution contained (in mm) 145 NaCl, 5.4 KCl, 5 HEPES, 1 MgCl2, and 1.8 CaCl2. The internal solution contained (in mm) 105 CsMeSO3, 10 CsF, 15 CsCl, 5 Cs4-BAPTA, 10 HEPES, 1 MgCl2, and 0.5 CaCl2 to which 60 μm spermine was added. The pH of both the external and internal solutions was adjusted to 7.3; their osmolarity was adjusted to 295 mOsm with sucrose. Before recording, we applied 0.3 mg/ml concanavalin A to individual cells for 4 min to attenuate desensitization. ATPA and (S)-5-iodowillardiine were gifts from Drs. P. Krogsgaard-Larsen (Royal Danish School of Pharmacy, Copenhagen, Denmark), J. C. Watkins, and D. E. Jane (University of Bristol, UK); additional samples of these drugs were purchased from Tocris (Ballwin, MO). BAPTA was purchased from Molecular Probes (Eugene, OR); all other reagents were purchased from Sigma (St. Louis, MO) or Aldrich (Milwaukee, WI). Recordings were made with an Axopatch-200B amplifier (Axon Instruments, Foster City, CA), using fire-polished thin-walled borosilicate glass pipettes (2–5MΩ). Series resistance (3–10MΩ) was compensated routinely by 90%. Records were stored on a Power Macintosh G3 computer with a 16 bit analog-to-digital converter (ITC-16; Instrutech, Elmont, NY) under the control of the data acquisition program Synapse (Synergy Research, Monrovia, MD). Agonists were applied via a stepper motor-based fast perfusion system, as described previously (Vyklicky et al., 1990).
Data analysis. Current–voltage (I–V) plots were generated with voltage ramps from −105 to +105 mV (0.42 V/sec). Procedures in the Igor program (WaveMetrics, Lake Oswego, OR) were used to generate and analyze conductance–voltage (G–V) plots. First, the reversal potential forI–V plots was estimated by using a fifth-order polynomial fit to the average of four leak-subtracted responses. ThenG–V plots were generated and fit with appropriate equations. For GluR5(Q) and GluR6(Q) homomeric receptors theG–V plots were fit with the Woodhull equation over the range from −100 to +20 mV:
| Equation 1 |
where Gmax is the conductance at a sufficiently hyperpolarized potential to produce full relief from block by polyamines; [Spm] is the cytoplasmic polyamine concentration (60 μm spermine was added to the internal solution); KD(0) is the dissociation constant for spermine at 0 mV membrane potential;Vm is the membrane potential;zθ is the valence and electrical distance for polyamine block. F, R, and T have their standard values. For GluR6(R) responses and for heteromeric receptors with outwardly rectifying G–V plots, the ratios of conductance values at +80 to −80 mV were used to describe the extent of rectification.
For responses with intermediate rectification recorded on the coexpression of the edited and unedited forms of GluRs, theG–V plots were fit with the sum of two Boltzmann functions over the range of −100 to +50 mV:
| Equation 2 |
where Gmax andVm have the same meaning as defined above; Vb is the membrane potential for a half-block of each component by polyamines;kb describes the voltage dependence of the block for each component; and C is a constant. Then theKD(0) values for polyamine block were calculated from the relationship:
| Equation 3 |
Values in the text are mean ± SEM unless noted differently. Statistical tests of differences between data sets were performed with Student’s t tests.
RESULTS
Selective activation of GluR5 by ATPA and (S)-5-iodowillardiine
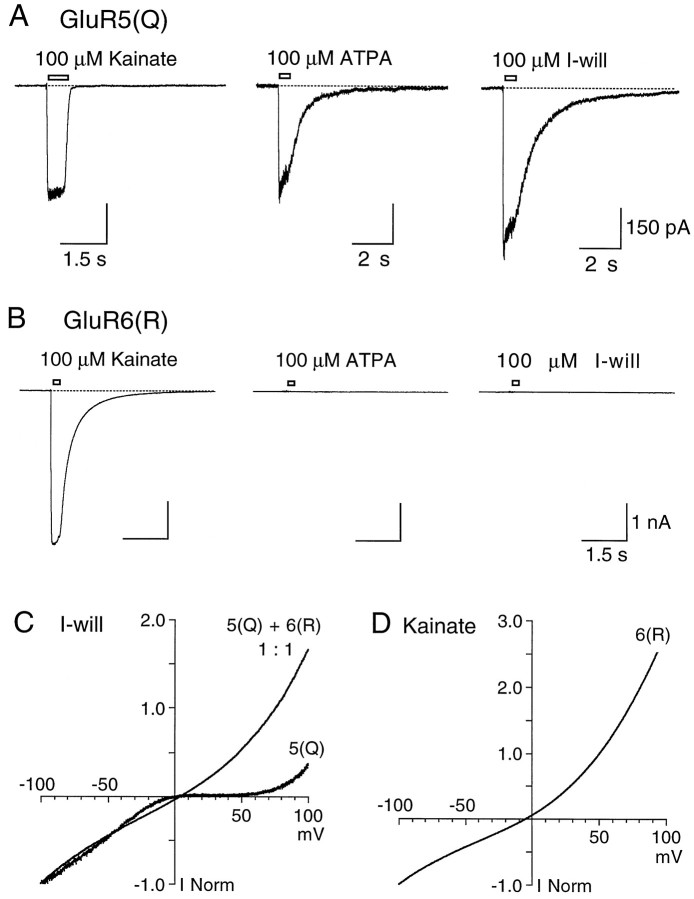
The design of our experiments required that we be able to distinguish responses mediated by heteromeric kainate receptors containing more than one type of subunit from those mediated by homomeric receptors. To do this, we used kainate receptor subtype selective agonists and the change in rectification that occurs on coassembly of the edited (R) and unedited (Q) forms of GluR subunits. To facilitate the analysis of rectification that used ramp changes in membrane potential, we attenuated the strong desensitization of kainate receptors by a treatment of HEK cells with 0.3 mg/ml concanavalin A for 4 min (Partin et al., 1993; Everts et al., 1999). We began our study by testing for the coassembly of GluR5 and GluR6. These experiments revealed that, although both GluR5(Q) and GluR6(R) were activated by 100 μm kainate as expected, the amplitude of responses for GluR5(Q), 354 ± 88 pA (n = 12) at −60 mV, was approximately one-fifth of that for GluR6(R), 2024 ± 540 pA (n = 7). Desensitization of kainate responses was attenuated to a similar extent for both subtypes, such that the ratio of peak responses to those measured at 200 msec was 1.08 ± 0.02 for GluR5(Q) and 1.02 ± 0.01 for GluR6(R). In contrast to the nonselective agonist action of kainate, 10 μmATPA and 100 μm I-will produced responses only for cells transfected with GluR5(Q), whereas for GluR6(R) these agonists were completely inactive (Fig.1A,B). On average, the amplitude of GluR5(Q) responses to 10 μm ATPA was 0.83 ± 0.05 (n = 6) of those to kainate recorded from the same cell, whereas for I-will the ratio was 1.29 ± 0.06 (n = 7). The attenuation of desensitization by concanavalin A was less for responses to ATPA and I-will than to kainate, but it was sufficient to generate large-amplitude equilibrium responses (Fig. 1A).
Fig. 1.
Selective activation of GluR5 by ATPA and I-will.A, Responses from a HEK cell transfected with GluR5(Q) to 100 μm kainate, 10 μm ATPA, and 100 μm I-will at −60 mV. B, When the same agonists were applied to a cell transfected with GluR6(R), only kainate produced inward current responses. C, RampI–V plots for responses to 100 μm I-will recorded from HEK cells transfected with GluR5(Q) or both GluR5(Q) and GluR6(R) at a cDNA ratio of 1:1. D, RampI–V plot for responses to 100 μm kainate recorded from a HEK cell transfected with GluR6(R). In all experiments the cells were treated with concanavalin A to attenuate desensitization.
Analysis of rectification was performed by using ramp changes in membrane potential from −105 to 105 mV (0.42 mV/msec) during equilibrium responses to selected agonists. As expected, theI–V plots for GluR5(Q) responses to I-will were birectifying because of a voltage-dependent polyamine block (Fig.1C), whereas I–V plots for GluR6(R) responses to kainate were outwardly rectifying (Fig. 1D). In contrast, when GluR5(Q) and GluR6(R) were coexpressed at a cDNA ratio of 1:1, the I–V plots for responses to 100 μm I-will were outwardly rectifying (Fig.1C) even though, as shown above, I-will is inactive at homomeric GluR6(R). This indicates that GluR5 and GluR6 can coassemble to form heteromeric kainate receptors.
Overexpression of GluR6 dominates heteromer formation with GluR5
To facilitate the analysis of the properties of heteromeric kainate receptors generated by the coassembly of GluR5 with GluR6, we used conductance–voltage plots (see Materials and Methods). When GluR5(Q) and GluR6(R) were cotransfected at a cDNA ratio of 1:1, this analysis revealed weak outward rectification for responses to both 10 μm ATPA (Fig.2A) and 100 μm I-will (Fig. 2B). In contrast, for homomeric GluR5(Q) the responses to these agonists showed strong biphasic rectification similar to that reported previously for the activation of unedited AMPA and kainate receptors (Bowie and Mayer, 1995). Surprisingly, when the cDNA ratios for GluR5(Q) to GluR6(R) were changed over the range of 1:1, 2:1, and 5:1, the responses remained outwardly rectifying (Fig. 2B), such that the ratio of the conductance at +80 to −80 mV was 1:1 = 1.53 ± 0.03 (n = 8), 2:1 = 1.73 ± 0.06 (n = 13), and 5:1 = 1.68 ± 0.04 (n = 5). Only when the cDNA ratio for GluR5(Q) to GluR6(R) was increased to 20:1 did we record responses with biphasic rectification intermediate between that for homomeric GluR5(Q) and GluR6(R) (Fig. 2C). The rectification of responses for the 20:1 ratio of cDNAs varied markedly from cell to cell (Fig.2C); such extreme variation was not observed for the GluR5 to GluR6 cDNA ratios of 1:1, 2:1, or 5:1.
Fig. 2.
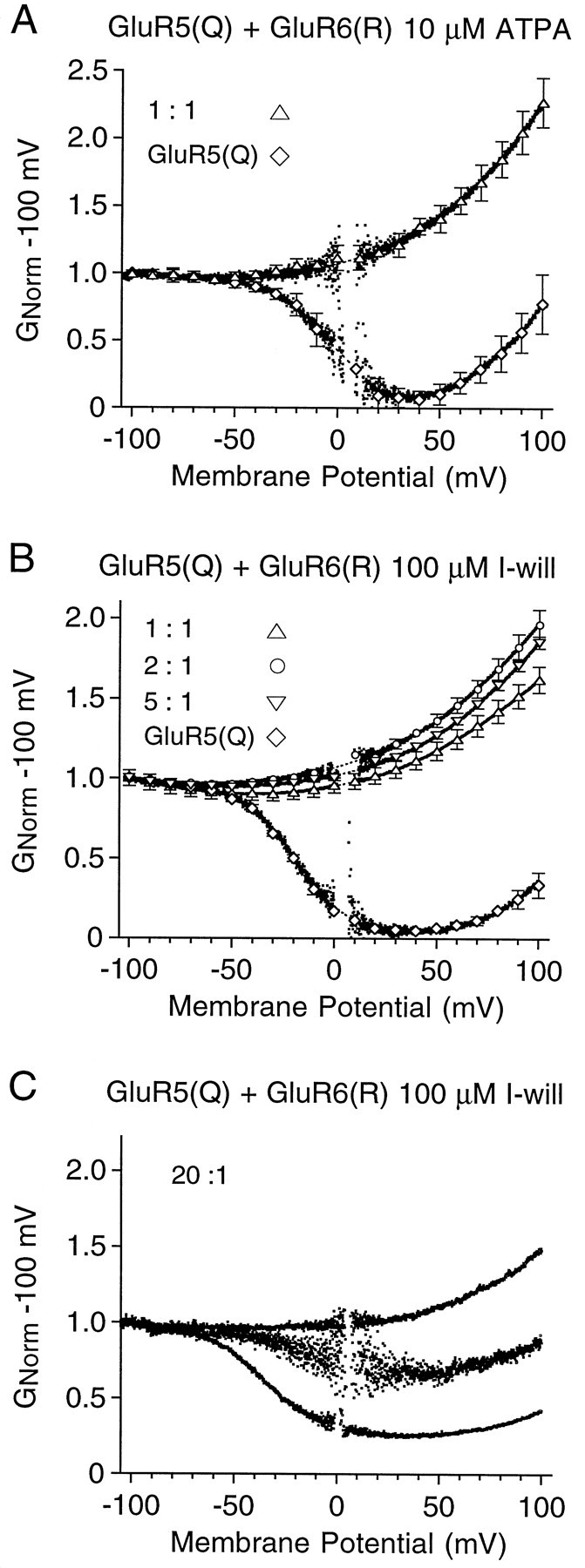
Functional overexpression of GluR6 dominates heteromer formation with GluR5. Shown are G–V plots for responses to 10 μm ATPA (A) and 100 μm I-will (B, C) recorded from HEK cells transfected with homomeric GluR5(Q) or GluR5(Q) plus GluR6(R) at cDNA ratios of 1:1, 2:1, 5:1, and 20:1, as indicated. In Aand B, the data points show the mean ± SEM for 4–12 cells per experiment; at cDNA ratios for GluR5(Q) to GluR6(R) of 1:1, 2:1, and 5:1 the responses to GluR5 selective agonists show a similar weak outward rectification. C,G–V plots for individual cells normalized to the conductance at −100 mV; at cDNA ratios of 20:1 the rectification varied from cell to cell.
Previous studies have shown that the single-channel conductance of GluR5(Q) is at least 50 to 100 times larger than that for GluR6(R) (Swanson et al., 1996). The fivefold larger amplitude of kainate responses for GluR6(R) versus GluR5(Q) described above suggests that in HEK cells the functional expression of GluR6 is considerably more efficient than that of GluR5 (see Fig. 1). We also observed a large (>40-fold) difference in response amplitudes for GluR6(Q) versus GluR5(Q) in Xenopus oocytes injected with equal amounts of cRNAs for these subunits, suggesting similar behavior in multiple heterologous expression systems. Replacing either the 5′-untranslated region (UTR) or both the 5′-UTR and signal peptide of GluR5 with that from GluR6 failed to increase the amplitude of GluR5 responses, suggesting either that the translation, assembly, cell surface expression, or stability of GluR5 is much less than that for GluR6 or that GluR5 opens with low probability as compared with GluR6. However, the latter possibility seems unlikely given similar maximum open probabilities for native kainate receptors in dorsal root ganglion (DRG) neurons (Huettner, 1990) and recombinant GluR6 (Traynelis and Wahl, 1997) and given results suggesting that kainate receptors in DRG neurons are generated by homomeric GluR5 (Partin et al., 1993; Swanson et al., 1998).
Recent experiments on polyamine block of heteromeric AMPA receptors suggest that the affinity for spermine varies with Q/R site stoichiometry (Washburn et al., 1997). It thus seemed likely that the strong attenuation of polyamine-mediated rectification observed for heteromeric kainate receptors generated by the coexpression of GluR5(Q) and GluR6(R) at cDNA ratios of 1:1, 2:1, and 5:1 (Fig.2A,B) reflects a constrained stoichiometry of assembly under these conditions rather than any functional dominance in heteromeric kainate receptors of edited versus unedited subunits. Evidence in support of this was obtained by examining changes in rectification produced by the cotransfection of GluR5(Q) with GluR5(R) or GluR6(Q) with GluR6(R). In single HEK cells we were unable to record responses for homomeric GluR5(R) because of the low amplitude of responses for this subunit (Sommer et al., 1992; Swanson et al., 1996); when expressed in Xenopus oocytes, the responses for GluR5(R) were large enough to permit reliable G–V plot analysis and revealed weak outward rectification indistinguishable from that recorded for homomeric GluR6(R). Subsequent experiments on HEK cells transfected with cDNAs for the Q and R forms of GluR5 at ratios of 1:1 and 5:1 revealed rectification intermediate between that for homomeric assemblies of edited or unedited GluR5 subunits (Fig.3A). Similar results were obtained on coexpression of GluR6(Q) and GluR6(R) (Fig. 3B).
Fig. 3.
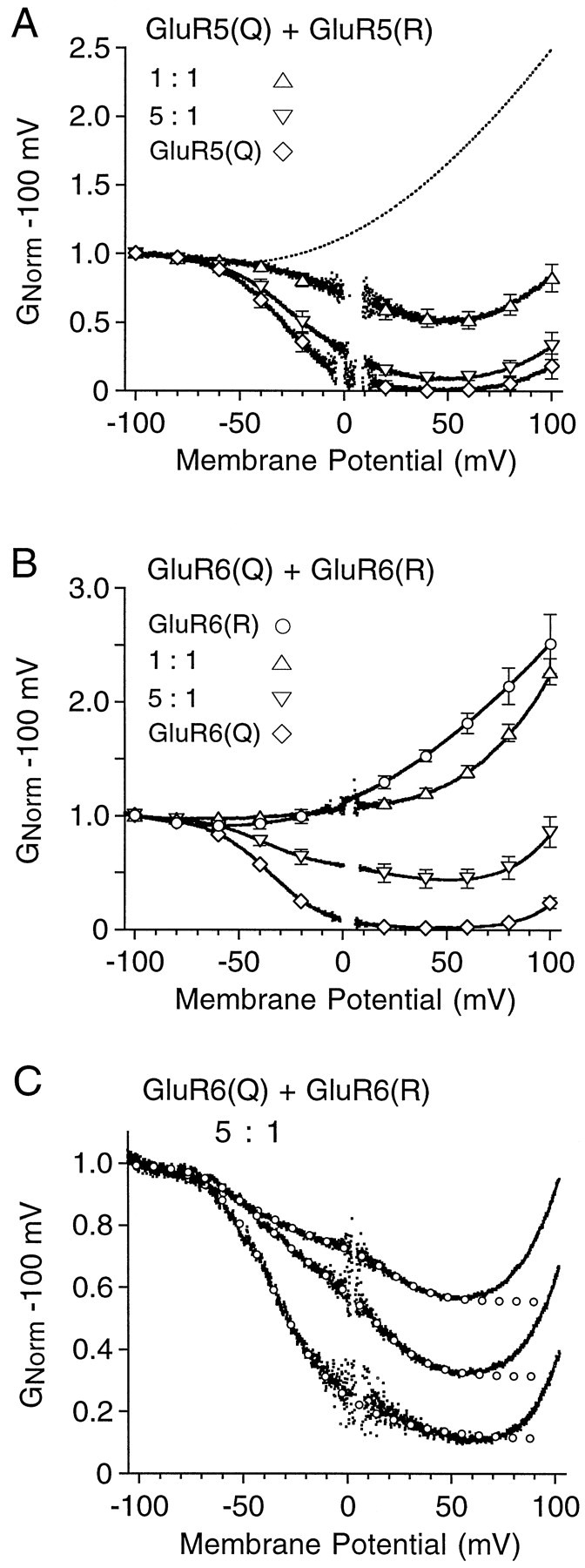
Coassembly of edited and unedited GluR5 or GluR6 generates multiple families of kainate receptors.A, G–V plots of the responses to 100 μm kainate for homomeric GluR5(Q) or GluR5(Q) plus GluR5(R) transfected at cDNA ratios of 1:1 and 5:1. B,G–V plots for homomeric GluR6(Q), homomeric GluR6(R), or GluR6(Q) plus GluR6(R) transfected at cDNA ratios of 1:1 and 5:1. The symbols show the mean ± SEM of responses to 100 μm kainate for 5–11 cells per experiment; thedotted line in A shows the responses for homomeric GluR6(R). C, Responses to 50 μmdomoate for three cells transfected with GluR6(Q) plus GluR6(R) at a cDNA ratio of 5:1; open circles indicate the fits of the sum of two Boltzmann functions over the range of −100 to +50 mV.
Although these results established that Q/R site stoichiometry in kainate receptors regulates polyamine affinity in a manner similar to that observed in previous studies on AMPA receptors (Washburn et al., 1997), we found that, when the ratio of cDNAs for the Q and R forms was increased from 1:1 to 5:1, there was considerable heterogeneity in the extent of rectification from cell to cell. Such variability resembled that described previously when GluR5 and GluR6 were coexpressed at a cDNA ratio of 20:1 (see Fig. 2). In many cases the G–Vplots for individual cells transfected with both the Q and R forms of either GluR5 or GluR6 clearly showed multiple components of polyamine block, visible as inflections on the descending limb of theG–V plot; this was particularly true for GluR6 (Fig.3C), although similar results were obtained in some cells for GluR5. As a result, in these cells the G–V plots were fit well by the sum of two Boltzmann functions, but not by a single Boltzmann (Fig. 3C). The simplest explanation for this would be the existence in single cells of multiple populations of kainate receptors with different KD values for polyamine block because of the incorporation of Q and R forms at different stoichiometry. Indeed, if the assembly of GluRs within a single gene family is dictated mainly by the relative concentrations of subunits within the endoplasmic reticulum, one would expect to observe multiple receptor populations with different sensitivity to polyamine block rather than the apparently homogeneous receptor population described in previous studies on heteromeric AMPA receptors expressed at different ratios of Q to R forms (Washburn et al., 1997). Thus, our results are explained best by the existence in individual cells of multiple receptor populations with different affinities for polyamines. For the coexpression of GluR6(Q) and GluR6(R) at a 5:1 ratio, theKD values for spermine block, calculated from fits of the sum of two Boltzmann functions (Eqs. 2, 3), were 4.5 ± 1.1 μm for the high-affinity component of the block, similar to previous estimates for homomeric GluR6(Q) (Bowie and Mayer, 1995; Bähring et al., 1997), and 238 ± 99 μm (mean ± SEM) for the low-affinity component of the block for the three cells shown in Figure3C, with the proportion of receptors with high and low affinity for spermine varying from cell to cell.
Responses to GluR5 selective agonists upregulated by coexpression with GluR6
The amplitude of responses to I-will appeared to be larger when GluR5 was coexpressed with GluR6 as compared with the responses that were recorded when GluR5 was expressed alone, suggesting that heteromeric kainate receptors assembled from GluR5 and GluR6 may have novel functional properties. This was true both for the coexpression of GluR5(R) with GluR6(Q) and for the coexpression of GluR5(Q) with GluR6(R).
In individual HEK cells we were unable to record responses for homomeric GluR5(R) larger than a few picoamps, in agreement with the results of previous studies (Sommer et al., 1992; Swanson et al., 1996). In contrast, when GluR5(R) was coexpressed with GluR6(Q) at a 1:1 ratio of cDNAs, outwardly rectifying responses were evoked reliably by the GluR5 selective agonist 100 μm I-will, with a mean amplitude of 165 ± 38 pA at −100 mV (Fig.4A,C). Although we could not quantify the effect, this suggests that functional upregulation must occur for GluR5(R) on coexpression with GluR6(Q). Strikingly, responses in the same cells to 100 μm kainate, a nonselective agonist that activates both GluR5 and GluR6, on average were 50 times larger than those to 100 μmI-will and displayed strong biphasic rectification, albeit somewhat weaker than that for homomeric GluR6(Q), suggesting that many receptors had failed to incorporate GluR5(R).
Fig. 4.
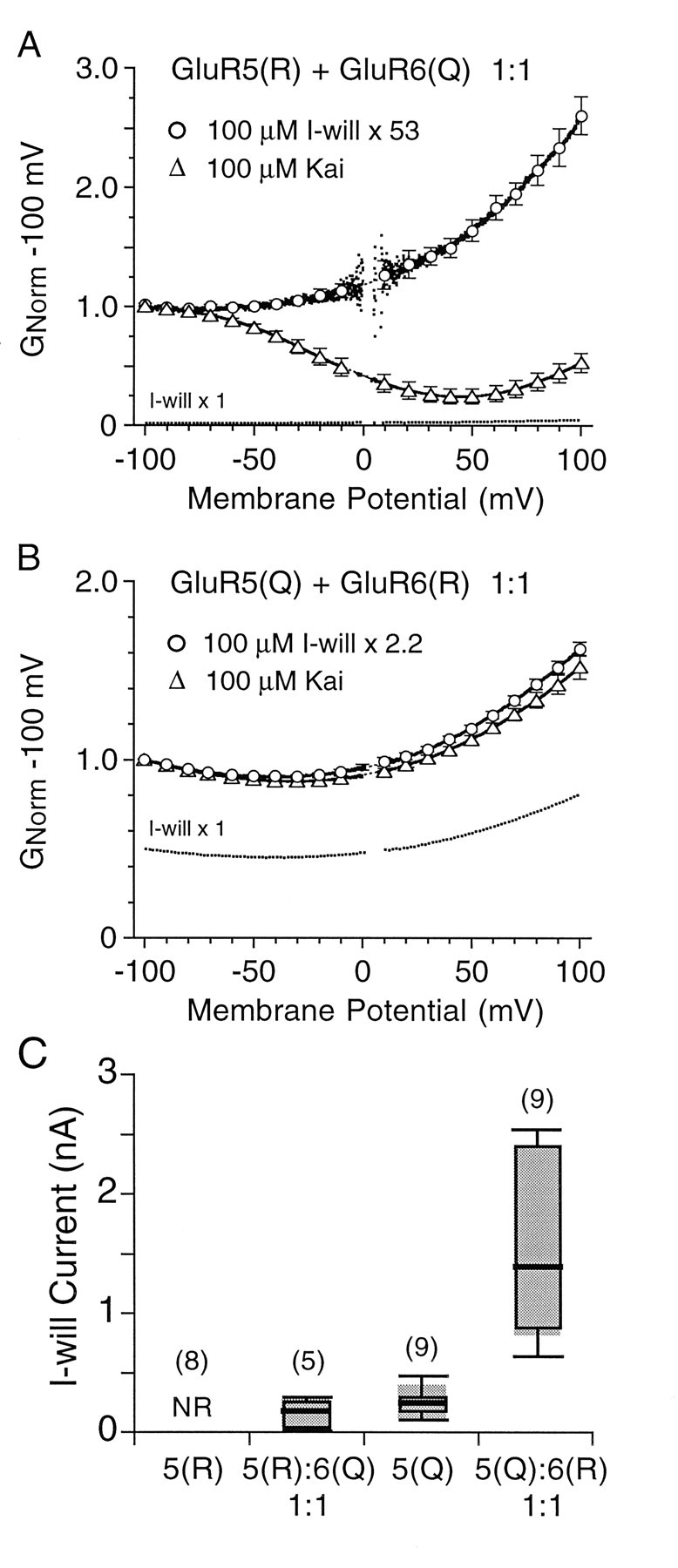
Coassembly of GluR5 and GluR6 upregulates the responses to GluR5 selective agonists. A,G–V plots for responses to 100 μm I-will and 100 μm kainate scaled to have the same amplitude at −100 mV for cells transfected with both GluR5(R) with GluR6(Q) at a cDNA ratio of 1:1; the symbols show the mean ± SEM of responses for six cells; the unscaled response to I-will is plotted as a dotted line. B,G–Vplots for responses to 100 μm I-will and 100 μm kainate normalized to have the same amplitude at −100 mV for cells transfected with both GluR5(Q) and GluR6(R) at a cDNA ratio of 1:1. The symbols show the mean ± SEM of responses for nine cells; the unscaled response to I-will is plotted as a dotted line. C, Box plots of the mean amplitude of I-will-evoked currents at −100 mV for the edited and unedited versions of GluR5 expressed alone or with GluR6 at a cDNA ratio of 1:1, as indicated; the cells were treated with concanavalin A to attenuate desensitization. The top and bottom boundaries of the boxes indicate 25 and 75% of the data, and the whiskers indicate 10 and 90% of the data. The median is indicated by a bold bar; theshaded areas indicate the mean ± SD. Note that the coexpression of GluR6 upregulates the amplitude of responses to I-will.
The amplitude of responses to the GluR5 selective agonist I-will also were upregulated when GluR5(Q) was coexpressed with GluR6(R) (Fig.4C). On average, after attenuation of desensitization by concanavalin A the responses to 100 μm I-will at −100 mV were 250 ± 46 pA (n = 9) for homomeric GluR5(Q), 933 ± 203 (n = 8) when GluR5(Q) was coexpressed with GluR6(R) at a 5:1 ratio, 1321 ± 620 pA (n = 10) at a 2:1 ratio, and 1609 ± 259 pA (n = 9) at a 1:1 ratio. When GluR5(Q) and GluR6(R) were coexpressed at a 1:1 cDNA ratio, the responses to both kainate and I-will showed weak outward rectification (Fig. 4B). The amplitude of responses to kainate was, on average, 2.2-fold larger than those to I-will recorded in the same cells. This suggests functional overexpression of GluR6(R) versus GluR5(Q), because after concanavalin A treatment the responses of homomeric GluR5(Q) to kainate and to I-will had similar steady-state amplitudes, whereas while GluR6(R) has a much smaller single-channel conductance than GluR5(Q) (Swanson et al., 1996). Alternatively, as discussed later, a larger single-channel conductance of GluR5(Q) plus GluR6(R) heteromers when all four subunits are activated by the nonselective agonist kainate, as compared with the response when only GluR5 subunits in the same GluR5 plus GluR6 heteromers are activated by I-will, also would be consistent with the results that were obtained.
Coexpression of GluR5 and GluR6 regulates kainate receptor desensitization
Previous studies have revealed that for homomeric GluR6 the responses to kainate desensitize rapidly and almost completely for Q/R site-edited and unedited forms (Partin et al., 1993; Swanson et al., 1997; Traynelis and Wahl, 1997). For homomeric GluR5(Q) and for native kainate receptors in DRG neurons, the responses to I-will also show strong desensitization with fast kinetics (Wong et al., 1994; Swanson et al., 1998). The present study revealed that, when GluR5 and GluR6 are coexpressed, the resulting heteromeric kainate receptors show reduced desensitization with an enhanced steady-state current in response to either I-will or kainate (Fig.5). For responses to 100 μmI-will the steady-state currents were 1.4 ± 0.4% of peak (n = 12) for homomeric GluR5(Q) but 10.7 ± 0.9% (n = 13) for GluR5(Q) plus GluR6(R) expressed at a cDNA ratio of 1:1. In these experiments there is no doubt that the change in desensitization results from the formation of heteromeric receptors, because GluR6(R) is unresponsive to I-will. On average, the steady-state amplitude of responses to 100 μmkainate was only 1.3 ± 0.2% of peak (n = 7) for homomeric GluR6(R), whereas for GluR5(Q) plus GluR6(R) coexpressed at a cDNA ratio of 1:1, 10 of 19 of cells showed much larger steady-state currents, on average 6.0 ± 1.0% (n = 10) of the peak response. Given the evidence for the functional overexpression of GluR6 (see Fig. 2), it is likely that in the remaining nine cells the responses to kainate were dominated by homomeric GluR6(R). Compared with responses for homomeric assemblies of GluR6(Q) or GluR6(R), the reduced desensitization observed on the coexpression of GluR5 and GluR6 brings to mind the enhanced steady-state response reported in some experiments on native kainate receptors in hippocampal neurons (Ruano et al., 1995; Wilding and Huettner, 1997; Paternain et al., 1998), suggesting that, at least in some cells, native kainate receptors may be heteromers containing GluR5 and GluR6, perhaps in combination with other subunits.
Fig. 5.
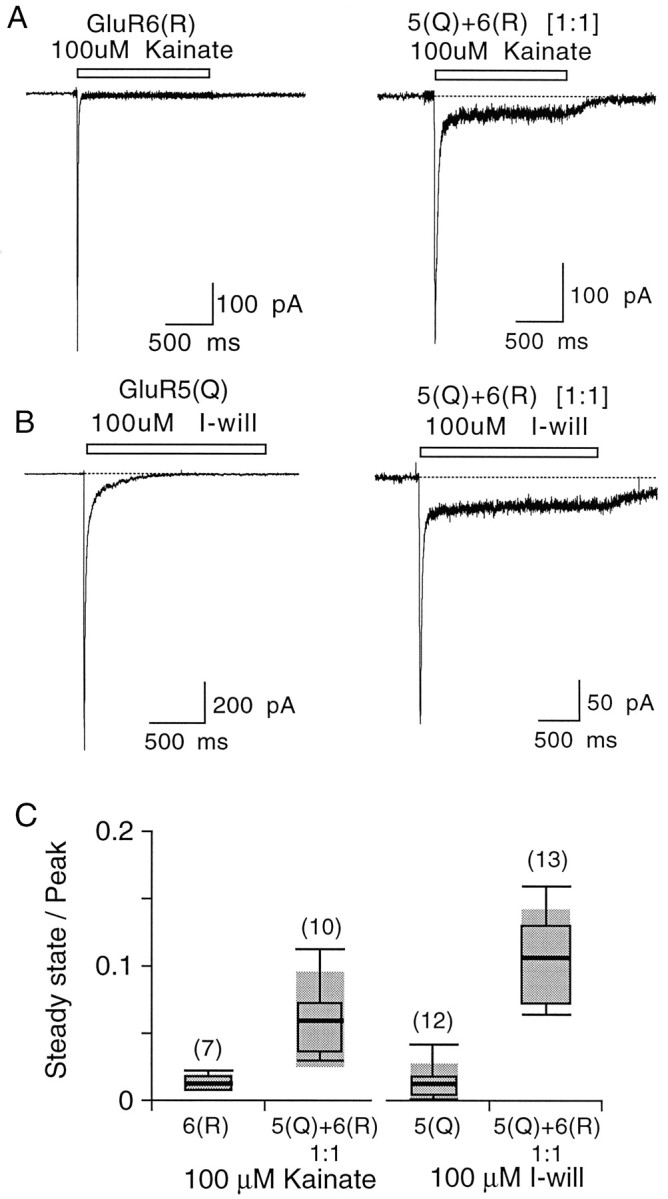
Coassembly of GluR5 and GluR6 reduces kainate receptor desensitization. A, Responses to 100 μm kainate recorded from HEK cells expressing homomeric GluR6(R) or GluR5(Q) plus GluR6(R) at a cDNA ratio of 1:1.B, Responses to 100 μm I-will recorded from HEK cells expressing homomeric GluR5(Q) or GluR5(Q) plus GluR6(R) at a cDNA ratio of 1:1. C, Desensitization, expressed as a steady-state/peak current, for homomeric GluR6(R), homomeric GluR5(Q), and GluR5(Q) plus GluR6(R) heteromers. The steady-state current amplitude was measured at 1 sec after the start of the application of the agonists. The box plots were generated by the method described in Figure 4C.
Although desensitization is reduced on the coexpression of GluR5(Q) and GluR6(R), the rate of onset of desensitization was not greatly affected. Previous studies found that homomeric GluR6 displays fast-desensitizing responses to kainate (Heckmann et al., 1996), whereas homomeric GluR5 shows profound heterogeneity in the rate of onset of desensitization (Swanson and Heinemann, 1998). When GluR5(Q) was coexpressed with GluR6(R), we found that responses to 100 μm kainate also exhibited inter-cell variability. We observed two types of desensitization: a “fast” type that can be well fit by a single exponential, τ =12.9 ± 0.8 msec (n = 9), and a “slow” type best fit by the sum of two exponentials, τ1 = 8.7 ± 0.8 msec (A1 = 74 ± 7.1%) and τ2 = 49.2 ± 10.4 msec (n= 10). The “fast” type of desensitization appeared not to differ from that for the responses of homomeric GluR6(R) to 100 μm kainate, τ = 12.5 ± 0.8 msec (n = 6). However, both the “fast” and “slow” types of desensitization developed with smaller time constants than those for responses of homomeric GluR5(Q) to 100 μm kainate: “fast” τ1 = 17.7 ± 2.7 msec (A141 ± 9.6%) and τ2 = 432 ± 85 msec (n = 7); “slow” τ1 = 117 ± 20 msec (A1 33.2 ± 8.0%) and τ2 = 452 ± 56 msec (n = 3). These results suggest that GluR6(R) is dominant over GluR5(Q) in controlling the rate of onset of desensitization. Similar to the results obtained for kainate, the responses to 100 μm I-will for homomeric GluR5(Q) also exhibited cell to cell heterogeneity in the rate of onset of desensitization. For 3 of 12 cells the responses were well fit by a single exponential, τ = 8.8 ± 1.3 msec; for 9 of 12 cells the onset of desensitization was fit by the sum of two exponentials, τ1 = 8.0 ± 1.1 msec (A1 = 78 ± 3.3%) and τ2 = 132 ± 19 msec. When GluR5(Q) was coexpressed with GluR6(R), the responses of individual cells to I-will also displayed “fast” and “slow” types of desensitization, with time constants similar to those for homomeric GluR5(Q).
For homomeric GluR5 expressed in HEK cells and for native kainate receptors expressed in DRG neurons, the recovery from desensitization evoked by I-will proceeds extremely slowly, with time constants of 2.5 and 4 min, respectively (Wong et al., 1994; Swanson et al., 1998). Rundown proceeds on the same time scale, making an accurate measurement of the kinetics of recovery from desensitization for I-will extremely difficult. To avoid this, we compared the extent of recovery from desensitization at a single 30 sec time point for homomeric GluR5(Q) and for GluR5(Q) plus GluR6(R) coexpressed at a cDNA ratio of 1:1 (Fig.6). With twin-pulse applications of 500 μm I-will separated by 30 sec, we observed 10.9 ± 1.7% (n = 5) recovery for homomeric GluR5(Q) but 48.2 ± 2.6% (n = 8) recovery for the 1:1 cotransfection of GluR5(Q) and GluR6(R). If we assume that recovery from desensitization follows single-exponential kinetics, the time constants given by these values would be 4.9 ± 1.0 min for homomeric GluR5(Q), in good agreement with previous estimates, and 0.78 ± 0.06 min for the 1:1 cotransfection of GluR5(Q) to GluR6(R). These experiments clearly reveal that the coexpression of GluR5(Q) with GluR6(R) speeds up recovery from desensitization, which might help to explain why hippocampal mossy fiber kainate receptor EPSCs do not inactivate during high-frequency presynaptic stimulation (Castillo et al., 1997; Vignes and Collingridge, 1997). We performed these experiments with the GluR5 selective agonist I-Will because, although knowledge of the changes in the kinetics of recovery from desensitization for glutamate is the physiologically relevant parameter that we need to measure to understand the behavior of EPSCs, the large population of homomeric GluR6 present when GluR5 and GluR6 are coexpressed (see Fig. 2) would interfere with the required observations.
Fig. 6.
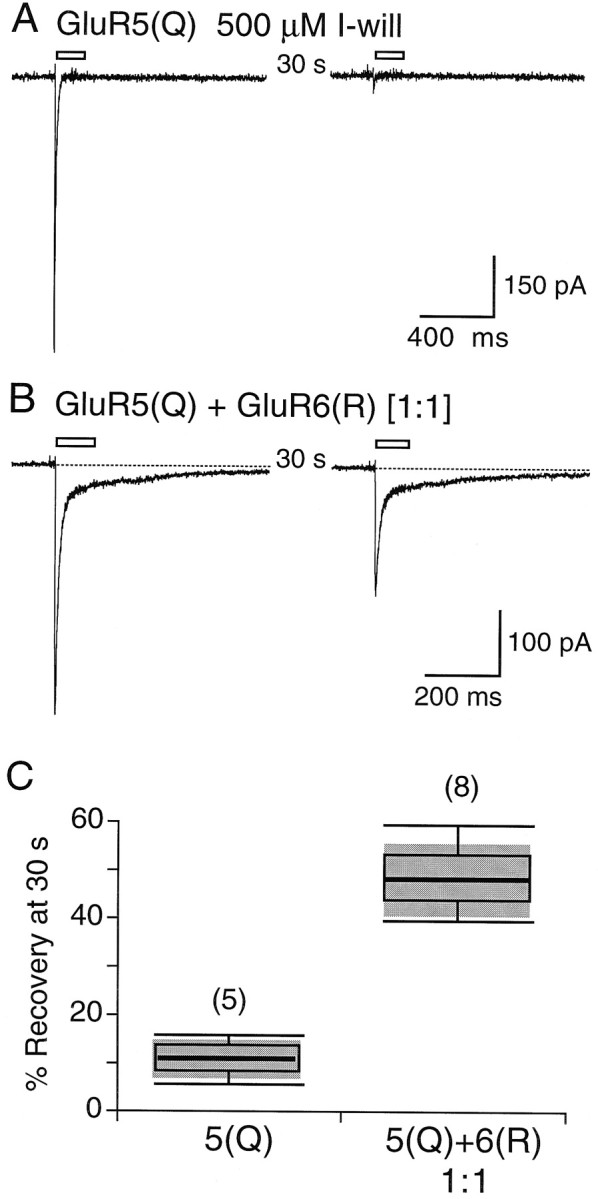
Coassembly of GluR5 and GluR6 speeds recovery from desensitization. A, Responses to paired applications of 500 μm I-will separated by 30 sec intervals reveal persistent desensitization for homomeric GluR5(Q). B, Recovery from desensitization is much faster when GluR5(Q) is coexpressed with GluR6(R) at a 1:1 ratio of cDNAs. C,Box plots for recovery from desensitization measured at 30 sec intervals between paired applications of I-will. The box plots were generated by the method described in Figure 4C.
Coassembly of GluR7 with GluR5 and GluR6
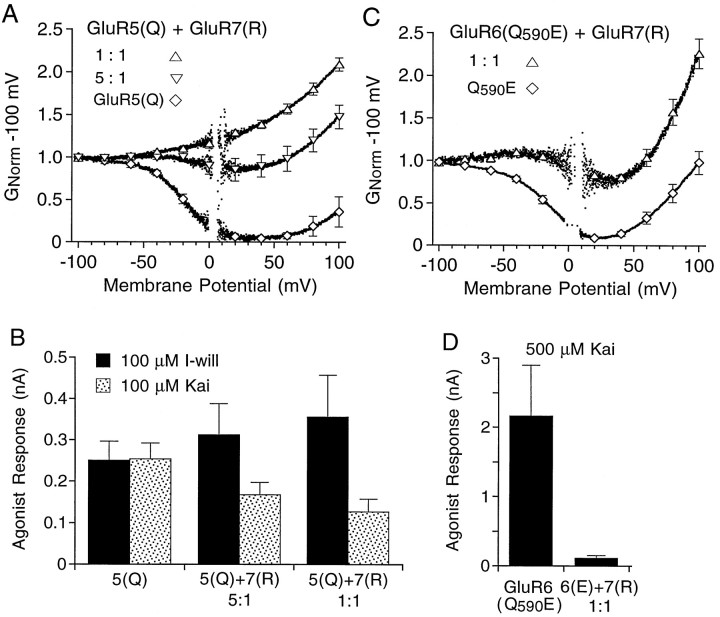
Because I-will does not activate homomeric kainate receptors assembled from GluR7 (Swanson et al., 1998), we were able to use the same strategy to test for the coassembly of GluR5 with GluR7 as was used in experiments with GluR6. Because GluR7 does not undergo Q/R site RNA editing (Lomeli et al., 1992), it was necessary first to generate the required construct by site-directed mutagenesis. When GluR5(Q) was coexpressed with GluR7(R) at cDNA ratios of 1:1 and 5:1, the responses to I-will recorded after concanavalin A treatment showed reduced biphasic rectification as compared with those for homomeric GluR5(Q) (Fig. 7A). This result indicates that GluR5 and GluR7 can coassemble to form heteromeric receptors. Two notable differences were observed in these experiments when compared with the results obtained for the coexpression of GluR5 with GluR6. First, biphasic rectification because of polyamine block was reduced to a much greater extent for the 1:1 cDNA ratio of GluR5(Q) to GluR7(R) than for the 5:1 cDNA ratio. This is similar to results obtained when the edited and unedited forms of GluR5 or GluR6 were coexpressed (see Fig. 3) but different from results obtained on coexpression of GluR5(Q) and GluR6(R) (see Fig. 2). Second, the amplitude of responses to I-will did not show clear evidence for upregulation on the coexpression of GluR5(Q) with GluR7(R). This was in marked contrast to results obtained for the coexpression of GluR5(Q) with GluR6(R). The mean amplitude of responses to 100 μm I-will was 250 ± 46 pA (n = 9) for homomeric GluR5(Q), 312 ± 76 pA (n = 8) for the 5:1 ratio of GluR5(Q) and GluR7(R), and 355 ± 100 pA (n = 4) for the 1:1 ratio; these values were not statistically different (p > 0.05). In the same cells the amplitude of responses to 100 μm kainate decreased slightly as the ratio of cDNAs for GluR5(Q) to GluR7(R) was decreased (Fig. 7B); the mean amplitudes were 254 ± 38 pA (n = 9) for homomeric GluR5(Q), 167 ± 29 pA (n = 8) for the 5:1 ratio of GluR5(Q) to GluR7(R), and 126 ± 31 pA (n = 4) for the 1:1 cDNA ratio. Although these values were also not statistically different (p > 0.05), the decline in the ratio of the amplitude of responses to kainate and I-will in individual cells was highly significant; the ratios were 1.09 ± 0.09 for homomeric GluR5(Q), 0.69 ± 0.10 (p < 0.01) for the 5:1 ratio of GluR5(Q) to GluR7(R), and 0.36 ± 0.02 (p < 0.01) for the 1:1 cDNA ratio (Fig. 7B).
Fig. 7.
Coassembly of GluR5 and GluR6 with GluR7.A, G–V plots of responses to 100 μm I-will for homomeric GluR5(Q) or GluR5(Q) and GluR7(R) transfected at cDNA ratios of 1:1 and 5:1; the symbolsindicate the mean ± SEM for three to five cells per experiment.B, Mean amplitude of responses to 100 μmkainate and 100 μm I-will recorded from cells transfected with homomeric GluR5(Q) or GluR5(Q) plus GluR7(R) at cDNA ratios of 5:1 and 1:1; the error bars indicate the mean ± SEM.C,G–V plots of responses to 500 μm kainate for homomeric GluR6(Q590E) or GluR7(R) and GluR6(Q590E) transfected at a cDNA ratio of 1:1; thesymbols indicate the mean ± SEM for five to six cells per experiment. D, Mean amplitude of responses to 500 μm kainate recorded from cells transfected with homomeric GluR6(Q590E) or GluR7(R) plus GluR6(Q590E); the error bars indicate the mean ± SEM. In all experiments the cells were treated with concanavalin A to attenuate desensitization.
The low affinity of GluR7(Q) for glutamate and kainate, rapid and complete desensitization to these agonists, and the lack of effect of concanavalin A on desensitization for GluR7(Q) (Schiffer et al., 1997) made it possible to test for the coassembly of GluR6 with GluR7 by recording responses to concentrations of kainate <1 mm,which are ineffective in activating GluR7 (Schiffer et al., 1997). In contrast to the results obtained for coexpression with GluR5(Q), when GluR7(R) was coexpressed with GluR6(Q) at cDNA ratios of 1:1, 2:1, and 5:1, extremely variable results were obtained. For some cells biphasic rectification was strongly attenuated, which indicates the formation of heteromers between GluR6(Q) and GluR7(R), whereas other cells showed little change in biphasic rectification as compared with the responses for wild-type GluR6(Q). The later case is probably attributable to a much larger macroscopic conductance for homomeric GluR6(Q) that in many cells most likely obscures the response of heteromers formed by the coassembly of GluR6(Q) and GluR7(R). At high (10:1) ratios of cDNAs for GluR7(R) to GluR6(Q), in many cells the responses to 500 μm kainate were too small to analyze accurately. These results suggest that coassembly with GluR7(R) downregulates the high levels of functional expression typical for homomeric GluR6(Q). When we repeated these experiments by using a mutant, GluR6(Q590E), which gives functional responses ∼100 times smaller than for wild-type GluR6(Q) but which has a threefold lower affinity for spermine than wild-type GluR6(Q) (Panchenko et al., 1999), more consistent results were obtained on coexpression with GluR7(R) at cDNA ratios of 1:1. For all of the cells that were tested, coexpression with GluR7(R) strongly attenuated the biphasic rectification characteristic of polyamine block for homomeric GluR6(Q590E) (Fig. 7C). In addition, these experiments revealed marked attenuation of the amplitude of responses to kainate on coexpression with GluR7(R) (Fig. 7D). The mean amplitudes of responses to 500 μm kainate were 2160 ± 740 pA (n = 6) for homomeric GluR6(Q590E) but only 110 ± 41 pA (n = 5) for the 1:1 ratio of GluR6(Q590E) to GluR7(R).
DISCUSSION
Our experiments were designed to address the issue of whether kainate receptor subunits from the GluR5–GluR7 gene family can coassemble to form heteromeric receptors with novel functional properties. Our results show clearly that they do. However, our results also raise a number of general issues related to the properties of heteromeric GluRs, ranging from the biophysical details of the mechanism of receptor gating to the signals mediating subtype-specific assembly of selected GluR subtypes and the likely subunit composition and role of kainate receptors in vivo.
Functional properties of heteromeric kainate receptors
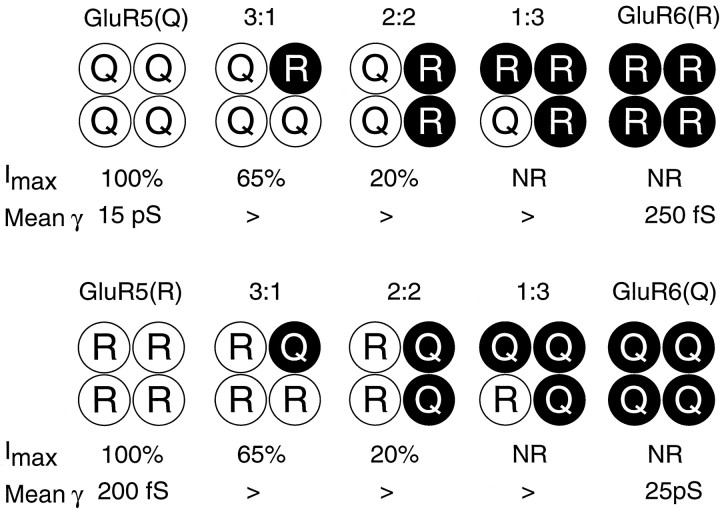
It would be predicted that, at saturating concentrations of GluR5 selective agonists, the maximal occupancy of heteromeric receptors generated by the assembly of GluR5 with GluR6 or GluR7 will be less than for homomeric GluR5 and will vary with subunit composition. The recent study of Rosenmund et al. (1998), which relates AMPA receptor single-channel conductance to the number of subunits occupied by an agonist, argued that GluRs were tetramers and that occupancy of a minimum of two agonist binding sites was required to allow the Q form of GluR3 to open to a subconductance state of 5 pS, with the binding of three and four agonist molecules producing openings to states of 15 and 23 pS. This scheme has the consequence that some of the subunit combinations likely to be generated under the conditions of our experiments will not generate currents in response to GluR5 selective agonists, even though an agonist has bound to the receptor, whereas other subunit combinations will produce much less than the maximum response possible when all agonist binding sites are occupied (Fig.8). When GluR5 is coexpressed with GluR6, according to the scheme of Rosenmund et al. (1998), we should not detect any response to GluR5 selective agonists for receptors of subunit composition 1GluR5:3GluR6, leaving only 2GluR5:2GluR6, 3GluR5:1GluR6, and homomeric GluR5 as possible functional combinations (Fig. 8). However, because of the functional overexpression of GluR6 when GluR5 and GluR6 are coexpressed at cDNA ratios of 1:1, 2:1, and even 5:1, the receptors of composition 2GluR5:2GluR6 are likely to be the major population that responds to GluR5 selective agonists rather than 3GluR5:1GluR6 or homomeric GluR5. If GluRs are pentamers and not tetramers, the quantitative details will be different but the result remains that single-channel conductance, and thus macroscopic current amplitude, will increase with the number of subunits occupied by an agonist. Such a scheme helps to explain the similar rectification for I-will responses when GluR5(Q) is coexpressed with GluR6(R) or when GluR5(R) is coexpressed with GluR6(Q) (see Figs. 2, 4), although we would expect functional overexpression of GluR6(R) in the first case and of GluR6(Q) in the second case. We suggest that, for coexpression of GluR5(Q) and GluR6(R), the dominant functional heteromer activated by GluR5 selective agonists will be 2GluR5(Q):2GluR6(R), whereas for coexpression of GluR5(R) and GluR6(Q) the dominant functional heteromer most likely will be 2GluR5(R):2GluR6(Q). Our results suggest that, no matter which form (Q or R) is activated by GluR5 selective agonists, heteromers of GluR5 and GluR6 containing 2R and 2Q subunits will exhibit similar outward rectification.
Fig. 8.
Molecular mechanisms for kainate receptor diversity. The scheme illustrates possible receptor combinations that could be formed by coassembly of the top row, GluR5(Q) with GluR6(R), and the bottom row, GluR5(R) with GluR6(Q), assuming a tetrameric stoichiometry.Imax indicates the maximum response that could be generated by a GluR5 selective agonist according to the values published by Rosenmund et al. (1998) for the activation of homomeric AMPA receptors at the appropriate receptor occupancies by GluR5 selective agonists; NR indicates that these forms would not be expected to respond to I-will or ATPA. Mean γ indicates the maximum values for single-channel conductance recorded bySwanson et al. (1996) for the Q and R forms of homomeric GluR5 and GluR6.
The results of experiments in which GluR5(Q) and GluR5(R) or GluR6(Q) and GluR6(R) were coexpressed (see Fig. 3) help to address the issue of the identity of the subunit composition of receptors with intermediate affinity for polyamine block and provide information for an interpretation of responses from heteromers containing more than one kainate receptor gene product. Although outward rectification that is independent of polyamine block (Bähring et al., 1997) limits the resolution of these experiments, in cells transfected at a 5:1 ratio of GluR6(Q) to GluR6(R) the G–V plots were well fit, assuming only two polyamine-sensitive receptor populations, expressed at different relative amounts (see Fig. 3C). One of these, with an affinity for spermine similar to that for homomeric GluR6(Q), most likely is a tetramer of unedited subunits. The second population, with 50-fold lower affinity, most likely represents the form 3Q:1R. If this is true, it seems reasonable that the forms 2Q:2R and 1Q:3R would have even lower affinity for polyamines and not be detectable as inflections on G–V plots under the conditions of our experiments. Returning to the case of heteromers formed by coassembly of GluR5 and GluR6 (see Figs. 2, 4), our results support the proposal that 2Q:2R heteromers, which we believe are not blocked by cytoplasmic concentrations of polyamines under the conditions of our experiments, underlie the weak outward rectification observed for I-will responses for both GluR5(Q):GluR6(R) and GluR5(R):GluR6(Q) expressed at 1:1 cDNA ratios.
The coexpression of GluR6 or GluR7 with GluR5 had different effects on the amplitude of equilibrium responses to agonists expected to activate GluR5 subunits selectively (see Figs. 4, 7). In the case of GluR6, which generates functional receptors with much higher efficiency than other AMPA or kainate receptor subunit combinations, it is possible that assembly or cell surface expression is facilitated, as compared with other GluR subunits, and that GluR6 acts as a chaperone when combined with GluR5. It is also possible that the upregulation of the amplitude of I-will responses is attributable to an effect of GluR6 on the gating of GluR5. Evidence consistent with such an effect is the observation that the coassembly of GluR5 with GluR6 reduces the extent of and speeds recovery from desensitization for GluR5 selective agonists. When GluR7 was coexpressed with GluR5, there was a reduction in the relative amplitude of responses to kainate versus I-will (see Fig. 7B). This indicates that GluR7 and GluR6 have different effects on GluR5. When GluR7 was coexpressed with GluR6, the amplitude of equilibrium responses to kainate was reduced dramatically (see Fig.7C). Although high concentrations of kainate are required to activate GluR7 channel gating (Schiffer et al., 1997), lower concentrations most likely produce desensitization; it is possible that, when GluR7 combines with GluR6, strong desensitization mediated by GluR7 interferes with the activation of GluR6 subunits even after treatment with concanavalin A. At the present time it is unknown whether the desensitization of GluRs occurs independently in individual subunits or whether this is a cooperative process in which individual subunits are coupled allosterically to their neighbors.
Subunit composition of native kainate receptors
Kainate receptor mRNAs are expressed widely throughout the CNS with levels that are higher at early developmental stages (Bettler et al., 1990; Wisden and Seeburg, 1993; Bahn et al., 1994; Bischoff et al., 1997). Although the brain region-specific expression patterns for GluR5, GluR6, and GluR7 are unique, they overlap in multiple regions. However, relatively few studies directly address whether single cells express more than one member of this gene family. Acutely isolated, purified cerebellar granule cells were shown by PCR to express RNA for both GluR5 and GluR6 (Belcher and Howe, 1997), with similar results obtained by single-cell PCR after short-term culture (Pemberton et al., 1998). The extent of RNA editing was different for the two kainate receptor subunits and increased during development but to different extents such that, by postnatal day 15, the most common forms were GluR5(Q) and GluR6(R). Although pharmacological approaches convincingly showed functional kainate receptors to be present in cerebellar granule cells (Pemberton et al., 1998), their physiological role and subunit composition remain unknown, but it is noteworthy that kainate receptors are expressed in the external germinal layer, before the excitatory synaptic innervation of granule cells (Ripellino et al., 1998).
In the case of hippocampal neurons, single-cell PCR of unidentified cells in short-term culture, which showed rapidly desensitizing responses to kainate, revealed the expression of GluR6 in 11of 11 cells that were examined, with coexpression of GluR5 in only 3 of the 11 cells (Ruano et al., 1995). However, as noted above, although mossy fiber kainate receptor-mediated EPSCs in CA3 neurons are absent in GluR6−/− mice, the functional properties of these synaptic currents are not well explained by those of homomeric GluR6. Most likely this is because additional subunits are required to generate or correctly target the postsynaptic kainate receptors in CA3 neurons. These additional subunits could be GluR5, KA1, or KA2, because the coassembly of GluR6 with either GluR5 or KA2 confers sensitivity to ATPA or I-will (Swanson et al., 1998). Despite this, experiments with GluR6−/− mice indicate that GluR6 plays a unique role in generating synaptic kainate receptors in CA3 neurons, perhaps by stabilizing or enhancing the functional response of other kainate receptor subunits. In contrast, trigeminal ganglion neurons (Sahara et al., 1997) and DRG neurons (Partin et al., 1993) express mRNA for GluR5 at much higher levels than for GluR6 or other kainate receptor subunits, suggesting considerable diversity of kainate receptor subunit composition in different areas of the CNS. This is reinforced by the results of in situ hybridization that reveal a large number of potential subunit combinations (Wisden and Seeburg, 1993; Bahn et al., 1994; Bischoff et al., 1997). Clearly, much remains to be learned about kainate receptors at many levels, from basic properties to their role in synaptic circuitry and behavior.
Footnotes
We thank Drs. P. Seeburg and S. Heinemann for cDNAs; Drs. J. C. Watkins, D. E. Jane, and P. Krogsgaard-Larsen for gifts of (S)-5-iodowillardiine and ATPA; Dr. V. Panchenko for Igor templates; Ms. Carla Glasser for technical assistance; and Dr. C. McBain for comments on this manuscript.
Correspondence should be addressed to Dr. M. L. Mayer, Building 49, Room 5A78, National Institutes of Health, 49 Convent Drive, Bethesda MD 20892-4495.
REFERENCES
- 1.Bahn S, Volk B, Wisden W. Kainate receptor gene expression in the developing brain. J Neurosci. 1994;14:5525–5547. doi: 10.1523/JNEUROSCI.14-09-05525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bähring R, Bowie D, Benveniste M, Mayer ML. Permeation and block of rat GluR6 glutamate receptor channels by internal and external polyamines. J Physiol (Lond) 1997;502:575–589. doi: 10.1111/j.1469-7793.1997.575bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belcher SM, Howe JR. Characterization of RNA editing of the glutamate-receptor subunits GluR5 and GluR6 in granule cells during cerebellar development. Brain Res Mol Brain Res. 1997;52:130–138. doi: 10.1016/s0169-328x(97)00252-0. [DOI] [PubMed] [Google Scholar]
- 4.Bettler B, Boulter J, Hermans-Borgmeyer I, O’Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron. 1990;5:583–595. doi: 10.1016/0896-6273(90)90213-y. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff S, Barhanin J, Bettler B, Mulle C, Heinemann S. Spatial distribution of kainate receptor subunit mRNA in the mouse basal ganglia and ventral mesencephalon. J Comp Neurol. 1997;379:541–562. doi: 10.1002/(sici)1096-9861(19970324)379:4<541::aid-cne6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 6.Bowie D, Mayer ML. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995;15:453–462. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 7.Brose N, Huntley GW, Stern-Bach Y, Sharma G, Morrison JH, Heinemann SF. Differential assembly of coexpressed glutamate receptor subunits in neurons of rat cerebral cortex. J Biol Chem. 1994;269:16780–16784. [PubMed] [Google Scholar]
- 8.Bureau I, Bischoff S, Heinemann SF, Mulle C. Kainate receptor-mediated responses in the CA1 field of wild-type and GluR6-deficient mice. J Neurosci. 1999;19:653–663. doi: 10.1523/JNEUROSCI.19-02-00653.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castillo PE, Malenka RC, Nicoll RA. Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature. 1997;388:182–186. doi: 10.1038/40645. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke VR, Ballyk BA, Hoo KH, Mandelzys A, Pellizzari A, Bath CP, Thomas J, Sharpe EF, Davies CH, Ornstein PL, Schoepp DD, Kamboj RK, Collingridge GL, Lodge D, Bleakman D. A GluR5 kainate receptor that regulates inhibitory synaptic transmission in the hippocampus. Nature. 1997;389:599–603. doi: 10.1038/39315. [DOI] [PubMed] [Google Scholar]
- 12.DeVries SH, Schwartz EA. Kainate receptors mediate synaptic transmission between cones and “OFF” bipolar cells in a mammalian retina. Nature. 1999;397:157–160. doi: 10.1038/16462. [DOI] [PubMed] [Google Scholar]
- 13.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–45. [PubMed] [Google Scholar]
- 14.Everts I, Petroski R, Kizelsztein P, Teichberg VI, Heinemann SF, Hollmann M. Lectin-induced inhibition of desensitization of the kainate receptor GluR6 depends on the activation state and can be mediated by a single native or ectopic N-linked carbohydrate side chain. J Neurosci. 1999;19:916–927. doi: 10.1523/JNEUROSCI.19-03-00916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frerking M, Malenka RC, Nicoll RA. Synaptic activation of kainate receptors on hippocampal interneurons [see comments]. Nat Neurosci. 1998;1:479–486. doi: 10.1038/2194. [DOI] [PubMed] [Google Scholar]
- 16.Heckmann M, Bufler J, Franke C, Dudel J. Kinetics of homomeric GluR6 glutamate receptor channels. Biophys J. 1996;71:1743–1750. doi: 10.1016/S0006-3495(96)79375-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herb A, Burnashev N, Werner P, Sakmann B, Wisden W, Seeburg PH. The KA-2 subunit of excitatory amino acid receptors shows widespread expression in brain and forms ion channels with distantly related subunits. Neuron. 1992;8:775–785. doi: 10.1016/0896-6273(92)90098-x. [DOI] [PubMed] [Google Scholar]
- 18.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 19.Huettner JE. Glutamate receptor channels in rat dorsal root ganglion neurons: activation by kainate and quisqualate, and blockade of desensitization by concanavalin A. Neuron. 1990;5:255–266. doi: 10.1016/0896-6273(90)90163-a. [DOI] [PubMed] [Google Scholar]
- 20.Keinänen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- 21.Köhler M, Burnashev N, Sakmann B, Seeburg PH. Determinants of Ca2+ permeability in both TM1 and TM2 of high-affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- 22.Lauridsen J, Honore T, Krogsgaard-Larsen P. Ibotenic acid ana-logues. Synthesis, molecular flexibility, and in vitro activity of agonists and antagonists at central glutamic acid receptors. J Med Chem. 1985;28:668–672. doi: 10.1021/jm50001a022. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Rogawski MA. GluR5 kainate receptor-mediated synaptic transmission in rat basolateral amygdala in vitro. Neuropharmacology. 1998;37:1279–1286. doi: 10.1016/s0028-3908(98)00109-9. [DOI] [PubMed] [Google Scholar]
- 24.Li P, Wilding TJ, Kim SJ, Calejesan AA, Huettner JE, Zhuo M. Kainate-receptor-mediated sensory synaptic transmission in mammalian spinal cord. Nature. 1999;397:161–164. doi: 10.1038/16469. [DOI] [PubMed] [Google Scholar]
- 25.Lomeli H, Wisden W, Köhler M, Keinänen K, Sommer B, Seeburg PH. High-affinity kainate and domoate receptors in rat brain. FEBS Lett. 1992;307:139–143. doi: 10.1016/0014-5793(92)80753-4. [DOI] [PubMed] [Google Scholar]
- 26.Mulle C, Sailer A, Perez-Otano I, Dickinson-Anson H, Castillo PE, Bureau I, Maron C, Gage FH, Mann JR, Bettler B, Heinemann SF. Altered synaptic physiology and reduced susceptibility to kainate-induced seizures in GluR6-deficient mice. Nature. 1998;392:601–605. doi: 10.1038/33408. [DOI] [PubMed] [Google Scholar]
- 27.Panchenko VA, Glasser CR, Partin KM, Mayer ML (1999) Amino acid substitutions in the pore of rat glutamate receptors at sites influencing block by polyamines. J Physiol (Lond), in press. [DOI] [PMC free article] [PubMed]
- 28.Partin KM, Patneau DK, Winters CA, Mayer ML, Buonanno A. Selective modulation of desensitization at AMPA versus kainate receptors by cyclothiazide and concanavalin A. Neuron. 1993;11:1069–1082. doi: 10.1016/0896-6273(93)90220-l. [DOI] [PubMed] [Google Scholar]
- 29.Paternain AV, Rodriguez-Moreno A, Villarroel A, Lerma J. Activation and desensitization properties of native and recombinant kainate receptors. Neuropharmacology. 1998;37:1249–1259. doi: 10.1016/s0028-3908(98)00098-7. [DOI] [PubMed] [Google Scholar]
- 30.Pemberton KE, Belcher SM, Ripellino JA, Howe JR. High-affinity kainate-type ion channels in rat cerebellar granule cells. J Physiol (Lond) 1998;510:401–420. doi: 10.1111/j.1469-7793.1998.401bk.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puchalski RB, Louis JC, Brose N, Traynelis SF, Egebjerg J, Kukekov V, Wenthold RJ, Rogers SW, Lin F, Moran T, Morrison JH, Heinemann SF. Selective RNA editing and subunit assembly of native glutamate receptors. Neuron. 1994;13:131–147. doi: 10.1016/0896-6273(94)90464-2. [DOI] [PubMed] [Google Scholar]
- 32.Ripellino JA, Neve RL, Howe JR. Expression and heteromeric interactions of non-NMDA glutamate receptor subunits in the developing and adult cerebellum. Neuroscience. 1998;82:485–497. doi: 10.1016/s0306-4522(97)00296-0. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Moreno A, Herreras O, Lerma J. Kainate receptors presynaptically downregulate GABAergic inhibition in the rat hippocampus. Neuron. 1997;19:893–901. doi: 10.1016/s0896-6273(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 34.Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- 35.Ruano D, Lambolez B, Rossier J, Paternain AV, Lerma J. Kainate receptor subunits expressed in single cultured hippocampal neurons: molecular and functional variants by RNA editing. Neuron. 1995;14:1009–1017. doi: 10.1016/0896-6273(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 36.Sahara Y, Noro N, Iida Y, Soma K, Nakamura Y. Glutamate receptor subunits GluR5 and KA-2 are coexpressed in rat trigeminal ganglion neurons. J Neurosci. 1997;17:6611–620. doi: 10.1523/JNEUROSCI.17-17-06611.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiffer HH, Swanson GT, Heinemann SF. Rat GluR7 and a carboxy-terminal splice variant, GluR7b, are functional kainate receptor subunits with a low sensitivity to glutamate. Neuron. 1997;19:1141–1146. doi: 10.1016/s0896-6273(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 38.Sommer B, Burnashev N, Verdoorn TA, Keinänen K, Sakmann B, Seeburg PH. A glutamate receptor channel with high affinity for domoate and kainate. EMBO J. 1992;11:1651–1656. doi: 10.1002/j.1460-2075.1992.tb05211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson GT, Heinemann SF. Heterogeneity of homomeric GluR5 kainate receptor desensitization expressed in HEK293 cells. J Physiol (Lond) 1998;513:639–646. doi: 10.1111/j.1469-7793.1998.639ba.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanson GT, Feldmeyer D, Kaneda M, Cull-Candy SG. Effect of RNA editing and subunit coassembly on single-channel properties of recombinant kainate receptors. J Physiol (Lond) 1996;492:129–142. doi: 10.1113/jphysiol.1996.sp021295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson GT, Gereau RW, Green T, Heinemann SF. Identification of amino acid residues that control functional behavior in GluR5 and GluR6 kainate receptors. Neuron. 1997;19:913–926. doi: 10.1016/s0896-6273(00)80972-1. [DOI] [PubMed] [Google Scholar]
- 42.Swanson GT, Green T, Heinemann SF. Kainate receptors exhibit differential sensitivities to (S)-5-iodowillardiine. Mol Pharmacol. 1998;53:942–949. [PubMed] [Google Scholar]
- 43.Traynelis SF, Wahl P. Control of rat GluR6 glutamate receptor open probability by protein kinase A and calcineurin. J Physiol (Lond) 1997;503:513–531. doi: 10.1111/j.1469-7793.1997.513bg.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vignes M, Collingridge GL. The synaptic activation of kainate receptors. Nature. 1997;388:179–182. doi: 10.1038/40639. [DOI] [PubMed] [Google Scholar]
- 45.Vignes M, Bleakman D, Lodge D, Collingridge GL. The synaptic activation of the GluR5 subtype of kainate receptor in area CA3 of the rat hippocampus. Neuropharmacology. 1997;36:1477–1481. doi: 10.1016/s0028-3908(97)00158-5. [DOI] [PubMed] [Google Scholar]
- 46.Vignes M, Clarke VR, Parry MJ, Bleakman D, Lodge D, Ornstein PL, Collingridge GL. The GluR5 subtype of kainate receptor regulates excitatory synaptic transmission in areas CA1 and CA3 of the rat hippocampus. Neuropharmacology. 1998;37:1269–1277. doi: 10.1016/s0028-3908(98)00148-8. [DOI] [PubMed] [Google Scholar]
- 47.Vyklicky L, Benveniste M, Mayer ML. Modulation of N-methyl-d-aspartic acid receptor desensitization by glycine in mouse cultured hippocampal neurones. J Physiol (Lond) 1990;428:313–331. doi: 10.1113/jphysiol.1990.sp018214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Washburn MS, Numberger M, Zhang S, Dingledine R. Differential dependence on GluR2 expression of three characteristic features of AMPA receptors. J Neurosci. 1997;17:9393–9406. doi: 10.1523/JNEUROSCI.17-24-09393.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wenthold RJ, Trumpy VA, Zhui WS, Petralia RS. Biochemical and assembly properties of GluR6 and KA2, two members of the kainate receptor family determined with subunit-specific antibodies. J Biol Chem. 1994;269:1332–1339. [PubMed] [Google Scholar]
- 50.Werner P, Voigt M, Keinänen K, Wisden W, Seeburg PH. Cloning of a putative high-affinity kainate receptor expressed predominantly in hippocampal CA3 cells. Nature. 1991;351:742–744. doi: 10.1038/351742a0. [DOI] [PubMed] [Google Scholar]
- 51.Wilding TJ, Huettner JE. Activation and desensitization of hippocampal kainate receptors. J Neurosci. 1997;17:2713–2721. doi: 10.1523/JNEUROSCI.17-08-02713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wisden W, Seeburg PH. A complex mosaic of high-affinity kainate receptors in rat brain. J Neurosci. 1993;13:3582–3598. doi: 10.1523/JNEUROSCI.13-08-03582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong LA, Mayer ML, Jane DE, Watkins JC. Willardiines differentiate agonist binding sites for kainate- versus AMPA-preferring glutamate receptors in DRG and hippocampal neurons. J Neurosci. 1994;14:3881–3897. doi: 10.1523/JNEUROSCI.14-06-03881.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]