Abstract
A developmentally regulated Homer/Vesl isoform, Cupidin (Homer 2a/Vesl-2Δ11), was isolated from postnatal mouse cerebellum using a fluorescent differential display strategy. The strongest expression of Cupidin was detected in the cerebellar granule cells at approximately postnatal day 7. Cupidin was enriched in the postsynaptic density fraction, and its immunoreactivity was concentrated at glomeruli of the inner granular layer when active synaptogenesis occurred. Cupidin protein could be divided into two functional domains: the N-terminal portion, which was highly conserved among Homer/Vesl family proteins, and the C-terminal portion, which consisted of a putative coiled-coil structure, including several leucine zipper motifs. The N-terminal fragment of Cupidin, which was able to associate with metabotropic glutamate receptor 1 (mGluR1), also interacted with F-actin in vitro. In keeping with this, F-actin immunocytochemically colocalized with Cupidin in cultured cerebellar granule cells, and a Cupidin–mGluR1–actin complex was immunoprecipitated from crude cerebellar lysates using an anti-Cupidin antibody. On the other hand, the C-terminal portion of Cupidin bound to Cdc42, a member of Rho family small GTPases, in a GTP-dependent mannerin vitro, and Cupidin functionally interacted with activated-Cdc42 in a heterologous expression system. Together, our findings indicate that Cupidin may serve as a postsynaptic scaffold protein that links mGluR signaling with actin cytoskeleton and Rho family proteins, perhaps during the dynamic phase of morphological changes that occur during synapse formation in cerebellar granule cells.
Keywords: Cupidin/Homer 2a/Vesl-2Δ11, cerebellar development, granule cell, mGluR, actin cytoskeleton, Rho family small GTPases
Neuronal migration, neurite extension, and synaptogenesis are three distinct steps that are essential for the maturation of a neuron and the development of the nervous system. In rodent cerebellar granule cells, in particular, migration and synaptogenesis are sequentially initiated and accomplished within the first 3 weeks after birth (Rakic, 1971; Hatten, 1997). During this period, postmitotic granule cells in the external germinal layer (EGL) start to migrate downward into the molecular layer (ML), crawling along processes of Bergmann glia, until they reach their final destination in the internal granular layer (IGL). In parallel with these events, the granule cells also extend their T-shaped axons called parallel fibers (PFs) in the ML. The PFs form synapses with the growing dendrites of Purkinje neurons. In the IGL, the axodendritic connections (glomeruli) are formed between the mossy fibers (MFs)–Golgi neurons and the dendrites of the granule cells (Altman and Bayer, 1996).
What are the critical cellular and molecular events in these developmental processes? One fundamental challenge lies in understanding the mechanisms of dynamic changes performed by the neurons in response to extracellular cues available at cell–cell and cell–matrix contacts and to various classes of diffusible factors and chemical neurotransmitters. It is believed that extracellular cues direct neuronal migration and specify synapse formation with the appropriate targets during axonal and dendritic outgrowth. Very little, however, is known about the intracellular signaling machinery that regulates the motility and shape of cerebellar neurons during these active neurogenetic stages. To elucidate the molecular basis of these neuronal events, we have begun to identify genes specifically expressed at a distinct stage of the postnatal cerebellar development using a fluorescence differential display (FDD) technique (Ito et al., 1994). We were especially interested in isolating genes whose expression peaked at approximately postnatal day 7 (P7), because this period matches with the time window when most of the dynamic cellular changes described above occur (Altman and Bayer, 1996).
Here, we report the isolation of a gene named Cupidin (CPD), which fulfills the criteria of developmentally regulated cerebellar gene. Cupidin was named after the Roman god of love, Cupid, because Cupidin mediated the association of several distinct classes of molecules thought to be remote: the actin-cytoskeleton, Rho family proteins, and metabotropic glutamate receptor 1α (mGluR1α). While this work was in progress, two other groups independently reported the isolation of an isoform of Homer (Brakeman et al., 1997) or Vesl (Kato et al., 1997) that was essentially identical to Cupidin, named Homer 2a (Xiao et al., 1998) or Vesl-2Δ11 (Kato et al., 1998). Homer/Vesl was initially characterized as an immediate early gene induced by synaptic-activity (Kato et al., 1997; Brakeman et al., 1997). Here, we show that Cupidin is developmentally regulated and is enriched at synapses on cerebellar granule cell dendrites. Furthermore, a potential link between mGluR and synaptic actin cytoskeleton via Cupidin is suggested.
MATERIALS AND METHODS
Fluorescence differential display. FDD was performed according to Ito et al. (1994). In brief, 2.5 μg of DNA-free total RNAs, prepared from cerebella of ICR mice (Nippon SLC, Shizuoka, Japan) at various developmental stages [embryonic day 18 (E18), P0, P3, P7, P12, P15, P21, and 8-week-old], were reverse-transcribed using a Superscript II reverse transcriptase (Life Technologies, Rockville, MD) in the presence of 2.5 μm anchored oligo-dT primers 5′-GT15A-3′. PCR was performed using an ExTaq polymerase kit (Takara Shuzo, Kyoto, Japan), the anchored primer, and arbitrary primers (10-mer), essentially as described by Ito et al. (1994). Clone 13–2 was generated using 5′-GTTTTCGCAG-3′ as an arbitrary primer. The PCR products were separated on a 6% polyacrylamide gel and analyzed by a computerized-fluorescent image analyzer FluorImager (Amersham Pharmacia Biotech, Piscataway, NJ) after staining with the fluorescent dye SYBR Green I (Takara Shuzo). Bands of interest were excised from FDD gels and were reamplified as described previously (Ito et al., 1994). Reamplified DNA fragments were cloned into the pT7Blue TA cloning vector (Novagen, Madison, WI) and were sequenced by the dideoxy chain termination method.
Reverse transcription-PCR analysis.A series of first strand cDNAs were produced by reverse-transcription (RT) from 20 ng of total cerebellar RNAs at the various developmental stages described above, using an oligo-dT primer. PCR was performed using a primer set corresponding to the end sequences of clone 13–2 (5′-CAGGGATGTTTAGATCTTCC-3′ and 5′-GTGGTTGACAATGTCATGTC-3′). Cycling conditions were as follows: 94°C for 3 min, 25 cycles of 94°C (15 sec), 55°C (2 min), and 72°C (1 min), and finally at 72°C for 5 min. The PCR products were analyzed as described above. To analyze tissue distribution, total RNAs prepared from various tissues of P7 mice were used for RT-PCR. RT-PCR for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed with primers 5′-GCCATCAACGACCCCTTCATTGACCTC-3′ and 5′-GCCATGTAGGCCATGAGGTCCACCAC-3′ as an internal control.
Northern blot hybridization analysis. A total of 10 μg of poly(A+) RNA for each sample was resolved by gel electrophoresis and transferred to a nylon membrane (Biodyne A; Pall BioSupport, East Hill, NY). The blot was hybridized with a random-primed [α-32P]dCTP-labeled full-length Cupidin cDNA probe.
In situ hybridization. Digoxigenin-labeled antisense or sense riboprobes were prepared from the nucleotide positions 647–1220 [amino acids (aa) 154–343 plus termination codon] of the Cupidin cDNA using a digoxigenin-dUTP labeling kit (Boehringer Mannheim, Indianapolis, IN). Cryosections of mouse brain (20-μm-thick) were fixed in 4% paraformaldehyde for 15 min, washed twice in PBS, and treated with freshly prepared 50 μg/ml proteinase K (Life Technologies) for 10 min at room temperature. After acetylation, sections were subjected to the digoxigenin-based hybridization procedures (Kondo et al., 1997). Briefly, sections were incubated in hybridization buffer containing 0.2 μg/ml digoxigenin-labeled riboprobes at 60°C overnight in a humid chamber. Hybridized sections were washed by successively immersing in 1× SSC (150 mm NaCl and 15 mm sodium citrate, pH 7.0) (60°C, 10 min, twice), 2× SSC (37°C, 10 min), 2× SSC containing 20 μg/ml RNaseA (37°C, 30 min), 2× SSC (37°C, 10 min), and 0.2× SSC (60°C, 30 min, twice). Hybridization signals were detected with the digoxigenin detection kit (Boehringer Mannheim).
Cloning of mouse Cupidin (Homer 2a/Vesl-2Δ11), Homer 1c/Vesl-1L, Homer 2b/Vesl-2, and Homer 3 cDNA. Clone 13–2 obtained from FDD was used as a probe to clone mouse Cupidin cDNA. Approximately 1 × 106 plaques of a cDNA library constructed from P6 cerebella of C57Bl/6J mice (a kind gift from Dr. M. E. Hatten, Rockefeller University, New York, NY) were screened using a 32P-labeled probe. Sequencing was performed in both directions by the dideoxy chain termination method using [α-32P] dCTP and the BcaBest dideoxy sequencing kit (Takara Shuzo). Homology search with the sequences in the GenBank database was performed using the BLAST and FASTA programs. Mouse Homer 1c/Vesl-1L, Homer 2b/Vesl-2, and Homer 3 cDNAs were cloned by PCR-based method with specific primer pairs corresponding to the reported cDNA sequences (Xiao et al., 1998). All cDNA sequences cloned were verified by sequencing.
Expression of glutathione S-transferase fusion proteins in Escherichia coli. Glutathione S-transferase (GST) fusion constructs were made by subcloning various parts of Cupidin cDNA into the GST fusion vector pGEX-KG, and the following constructs were obtained: pGEX-CPD (the full length; aa 1–343), pGEX-CPD/N (the N-terminal 111 aa; aa 1–111), and pGEX-CPD/C (the C-terminal 232 aa; aa 112–343). The full-length cDNA of other Homer isoforms were also subcloned into pGEX-KG. GST fusion proteins were expressed in E. coli JM109 and affinity-purified with glutathione Sepharose 4B (Amersham Pharmacia Biotech). For cosedimentation assay with F-actin, GST fusion proteins were further purified through a Mono Q anion exchange column (Amersham Pharmacia Biotech). Concentrations of proteins were determined using a protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin (BSA) as a standard.
Coprecipitation of mGluR1α and GST-CPD/N. P7 mouse cerebra and cerebella were homogenized in an ice-chilled glass Teflon Potter homogenizer containing 9 vol of homogenizing buffer (0.32m sucrose, 5 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.1 mmPMSF, 10 μm pepstatin A, and 10 μm leupeptin). The homogenates were centrifuged at 1,000 × g for 15 min at 4°C to obtain nuclei-free S1 fractions. After adding Trition X-100 to give a final concentration of 0.1%, the S1 fractions were centrifuged at 100,000 ×g for 4°C for 1 hr. A 2 ml sample of the lysates (1 mg/ml protein concentration) was incubated with 50 μg of GST-CPD/N for 1 hr at 4°C. After adding 50 μl of glutathione Sepharose beads (50% slurry) (Amersham Pharmacia Biotech), the mixtures were incubated for another 1 hr. The beads were extensively washed with wash buffer (20 mm Tris-HCl, pH 8.0, 0.1 mNaCl, 0.1% Triton X-100, and 1 mm2-mercaptoethanol). Bound proteins were dissolved in SDS-PAGE sample buffer and were subjected to Western blot analysis with an anti-mGluR1α antibody (Ryo et al., 1993).
Cosedimentation assay with F-actin. Actin was isolated from acetone powders prepared from chicken skeletal muscle, as described byPardee and Spudich (1982). To obtain F-actin, G-actin was allowed to polymerize for 30 min at room temperature by addition of MgCl2 and KCl to final concentrations of 2 and 10 mm, respectively. GST and GST fusion proteins, purified through Mono Q column chromatography, were precleared by centrifugation at 140,000 × g for 20 min at room temperature before use. F-actin and GST fusion proteins were mixed at concentration of 500 and 280 μg/ml, respectively, in a polymerization buffer (10 mm Tris-HCl, pH 7.0, 50 mm KCl, 1 mmMgCl2, and 1 mm ATP). After incubation for 30 min at room temperature, the mixtures were applied to Ultra-Clear tubes (Beckman Instruments, Fullerton, CA) containing a 30% sucrose cushion in the polymerization buffer and were centrifuged at 140,000 × g for 20 min at room temperature. The resulting pellets were resuspended to the initial assay volume of the polymerization buffer. Equal amounts of both the supernatant and the resuspended pellet were then analyzed by 10% SDS-PAGE.
Preparation of anti-Cupidin antibody and coimmunoprecipitation assay. A rabbit polyclonal antibody against Cupidin was raised against a His-tagged CPD/C (aa 112–343) that was bacterially expressed. After preabsorption with GST-Homer 1c immobilized on Sepharose 4B (Amersham Pharmacia Biotech), the residual antisera were affinity-purified against the GST-CPD/C covalently coupled to CNBr-activated Sepharose 4B. P7 mouse cerebella (2 gm, wet weight) were homogenized in 18 ml of 10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 50 mm KCl, 1% NP-40, 5 mm MgCl2, 1 mm EGTA, 0.1 mm PMSF, 10 μm pepstatin A, and 10 μm leupeptin. The homogenate was centrifuged at 16,000 × g for 20 min at 4°C, and the supernatant were precleared with Protein A/G resin (50% slurry) (Cytosignal Inc., Irvine, CA). Aliquots of the supernatant (125 μl) were incubated with either the affinity-purified anti-Cupidin polyclonal antibody or the preimmune serum for 4 hr at 4°C and then with an equal amount of Protein A/G resin (50% slurry) for 30 min at 4°C. After washing the resin with homogenizing buffer five times, bound proteins were analyzed by Western blotting with the anti-Cupidin, anti-actin (Sigma, St. Louis, MO), or anti-mGluR1α antibodies.
Ligand overlay assay with Rho small GTPases. Bacterially expressed GST-Rho family protein fusions (GST-RhoA, GST-RhoAA37, GST-Rac1, and GST-Cdc42) and GST-Citron were purified through glutathione Sepharose column chromatography according to the procedure described previously (Fujisawa et al., 1998). A 1 μg sample of GST-CPD, GST-CPD/C, and GST-Citron and 3 μg of GST and GST-CPD/N were separated by 10% SDS-PAGE and blotted onto nitrocellulose membrane (Hybond-ECL; Amersham Pharmacia Biotech). Ligand overlay assay was performed as described previously (Matsui et al., 1996). Briefly, after the GST-fusion proteins on the blots were renatured, the protein blots were probed by incubating with each GST-Rho family fusion protein loaded with either [35S]GTPγS or [35S]GDPβS at an equal specific activity. After washing three times, the ligand-bound blots were air-dried, and the radioactivities were analyzed with a Bioimaging analyzer BAS2000 (Fujix, Tokyo, Japan).
Coimmunoprecipitation of Cupidin and GST-Rho family proteins. Cerebella (0.5 gm, wet weight) from P7 mice were homogenized in 5 ml of a buffer (10 mm HEPES-KOH, pH 7.4, 100 mm NaCl, 50 mmKCl, 5 mm MgCl2, 1 mm EGTA, 1% NP-40, 0.1 mmPMSF, 10 μm pepstatin A, and 10 μm leupeptin) and then centrifuged at 16,000 × g for 20 min at 4°C, and the supernatant was precleared with Protein G Sepharose beads (Amersham Pharmacia Biotech) for 1 hr at 4°C. Five micrograms of GST, GST-RhoA, GST-Rac1, or GST-Cdc42 expressed in E. coli were incubated with a nucleotide-loading buffer (25 mm Tris-HCl, pH 8.0, 2 mm EDTA, 100 mmNaCl, 1 mm MgCl2, 0.05% Tween 20, 5 mm DTT, and 100 μm GDP or GTPγS) for 20 min at 30°C, and the reaction was stopped by adding a quarter volume of stop solution (25 mm Tris-HCl, pH 8.0, 20 mm MgCl2, and 100 μm GDP or GTPγS). The nucleotide-loaded GST proteins were sequentially incubated with the cerebellar supernatants for 1 hr at 4°C with either the affinity-purified anti-Cupidin polyclonal antibody or the preimmune serum for 1 hr at 4°C and then with Protein G Sepharose beads (50% slurry) for 30 min at 4°C. After washing the beads with homogenizing buffer five times, proteins bound to the beads were analyzed by Western blotting using an anti-GST antibody (Amersham Pharmacia Biotech).
Cross-linking assay of Cupidin with Rho family proteins. GST or GST-Rho family proteins expressed in E. coli were loaded with either GTPγS or GDP as described above. GST-CPD proteins were digested with thrombin, and then GST-free CPD proteins were purified through Mono Q column. Each protein sample was finally dialyzed against a cross-linking buffer (10 mm HEPES-NaOH, pH 7.5, 2 mm EDTA, 1 mmMgCl2, 0.05% Tween 20, 5 mm DTT, and 1 mm GDP). CPD protein (25 μg/ml) and each GST-Rho family protein (25 μg/ml) were mixed and incubated for 1 hr at room temperature. Dimethyl pimalidate (DMP) (Pierce, Rockford, IL) was added to give a final concentration of 10 mm and incubated for another 1 hr at room temperature. Equal amount of the DMP-treated protein mixtures was analyzed by Western blotting using the anti-GST antibody.
Transient expression of Cupidin in mammalian cells. Cupidin cDNA was subcloned into the pEF-BOS plasmid (Mizushima and Nagata, 1990). Green fluorescent protein (GFP)-fused Cupidin (GFP-CPD), GFP-CPD/N, and GFP-CPD/C expression plasmids were constructed by subcloning the corresponding parts of Cupidin cDNA into the pEGFP-C3 (Clontech, Palo Alto, CA). Madin–Darby canine kidney (MDCK) cells were transiently transfected with GFP-CPD expression plasmids by lipofection using Lipofectamine Plus (Life Technologies). HeLa cells were transiently transfected with pEF-BOS-CPD, pEF-BOS-myc-tagged Cdc42V12, or both. After 30 hr incubation, the cells were fixed in 4% paraformaldehyde, and GFP-fluorescence was observed. For visualization of intracellular F-actin organization, the cells were probed with Texas Red-conjugated phalloidin. For detection of nontagged CPD, the samples were incubated with the anti-CPD antibody, followed by an FITC-conjugated secondary antibody. Fluorescent images were visualized using a confocal microscope (MRC1024; Bio-Rad). Images were acquired digitally or on films, and pseudocoloring was performed using Photoshop 3.5 (Adobe, San Jose, CA).
Subcellular fractionation. Subcellular fractionation of P7 mouse cerebella was performed exactly as described previously (Huttner et al., 1983; Carlin et al., 1980).
Immunohistochemistry. Cerebellar granule cells were prepared from P2 ICR mouse cerebella as described previously (Yuzaki and Mikoshiba, 1992). The culture was highly enriched in granule cells, and after 14 d in vitro (DIV), only a few calbindin-positive Purkinje cells (2–5 Purkinje cells per 105 plating cells) were detected. In brief, the cerebella from P2 mice were treated with 0.1% trypsin (Sigma) and 0.05% DNase I (Boehringer Mannheim) in Ca2+–Mg2+-free HBSS (Sigma) for 13 min at 37°C. The cells were washed with Ca2+–Mg2+-free HBSS, dissociated by repeated passage through a micropipette tip (200 μl size) in Ca2+–Mg2+-free HBSS containing 0.05% DNase I and 12 mmMgSO4, and then rinsed with the culture medium. Dispersed cells were plated at a density of 2 × 105 cells onto poly-l-lysine (Sigma)-coated glass coverslips (12 mm diameter; Matsunami, Tokyo, Japan) in serum-free defined medium: Eagle’s medium supplemented with 1 mg/ml BSA, 10 μg/ml insulin, 0.1 nM l-thyroxine, 0.1 mg/ml transferrin, 1 μg/ml aprotinin (all from Sigma), 30 nm selenium (Merck, Darmstadt, Germany), 0.25% (w/v) glucose, 2 mm glutamin, 2 mg/ml sodium bicarbonate, 0.1 mg/ml streptomycin (Meiji, Tokyo, Japan), and 100 U/ml penicillin (Banyu Pharmaceutical Co., Tokyo, Japan). The cultures were maintained in a humidified atmosphere of 5% CO2 in air at 37°C. After 7, 14, and 21 DIV, respectively, the cells were fixed with 4% paraformaldehyde and treated as described below. For native tissues, ICR mice (P4, P7, P21, and adult) were transcardially perfused with 4% paraformaldehyde in PBS(−), and excised brains were immersed for 2 hr in the same fixative and cryosectioned (8-μm-thick). For immunoreaction, fixed cultured cells or brain sections were preincubated with 2% normal goat serum and 2% normal horse serum in PBS(−) for 1 hr and then incubated with the affinity-purified anti-Cupidin (1.3 μg/ml) and mouse anti-synaptophysin (Sigma) antibodies for 1 hr at room temperature. After washing with PBS(−), the samples were incubated with the FITC-conjugated goat anti-rabbit IgG antibody (Vector Laboratories, Burlingame, CA) for Cupidin staining and with Texas Red-conjugated horse anti-mouse IgG antibody (Vector Laboratories) for synaptophysin staining. To visualize F-actin, samples were incubated with Texas Red-conjugated phalloidin. Immunofluorescence was observed using a Zeiss (Oberkochen, Germany) Axiophot epifluorescence microscope. Conventional immunodetection was also performed by using diaminobenzidine (DAB) and horse radish peroxidase-conjugated secondary antibody. Immunospecificity to Cupidin was confirmed by parallel experiments using the preimmune serum or anti-Cupidin antibody preincubated with excess amounts of the antigenic polypeptide.
RESULTS
Identification of clone 13–2, a transcript transiently induced during the development of cerebellar cortex
In the course of a systematic FDD study of genes expressed in the developing mouse cerebellum, we identified a PCR product of 323 base pairs, named clone 13–2, with a characteristic expression pattern: the 13–2 transcript was detected from as early as E18 up to P21, with a peak at P7 (Fig.1A,arrowhead). This time profile coincided with important events in the development of cerebellar cortex, such as migration of granule cells, dendrogenesis of Purkinje cells, and formation of synapses. RT-PCR analysis using a primer set corresponding to both ends of clone 13–2 confirmed this characteristic developmental expression pattern (Fig. 1B), which was further supported by a Northern analysis with a full-length cDNA probe (Fig. 1C,arrowhead). RT-PCR using RNAs prepared from P7 mice showed that the expression of clone 13–2 was widespread and found in various tissues at different levels (Fig. 1D).
Fig. 1.
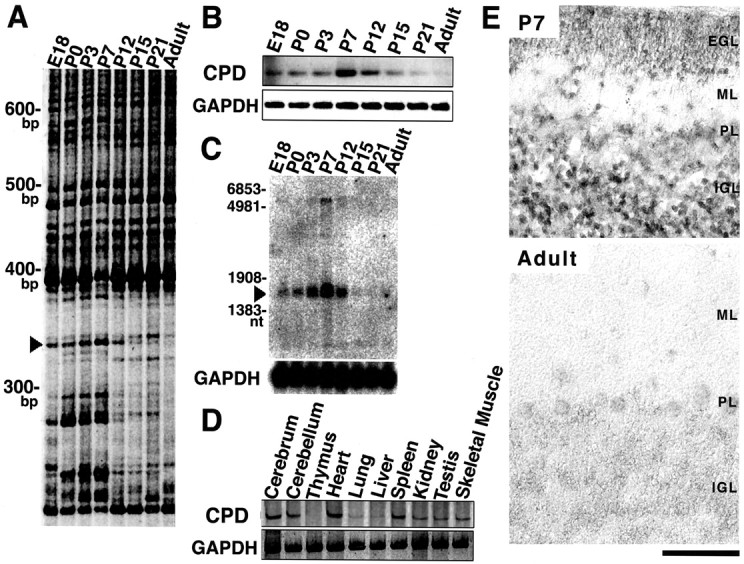
Isolation, detection, and expression pattern of clone 13–2/Cupidin during cerebellar development and in various tissues. A, FDD analysis. Cupidin was first detected as a developmentally induced band named clone 13–2, indicated by anarrowhead. B, RT-PCR analysis in the developing cerebellum. RT-PCR for Cupidin was performed using an inner primer set corresponding to the internal sequences of the cloned FDD band. GAPDH was used as an RT-PCR control. C, Poly(A)+ RNA (10 μg) was transferred onto a nylon membrane. 32P-labeled full-length Cupidin cDNA was used as a probe. As a control, a 32P-labeled GAPDH cDNA was used.D, RT-PCR analysis in various tissues of P7 mice.E, In situ localization of clone 13–2/Cupidin mRNA in the mouse cerebellum. Parasagittal sections of cerebella at P7 and adult stage were probed with the digoxigenin-labeled antisense cRNA probe prepared using a fragment (647–1220 nt) of Cupidin cDNA as a template. The antisense probe recognized two splicing variant forms of Cupidin. Specificity of labeling signals was confirmed using the sense probe (data not shown).PL, Purkinje cell layer. Magnification, 400×. Scale bar, 100 μm.
Cellular localization of clone 13–2 was examined in the developing mouse cerebellum by in situ hybridization analysis (Fig.1E). The cRNA probe used was able to recognize two transcripts of 1.8 and 6 kb in size, presumably produced by alternative splicing, both of which were predominantly expressed at P7, whereas the signal of 6 kb transcript was slightly detected at adult (data not shown). At P7, a strong signal was observed in the majority of differentiating granule cells in the IGL in addition to migrating granule cells within the ML. In adult, the signals in the IGL were greatly reduced, and a very faint signal, probably caused by the 6 kb transcript, was observed in the Purkinje cells (Fig.1E). Together, these results indicated that the expression of clone 13–2 in the granule cells was induced at either the migrating stage in the ML or the differentiating stage in the IGL, suggesting that its expression was spatiotemporally regulated during the development of the cerebellar cortex.
Clone 13–2 is derived from a transcript encoding a multimodular protein, Cupidin
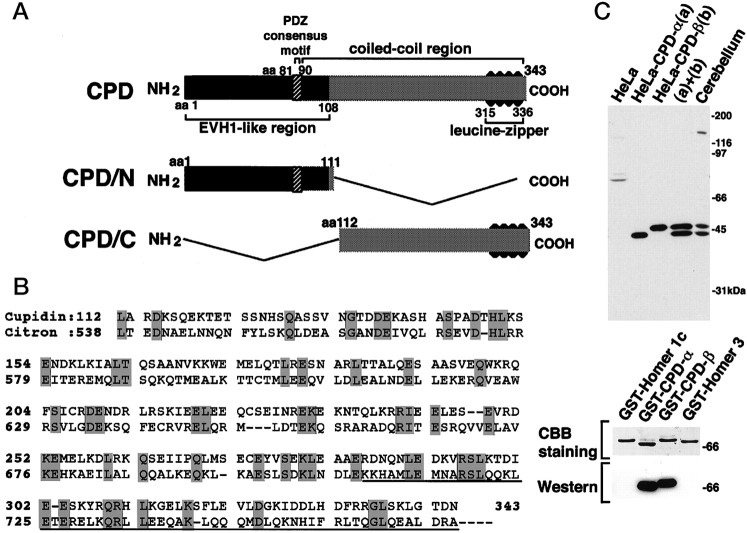
The cDNA sequence of clone 13–2 obtained from FDD showed no homology with any DNA sequence available on the GenBank database. Because clone 13–2 was likely to represent a 3′-noncoding sequence adjacent to the poly(A+) tail, we isolated a full-length cDNA from a P6 mouse cerebellar cDNA library. The entire sequence of the isolated cDNA consisted of 1761 nucleotides, and a putative open reading frame of 343 aa (calculated molecular mass of 39,461 Da) was found upstream of the nucleotide sequence for clone 13–2. From the hydropathy and secondary structure prediction, it was suggested that this protein was likely to be a soluble protein with a long α-helical stretch spanning from Ser-90 to near the C-terminal end. This protein contained Arg-81 and 87-GlyLeuGlyPhe-90 sequence, reminiscent of a minimal PDZ [postsynaptic density (PSD)/Discs large/zona occludens-1] domain consensus sequence (Fig.2A). The putative C-terminal α-helical region included several leucine zipper motifs and was predicted to form a coiled-coil structure. Homology search revealed that this protein was a close relative of Homer (Brakeman et al., 1997; Kato et al., 1997) and was subsequently found to be identical to Homer 2a/Vesl-2Δ11 (Kato et al., 1998; Xiao et al., 1998). All members of the Homer family share a highly conserved N-terminal 120 aa region homologous to the EVH1 domain [Enabled (Ena)/vasodilator-stimulated phosphoprotein (VASP) homology] (31% aa identity). Interestingly, we found that the C-terminal region of this protein from Ser-90 to the C terminus revealed a weak identity (22%) with a portion of Citron, which includes the Rho/Rac binding region and a part of the leucine zipper motif (Madaule et al., 1995) (Fig.2B).
Fig. 2.
Schematic structure of Cupidin and the specificity of anti-Cupidin antibody. A, Schematic representation of the structure of Cupidin and its truncation mutants. CPD is characterized by two major regions: an EVH1-like region, including a single PDZ consensus motif, and a Citron-like coiled-coil region, including leucine zippers. CPD/N (aa 1–111) and CPD/C (aa 112–343) were constructed by deleting the C-terminal portion and the N-terminal portion, respectively. B, Sequence alignment of Cupidin with Citron, a putative target protein for Rho GTPase. Identical amino acids are shaded. The Rho-binding region of Citron isunderlined. The C-terminal region of Cupidin (Leu-112 to Leu-336) exhibited a weak identity (22%) with a portion of Citron, including its Rho-binding region. C, Affinity-purified anti-Cupidin antibody specifically recognized a doublet band (44 and 46 kDa) in lysates of P7 mouse cerebellum (20 μg). Thebottom 44 kDa band comigrated with Cupidin (CPD-α) expressed in HeLa cells. The top 46 kDa band comigrated with a splice variant of Cupidin (CPD-β) expressed in HeLa cells. The antibody did not recognized other Homer family members, Homer 1c and Homer 3.
This identified protein thus contained multiple putative protein–protein interaction motifs and appeared to be a good candidate as an adapter molecule that mediates the association and binding of distinct classes of proteins. We hereafter refer to this protein as Cupidin (Homer 2a/Vesl-2Δ11).
Interaction with the Rho family small GTPase proteins through the C-terminal region
To examine the relevance of the several putative protein–protein interaction domains, we constructed two deletion mutants of Cupidin, CPD/N and CPD/C, comprising the N-terminal first 111 aa and the C-terminal 232 aa (residues 112–343), respectively (Fig.2A).
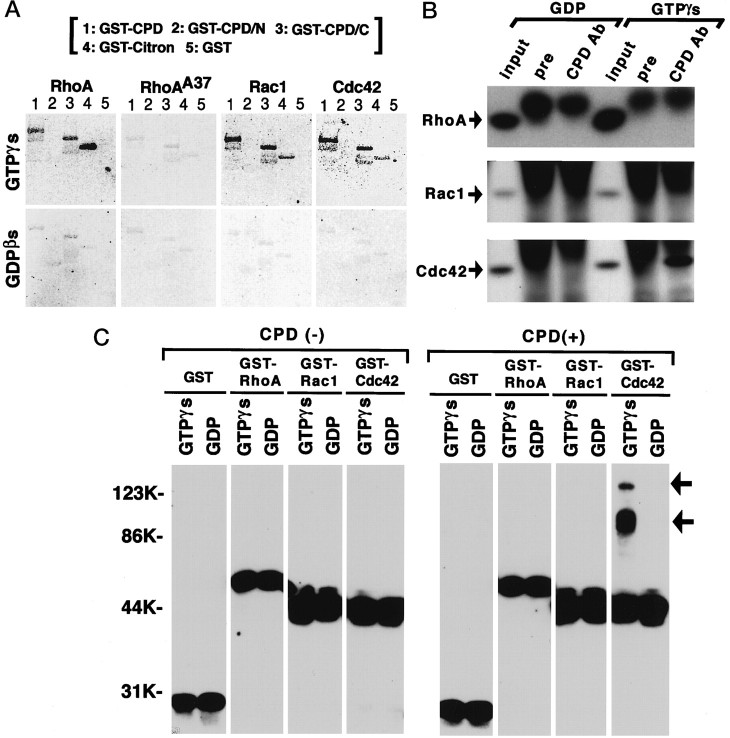
We investigated a possible interaction of Cupidin with Rho family GTPases, because the C-terminal region of Cupidin showed some homology to the Rho binding site of Citron (Madaule et al., 1995) (Fig.2B). Using a filter-overlay assay, filters, blotted with GST-CPD, GST-CPD/N, GST-CPD/C, GST-Citron (GST fused to Citron Rho-binding region), and GST were probed with GST-RhoA, GST-RhoAA37 (GST fused to an effector-binding incompetent mutant of RhoA) (Matsui et al., 1996), GST-Rac1 (GST fused to Rac1), and GST-Cdc42 (GST fused to Cdc42), in the presence of either [35S]GTPγS or [35S]GDPβS. As shown in Figure3A, GST-CPD (lane 1) and GST-CPD/C (lane 3) were intensely labeled with the [35S]GTPγS-bound form of GST-RhoA, GST-Rac1, and GST-Cdc42, but only weakly with that of GST-RhoAA37. In contrast, the labeling intensity of GST-Citron (lane 4) was much stronger with [35S]GTPγS-bound GST-RhoA compared with the other [35S]GTPγS-bound probes, which appears to be consistent with the previous report (Madaule et al., 1995). For GST-CPD, GST-CPD/C, and GST-Citron, labeling intensities detected with [35S]GDPβS-bound probes were inferior to those with all [35S]GTPγS-bound forms. Neither GST-CPD/N (lane 2) nor GST alone (lane 5) showed any significant labeling with any probes used. These results indicated that the C-terminal region of Cupidin, which shares a weak homology to the Rho/Rac-binding domain of Citron, was able to interact with Rac1 and Cdc42, as well as RhoA, in a GTP-dependent manner in vitro. A similar result was obtained with the other isoforms of CPD (Homer 1c, Homer 2b, and Homer 3) (data not shown).
Fig. 3.
Cupidin interacts with Rho family small GTPases. A, Direct binding of RhoA, Rac1, and Cdc42 to GST-fused Cupidin in a ligand overlay assay. GST-CPD (lane 1), GST-CPD/N (lane 2), GST-CPD/C (lane 3), GST-Citron (lane 4), and GST (lane 5) were separated by SDS-PAGE, blotted onto nitrocellulose membranes, and probed with [35S]GTPγS-bound (top) or [35S]GDPβS-bound (bottom) forms of each GST-fused Rho family GTPases (RhoA, RhoAA37, Rac1, Cdc42), as indicated at thetop. B, In vitro binding of Cupidin to Cdc42 in a coimmunoprecipitation assay. GST, GST-RhoA, GST-Rac1, and GST-Cdc42, loaded with either GTPγS or GDP, were mixed with P7 mouse cerebellar lysates. Each aliquot was immunoprecipitated with either the anti CPD antibody (CPD Ab) or preimmune serum (pre) and was analyzed by Western blot using the anti-GST antibody. Loaded nucleotides and composition of the analyzed mixtures are indicated at the top of each lane.Input, The equivalent mixture without being subjected to immunoprecipitation. C, A cross-link assay of Cupidin. GST, GST-RhoA, GST-Rac1, and GST-Cdc42, loaded with either GTPγS or GDP, were mixed with GST-free Cupidin protein. Each aliquot was treated with the cross-linker DMP and analyzed by Western blotting using the anti-GST antibody. Loaded nucleotides and the compo-sition of the analyzed mixture are indicated at the top of eachlane. Note that, by cross-linking in the presence of CPD, a part of Cdc42 loaded with GTPγS (GTPγS-Cdc42) enters a higher molecular weight complex, indicated byarrows.
To confirm the interaction between Cupidin and Rho family small GTPases, we performed two other sets of in vitro binding experiments. First, we tested whether exogenously added GDP- or GTP- loaded Rho family proteins were able to be coprecipitated with endogenous CPD proteins from P7 mouse crude cerebellar lysates. We raised a rabbit polyclonal anti-CPD antibody using a recombinantly expressed C-terminal portion of Cupidin as an antigen. The C terminus showed a very limited homology among Homer/Vesl-family proteins. The affinity purified anti-CPD antibody recognized two splice variants of Cupidin (CPD-α, Homer 2a/Vesl-2Δ11; CPD-β, Homer 2b/Vesl-2) (Fig.2C, top) but did not significantly cross-react with either recombinant Homer 1c or Homer 3 bacterially expressed (Fig.2C, bottom). The anti-CPD antibody immunoprecipitated both CPD-α and CPD-β from P7 cerebellar lysates. Addition of GDP- or GTPγS-loaded Rho family proteins did not change the efficiency of immunoprecipitation of cerebellar Cupidin protein with the antibody. Under this condition, the anti-CPD antibody could coprecipitate GST-Cdc42 in the presence of GTPγS but not of GDP (Fig.3B). Furthermore, we studied the direct interaction between Cupidin and Rho family proteins using DMP, a protein cross-linker with a 9.2 Å spacer arm. After cross-linking without Cupidin, GST-Rho family proteins remained monomeric, irrespective of whether GDP or GTPγS was loaded, as judged by their respective mobility on SDS-PAGE (Fig. 3C, left). When Cupidin was coincubated in these cross-linking reaction mixture, only GTPγS-loaded Cdc42 could form a complex with Cupidin, which represented the mobility shift to a higher molecular weight band calculated as ∼100 and ∼150 kDa (Fig.3C, right). These results suggested that Cdc42 could directly interact with Cupidin in a GTP-dependent manner. Although the detection is limited under experiment shown above, an interaction between Cupidin and RhoA or Rac1 cannot be excluded and remains to be studied.
It is known that the activation of Cdc42 contributes the formation of filopodial protrusions at the cell periphery (Kozma et al., 1995). To determine whether Cupidin could interact with Cdc42 in vivo, we examined the functional effect of overexpressing Cupidin on Cdc42 phenotype of HeLa cells expressing Cdc42V12, a constitutively active mutant of Cdc42 (Takaishi et al., 1997). Under our experimental conditions, 58% of HeLa cells expressing Cdc42V12indeed formed filopodia-like microspike structures (Fig.4, n = 300). Eighteen percent of the remaining cells showed atypical aggregation of F-actin in the cytoplasm, whereas 24% displayed other phenotypes, such as thickening of cortical actin filaments and blebbings at the cell surface (data not shown). The latter presumably represented the agonal state of the cells because of the prolonged expression of Cdc42V12. In contrast, the cells overexpressing Cupidin alone showed no obvious change in cell shape (Fig. 4). By coexpressing both Cupidin and Cdc42V12, however, cells with the proportion showing the typical microspikes drastically decreased (Fig.4); only 2% of the cells (n = 300) showed microspike formation, 33% was in agonal state, and 65% was as the control with no obvious peripheral change. These results indicated that Cupidin somehow interfered Cdc42-induced microspike formation.
Fig. 4.
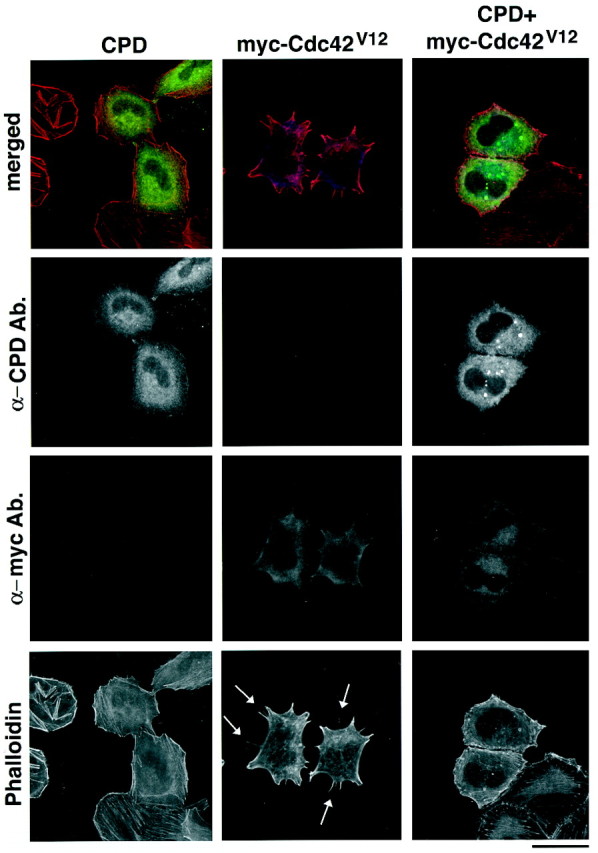
Cupidin overexpression affects Cdc42V12 phenotype in HeLa cells. Overexpression of CPD alone (left panels), myc tagged-Cdc42V12 alone (middle panels), or both (right panels) in HeLa cells. Transfected cells were stained using the anti CPD antibody (FITC), anti-myc antibody (Cy5), and Texas Red-phalloidin. Top panels are superimposed composite images using three pseudocolors (green for CPD, redfor phalloidin, and blue for myc tagged-Cdc42V12). Note that filopodia-like microspike structures were visible in Cdc42V12-overexpressed cells, as indicated byarrows. However, few such structures were induced in cells overexpressing either CPD alone or CPD and Cdc42V12. Scale bar, 60 μm.
Cupidin interacts with F-actin and mGluR1α through its N-terminal region
Next, we asked whether the EVH1-like domain present in Cupidin could interact in some way with F-actin, because the VASP/Ena family proteins containing the EVH1 domain are known to be associated with actin filaments, either directly or indirectly (Gertler et al., 1996). As shown in Figure 5A, GST-CPD/N and GST-CPD/C were subjected to an in vitrocosedimentation assay with F-actin. In the presence of F-actin, a substantial fraction of GST-CPD/N (43%) was recovered into the pellet fraction after high-speed centrifugation. However, neither GST-CPD/C nor GST alone was coprecipitated with F-actin. The full-length Cupidin fused to GST was also coprecipitated with F-actin (data not shown). These results indicated that F-actin could interact with Cupidin through the N-terminal first 111 aa.
Fig. 5.
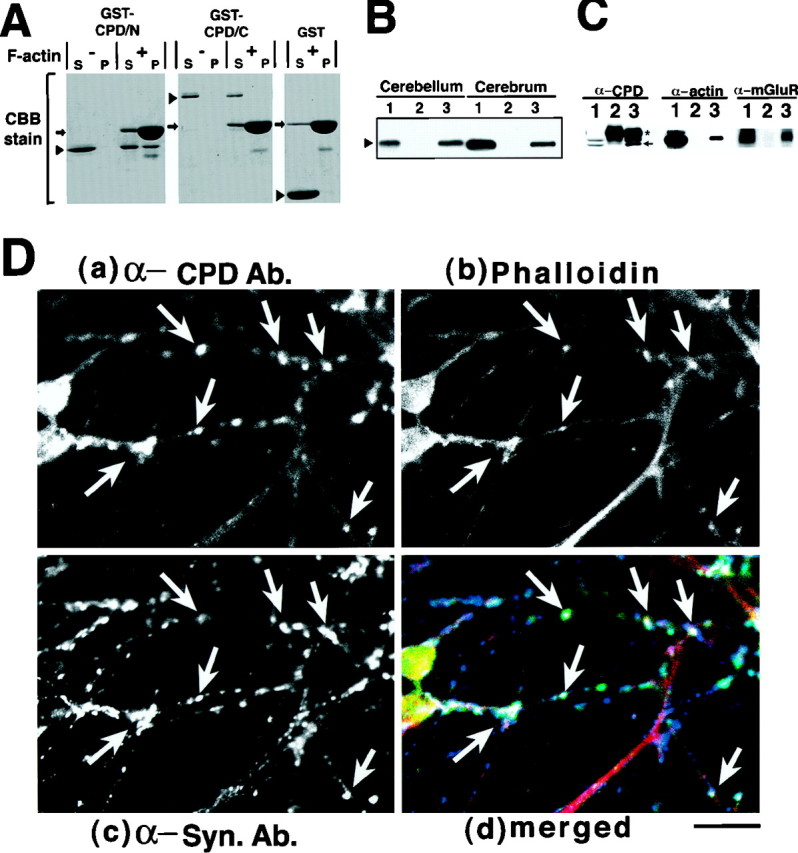
Cupidin interacts with F-actin and mGluR1α. A, F-actin binding to Cupidin in a cosedimentation assay. GST, GST-CPD/N, and GST-CPD/C were incubated with (+) or without (−) F-actin prepared from chicken skeletal muscles and then centrifuged. Equivalent protein amounts of the supernatant (S) and pellet (P) fractions were separated by SDS-PAGE and stained with Coomassie Brilliant Blue (CBB). A representative result of five independent experiments is shown here. The arrowsindicate the position of actin, and the arrowheadsrepresent each GST-fusion protein. B, Binding of Cupidin to mGluR1α in a pull down assay. The S1 fraction prepared from P7 mouse cerebellum or cerebrum was incubated with GST-CPD/N protein and then immobilized onto glutathione-Sepharose beads. After extensive washing, GST-CPD/N-bound proteins were extracted with SDS-PAGE sample buffer and were analyzed by Western blotting using an anti-mGluR1α polyclonal antibody. Lane 1, Input (the same amounts of lysates used for the assay were loaded); lane 2, eluate from a GST-bound column; lane 3, eluate from a GST-CPD/N bound column. C, Coimmunoprecipitation of both mGluR1 and actin from P7 mouse cerebellar lysates using the anti-CPD antibody. The immunoprecipitates obtained with either the preimmune serum (lane 2) or the affinity-purified anti-CPD polyclonal antibody were examined by Western blotting with the indicated antibodies (anti-CPD, anti-actin, and anti-mGluR1α). Lane 1 is the detergent extract of P7 mouse cerebellum. Thearrow indicates Cupidin signal, and theasterisk indicates the heavy chain of IgG.D, Primary-cultured mouse cerebellar granule neurons at 7 DIV were triple-stained with an anti-CPD polyclonal antibody (FITC;a), Texas Red-phalloidin (b), and an anti-synaptophysin monoclonal antibody (Cy5; c) and observed by confocal microscopy.d is a superimposed composite images ofa–c using three pseudocolors (green for CPD, red for phalloidin, and blue for synaptophysin).Arrows indicate representative positions at which the three pseudocolors overlapped. Scale bar, 10 μm.
All members of the Homer/Vesl family have been shown to interact with the group I mGluRs through the N-terminal region: the N-terminal 131 aa of Homer 1/Vesl-1 and the N-terminal 2–141 aa of Homer 2/Vesl-2 (Kato et al., 1998). We showed that GST-CPD/N could pull down mGluR1α from lysates of both cerebellum and cerebrum of P7 mice (Fig. 5B,lane 3), indicating that the N-terminal 111 aa are sufficient not only for F-actin interaction but also for mGluR1α binding.
We further examined whether Cupidin was indeed able to form a protein complex with F-actin and mGluR1α in the developing cerebellum. Anti-Cupidin antibody, recognizing both splicing forms of Cupidin (Fig.2C), could coimmunoprecipitate both actin and mGluR1α from the detergent extracts of P7 mouse cerebellar lysates (Fig.5C), thus supporting the possibility as to formation of a Cupidin–mGluR–actin complex, to some part of which CPD-β may also contribute.
Subcellular localization of Cupidin
To address the relevance of Cupidin–actin interaction, we examined the subcellular localization of Cupidin in primary-cultured cerebellar granule cells. Cupidin immunoreactivity was observed in a patchy and punctate pattern along the dendrite of granule cells (Fig.5D, a). Texas Red-conjugated phalloidin labeled the F-actin in the neurites in a punctate manner (Fig. 5D,b). Interestingly, many F-actin-positive puncta appeared to overlap with Cupidin-positive ones. The immunoreactivity for synaptophysin, a presynaptic marker protein, was simultaneously visualized to clarify synaptic sites (Fig. 5D,c). The merged image of the triple staining revealed that a large number of the Cupidin-positive and F-actin-positive puncta were in close proximity to the synaptophysin-positive synaptic structures (Fig. 5D, d), suggesting that Cupidin may colocalize with F-actin at granule cell synapses.
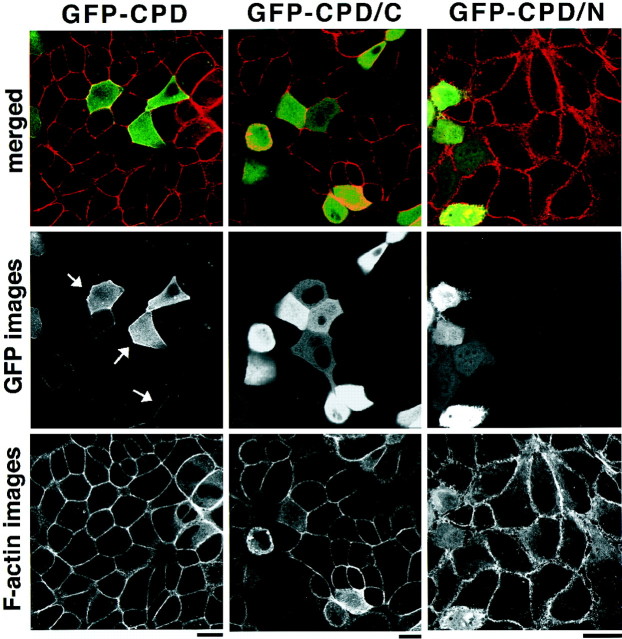
We next examined the subcellular localization of overexpressed GFP-CPD in MDCK cells, a canine epithelial cell line. The GFP fluorescence for Cupidin was diffusely localized throughout the cytoplasm and was occasionally seen in a punctate pattern that had the appearance of a stack in Golgi-like structures. Notably, the strong intensity of GFP fluorescence was remarkable at regions of cell–cell contacts at which Texas-Red fluorescence for F-actin was also the highest (Fig.6, left panels). A similar prominence of GFP fluorescence at cell–cell contact sites was observed in HeLa cells overexpressing GFP-CPD (data not shown). Interestingly, GFP fusions with either truncated form of Cupidin, GFP-CPD/N or GFP-CPD/C, overexpressed in MDCK cells were no longer localized at the cell–cell contact sites, suggesting that both the N-terminal and the C-terminal domains of CPD are required for this localization. Together with the in vitro binding to F-actin (Fig.5A,C), both types of subcellular localizations in cerebellar granule cells and in MDCK cells are consistent with the idea that Cupidin may accumulate at structures in which the actin cytoskeleton concentrates.
Fig. 6.
Localization of Cupidin overexpressed in MDCK cells at sites of cell–cell contact. MDCK cells overexpressing GFP-CPD (left panels), GFP-CPD/C (middle panels), and GFP-CPD/N (right panels) were stained with Texas Red-phalloidin. Middle panels show GFP images,bottom panels show F-actin images overlapping cell–cell contact sites, and top panels show the merged images. Note that overexpressed GFP-CPD specifically concentrated at cell–cell contact sites, as indicated by arrows, whereas both overexpressed GFP-CPD/N and GFP-CPD/C widely localized throughout the cytoplasm. Scale bars, 10 μm.
Cupidin is localized at sites undergoing active synaptogenesis in the developing mouse cerebellum
To gain further insight into the possible role of Cupidin in the developing cerebellum, we next attempted to characterize the distribution of Cupidin in mouse cerebellum using both immunohistochemical and biochemical approaches.
An immunohistochemical study with the anti-Cupidin antibody showed that the immunoreactive signals were predominantly localized in the ML and the IGL of P7 mouse cerebellum (Fig.7A). Most of the immunoreactivity in the IGL was observed as puncta with strong signal intensity in small clear spaces surrounded by granule cell soma. The localization of presynaptic synaptophysin immunoreactivity seemed to substantially overlap with that of the Cupidin-positive punctate structures. This result raised the possibility that the Cupidin signals in the IGL might correspond to glomeruli formed at the synaptic junctions between the MF–Golgi neuron terminals and the granule cell dendrites (Fig. 7, insets A′, B′). Subcellular fractionation of P7 cerebellar extracts (Fig. 7C) demonstrated that Cupidin, together with its alternative splicing variant CPD-β, was abundant in the crude synaptosomal fraction (P2′). Further subfractionation revealed that Cupidin, along with PSD-95, was enriched in the synaptic heavy membrane (LP1) and in the PSD fractions. These data indicated that, at P7, Cupidin exists in the postsynapses, most likely in the PSD area.
Fig. 7.
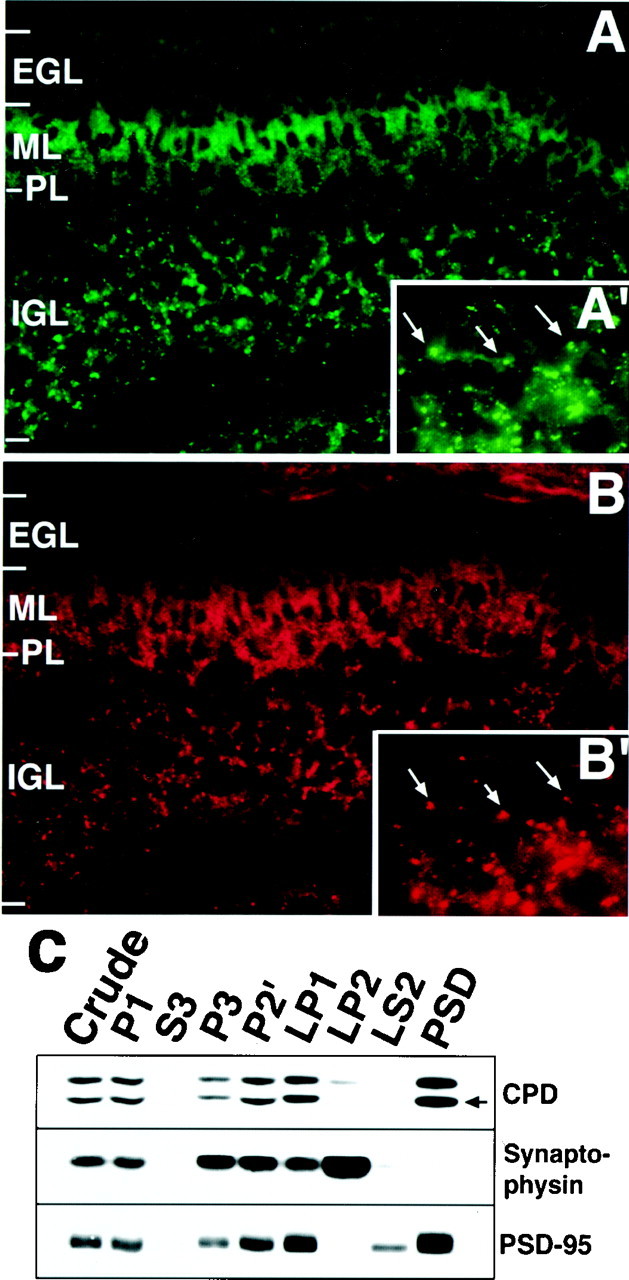
The cellular and subcellular distribution of Cupidin. (A, A′, B,B′) A sagittal cerebellar section of P7 mouse was immunostained with an anti-Cupidin antibody (A,A′) and an anti-synaptophysin antibody (B, B′). Magnification: A, B, 400×; A′, B′, 1000× of IGL region. Examples that Cupidin signals could be overlapped with synaptophysin signals are indicated by arrows.PL, Purkinje cell layer. C, Crude synaptosomes from P7 mouse cerebellum were prepared by differential centrifugation (P2′), lysed hypotonically, and fractionated into heavy membranes (LP1), synaptic vesicles (LP2), and cytosol (LS2). PSD fractions were prepared from LP1 fractions. Proteins (25 μg) from each fraction were subjected to Western blot using the antibodies against the indicated proteins. An arrow indicates the signal of Cupidin.
The developmental profile of synaptic expression of Cupidin was further examined. In line with our data obtained from the FDD, RT-PCR, and Northern analysis, the expression of 44 kDa Cupidin (CPD-α) was most prominent at approximately P7 in the developing mouse cerebellum (Fig.8A). On the other hand, a 46 kDa long splicing isoform (probably CPD-β) was upregulated until P7, and its expression level stayed relatively constant from then onward (Fig. 8A). The immunohistochemical study using cerebellar sections at various stages (Fig. 8B) and the immunocytochemical analysis using cultured cerebellar granule neurons (Fig. 8C) confirmed a sharp decline in the Cupidin immunoreactivity of granule cells after P7 and 7 DIV, respectively. The Cupidin-immunoreactive signals in the IGL of P7 cerebellum seemed to be concentrated in glomeruli in which active synaptogenesis was undergoing. In adult, most immunoreactivity was weakly detected in the Purkinje cell dendrites and soma. Considering the developmental expression profile seen in Figure 8A, the signals seen in the adult cerebellum were probably attributable to the expression of the 46 kDa CPD-β. The striking temporal profile, along with a transient concentration at the cerebellar granule cell synapses, indicates that Cupidin gene expression may be tightly coupled with the formation of new synapses in the cerebellar cortex.
Fig. 8.
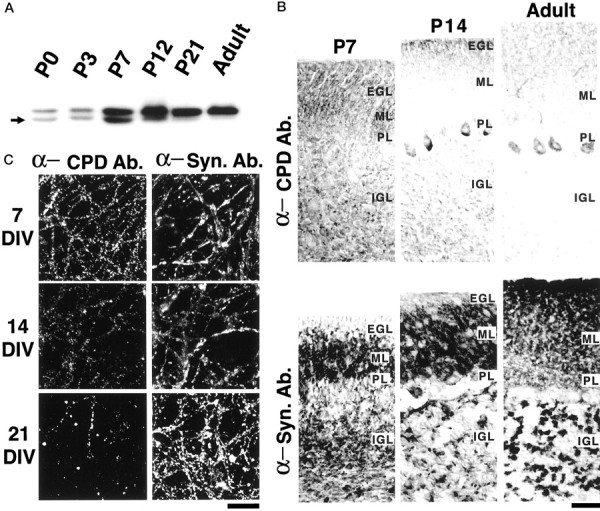
Synaptic expression of Cupidin in granule cells during the development of the cerebellar cortex. A, Western blot analysis for developmental expression of Cupidin in the cerebellum. Whole lysates of mouse cerebellum at various stages were examined by Western blotting using the anti-CPD antibody. The expression of the top 46 kDa signal persisted even after P7, whereas the expression of the bottom 44 kDa Cupidin was peaked at approximately P7 and then was hardly detectable in adult lysates. B, Sagittal sections of P7, P14, and adult mouse cerebella were immunostained with the anti-CPD antibody (α-CPD Ab.; top panels) and anti-synaptophysin antibody (α-Syn. Ab.; bottom panels), followed by HRP-based DAB–H2O2 reaction. Apparently, a high intensity of CPD signals in the IGL was prominent at P7 and then diminished onwards. In contrast, synaptophysin signals in IGL were increased both in size and intensity, indicating the maturation of glomeruli. PL, Purkinje cell layer. Magnification, 400×. Scale bar, 40 μm. C, Primary cultured mouse cerebellar granule neurons at 7, 14, and 21 DIV were costained with the anti-CPD antibody (α-CPD Ab.; left panels) and the anti-synaptophysin antibody (α-Syn. Ab.; right panels) and observed using confocal microscopy. Note that a large number of punctate CPD-signals again peaked at 7 DIV, in contrast to the lasting increase in synaptophysin-signals until 21 DIV. Scale bar, 50 μm.
DISCUSSION
Isolation of Cupidin, a Homer/Vesl isoform, as a developmentally regulated gene in cerebellar cortex
In the course of a systematic study of genes that are developmentally regulated in the mouse cerebellum, we have identified a clone encoding a multimodular protein, Cupidin. Cupidin was subsequently found to be identical to Homer 2a/Vesl-2Δ11 (Kato et al., 1998; Xiao et al., 1998). Our study indicates that Cupidin gene expression is highly regulated in the postnatal developing cerebellar granule cells. Although there was no isoform-specific probes that could distinguish Cupidin (CPD-α) from splicing isoform CPD-β (Homer 2b/Vesl-2), our data clearly demonstrates that both Cupidin mRNA and protein expressions are strongest at P7 in cerebellar granule cells while these neurons were undergoing active synaptogenesis. The Cupidin immunoreactivity decreased from the IGL afterwards, and CPD-β might also show the same expression profile in granule cells. However, CPD-β expression in whole cerebellum drastically increased at approximately P7 and continued to adult stage. In adult cerebellum, CPD-β appeared to localize in Purkinje cell dendrites and soma. These data suggest that the alternative splicing of Cupidin and CPD-β is highly development-specific and that Cupidin and CPD-β may be involved in a cell type-specific regulation.
Cupidin as a scaffold protein linking mGluR with actin cytoskeleton
Cupidin was structurally marked by two regions: the N-terminal region (the N terminus to residue 111) and the C-terminal region (residue 112 to the C terminus). We showed that the N-terminal region could bind both mGluR1α and F-actin, although we could not rule out the possibility that the latter interaction may involve other intermediate molecules. Although the N-terminal region of Cupidin contains a single Gly-Leu-Gly-Phe sequence preceded with Arg (residues 81–90), just as seen in several typical PDZ domains some of which are known to associate with various kinds of plasma membrane proteins (Doyle et al., 1996; Sheng, 1996), the flanking sequence, especially the sequence after the Gly-Leu-Gly-Phe, has no aa homology with any other PDZ domain proteins.
Although mGluR1α is predominantly expressed in Purkinje cells of adult cerebellum, it was reported that both mGluR1α mRNA (Shigemoto et al., 1992; Bessho et al., 1993) and protein (Baude et al., 1993; Ryo et al., 1993) were detected in the granule cells at a low level. It was also reported that mGluR1α immunoreactivity was localized in the glomerulus (Baude et al., 1993), a structure in which synapses are formed between the granule cells and MFs–Golgi neurons. In the present study, we have shown that Cupidin probably forms a complex with mGluR1α in the P7 mouse cerebellum. Thus, in the developing granule cells, Cupidin may play a role in mGluR1α signaling.
Ionotropic glutamate receptors (iGluRs) bind to PDZ domain-based adapter proteins such as glutamate receptor-interacting protein (GRIP) (for certain AMPA receptor subunits) and PSD-95 (for several NMDA receptor subunits), and it is known that PSD-95 can anchor iGluRs to dendritic F-actin via actin-binding protein actinin (Allison et al., 1998). The present study suggests that Cupidin may provide a physical link between mGluRs and the synaptic actin cytoskeleton. The N-terminal region of Cupidin has a weak overall homology to the EVH1 domain of the VASP/Ena family, which includes VASP (Reinhard et al., 1992),Drosophila Enabled/mouse Ena homolog (Mena) (Gertler et al., 1996), and Wiscott–Aldrich syndrome protein (WASP) (Symons et al., 1996). One of the common features of this protein family is thought to be the regulation of actin cytoskeleton, perhaps through the shared EVH1 domain present in the N-terminal region. We have found that Cupidin appears to have the ability to form a stable complex with F-actin via its N-terminal region containing the EVH1-like domain. The immunocytochemical studies and coimmunoprecipitation experiments also clearly indicated that Cupidin is likely to interact with the actin cytoskeleton in developing granule cells. In a quantitative point of view, only a limited fraction of Cupidin could bind to F-actin by ourin vitro F-actin binding experiments. This low efficiency in the in vitro Cupidin–F-actin interaction suggests that other accessory or intermediate protein(s) may be involved in the interaction between Cupidin and actin filaments.
The C-terminal two-thirds of Cupidin has been predicted to be an α-helical, coiled-coil structure containing several leucine zipper motifs (Fig. 2A). Isoforms of Homer/Vesl have been proposed to multimerize through their C-terminal ends (Kato et al., 1998; Xiao et al., 1998). In the present study, we have further identified the C-terminal region as the possible interaction domain with Rho family small GTPase proteins, a well-established class of cytoskeletal regulators. Together with the synaptic localization, Cupidin may act as a scaffold protein that links multiple signaling molecules, including G-protein-coupled cell surface receptor such as mGluR1, the actin cytoskeleton, and Rho-family proteins, especially at sites making synaptic contacts in the developing granule cells.
A possible interaction between Cupidin protein complex and Rho family small GTPases
Members of the Rho small GTPase family play important roles in regulation of the actin cytoskeleton in a wide variety of organisms, ranging from yeast to mammals (for review, see Narumiya et al., 1997) and have been implicated in neuronal migration, dendrite outgrowth, and axon guidance (Luo et al., 1996; Threadgill et al., 1997). Recently, the molecular interaction between a Rho-target protein, Citron, and PSD-95 has been demonstrated (Furuyashiki et al., 1999).We here have found that, in ligand overlay assay, Cupidin could potentially interact with RhoA, Rac1, and Cdc42 in a GTP-dependent manner through the C-terminal two-thirds homologous to the Rho binding site of Citron. It should be noted that the three different in vitro studies we performed to assess these possible interactions always revealed that Cupidin interacted with Cdc42. The heterologous expression studies revealed that Cupidin overexpression inhibited the active Cdc42 induced microspike formation in HeLa cells. Although the most likely explanation for this inhibition may be the displacement of Cdc42 by overexpressed Cupidin, it remains possible that Cupidin may function as a regulator of Cdc42. Whether Cupidin may be involved in the regulation of cell shape during synaptogenesis, possibly via Rho family GTPases, will be the subject of further studies.
The heterologous expression experiments in MDCK cells showed that Cupidin appeared to be colocalized with certain actin-associated structures at the cell–cell junctions. Indeed, it was reported that Rho family proteins may play a regulatory role in cell–cell adhesion in MDCK cells (Takaishi et al., 1997) and that GTPase-activating protein-containg Ile-Gln motifs, a target for Cdc42 and Rac1, is localized at cell–cell contact regions (Kuroda et al., 1996). Thus, the binding of Rho family members may be important in regulating the cellular localization of Cupidin or its association with other target proteins.
A Cupidin-linked signaling complex in the developing cerebellum
The present study suggests that Cupidin interacts with several signaling molecules, including mGluR1α, F-actin, and perhaps Rho family small GTPases. We have also shown that Cupidin protein is predominantly expressed in punctate structures in the IGL and the ML of P7 mouse cerebellum. Furthermore, Cupidin-positive structures seems to correspond to the MF–granule cell synapses, glomeruli, and the PF–Purkinje cell dendrite synapses during the period when active synaptogenesis is occurring. In the primary-cultured granule cell neurites, Cupidin protein substantially colocalizes with F-actin in close vicinity to the synaptophysin-positive punctate structures.
A recent study noted that Homer protein appeared to bind the inositol 1,4,5-trisphosphate receptor (IP3R) (Tu et al., 1998), an IP3-gated Ca2+ release channel downstream of the mGluR1 signaling cascade (Furuichi and Mikoshiba, 1995). Because Cupidin is identical to the Homer 2a isoform and its expression is tightly regulated in the developing cerebellar cortex, Cupidin may be a physical link between mGluR1 and IP3R. Whether Cupidin is implicated in a mGluR1-regulated Ca2+ signaling events during cerebellar development remains to be established.
In migrating granule cells, actin filament was observed in a subcortical rim of soma and in lamellipodial and filopodial extensions of the leading process, whereas in the granule cell-extending neurites, it was distributed in the periphery of the growth cone but was not enriched in the leading processes (Rivas and Hatten, 1995). Because Cupidin binds F-actin and is expressed in migrating and neurite-extending granule cells, one might speculate a possible role for Cupidin in the cytoskeletal regulation required for cell motility and/or dynamic changes in cell shape during the development of granule cells.
In conclusion, we have uncovered the intriguing molecular and developmental properties of a Homer/Vesl isoform, Cupidin. Cupidin appears to act as a scaffold protein linking multiple signaling molecules, including mGluR1, F-actin, and perhaps Rho family small GTPases. Because Cupidin expression is prominent in the early differentiating granule cells and enriched at the synapses, a specific involvement of Cupidin in regulating granule cell synaptogenesis is suggested.
Footnotes
This research was supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan, the Ministry of Health and Welfare of Japan, the Asahi Glass Foundation, and the Institute of Physical and Chemical Research (RIKEN). We thank Dr. M. E. Hatten (Rockefeller University) for a cerebellar cDNA library, Dr. K. Kaibuchi (Nara Institute of Science and Technology) for the plasmid construct of RhoAA37 and for sharing the GST-Rho overlay assay protocol, Drs. K. Inokuchi (Mitsubishi Chemicals Institute of Life Science) and A. Kato (Kyushu University) for communicating unpublished results, Dr. T. Furuyashiki (Kyoto University) for sharing the GST-Rho pull-down assay protocol, and Dr. N. Watanabe (Kyoto University) for discussions.
Correspondence should be addressed to Dr. Teiichi Furuichi, Laboratory for Molecular Neurogenesis, Brain Science Institute, RIKEN, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan.
REFERENCES
- 1.Allison DW, Gelfand VI, Spector I, Craig AM. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman J, Bayer SA. Section E. The third stage of cerebellar development: maturation of the cerebellar system. Development of the cerebellar system, pp 324–524. CRC; Boca Raton, FL: 1996. [Google Scholar]
- 3.Baude A, Nusser Z, Roberts JDB, Mulvihill E, McIlhinney RAJ, Somogyi P. The metabotropic glutamate receptor (mGluR1α) is concentrated at perisynaptic membrane of neuronal subpopulations as detected by immunogold reaction. Neuron. 1993;11:771–787. doi: 10.1016/0896-6273(93)90086-7. [DOI] [PubMed] [Google Scholar]
- 4.Bessho Y, Nawa H, Nakanishi S. Glutamate and quisqualate regulate expression of metabotropic glutamate receptor mRNA in cultured cerebellar granule cells. J Neurochem. 1993;60:253–259. doi: 10.1111/j.1471-4159.1993.tb05845.x. [DOI] [PubMed] [Google Scholar]
- 5.Brakeman PR, Lanahan AA, O’Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 6.Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86:831–843. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 8.Fujisawa K, Madaule P, Ishizaki T, Watanabe G, Bito H, Saito Y, Hall A, Narumiya S. Different regions of Rho determine Rho-selective binding of different classes of Rho target molecules. J Biol Chem. 1998;273:18943–18949. doi: 10.1074/jbc.273.30.18943. [DOI] [PubMed] [Google Scholar]
- 9.Furuichi T, Mikoshiba K. Inositol 1,4,5-trisphosphate receptor-mediated Ca2+ signaling in the brain. J Neurochem. 1995;64:953–960. doi: 10.1046/j.1471-4159.1995.64030953.x. [DOI] [PubMed] [Google Scholar]
- 10.Furuyashiki T, Fujisawa K, Fujita A, Madaule P, Uchino S, Mishina M, Bito H, Narumiya S. Citron, a Rho-target, interacts with PSD-95/SAP-90 at glutamatergic synapses in the thalamus. J Neurosci. 1999;19:109–118. doi: 10.1523/JNEUROSCI.19-01-00109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- 12.Hatten ME, Alder J, Zimmerman K, Heintz N. Genes involved in cerebellar cell specification and differentiation. Curr Opin Neurobiol. 1997;7:40–47. doi: 10.1016/s0959-4388(97)80118-3. [DOI] [PubMed] [Google Scholar]
- 13.Huttner WB, Schiebler W, Greengard P, De Camilli P. Synapsin I (Protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicles preparation. J Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito T, Kito K, Adati N, Mitsui Y, Hagiwara H, Sakaki Y. Fluorescent differential display; arbitrarily primed RT-PCR fingerprinting on an automated DNA sequencer. FEBS Lett. 1994;351:231–236. doi: 10.1016/0014-5793(94)00867-1. [DOI] [PubMed] [Google Scholar]
- 15.Kato A, Ozawa F, Saitoh Y, Hirai K, Inokuchi K. vesl, a gene encoding VASP/EVH1 family related protein, is upregulated during seizure, long-term potentiation and synaptogenesis. FEBS Lett. 1997;412:183–189. doi: 10.1016/s0014-5793(97)00775-8. [DOI] [PubMed] [Google Scholar]
- 16.Kato A, Ozawa F, Saitoh Y, Fukazawa Y, Sugiyama H, Inokuchi K. Novel members of the Vesl/Homer Family of PDZ proteins that bind metabotropic glutamate receptors. J Biol Chem. 1998;273:23969–23975. doi: 10.1074/jbc.273.37.23969. [DOI] [PubMed] [Google Scholar]
- 17.Kondo M, Sumino R, Okado H. Combinations of AMPA receptor subunit expression in individual cortical neurons correlate with expression of specific calcium-binding proteins. J Neurosci. 1997;17:1570–1581. doi: 10.1523/JNEUROSCI.17-05-01570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozma R, Ahmed S, Best A, Lim L. The ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroda S, Fukata M, Kobayashi K, Nakafuku M, Nomura N, Iwamatsu A, Kaibuchi K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J Biol Chem. 1996;271:23363–23367. doi: 10.1074/jbc.271.38.23363. [DOI] [PubMed] [Google Scholar]
- 20.Luo L, Hensch TK, Ackerman L, Barbel S, Jan LJ, Jan YN. Different effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- 21.Madaule P, Furuyashiki T, Reid T, Ishizaki T, Watanabe G, Morii N, Narumiya S. A novel partner for the GTP-bound forms of rho and rac. FEBS Lett. 1995;377:243–248. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]
- 22.Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J. 1996;15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5332. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narumiya S, Ishizaki T, Watanabe N. Rho effectors and reorganization of actin cytoskeleton. FEBS Lett. 1997;410:68–72. doi: 10.1016/s0014-5793(97)00317-7. [DOI] [PubMed] [Google Scholar]
- 25.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 26.Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electron microscopic study in Macacus rhesus. J Comp Neurol. 1971;141:283–312. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- 27.Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivas RJ, Hatten ME. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J Neurosci. 1995;15:981–989. doi: 10.1523/JNEUROSCI.15-02-00981.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryo Y, Miyawaki A, Furuichi T, Mikoshiba K. Expression of metabotropic glutamate receptor mGluR1α and the ionotropic glutamate receptor GluR1 in the brain during the postnatal development of normal mouse and in the cerebellum from mutant mice. J Neurosci Res. 1993;36:19–32. doi: 10.1002/jnr.490360104. [DOI] [PubMed] [Google Scholar]
- 30.Sheng M. PDZs and receptor/channel clustering: rounding up the latest suspects. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 31.Shigemoto R, Nakanishi S, Mizuno N. Distribution of the mRNA for a metabotropic glutamate receptor (mGluR1) in the central nervous system: an in situ hybridization study in adult and developing rat. J Comp Neurol. 1992;322:121–135. doi: 10.1002/cne.903220110. [DOI] [PubMed] [Google Scholar]
- 32.Symons M, Derry JMJ, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott–Aldrich syndrome protein, a novel effector for the GTPase Cdc42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 33.Takaishi K, Sasaki T, Kotani H, Nishioka H, Takai Y. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J Cell Biol. 1997;139:1047–1059. doi: 10.1083/jcb.139.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 35.Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 36.Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of Homer-related, synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 37.Yuzaki M, Mikoshiba K. Phrmacological and immunocytochemical characterization of metabotropic glutamate receptors in cultured Purkinje cells. J Neurosci. 1992;12:4253–4263. doi: 10.1523/JNEUROSCI.12-11-04253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]