Abstract
The site for interactions between the nervous system and much of the chemical world is in the olfactory sensory neuron (OSN). Odorant receptor proteins (ORPs) are postulated to mediate these interactions. However, the function of most ORPs has not been demonstrated in vivo or in vitro. For this and other reasons, we created a conditionally immortalized cell line derived from the OSN lineage, which we term odora. Odora cells, under control conditions, are phenotypically similar to the OSN progenitor, the globose basal cell. After differentiation, odoracells more closely resemble OSNs. Differentiated odoracells express neuronal and olfactory markers, including components of the olfactory signal transduction pathway. Unlike other cell lines, they also efficiently target exogenous ORPs to their surface. Strikingly, differentiated odora cells expressing ORPs respond to odorants, as measured by an influx of calcium. In particular, cells expressing one ORP demonstrate a specific response to only one type of tested odorant. Odora cells, therefore, are ideal models to examine the genesis and function of olfactory sensory neurons.
Keywords: olfactory epithelium, globose basal cell, olfactory sensory neuron, olfactory receptor protein, odorant signal transduction, conditional immortalization
The mature sensory neurons of the mammalian olfactory epithelium (OE) are the primary transducers of odorant signals from the external world to the CNS. Olfactory sensory neurons (OSNs) receive and transmit their stimuli directly: their dendritic termini lie in the nasal cavity, directly apposed to the environment; their axonal termini, unlike those of other sensory neurons, synapse directly onto second-order neurons within the forebrain (Graziadei and Metcalf, 1971; Moulton, 1974; Costanzo and Morrison, 1989). At least partly because of their constant exposure to environmental insults, OSNs die and are replenished throughout life (Farbman, 1990; Crews and Hunter, 1994); this property makes them unlike most other neurons, which, under normal conditions, show limited regeneration in mature mammals (Brustle and McKay, 1996; Weiss et al., 1996; Kuhn et al., 1997; Fawcett and Geller, 1998).
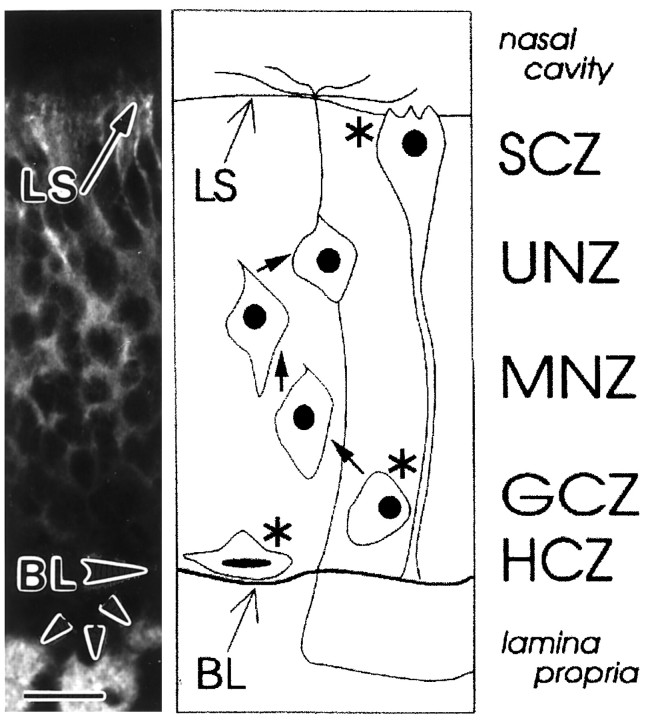
There are three dividing cell types within the mature olfactory epithelium, only one of which gives rise to OSNs (Fig.1; Caggiano et al., 1994). These are: (1) the horizontal basal cell (HBC), whose nucleus resides in the horizontal cell zone (HCZ); (2) the sustentacular cell (SC), whose nucleus resides in the sustentacular cell zone (SCZ); and (3) the globose basal cell (GBC), whose nucleus resides in the globose cell zone (GCZ). Of these, the GBC is the most prolific in vivo(Graziadei and Monti Graziadei, 1979; Schwartz Levey et al., 1991;Caggiano et al., 1994); for example, retroviral lineage tracing suggests that at least 50% of all cell division occurring just after birth in the rat OE is within the GCZ (Caggiano et al., 1994).
Fig. 1.
Major cellular elements of the olfactory epithelium. The olfactory epithelium contains the primary cell in the olfactory pathway, the OSN, which expresses general neuronal markers, including neurotubulin (left panel). In addition, there are several other cell types within the epithelium; the nuclei of all types are present in cell type-specific zones (right panel; cell sizes are exaggerated). Included are those of the three dividing progenitors (asterisks) that give rise to the cells of the mature olfactory epithelium. Two of these, the horizontal basal cell within the horizontal cell zone (HCZ) and the sustentacular cell within the sustentacular cell zone (SCZ), give rise only to themselves. The third, the globose basal cell within the globose cell zone (GCZ), gives rise to itself, as well as to immature OSNs within the middle neuron zone (MNZ;arrow), which in turn give rise to mature OSNs in the upper neuron zone (UNZ; arrow). Together, these zones span the width of the epithelium from the basal lamina (BL), which separates the epithelium from the underlying lamina propria in which OSN axons extend in bundles (small arrowheads), to the nasal cavity, in which odorants are presented to OSN dendrites located at the lumenal surface (LS). Adapted from Caggiano et al. (1994). Scale bar, 20 μm.
It has been difficult to recapitulate GBC turnover and differentiationin vitro, both in primary cultures (Calof and Chikaraishi, 1989; Pixley, 1992; Mahanthappa and Schwarting, 1993) and in cell lines (Goldstein et al., 1997; Coronas et al., 1997a). In addition, primary culture of OSNs is frequently inefficient and cumbersome (Bozza and Kauer, 1998). Because of these technical obstacles, many questions about the turnover and function of OSNs have remained unanswered. Perhaps the most vexing has been the role of the family of genes encoding putative odorant receptor proteins (ORPs; Buck and Axel, 1991); the lack of robust in vitro models has particularly hindered any direct functional investigations into the coupling by specific members of this family to a physiological odorant response. Although ORPs can be expressed intracellularly via baculoviral infection of insect cells (Raming et al., 1993; Nekrasova et al., 1996), chimeric ORPs can be expressed in heterologous cells (Krautwurst et al., 1998), and at least two receptors have been successfully introduced to OSNs in vivo (Zhao et al., 1998; Touhara et al., 1999); functional surface expression of ORPs in intact OSNs has not been achieved in vitro. We, therefore, have created a cell line that we propose will be useful in defining: (1) the role of ORPs in odorant detection; (2) the transduction apparatus within OSNs; (3) signals that influence trafficking of ORPs and other olfactory proteins within OSNs; and (4) factors that influence GBC turnover, differentiation, and maturation.
MATERIALS AND METHODS
Creation of the odora line. A temperature-sensitive mutant (tsA58) (Jat et al., 1986) of the SV40 large T antigen (Southern and Berg, 1982) in a retroviral backbone (Cepko et al., 1984) was used to create cell lines from the olfactory epithelium. Postnatal day 3 rats were killed according to protocols approved by Tufts University, the Society for Neuroscience, and the National Institutes of Health. Olfactory epithelia from eight pups were dissociated using a method (Hunter et al., 1992) we had developed for dissociation of retinae, and were incubated at 33°C (at which tsA58 should be active) in normal medium (DMEM; Bio-Whittaker, Walkersville, MD) containing 10% fetal bovine serum (FBS; Hyclone, Logan, UT), 100 U/ml penicillin (Irvine Scientific, Santa Ana, CA) and 100 μg/ml streptomycin (Irvine Scientific). During plating, 30 μl of a tsA58-Tag viral concentrate (Jat and Sharp, 1989) (∼106 pfu/ml, prepared as in Caggiano et al., 1994) was added to each dish; after 2.5 hr, an additional 30 μl was added. After 2 d, the medium was supplemented with 400 μg/ml G418 (active drug concentration; Life Technologies, Gaithersburg, MD). After 5 weeks, eighteen colonies had formed. All were isolated in cloning rings, then replated in G418-containing medium. After reaching confluency, all lines were frozen and stored in liquid nitrogen. After thawing, only five remained viable; all have similar characteristics. None was able to survive our attempts at single-cell cloning in 96-well plates; we presume that this reflects a requirement for autocrine factors. Interestingly, all adhere tenaciously to both plastic and glass, suggesting that these cells produce strong adhesion molecules. The cells cannot be removed easily by standard trypsinization (0.05% in DMEM), even when incubated at 37°C for 1 hr; however, cells readily detach when divalent cations are chelated, suggesting that they adhere via a calcium- (perhaps cadherin-) mediated mechanism. One of these lines we now call odora.
Odora culture. Odora cells were grown at 33°C to ∼30% confluency, then shifted to 39°C (at which tsA58-Tag should be degraded; see Fig. 3) and incubated for 5–7 d in normal medium (see above) supplemented with 1 μg/ml insulin, which is important in maintaining an independent olfactory cell line (Goldstein et al., 1997), and 20 μm dopamine and 100 μm ascorbic acid (both from Sigma, St. Louis, MO), which can elicit differentiation in yet another cell line (Coronas et al., 1997a). Together, these changes resulted in differentiation ofodora cells. Control cells remained in unsupplemented normal medium and at 33°C. For assessment of proliferation, odoracells were cultured with a combination of fluoro- and bromo-deoxyuridine (cell proliferation labeling reagent; Amersham, Arlington Heights, IL) during the last 18 hr of culture. Cells were then rinsed, fixed in absolute methanol at −20°C, and processed for BrdU immunohistochemistry. The average number of cells from 14 random fields taken from three individual coverslips was determined, and the percent that had incorporated BrdU was calculated. The total number of labeled cells present at 33°C was 4.8-fold higher than that at 6 d after the shift to 39°C, reflecting the more rapid proliferation seen when tsA58-T antigen is active.
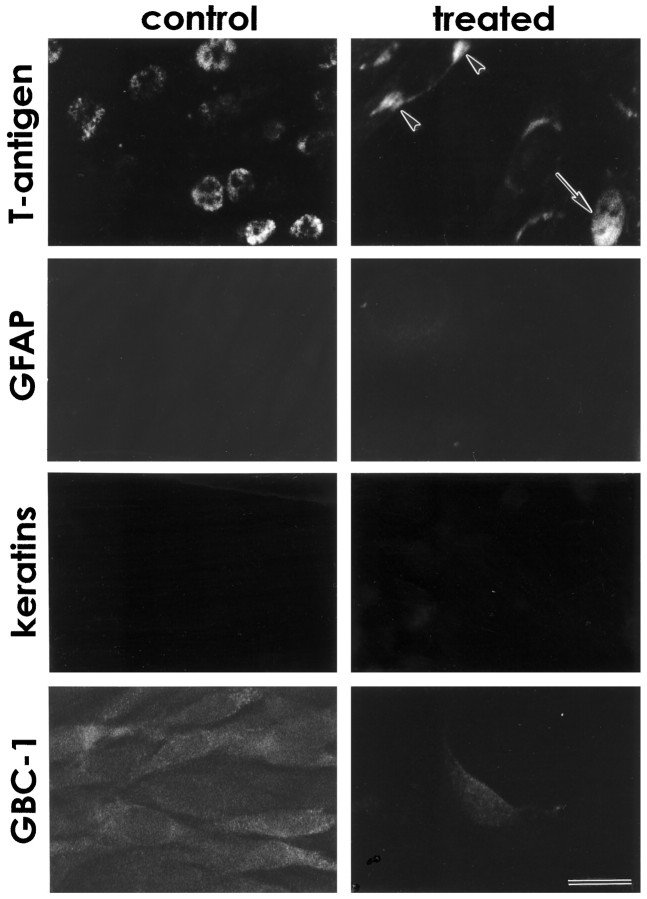
Fig. 3.
Odora cells express markers of the olfactory sensory neuron lineage, but not of the other two lineages within the mature olfactory epithelium. T-antigen, the immortalizing oncogene, is expressed in the nuclei of odora cells at the permissive temperature (control), but only infrequently after a shift to the nonpermissive temperature (treated) in nuclei (arrow), and occasionally in perinuclear regions (arrowheads). Markers of glia (GFAP), horizontal basal cells (keratins), and sustentacular cells (SUS-4; data not shown) are not expressed in either condition. Importantly,odora cells express a marker of globose basal cells,GBC-1, under control conditions; this expression decreases dramatically with differentiation, as it does in OSNs. Scale bar, 20 μm.
Immunohistochemistry and immunofluorescence.Immunohistochemistry was performed as previously described (Libby et al., 1996). For sections, the rostral portion of the entire head of a postnatal day 1 to 3 rat, frozen in liquid nitrogen-cooled isopentane, was cut at 10–12 μm and placed onto ethanol-cleaned glass slides. After drying, the sections were either left unfixed or fixed with 4% paraformaldehyde (Polysciences, Warrington, PA) in PBS (in mm: 137 NaCl, 2.7 KCl, 10 Na2HPO4, and 1.8 KH2PO4, pH 7.2–7.3) for 5 min at ambient temperature. Odora cells on 12 mm round glass coverslips were washed with PBS, then fixed in 4% paraformaldehyde in PBS for 5 min at ambient temperature. The primary antibodies used are given in Table 1. Most have been shown by other laboratories to react specifically with their stated antigen in tissue homogenates; we, and our collaborators (B. Talamo, unpublished observations), have additionally demonstrated that the antibodies recognizing the signal transduction components (Gαolf, ACIII, and oCNGβ) react with appropriately sized species on protein transfer (Western) blots of extracts of odora cells (see Fig. 6). Secondary antibodies coupled to fluorescein, rhodamine, and R-phycoerythrin were obtained from Sigma and Molecular Probes (Eugene, OR). All antibodies were diluted in a potassium Ringer’s buffer (KRB; in mm: 140 NaCl, 5 KCl, 5 NaHCO3, 2 MgCl2, 1 CaCl2, 10 glucose, and 10 HEPES, pH 7.4) containing 2% bovine serum albumin (Sigma), and 0.01% Triton X-100 (Sigma); the wash solution was KRB. For an assessment of percentage of cells expressing a given marker, the total numbers of cells in random fields (assessed by contrast or with a nuclear stain) were counted, and the numbers of cells within those fields expressing the marker were calculated. Data are expressed as average percent of cells (± SD) expressing detectable levels of the marker in the given conditions.
Table 1.
Sources and reactivity of antibodies used in this study (in order of appearance)
| Antigen | Antibody | Source; Reactivity in OE |
|---|---|---|
| Neurotubulin (β-III tubulin) | Mouse monoclonal, “J1” | A. Frankfurter; Talamo et al., 1989 |
| T-antigen | Rabbit polyclonal | D. Hanahan; this report |
| Glial fibrillary acidic protein (GFAP) | Rabbit polyclonal | DAKO; Calof and Chikaraishi, 1989 |
| Sustentacular cell antigen | Mouse monoclonal, “SUS-4” | J.E. Schwob; Goldstein and Schwob, 1996 |
| Globose basal cell antigen | Mouse monoclonal, “GBC-1” | J.E. Schwob; Goldstein and Schwob, 1996 |
| Neural cell adhesion molecule (NCAM) | Rabbit polyclonal | Sigma; Caggiano et al., 1994 |
| Microtubule-associated protein 5 (MAP5) | Mouse monoclonal | Sigma; Coronas et al., 1994 |
| Growth-associated protein 43 (GAP-43) | Mouse monoclonal | Sigma; Goldstein and Schwob, 1996 |
| Olfactory G-protein (Gαolf) | Rabbit polyclonal | Santa Cruz; Jones and Reed, 1989 |
| Adenylyl cyclase type III (ACIII) | Rabbit monoclonal | Santa Cruz; Juilfs et al., 1997 |
| Olfactory cyclic nucleotide-gated channel (oCNGβ) | Mouse monoclonal | K. Zinn; this report |
| Olfactory marker protein (OMP) | Goat polyclonal | F.L. Margolis; Margolis, 1972 |
| Hemagglutinin | Mouse monoclonal | BAbCo; this report |
Commercial antibody suppliers: DAKO, Carpinteria, CA; Sigma, St. Louis, MO; Santa Cruz Biotechnologies, Santa Cruz, CA; and Berkeley Antibody Company (BAbCo), Berkeley, CA.
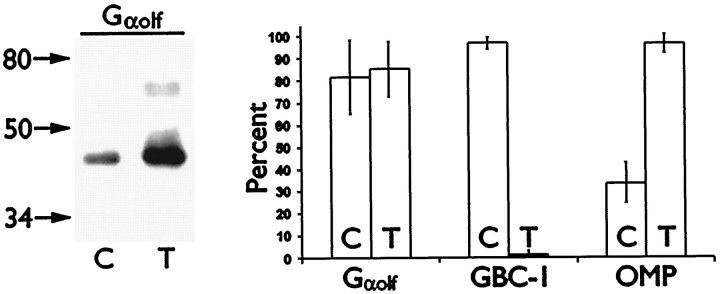
Fig. 6.
Gαolf, OMP, and GBC-1 levels change after differentiation of odora cells. Protein transfer blots (left) of control (C) and treated (T) odora cell extracts reveal an increase in overall Gαolf expression after differentiation. However, the percentage of cells (right, average ± SD) expressing immunohistochemically detectable levels of Gαolf does not increase after treatment, suggesting an increase in Gαolflevels in each expressing cell. In contrast, the percent of cells expressing detectable levels of OMP increases, and the percent expressing GBC-1 decreases after differentiation.
Protein transfer blot. Control and differentiatedodora cells, grown as described above, were solubilized in 10 mm Tris-HCl, pH 7.5, containing 1% SDS, 1 mm sodium orthovanadate, 1 mm sodium pyrophosphate, 5 μg/ml leupeptin, 5 μg/ml pepstatin, 5 μg/ml aprotinin, 5 μg/ml chymostatin, and 100 μg/ml pefabloc. After collection and sonication, two volumes of sample buffer (192 mm Tris-HCl, pH 6.8, containing 9% SDS, 15% glycerol, and 2% 2-mercaptoethanol) were added. Lysates were boiled and separated by electrophoresis on a denaturing 10% polyacrylamide gel, then transferred to Immobilon-P (Millipore, Bedford, MA). The membrane was blocked in 5% (w/v) nonfat milk in 200 mm NaCl, 0.1% Tween 20, 50 mm Tris, pH 7.4 for 1 hr, incubated in primary antibody for 2 hr and peroxidase-conjugated secondary antibody (New England Biolabs, Beverly, MA) for 1 hr, all at ambient temperature. After washing, the membrane was incubated in a chemiluminescent substrate (New England Nuclear, Boston, MA) for 1 min and exposed to x-ray film. Samples were normalized by nucleic acid content in order to load extracts from approximately equal numbers of cells per lane.
RT-PCR. RNA was isolated from dissected adult rat olfactory epithelium and retina using RNAzol B (Biotecx, Houston, TX) and from odora cells using Ultraspec (Biotecx) following the manufacturer’s recommendations. For oCNGα, oCNGβ, and rCNGβ-t, 4 μg of RNA was reverse-transcribed using SuperScript II (Life Technologies) primed with random hexamers following the manufacturer’s recommendations. In addition, for oCNGβ, 100 ng of RNA was similarly reverse-transcribed using a specific oligonucleotide primer (5′-TACATCTCTCGGCCAATGTC-3′). The resultant cDNAs (5% of the randomly primed, or all of the specifically primed) were amplified in PCRs using Platinum Taq High Fidelity (Life Technologies) in 2 mm MgSO4. Oligonucleotides used were: oCNGα, forward, 5′-GTCATCATCCACTGGAATGCTTG-3′ and reverse, 5′-ATCAGCTACCACTGCCAACTTGCCC-3′ (Kingston et al., 1996); oCNGβ, forward, 5′-ACCATGCGCTGGTAAAGAAG-3′ and reverse, 5′-TACATCTCTCGGCCAATGTC-3′ (sequences obtained from the Mombaerts laboratory); and rCNGβ-t, forward, 5′-TCCATGCTGTGCCAATCACA-3′ and reverse, 5′-CTGGTCCACATCAGCCTGCA-3′ (Sautter et al., 1998). Cycling protocol was: 94°C, 2 min; 35–40 cycles of 94°C, 45 sec; 52°C, 45 sec; 72°C, 2 min; and 10 min at 72°C. Fifty percent of each reaction was analyzed on a 1% agarose/1% NuSieve (FMC Bioproducts, Rockland, ME) gel. The primers used for oCNGα and rCNGβ-t have been shown previously not to amplify sequences from related CNG cDNAs. In addition, the primers used for oCNGα and oCNGβ span introns in genomic DNA; both sets would, therefore, detect contaminating genomic DNA in our RNA samples. We did not detect these larger products derived from genomic DNA in our samples, demonstrating a lack of contamination.
Calcium imaging. Differentiated cells were loaded at 39°C with the ratiometric calcium imaging dye, fura-2 AM (4 μm; also contains 0.01% Pluronic F-127; both from Molecular Probes) on 31 mm round glass coverslips in odorant Ringer’s solution (ORS; in mm: 140 NaCl, 5 KCl, 10 glucose, 1 Na pyruvate, 1 CaCl2, 1 MgCl2, and 10 HEPES, pH 7.2) as previously described for primary cultures of OSNs (Bozza and Kauer, 1998). After 1 hr, cells were rinsed and placed into a perfusion chamber through which odorants (Table2; final concentrations of 50 μm each in ORS that had been stored in a polypropylene tube) were presented for 16–24 sec. Two controls were used: “Ringer’s”, which was a pulse of Ringer’s stored in polystyrene, and “no odorant”, which was a pulse of Ringer’s stored in polypropylene, which has an intrinsically different odor than that of polystyrene. No differences were ever seen between the two controls. Unless otherwise noted, data were collected at 340 and 380 nm every 4 sec, stored, and analyzed and plotted using Quattro Pro versions 5 and 7 (Corel, Ottawa, Canada). For simplicity, the signals are shown as the ratios of the fluorescence intensity at 340 nm divided by that at 380 nm; this roughly reflects the concentration of intracellular calcium (Bozza and Kauer, 1998, their references). Note that the baseline ratio varies with each experiment, presumably reflecting such parameters as length and efficiency of dye loading, as well as the ambient temperature during the experiment (which varied from 26 to 33°C).
Table 2.
Odorant mixes used in this study
| Mix A | Mix B | Mix C | Mix D | Mix E | Mix F |
|---|---|---|---|---|---|
| Geraniol | N-valeric acid | (+)-carvone | Ethyl-N-butyrate | Phenyl ethyl alcohol | Lyral |
| N-amyl alcohol | N-enanthic acid | Eugenol | Hexylacetate | Eugenol | Phenyl ethyl amine |
| N-butyl acetate | N-pelargonic acid | Cinnamaldehyde | Citralva | Acetophenone | Vanillin |
| Propionic acid | N-heptanol | (−)-limonene | Cineole | Citralva | Isovaleric acid |
| Benzaldehyde | N-hexanol | Citral | (1R)-(+)-(α)-pinene | Geraniol | |
| Acetophenone | N-butanol | 2-ethyl fenchol | Isobutyraldehyde | ||
| Benzyl alcohol |
All odorants were at 50 μm. Some mixes were presented simultaneously; these are noted, for example, as “CD”, for a presentation of mixes C and D.
Transfection of odora cells. Odoracells, grown on 31 mm round glass coverslips in individual 35 mm tissue culture dishes, were differentiated as described above. Five days after differentiation was begun, odora cells were transfected for 2 hr with 0.625 μg of a plasmid containing the β2-adrenergic receptor or an ORP construct (U131 or OR5) and a hemagglutinin tag (McClintock et al., 1997), using 3.75 μl of the SuperFect reagent (Qiagen, Chatsworth, CA) per dish, then were washed and returned to the appropriate medium for 2 d. SuperFect was by far the most efficient transfection reagent tested, resulting in 50–80% transfection efficiency. To assess surface expression, cells were removed to 0°C to prevent receptor and antibody internalization and incubated for 2 hr with a monoclonal mouse anti-HA antibody (BAbCo, Berkeley, CA), then fixed in 4% paraformaldehyde in PBS. Cells were washed and incubated in a secondary antibody, coupled to R-phycoerythrin (Molecular Probes) for 45–60 min at ambient temperature and coverslipped in ProLong (Molecular Probes). Intracellular calcium responses were assayed (see above) in the presence of the β2-adrenergic receptor agonist, isoproterenol (25 mm; Sigma) in cells transfected with the β2-adrenergic receptor; cells transfected with ORP constructs were assayed for responses to our odorant mixtures (see above).
RESULTS
Creation of an olfactory sensory neuron line
We chose to immortalize cells derived from the OE of perinatal rats; at this age, the OE has achieved a mature morphology, but is still undergoing massive proliferation (Caggiano et al., 1994). Because of the high degree of division occurring in the basal cell populations at this age, a large number of dividing progenitors (HBC, GBC, and SC; Fig. 1) are present. We have previously shown that ∼50% of the mitotically active cells present just after birth are HBCs, and ∼50% are GBCs or their immediate progeny (Caggiano et al., 1994); an immortalizing oncogene could, therefore, be transmitted to these dividing populations via infection with a replication-incompetent retrovirus. After infection of a cell, the oncogene will be incorporated into the DNA of one of its daughter cells and subsequently passed onto that daughter’s progeny. We chose a temperature-sensitive oncogene in the hope that cells that were proliferating at the permissive temperature of the oncogene would cease proliferation and differentiate when the cells were removed to a nonpermissive temperature.
Cells from freshly dissociated perinatal rat OE were infected with a retrovirus carrying a neomycin resistance gene and a temperature-sensitive mutant of an oncogene, large T antigen (tsA58-Tag), driven by the murine moloney leukemia virus long terminal repeat (MMLV-LTR) (Southern and Berg, 1982; Cepko et al., 1984; Jat et al., 1986; Jat and Sharp, 1989). After selection at the permissive temperature (33°C), 18 colonies were isolated; of these, five remained viable after freezing and replating. Of these five, we have chosen one for further study; we now call these odora cells, for olfactory- derived, odorant receptor activatable cells. Surprisingly, the other four lines are qualitatively similar in growth properties, gross morphology, and antigen expression (data not shown), suggesting that they were derived from similar progenitor cells. Thus, one population of progenitors appears to be more easily immortalized.
Characterization of odora cells
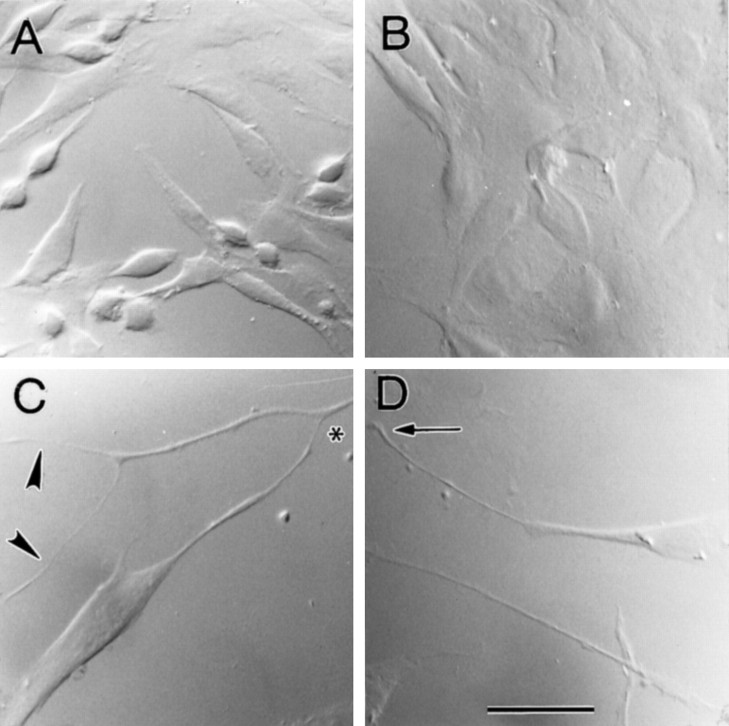
Odora cells grow rapidly, attach readily to uncoated glass, and are easily dissociated when grown at 33°C in normal medium (see Materials and Methods). This line, which was originally created in 1995, has now been in continuous culture for 2 years; over that time, the gross characteristics of the cells have not changed. Briefly, the cells achieve an epithelial morphology quickly, only rounding up occasionally (Fig. 2A). After differentiation [shift to the nonpermissive temperature (39°C) and addition of insulin and dopamine; see Materials and Methods],odora cells become somewhat flatter and larger (Fig.2B); in addition, they begin to attain a bipolar morphology, frequently extending long processes across many tens of micrometers (Fig. 2C, D). Their division rate also decreases dramatically: after an 18 hr pulse of the thymidine analog bromodeoxyuridine (BrdU), >90% of the nuclei of the control cells have incorporated BrdU, whereas only 20% of the nuclei of the differentiated cells have (see Materials and Methods).
Fig. 2.
Morphology of odora cells.Odora cells, immortalized by infection with a temperature-sensitive variant of large T antigen (tsA58-Tag), exhibit an epithelial morphology when grown at 33°C in normal medium (A). Dividing cells, which normally round up from the epithelial sheet, are frequent. In contrast, after differentiation, cells remain largely flattened (B); when sparse, their bipolar morphology is apparent. Differentiated cells occasionally extend long, sometimes branched, processes (arrowheads) over many tens of micrometers (C, D). These processes occasionally contact those from other cells (asterisk) or terminate in a filopodium (arrow). Scale bar, 40 μm.
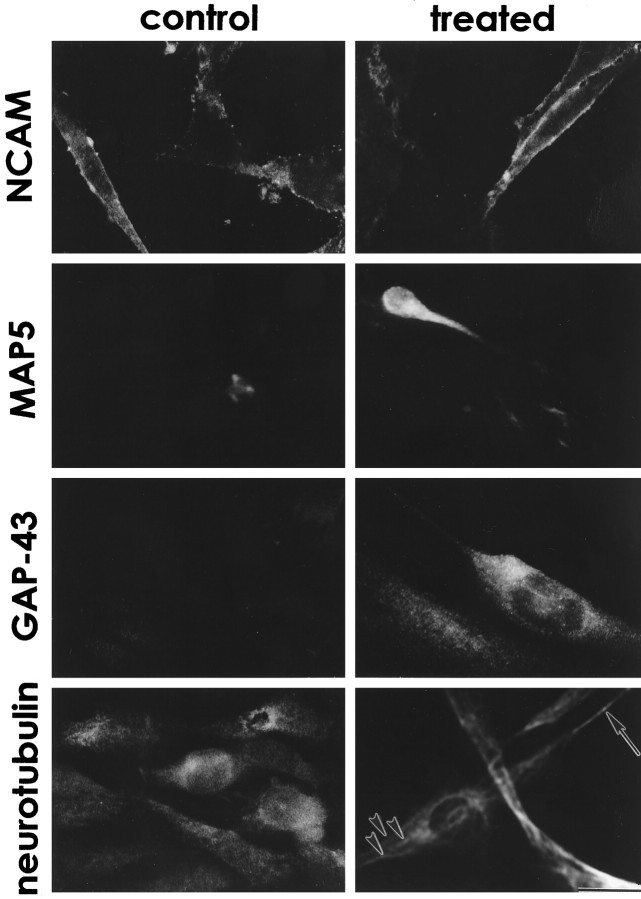
Immunohistochemically, control odora cells express nuclear T-antigen (Fig. 3), and do not express markers of glia (GFAP; Fig. 3), of horizontal basal cells (keratins; Fig. 3), or of sustentacular cells (SUS-4; data not shown). However, control odora cells do express a marker of the globose basal cell (GBC-1; Fig. 3), as well as the neural cell adhesion molecule, NCAM (as do GBCs; Caggiano et al., 1994; Fig.4). Together, these data suggest thatodora cells were derived from infection of a dividing globose basal cell. Odora cells do not express appreciable quantities of several neuronal markers, including the microtubule-associated protein MAP5 and the growth-associated protein GAP-43; they also do not appear to assemble neurotubulin into microtubules (Fig. 4). Thus, control odora cells share some characteristics with the undifferentiated OSN progenitor, the GBC. As noted above, our other lines appear qualitatively similar; this suggests that cells within the GBC lineage are more easily immortalized, or more easily cultured, than those within the HBC lineage.
Fig. 4.
Odora cells express neuronal markers. Under control conditions, odora cells, like globose basal cells, express NCAM, but little MAP5, and no GAP-43. After differentiation, all three markers are expressed. Neurotubulin, although expressed in control conditions, is not assembled into discernible microtubules until the cells are differentiated, where it is present both in long processes (arrow) and in bundles within the cytoplasm (arrowheads). Scale bar, 20 μm.
Differentiated odora cells largely do not express nuclear T-antigen (Fig. 3), consistent with their slowing in division (cf. Jat and Sharp, 1989). They also have an antigenic profile similar to that for OSNs: they largely cease expression of GBC-1 (Fig. 3), maintain expression of NCAM (Fig. 4), and begin to express MAP5 and GAP-43 (Fig.4). In addition, they appear to assemble neurotubulin into discernible microtubules within their somata and processes (Fig. 4). Thus, in many respects, differentiated odora cells are similar to olfactory sensory neurons.
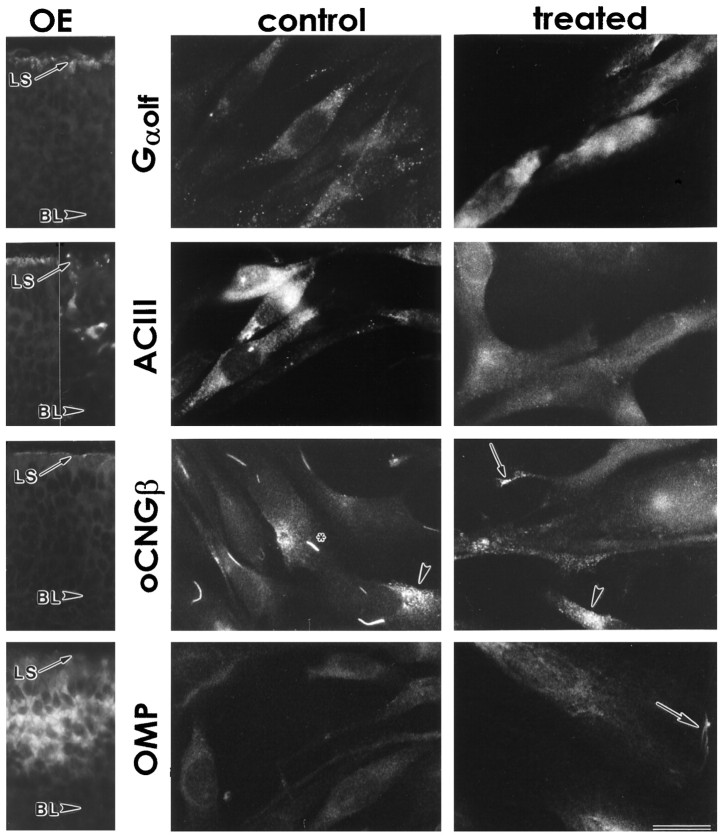
Odora cells also express markers that are more selective for OSNs. These include components of the olfactory signal transduction machinery: the olfactory G-protein (Gαolf), adenylyl cyclase type III (ACIII), and the β subunit of the olfactory cyclic nucleotide-gated channel (oCNGβ), as well as olfactory marker protein (OMP). In tissue sections of the olfactory epithelium, the components of the transduction machinery are located within the dendrites at the lumenal surface, whereas OMP is throughout the somata of OSNs (Fig. 5). In odoracells, all of these markers are expressed to some degree before differentiation (Fig. 5); in this regard, controlodora cells appear to be more differentiated than GBCs.
Fig. 5.
Odora cells express markers that are expressed by olfactory sensory neurons in vivo, including components of the olfactory signal transduction machinery. Olfactory cells, both in situ and in vitro, express the olfactory G-protein (Gαolf), adenylyl cyclase type III (ACIII), the β subunit of the olfactory cyclic nucleotide-gated channel (oCNGβ), and olfactory marker protein (OMP). In situ, mature sensory neurons express all four (OE; oCNGβ section was fixed in 4% paraformaldehyde; others were unfixed). The basal lamina (BL) and lumenal surface (LS) are noted for orientation (compare Fig. 1); for ACIII, two sections are shown, because the expression within the somata is variable. Note that in control odora cells, oCNGβ appears trapped in perinuclear compartments, some of which are strikingly bar- or crescent-shaped (asterisk), whereas others are more diffuse (arrowhead); after differentiation, the channel largely moves away from these sites, including to the distal ends of cellular processes (arrow), although some remains diffusely perinuclear (arrowhead). OMP also appears to be distributed to additional, peripheral, locations after differentiation (arrow). Scale bar, 40 μm for tissue sections; 20 μm for odora cells.
However, the expression of Gαolf and OMP increases after differentiation. For Gαolf, the protein level of each expressing cell increases: although the percent of cells that produce immunohistochemically detectable levels of Gαolf does not change with differentiation (Figs. 5, 6), the overall amount of Gαolf does increase after differentiation, as assayed by protein transfer (Western) blot (Fig.6). In contrast, for OMP, this change appears to be in the number of cells that express immunohistochemically detectable amounts of OMP: although some cells produce detectable levels before differentiation, nearly all cells produce detectable OMP after differentiation (Figs. 5,6). This increase in OMP expression coincides with the dramatic decrease in numbers of cells producing detectable levels of GBC-1 after differentiation (Figs. 3, 6).
In addition, the distributions of oCNGβ and OMP change after differentiation (Fig. 5). Both shifts are consistent with the differentiation of odora cells to a phenotype more like that of an OSN. Strikingly, the altered distribution of oCNGβ after differentiation suggests a novel regulation of subcellular distribution within OE-derived cells that may be partly responsible for localizing olfactory-specific components of OSNs (including, potentially, ORPs; see below) to the plasma membrane.
We have shown that odora cells produce the β subunit of the olfactory cyclic nucleotide-gated channel (Fig. 5). However, the cyclic nucleotide-gated channel in OSNs is thought to consist of at least two subunits, oCNGα (Dhallan et al., 1990) (also known as oCNC1, CNG2, or CNCα3) and oCNGβ (Bradley et al., 1994; Liman and Buck, 1994) (also known as oCNC2, CNG5, or CNCα4). In addition, the olfactory epithelium specifically produces a further potential subunit, rCNGβ-t (also known as CNG4.3 or CNCβ1b), which is produced from an alternative transcript of the retinal rod β subunit (Sautter et al., 1998; Bönigk et al., 1999). The exact combination of subunits that is used in native OSNs is not known, but coexpression of all three cDNAs results in a channel similar to that found in OSNs (Sautter et al., 1998; Bönigk et al., 1999), and all three polypeptides are expressed in the OE (Bönigk et al., 1999), suggesting that the native channel is a heterotrimer.
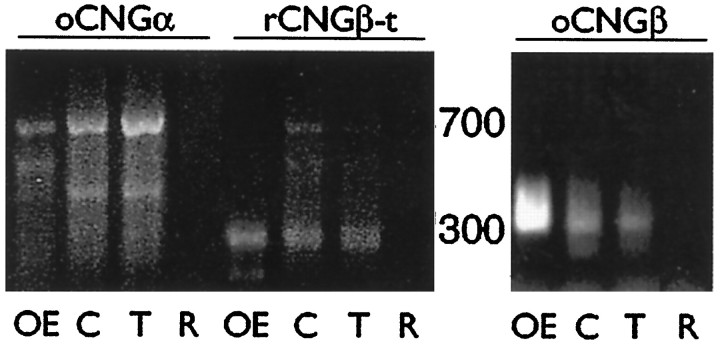
We used RT-PCR to demonstrate the presence of RNAs encoding all three of these potential subunits. Using primers for oCNGα that have been shown to amplify that subunit specifically (Kingston et al., 1996), we find that odora cells, like OE (but not retina), produce the oCNGα RNA, and that differentiation of odora cells appears to increase the amount of oCNGα RNA (Fig.7). Using primers for rCNGβ-t that have similarly been shown to be specific (Sautter et al., 1998), we also find that odora cells, like OE (but not retina), produce the rCNGβ-t RNA (Fig. 6). Finally, using primers for oCNGβ, we find that odora cells, like OE (but not retina) produce the oCNGβ RNA; however, this RNA appears to be in low abundance inodora cells relative to OE, because it is difficult to amplify from odora-derived cDNA that has been randomly primed (requiring additional cycles of amplification), or requires specific priming in order to produce substantial amplification (Fig.7). The products we amplified were not derived from genomic DNA, as at least two of the primer sets (those for oCNGα and oCNGβ) span introns, and would have resulted in larger products than those we amplified in these reactions. Together, these RT-PCR data suggest thatodora cells, like OSNs, produce three potential cyclic nucleotide-gated channel subunits, and are, therefore, likely to assemble an OSN-like channel. Although we have shown immunohistochemically that the oCNGβ protein is present inodora cells and in OSNs (Fig. 5), the precise combination of proteins that is produced and used in odora cells and in OSNs awaits further characterization of the native channels.
Fig. 7.
Odora cells express RNAs encoding three potential subunits of a cyclic nucleotide-gated channel. RNAs were reverse-transcribed and amplified in PCRs as described in Materials and Methods. Products can be amplified in 35 cycles for oCNGα (674 nt) and rCNGβ-t (293 nt) using randomly primed cDNAs, and for oCNGβ (313 nt) using specifically primed cDNAs, from olfactory epithelium (OE), control odoracells (C), and treated odora cells (T). Products were not derived from genomic DNA, because two of the primer sets (those for oCNGα and oCNGβ) span introns and would have resulted in larger products than those we amplified in these reactions. No products were amplified when cDNAs from retina (R) were used in parallel reactions.
Responses to stimuli in odora cells
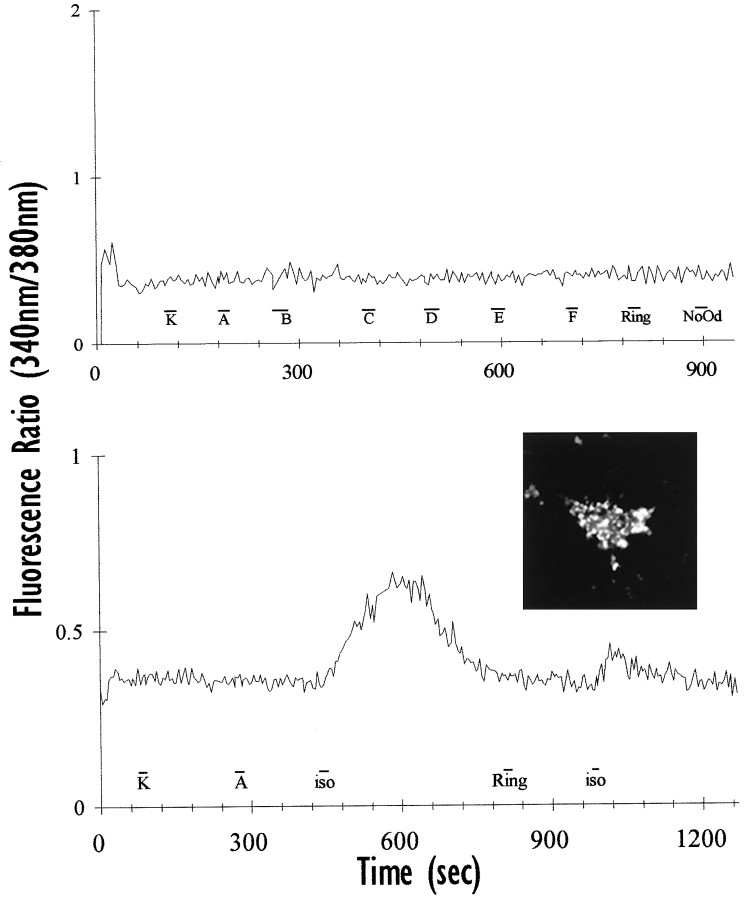
As differentiated odora cells contain many of the components of the putative signal transduction machinery, we next asked whether these cells can respond to odorants. These cells, unlike many neurons, frequently did not respond (as assayed by changes in intracellular calcium levels; see Materials and Methods) to a potentially depolarizing concentration of potassium (100 mm; Fig. 8). The reasons that they may not always respond to a high concentration of potassium are many; for example, they may not express voltage-activated calcium channels, or they may have an unusually high resting potential. An accurate description of this phenomenon awaits a full characterization of the electrophysiological properties ofodora cells.
Fig. 8.
Odora cells can target functional exogenous proteins to their plasma membrane. Top,Untransfected odora cells do not respond to pulses (16–24 sec, displayed as bars below the response traces) of our odorant mixtures (data from one cell shown; compare Table 2), or to a pulse of Ringer’s (Ring) or odorant diluent (NoOd). Odora cells frequently do not respond to 100 mm potassium (K). Bottom, Odora cells, when transfected with a plasmid containing the β2-adrenergic receptor and an HA tag, properly target the exogenous protein their surface, as detected by immunoreactivity for the HA tag in live (non-permeabilized) cells (inset). Transfected odora cells respond to isoproterenol (iso) but not to odorant mixtures (A). Response for one cell shown.
We have not yet determined whether odora cells express an endogenous ORP; however, because there are hundreds of members of the odorant receptor gene family (Buck and Axel, 1991), the likelihood of testing the cells with an odorant that happened to stimulate their endogenous receptors seems slight. We have used six mixtures of odorants (containing a total of 30 odorants; Table 2), which, because of the broad odorant responsivity in individual cells (Bozza and Kauer, 1998; Malnic et al., 1999), is likely to stimulate, at least partially, many more than 30 different ORPs. Differentiated odora cells never responded to these mixes (Fig. 8, top; Table3); although this is consistent with a lack of endogenous ORP expression, it could just as easily reflect our relatively small odorant profile.
Table 3.
Responses to odorant mix B and selected components (Comp), including those cells that responded to: EA (total; includes 34 that responded only to EA); PA (only); and both EA and PA
| # Slips | #Cells | Response | |||||
|---|---|---|---|---|---|---|---|
| B | VA | EAtotal | PAonly | EA and PA | |||
| No DNA | 2 | 30 | 0 | ND | ND | ND | ND |
| U131 | |||||||
| Mixes | 18 | 303 | 98 | ND | ND | ND | ND |
| Components | 11 | 225 | 65 | 0 | 48 | 0 | 14 |
Note that none of the cells listed (0/558) responded to any of the other odorant mixes texted. ND, Not determined.
Others have shown that the β2-adrenergic receptor can activate the olfactory G-protein (Gαolf) (Jones et al., 1990), resulting in a transduction cascade similar to that after odorant exposure. Because we had never elicited a response to odorants in differentiatedodora cells (see above), and because we knew thatodora cells could produce, and distribute, many components of the transduction machinery (see above), we transfected cells with a plasmid containing the β2-adrenergic receptor (see Materials and Methods). In this plasmid, the β2-adrenergic receptor is coupled to a hemagglutinin (HA) tag (McClintock et al., 1997), which can subsequently be detected in living cells with an antibody directed against HA. Unlike many neuronally-derived cells, odoracells are readily transfected: we typically achieve 50–80% transfection efficiency (as assayed by expression of the β2-adrenergic receptor in live cells; Fig. 8,inset) in transient transfections.
Odora cells transfected with the β2-adrenergic receptor are able to respond with an increase in intracellular calcium to a saturating concentration of an appropriate agonist (isoproterenol, 6 of 12 cells; Fig. 8). Importantly, when presented with our odorant mixtures before isoproterenol, these transfected cells did not respond (0 of 12 on two coverslips; Fig. 8), demonstrating that expression of a non-ORP receptor is not sufficient to confer an odorant response. Althoughodora cells, like OSNs, appear to produce some Gαs (data not shown), these data, nevertheless, suggest that odora cells can couple ligand activation of a receptor (in this case, the β2-adrenergic receptor) to an increase in intracellular calcium, presumably via a stimulation of adenylyl cyclase by Gαolf (or Gαs). The resultant increase in cAMP is likely to result in opening of cyclic nucleotide-gated channels within the plasma membrane, as is thought to occur in OSNs.
Expression and function of ORPs in odora cells
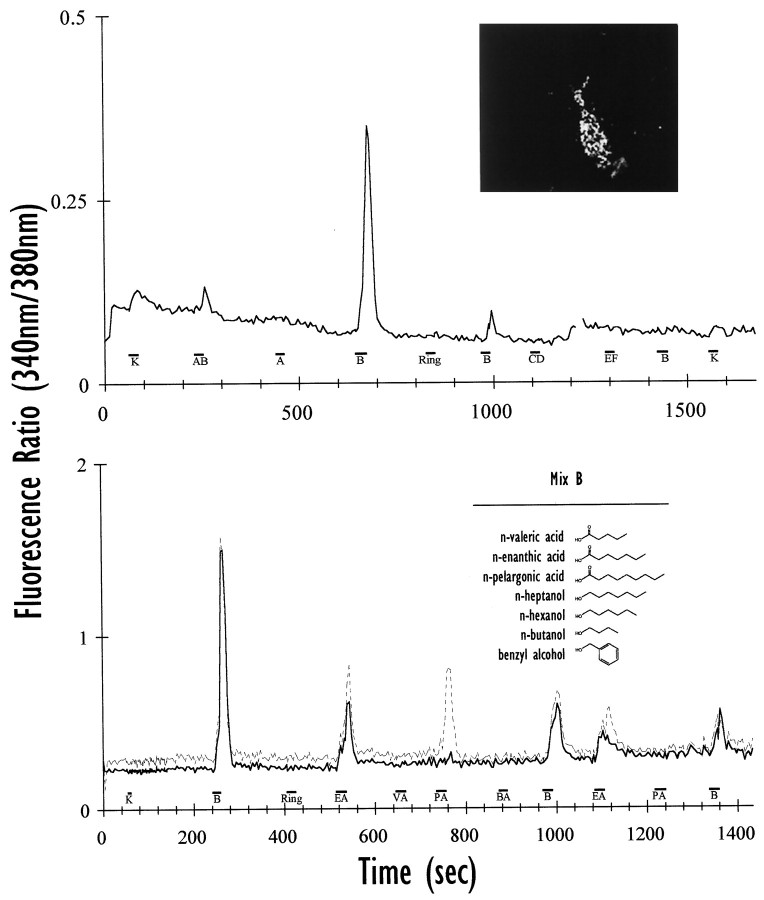
In marked contrast to the inability of other cell lines to direct ORPs to the plasma membrane (McClintock et al., 1997), odoracells properly and efficiently target exogenous ORPs to their surface in a punctate pattern (Fig. 9,inset). This pattern was seen with both ORP constructs tested (U131, Fig. 9; OR5, data not shown), and was similar to that seen after expression of the β2-adrenergic receptor in odora cells (compare Fig. 8). Odoracells are, therefore, unique in their ability to direct ORPs to their surface. As these ORP-transfected odora cells express (1) an ORP, (2) Gαolf, (3) ACIII, and (4) subunits of the cyclic nucleotide-gated channel, we reasoned that these cells should respond to stimuli for the exogenous ORP. We have assayed responses by detecting changes in intracellular calcium by ratiometric imaging with fura-2 AM (see Materials and Methods), focusing on those cells that were transfected with the rat ORP, U131. This ORP was isolated in 1997 (McClintock et al., 1997), but its function has not been previously characterized.
Fig. 9.
Odora cells can express functional exogenous odorant receptor proteins. Top, Odora cells, when transfected with a plasmid containing an ORP (U131) and an HA tag, properly target the exogenous ORP to their surface (inset). The transfected odora cells respond to a mix of odorants (B), but not to others (A, CD, EF); therefore, expression of U131 confers odorant responsiveness. Cells occasionally respond to 100 mm potassium (K). The response to mix B desensitizes: the second response is markedly smaller than the first, and the third pulse elicited no response. The response to a mixture of A and B (AB) is smaller than that for B alone; this may, in part, reflect the dilution inherent in mixing A and B, but could also be caused by more complicated interactions among the various components of A and B. The data shown are the average of the responses from four cells; the break in the line reflects a sampling error at 380 nm. Bottom, Cells that responded to mix B were divided into two categories: those that responded ton-enanthic acid (EA; solid line; average of five cells), and those that responded both to EA and n-pelargonic acid (PA;dotted line; average of five cells); responses to PA are never seen in cells that do not respond to EA. Importantly, expression of U131 does not confer response to compounds with structures that are similar to EA, including n-valeric acid (VA), benzyl alcohol (BA), and heptanol (Table 2). The responses desensitize, both to mix B (presented three times in 20 min) and to the individual components (each presented twice in 10 min). Together, these data suggest that U131 acts as a receptor for seven- and nine-carbon saturated fatty acids, and thatodora cells can appropriately couple this receptor to a physiological response. We did not detect a response to 100 mm potassium (K) in the differentiated odora cells, even when data were collected every 880 msec during and after the K pulse (data collection returned to every 4 sec after 92 sec).
We have used, as noted above, six mixtures of odorants (Table 2) to test for potential odorant responses. As also noted above, the relatively broad responsivity displayed by OSNs (Bozza and Kauer, 1998;Malnic et al., 1999) suggests that our mixes might contain a ligand that would interact with the transfected ORP, particularly since expression of transfected constructs can be quite high.
Differentiated odora cells that had been transiently transfected with an ORP, U131, respond to one of our six mixes of odorants (mix B), but not to the five others (mixes A, C–F; Fig. 9). This increase in intracellular calcium is completely blocked when extracellular calcium is chelated (G. Liu and B. Talamo, personal communication), suggesting that the increase results from calcium influx rather than from release of calcium from intracellular stores. On 18 coverslips, 32% of the cells responded to odorant mix B (Table 3), suggesting that a large fraction (∼50%) of the transfected cells (themselves 50–80% of the total number of cells) displayed an odorant response. Those cells that were transfected but did not respond to mix B may express lower levels of transfected ORP, may have been incapable of responding because of changes in other components of the transduction pathway, or may have had inadequate access to the odorant. Nevertheless, after transient transfection with a specific ORP, a relatively large number of cells produced responses to one specific odorant mixture.
Intriguingly, odora cells transfected with U131 display odorant desensitization, a phenomenon that has been described in OSNs (Bozza and Kauer, 1998, their references). We routinely observe a diminished response to successive challenges of odorant (Fig. 9), even when those presentations are minutes apart. Remarkably, within a given experiment, there is little difference among individual cells, both in initial amplitude of response and subsequent desensitization.Odora cells present a rich, relatively uniform, system in which to study this phenomenon in detail.
Which of the components of the stimulating mix can elicit a response? When tested individually, only two, the seven-carbon saturated fatty acid (enanthic acid; EA) and nine-carbon satu- rated fatty acid (pelargonic acid; PA), ever elicited a response in odoracells (Fig. 9; Table 3). Strikingly, the five-carbon saturated fatty acid (valeric acid; VA) and the seven-carbon saturated alcohol (heptanol) gave no measurable response (0 of 225 cells). Of those cells that responded to odorant mix B, 73% responded to EA (Table 3); of those that responded to EA, 29% also responded to PA. The only cells that responded to EA or PA were among those that had responded to odorant mix B; similarly, no cells ever responded to PA that did not respond to EA. Remarkably, as noted earlier with odorant mixtures, the amplitude and duration of responses among cells within each of the groups were consistent within a given experiment, even in the degree of desensitization. A more exhaustive characterization of the response profile awaits additional experiments; nevertheless, it is clear that U131 provides odora cells with the capacity to respond to seven- and nine-carbon saturated fatty acids.
DISCUSSION
The nature of the odora progenitor
The postnatal rodent olfactory epithelium contains three major dividing cell types: sustentacular cell, horizontal basal cell, and globose basal cell. We have previously shown, under the conditions we have studied, that these three cell types appear to be in independent lineages, and that olfactory sensory neurons appear to be exclusively derived from GBCs (Caggiano et al., 1994). Similar results have been reported by others (Schwob et al., 1994). However, these lineal relationships are not absolute: under some experimental conditions, such as exposure to methyl bromide gas, a single, HBC-like progenitor is competent to give rise to all cell types within the olfactory epithelium (Huard et al., 1998). Thus, there is some plasticity within the progenitors which, under most conditions, is dampened.
It is possible that factors present in the resting olfactory epithelium act to repress transdifferentiation among the three lineages. Indeed, within the OSN lineage itself, it is thought that some aspects of the mature OSN phenotype are transcriptionally repressed in the progenitor cells (Tsai and Reed, 1997; Wang et al., 1997). It is conceivable that some of these controls are lost after transfer of cells ex vivo. The loss of these controls would, at least in part, explain the difficulty that many laboratories have had in recapitulating OSN generation in vitro, and may also partly account for the partially differentiated phenotype that we observe in controlodora cells.
Within the usual OSN lineage, i.e., from GBC to OSN, cells move through several stages of development (for review, see Calof et al., 1998). During the initial stages, the cell is still mitotically active; as it progresses through the subsequent stages, mitosis ceases. We could have infected any of these dividing cells within the early stages of OSN production with our retrovirus containing the immortalizing oncogene, thereby obtaining cell lines with a phenotype between GBC and OSN. Our immunohistochemical results with the odora line are consistent with our immortalization of a slightly differentiated OSN progenitor.
Dopamine as a differentiation agent
Olfactory sensory neurons express D2 dopamine receptor RNA and protein within the olfactory epithelium (Coronas et al., 1997b), as well as at their target sites in the glomeruli of the olfactory bulb (Nickell et al., 1991; Coronas et al., 1997b). One synaptic target of OSNs is the dopaminergic periglomerular cell, which may, therefore, provide the neurotransmitter that acts on D2 receptors expressed by OSNs. It is conceivable that one form of communication between an OSN and its target is the detection of dopamine by presynaptic receptors on OSN terminals. Thus, dopamine is ideally situated as a potential modulator of OSN function. Indeed, dopamine has been shown to modulate adenylyl cyclase levels in olfactory epithelium (Mania-Farnell et al., 1993) and to modulate an inwardly rectifying hyperpolarization-activated current in OSNs (Vargas and Lucero, 1999).
Dopamine may also modulate OSN maturation. Others have shown that activation of a D2 receptor in an olfactory epithelium-derived cell line can lead to differentiation of those cells (Coronas et al., 1997a). Similarly, we have shown here that dopamine elicits differentiation in odora cells; we have also found thatodora cells express D2 receptors (J. R. Murrell and D. D. Hunter, unpublished observations). Thus, dopamine, derived from targets within the olfactory bulb, and acting on D2 receptors in OSNs, may act as a differentiation signal for some of the final aspects of OSN maturation.
The cyclic nucleotide-gated channel
One component of the odorant signal transduction system that is thought to be vital is the cyclic nucleotide-gated channel. Animals in which one subunit of the channel has been disrupted by homologous recombination display anosmia to all tested odorants (Brunet et al., 1996), suggesting that all of these compounds use activation of this channel as part of the signal transduction process. However, the exact nature of the channel as it exists in OSNs is unclear. The olfactory channel consists of at least two subunits, oCNGα (Dhallan et al., 1990; also known as oCNC1, CNG2, or CNCα3) and oCNGβ (Bradley et al., 1994; Liman and Buck, 1994; also known as oCNC2, CNG5, or CNCα4), and perhaps a third, rCNGβ-t (Sautter et al., 1998;Bönigk et al., 1999; also known as CNG4.3 or CNCβ1b). It seems plausible that the native channel is a heterotrimer of all three subunits (Sautter et al., 1998; Bönigk et al., 1999). We have shown that odora cells produce RNAs encoding all three potential subunits of the olfactory cyclic nucleotide-gated channel. Therefore, odora cells are likely to serve as a good model for studying this channel.
Activation of the olfactory cyclic nucleotide-gated channel can lead to an influx of calcium through the OSN membrane, which could subsequently activate a variety of channels within the plasma membrane (for review, see Schild and Restrepo, 1998). Whether a similar cascade exists withinodora cells is unclear; however, the influx in calcium we measure in response to odorants suggests that at least some portion of this pathway is operating as it does in OSNs.
Odorant receptor proteins and odor codes
We have, for the first time, demonstrated the expression of full-length exogenous odorant receptor proteins in a cell line. The ready availability of large numbers of cells expressing the same odorant receptor will allow for a careful analysis of the odorant response profiles conferred by a given odorant receptor protein. Here, we have shown a moderate degree of flexibility, but also a fair amount of selectivity, in the chemical nature of the odorant that stimulates a given odorant receptor protein. Similar results have been obtained in OSNs that were infected with a single odorant receptor (I7, Zhao et al., 1998; MOR23, Touhara et al., 1999), and are consistent with the many reports that the primary reception event is somewhat broadly tuned (Bozza and Kauer, 1998; Malnic et al., 1999; Duchamp-Viret et al., 1999).
Where, then, is odorant selectivity achieved? The bulk of the data generated within the olfactory system in the last several decades has led to the concept that much of the selectivity is achieved via interactions among the neurons of the network within the olfactory bulb (Schild, 1988; Kauer, 1991). An understanding of the relationships among individual odorants and their receptors will, therefore, only lay a partial groundwork for an understanding of odor perception. Nevertheless, it will be important to show carefully whether reproducible response profiles contribute to the combinatorial events that occur within the olfactory bulb and result in odorant discrimination. Such profiles can be more easily generated in the defined, consistent system provided by odora cells.
Summary
Our creation and characterization of odora cells is a major step forward: although other lines have been created (Goldstein et al., 1997; Coronas et al., 1997a), none has been as extensively characterized as this line, none appears to follow the course of differentiation (similar to that of GBCs) displayed by this line, and, in particular, none shows the capacity to respond to odorants when expressing a known ORP that we have shown here. Together, these properties of odora cells should allow for an extensive characterization of the generation and function of odorant-responsive cells, a task that has been limited in the past by the necessity to study OSNs in vivo or in relatively inefficient primary culture systems.
The presence of a family of G-protein-coupled odorant receptors was originally proposed in 1991 (Buck and Axel, 1991), based on the initial identification of a multitude of RNAs that could potentially encode these proteins. A major problem in previous experiments has been a lack of correct targeting of heterologously expressed ORPs; the shift in targeting that occurs when odora cells are differentiated provides us with an ideal model system for studying trafficking of membrane proteins, including ORPs. In addition, we are now in a position that allows for a thorough analysis of the issues surrounding the contribution that individual ORPs may make to odorant sensitivity and selectivity. Similarly, the ready isolation of large quantities of the signal transduction components from a cell line represents a unique opportunity for biochemical elucidation of many aspects of this cascade which have, for now, remained a mystery, including such issues as odorant desensitization. In summary, as models of the OSN lineage,odora cells will now allow us to study the genesis, maturation, function, death, and replenishment of OSNs in a controlled environment, enabling us to ask fundamental questions concerning the olfactory system that could previously not be asked.
Footnotes
This work was supported by funds provided by Tufts University. We particularly thank Tom Bozza for many helpful discussions during the course of this work, Tim McClintock for providing advice and receptor constructs, and members of the laboratory of Barbara Talamo for comments and sharing unpublished data, as well as Atsuko Polzin for help in performing some of the original infections, Joel White and members of the laboratory of Jim Wang for comments, and Dona Chikaraishi for encouragement during the beginning of this project. We also thank William Brunken for many hours of support and advice.
Correspondence should be addressed to Dr. Dale D. Hunter, Department of Neuroscience SC-6, Tufts University School of Medicine, 136 Harrison Avenue, Boston, MA 02111.
REFERENCES
- 1.Barber PC, Lindsay RM. Schwann cells of the olfactory nerves contain glial fibrillary acidic protein and resemble astrocytes. Neuroscience. 1982;7:3077–3090. doi: 10.1016/0306-4522(82)90231-7. [DOI] [PubMed] [Google Scholar]
- 2.Bönigk W, Bradley J, Müller F, Sesti F, Boekhoff I, Ronnett GV, Kaupp UB, Frings S. The native rat olfactory cyclic nucleotide-gated channel is composed of three distinct subunits. J Neurosci. 1999;19:5332–5347. doi: 10.1523/JNEUROSCI.19-13-05332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozza TC, Kauer JS. Odorant response properties of convergent olfactory receptor neurons. J Neurosci. 1998;18:4560–4569. doi: 10.1523/JNEUROSCI.18-12-04560.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley J, Li J, Davidson N, Lester HA, Zinn K. Heteromeric olfactory cyclic nucleotide-gated channels: a subunit that confers increased sensitivity to cAMP. Proc Natl Acad Sci USA. 1994;91:8890–8894. doi: 10.1073/pnas.91.19.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet LJ, Gold GH, Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17:681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 6.Brustle O, McKay RD. Neuronal progenitors as tools for cell replacement in the nervous system. Curr Opin Neurobiol. 1996;6:688–695. doi: 10.1016/s0959-4388(96)80104-8. [DOI] [PubMed] [Google Scholar]
- 7.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 8.Caggiano ME, Kauer JS, Hunter DD. Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication-incompetent retrovirus. Neuron. 1994;13:339–352. doi: 10.1016/0896-6273(94)90351-4. [DOI] [PubMed] [Google Scholar]
- 9.Calof AL, Chikaraishi DM. Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron. 1989;3:115–127. doi: 10.1016/0896-6273(89)90120-7. [DOI] [PubMed] [Google Scholar]
- 10.Calof AL, Mumm JS, Rim PC, Shou J. The neuronal stem cell of the olfactory epithelium. J Neurobiol. 1998;36:190–205. doi: 10.1002/(sici)1097-4695(199808)36:2<190::aid-neu7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 11.Cepko CL, Roberts BE, Mulligan RC. Construction and applications of a highly transmissible murine retrovirus shuttle vector. Cell. 1984;37:1053–1062. doi: 10.1016/0092-8674(84)90440-9. [DOI] [PubMed] [Google Scholar]
- 12.Coronas V, Feron F, Hen R, Sicard G, Jourdan F, Moyse E. In vitro induction of apoptosis or differentiation by dopamine in an immortalized olfactory neuronal cell line. J Neurochem. 1997a;69:1870–1881. doi: 10.1046/j.1471-4159.1997.69051870.x. [DOI] [PubMed] [Google Scholar]
- 13.Coronas V, Srivastava LK, Liang JJ, Jourdan F, Moyse E. Identification and localization of dopamine receptor subtypes in rat olfactory mucosa and bulb: a combined in situ hybridization and ligand binding radioautographic approach. J Chem Neuroanat. 1997b;12:243–257. doi: 10.1016/s0891-0618(97)00215-9. [DOI] [PubMed] [Google Scholar]
- 14.Costanzo RM, Morrison EE. Three-dimensional scanning electron microscopic study of the normal hamster olfactory epithelium. J Neurocytol. 1989;18:381–391. doi: 10.1007/BF01190841. [DOI] [PubMed] [Google Scholar]
- 15.Crews L, Hunter DD. Neurogenesis in the olfactory epithelium. Perspect Dev Neurobiol. 1994;2:151–161. [PubMed] [Google Scholar]
- 16.Dhallan RS, Yau KW, Schrader KA, Reed RR. Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature. 1990;347:184–187. doi: 10.1038/347184a0. [DOI] [PubMed] [Google Scholar]
- 17.Duchamp-Viret P, Chaput MA, Duchamp A. Odor response properties of rat olfactory receptor neurons. Science. 1999;284:2171–2174. doi: 10.1126/science.284.5423.2171. [DOI] [PubMed] [Google Scholar]
- 18.Farbman AI. Olfactory neurogenesis: genetic or environmental controls? Trends Neurosci. 1990;13:362–365. doi: 10.1016/0166-2236(90)90017-5. [DOI] [PubMed] [Google Scholar]
- 19.Fawcett JW, Geller HM. Regeneration in the CNS: optimism mounts. Trends Neurosci. 1998;21:179–180. doi: 10.1016/s0166-2236(98)01241-7. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein BJ, Schwob JE. Analysis of the globose basal cell compartment in rat olfactory epithelium using GBC-1, a new monoclonal antibody against globose basal cells. J Neurosci. 1996;16:4005–4016. doi: 10.1523/JNEUROSCI.16-12-04005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein BJ, Wolozin BL, Schwob JE. FGF2 suppresses neuronogenesis of a cell line derived from rat olfactory epithelium. J Neurobiol. 1997;33:411–428. [PubMed] [Google Scholar]
- 22.Graziadei PPC, Metcalf JF. Autoradiographic and ultrastructural observations on the frog’s olfactory mucosa. Z Zellforsch. 1971;116:305–318. doi: 10.1007/BF00330630. [DOI] [PubMed] [Google Scholar]
- 23.Graziadei PPC, Monti Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- 24.Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol. 1998;400:469–486. [PubMed] [Google Scholar]
- 25.Hunter DD, Murphy MD, Olsson CV, Brunken WJ. S-laminin expression in adult and developing retinae: a potential cue for photoreceptor morphogenesis. Neuron. 1992;8:399–413. doi: 10.1016/0896-6273(92)90269-j. [DOI] [PubMed] [Google Scholar]
- 26.Jat PS, Cepko CL, Mulligan RC, Sharp PA. Recombinant retroviruses encoding simian virus 40 large T antigen and polyomavirus large and middle T antigens. Mol Cell Biol. 1986;6:1204–1217. doi: 10.1128/mcb.6.4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jat PS, Sharp PA. Cell lines established by a temperature-sensitive simian virus 40 large-T-antigen gene are growth restricted at the nonpermissive temperature. Mol Cell Biol. 1989;9:1672–1681. doi: 10.1128/mcb.9.4.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones DT, Reed RR. Golf: an olfactory neuron specific G protein involved in odorant signal transduction. Science. 1989;244:790–795. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- 29.Jones DT, Masters SB, Bourne HR, Reed RR. Biochemical characterization of three stimulatory GTP-binding proteins. The large and small forms of Gs and the olfactory-specific G-protein, Golf. J Biol Chem. 1990;265:2671–2676. [PubMed] [Google Scholar]
- 30.Juilfs DM, Fulle HJ, Zhao AZ, Houslay MD, Garbers DL, Beavo JA. A subset of olfactory neurons that selectively express cGMP-stimulated phosphodiesterase (PDE2) and guanylyl cyclase-D define a unique olfactory signal transduction pathway. Proc Natl Acad Sci USA. 1997;94:3388–3395. doi: 10.1073/pnas.94.7.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kauer JS. Contributions of topography and parallel processing to odor coding in the vertebrate olfactory pathway. Trends Neurosci. 1991;14:79–85. doi: 10.1016/0166-2236(91)90025-p. [DOI] [PubMed] [Google Scholar]
- 32.Kingston PA, Zufall F, Barnstable CJ. Rat hippocampal neurons express genes for both rod retinal and olfactory cyclic nucleotide-gated channels: novel targets for cAMP/cGMP function. Proc Natl Acad Sci USA. 1996;93:10440–10445. doi: 10.1073/pnas.93.19.10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn HG, Winkler J, Kempermann G, Thal LJ, Gage FH. Epidermal growth factor and fibroblast growth factor-2 have different effects on neural progenitors in the adult rat brain. J Neurosci. 1997;17:5820–5829. doi: 10.1523/JNEUROSCI.17-15-05820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Libby RT, Hunter DD, Brunken WJ. Developmental expression of laminin β2 in rat retina: further support for a role in rod morphogenesis. Invest Ophthalmol Vis Sci. 1996;37:1651–1661. [PubMed] [Google Scholar]
- 36.Liman ER, Buck LB. A second subunit of the olfactory cyclic nucleotide-gated channel confers high sensitivity to cAMP. Neuron. 1994;13:611–621. doi: 10.1016/0896-6273(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 37.Mahanthappa NK, Schwarting GA. Peptide growth factor control of olfactory neurogenesis and neuron survival in vitro: roles of EGF and TGF-βs. Neuron. 1993;10:293–305. doi: 10.1016/0896-6273(93)90319-m. [DOI] [PubMed] [Google Scholar]
- 38.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 39.Mania-Farnell BL, Farbman AI, Bruch RC. Bromocriptine, a dopamine D2 receptor agonist, inhibits adenylyl cyclase activity in rat olfactory epithelium. Neuroscience. 1993;57:173–180. doi: 10.1016/0306-4522(93)90119-z. [DOI] [PubMed] [Google Scholar]
- 40.Margolis FL. A brain protein unique to the olfactory bulb. Proc Natl Acad Sci USA. 1972;69:1221–1224. doi: 10.1073/pnas.69.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClintock TS, Landers TM, Gimelbrant AA, Fuller LZ, Jackson BA, Jayawickreme CK, Lerner MR. Functional expression of olfactory-adrenergic receptor chimeras and intracellular retention of heterologously expressed olfactory receptors. Brain Res Mol Brain Res. 1997;48:270–278. doi: 10.1016/s0169-328x(97)00099-5. [DOI] [PubMed] [Google Scholar]
- 42.Moulton DG. Dynamics of cell populations in the olfactory epithelium. Ann NY Acad Sci. 1974;237:52–61. doi: 10.1111/j.1749-6632.1974.tb49843.x. [DOI] [PubMed] [Google Scholar]
- 43.Nekrasova E, Sosinskaya A, Natochin M, Lancet D, Gat U. Overexpression, solubilization and purification of rat and human olfactory receptors. Eur J Biochem. 1996;238:28–37. doi: 10.1111/j.1432-1033.1996.0028q.x. [DOI] [PubMed] [Google Scholar]
- 44.Nickell WT, Norman AB, Wyatt LM, Shipley MT. Olfactory bulb DA receptors may be located on terminals of the olfactory nerve. NeuroReport. 1991;2:9–12. doi: 10.1097/00001756-199101000-00002. [DOI] [PubMed] [Google Scholar]
- 45.Pixley SK. CNS glial cells support in vitro survival, division, and differentiation of dissociated olfactory neuronal progenitor cells. Neuron. 1992;8:1191–1204. doi: 10.1016/0896-6273(92)90139-5. [DOI] [PubMed] [Google Scholar]
- 46.Raming K, Krieger J, Strotmann J, Boekhoff I, Kubick S, Baumstark C, Breer H. Cloning and expression of odorant receptors. Nature. 1993;361:353–356. doi: 10.1038/361353a0. [DOI] [PubMed] [Google Scholar]
- 47.Sautter A, Zong X, Hofmann F, Biel M. An isoform of the rod photoreceptor cyclic nucleotide-gated channel β subunit expressed in olfactory neurons. Proc Natl Acad Sci USA. 1998;95:4696–4701. doi: 10.1073/pnas.95.8.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schild D. Principles of odor coding and a neural network for odor discrimination. Biophys J. 1988;54:1001–1011. doi: 10.1016/S0006-3495(88)83038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schild D, Restrepo D. Transduction mechanisms in vertebrate olfactory receptor cells. Physiol Rev. 1998;78:429–466. doi: 10.1152/physrev.1998.78.2.429. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz Levey M, Chikaraishi DM, Kauer JS. Characterization of potential precursor populations in the mouse olfactory epithelium using immunocytochemistry and autoradiography. J Neurosci. 1991;11:3556–3564. doi: 10.1523/JNEUROSCI.11-11-03556.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwob JE, Huard JM, Luskin MB, Youngentob SL. Retroviral lineage studies of the rat olfactory epithelium. Chem Senses. 1994;19:671–682. doi: 10.1093/chemse/19.6.671. [DOI] [PubMed] [Google Scholar]
- 52.Southern PJ, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 53.Talamo BR, Rudel R, Kosik KS, Lee VM, Neff S, Adelman L, Kauer JS. Pathological changes in olfactory neurons in patients with Alzheimer’s disease. Nature. 1989;337:736–739. doi: 10.1038/337736a0. [DOI] [PubMed] [Google Scholar]
- 54.Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc Natl Acad Sci USA. 1999;96:4040–4045. doi: 10.1073/pnas.96.7.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai RY, Reed RR. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J Neurosci. 1997;17:4159–4169. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vargas G, Lucero MT. Dopamine modulates inwardly rectifying hyperpolarization-activated current (Ih) in cultured rat olfactory receptor neurons. J Neurophysiol. 1999;81:149–158. doi: 10.1152/jn.1999.81.1.149. [DOI] [PubMed] [Google Scholar]
- 57.Wang SS, Tsai RYL, Reed RR. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss S, Reynolds BA, Vescovi AL, Morshead C, Craig CG, van der Kooy D. Is there a neural stem cell in the mammalian forebrain? Trends Neurosci. 1996;19:387–393. doi: 10.1016/s0166-2236(96)10035-7. [DOI] [PubMed] [Google Scholar]
- 59.Zhao H, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein S. Functional expression of a mammalian odorant receptor. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]