Abstract
The myelin proteolipid protein (PLP) gene (i.e., the PLP/DM20 gene) has been of some interest because of its role in certain human demyelinating diseases, such as Pelizaeus-Merzbacher disease. A substantial amount of evidence, including neuronal pathology in knock-out and transgenic animals, suggests the gene also has functions unrelated to myelin structure, but the products of the gene responsible for these putative functions have not yet been identified. Here we report the identification of a new exon of the PLP/DM20 gene and at least two new products of the gene that contain this exon. The new exon, located between exons 1 and 2, is spliced into PLP and DM20 mRNAs creating a new translation initiation site that generates PLP and DM20 proteins with a 12 amino acid leader sequence. This leader sequence appears to target these proteins to a different cellular compartment within the cell bodies of oligodendrocytes and away from the myelin membranes. Furthermore, these new products are also expressed in a number of neuronal populations within the postnatal mouse brain, including the cerebellum, hippocampus, and olfactory system. We term these products somal-restricted PLP and DM20 proteins to distinguish them from the classic PLP and DM20 proteolipids. They represent putative candidates for some of the nonmyelin-related functions of the PLP/DM20 gene.
Keywords: myelin proteins, protein targeting, gene structure, myelin protein genes, neuronal genes, gene expression
The gene encoding the myelin proteolipid protein (PLP) is alternatively spliced to produce two mRNA products that encode two transmembrane proteins (i.e., PLP and DM20). These proteolipids constitute ∼50% of the protein in the myelin sheath and, as such, are among the most abundant proteins in the CNS. Peptide mapping (Trifilieff et al., 1986) and molecular biological studies (Nave et al., 1987) established that DM20 differs from PLP by an internal deletion of 35 amino acids encoded by exon 3B of the gene. The complete amino acid sequences of both proteolipids have been deduced from their corresponding cDNAs in many species, and their strong conservation among species has been noted by many investigators (Nave and Milner, 1989; Hudson and Nadon, 1992;Macklin et al., 1990; Griffiths et al., 1995, 1998a;Griffiths, 1996).
Expression of the PLP/DM20 gene was thought to be confined to myelin-forming cells in the CNS. In recent years expression of the gene has been observed in the PNS, embryonic CNS, and the heart (Puckett et al., 1987; Campagnoni et al., 1992; Ikenaka et al., 1992; Kamholz et al., 1992; Timsit et al., 1992, 1995). Furthermore it appears that expression of both isoforms, PLP and DM20, can occur in the thymus, spleen, and lymph nodes of the immune system (Pribyl et al., 1996a,b). Thus, the expression of the PLP/DM20 gene in so many nonmyelinating cells has strongly suggested, although indirectly, a role for PLP and/or DM20 beyond that of a myelin structural protein.
It has been difficult to define the nonmyelin role of the PLP/DM20 gene or even to assess whether this role was performed exclusively by the known gene products or perhaps by, as yet, undefined products of the gene. As part of our investigations into the expression of the MBP gene in the immune system, we found that the PLP/DM20 gene was also expressed in these tissues (Pribyl et al., 1996a,b). Subsequent reverse transcription (RT)-PCR analysis of the PLP-related products in mouse thymus revealed the existence of a DM20 isoform with a structure predicting a new exon of the gene. Here we report the isolation of cDNAs encoding a novel set of PLP and DM20 transcripts from the brain that define this new exon, and we map it to a genomic segment between exons 1 and 2. Of additional interest is the observation that these novel PLP and DM20 isoforms are expressed in neurons as well as in oligodendrocytes (OLs) in the CNS. These findings reinforce the possibility of a nonmyelin role for the PLP/DM20 gene and suggest that a more complex array of products is expressed by the gene, which may be partly responsible for that role.
MATERIALS AND METHODS
Cell culture. Primary glial cell cultures and enriched OLs were prepared as described previously (Bongarzone et al., 1996). Cultures of cerebellar granular neurons were prepared from postnatal day 7 (P7) cerebellar cortices as described elsewhere (Gao et al., 1991). The LBRM T-cell line (a gift of Dr. J. Merrill) and the CN1.4 neuronal line (Bongarzone et al., 1998b) were grown in 10% FCS-DMEM for 4 d before collection.
Extraction of RNA, RT-PCR, and cloning. Total RNA was extracted from brain tissue or cell pellets using Trizol reagent (Life Technologies, Gaithersburg, MD). Poly(A+) selection was performed as described in Sambrook et al. (1989). RT-PCR was performed essentially as described previously (Campagnoni et al., 1992; Bongarzone et al., 1998a) using the following primer pairs: (1) 114S and 3NPLP to amplify between exons 1.1 and 7, (2) 114S and 114A to amplify exon 1.1 only, and (3) 114S and 129A to amplify between exons 1.1 and 2.
Primer 114S: 5′-ACATGGCATTTAACTGTATTAACCCCTT
Primer 3NPLP: 5′-TCAGAACTTGGTGCCTCGGCCCATG
Primer 114A: 5′-AAGCCTGTGTGCATTTTCCAAAGGTCTGG
Primer 129A: 5′-CCTACCAGACATCTAGCACAA
PCR conditions were as follows: 94OC for 4 min; 40 cycles of 94OC for 1.25 min, 60OC for 1.25 min, and 72OC for 1.50 min; and 72OC for 7 min. Products of the PCR reaction were subcloned using the pGEM-T Easy Vector System according to the manufacturer’s instructions (Promega, Madison, WI) and subcloned further into pSPORT (Life Technologies). Sequence analysis was performed on representative samples of the PCR products.
Northern blot and nuclease protection assay. Northern blot (10 μg of total RNA) analysis was performed essentially as described by Sambrook et al. (1989) using a riboprobe to detect the mRNAs. A pSPORT (Life Technologies) clone generated by PCR amplification between primers 114S and 114A (see above), containing exon 1.1, was linearized with SalI, and a 182 nucleotide antisense probe was synthesized from the T7 promoter. This riboprobe was hybridized at 65°C to Northern blots, which were then washed at 37 and 60°C. Sizes of bands were assigned according to a Gibco-BRL RNA ladder (Life Technologies).
The same probe was used for the ribonuclease protection assay (RPA) (see Fig. 3A) using a commercially available RNase assay kit from Ambion (Austin, TX). A clone (see Fig. 3B) containing exon 1.1 plus the first 34 nucleotides of exon 2 in pGEM-T Easy (Promega) was linearized with SalI, and a 225 nucleotide antisense riboprobe was generated from the T7 promoter and used in RPA assays. For RNase protection, either 10 μg of total RNA or 3 μg of poly(A+) RNA was hybridized to the probe at 45°C.
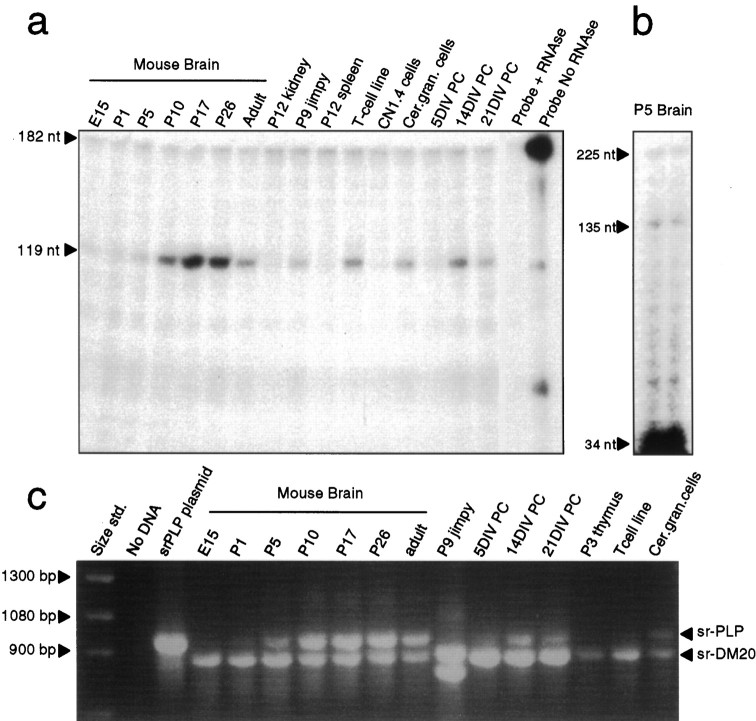
Fig. 3.
Distribution of srPLP and srDM20 in various tissues and cells. a, The expression of srPLP and srDM20 was examined by RPA using an exon 1.1–specific sequence. In mouse brain, peak expression of the srPLP and srDM20 mRNAs was observed between P17 and P26, closely resembling the developmental expression of the classic PLP and DM20 mRNAs. b, The steady levels of both sr and classic mRNAs in P5 mouse brain were examined by RPA using a riboprobe that contained 101 nt of exon 1.1 and 34 nt of exon 2. The level of protection of the classic fragment (34 nt) was significantly higher than that of the sr fragment (135 nt).c, The expression of either srPLP or srDM20 mRNAs was analyzed by RT-PCR using primers 114S and 3NPLP. The expression of srDM20 was found to precede that of srPLP during the development of the mouse brain and in mouse brain mixed glial primary cultures (PC). Truncated forms of srPLP and srDM20 mRNAs, reflecting the deletion of exon 5, were expressed in jimpy brains. Interestingly, the srPLP and srDM20 mRNAs were also expressed in cerebellar granule neurons as well as in thymus and a mouse T-cell line (LBRM). Cer. gran., Cerebellar granule;CN1.4, a conditionally immortalized cortical neuronal cell line; DIV, days in vitro;nt, nucleotide.
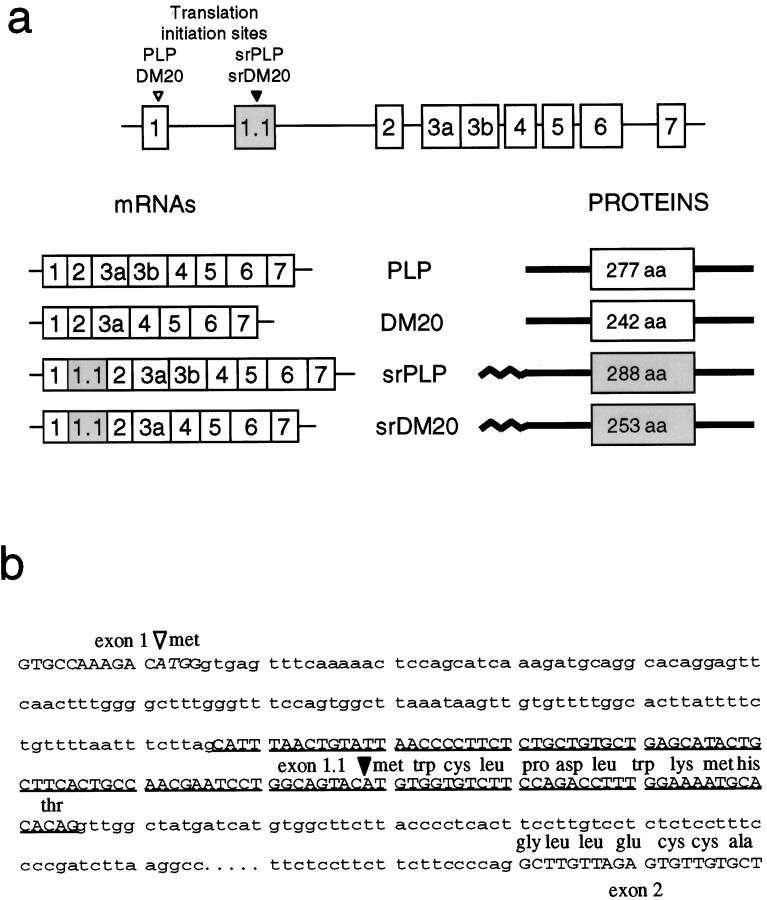
Mapping of exon 1.1 to the first intron of the gene. Eight clones were isolated from a screen of a mouse lambda genomic library (Stratagene 946308, La Jolla CA) with the somal-restricted (sr)PLP cDNA. These were digested with EcoRI, BamHI, andHindIII and with all possible combinations of these enzymes. From a comparison of the resulting Southern blots probed with a full-length PLP cDNA and a map published by Macklin et al. (1987), we identified a lambda clone that contained both exons 1 and 2 of the PLP gene. A comparison of the same blot stripped and reprobed with srPLP cDNA detected no restriction fragments that uniquely reacted with the novel sequence in srPLP. However, an ∼2 kbBamHI–EcoRI fragment that contained only 83 bp of the 3′ end of exon 1 appeared much more heavily labeled in the blots probed with srPLP. We subcloned an ∼2.2 kbHindIII–EcoRI fragment calculated to contain all of exon 1, sequenced the ends to confirm the presence of exon 1, and then sequenced with a primer specific to the novel srPLP cDNA sequence to confirm the presence of the new exon. We deleted the clone from its 5′ end and sequenced the only two deletion clones that still reacted with the srPLP probe. One deletion began in exon 1 and bridged the gap between exon 1 and the new exon (121 bp). A complete sequence for intron 1 can be found in the GenBank (accession number, AF003838) (Wight and Dobretsova, 1997).
In vitro transcription–translation.35S-Proteins were synthesized in the TNT T7-Coupled Reticulocyte Lysate System (Promega) and analyzed in a 12.5% SDS-polyacrylamide gel.
In situ hybridization. Sense and antisense33P-riboprobes corresponding to exon 1.1 were synthesized, and in situ hybridization was performed essentially as described (Landry et al., 1998) except that prehybridization and hybridization were performed at 55°C. Slides were exposed to Hyperfilm β-Max (Amersham, Arlington Heights, IL) and then dipped in photographic emulsion (Kodak NTB-2; Eastman Kodak, Rochester, NY) and stored at 4°C for 4 weeks.
Production of antibodies. A synthetic peptide containing the first 16 amino acids of the srPLP and srDM20 proteins was synthesized, coupled to a multiple antigen protein (Research Genetics, Huntsville, AL), and injected into female New Zealand White rabbits. After a primary immunization in complete Freund’s adjuvant, a series of four boosts in incomplete adjuvant were performed before final bleeding. Antibody titer was monitored by ELISA.
Western blots. Total proteolipid proteins were extracted from mouse brain by the method of Gardinier and Macklin (1988); dissolved in 8 m urea, 125 mm Tris, pH 6.8, 4% SDS, and 3% DTT; and loaded on a 15% gel. After transfer to Immobilon-P (Millipore, Bedford, MA), antibody-reactive products were detected using the Vectastain Elite avidin–horseradish peroxidase–coupled system (Vector Laboratories, Burlingame, CA) and SuperSignal chemiluminescent substrate (Pierce, Rockford, IL). Mobilities were compared with Bio-Rad Kaleidoscope and Prestained Molecular Weight Standards (Hercules, CA).
Immunofluorescence cytochemistry. Fixed cells were treated with 0.1% Triton X-100 and blocked with 10% normal goat serum. The following primary antibodies were used: rabbit polyclonal anti-srPLP and -srDM20 (1:2000); rat monoclonal anti-PLP and -DM20 (AA3; 1:200) (gift of Dr. K. Ikenaka); and mouse monoclonal antibodies A2B5 (1:100) (obtained from American Type Culture Collection, Rockville, MD), galactocerebroside (O1; 1:100), and O4 (1:100) (gifts of Drs. M. Schachner, J. Trotter, and A. Gard). Samples were analyzed with a Leica DM RXA microscope (Nussloch, Germany) and a Zeiss LSM 310 confocal microscope.
Immunohistochemistry. P14 mouse brains were fixed in 4% paraformaldehyde, and sagittal sections (50 μm) were processed using the free-floating technique. Sections were incubated with anti-srPLP and -srDM20 antibodies (1:2000 in 0.1% Triton X-100 and 0.1% casein PBS) and processed with the appropriate Vectastain secondary system.
RESULTS
Identification of a new exon in the PLP/DM20 gene
As part of our studies on the expression of the myelin protein genes in the immune system, we performed RT-PCR experiments to determine which isoforms of the PLP/DM20 gene were expressed in P0 thymus. After subcloning and sequencing many of these products, we identified, in addition to DM20 and PLP mRNAs, an unusual PLP/DM20 clone that we called clone 114. It was found to be derived from a DM20 variant mRNA with 109 novel base pairs inserted between sequences encoded by exons 1 and 2 of the PLP/DM20 gene, indicating the existence of a new exon of the gene. Two examples were isolated from a screen of 44 RT-PCR clones from P0 mouse thymus using a probe specific for the new sequence, and one clone was obtained from a screen of 58 RT-PCR clones from embryonic day 14 (E14) mouse brain. An additional screen of ∼250,000 clones of a mouse oligodendrocyte cDNA library yielded a clone containing the complete open reading frame for the srPLP and one for the srDM20. These results indicate that (1) the new isoforms are expressed in both the immune system and the nervous system and (2) the new exon can be spliced into either DM20 or PLP mRNAs.
Using the novel cDNA sequence, we mapped the location of this new exon (designated exon 1.1) between exons 1 and 2 of the PLP/DM20 gene in the mouse (Fig. 1a). The new sequence was located in a 2.2 kb genomic fragment, which also contained exon 1. This fragment was sequenced; the new exon 1.1 was found to be 121 bp downstream of exon 1, and it was flanked by appropriate splice donor/acceptor consensus sequences (see Fig. 1b).
Fig. 1.
Organization of the myelin proteolipid protein gene and the sequence of the new exon and the N-terminal peptide encoded by this exon. a, Gene structure showing the location of the new exon 1.1 between exons 1 and 2.Below the gene to the left are shown the exonic compositions of the srPLP and srDM20 mRNAs containing exon 1.1 compared with those of the classic PLP and DM20 mRNAs. The translation initiation sites for the classic PLP and DM20 and for the srPLP and srDM20 proteins are indicated in the gene structure within exons 1 and 1.1, respectively. To the right of the mRNAs arediagrams of the protein products corresponding to each of the mRNAs and the number of amino acids (aa) in the protein. The 12 amino acid N-terminal sequences encoded by exon 1.1 are indicated on thediagrams for srPLP and srDM20 (wavylines). The sizes and locations of the exons are not drawn to scale. b, The sequence of the new 109 bp exon 1.1, underlined in capitalletters, given along with the complete intronic sequence between exons 1 and 1.1 as well as some intronic sequence between exons 1.1 and 2 (in lowercase). The translation start site (filledarrowhead) for srPLP and srDM20 proteins and the new N-terminal amino acid sequence contributed by exon 1.1 are shown above the sequence. The translation initiation site used in the classic PLP and DM20 mRNAs (openarrowhead) at the 3′ end of exon 1 is shown in italics.
Identification of new PLP and DM20 protein variants
The largest open reading frame in the new PLP or DM20 clones predicted a translated protein sequence that was identical to PLP or DM20 except for an additional 12 amino acids at the N terminals of the proteins, which is reminiscent of a targeting sequence. In the “classic” PLP and DM20 mRNAs, the translation initiation site is at the 3′ end of exon 1. In the new clones, splicing of exon 1 into exon 1.1 produces a termination signal in-frame with the classic translation start site not far downstream so that this initiation site is unlikely to be used. However, there is a second translation initiation site within exon 1.1 that defines the largest open reading frame in the new mRNAs. The predicted protein from this initiation site would be translated, in-frame, into exon 2 to produce DM20 or PLP proteins with a 12 amino acid peptide linked to their N terminals (shown in Fig. 1). We termed these products srPLP or srDM20 (for “somal-restricted” PLP or DM20 in reference to their subcellular localization to be described). To determine whether this second initiation site was actually used, we translated the srDM20 cDNA in an in vitrotranscription–translation system. Figure2a shows selected lanes of a gel in which the translation of the srDM20 cDNA is compared with the products synthesized by either DM20 or PLP cDNA. The major srDM20 product of the translation migrates between the PLP and DM20 controls (shown by hatched and open arrowheads, respectively, in Fig. 2) with the expected mobility predicted by the open reading frame. The higher molecular weight bands observed in the analysis of the total products are probably aggregates of PLP, DM20, and srDM20, a phenomenon observed by many investigators (Chan and Lees, 1974; Sakura, 1981; Bizzozero et al., 1982; Trifilieff et al., 1986).
Fig. 2.
Invitrotranscription–translation of srDM20 and Northern blot of srPLP and srDM20 mRNAs. a, Plasmids containing the coding regions of srDM20 and the classic DM20 and PLP isoforms were translated in anin vitro transcription–translation system and analyzed by SDS-PAGE. Autoradiograms of the total products of synthesis indicated that the principal radiolabeled bands corresponded in size to the new srDM20 (filledarrowhead,middlelane) and the classic DM20 and PLP (open and hatchedarrowheads, left and rightlanes, respectively). The individuallanes were part of the same transcription–translation experiment, analyzed in the same gel. Molecular weight standards were also included in the gel to assign the size for each band but are not shown. b, Total RNA from P18 mouse brain (Br) and from enriched OL cultures (OL) was analyzed by Northern blot with a 32P-riboprobe specific to exon 1.1. A 3.4 kb band was identified in the brain sample. In enriched OL RNA, the 3.4 kb band appeared as a doublet with a minor band at ∼3.5 kb. In both samples, a 12 kb band, presumably corresponding to heterogeneous nuclear RNA, was observed. Migration of molecular weight standards (data not shown) was used to assign the size for each band.
srPLP and srDM20 mRNAs are expressed in several tissues and cell types
Northern blot analysis of brain RNA indicated that the size of the srPLP and srDM20 mRNAs was ∼3.4 kb (Fig. 2b), slightly larger than the classic PLP and DM20 mRNAs (Sorg et al., 1987). Analysis of mouse OL RNA revealed two bands between 3.4 and 3.5 kb. Figure 3a shows the expression of the srPLP and srDM20 mRNAs by RPA using a probe specific for exon 1.1. In the developing mouse brain the pattern of expression of srPLP and srDM20 mRNAs was quite similar to that of classic PLP and DM20 mRNA expression, peaking around the age of maximal myelination (P17) and declining somewhat in the adult. The srPLP and srDM20 mRNAs were expressed in a mouse T-cell line (LBRM) and in mixed glial cultures. The srPLP and srDM20 mRNAs also were found to be expressed in primary cultures of cerebellar granule cells. Figure 3bshows an RPA assay designed to illustrate the difference in the levels of classic and srPLP and srDM20 mRNAs using a riboprobe that protects exon 1.1 and the first 34 nucleotides of exon 2. The 135 nt fragment is derived from the srPLP and srDM20 mRNAs, and the 34 nt fragment is derived from the classic PLP and DM20.
Because RPA analyses do not indicate which of the srPLP and srDM20 mRNA isoforms are expressed, we conducted RT-PCR experiments to determine this (Fig. 3c). In the brain, the developmental pattern of expression of the sr isoforms was identical to that of the classic isoforms. Expression of srDM20 mRNA preceded that of srPLP mRNA. The srDM20 mRNA was expressed in the embryonic brain, and the srPLP mRNA was not detected until approximately P5 and then increased to a maximum around the age of peak myelination. The developmental expression in mixed glial cultures followed that of the in vivo results in that srDM20 expression occurred before srPLP expression. In the jimpy brain, both isoforms were detected, but they were smaller than the wild-type isoforms because the jimpy isoforms are missing exon 5 because of a mutation in the acceptor splice site of exon 5. The srDM20 isoform was expressed in P3 thymus and in the LBRM T-cell line, and both srPLP and srDM20 were expressed in cultured cerebellar granule cells.
Expression of srPLP and srDM20 proteins in OLs and neurons
Rabbit polyclonal antisera against the peptide unique to the srPLP and srDM20 proteins were prepared. In Western blots these antisera recognized bands at ∼27 and ∼21 kDa, corresponding in size to srPLP and srDM20 (Fig. 4a) along with other lower and higher molecular weight bands reminiscent of blots obtained by others with PLP antisera (Fig. 4b). These blots were stripped and reprobed with AA3, a rat monoclonal antibody to the C-terminal region of PLP. The srPLP and srDM20 migrated slightly slower than the classic PLP (∼25 kDa) and DM20 (∼20 kDa) because they are of slightly higher molecular weight because of the extra N-terminal sequence. In PLP and DM20 Western blots of brain, the detection of higher and lower molecular weight products was somewhat variable. The higher molecular weight bands are generally believed to be aggregates of PLP and/or DM20 (Chan and Lees, 1974; Sakura, 1981; Bizzozero et al., 1982; Trifilieff et al., 1986), and the lower molecular weight bands have not yet been identified, although partial peptide sequences have been obtained for the bovine proteins (Lepage et al., 1986). Our results would suggest that some of these bands contain the peptide encoded by exon 1.1.
Fig. 4.
Detection of srPLP and srDM20 proteins in proteolipid extracts by Western blot. Extracts of total proteolipid proteins were prepared from mouse P14 brains, separated by SDS-PAGE, and analyzed on Western blots with a polyclonal antiserum specific for the N terminal of srPLP and srDM20. The anti-srPLP and -srDM20 recognized several bands, the most intense of which migrated with an apparent molecular weight of 27 kDa (lanea). Other lower molecular weight bands were evident at 21, 20, and 11 kDa. The blot was stripped and reprobed with AA3, a monoclonal antibody that recognizes the C terminal of PLP and DM20 (lane b). Very heavy bands of PLP and DM20 were evident, and these migrated with apparent molecular weights of 25 and 20 kDa. Because of the differences in the levels of srPLP and srDM20 and of classic PLP and DM20, laneb is an adjacent lane on the reprobed blot, loaded with less protein. Molecular weight standards were included to assign the relative sizes (data not shown). On the basis of the relative sizes and mobilities of the bands in both preparations, the srPLP and srDM20 were assigned to the 27 and 21 kDa bands, respectively.
Expression in OLs
Expression of srPLP and srDM20 proteins was readily detected in OLs in vitro. In double-labeling experiments we observed coexpression of srPLP and srDM20 in cells that were A2B5+ (Fig.5, top left panel), O4+ (Fig. 5, top middle panel), O1+ (Fig. 5, top right panel, bottom panels), or PLP+ (Fig. 5, middle panels), suggesting expression of these products over a long period of OL development. In all cases we saw that labeling of srPLP and srDM20 was restricted to the cell bodies and to some processes, but we never observed labeling of the sheets elaborated by OLs in culture (e.g., see Fig. 5, right panels). These results indicate that the srPLP and srDM20 proteins are not localized primarily in the myelin sheath in the OL and suggest that the 12 amino acid leader in srPLP and srDM20 might serve as a targeting sequence. Interestingly, purified OLs, after shaking off and separation from microglia and astrocytes, exhibit very intense staining with the srPLP and srDM20 antibody, significantly higher than that observed in OLs in the mixed cultures.
Fig. 5.
Expression of srPLP and srDM20 in OLs is localized in the cell bodies but not in the myelin-like membranes. Top Panels, Cells within the OL lineage were found to coexpress srPLP and srDM20 (redfluorescence) with markers of different oligodendroglial developmental stages (greenfluorescence) including A2B5 (precursors), O4 (immature/mature), and O1 (mature). The combined immunofluorescence for srPLP and srDM20 with these markers is shown.Middle,Bottom Panels, Single confocal sections of mature, enriched OLs double stained for srPLP and srDM20 (redfluorescence) and either O1, an antibody that stains primarily galactocerebrosides, or AA3, an antibody that stains classic PLP and DM20 (greenfluorescence), are shown. In all cases examined the srPLP and srDM20 staining was localized primarily in the cell soma and major processes but not in the membranes elaborated by the OLs.
In situ hybridization with an srPLP- and srDM20-specific probe at a gross level indicates that expression is primarily in white matter–enriched regions. Figure 6,a and b, shows a sagittal section of P14 brain hybridized to 33P-antisense and sense exon 1.1–specific cRNA, respectively. There was strong labeling in the white matter regions of the cerebellum, the corpus callosum, and the internal capsule. Higher magnification of these regions shows punctate cellular staining, presumably within OLs of the corpus callosum (Fig. 6c) and the deep cerebellar white matter (Fig.6e). Analogous regions immunostained for srPLP and srDM20 clearly show staining of interfascicular OLs within the corpus callosum (Fig. 6d, black arrows) and some satellite OLs in cerebellar white matter (Fig. 6f, red arrows). In marked contrast with PLP and DM20 immunostaining of the myelin sheaths, the srPLP and srDM20 staining is localized to the cell bodies, again consistent with thein vitro data for a different subcellular localization of these proteolipids.
Fig. 6.

Expression of srPLP and srDM20 is primarily in white matter regions of the mouse brain but is present in some neuronal populations. a,In situ hybridization of a sagittal section of a P14 mouse brain with a33P-riboprobe specific for exon 1.1 showing expression of the srPLP and srDM20 mRNAs. b,In situ hybridization of a section adjacent toa using a sense control. c, Higher magnification of a different section (counterstained with cresyl violet) showing portions of the corpus callosum and hippocampus and illustrating the punctate labeling throughout the corpus callosum and in the hippocampus below it. d, Immunohistochemical detection of srPLP and srDM20 in the corpus callosum and the hippocampus. Arrows in c andd point to cells of the size, location, and morphology of OLs that contain srPLP and srDM20 mRNAs and proteins, respectively. Unlike classic PLP and DM20 staining in the corpus callosum, srPLP and srDM20 labeling is confined to the cell somas. Also labeled are the CA2 and CA3 regions of the hippocampus.e, Higher magnification of a cerebellar region showing punctate labeling of srPLP and srDM20 mRNAs in cells throughout the white matter (redarrows) and the internal granular cell layer (whitearrows) as well as the Purkinje cells (yellowarrows). f,Immunohistochemical detection of srPLP and srDM20 in a region of the cerebellum similar to that in e. Cell bodies of OLs are indicated with redarrows, whereas Purkinje cells are indicated with blackarrows. Granule neurons located within theIGL were readily immunostained with the antibody.g, Higher magnification of the immunohistochemical detection of srPLP and srDM20 in the internal granule cell layer (openarrows) and Purkinje cells (filledarrows) from the mouse cerebellum. h, Immunohistochemical detection of srPLP and srDM20 proteins in medium-sized neurons within the anterior olfactory nucleus of the P14 mouse brain. i,Immunohistochemical control for background staining in the cerebellum with preimmune serum to the srPLP and srDM20 antibody.j–l, Primary granular neuron cultures doubly stained for srPLP and srDM20 (j;redfluorescence) and for tubulin III (k;green). l is an overlay showing that srPLP and srDM20 proteins remain associated with the neuronal cell bodies. CC, Corpus callosum; DCWM, deep cerebellar white matter; Hp, hippocampus;IC, internal capsule; IGL, internal granule cell layer.
Expression in neurons
The in situ hybridization data suggested that the srPLP and srDM20 proteins might be expressed in certain neuronal populations within the cerebellum, hippocampus, and olfactory system. Light, but distinct, labeling of neuronal regions within the hippocampus was evident (Fig. 6a,c). In the hippocampus there was significant labeling of pyramidal neurons in the CA1 (data not shown), CA2, and CA3 (Fig. 6d) regions as well as in hippocampal granule cells in the dentate gyrus (data not shown). Labeling of srPLP and srDM20 mRNA was also evident in neurons within the cerebellar granule cell layer (Fig. 6e,white arrows) as well as in Purkinje cells (Fig.6e, yellow arrows). Immunohistochemical studies of the same regions showed labeling of the cerebellar granule cell layer (Fig. 6f) and Purkinje cells (Fig. 6f, black arrows; see higher magnification in Fig. 6g).
To confirm these in vivo results, we examined cerebellar granule cell cultures for the expression of srPLP and srDM20. As shown in Figure 6j–l, most of the cells in these preparations stained robustly, along with some that were more lightly stained. As in OLs, the immunostaining of srPLP and srDM20 was confined to cerebellar granule cell bodies.
Olfactory neurons were another class of neurons that expressed srPLP and srDM20. Immunohistochemical analyses of the myelinating mouse brain with the srPLP and srDM20 antibody showed clear labeling of medium-sized neurons in the olfactory regions, a field of which is illustrated in Figure 6h.
DISCUSSION
In this paper we report the identification of a new exon (exon 1.1) of the PLP/DM20 gene and two new products of the gene that we call srDM20 and srPLP. Exon 1.1 was mapped between exons 1 and 2, thereby increasing the number of exons identified in the gene to eight. This exon is flanked by appropriate splice consensus sequences and, when spliced into PLP and DM20 mRNAs, increases the size of the messages by 109 bp.
The splicing of exon 1.1 in the mRNA also creates a new translation initiation site 72 bp downstream of the beginning of exon 1.1. The existence of the upstream translation start site followed relatively closely by an in-frame termination signal is frequently associated with poorly translated mRNAs and might reduce the translational efficiency of the new srPLP and srDM20 mRNA variants. In spite of this, the mRNAs were translated readily in vitro, and the presence of these isoforms was evident both on Western blots and by immunocytochemistry.
The protein products of srPLP and srDM20 mRNAs appear to be identical to their respective classic counterparts except for an additional 12 amino acid sequence at their N terminals. The distribution of the srPLP and srDM20 proteins in the OLs in vivo and in vitro indicates that they are targeted primarily to sites within the cell bodies and not to the myelin sheath. Analysis of the 12 amino acid sequence found in srPLP and srDM20 in the data base revealed a 55% homology to targeting sequences of proteins localized in the endoplasmic reticulum (ER). This suggests that the variant isoforms might be associated with ER/Golgi membranes rather than with components of the myelin membrane. The antiserum generated against the unique peptide region in the sr proteolipids may prove to be useful for immunohistochemical studies because it stains OL cell bodies and not the myelin sheaths in tissue sections.
The presence of the additional N-terminal sequence in srPLP and srDM20 does not greatly alter the accepted theoretical topography from that of the classic PLP and DM20 proteins (Weimbs and Stoffel, 1992) because hydrophilicity plots show no significant differences. The N terminals of the new isoforms remain hydrophilic and would be found on the cytoplasmic side of the tetraspan protein in the membrane without altering the hydrophobic domains of the protein.
Finding the expression of the srPLP and srDM20 variants in OLs and in the thymus is consistent with the known expression of PLP and DM20 in these tissues. On the other hand, finding immunoreactivity associated with neurons was surprising. The N-terminal sequence in the sr proteolipids has no homology at all to the M6 proteins related to DM20 described by Yan et al. (1993, 1996), and the antibody should not cross-react with these proteins. Until now, there have been no reports of PLP immunoreactivity in neurons. It is possible that the 50- to 100-fold higher abundance of PLP and DM20 proteins in white matter overshadows the signal from brain regions in which the sr proteolipids are expressed, which we found to be the case with detection of the golli products of the MBP gene with MBP antibodies. It is also possible that conformational changes in the srPLP and srDM20 mask detection with PLP antibodies as is the case with the 010 antibody that is sensitive to PLP and DM20 conformation (Jung et al., 1996). We also have found that the reaction of golli proteins with MBP antibodies is substantially less sensitive than that with the golli antibodies (Landry et al., 1996), possibly because of conformational differences.
Our finding that these new products of the PLP/DM20 gene are expressed in neurons is in keeping with a number of recent studies reporting neuronal abnormalities in PLP–knock-out or –overexpressing mice. For example, recent studies have shown that in the knock-out mice at 6–8 weeks axonal swellings of small diameter axons with subsequent fiber degeneration were detected throughout the white and gray matter (Griffiths et al., 1998b). At later ages (∼1 year), larger diameter axons became affected, accompanied by fiber degeneration. This phenomenon appeared to be caused by the PLP/DM20 gene ablation because similar swellings were absent in the shiverer mutant with a disrupted MBP gene. Similarly, in older animals of the PLP-overexpressing lines generated by Readhead et al. (1994), late-onset neurodegeneration has been observed (Anderson et al., 1998). Very recently, Coetzee et al. (1999) have reported abnormalities of the cerebellar granule cell layer in UDPgalactose:ceramide galactosyltransferase–PLP double knock-out mice, a finding consistent with our observations of the expression of the srPLP and srDM20 in cerebellar granule cells. The extent to which expression of these new isoforms in neuronal populations might contribute to the abnormalities observed in these mice remains to be determined, but they are certainly attractive candidates for this.
The notion that products of the PLP/DM20 gene may be involved in processes other than myelination first came from cell biological studies on the jimpy mutant. In this mouse there are a number of developmental abnormalities that occur before active myelin synthesis (Knapp, 1996). For example, premature OL celldeath as well as increased OL proliferation has been reported in the jimpy mouse (Skoff, 1995). This leads to normal numbers of OLs in the mutant, but they are immature. Interestingly, many aspects of the dysmyelinating phenotype in jimpy are maintained in cells grown in primary culture (Skoff and Knapp, 1990). These results were difficult to reconcile with a single structural role for PLP and/or DM20 in myelin and suggested that the gene must encode products with biological roles beyond that of myelin structural proteins, because of the apparently unrelated effects of the mutation on the cell biology and differentiation of the OL. These roles have been attributed to DM20 because its expression seems to be more promiscuous than that of PLP. However, concrete evidence of an alternative function for DM20 has not yet been found and does not exclude the possibility of this role being assumed by other products of the PLP/DM20 gene.
The developmental expression and subcellular localization of srPLP and srDM20 suggest several possible functions for these proteins in OLs. The timing of expression of the sr proteolipids is consistent with a role before myelination in the OL precursor and immature OL. For example, in vitro we found colocalization of the srPLP and srDM20 with markers of cells at many stages of development from precursors to mature OLs, suggesting a long-term expression of srPLP and srDM20 in the OL lineage. It is possible that the sr proteolipids play a role in the survival or death of immature OLs. The localization of the sr proteolipids primarily within cell somas, perhaps as part of the ER/Golgi system, suggests that they could play some role in the trafficking of molecules, such as the classic PLP and DM20, within that network. Although the role of these proteins is, as yet, uncertain, they would also be mutated in the PLP mutants. If they have some general role in OLs and neurons, then that might account for some of the nonmyelin-related cell biological aberrations observed in the PLP mutants.
Footnotes
This research was supported by National Institutes of Health Grants NS23022 and NS33091 and the National Multiple Sclerosis Society Grant PP0556. We wish to thank Robert Woodhall for technical assistance and Drs. Marjorie Lees, Kazuhiro Ikenaka, and Steve Pfeiffer for providing us with the AA3 PLP monoclonal antibodies.
Correspondence should be addressed to Dr. Anthony T. Campagnoni, Neuropsychiatric Institute, Room 47-448, University of California, Los Angeles, Medical School, 760 Westwood Plaza, Los Angeles, CA 90024-1759.
E.R.B. and C.W.C. contributed equally to this work.
REFERENCES
- 1.Anderson TJ, Schneider A, Barrie JA, Klugmann M, McCulloch MC, Kirkham D, Kyriakides E, Nave KA, Griffiths IR. Late-onset neurodegeneration in mice with increased dosage of the proteolipid protein gene. J Comp Neurol. 1998;394:506–519. doi: 10.1002/(sici)1096-9861(19980518)394:4<506::aid-cne8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 2.Bizzozero OA, Besio-Moreno M, Pasquini JM, Soto EF, Gomez CJ. An electrophoretic analysis of proteolipids from different rat brain subcellular fractions. Biochim Biophys Acta. 1982;691:281–292. doi: 10.1016/0005-2736(82)90417-5. [DOI] [PubMed] [Google Scholar]
- 3.Bongarzone ER, Foster LM, Byravan S, Verity AN, Landry CF, Schonmann V, Amur-Umarjee S, Campagnoni AT. Conditionally immortalized neural cell lines: potential models for the study of neural cell function. Methods. 1996;10:489–500. doi: 10.1006/meth.1996.0126. [DOI] [PubMed] [Google Scholar]
- 4.Bongarzone ER, Howard SG, Schonmann V, Campagnoni AT. Identification of the dopamine D3 receptor in oligodendrocyte precursors: potential role in regulating differentiation and myelin formation. J Neurosci. 1998a;18:5344–5353. doi: 10.1523/JNEUROSCI.18-14-05344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bongarzone ER, Foster L, Byravan S, Casaccia-Bonnefil P, Schonmann V, Campagnoni AT. Two neuronal cell lines expressing the Golli-MBP gene display differences in their in vitro survival and in their interaction with glia. J Neurosci Res. 1998b;54:309–319. doi: 10.1002/(SICI)1097-4547(19981101)54:3<309::AID-JNR2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Campagnoni CW, Garbay B, Micevych P, Pribyl T, Kampf K, Handley VW, Campagnoni AT. DM20 mRNA splice product of the myelin proteolipid protein gene is expressed in the murine heart. J Neurosci Res. 1992;33:148–155. doi: 10.1002/jnr.490330119. [DOI] [PubMed] [Google Scholar]
- 7.Chan DS, Lees MB. Gel electrophoresis studies of bovine brain white matter proteolipid and myelin proteins. Biochemistry. 1974;13:2704–2712. doi: 10.1021/bi00710a008. [DOI] [PubMed] [Google Scholar]
- 8.Coetzee T, Suzuki K, Nave KA, Popko B. Myelination in mice lacking galactolipids and proteolipid protein. J Neurochem. 1999;72[Suppl]:S45. doi: 10.1006/mcne.1999.0768. [DOI] [PubMed] [Google Scholar]
- 9.Gao WO, Heintz N, Hatten ME. Cerebellar granule cell neurogenesis is regulated by cell-cell interactions in vitro. Neuron. 1991;6:705–715. doi: 10.1016/0896-6273(91)90168-y. [DOI] [PubMed] [Google Scholar]
- 10.Gardinier MV, Macklin WB. Myelin proteolipid protein gene expression in jimpy and jimpy(msd) mice. J Neurochem. 1988;51:360–369. doi: 10.1111/j.1471-4159.1988.tb01047.x. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths I, Klugmann M, Anderson T, Thomson C, Vouyiouklis D, Nave KA. Current concepts of PLP and its role in the nervous system. Microsc Res Tech. 1998a;41:344–358. doi: 10.1002/(SICI)1097-0029(19980601)41:5<344::AID-JEMT2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths I, Klugmann M, Anderson T, Yool D, Thomson C, Schwab MH, Schneider A, Zimmermann F, McCulloch M, Nadon N, Nave KA. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science. 1998b;280:1610–1613. doi: 10.1126/science.280.5369.1610. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths IR. Myelin mutants: model systems for the study of normal and abnormal myelination. Bioessays. 1996;18:789–797. doi: 10.1002/bies.950181005. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths IR, Montague P, Dickinson P. The proteolipid protein gene. Neuropathol Appl Neurobiol. 1995;21:85–96. doi: 10.1111/j.1365-2990.1995.tb01034.x. [DOI] [PubMed] [Google Scholar]
- 15.Hudson LD, Nadon NL. Amino acid substitutions in proteolipid protein that cause dysmyelination. In: Martenson RE, editor. Myelin: biology and chemistry. CRC; Boca Raton, FL: 1992. pp. 677–702. [Google Scholar]
- 16.Ikenaka K, Kagawa T, Mikoshiba K. Selective expression of DM-20, an alternatively spliced myelin proteolipid protein gene product, in developing nervous system and in nonglial cells. J Neurochem. 1992;58:2248–2253. doi: 10.1111/j.1471-4159.1992.tb10970.x. [DOI] [PubMed] [Google Scholar]
- 17.Jung M, Sommer I, Schachner M, Nave KA. Monoclonal antibody O10 defines a conformationally sensitive cell-surface epitope of proteolipid protein (PLP): evidence that PLP misfolding underlies dysmyelination in mutant mice. J Neurosci. 1996;16:7920–7929. doi: 10.1523/JNEUROSCI.16-24-07920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamholz J, Sessa M, Scherer S, Vogelbacker H, Mokuno K, Baron P, Wrabetz L, Shy M, Pleasure D. Structure and expression of proteolipid protein in the peripheral nervous system. J Neurosci Res. 1992;31:231–244. doi: 10.1002/jnr.490310204. [DOI] [PubMed] [Google Scholar]
- 19.Knapp PE. Proteolipid protein: is it more than just a structural component of myelin? Dev Neurosci. 1996;18:297–308. doi: 10.1159/000111420. [DOI] [PubMed] [Google Scholar]
- 20.Landry CF, Ellison JA, Pribyl TM, Campagnoni C, Kampf K, Campagnoni AT. Myelin basic protein gene expression in neurons: developmental and regional changes in protein targeting within neuronal nuclei, cell bodies, and processes. J Neurosci. 1996;16:2452–2462. doi: 10.1523/JNEUROSCI.16-08-02452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landry CF, Pribyl TM, Ellison JA, Givogri MI, Kampf K, Campagnoni CW, Campagnoni AT. Embryonic expression of the myelin basic protein gene: identification of a promoter region that targets transgene expression to pioneer neurons. J Neurosci. 1998;18:7315–7327. doi: 10.1523/JNEUROSCI.18-18-07315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lepage P, Helynck G, Chu JY, Luu B, Sorokine O, Trifilieff E, Van Dorsselaer A. Purification and characterization of minor brain proteolipids: use of fast atom bombardment-mass spectrometry for peptide sequencing. Biochimie. 1986;68:669–686. doi: 10.1016/s0300-9084(86)80161-4. [DOI] [PubMed] [Google Scholar]
- 23.Macklin WB, Campagnoni CW, Deininger PL, Gardinier MV. Structure and expression of the mouse myelin proteolipid protein gene. J Neurosci Res. 1987;18:383–394. doi: 10.1002/jnr.490180302. [DOI] [PubMed] [Google Scholar]
- 24.Macklin WB, Gardinier MV, Obeso Z. Structure and expression of the mouse myelin proteolipid protein gene. Ann NY Acad Sci. 1990;605:183–193. doi: 10.1111/j.1749-6632.1990.tb42392.x. [DOI] [PubMed] [Google Scholar]
- 25.Nave KA, Milner RJ. Proteolipid proteins: structure and genetic expression in normal and myelin-deficient mutant mice. Crit Rev Neurobiol. 1989;5:65–91. [PubMed] [Google Scholar]
- 26.Nave KA, Lai C, Bloom FE, Milner RJ. Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc Natl Acad Sci USA. 1987;84:5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pribyl TM, Campagnoni C, Kampf K, Handley VW, Campagnoni AT. The major myelin protein genes are expressed in the human thymus. J Neurosci Res. 1996a;45:812–819. doi: 10.1002/(SICI)1097-4547(19960915)45:6<812::AID-JNR18>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 28.Pribyl TM, Campagnoni CW, Kampf K, Kashima T, Handley VW, McMahon J, Campagnoni AT. Expression of the myelin proteolipid protein gene in the human fetal thymus. J Neuroimmunol. 1996b;67:125–130. doi: 10.1016/0165-5728(96)00058-6. [DOI] [PubMed] [Google Scholar]
- 29.Puckett C, Hudson L, Ono K, Friedrich V, Benecke J, Dubois-Dalcq M, Lazzarini RA. Myelin-specific proteolipid protein is expressed in myelinating Schwann cells but is not incorporated into myelin sheaths. J Neurosci Res. 1987;18:511–518. doi: 10.1002/jnr.490180402. [DOI] [PubMed] [Google Scholar]
- 30.Readhead C, Schneider A, Griffiths IR, Nave KA. Premature arrest of myelin formation in transgenic mice with increased proteolipid protein gene dosage. Neuron. 1994;12:583–595. doi: 10.1016/0896-6273(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 31.Sakura JD. Physical properties of bovine white matter proteolipid apoprotein-sodium dodecyl sulfate complexes. J Neurosci Res. 1981;6:689–700. doi: 10.1002/jnr.490060602. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Frisch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 33.Skoff RP. Programmed cell death in the dysmyelinating mutants. Brain Pathol. 1995;5:283–288. doi: 10.1111/j.1750-3639.1995.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 34.Skoff RP, Knapp PE. Expression of the Jimpy phenotype in relation to proteolipid protein appearance. Ann NY Acad Sci. 1990;605:122–134. doi: 10.1111/j.1749-6632.1990.tb42387.x. [DOI] [PubMed] [Google Scholar]
- 35.Sorg BA, Smith M, Campagnoni AT. Developmental expression of the myelin proteolipid protein and basic protein mRNAs in normal and dysmyelinating mutant mice. J Neurochem. 1987;49:1146–1154. doi: 10.1111/j.1471-4159.1987.tb10005.x. [DOI] [PubMed] [Google Scholar]
- 36.Timsit S, Martinez S, Allinquant B, Peyron F, Puelles L, Zalc B. Oligodendrocytes originate in a restricted zone of the embryonic ventral neural tube defined by DM-20 mRNA expression. J Neurosci. 1995;15:1012–1024. doi: 10.1523/JNEUROSCI.15-02-01012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timsit SG, Bally-Cuif L, Colman DR, Zalc B. DM-20 mRNA is expressed during the embryonic development of the nervous system of the mouse. J Neurochem. 1992;58:1172–1175. doi: 10.1111/j.1471-4159.1992.tb09378.x. [DOI] [PubMed] [Google Scholar]
- 38.Trifilieff E, Luu B, Nussbaum JL, Roussel G, Espinosa de los Monteros A, Sabatier JM, Van Rietschoten J. A specific immunological probe for the major myelin proteolipid. Confirmation of a deletion in DM-20. FEBS Lett. 1986;198:235–239. doi: 10.1016/0014-5793(86)80412-4. [DOI] [PubMed] [Google Scholar]
- 39.Weimbs T, Stoffel W. Proteolipid protein (PLP) of CNS myelin: positions of free, disulfide-bonded, and fatty acid thioester-linked cysteine residues and implications for the membrane topology of PLP. Biochemistry. 1992;31:12289–12296. doi: 10.1021/bi00164a002. [DOI] [PubMed] [Google Scholar]
- 40.Wight PA, Dobretsova A. The first intron of the myelin proteolipid protein gene confers cell type-specific expression by a transcriptional repression mechanism in non-expressing cell types. Gene. 1997;201:111–117. doi: 10.1016/s0378-1119(97)00435-6. [DOI] [PubMed] [Google Scholar]
- 41.Yan Y, Lagenaur C, Narayanan V. Molecular cloning of M6: identification of a PLP/DM20 gene family. Neuron. 1993;11:423–431. doi: 10.1016/0896-6273(93)90147-j. [DOI] [PubMed] [Google Scholar]
- 42.Yan Y, Lagenaur C, Narayanan V. Expression of members of the proteolipid protein gene family in the developing murine central nervous system. J Comp Neurol. 1996;370:465–478. doi: 10.1002/(SICI)1096-9861(19960708)370:4<465::AID-CNE4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]