Abstract
Objective
To explore the regulatory effects of circZNF609 on proliferative and migratory capacities of gastric cancer (GC) and its underlying mechanism.
Methods
Expression level of circZNF609, CDK6 and miR-483-3p in GC tissues and cells were detected qRT-PCR verification. CCK-8 and transwell assay were conducted the cell viability and migratory capacities of GC cells. Dual luciferase assay was enrolled to confirm the interaction among circZNF609, CDK6 and miR-483-3p. Western blot was used to detect the protein level of CDK6.
Results
Expression levels of circZNF609 were higher in GC patients by qRT-PCR.GC patients with higher expression of circZNF609 were expected to have a higher TNM stage and lower 5-year survival than those with lower expression. ROC curves showed a well diagnostic value of circZNF609 in GC. Treatment of RNase R in GC cells downregulated the expression of ZNF609, whereas circZNF609 expression did not change. Furthermore, cytoplasmic expression of circZNF609 was higher than those of nuclear expression. Besides, biological experiments indicated that overexpression of circZNF609 promoted the proliferative and migratory capacities of GC cells. To demonstrate the underlying mechanism of circZNF609, we found that circZNF609 bound to miR-483-3p, which presented a lower expression in GC tissues than that of paracancerous tissues. Both circZNF609 and miR-483-3p could bind to Ago2, suggesting that circZNF609 may act as a sponge of miR-483-3p. In addition, the effect of overexpressed circZNF609 on cellular behaviors of GC cells were partly reversed by overexpression of miR-483-3p. Bioinformatics suggested that CDK6 has a potential binding site with miR-483-3p. The expression of CDK6 markedly increased in GC tissues and cells, which was negatively correlated with miR-483-3p expression. Dual-luciferase reporter gene results indicated that miR-483-3p could bind to the 3’-UTR of CDK6. Moreover, miR-483-3p downregulated CDK6 at both mRNA and protein levels. Overexpression of miR-483-3p inhibited proliferative and migratory capacities of GC cells, which were reversed by CDK6 overexpression.
Conclusion
In summary, the expression of circZNF609 is upregulated in GC. CircZNF609 can be used as the sponge of miR-483-3p to regulate the expression level of CDK6, thus participating in the progression of GC by regulating the proliferative and migratory capacities of GC cells.
Keywords: circZNF609, MiR-483-3p, CDK6, proliferation, migration, gastric cancer
Introduction
Gastric cancer (GC) is one of the most common malignant tumors of the digestive system in the world. It is the second leading cause of death from cancer worldwide. GC manifests as low survival, low cure rate, high recurrent rate and poor prognosis, seriously threatening human health.1 Pathological examination is currently the major diagnostic approach for GC. Unfortunately, a large number of GC patients have already progressed in the advanced stage at the first time of diagnosis since efficient hallmark in early diagnosis is lacking.2 Effective interventions for GC are urgently required. The occurrence of GC usually involves the interaction of multiple factors, showing genetic complexity and heterogeneity.3 The molecular mechanisms involved in the development and progression of GC are still not comprehensively elucidated. Therefore, it is particularly significant to find molecular biomarkers for early diagnosis, prognosis and treatment of GC.
In recent years, a large number of studies have shown that noncoding RNA (ncRNA) is able to regulate gene expressions in tumor progression.4 miRNAs regulate crucial biological processes, such as cell division, differentiation and apoptosis. In particular, some miRNAs are closely related to the diagnosis and prognosis of GC, serving as oncogenes or tumor-suppressor genes.5 Circular RNA (circRNA) is recently discovered ncRNA with specific functions. It is abundantly present in the cytoplasm of eukaryotic cells and highly conserved. Besides, circRNA is differentially expressed in human tissues.6 circRNA contains a covalently closed circular structure. Serum level of circRNA is expected to become a hallmark for diagnosing, monitoring and evaluating GC, showing a clinical significance.7,8 For example, the expression level of hsa_circ_0003159 was found to be negatively correlated with the TNM stage and distant metastasis of GC patients.9
circRNA exerts multiple regulatory functions by binding to RNA-binding protein (RBP) or other RNAs through base pairing.10 In addition, circRNA can competitively bind to intracellular miRNA as a miRNA sponge and block the inhibitory effects of miRNA on its target genes, thus participating in biological processes such as cell proliferation, apoptosis and senescence.11 It is found that hsa_circ_0007534 circRNA is highly expressed in breast cancer. Downregulation of hsa_circ_0007534 can inhibit proliferative and invasive capacities of breast cancer cells by targeting miR-593/MUC19 axis.12 circFBLIM1 exerts regulatory functions in hepatocellular carcinoma by competitively binding to miR-346 as a competing endogenous RNA.13 circZNF609 shows an essential role in promoting myoblast proliferation.14 miR-483-3p inhibits the progression of breast cancer by inhibiting circCCNE1 expression.15 However, the roles of circZNF609 and miR-483-3p in GC have not been reported yet.
Methods and materials
Clinical samples
80 paired GC tissues and paracancerous tissues were collected in Danyang People’s Hospital from 2003 to 2016. The basic information of the patients was included in Supplementary table 1. Tissues were preserved in liquid nitrogen. Enrolled patients were pathologically diagnosed as GC. Informed consent was signed before the study. The Ethics Committee of Danyang People’s Hospital approved this study. All population-related studies were conducted in strict compliance with the Ethics Committee of Danyang People’s Hospital, and in accordance with the 1964 Helsinki declaration and its later amendments.
Cell culture and transfection
Gastric mucosal cell line (GES-1) and GC cell lines (SGC-7901, HGC-27, AGS, MKN-45 and BGC-823) were obtained from ATCC (American Type Culture Collection). Cells were cultured in DMEM containing 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin (Hyclone, South Logan, UT, USA). Cells were incubated in a 5% CO2 incubator at 37°C. Cell passage was performed using trypsin until 80%–90% of confluence.
Cells were transfected with corresponding plasmids following the instructions of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Culture medium was replaced 4 hrs later. Transfection plasmids were constructed by GenePharma, Shanghai, China.
Chromatin fractionation
Cells were fully lysed in cell fractionation buffer, incubated on ice for 20 mins and centrifuged at 3000 r/min for 15 mins. The supernatant contained cytoplasmic proteins. The precipitant was washed with RLA for three times and lysed with RIPA for incubation on ice for a total of 20 mins. The mixture underwent vortex oscillation every 5 mins for 30 s. Finally, the mixture was centrifuged at 12,500 rpm/min for 15 mins, and the supernatant contained nuclear proteins.
QRT-PCR
Total RNA in treated cells or tissues was extracted using TRIzol method for reverse transcription according to the instructions of PrimeScript RT reagent Kit (Takara, Tokyo, Japan). RNA concentration was detected using a spectrometer. QRT-PCR was then performed based on the instructions of SYBR Premix Ex Taq TM (Takara). The relative gene expression was calculated using 2−△Ct method. Primers used in the study were as follows: miR-483-3p, F: 5ʹ- CTCTCGTAACTGGCTAGTCAAGAGAGTCATTTGACTCGCA-3ʹ, R: 5ʹ- CTGTACTCATGAGGTGGTTGACGCAGGAGG-3ʹ; CDK6, F: 5ʹ- ACTTGCACATCTCTGGTAGC-3ʹ, R: 5ʹ-GCTTGACTGATTCGTACTGC-3ʹ; GAPDH, F: 5ʹ-CACCCACTCCTCCACCTTTG-3ʹ, R: 5ʹ- GCTCATTCAACGGATAAGTC-3ʹ; CircZNF609, F: 5ʹ- CATGCTGATTACGCTTTGACTC-3ʹ, R: 5ʹ-GTTTCGTTCATCTGCTACTT-3ʹ.
Western blot
Cells were lysed with RIPA lysis buffer in the presence of a protease inhibitor (Sigma-Aldrich, St. Louis, MO, USA) to harvest total cellular protein. The protein concentration of each cell lysate was quantified using the BCA (bicinchoninic acid) protein assay kit (Pierce, Rockford, IL, USA). An equal amount of protein sample was loaded onto a 10% SDS-PAGE gel and then transferred to a PVDF (polyvinylidene fluoride) membrane after separation. After blockage with skim milk, membranes were incubated with primary and secondary antibody (Cell Signaling Technology, Danvers, MA, USA). Finally, an image of the protein band was captured by the Tanon detection system using ECL reagent (Thermo, Waltham, MA, USA).
CCK-8 assay
Transfected cells were seeded into 96-well plates at a density of 3×105/μL. 10 μL of CCK-8 solution (cell counting kit-8, Dojindo, Kumamoto, Japan) was added in each well after cell culture for 0, 24 hrs, 48 hrs, 72 hrs and 96 hrs, respectively. The absorbance at 450 nm of each sample was measured by a microplate reader (Bio-Rad, Hercules, CA, USA).
Transwell assay
After 24 hrs of transfection, cells were digested and resuspended in serum-free medium. Cell density was adjusted to 2.0×108/mL. Transwell chambers precoated with Matrigel were placed in 24-well plates. 100 μL of cell suspension and 500 μL of medium containing 10% FBS were added in the upper and lower chambers, respectively. After cell culture for 48 hrs, cells were fixed with 4% paraformaldehyde for 15 mins and stained with crystal violet for 10 min. Inner cells were carefully cleaned. Penetrating cells were captured in 5 randomly selected fields of each sample.
Dual-luciferase reporter gene assay
StarBase v2.0 (http://starbase.sysu.edu.cn), Pctar and miRanda were used for predicting the target gene of miR-483-3p. GC cells were first seeded in the 96-well plate with 2×103 per well. Co-transfection of miR-483-3p mimics and wild-type or mutant type of CDK6 or circZNF609 was performed until 70% of cell confluence. Each group set 5 replicates. Luciferase activity was determined 48 hrs later. 100 μL of LARII and 20 μL of PLB were added in the fluorescent photoluminescence tube for determining firefly fluorescence signal. Subsequently, 100 μL of Stop&Glo was added to terminate the reaction and Renilla fluorescence signal was finally detected. The original data of dual-luciferase reporter were included in Supplementary table 2.
RNA-binding protein immunoprecipitation (RIP)
RIP was performed using the Magna RIP RNA Binding kit (Merck Millipore, USA). Cells were washed and cross-linked with 0.01% formaldehyde for 15 mins. After centrifugation and cell lysis, cell extraction was incubated with RIP buffer containing protein A/G magnetic beads coated with anti-Ago2 or negative control anti-IgG antibody. After overnight incubation at 4°C, cells were incubated with Protein A Agarose for 1 hr at 4°C, followed by the isolation of RNA. Expression levels of circZNF609 and miR-483-3p were detected by qRT-PCR, whereas Ago2 expression was determined by Western blot.
Statistical analysis
SPSS 22.0 statistical software was used for data analysis. Data were expressed as mean ± SD(x ® ± s). Differences between the two groups were compared using the Student's t-test. Pearson correlation coefficient analysis was introduced for evaluating relationship in target genes. Analysis of survival data was performed using Kaplan–Meier method. Receiver operating characteristic curve) curves were introduced for determining diagnostic sensitivity and specificity of miRNAs. P<0.05 considered the difference was statistically significant.
Results
circZNF609 expression was pronounced in GC
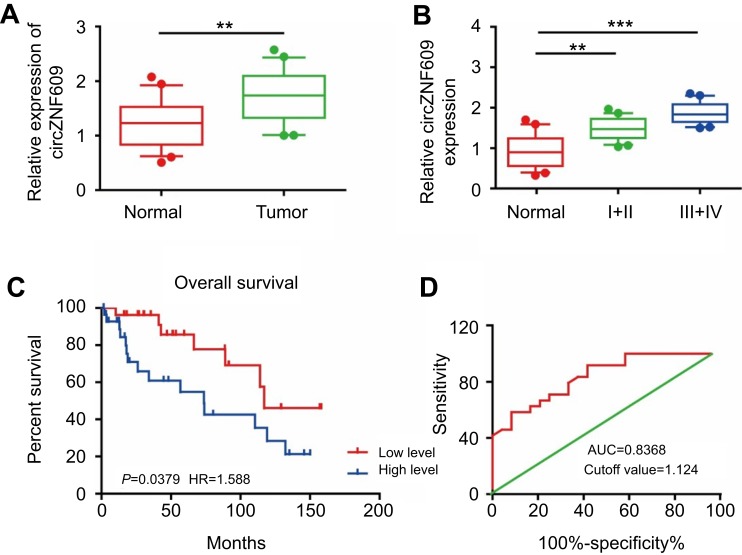
Previous studies have demonstrated the role of circZNF609 in other diseases,16,17 so we first detected circZNF609 expression in 80 paired GC and paracancerous tissues by qPCR. As the data showed, circZNF609 was highly expressed in GC tissues compared with that of paracancerous tissues (Figure 1A). We observed that circZNF609 expression increased with the aggravation of tumor stage in GC patients, suggesting the potential effect of circZNF609 on GC development (Figure 1B). Furthermore, the 5-year survival of GC patients showing higher circZNF609 expression was expected to be lower than those with lower circZNF609 expression (P=0.0379, HR=1.588, Figure 1C). ROC analysis indicated a certain diagnostic value of circZNF609 in GC (AUC=0.8368, cutoff value=1.124, Figure 1D).
Figure 1.
circZNF609 was highly expressed in GC and promoted the proliferative and migratory capacities of GC cells. (A) circZNF609 was highly expressed in GC tissues than that of paracancerous tissues (P<0.01). (B) circZNF609 expression in GC tissues with different tumor stages (P<0.01). (C) The 5-year survival of GC patients with high or low expression of miR-483-3p (P=0.0379, HR=1.588). (D) Survival curves of miR-483-3p expression in GC patients (AUC=0.8368, cutoff value=1.124). ** P<0.01, *** P<0.001.
Abbreviations: circZNF609, circular RNA ZNF609; GC, gastric cancer.
CircZNF609 promoted proliferative and migratory rates of GC cells
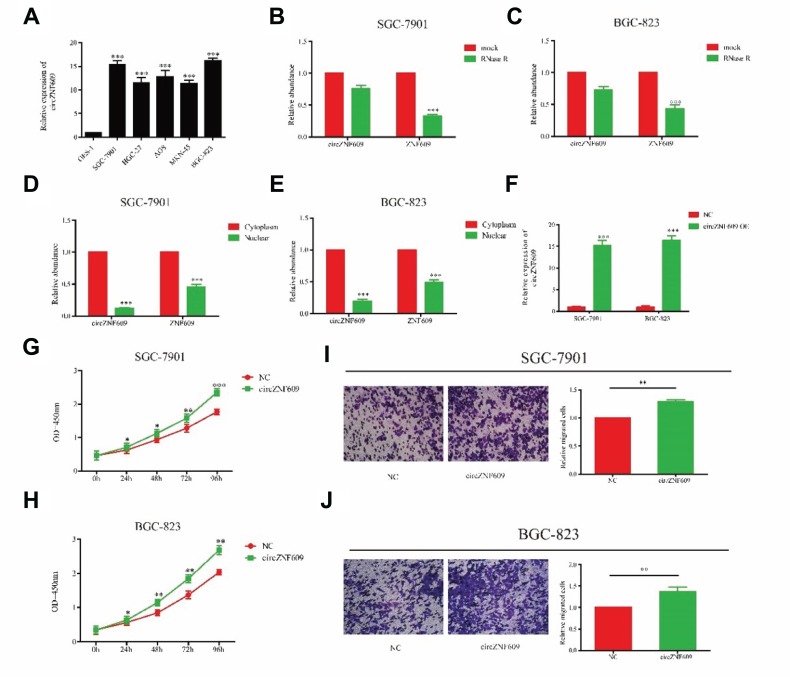
We speculated that circZNF609 may stimulate GC cell growth due to its high expression in GC tissues. First of all, circZNF609 expression was verified to be highly expressed in GC cell lines than that of gastric epithelial cell line GES-1 (Figure 2A). Particularly, SGC-7901 and BGC-823 cells showed a relatively high expression of circZNF609, which were selected for the subsequent experiments. It is found that circZNF609 expression was not altered after RNase R induction, whereas ZNF609 expression was markedly downregulated in SGC-7901 and BGC-823 cells, confirming the circRNA characteristics of circZNF609 (Figure 2B and C). Through chromatin fractionation assay, circZNF609/ZNF609 was found to be mainly distributed in the cytoplasm, suggesting its biological function at post-transcriptional level (Figure 2D and E). To verify the influence of circZNF609 on GC cell growth, we first verified the transfection efficacy of circZNF609 overexpression plasmid in SGC-7901 and BGC-823 cells (Figure 2F). The data showed promoted proliferative rate of GC cells by circZNF609 overexpression (Figure 2G and H). Similarly, circZNF609 overexpression markedly accelerated the migratory rate of GC cells (Figure 2I and J). The above data demonstrated the promotive effect of circZNF609 on proliferative and migratory rates of GC cells.
Figure 2.
Overexpression of circZNF609 promoted the migratory and proliferative ability in GC. (A) circZNF609 expression in GC cell lines (P<0.01). B and C. Expression levels of circZNF609 and ZNF609 in SGC-7901 (B) and BGC-823 (C) cells treated with RNase R. (D and E) Cytoplasmic and nuclear levels of circZNF609/ZNF609 in SGC-7901 (D) and BGC-823 (E) cells. (F) The effect of overexpression of circCNF609 in SGC-7901 and BGC-823. (G and H) CircZNF609 overexpression promoted proliferative rate of SGC-7901 (G) and BGC-823 (H) cells. (I and J) circZNF609 overexpression promoted migratory rate of SGC-7901 (I) and BGC-823 (J) cells. *P<0.05, **P<0.01, ***P<0.001.
Abbreviation: GC, gastric cancer.
CircZNF609 sponged miR-483-3p
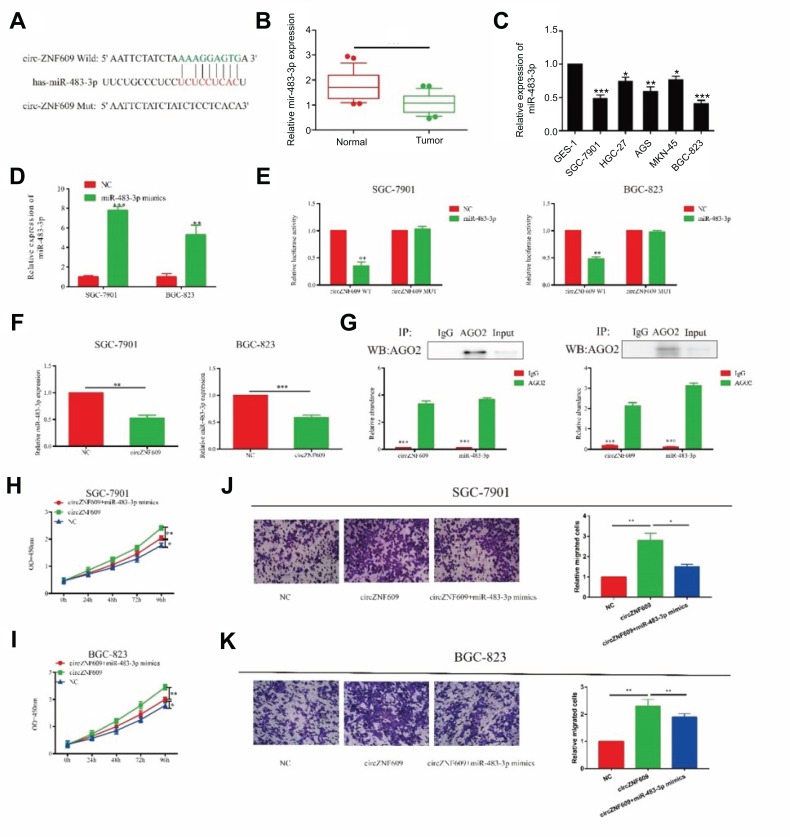
Previous studies have shown that circRNA exerts its function by sponging target miRNA.18 Bioinformatics analysis revealed a potential binding site for miR-483-3p and circZNF609 (Figure 3A). QRT-PCR results showed a low expression of miR-483-in GC (Figure 3B). Compared with GES-1 cells, miR-483-3p expression also decreased in GC cell lines (Figure 3C). To verify that circZNF609 could bind to miR-483-3p, we constructed a reporter plasmid. Transfection efficiency of miR-483-3p mimics in GC cells was verified as shown in Figure 3D. Dual-luciferase reporter gene results further verified the binding between miR-483-3p and circZNF609 (Figure 3E). Overexpression of circZNF609 downregulated miR-483-3p expression in GC cells, indicating that circZNF609 could negatively regulate miR-483-3p in GC (Figure 3F). Since circRNA exerted its function by a silencing complex of circRNA/miRNA binding to Ago2, we thereafter detected the expression level of circZNF609/miR-483-3p binding to Ago2 by RIP assay. Compared with IgG, circZNF609/miR-483-3p was primarily associated with Ago2 (Figure 3G). To demonstrate whether circZNF609 exerted its ceRNA function by sponging miR-483-3p, we co-overexpressed circZNF609 and miR-483-3p in GC cells. Promoted proliferative and migratory rates of GC cells induced by circZNF609 overexpression were partially reversed by miR-483-3p overexpression (Figure 3H–K). Based on the above results, we speculated that circZNF609 exerted its function in GC by sponging miR-483-3p.
Figure 3.
circZNF609 regulated proliferative and invasive capacities of GC cells through sponging miR-483-3p. (A) miR-483-3p was the potential target of circZNF609. (B and C) miR-483-3p was downregulated in GC (B) and GC cell lines (C). (D) The overexpression efficiency of miR-493-3p in SGC-7901 and BGC-823. (E and F) Dual-luciferase reporter gene assay showed the binding between circZNF609 and miR-483-3p. (F) CircZNF609 overexpression downregulated miR-483-3p overexpression in SGC-7901 and BGC-823 cells (P<0.01). (G) Abundances of circZNF609 and miR-483-3p in SGC-7901 and BGC-823 cells detected by AGO2 (RIP) assay. Ago2 was detected using IP-Western blot (up panel), and expression levels of circZNF609 and miR-483-3p were detected using qRT-PCR (down panel). (H and I) Promoted proliferative rate induced by circZNF609 overexpression was reversed after miR-483-3p overexpression in SGC-7901 (H) and BGC-823 (I) cells. (J and K) Accelerated migratory rate induced by circZNF609 overexpression was reversed after miR-483-3p overexpression in SGC-7901 (J) and BGC-823 (K) cells. *P<0.05, **P<0.01, ***P<0.001.
CDK6 was the target gene of miR-483-3p
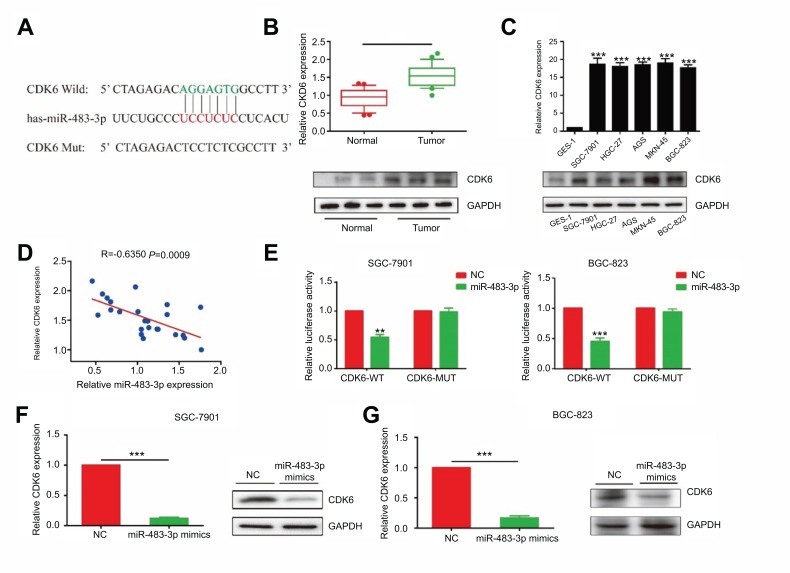
Biological function of miRNA was achieved by targeting on some certain downstream genes. We found a potential binding site between miR-483-3p and CDK6 through online prediction and analysis (Figure 4A). Besides, CDK6, as a cell cycle regulator, participates in a lot of biological processes in cancer development.19 CDK6 expression was found to be highly expressed in GC tissues and cell lines (Figure 4B and C). Pearson correlation coefficient revealed a negative correlation between miR-483-3p and CDK6 (R=−0.635, P=0.0009, Figure 4D). Furthermore, dual-luciferase reporter gene assay confirmed the binding condition between miR-483-3p and CDK6 (Figure 3E). By overexpression of miR-483-3p in GC cells, both mRNA and protein levels of CDK6 were reduced (Figure 4F and G). The above all demonstrated CDK6 was the target gene of miR-483-3p.
Figure 4.
miR-483-3p-regulated proliferative and invasive capacities of GC cells through targeting CDK6. (A) CDK6 is the target of miR-483-3p. (B) CDK6 was highly expressed in GC tissues than that of paracancerous tissues (P<0.01). (C) CDK6 expression in GC cell lines (P<0.01). (E) Pearson correlation analysis showed a negative correlation between expressions of miR-483-3p and CDK6 (R=−0.635, p=0.0009). (E) Dual-luciferase reporter gene assay showed the binding between CDK6 and miR-483-3p. (F and G) The mRNA and protein levels of CDK6 in SGC-7901 (F) and BGC-823 (G) cells transfected with miR-483-3p mimics. **P<0.01; ***P<0.001.
MiR-483-3p exerted its function by targeting CDK6
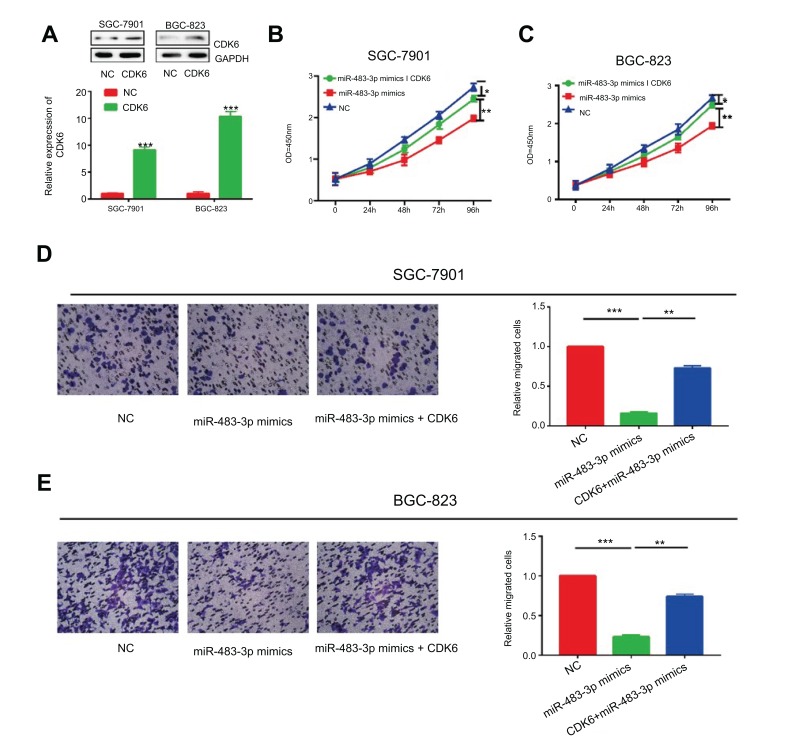
Since CDK6 the target gene of miR-483-3p, we hypothesized that miR-483-3p exerted its biological function through CDK6. Firstly, both mRNA and protein levels of CDK6 upregulated after transfection of CDK6 overexpression plasmid in GC cells (Figure 5A). Co-overexpression of miR-483-3p/CDK6 in SGC-7901 and BGC-823 cells reversed the inhibited proliferative rate by miR-483-3p overexpression (Figure 5B and C). Identically, CDK6 overexpression also reversed the inhibited migratory rate of GC cells overexpressing miR-483-3p (Figure 5D and E). To sum up, our results demonstrated a potential role of circZNF609/miR-483-3p/CDK6 axis in the occurrence and development of GC.
Figure 5.
The effect of CDK6 was partly abolished by miR-483-3p. (A) The overexpression efficiency of CDK6 in SGC-7901 and BGC-823. (B and C) Inhibited proliferative rate induced by miR-483-3p overexpression was reversed after CDK6 overexpression in SGC-7901 (B) and BGC-823 (C) cells. (D and E) Inhibited migratory rate induced by miR-483-3p overexpression was reversed after CDK6 overexpression in SGC-7901 (D) and BGC-823 (E) cells. *P<0.05, **P<0.01, ***P<0.001.
Discussion
GC is a common type of malignancies throughout the world. Its morbidity and mortality remain high. Due to the low early diagnosis rate and 5-year survival rate, it is necessary to find effective biomarkers in early diagnosis and therapeutic targets for GC.
Numerous studies have shown that ncRNAs are closely related to the regulation of various pathophysiological processes in tumors.20 In the present study, circZNF609 was differentially expressed in GC tissues and paracancerous tissues. Its expression level was closely related to the clinical TNM stage and prognosis of GC patients, suggesting the potential diagnostic and prognostic values of circZNF609 in GC. Hence, circZNF609 is expected to be a novel hallmark for tumor diagnosis and evaluation. In vitro assay demonstrated the high expression of circZNF609 in GC, which was able to promote proliferative and migratory capacities of GC cells. Besides, numerous miRNAs are involved in tumor development by regulating cell proliferation and migration.21,22 For example, miR-647 is lowly expressed in GC. Overexpression of miR-647 promotes proliferation and migration of tumor cells by regulating expression levels of ANK2, FAK, MMP2, MMP12 and CD44.23 Previous studies have shown that miR-483-3p inhibits the progression of breast cancer by inhibiting CCNE1 expression.15 miR-483-3p reverses EMT and inhibits gefitinib resistance by directly targeting on integrin β3 through inhibiting downstream FAK/ERK pathway.24 In the present study, circZNF609 was confirmed for its sponge of miR-483-3p through dual-luciferase reporter gene assay and RIP assay. Overexpression of miR-483-3p could reverse the promoted cell proliferation and migration induced by the overexpressed circZNF609, suggesting that the regulatory effect of circZNF609 on GC cells was mainly through absorbing miR-483-3p.
As a cell cycle-promoting factor, CDK6 is involved in tumor pathogenesis. CDK6 activity markedly alters in ovarian and bladder cancer.25,26 miR-105 is capable of inhibiting the growth of prostate cancer by downregulating CDK6 expression.27 It is reported that circZEB1.33 promotes the proliferation of liver cancer cells by upregulating CDK6 as a sponge of miR-200a-3p, thus promoting the progression of liver cancer.28 This study found that CDK6 expression was negatively correlated to miR-483-3p expression in GC. Furthermore, dual-luciferase reporter gene assay confirmed that CDK6 was the target gene of miR-483-3p, indicating that miR-483-3p could regulate biological performances of GC cells through targeting CDK6. In vitro overexpression of CDK6 reversed the inhibitory effects of miR-483-3p on the proliferative and migratory capacities of GC cells, further confirming the above conclusions.
This is the first time to uncover the role of circZNF609 in GC. However, there are still some shortcomings in this study. First of all, the colocalization between circZNF609 and miR-483-3p is not elucidated. Second, whether circZNF609 can regulate in vivo proliferation and metastasis is needed for further exploration.
In summary, the expression of circZNF609 is upregulated in GC. It can be used as a sponge of miR-483-3p to regulate the expression level of CDK6, thus participating in the progression of GC by regulating the proliferative and migratory capacities of GC cells.
Acknowledgments
We would like to express our sincere thanks to all those patients who donated to this work. This work was supported by National Natural Science Foundation of China (31700444), Social Development Guide Project of Zhenjiang (FZ2015074 and FZ2015075), and Science and Technological Development Special Foundation of Danyang (SF201510). Weidong Wu and Ningxian Wei are co-first authors for this study.
Ethics approval and consent to participate
Sample collection was obtained from the written informed consent of patients and approved by the ethics committee of Danyang People’s Hospital of Jiangsu Province.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lordick F, Allum W, Carneiro F, et al. Unmet needs and challenges in gastric cancer: the way forward. Cancer Treat Rev. 2014;40:692–700. doi: 10.1016/j.ctrv.2014.03.002 [DOI] [PubMed] [Google Scholar]
- 2.Seidlitz T, Merker SR, Rothe A, et al. Human gastric cancer modelling using organoids. Gut. 2019;68(2):207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen DJ, Leichman L. Controversies in the treatment of local and locally advanced gastric and esophageal cancers. J Clin Oncol. 2015;33:1754–1759. doi: 10.1200/JCO.2014.59.7765 [DOI] [PubMed] [Google Scholar]
- 4.Lin T, Fu Y, Zhang X, et al. A seven-long noncoding rna signature predicts overall survival for patients with early stage non-small cell lung cancer. Aging (Albany NY). 2018. doi: 10.18632/aging.101550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Wang F, Wang X, He Z, Zhu X. Mirna-543 promotes cell migration and invasion by targeting spop in gastric cancer. Onco Targets Ther. 2018;11:5075–5082. doi: 10.2147/OTT.S161316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Y, Shao Y, Sun W, et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12:11–20. [DOI] [PubMed] [Google Scholar]
- 7.Shen Y, Zhang J, Fu Z, et al. Gene microarray analysis of the circular rnas expression profile in human gastric cancer. Oncol Lett. 2018;15:9965–9972. doi: 10.3892/ol.2018.8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular rna hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23:6330–6338. doi: 10.3748/wjg.v23.i34.6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian M, Chen R, Li T, Xiao B. Reduced expression of circrna hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32. doi: 10.1002/jcla.22281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou C, Tan DM, Chen L, et al. Effect of mir-212 targeting tcf7l2 on the proliferation and metastasis of cervical cancer. Eur Rev Med Pharmacol Sci. 2017;21:219–226. [PubMed] [Google Scholar]
- 11.Wu Z, Huang W, Wang X, et al. Circular rna cep128 acts as a sponge of mir-145-5p in promoting the bladder cancer progression via regulating sox11. Mol Med. 2018;24:40. doi: 10.1186/s10020-018-0039-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song L, Xiao Y. Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting mir-593/muc19 signal pathway. Biochem Biophys Res Commun. 2018;503:2603–2610. doi: 10.1016/j.bbrc.2018.08.007 [DOI] [PubMed] [Google Scholar]
- 13.Bai N, Peng E, Qiu X, et al. Circfblim1 act as a cerna to promote hepatocellular cancer progression by sponging mir-346. J Exp Clin Cancer Res. 2018;37:172. doi: 10.1186/s13046-018-0838-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legnini I, Di Timoteo G, Rossi F, et al. Circ-znf609 is a circular rna that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, Lyu J. Tumor suppressor function of microRNA-483-3p on breast cancer via targeting of the cyclin e1 gene. Exp Ther Med. 2018;16:2615–2620. doi: 10.3892/etm.2018.6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JJ, Liu C, Shan K, et al. Circular rna-znf609 regulates retinal neurodegeneration by acting as mir-615 sponge. Theranostics. 2018;8:3408–3415. doi: 10.7150/thno.25156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng L, Chen G, Zhu Z, et al. Circular rna znf609 functions as a competitive endogenous rna to regulate akt3 expression by sponging mir-150-5p in hirschsprung’s disease. Oncotarget. 2017;8:808–818. doi: 10.18632/oncotarget.13656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circrna_103809 promotes lung cancer progression via facilitating znf121-dependent myc expression by sequestering mir-4302. Biochem Bioph Res Co. 2018;4:846–851. doi: 10.1016/j.bbrc.2018.04.172 [DOI] [PubMed] [Google Scholar]
- 19.Zhang WY, Liu YJ, He Y, Chen P. Down-regulation of long non-coding rna anril inhibits the proliferation, migration and invasion of cervical cancer cells. Cancer Biomark. 2018. doi: 10.3233/CBM-181467 [DOI] [PubMed] [Google Scholar]
- 20.Shimizu D, Kanda M, Kodera Y. Review of recent molecular landscape knowledge of gastric cancer. Histol Histopathol. 2018;33:11–26. doi: 10.14670/HH-11-898 [DOI] [PubMed] [Google Scholar]
- 21.Azarbarzin S, Feizi M, Safaralizadeh R, Kazemzadeh M, Fateh A. The value of mir-383, an intronic mirna, as a diagnostic and prognostic biomarker in intestinal-type gastric cancer. Biochem Genet. 2017;55:244–252. doi: 10.1007/s10528-017-9793-x [DOI] [PubMed] [Google Scholar]
- 22.Shen YH, Xie ZB, Yue AM, et al. Expression level of microrna-195 in the serum of patients with gastric cancer and its relationship with the clinicopathological staging of the cancer. Eur Rev Med Pharmacol Sci. 2016;20:1283–1287. [PubMed] [Google Scholar]
- 23.Cao W, Wei W, Zhan Z, Xie D, Xie Y, Xiao Q. Role of mir-647 in human gastric cancer suppression. Oncol Rep. 2017;37:1401–1411. doi: 10.3892/or.2017.5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue J, Lv D, Wang C, et al. Epigenetic silencing of microRNA-483-3p promotes acquired gefitinib resistance and emt in egfr-mutant nsclc by targeting integrin beta3. Oncogene. 2018;37:4300–4312. doi: 10.1038/s41388-018-0276-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Ying Y, Xie H, et al. Dual regulatory role of ccna2 in modulating cdk6 and met-mediated cell-cycle pathway and emt progression is blocked by mir-381-3p in bladder cancer. Faseb J. 2019;33;1374–1377. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, Guo X, Shen J, Hua D. Alpinetin inhibits proliferation and migration of ovarian cancer cells via suppression of stat3 signaling. Mol Med Rep. 2018;8: 4030–4036. [DOI] [PubMed] [Google Scholar]
- 27.Honeywell DR, Cabrita MA, Zhao H, Dimitroulakos J, Addison CL. Mir-105 inhibits prostate tumour growth by suppressing cdk6 levels. PLoS One. 2013;8:e70515. doi: 10.1371/journal.pone.0070515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong Y, Mao J, Wu D, et al. Circ-zeb1.33 promotes the proliferation of human hcc by sponging mir-200a-3p and upregulating cdk6. Cancer Cell Int. 2018;18:116. doi: 10.1186/s12935-018-0602-3 [DOI] [PMC free article] [PubMed] [Google Scholar]