ABSTRACT
Epithelial growth factor-like 7 (EGFL7) is a secretory protein with a well-characterized role in angiogenesis and the oncogenesis of certain solid tumors. Overexpression of EGFL7 is associated with adverse prognosis in patients with cytogenetically normal acute myeloid leukemia (CN-AML). However, whether this association persists after allogeneic hematopoietic stem cell transplantation (allo-HSCT) remains unclear. To further clarify the prognostic role of EGFL7, seventy-one AML patients with EGFL7 expression data who underwent allo-HSCT from The Cancer Genome Atlas database were included and divided into either EGFL7high or EGFL7low group based on the median EGFL7 expression level. Two groups had similar clinical and molecular characteristics except that the EGFL7high group had less frequent NPM1 mutations (P= .001). Kaplan-Meier survival curves showed that high EGFL7 expressers had shorter OS than the low expressers (P= .040). Univariate analysis showed that high EGFL7 expression, MLL-PTD, RUNX1 and TP53 mutations were associated with short OS (all P< .05). Multivariate analysis indicated that high EGFL7 expression, FLT3-ITD, RUNX1 and TP53 mutations were independent risk factors for OS (all P< .05). Collectively, our study suggested that EGFL7, like the other widely-used risk stratification factors, could serve as a prognostic tool and therapeutic target in AML, even after allo-HCST.
KEYWORDS: Acute myeloid leukemia, EGFL7, allogeneic hematopoietic stem cell transplantation, prognosis, overall survival
Introduction
The clinical and genetic heterogeneity of acute myeloid leukemia (AML) has made pre-treatment genetic mutation and expression profiling a pre-requisite for individualized prognostication and treatment.1,2 For example, DNMT3A and TP53 mutations are poor-prognostic factors,3,4 while NPM1 mutation is associated with favorable prognosis.5 With the advances in molecular diagnostics, not only the mutations, but also the abnormal expression of certain genes can be utilized for refined risk stratification of AML. Over-expressions of DOK4/5, PDK2/3, FHL2, and iASPP are associated with poor prognosis, whereas overexpression of DOK7 correlates with good prognosis.6–8 Due to the complexity of AML leukemogenesis and treatment, it is in dire need to find more and better risk stratification markers in the foreseeable future.
Epithelial growth factor-like 7 (EGFL7) is a secretory endothelial cell protein that contains two epidermal growth factor-like domains and plays an important role in regulating vasculogenesis.9 A previous study showed that EGFL7 was highly expressed in human epithelial tumor tissues, including lung cancer, hepatocellular carcinoma, gastric cancer, esophageal cancer, and renal cancer.10 Some studies have indicated that high EGFL7 expression was associated with poor prognosis and advanced stages in multiple types of human cancer, including epithelial ovarian cancer, pancreatic cancer, and gastric cancer.11–13 Another recent study demonstrated that high expression of EGFL7 was correlated with lower complete remission (CR) rates, shorter event-free survival (EFS) and overall survival (OS) in patients with cytogenetically normal AML (CN-AML).14
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) significantly reduces leukemia residual disease and has conferred cure on many AML patients.15 However, it remains unclear whether the prognostic effect of EGFL7 will persist after allo-HSCT. We aim to answer this question by studying whether high EGFL7 expression predicts poor prognosis in AML patients who have undergone allo-HSCT.
Patients and methods
Patients
From The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/), a total of seventy-one AML patients who underwent allo-HSCT and had EGFL7 expression data were included in this study.16 All patients were between ages 18 and 72. Clinical characteristics at diagnosis were available in the database, including peripheral blood (PB) white blood cell count (WBC), PB and bone marrow (BM) blast percentages, French-American-British (FAB) subtypes, and the frequencies of other genetic abnormalities. Overall survival (OS) was the primary endpoint of this study. OS was defined as the time from diagnosis to death of any cause, or was censored at the last follow-up. All patients provided informed consent, and the study protocol was approved by the Washington University Human Studies Committee.
Statistical analysis
Descriptive statistics were used to summarize the clinical and molecular characteristics of the patients. Data sets were described by median and/or range. As the numerical data were not normally distributed, we used the Wilcoxon-Mann-Whitney test to compare the two groups. Categorical data were compared using the chi-square test. Survival was estimated using the Kaplan-Meier methods and log-rank test. Univariate and multivariate Cox proportional hazard models were constructed for OS, using a limited backward elimination procedure. A two-tailed P< .05 was considered statistically significant. All statistical analyses were performed by SPSS software 25.0 and GraphPad Prism software 7.0.
Results
Differences in the clinical and molecular characteristics between the EGFL7high and EGFL7low groups
All patients were divided into two groups based on the median EGFL7 expression level. Comparison of the two groups’ clinical and molecular characteristics were showed in Table 1. EGFL7high group had less frequent NPM1 mutations (P= .001). No significant differences were found in age, gender distribution, WBC count, BM blasts, PB blasts, FAB subtypes, karyotype, and risk-group distribution. The two groups had similar mutation frequencies in FLT3-ITD, DNMT3A, IDH1/IDH2, RUNX1, NRAS/KRAS, TET2, TP53, and MLL-PTD. Relapse rate and HSCT donor type were similar in the two groups (all P> .05).
Table 1.
Clinical and molecular characteristics of the patients.
| Characteristics | EGFL7high (n = 35) | EGFL7low (n = 36) | P |
|---|---|---|---|
| Age/years, median (range) | 54 (22–72) | 48 (18–63) | .102* |
| Age group/n (%) | .381§ | ||
| <60 years | 24 (68.6) | 28 (77.8) | |
| ≥60 years | 11 (31.4) | 8 (22.2) | |
| Gender/n (%) | .705§ | ||
| Male | 21 (60.0) | 20 (55.6) | |
| Female | 14 (40.0) | 16 (44.4) | |
| WBC/×109/L, median (range) | 29.4 (0.6–223.8) | 29.6 (0.8–202.7) | .505* |
| BM blast/%, median (range) | 67 (30–99) | 78 (34–100) | .066* |
| PB blast/%, median (range) | 49 (0–94) | 48 (0–96) | .729* |
| FAB subtypes/n (%) | |||
| M0 | 5 (14.3) | 4 (11.1) | .735§ |
| M1 | 13 (37.1) | 10 (27.8) | .399§ |
| M2 | 11 (31.4) | 7 (19.4) | .246§ |
| M3 | 0 (0.0) | 1 (2.8) | 1.000§ |
| M4 | 4 (11.4) | 9 (25.0) | .139§ |
| M5 | 0 (0.0) | 4 (11.1) | .115§ |
| M6 | 1 (2.9) | 0 (0.0) | .493§ |
| M7 | 1 (2.9) | 1 (2.8) | 1.000§ |
| Cytogenetics/n (%) | |||
| Normal | 14 (40.0) | 19 (52.8) | .280§ |
| Complex karyotype | 5 (14.3) | 6 (16.7) | .782§ |
| 8 Trisomy | 4 (11.4) | 2 (5.6) | .429§ |
| inv(16)/CBFβ-MYH11 | 4 (11.4) | 1 (2.8) | .199§ |
| 11q23/MLL | 0 (0.0) | 3 (8.3) | .239§ |
| −7/7q- | 3 (8.6) | 0 (0.0) | .115§ |
| t(15;17)/PML-RARA | 0 (0.0) | 1 (2.8) | 1.000§ |
| t(9;22)/BCR-ABL1 | 2 (5.7) | 0 (0.0) | .239§ |
| t(8;21)/RUNX1-RUNX1T1 | 1 (2.9) | 0 (0.0) | .493§ |
| Others | 2 (5.7) | 4 (11.1) | .674§ |
| Risk/n (%) | |||
| Good | 5 (14.7) | 2 (5.6) | .253§ |
| Intermediate | 16 (47.1) | 24 (60.0) | .098§ |
| Poor | 13 (38.2) | 10 (27.8) | .352§ |
| FLT3-ITD/n (%) | .832§ | ||
| Positive | 8 (22.9) | 9 (25.0) | |
| Negative | 27 (77.1) | 27 (75.0) | |
| NPM1/n (%) | .001§ | ||
| Mutation | 3 (8.6) | 15 (41.7) | |
| Wild type | 32 (91.4) | 21 (58.3) | |
| DNMT3A/n (%) | .832§ | ||
| Mutation | 8 (22.9) | 9 (25.0) | |
| Wild type | 27 (77.1) | 27 (75.0) | |
| IDH1/IDH2/n (%) | .368§ | ||
| Mutation | 10 (28.6) | 7 (19.4) | |
| Wild type | 25 (71.4) | 29 (80.6) | |
| RUNX1/n (%) | .710§ | ||
| Mutation | 3 (8.6) | 5 (13.9) | |
| Wild type | 32 (91.4) | 31 (86.1) | |
| NRAS/KRAS/n (%) | .710§ | ||
| Mutation | 4 (11.4) | 3 (8.3) | |
| Wild type | 31 (88.6) | 33 (91.7) | |
| TET2/n (%) | .614§ | ||
| Mutation | 1 (2.9) | 3 (8.3) | |
| Wild type | 34 (97.1) | 33 (91.7) | |
| TP53/n (%) | 1.000§ | ||
| Mutation | 2 (5.7) | 2 (5.6) | |
| Wild type | 33 (94.3) | 34 (94.4) | |
| MLL-PTD/n (%) | 1.000§ | ||
| Positive | 2 (5.7) | 2 (5.6) | |
| Negative | 33 (94.3) | 34 (94.4) | |
| Relapse/n (%) | .932§ | ||
| Yes | 23 (65.7) | 24 (66.7) | |
| No | 12 (34.3) | 12 (33.3) | |
| HSCT | .903§ | ||
| Haplo | 1 (2.9) | 1 (2.8) | |
| Sib allo | 14 (40.0) | 16 (44.4) | |
| MUD | 20 (57.1) | 19 (52.8) |
Abbreviations: WBC, white blood cell; BM, bone marrow; PB, peripheral blood; FAB, French American British; HSCT, hematopoietic stem cell transplantation; Haplo, haploidentical; Allo, allogeneic; MUD, matched unrelated donor.
‘*’ denotes Mann-Whitney U test; ‘§’ denotes chi-square test.
Prognostic value of EGFL7 expression
Kaplan-Meier analysis demonstrated that high EGFL7 expressers had shorter OS than the low expressers (P= .040, Figure 1). To evaluate the prognostic significance of EGFL7 expression level and other clinical or molecular variables, univariate and multivariate Cox proportional hazard models were constructed. The variables included expression levels of EGFL7 (high vs. low), age (≥60 vs. <60 years), WBC count (≥15 × 109/L vs. <15 × 109/L), BM blasts (≥70% vs. <70%), PB blasts (≥20% vs. <20%), FLT3-ITD (positive vs. negative), MLL-PTD (positive vs. negative), and other common AML mutations (NPM1, DNMT3A, RUNX1, and TP53; mutated vs. wild). Results were shown in Tables 2 and 3.
Figure 1.
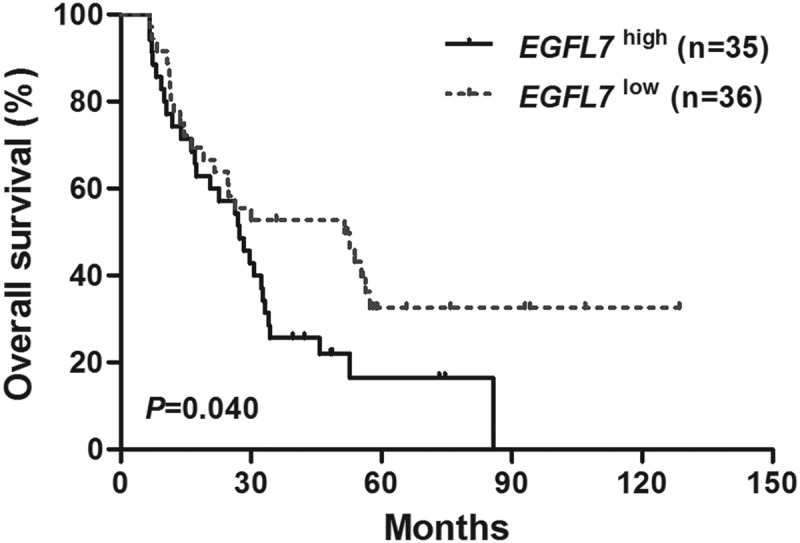
Kaplan-Meier curves of overall survival (OS) in the patients. Patients with high EGFL7 expression had shorter OS than those with low expression.
Table 2.
Univariate analysis of OS in the patients.
| Variables | OS |
|
|---|---|---|
| HR (95%CI) | P-value | |
| EGFL7 (high vs. Low) | 1.789 (1.020–3.137) | .042 |
| Age (≥60 vs. <60 years) | 1.406 (0.769–2.571) | .268 |
| WBC (≥15 vs. <15 × 109/L) | 1.161 (0.665–2.028) | .600 |
| BM blasts (≥70 vs. <70%) | 0.840 (0.487–1.449) | .530 |
| PB blasts (≥20 vs. <20%) | 1.121 (0.607–2.071) | .716 |
| FLT3-ITD (positive vs. negative) | 1.666 (0.884–3.139) | .114 |
| NPM1 (mutated vs. wild) | 0.805 (0.422–1.536) | .510 |
| DNMT3A (mutated vs. wild) | 1.259 (0.668–2.374) | .477 |
| RUNX1 (mutated vs. wild) | 2.437 (1.127–5.270) | .024 |
| TP53 (mutated vs. wild) | 4.270 (1.437–12.688) | .009 |
| MLL-PTD (positive vs. negative) | 3.307 (1.171–9.334) | .024 |
Abbreviations: OS, Overall survival; HR, hazard ratio; CI, confidence interval; WBC, white blood cell; BM, bone marrow; PB, peripheral blood.
Table 3.
Multivariate analysis of OS in the patients.
| Variables | OS |
|
|---|---|---|
| HR (95%CI) | P-value | |
| EGFL7 (high vs. Low) | 2.307 (1.151–4.625) | .019 |
| Age (≥60 vs. <60 years) | 1.229 (0.614–2.459) | .560 |
| WBC (≥15 vs. <15 × 109/L) | 1.325 (0.674–2.602) | .415 |
| BM blasts (≥70 vs. <70%) | 0.833 (0.419–1.659) | .604 |
| PB blasts (≥20 vs. <20%) | 1.589 (0.753–3.354) | .224 |
| FLT3-ITD (positive vs. negative) | 2.406 (1.086–5.329) | .031 |
| NPM1 (mutated vs. wild) | 1.388 (0.562–3.430) | .477 |
| DNMT3A (mutated vs. wild) | 1.158 (0.562–2.383) | .691 |
| RUNX1 (mutated vs. wild) | 3.165 (1.332–7.521) | .009 |
| TP53 (mutated vs. wild) | 12.175 (3.161–46.895) | .000 |
| MLL-PTD (positive vs. negative) | 2.425 (0.776–7.577) | .128 |
Abbreviations: OS, Overall survival; HR, hazard ratio; CI, confidence interval; WBC, white blood cell; BM, bone marrow; PB, peripheral blood.
Univariate analysis showed that high EGFL7 expression was associated with short OS (P= .042). RUNX1 mutations, TP53 mutations, and MLL-PTD were unfavorable for OS as well (P= .024, P= .009, P= .024, respectively). Multivariate analysis showed that high EGFL7 expression was an independent risk factor for OS (P= .019), as well as FLT3-ITD, mutations in RUNX1 and TP53 (P= .031, P= .009, P< .001, respectively).
Discussion
Our findings suggested that high EGFL7 expression was an independent risk factor for OS in AML patients who had undergone allo-HSCT, which not only concurred with the previous finding that high EGFL7 expression was associated with worse outcome in CN-AML,14 but also indicated that EGFL7 could be a prognostic indicator unaffected by treatment.
The outcome of allo-HSCT in AML is influenced by a variety of factors. The degree of remission before transplantation is a strong predictor of the relapse risk after transplantation. Patients with morphological disease at the time of transplantation have the highest risk of relapse, followed by patients in high-risk remission (CR-HR), while patients in first complete remission (CR1) or second complete remission (CR2) at the time of transplantation have the lowest risk of relapse rate.17 Studies have shown that NPM1 and FLT3 mutational status before transplant are associated with higher relapse risk and poorer outcome in transplant patients, and may serve as useful markers for minimal residual disease (MRD).17,18 The overexpression of WT1 before transplant may also represent an MRD tool for risk stratification, and predicts poor survival in AML patients.19 In addition, pre-transplant bone marrow status is an important prognostic factor for AML patients, as patients with higher marrow blast percentage at transplant usually have higher relapse risk and shorter survival.20 Some post-transplant factors also influence the prognosis of AML patients undergoing allo-HSCT. The existence of acute and chronic graft versus host disease (GVHD) is associated with higher survival and lower relapse rate in leukemia patients post allo-HSCT, due to the graft versus leukemia (GVL) effect.21 On the other hand, commonly used chemotherapy drugs for treating AML and transplant conditioning confers increased risk of cardiovascular events (CVEs) after transplant, thereby increasing the risk of death.22 In our study, we could not control for the above factors and the data were not available from the database. Nonetheless, the two groups, except the difference in EGFL7 expression levels, were essentially balanced with respect to most of the clinical and molecular characteristics. Both univariate and multivariate analyses indicated that high EGFL7 expression was an indicator of poor prognosis in transplanted AML patients. This suggests that high EGFL7 expression may also be a negative prognostic indicator of AML, the same as FLT3-ITD and TP53 mutations.
The role of EGFL7 in tumorigenesis, particularly leukemogenesis, is being defined. A study showed that overexpression of EGFL7 could promote epidermal growth factor receptor (EGFR) and protein kinase B (AKT) phospho-activation and induce the migration of gastric cancer cells. EGFL7 knockdown reversed the morphology of epithelial-mesenchymal transition (EMT), decreased vimentin and Snail expression in gastric cancer cell lines.13 In addition, high expression of EGFL7 can also promote the migration of human pancreatic cancer cells and act through transcription factors Snail and Slug to induce EMT.23 These findings suggest that EGFL7 promotes solid tumor cell metastasis by activating EMT through an EGFR-AKT-Snail signaling pathway. In AML, a study indicated that the AML blasts can synthesize and secrete EGFL7 protein and promote the leukemic cell growth in an autocrine fashion, suggesting that targeting EGFL7 may be an effective therapeutic option.14
In univariate and multivariate analyses, we also found that RUNX1 and TP53 mutations were associated with poor OS, consistent with former findings that somatic mutations in TP53 and RUNX1 are indicators of inferior survival in patients with myelodysplastic syndrome, and that mutations in TP53 and RUNX1 predict poor outcomes in AML.24,25 In this study, FLT3-ITD was also an independent risk factor for OS, in line with the fact that FLT3-ITD is associated with increased risk of relapse in AML.26 However, MLL-PTD, NPM1 and DNMT3A mutations had no effects on OS in our study. This may be due to the small sample size and the unpredictable interaction of multiple gene mutations. Despite the limitations, EGFL7 has the potential to become a useful tool in AML risk stratification, especially in patients undergoing allo-HSCT.
In conclusion, our results suggest that high EGFL7 expression, similar to FLT3-ITD and TP53 mutations, may also predict poor prognosis in AML patients undergoing allo-HSCT. Larger prospective studies are needed to further validate our results.
Funding Statement
This study was funded by the National Natural Science Foundation of China [81500118]; the National Natural Science Foundation of China [61501519].
Abbreviations
- AML
acute myeloid leukemia
- EGFL7
epithelial growth factor-like 7
- CR
complete remission
- EFS
event-free survival
- OS
overall survival
- Allo-HSCT
allogeneic hematopoietic stem cell transplantation
- BM
bone marrow
- PB
peripheral blood
- WBC
white blood cell count
- MRD
minimal residual disease
- GVHD
graft versus host disease
- CVE
cardiovascular event
- EMT
epithelial-mesenchymal transition
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Ethical approval
All data in this study were downloaded from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). We did not involve direct interaction with patients. So, this article does not contain any studies with human participants performed by any of the authors.
References
- 1.Kreso A, Dick JE.. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Mrózek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD.. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar D, Mehta A, Panigrahi MK, Nath S, Saikia KK. DNMT3A (R882) mutation features and prognostic effect in acute myeloid leukemia in Coexistent with NPM1 and FLT3 mutations. Hematol Oncol Stem Cell Ther. 2018;11:82–89. doi: 10.1016/j.hemonc.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Bullinger L, Döhner K, Döhner H. Genomics of acute myeloid leukemia diagnosis and pathways. J Clin Oncol. 2017;35:934–946. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- 5.Heath EM, Chan SM, Minden MD, Murphy T, Shlush LI, Schimmer AD. Biological and clinical consequences of NPM1 mutations in AML. Leukemia. 2017;31:798–807. doi: 10.1038/leu.2017.30. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Li R, Hu K, Dai Y, Pang Y, Jiao Y, Liu Y, Cui L, Shi J, Cheng Z, et al. Prognostic role of DOK family adapters in acute myeloid leukemia. Cancer Gene Ther. 2018. doi: 10.1038/s41417-018-0052-z. [DOI] [PubMed] [Google Scholar]
- 7.Cui L, Cheng Z, Liu Y, Dai Y, Pang Y, Jiao Y, Ke X, Cui W, Zhang Q, Shi J, et al. Overexpression of PDK2 and PDK3 reflects poor prognosis in acute myeloid leukemia. Cancer Gene Ther. 2018. doi: 10.1038/s41417-018-0071-9. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Z, Dai Y, Pang Y, Jiao Y, Zhao H, Zhang Z, Qin T, Hu N, Zhang Y, Ke X, et al. Enhanced expressions of FHL2 and iASPP predict poor prognosis in acute myeloid leukemia. Cancer Gene Ther. 2019;26:17–25. doi: 10.1038/s41417-018-0027-0. [DOI] [PubMed] [Google Scholar]
- 9.Bambino K, Lacko LA, Hajjar KA, Stuhlmann H. Epidermal growth factor-like domain 7 is a marker of the endothelial lineage and active angiogenesis. Genesis. 2014;52:657–670. doi: 10.1002/dvg.22781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan C, Yang LY, Wu F, Tao YM, Liu LS, Zhang JF, He YN, Tang LL, Chen GD, Guo L. The expression of Egfl7 in human normal tissues and epithelial tumors. Int J Biol Markers. 2013;28:71–83. doi: 10.5301/JBM.2013.10568. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L, Li J, Zhao YP, Guo JC, Cui QC, Zhou WX, Zhang TP, Wu WM, You L, Shu H. Prognostic significance of epidermal growth factor-like domain 7 in pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2014;13:523–528. PMID: 25308363. [DOI] [PubMed] [Google Scholar]
- 12.Oh J, Park SH, Lee TS, Oh HK, Choi JH, Choi YS. High expression of epidermal growth factor-like domain 7 is correlated with poor differentiation and poor prognosis in patients with epithelial ovarian cancer. J Gynecol Oncol. 2014;25:334–341. doi: 10.3802/jgo.2014.25.4.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo BH, Xiong F, Wang JP, Li JH, Zhong M, Liu QL, Luo GQ, Yang XJ, Xiao N, Xie B, et al. Epidermal growth factor-like domain-containing protein 7 (EGFL7) enhances EGF receptor-AKT signaling, epithelial-mesenchymal transition, and metastasis of gastric cancer cells. PLoS ONE. 2014;9:e99922. doi: 10.1371/journal.pone.0099922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papaioannou D, Shen C, Nicolet D, McNeil B, Bill M, Karunasiri M, Burke MH, Ozer HG, Yilmaz SA, Zitzer N, et al. Prognostic and biological significance of the proangiogenic factor EGFL7 in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2017;114:E4641–E4647. doi: 10.1073/pnas.1703142114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta V, Tallman MS, He W, Logan BR, Copelan E, Gale RP, Khoury HJ, Klumpp T, Koreth J, Lazarus HM, et al. Comparable survival after HLA-well-matched unrelated or matched sibling donor transplantation for acute myeloid leukemia in first remission with unfavorable cytogenetics at diagnosis. Blood. 2010;116:1839–1848. doi: 10.1182/blood-2010-04-278317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ Jr, Laird PW, Baty JD, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaballa S, Saliba R, Oran B, Brammer JE, Chen J, Rondon G, Alousi AM, Kebriaei P, Marin D, Popat UR, et al. Relapse risk and survival in patients with FLT3 mutated acute myeloid leukemia undergoing stem cell transplantation. Am J Hematol. 2017;92:331–337. doi: 10.1002/ajh.24632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayser S, Benner A, Thiede C, Martens U, Huber J, Stadtherr P, Janssen JW, Röllig C, Uppenkamp MJ, Bochtler T, et al. Pretransplant NPM1 MRD levels predict outcome after allogeneic hematopoietic stem cell transplantation in patients with acute myeloid leukemia. Blood Cancer J. 2016;6:e449. doi: 10.1038/bcj.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frairia C, Aydin S, Audisio E, Riera L, Aliberti S, Allione B, Busca A, D’Ardia S, Dellacasa CM, Demurtas A, et al. Post-remissional and pre-transplant role of minimal residual disease detected by WT1 in acute myeloid leukemia: a retrospective cohort study. Leuk Res. 2017;61:10–17. doi: 10.1016/j.leukres.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Lee SE, Yoon JH, Shin SH, Yahng SA, Cho BS, Eom KS, Kim YJ, Min CK, Lee S, Cho SG, et al. Impact of pre-transplant marrow blasts on survival of allogeneic stem cell transplantation in adult acute myeloid leukemia. Int J Hematol. 2013;97:640–649. doi: 10.1007/s12185-013-1312-1. [DOI] [PubMed] [Google Scholar]
- 21.Rubio MT, Labopin M, Blaise D, Socié G, Contreras RR, Chevallier P, Sanz MA, Vigouroux S, Huynh A, Shimoni A, et al. The impact of graft-versus-host disease prophylaxis in reduced-intensity conditioning allogeneic stem cell transplant in acute myeloid leukemia: a study from the acute leukemia working party of the European Group for Blood and Marrow Transplantation. Haematologica. 2015;100:683–689. doi: 10.3324/haematol.2014.119339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fossard G, Blond E, Balsat M, Morisset S, Giraudier S, Escoffre-Barbe M, Labussière-Wallet H, Heiblig M, Bert A, Etienne M, et al. Hyperhomocysteinemia and high doses of nilotinib favor cardiovascular events in chronic phase Chronic Myelogenous Leukemia patients. Haematologica. 2016;101:e86–e90. doi: 10.3324/haematol.2015.135103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang YL, Dong FL, Yang J, Li Z, Zhi QM, Zhao X, Yang Y, Li DC, Shen XC, Zhou J. Suppression of the epidermal growth factor-like domain 7 and inhibition of migration and epithelial-mesenchymal transition in human pancreatic cancer PANC-1 cells. Asian Pac J Cancer Prev. 2015;16:4065–4069. PMID: 25987088. doi: 10.7314/apjcp.2015.16.9.4065. [DOI] [PubMed] [Google Scholar]
- 24.Schnittger S, Dicker F, Kern W, Wendland N, Sundermann J, Alpermann T, Haferlach C, Haferlach T. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2010;117:2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 25.Stengel A, Kern W, Haferlach T, Meggendorfer M, Fasan A, Haferlach C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL-an analysis of 3307 cases. Leukemia. 2016;31:705–711. doi: 10.1038/leu.2016.263. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Z, Dai Y, Pang Y, Jiao Y, Zhao H, Wu S, Zhang L, Zhang Y, Wang X, Wang L, et al. Clinical and biological implications of mutational spectrum in acute myeloid leukemia of FAB subtypes M0 and M1. Cell Physiol Biochem. 2018;47:1853–1861. doi: 10.1159/000491065. [DOI] [PubMed] [Google Scholar]


