ABSTRACT
Limited studies are available on the molecular pathogenesis of Epstein-Barr virus (EBV)-associated T or natural killer (NK) cell lymphoproliferative disorders (EBV+T/NK-LPD). In this retrospective study, we aim to elucidate the mutation profile of EBV+T/NK-LPD using capture-based targeted sequencing with a panel consisting of 64 lymphoma-related genes to identify driver genes associated with the development of EBV+T/NK-LPD. Targeted sequencing of 169 EBV+T/NK-LPD cases was performed using a panel of 64 lymphoma-related genes. Of the 169 EBV+T/NK-LPD cases, 123 had extra-nodal NK/T-cell lymphoma (ENKTL), 12 had aggressive NK-cell leukemia (ANKL) and 34 had EBV+ T-cell lymphoma of childhood (EBV+TL). The mutation profile revealed that all three subtypes of EBV+T/NK-LPDs had high mutation rates in STAT3, KMT2D, DDX3X, NOTCH1 and TET2. Target sequencing revealed that ENKTL, ANKL and EBV+TL were molecularly distinct, the mutation in nasal-ENKTL and extra-nasal-ENKTL are also different. Survival analysis revealed that ENKTL patients with gene mutations or loss of protein expression in either KMT2D or TET2 were significantly correlated with shorter overall survival. And although the EBV+TL and ANKL groups were too small to confirm survival disadvantage, the adverse prognosis trends of KMT2D or TET2 were showed in these two groups. We conclude that EBV+T/NK lymphoproliferative disorders have very distinct molecular profiles. Our findings also suggest the likely involvement of KMT2D and TET2 in the development of ENKTL, and possibly EBV+T/NK-LPDs in general.
KEYWORDS: Epstein-Barr virus, lymphoproliferative disorder, extra-nodal NK/T-cell lymphoma, aggressive NK-cell leukemia, EBV+ T-cell lymphoma, KMT2D, TET2
Introduction
Epstein-Barr virus (EBV)-associated mature T-cells (T) or natural killer cells (NK) lymphoproliferative disorders (LPD) (EBV+T/NK-LPD) are rare systemic diseases characterized by clonal malignant proliferation of EBV-positive T/NK cells. Despite treatment with chemotherapy and/or radiotherapy, the prognosis for patients with EBV+T/NK-LPD remains poor1–3 EBV+T/NK-LPDs are more prevalent in indigenous populations of Central and South America and Asia. China is one of countries with the highest prevalent rate.4–7 In the 2016 update of the World Health Organization (WHO) diagnostic classification of lymphoid neoplasms, T/NK neoplasms associated with EBV infection included extra-nodal NK/T cell lymphoma (ENKTL), aggressive natural killer cell leukemia (ANKL) and systemic EBV-positive T-cell lymphoma of childhood (EBV+TL).3 ENKTL is the most common type and occurs predominantly in the upper aerodigestive track (nasal-ENKTL; i.e. nasal cavity, nasopharynx, paranasal sinus and palate) but are also found in extra-nasal sites (extra-nasal-ENKTL) such as the skin, soft tissue, gastrointestinal tract, testis and less frequently in the liver, lung, brain, breast, ovaries, adrenal glands and spleen.5,8 Albeit having varied phenotypic characteristics and site of origin, certain overlapping clinicopathologic features exists among ENKTL, ANKL and EBV+TL, thus making differential diagnosis difficult.5,9–11 EBV infection has been established to be an early event in the disease development and additional somatic genetic alterations are necessary to induce carcinogenesis.12 However, the underlying molecular mechanisms in the malignant proliferation associated with EBV+T/NK-LPD remain elusive.
Next-generation sequencing (NGS) has already become a widely used technology in detecting genomic alterations in the clinical settings due to its requirement for small sample volume and ability to simultaneously interrogate multiple genes. Thus, NGS has led to the understanding of the molecular landscape and the discovery of therapeutic targets in different cancer types.13 In efforts to understand the molecular mechanism and identify potential drug targets, mutational profile of ENKTL patients has been elucidated by several groups.14–18 However, limited studies have investigated the molecular landscape of other subtypes of EBV+T/NK-LPDs. In this study, we aim to elucidate the mutation profile of all three subtypes of EBV+T/NK-LPD using capture-based targeted sequencing with a panel consisting of 64 lymphoma-related genes to identify other driver genes associated with the development of EBV+T/NK-LPD.
Results
Mutational profile of EBV+T/NK-LPD cases
Among the 123 ENKTL cases, we identified 253 mutations in 51 genes in 106 cases. Seventeen patients had no mutation detected from this panel, resulting in a positive detection rate of 80.3% (106/123). The most common mutated genes in ENKTL cases were STAT3, DDX3X, KMT2D, TET2, EP300 and BCOR (Figure 1a-b).
Figure 1.
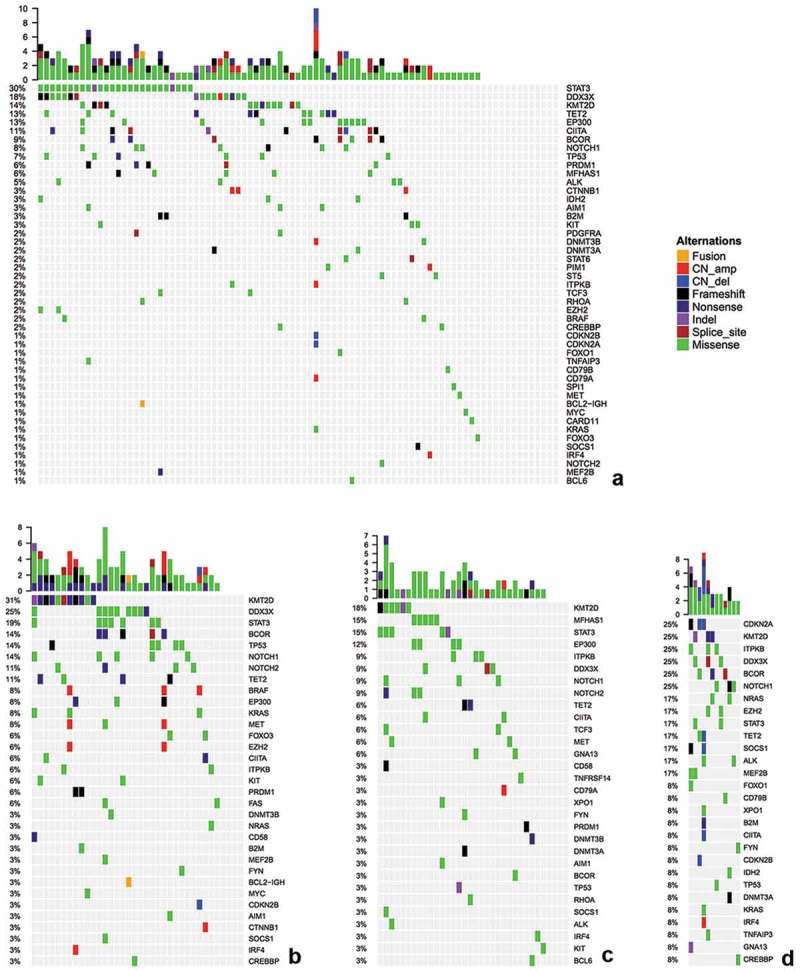
Targeted sequencing results of nasal-ENKTL (a), extra-nasal-ENKTL (b), EBV+TL (c) and ANKL (d). Each colmn represents a patient and each row represents a gene. Top plot represents the overall number of mutations a patient carried. Side bars represent the percentage of patients with a certain mutation. Different colors denote different types of mutation. Negative denotes the absence of any mutation.
Of the 123 ENKTL cases, 87 cases had nasal-ENKTL. Among the nasal-ENKTL cases, 175 mutations from 47 genes were detected in 74 cases, resulting in a positive detection rate of 85.1% (74/87) (Figure 1a). The remaining 13 cases had no mutation detected from our gene panel. STAT3 was the most frequently mutated gene, occurring in 29.9% (26/87) of the cases, followed by DDX3X and KMT2D, occurring in 18.4% (16/87) and 13.8% (12/87) of the cases, respectively. Other frequently mutated genes included TET3 (12.6%), EP300 (12.6%), CIITA (11.5%), BCOR (9.2%), NOTCH1 (8.0%) and TP53 (6.9%).
Among the remaining 36 cases with extra-nasal-ENKTL, 96 mutations from 33 genes were detected in 88.9% (32/36) of the cases (Figure 1b). KMT2D was the most commonly mutated gene in extra-nasal-ENKTL and was detected in 30.6% of the patients (11/36), followed by DDX3X and STAT3, occurring in 25% (9/36) and 19.4% (7/36) of the cases, respectively. Other frequently mutated genes included BCOR (13.9%), TP53 (13.9%), NOTCH1 (13.9%), NOTCH2 (11.1%) and TET2 (11.1%). Moreover, mutations in KMT2D showed mutually exclusive from mutations in DDX3X (OR = 0.21) and STAT3 (OR = 0.31) (Figure 1b).
On the other hand, among the 34 EBV+TL cases, 60 mutations from 27 genes (42.2%) were detected in 88.2% (30/34) of the cases (Figure 1c). KMT2D was the most frequently mutated gene, occurring in 17.6% (6/34) of the cases, followed by MFHAS1 (14.7%, 5/34), STAT3 (14.7%, 5/34), EP300 (11.8%, 4/34), ITPKB (8.8%, 3/34), DDX3X (8.8%, 3/34), NOTCH1 (8.8%, 3/34) and NOTCH2 (8.8%, 3/34). Interestingly, mutations in KMT2D showed mutually exclusive from mutations in MFHAS1 (OR = 0) (Figure 1c).
Among the 12 patients with ANKL, 47 mutations from 28 genes (43.8%) were detected in all the cases (Figure 1d). Mutations in CDKN2A, KMT2D, ITPKB, DDX3X, BCOR and NOTCH1 were detected in 25% (3/12) of the cases, respectively.
Collectively, the most frequently mutated genes in all 169 EBV+T/NK-LPD cases in our cohort were STAT3 and KMT2D with a mutation frequency of more than 10%, followed by DDX3X, TET2, EP300 and NOTCH1 with mutation frequencies of greater than 6%. However, mutation rates in ITPKB (p = .01), CDKN2A (p < .001), MEF2B (p = .041), SOCS1 (p = .041) and NRAS (p = .011) were significantly higher among ANKL cases than in ENKTL cases (Table 2). On the other hand, mutations in MFHAS1 was more common in EBV+TL than ENKTL (p = .025). Among the three subtypes of EBV+T/NK-LPDs, mutations in PRDM1, KIT, BRAF, AIM1 and CTNNB1 were exclusively detected in ENKTL (Figure 2). However, the mutation rates of these 5 genes were only between 3.3% and 5.7%. In addition, mutations in CD79B, CDKN2B, CREBBP, DNMT3A, FOXO1, GNA13, IDH2, IRF4, TNFAIP3, XPO1, MEF2B, NRAS, and CDKN2A were only detected in ANKL cases. Meanwhile, TCF3 as only found in EBV+TL cases.
Table 2.
Chi-square analysis of mutated genes in patients with ENKTL (both nasal and extra-nasal), ANKL and EBV+TL.
| Gene | ENKTLs | ANKLs | EBV+TLs |
P 1 |
P 2 |
|---|---|---|---|---|---|
| (ENKTL vs ANKL) | (ENKTL vs EBV+TL) | ||||
| STAT3 | 33 | 2 | 5 | 0.763 | 0.144 |
| KMT2D | 23 | 3 | 6 | 0.885 | 0.889 |
| DDX3X | 25 | 3 | 3 | 0.993 | 0.121 |
| TET2 | 15 | 2 | 2 | 1.000 | 0.461 |
| EP300 | 14 | 0 | 4 | 0.460 | 1.000 |
| NOTCH1 | 12 | 3 | 3 | 0.262 | 1.000 |
| BCOR | 13 | 3 | 1 | 0.313 | 0.298 |
| CIITA | 12 | 1 | 2 | 1.000 | 0.718 |
| TP53 | 11 | 1 | 1 | 1.000 | 0.423 |
| MFHAS1 | 5 | 0 | 5 | 1.000 | 0.025 |
| ITPKB | 4 | 3 | 3 | 0.010 | 0.356 |
| PRDM1 | 7 | 0 | 1 | 0.868 | 0.838 |
| NOTCH2 | 5 | 0 | 3 | 1.000 | 0.499 |
| ALK | 4 | 2 | 1 | 0.156 | 1.000 |
| EZH2 | 5 | 2 | 0 | 0.231 | 0.520 |
| CDKN2A | 1 | 3 | 0 | <0.001 | 1.000 |
| MEF2B | 2 | 2 | 0 | 0.041 | 1.000 |
| SOCS1 | 2 | 2 | 2 | 0.041 | 0.436 |
| NRAS | 1 | 2 | 0 | 0.011 | 1.000 |
Figure 2.
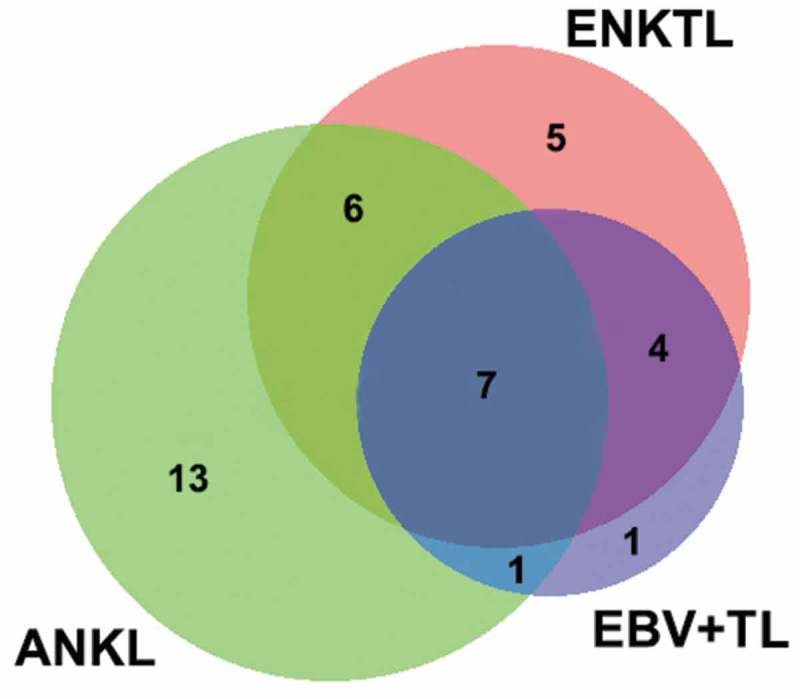
Venn diagram depicting the number of genes exclusive or shared among ENKTL, EBV+TL and ANKL.
Moreover, extra-nasal-ENKTL had significantly more mutations in KMT2D and NOTCH2 as compared to nasal-ENKTL (Table 3). Additionally, mutations in MFHAS1, ALK, AIM1, B2M, CTNNB1 and IDH2 were exclusively detected in nasal-ENKTL, while mutations in NOTCH2, BRAF, KRAS, MET, ITPKB, FOXO3 and FAS were exclusively found in extra-nasal-ENKTL (Supplementary Figure 1).
Table 3.
Chi-square analysis of mutated genes in patients with nasal-ENKTL and extra-nasal-ENKTL.
| Gene | Nasal-ENKTL | Extra-nasal-ENKTL | p |
|---|---|---|---|
| STAT3 | 26 | 7 | 0.234 |
| DDX3X | 16 | 9 | 0.497 |
| KMT2D | 12 | 11 | 0.030 |
| TET2 | 11 | 4 | 1.000 |
| EP300 | 11 | 3 | 0.709 |
| BCOR | 8 | 5 | 0.441 |
| NOTCH1 | 7 | 5 | 0.320 |
| CIITA | 10 | 2 | 0.499 |
| NOTCH2 | 1 | 4 | 0.041 |
Furthermore, analysis of the distribution of the mutation sites in STAT3 revealed clustering at the Src homology 2 (SH2) domain, with N647I, Y640F, D661Y, S614R and E616G identified as hotspot mutations (Figure 3). On the other hand, mutations in DDX3X were predominantly distributed either the helicase ATP binding domain or the helicase C-terminal domain (Figure 3). However, mutations in KMT2D, TET2, EP300 and NOTCH1 were distributed throughout the length of the gene with no particular hotspot (Figure 3). Mutations in KMT2D and TET2 were mainly nonsense mutations and small insertion-deletion mutations resulting in frameshifts, which can both lead to protein loss of function (Figure 1a-d). However, mutations in DDX3X, EP300 and NOTCH1 were primarily missense mutations that can affect the protein structure (Figure 1a-d). Sanger sequencing results were consistent with the mutations identified by targeted sequencing in KMT2D, TET2, EP300 and NOTCH1 (results of 4 representative mutations in each gene was depicted in Supplementary Figure 2).
Figure 3.
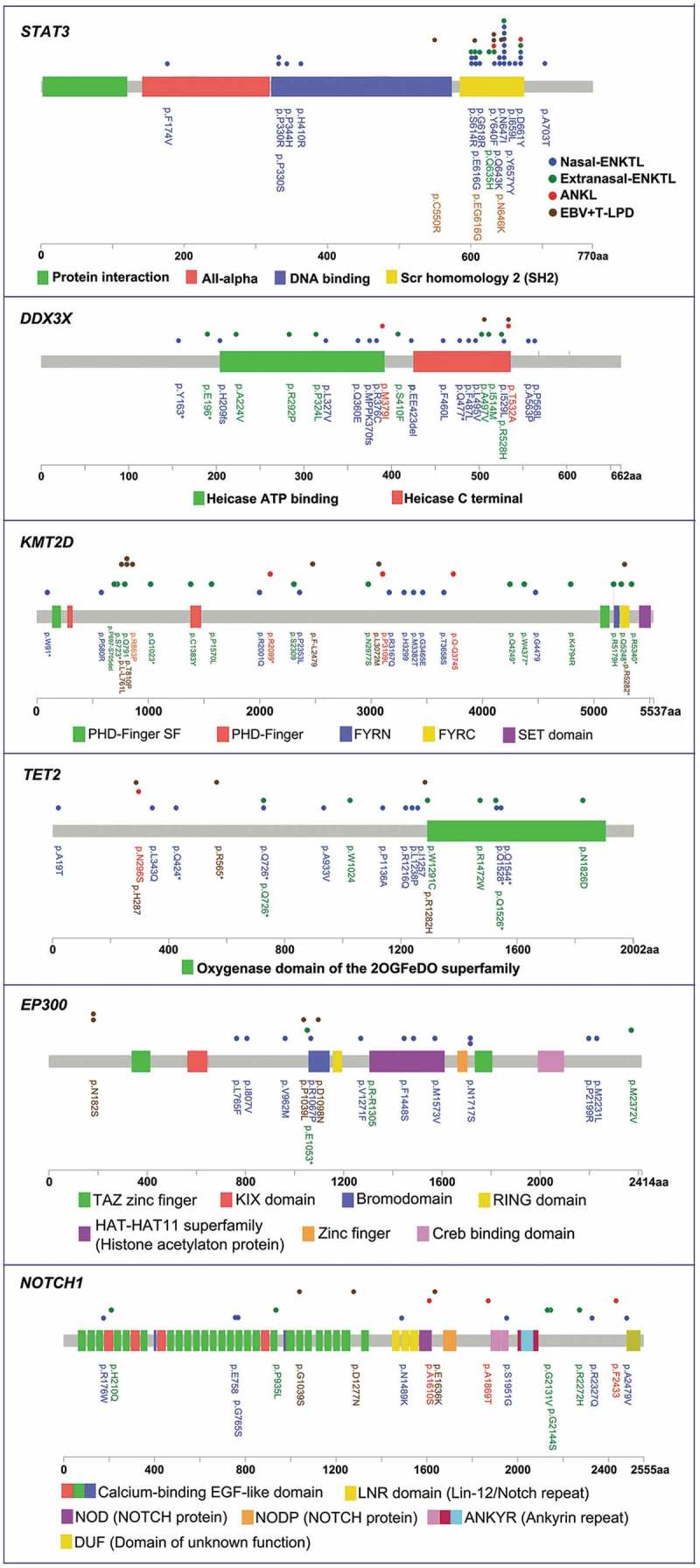
The distribution of mutations in STAT3, DDX3X, KMT2D, TET2, EP300 and NOTCH1. Genomic open reading frame were labelled with amino acid positions. Colored boxes depict the different functional domains along the gene. The location of mutations is denoted by colored circles. Colors of the circles denote the EBV+T/NK-LPD subtype.
Characteristics of ENKTL patients
ENKTL was the most common disorder among the three subtypes of EBV+T/NK-LPD. In our cohort, ENKTL cases accounted for 72.7% (123/169), with a male to female ratio of 2.32:1. The median age of the 123 ENKTL patients was 46 years. Among the 95 evaluable ENTKL patients, the median overall survival was 489 days, ranging from 46 to 1308 days.
Among the 87 nasal-ENKTL patients, a majority were males (75.9%, 66/87), with a median age of onset of 47 years, ranging from 17 to 83 years. The patients were predominantly diagnosed with stage I-II (87.4%, 76/87), while the remaining 11 patients were diagnosed with stage III-IV according to the Ann Arbor staging system. The median overall survival of the 52 evaluable nasal-ENKTL patients was 511 days, ranging from 47 to 930 days.
On the other hand, among the 36 extra-nasal-ENKTL patients, 55.6% (20/36) were males, while 44.4% (16/36) were females, with a median age of onset of 46 years, ranging from 7 to 72 years. Forty-two percent (41.7%, 15/36) of the patients were diagnosed with stage Ⅰ-Ⅱ, while 58.3% (21/36) were diagnosed with stage Ⅲ-Ⅳ according to the Ann Arbor staging classification. The extra-nasal sites of the primary lesions detected among the patients included 47.2% (17/36) in skin, 33.3% (12/36) in intestines, 11.1% (4/36) in lymph node, 5.6% (2/36) in soft tissues surrounding the kidney and 2.8% (1/36) in testis. The median overall survival of the 29 valuable extra-nasal-ENKTL patients was 435 days, ranging from 46 to 706 days.
GO and KEGG analysis in ENKTL cases
We performed Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis to further understand the biological process, cellular component, molecular function, or pathway affected by gene mutations detected among the ENKTL patients of both nasal and extra-nasal types. The GO enrichment analysis revealed that gene mutations detected in ENKTL patients were involved in myeloid progenitor cell differentiation, positive regulation of gene expression and positive regulation of transcription from RNA polymerase II promoter in terms of biological process (Supplementary Figure 3a); involved in MHC class I protein complex, nuclear chromatin and transcription factor complex in terms of cellular component (Supplementary Figure 3b); and involved in chromatin DNA binding, transmembrane receptor protein tyrosine kinase activity and transcription regulation DNA binding in terms of molecular function (Supplementary Figure 3c). In addition, KEGG analysis showed that the mutated genes were mainly involved in micro-RNA in cancer, thyroid hormone signaling pathway and pathways in cancer (Supplementary Figure 3d).
Analysis of KMT2D, TET2, EP300 and NOTCH1 in ENKTL cases
Mutations in STAT3, DDX3X, KMT2D, TET2, EP300 and NOTCH1 were frequently mutated among the ENKTL cases. STAT3 and DDX3X have already been implicated in the development of ENKTL.14,16,19–22 However, the association of mutations in KMT2D, TET2, EP300 and NOTCH1 in the genesis and development of ENKTL has not been elucidated.
In general, mutations in KMT2D, TET2, EP300 and NOTCH1 were detected in 18.7% (23/123), 12.2% (15/123), 11.4% (14/123) and 9.8% (12/123) of ENKTL cases, respectively. Immunohistochemistry staining revealed that 27.6% (34/123), 61.8% (76/123), 29.3% (36/123) and 72.4% (89/123) of ENKTL cases were negative for KMT2D, TET2, EP300 and NOTCH1 (Representative figure is depicted in Figure 4a-4b, Supplementary Figure 4a-4b), respectively, suggesting the loss of these proteins might be involved in the development of ENKTL. Correlation analysis revealed that gene mutation in KMT2D (p < .01) and TET2 (p = .034) were associated with the loss of protein expression (Table 4). Furthermore, survival analysis of ENKTL patients with gene mutations in either KMT2D (p = .002) or TET2 (p = .001) or loss of protein expression in either KMT2D (p = .024) or TET2 (p < .001) revealed significantly shorter overall survival (Table 5, Figure 4c), and the there was no difference between the nasal and extranasal ENKTL subgroups. On the contrary, no correlation was found between the gene mutation and loss of protein expression in EP300 (p = .951) and NOTCH1 (p = .252). Consistently, survival analysis of ENKTL patients with gene mutations in either EP300 (p = .727) or NOTCH1 (p = .600) or loss of protein expression in either EP300 (p = .837) or NOTCH1 (p = .947) did not show any significant difference in overall survival (Table 5, Supplementary Figure 4c). These findings implicate gene mutations or loss of protein expression in KMT2D or TET2 in the poor prognosis observed in ENKTL patients. These observations strongly suggest that KMT2D and TET2 are involved in driving carcinogenesis in ENKTL. In addition, although the EBV+TL and ANKL groups were too small to confirm survival disadvantage, the adverse prognosis trends of KMT2D or TET2 were showed in these two groups.
Figure 4.
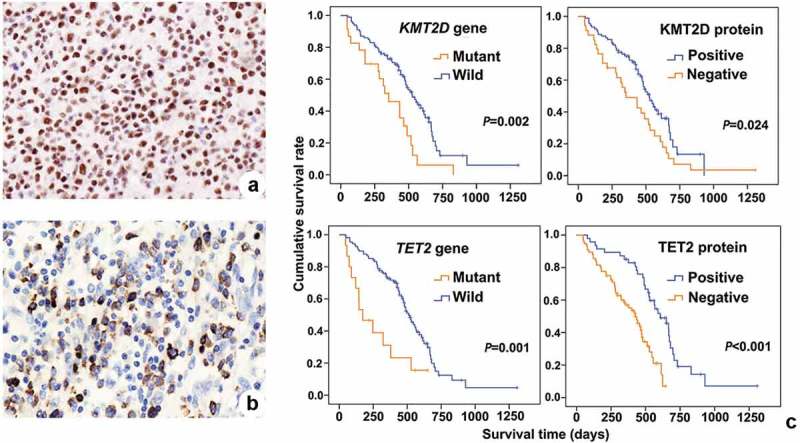
Immunohistochemistry and survival curves of KMT2D and TET2 in ENKTL patients. (a), Immunohistochemistry of KMT2D. (b), Immunohistochemistry of TET2. (c), survival curves of ENKTL patients according to KMT2D and TET gene mutation or protein loss of function.
Table 4.
Correlation analysis between protein loss and gene mutation.
| Genes | Protein detection | Gene detection |
χ2 | P | |
|---|---|---|---|---|---|
| Wild-type | Mutant | ||||
| KMT2D | Positive | 82 | 7 | 25.859 | <0.01 |
| Negative | 18 | 16 | |||
| TET2 | Positive | 45 | 2 | 4.478 | 0.034 |
| Negative | 63 | 13 | |||
| EP300 | Positive | 77 | 10 | 0.004 | 0.951 |
| Negative | 32 | 4 | |||
| NOTCH1 | Positive | 29 | 5 | 1.308 | 0.252 |
| Negative | 82 | 7 | |||
Table 5.
The prognostic significance of gene mutation, protein expression in ENKTL.
| Gene mutant cases | % | P | Protein negative cases | % | P | |
|---|---|---|---|---|---|---|
| KMT2D | 23 | 18.7 | 0.002 | 34 | 27.7 | 0.024 |
| TET2 | 15 | 12.2 | 0.001 | 76 | 61.8 | <0.001 |
| EP300 | 14 | 11.4 | 0.727 | 36 | 29.3 | 0.837 |
| NOTCH1 | 12 | 9.8 | 0.600 | 89 | 72.4 | 0.947 |
Discussion
To our knowledge, this is the largest sample study of next generation sequencing on EBV+T/NK LPD that interrogated 64 lymphoma-related genes in a cohort of 169 EBV+T/NK-LPD cases, including 123 ENKTL cases, 12 ANKL cases and 34 EBV+TL cases. Studies on EBV+T/NK LPD are limited due to the rarity of the disease and the difficulty in obtaining samples.
Target sequencing revealed that ENKTL, ANKL and EBV+TL were molecularly distinct. Collectively, mutations in STAT3, KMT2D, DDX3X, NOTCH1 and TET2 were found to be frequent among the three subtypes of EBV+T/NK-LPD. However, certain gene mutations were exclusively detected in particular subtype; such as PRDM1, KIT, BRAF, AIM1 and CTNNB1, which were only found among ENKTL cases. Conversely, certain gene mutations were more frequent in particular types; such as ITPKB, CDKN2A, MEF2B, SOCS1 and NRAS were more frequent in ANKL than in ENKTL and MFHAS1 mutation was more common in EBV+TL than ENKTL.
Consistent with other reports on molecular profiling of ENKTL cases,14–18 STAT3, DDX3X, KMT2D, TP53, TET2, EP300, BCOR, NOTCH1 and CIITA had the highest mutation rates among the ENKTL cases in our cohort. However, the mutation rates varied among the studies. The variation in mutation rates possibly arose from differences in geographical distribution of the population studied (i.e. Chinese, Caucasian, Korean, Japanese vs. Chinese), the size of the population (i.e. 25 to 105 vs. 123 ENKTL cases in our cohort), the technique used (i.e. PCR-SSCP followed by direct sequencing, transcriptome or targeted sequencing) and region of gene sequenced. The mutation rates of the genes from our cohort and the previously reported mutation rates from other populations were summarized in Table 6.
Table 6.
Comparison of mutant genes of ENKTL in our study and reported in the literatures.
| Study | This study | Kucuk, et.al. [12] | Lee, et.al. [13] | Jiang, et.al. [14] |
Dobashi, et.al. [15] |
|---|---|---|---|---|---|
| Year/Country | 2017/China | 2015/America | 2015/South Korea | 2015/China | 2016/Japan |
| Total Sample | 123 | 51 | 34 | 105 (3 Extra-nasal- ENKTL) |
25 (1 Extra-nasal- ENKTL) |
| Whole exon sequencing | 0 | 1 | 9 | 25 | 0 |
| Targeted sequencing | 123 | 0 | 21 | 80 | 25 |
| RNA sequencing | 0 | 30 | 0 | 0 | 0 |
| Sanger sequencing | 0 | 30 | 0 | 0 | 0 |
| STAT3 | 26.8% | 5.9% (3/51) | 26.4% | 10.4% | 8% |
| KMT2D | 18.7% | 5% (1/20) | 17.6% | 6.7% | 8% |
| DDX3X | 20.3% | 5.9% (1/17) | - | 20% | 12% |
| TP53 | 8.9% | 23.8% (5/21) | 11.8% | 13.3% | 16% |
| TET2 | 12.2% | 5.9% (1/17) | 0 | 0 | 8% |
| EP300 | 11.4% | 15% (3/20) | - | 3.8% | 0 |
| BCOR | 10.6% | 17.6% (3/17) | 20.6% | 0 | 32% |
| NOTCH1 | 9.8% | 5.9% (1/17) | - | 0 | 8% |
| CIITA | 9.8% | 11.8% (2/17) | 0 | 2.9% | 0 |
- Not tested
Among all the genes in the panel used for targeted sequencing, STAT3 had the highest mutation frequency in all the three subtypes of EBV+T/NK-LPD. Mutations in STAT3 were predominantly localized in the SH2 domain, which is critical for STAT activation.20 Consistent with other reports,14,15 hot spot mutations including N647I, Y640F, D661Y, S614R and E616G were also detected in our cohort, with N647I being the most recurrent among the ENKTL cases in our cohort. On the other hand, the mutations in DDX3X, KMT2D, TET2, EP300 and NOTCH1 were distributed randomly throughout the length of the gene with no particular hotspots. Since gene mutations in EBV+T/NK-LPD did not have particular hotspot except for STAT3, the use of traditional methods such as Sanger sequencing and restriction fragment polymorphism analysis would be laborious and time-consuming. Hence, targeted sequencing is more suitable in the molecular profiling of EBV+T/NK-LPD. Conversely, the types of mutations in KMT2D and TET2 were primarily nonsense and frameshift mutations that could result in premature termination, translation of truncated proteins and degradation, ultimately leading to the loss of protein function. Consistently, gene mutations in KMT2D and TET2 were found to be strongly associated with the loss of protein expression (KMT2D, p < .01; TET2, p = .034). On the other hand, the mutations in DDX3X, EP300 and NOTCH1 were mainly missense mutations that can affect the protein structure.
Interestingly, four of the highly mutated genes in the three subtypes of EBV+T/NK-LPD, KMT2D, TET2, EP300 and BCOR, are related to epigenetic modification. Epigenetic dysregulation represents an emerging component of cancer genomics.23 KMT2D, also referred to as MLL2 or MLL4, encodes histone-lysine N- methyltransferase involved in transcription activation by methylating histone H3.24 TET2 encodes methylcytosine dioxygenase, which is associated with DNA demethylation.25 EP300 and BCOR encode histone acetyltransferase P300 and BCL-6 co-inhibitory factors, respectively, which regulate histone acetylation and deacetylation.17,26 We postulate that abnormal epigenetic regulation brought about by mutations in these genes may play important roles in driving carcinogenesis in EBV+T/NK-LPDs.
Since STAT3, DDX3X and BCOR have already been implicated in the development of ENKTL,14–17,19,21,22,27 we further our study to understand the role of KMT2D, TET2, EP300 and NOTCH1 in ENKTL. Based on the analysis of survival data of ENKTL patients with either gene mutation or loss of protein expression of EP300 or NOTCH1, both EP300 and NOTCHI1 did not affect prognosis. On the other hand, ENKTL patients with mutations in either KMT2D (p = .002) or TET2 (p = .001) or loss of protein expression in KMT2D (p = .024) or TET2 (p < .001) had inferior overall survival, indicating poor prognosis of this subset of patients. Interestingly, there are mutually exclusive phenomenons between mutational in KMT2D and TET2 (OR = 1.71). Moreover, mutations in KMT2D and TET2 were also showed mutually exclusive from STAT3 (KMT2D vs. STAT3 OR = 0.72; TET2 vs. STAT3 OR = 0.99) or DDX3X (KMT2D vs. DDX3X OR = 0.15; TET2 vs. DDX3X OR = 0.57) Further suggesting that carcinogenesis in a certain subset of ENKTL is driven by mutations in either KMT2D or TET2. From these evidences, we hypothesize that KMT2D and TET2 likely play critical roles in the development of ENKTL, and could be potential therapeutic targets. Further studies are required to explore the functional roles of KMT2D and TET2 in the development of EBV+T/NK-LPD.
Prognosis for patients with EBV+T/NK-LPD remains poor with no optimal treatment available. Further studies to explore novel therapeutic approaches are required to improve the prognosis of these patients including more clinical trials on therapeutic agents available for the clinic (e.g. chemotherapy, immunotherapy), pre-clinical and clinical studies on small molecules designed for JAK/STAT pathway (i.e. JAK3 inhibitor tofacitinib, STAT3 inhibitor WP1066 and DDX3X inhibitor RK-33) and development of new agents targeting EBV or specific signaling pathways such as inhibitors for epigenetic modifiers (i.e. BCOR, KMT2D, TET2).
we have elucidated the molecular profile of the three subtypes of EBV+T/NK LPDs. Our findings also implicate KMT2D and TET2 in the development of ENKTL, and possibly EBV+T/NK-LPDs in general.
Materials and methods
Materials
Formalin-fixed, paraffin-embedded (FFPE) tissue samples from 169 EBV+T/NK LPD patients from January 2010 to December 2017 were retrieved from the Department of Pathology of the West China Hospital of Sichuan University. Tumor histology was reviewed by trained pathologists according to the WHO classification for tumors of hematopoietic and lymphoid tissues.3 The study had been approved by the relevant Institutional Review Board. Prior written informed consent was acquired from each of the recruited patients for the use of their clinicopathological data and tissue samples.
Tissue DNA isolation
Genomic DNA from FFPE tissue samples were purified using QIAamp DNA FFPE Tissue Kit (Qiagen Inc., Valencia, CA).
Targeted sequencing
A minimum of 50ng of DNA is required for NGS library construction. Tissue DNA was sheared using Covaris M220, followed by end repair, phosphorylation, and adaptor ligation. Fragments of size 200–400bp were selected by bead (Agencourt AMPure XP Kit; Beckman Coulter, CA, USA), followed by hybridization with capture probes baits, hybrid selection with magnetic beads and PCR amplification. a gene panel including all exons and selected introns of 64 genes involved in the development of lymphoma previously reported in literature (Table 1). The quality and the size of the fragments were assessed using a Qubit 2.0 Fluorimeter with the dsDNA high sensitivity assay kit (Life Technologies; Thermo Fisher Scientific, CA, USA). Indexed samples were sequenced on Nextseq500 (Illumina, Inc., CA, USA) with paired-end reads.
Table 1.
Functional classification of the 64 genes used for targeted sequencing.
| Functional classification | Genes |
|---|---|
| Oncogenes | BRAF, FYN, KIT, KRAS, MET, MFHAS1, MYC, NRAS, PDGFRA, PIM1, SPI1 |
| Anti-oncogene | AIM1, PRDM1, PTEN, ST5, TP53 |
| Apoptosis | BCL2, CARD11, FAS, TNFAIP3, TNFRSF14 |
| Cell cycle | CCND1, CDKN2A, CDKN2B, XPO1 |
| Immune response | CD58, CD79A, CD79B, CXCR4, IGHD, IGHJ, KIR2DL4, KIR3DL2 |
| Transcriptional regulation | BCL6, ID3, IRF4, TCF3, DDX3X |
| DNA methylation | DNMT3A, DNMT3B, TET2 |
| Histone acetylation | BCOR, CREBBP, EP300 |
| Histone methylation | KMT2D, EZH2 |
| Signaling pathways | NOTCH1, NOTCH2, STAT3, STAT6, CTNNB1, SOCS1, GNA13 |
| Receptor tyrosine kinase | ALK |
| Autophagy | ATG5 |
| Homeostasis | FOXO1, FOXO3 |
| Others | IDH2, ITPKB, MEF2B, MYD88, RHOA, B2M, CIITA |
Sequence data analysis
Sequence data were mapped to the reference human genome (hg19) using Burrows-Wheeler aligner 0.7.10. Local alignment optimization, variant calling and annotation were performed using Genome Analysis Tool Kit 3.2, MuTect, and VarScan. Variants were filtered using the VarScan fpfilter pipeline, loci with depth less than 100 were filtered out. Base calling in tissue samples required at least 5 and 8 supporting reads for small insertion-deletions (INDELs) and single nucleotide variants (SNVs), respectively. INDELs and SNVs with population frequency over 0.1% in the ExAC, 1000 Genomes, dbSNP or ESP6500SI-V2 databases were grouped as SNP and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff v3.6. Analysis of DNA translocation was performed using both Tophat2 and Factera 1.4.3.
Sanger sequencing and data analysis
EBV+T/NK-LPD patients with mutations in KMT2D, TET2, EP300 and NOTCH1 detected by NGS were validated with Sanger sequencing. Primer sequences were listed in Supplementary Table 1 to 4. Every amplicon was confirmed by Tris-acetate-EDTA agarose gel electrophoresis. QIAquick PCR purification kit (Qiagen Inc., CA, USA) was used to purify the PCR-amplified fragments. The forward and reverse primers used for PCR amplifications were also used for Sanger sequencing of the amplicons with Applied Biosystems ABI 3730 48-capillary electrophoresis DNA analyzer (Applied Biosystems; Thermo Fisher Scientific, CA, USA). The sequencing results were analyzed by aligning the sequences with the National Center for Biotechnology Information (NCBI) KMT2D, TET2, EP300 and NOTCH1 messenger RNA Reference Sequence (NM_003482.3, NM_001127208.2, NM_001362843.1 and NM_017617.5, respectively) using Vector NTI 10.3.0 software (Invitrogen; Thermo Fisher Scientific, CA, USA). The sequencing electropherograms were visually analyzed using Sequence Scanner v2 software (Applied Biosystems; Thermo Fisher Scientific, CA, USA).
Immunohistochemistry
The EnVision method28 was used for immunohistochemical (IHC) staining of KMT2D (Polyclonal; Sigma), EP300 (Polyclonal; Sigma) and NOTCH1 (OTI3E12; Zhongshan). Similarly, the EliVision IHC kit (Wuhan Boster Biological Engineering Co., Ltd.) was used for TET2 (Polyclonal; Darmstadt) staining.
Statistical analysis
Chi-square test was performed to analyze the correlation between protein expression and gene mutation. Chi-square test and Venn analysis were used to analyze the differences of mutational genes among different diseases. Chi-square test and Fisher’s exact test were performed to analyzed the mutational mutually exclusive. Survival data were analyzed by Kaplan–Meier and log-rank test was used to compare the difference between survival groups. For all statistical tests, p < .05 was considered statistically significant.
The study was approved by the institutional review board (IRB) at the West china hospital of Sichuan University.
Funding Statement
The authors were supported by the national natural science foundation of china (81272626) and project foundation of Sichuan provincial health and family planning commission (16ZD009).
Acknowledgments
We would like to give our sincere appreciation to the reviewers for their helpful comments on this article.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Competing interests
The authors declare that they have no competing interests.
Supplementary material
Supplementary data for this article can be accessed here
References
- 1.Kwong YL, Anderson BO, Advani R, Kim WS, Levine AM, Lim ST.. Management of T-cell and natural-killer-cell neoplasms in Asia: consensus statement from the Asian oncology summit 2009. Lancet Oncol. 2009;10:1093–1101. doi: 10.1016/S1470-2045(09)70265-7. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi M, Suzuki R, Oguchi M. Advances in the treatment of extranodal NK/T-cell lymphoma, nasal type. Blood. 2018;131:2528–2540. doi: 10.1182/blood-2017-12-791418. [DOI] [PubMed] [Google Scholar]
- 3.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J, Yang Q, Lu Z, He M, Gao L, Zhu M, Sun L, Wei L, Li M, Liu C, et al. Distribution of lymphoid neoplasms in China: analysis of 4,638 cases according to the World Health Organization classification. Am J Clin Pathol. 2012;138:429–434. doi: 10.1309/AJCP7YLTQPUSDQ5C. [DOI] [PubMed] [Google Scholar]
- 5.Park S, Ko YH. Epstein-Barr virus-associated T/natural killer-cell lymphoproliferative disorders. J Dermatol. 2014;41:29–39. doi: 10.1111/1346-8138.12322. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Pol S, Silva O, Natkunam Y. Defining the elusive boundaries of chronic active Epstein-Barr virus infection. Haematologica. 2018;103:924–927. doi: 10.3324/haematol.2018.193714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang QP, Zhang WY, Yu JB, Zhao S, Xu H, Wang WY, Bi CF, Zuo Z, Wang XQ, Huang J, et al. Subtype distribution of lymphomas in Southwest China: analysis of 6,382 cases using WHO classification in a single institution. Diagn Pathol. 2011;6:77. doi: 10.1186/1746-1596-6-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanavaros P, De Bruin PC, Briere J, Meijer CJ, Gaulard P. Epstein-Barr virus (EBV) in extranodal T-cell non-Hodgkin’s lymphomas (T-NHL). Identification of nasal T-NHL as a distinct clinicopathological entity associated with EBV. Leuk Lymphoma. 1995;18:27–34. doi: 10.3109/10428199509064919. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki R, Suzumiya J, Nakamura S, Aoki S, Notoya A, Ozaki S, Gondo H, Hino N, Mori H, Sugimori H, et al. Aggressive natural killer-cell leukemia revisited: large granular lymphocyte leukemia of cytotoxic NK cells. Leukemia. 2004;18:763–770. doi: 10.1038/sj.leu.2403262. [DOI] [PubMed] [Google Scholar]
- 10.Ryder J, Wang X, Bao L, Gross SA, Hua F, Irons RD. Aggressive natural killer cell leukemia: report of a Chinese series and review of the literature. Int J Hematol. 2007;85:18–25. doi: 10.1532/IJH97.A10612. [DOI] [PubMed] [Google Scholar]
- 11.Gao LM, Zhao S, Liu WP, Zhang WY, Li GD, Kucuk C, Hu X-Z, Chan WC, Tang Y, Ding W-S, et al. Clinicopathologic characterization of aggressive natural killer cell leukemia involving different tissue sites. Am J Surg Pathol. 2016;40:836–846. doi: 10.1097/PAS.0000000000000634. [DOI] [PubMed] [Google Scholar]
- 12.Kanavaros P, Briere J, Emile JF, Gaulard P. Epstein-Barr virus in T and natural killer (NK) cell non-Hodgkin’s lymphomas. Leukemia. 1996;10(Suppl 2):s84–7. [PubMed] [Google Scholar]
- 13.Serratì S, De Summa S, Pilato B, Petriella D, Lacalamita R, Tommasi S, Pinto R. Next-generation sequencing: advances and applications in cancer diagnosis. Onco Targets Ther. 2016;9:7355–7365. doi: 10.2147/OTT.S99807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kucuk C, Jiang B, Hu X, Zhang W, Chan JK, Xiao W, Lack N, Alkan C, Williams JC, Avery KN, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun. 2015;6:6025. doi: 10.1038/ncomms7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S, Park HY, Kang SY, Kim SJ, Hwang J, Lee S, Kwak SH, Park KS, Yoo HY, Kim WS, et al. Genetic alterations of JAK/STAT cascade and histone modification in extranodal NK/T-cell lymphoma nasal type. Oncotarget. 2015;6:17764–17776. doi: 10.18632/oncotarget.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY, Zhang ZG, Pan C-M, Hu Y, Cai C-P, Dong Y, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015;47:1061–1066. doi: 10.1038/ng.3358. [DOI] [PubMed] [Google Scholar]
- 17.Dobashi A, Tsuyama N, Asaka R, Togashi Y, Ueda K, Sakata S, Baba S, Sakamoto K, Hatake K, Takeuchi K. Frequent BCOR aberrations in extranodal NK/T-cell lymphoma, nasal type. Genes Chromosomes Cancer. 2016;55:460–471. doi: 10.1002/gcc.v55.5. [DOI] [PubMed] [Google Scholar]
- 18.Choi S, Go JH, Kim EK, Lee H, Lee WM, Cho CS, Han K. Mutational analysis of extranodal NK/T-cell lymphoma using targeted sequencing with a comprehensive cancer panel. Genomics Inform. 2016;14:78–84. doi: 10.5808/GI.2016.14.3.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolskee E, Jobanputra V, Jain P, Chen J, Ganapathi K, Nahum O, Levy B, Morscio J, Murty V, Tousseyn T, et al. Genetic landscape of T- and NK-cell post-transplant lymphoproliferative disorders. Oncotarget. 2016;7:37636–37648. doi: 10.18632/oncotarget.9400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koo GC, Tan SY, Tang T, Poon SL, Allen GE, Tan L, Chong SC, Ong WS, Tay K, Tao M, et al. Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov. 2012;2:591–597. doi: 10.1158/2159-8290.CD-12-0028. [DOI] [PubMed] [Google Scholar]
- 21.Bouchekioua A, Scourzic L, de Wever O, Zhang Y, Cervera P, Aline-Fardin A, Mercher T, Gaulard P, Nyga R, Jeziorowska D, et al. JAK3 deregulation by activating mutations confers invasive growth advantage in extranodal nasal-type natural killer cell lymphoma. Leukemia. 2014;28:338–348. doi: 10.1038/leu.2013.157. [DOI] [PubMed] [Google Scholar]
- 22.Guo Y, Arakawa F, Miyoshi H, Niino D, Kawano R, Ohshima K. Activated janus kinase 3 expression not by activating mutations identified in natural killer/T-cell lymphoma. Pathol Int. 2014;64:263–266. [DOI] [PubMed] [Google Scholar]
- 23.Ntziachristos P, Abdel-Wahab O, Aifantis I. Emerging concepts of epigenetic dysregulation in hematological malignancies. Nat Immunol. 2016;17:1016–1024. doi: 10.1038/ni.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo C, Chen LH, Huang Y, Chang CC, Wang P, Pirozzi CJ, Qin X, Bao X, Greer PK, McLendon RE, et al. KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget. 2013;4:2144–2153. doi: 10.18632/oncotarget.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tessadori F, Giltay JC, Hurst JA, Massink MP, Duran K, Vos HR, van Es RM, Scott RH, van Gassen KLI, Bakkers J, et al. Germline mutations affecting the histone H4 core cause a developmental syndrome by altering DNA damage response and cell cycle control. Nat Genet. 2017;49:1642–1646. doi: 10.1038/ng.3956. [DOI] [PubMed] [Google Scholar]
- 27.Sim SH, Kim S, Kim TM, Jeon YK, Nam SJ, Ahn YO, Keam B, Park HH, Kim D-W, Kim CW, et al. Novel JAK3-activating mutations in extranodal NK/T-cell lymphoma, nasal type. Am J Pathol. 2017;187:980–986. doi: 10.1016/j.ajpath.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Kammerer U, Kapp M, Gassel AM, Richter T, Tank C, Dietl J, Ruck P. A new rapid immunohistochemical staining technique using the EnVision antibody complex. J Histochem Cytochem. 2001;49:623–630. doi: 10.1177/002215540104900509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


