Abstract
Background:
Kinesin family member C1 (KIFC1), a C-type kinesin motor protein, plays important roles in centrosome assembly and intracellular transport. Numerous studies have focused on the prognostic value of KIFC1 in malignant tumors and the relationship between KIFC1 expression and clinicopathological traits of cancer patients, but the studies remain controversial. And no meta-analysis has yet shown the association between KIFC1 and various cancers.
Methods:
Systematic retrieval was carried out within several databases, including PubMed, Embase, Web of Science, Wanfang and China National Knowledge Infrastructure (CNKI). In addition, hazard ratios (HR) and relative risks (RR) with 95% confidence intervals (CIs) were calculated to examine the risk or hazard correlation by Stata SE15.1.
Results:
Eleven studies with the overall 2424 participants were included in this research. High KIFC1 expression was remarkably correlated with worse OS (HR = 1.33, 95% CI = 1.07–1.60) and poorer relapse-free survival (HR = 2.28, 95% CI = 1.75–2.80). In subgroup analysis, high KIFC1 expression was a negative predictor for OS in patients with ovarian cancer (P < .001), breast cancer (P < .001), hepatocellular carcinoma (P < .001), and non-small cell lung cancer (P < .001), but not for esophageal squamous cell carcinoma (P = .246). Moreover, high levels of KIFC1 were related with positive lymph node metastasis (RR = 1.23, 95% CI = 1.01–1.50, P = .041) and advanced tumor node metastasis (TNM) stage (RR = 1.55, 95% CI = 1.27–1.89, P < .001).
Conclusions:
KIFC1 overexpression indicates poor prognosis and more serious clinicopathological characteristics in kinds of malignancies. Thus, we conclude that KIFC1 could be a target for clinical diagnosis and treatment of various cancers.
Keywords: cancers, KIFC1, meta-analysis, prognosis
1. Introduction
Cancer is a leading cause of death worldwide and a major problem affecting public health.[1] In spite of the great progress of diagnostic methods and treatments, accurate cancer prognosis remains challenging.[2] The possible causes are the recurrence of cancers and the difficulty of early diagnosis. Effective biomarkers are powerful helpers in predicting cancer prognosis and monitoring cancer development. Therefore, clinicians and researchers are devoting great efforts to finding optimal prognostic biomarkers for cancer.
Kinesin family member C1 (KIFC1), namely HSET, is a minus end-directed motor protein[3] and a member of the kinesin-14 family.[4] It facilitates the crosslinking and sliding of microtubules, spindle pole focusing and vesicle transport.[5,6] KIFC1 also plays a vital role in centrosome clustering in cancer cells.[7] Centrosome amplification is a hallmark of cancers and contributes to genomic instability.[8] However, the extra centrosomes could lead to missegregation and aneuploidy eventually leading to cell death. KIFC1 may rescue cancer cells via centrosome clustering to limit negative effects of multipolar mitosis, which supports their survival.[9] By contrast, KIFC1 is nonessential for bipolar spindle assembly in normal somatic cells.[10] It has been testified that KIFC1 expression is upregulated in various human cancers, such as ovarian cancer, lung cancer, breast cancer, and hepatocellular carcinoma.[10–13] Furthermore, High KIFC1 expression induces resistance to docetaxel-mediated apoptosis in breast cancer cells[14] and its expression levels predict the risk of brain metastasis in lung cancer.[15] It is speculated that KIFC1 may play a vital role in the occurrence and development of various cancers. And further research on KIFC1 will provide new ideas for molecular therapy and targeted therapy of cancers. However, the present researches remain disputed and there is no correlated meta-analysis to evaluate the diagnostic accuracy of preoperative KIFC1 in malignancies. In this research, we systematically collected published evidences to reveal the relationship between KIFC1 expression and overall survival (OS), relapse-free survival (RFS) and clinicopathological characteristics of cancer patients. Afterwards, we could possibly identify a new biomarker for cancer prognosis and provide a reference for clinical diagnosis and risk assessment.
2. Methods
2.1. Literature retrieval strategy
A systematically comprehensive search was performed in different databases, including PubMed, Embase, Web of Science, Wanfang, and CNKI. The retrieval keywords were as follows: (“KIFC1” OR “HSET” OR “kinesin family member C1”) AND (“cancer” OR “tumor” OR “neoplasm” OR “carcinoma” OR “malignancy”) AND “prognosis.” In addition, manual examination was also performed in references from the retrieved literatures to further identify potentially relevant studies.
2.2. Selection criteria
Eligible materials were collected into this meta-analysis based on the following criteria:
-
(1)
KIFC1 expression was detected in cancers;
-
(2)
illuminated the relationship between the level of KIFC1 and OS or RFS; and
-
(3)
hazard ratios (HR) and 95% confidence intervals (CI) can be accessed directly or estimated.
Exclusion criteria were:
-
(1)
non-English publication;
-
(2)
studies with insufficient data;
-
(3)
case reports, letters, reviews, and expert opinions.
2.3. Date extraction and quality assessment
The extracted data and information were following: the first author's name, year of publication, country, cancer type, number of patients, gender, method of assessing KIFC1 expression, cut-off value, follow-up times (months), outcome measures, analysis type, clinicopathological characteristics, HRs with 95% CI, prognostic value (OS, RFS), and P-values. For studies that only provide the survival curve in figures, the approximated survival data were extracted by Engauge Digitizer.
2.4. Statistical analysis
All related statistical analyses were carried out using Stata SE15.1. The HR with 95% CI for OS and RFS were computed, as well as the relative risks (RRs) for clinical pathology parameters. Chi-squared tests and I2 statistics were used to assess heterogeneity among studies. I2 > 50% and Pheterogeneity ≤ .10 were used as criteria for statistically significant heterogeneity. Studies with markedly heterogeneity adopted the random-effect model. Otherwise (I2 ≤ 50%, Pheterogeneity > .10), a fixed-effects model was used. In addition, the publication bias was assessed with a funnel plot and the Egger test, in which case P < .05 was recognized as statistically significant.
3. Results
3.1. Characteristics of the included studies
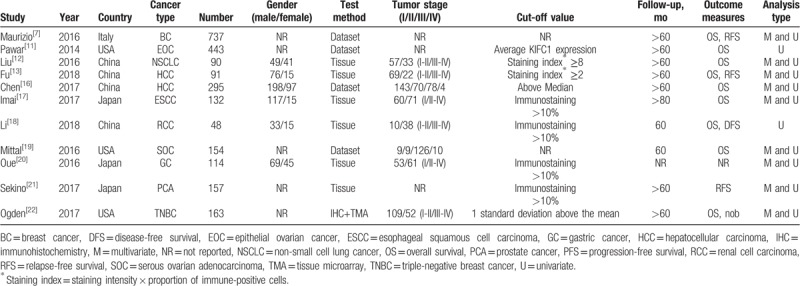
The above-described keyword search in the PubMed, Web of Science, Embase, Wanfang, and CNKI databases yielded 389 potentially relevant studies. After screening the titles and abstracts, 378 studies were excluded, and 11 studies[7,11–13,16–22] were included in this meta-analysis, encompassing a total of 2424 patients (Fig. 1). Table 1 lists the main information of the 11 eligible studies. All included studies were published between 2014 and March 2018, and the sample sizes ranged from 48 to 737. Four researches were from China, 3 from Japan, 3 from the USA and 1 from Italy. A variety of cancers were investigated, including hepatocellular carcinoma, esophageal squamous cell carcinoma, renal cell carcinoma, non-small cell lung cancer, ovarian cancer, triple-negative breast cancer, gastric cancer, and prostate cancer. The cut-off values used in these studies were not uniform. Four studies had a KIFC1 cut-off value of immunostaining >10%, 1 study applied 1 standard deviation above the mean, and 2 studies used staining indices greater than 8 and 2, respectively. The remaining 4 studies obtained data from datasets, each with different cut-off values, which were provided in Table 1. Among the 11 studies, 8 were available to appraise HR of OS for various cancers, and 3 of these directly reported HRs with the corresponding 95% CIs, while 5 studies only provided Kaplan–Meier curves.
Figure 1.
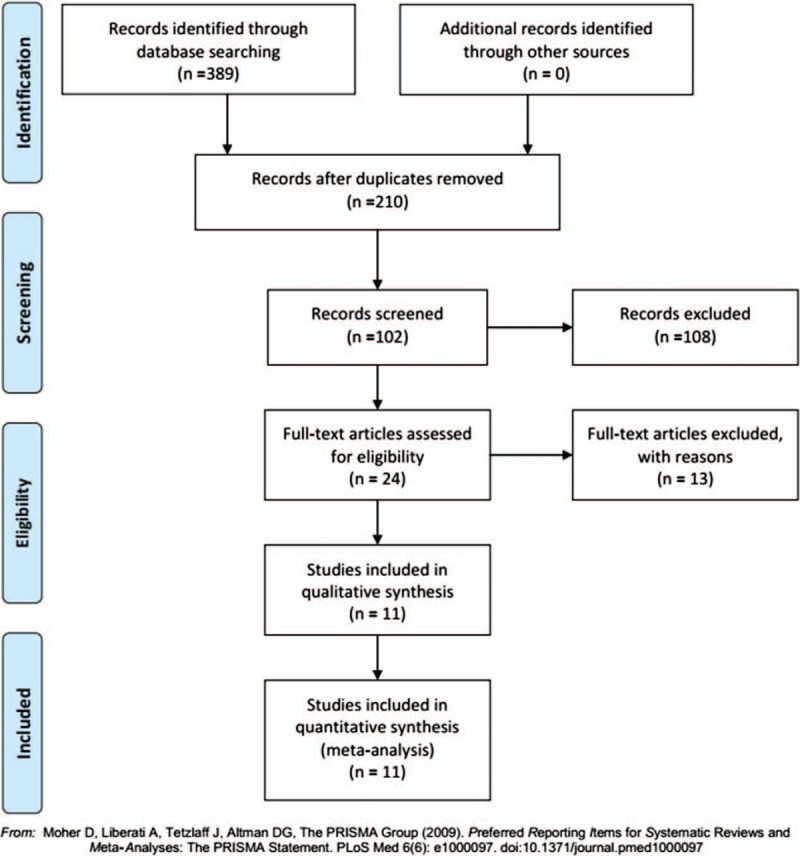
Flow chart of the literature retrieval and selection process.
Table 1.
Characteristics of enrolled studies in the meta-analysis.
3.2. Correlation between KIFCI expression levels and OS
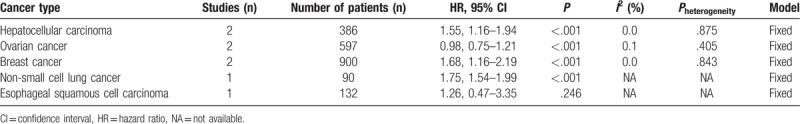
There were 8 studies including 2105 patients showed HRs for OS in this meta-analysis. A random-effect model was adopted in the analysis of data between KIFCI expression level and OS with significant heterogeneity (I2 = 57.1%, Pheterogeneity = .004). The outcome indicated that a shorter OS in the patients with high expression of KIFCI than in the low KIFCI expression (HR = 1.33, 95% CI = 1.07–1.60, P < .001; Fig. 2). Subgroup analyses classified by cancer type and analysis type were processed to further investigate the correlation between high KIFC1 expression and OS. A significant association between KIFC1 expression and OS remained in hepatocellular carcinoma (P < .001), ovarian cancer (P < .001), breast cancer (P < .001), non-small cell lung cancer (P < .001), but not in esophageal squamous cell carcinoma (P = .246) (Table 2). In terms of analysis type, significant correlation was found in univariate analysis (HR = 1.39, 95% CI = 1.10–1.68), but not in multivariate analysis (HR = 0.96, 95% CI = 0.40–1.53; Fig. 2).
Figure 2.
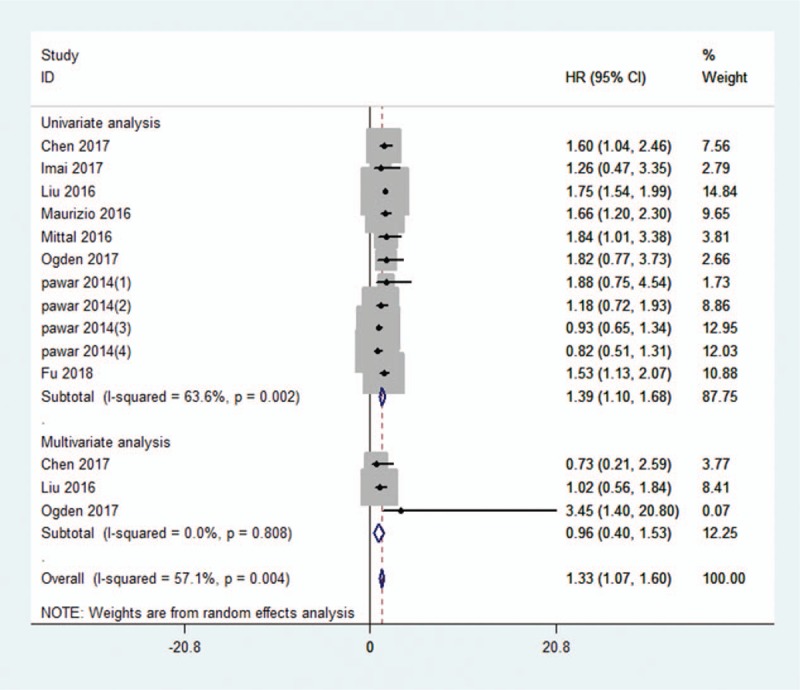
Forest plot of the pooled HRs for OS in cancer patients. Eight studies were included and the random-effect model was adopted. The pooled HR was 1.33 (95% CI = 1.07–1.60, P < .001). CI = confidence interval, HR = hazard ratio, OS = overall survival.
Table 2.
The subgroup analysis of HRs for overall survival.
3.3. Correlation between KIFC1 expression level and RFS
The HRs for RFS from 3 studies involving 1162 patients was available. A fixed-effect model was used on account of no significant heterogeneity (I2 = 0.0%, Pheterogeneity = .898). The pooled HR for RFS was 2.28 (95% CI = 1.75–2.80, P < .001), which indicated a significant association between high KIFC1 levels and poorer RFS, as shown in Figure 3. A subgroup analysis based on the analysis model revealed that increased expression of KIFCI was a negative predictor for RFS in both univariate and multivariate analysis.
Figure 3.
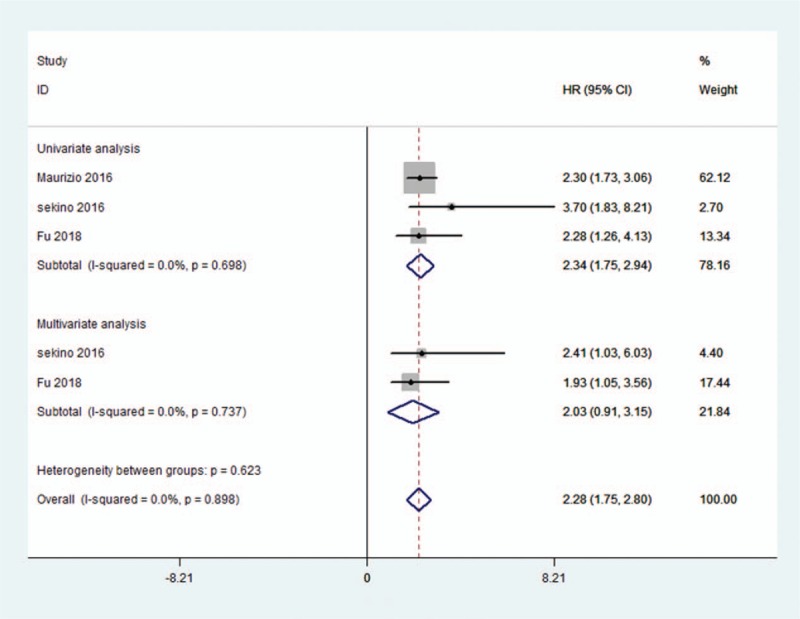
Forest plot of the pooled HRs for RFS in cancer patients. Three studies were available and the fixed-effect model was used. The pooled HR was 2.28 (95% CI = 1.75–2.80, P < .001). CI = confidence interval, HR = hazard ratio, RFS = relapse-free survival.
3.4. Correlation between KIFCI expression levels and clinicopathological characteristics
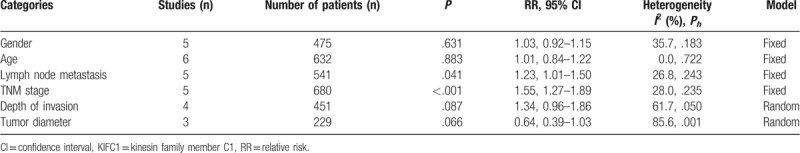
Associations between high expression of KIFCI and clinicopathological characteristics were summarized as pooled RRs with 95% CIs, as shown in Table 3. The results suggested that elevated KIFC1 expression levels were associated with advanced tumor node metastasis (TNM) stage (RR = 1.55, 95% CI = 1.27–1.89, P < .001) and positive lymph node metastasis (RR = 1.23, 95% CI = 1.01–1.50, P = .041). Whereas, insignificant association was discovered between high expression of KIFC1 and gender (RR = 1.03, 95% CI = 0.92–1.15, P = .631), age (RR = 1.01, 95% CI = 0.84–1.22, P = .883), depth of invasion (RR = 1.34, 95% CI = 0.96–1.86, P = .087), or tumor diameter (RR = 0.64, 95% CI = 0.39–1.03, P = .066). Currently, knowledge about the relation between high KIFC1 expression and other clinicopathological characteristics is still limited.
Table 3.
Meta-analysis of the correlation between overexpressed KIFC1 and clinicopathological characteristics.
3.5. Publication bias and sensitivity analysis
The funnel plot for publication bias showed symmetry (Fig. 4). And no publication bias was detected by the Begg test (P = .827) and Egger test (P = .310) for OS, respectively. Additionally, the sensitivity analysis results showed that omitting singe 1 study had no significant influence on pooled HRs, confirming the robustness of the result (Fig. 5).
Figure 4.
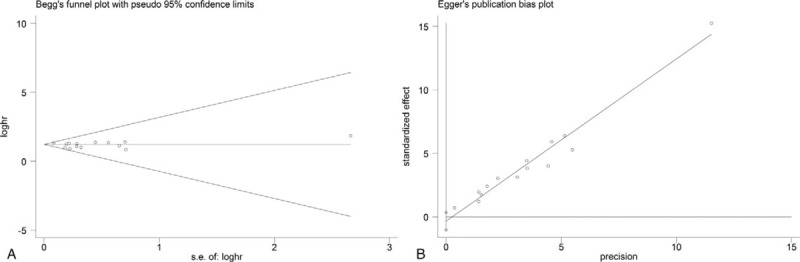
Begg funnel plot (A) and Egger test (B) for publication bias of the correlation between KIFC1 expression and OS. The P-value of Begg test was .827 and .310 for Egger test. OS = overall survival.
Figure 5.
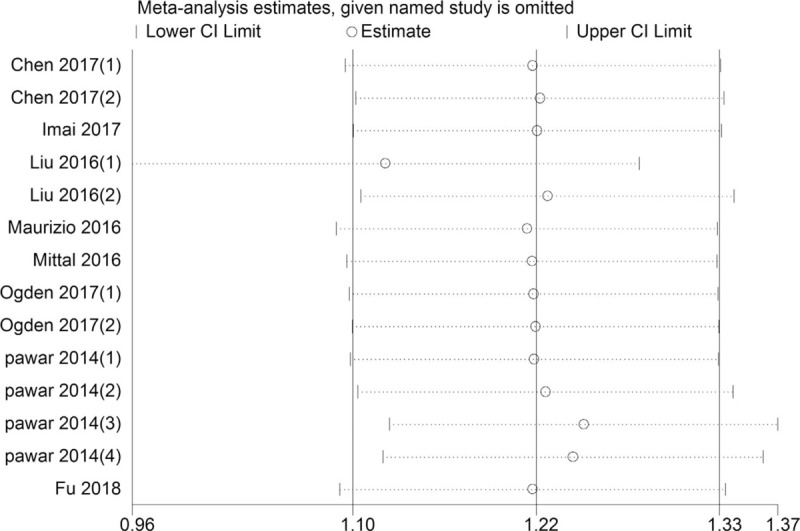
Sensitivity analysis for OS. OS = overall survival.
4. Discussion
In spite of its high incidence, cancer is usually discovered comparatively late, even though it is much easier to treat if diagnosed at early stages. Thus, finding optimal biomarkers with high prognostic value would greatly aid cancer therapy. KIFC1 is a C-type kinesin motor protein that is encoded by the KIFC1 gene located on chromosome 6 in humans.[4] It not only assists the motility of microtubules, but is also considered a promising chemotherapy target due to its multi-centrosome clustering activity in multi-centrosome cancer cells.[9] KIFC1 is absent or shows low-level expression in normal tissues but is overexpressed in cancer cells.[10,11] Several studies have shown that KIFC1 knockdown significantly inhibits cancer cell proliferation,[18] sphere formation,[17,20,21] and cell migration and invasion,[13,18] as well as causes G2/M cell cycle arrest.[12,18] Moreover, depletion of KIFC1 produces multiple microtubule organizing centers (MTOCs) and delays early mitosis, while KIFC1 silencing suppresses cell proliferation and delays cyclin A degradation in the human primary fibroblast cell line IMR-90, suggesting that it plays an essential role in bipolar MTOC formation and maintaining chromosomal stability during mitosis.[23] Thus, it is possible that KIFC1 serves as an oncogene in human tumorigenesis. On the other hand, KIFC1 can increase the probability of genetic instability to enhance the degree of tumor malignancy in breast cancer[10] and ovarian cancer.[11] Consequently, extensive attention has been paid to the potential prognostic value of KIFC1 expression. However, the present studies remain controversial and no meta-analysis has been conducted to evaluate the prognostic role of high KIFC1 expression in human malignancies. Therefore, the first meta-analysis was completed by us to assess whether KIFCI expression influenced cancer prognosis.
The pooled results of this meta-analysis showed that KIFCI overexpression was significantly correlated with worse OS in univariate analysis and poorer RFS in both univariate and multivariate analysis. This implied that KIFC1 is a potential prognostic biomarker of multiple cancers. However, multivariate analysis showed no significant correlation between high KIFCI expression levels and OS. The potential reason could be the small sample size. Only 3 studies reported multivariate analysis in hepatocellular carcinoma, non-small cell lung cancer, and breast cancer, respectively. Second, the complex mechanism by which KIFC1 acts may lead to tumor type-specific roles of this molecule. In terms of cancer type, high KIFC1 expression predicted poorer OS in patients with hepatocellular carcinoma, ovarian cancer, breast cancer, and non-small cell lung cancer, but not esophageal squamous cell carcinoma. The studies included in this meta-analysis did not cover all types of cancer and the subgroup analysis was only data obtained from 1 or 2 studies. Therefore, further studies are needed to explore the prognostic significance of KIFC1 in other cancer types.
In terms of clinicopathological characteristics, high levels of KIFC1 were related with advanced TNM stage (RR = 1.55, 95% CI = 1.27–1.89, P < .001) and positive lymph node metastasis (RR = 1.23, 95% CI = 1.01–1.50, P = .041). This indicated that KIFC1 could promote tumor progression, deepen tumor malignancy and enhance the mobility of cancer cells. Hence, high KIFC1 expression reduced the survival of the patients compared to the low expression group. However, gender, age, depth of invasion, and tumor diameter were not significantly related to high KIFC1 expression
As far as we know, this is the first meta-analysis to systematically review the prognostic value of KIFC1 in various cancers. However, several limitations still existed. First of all, only 11 studies including 2424 patients could be included in this meta-analysis. Hence, the sample size was not big enough, which may lead to bias. Second, the technique for detecting KIFC1 expression and the definition of cut-off value was not uniform in the studies, and these differences could contribute to heterogeneity. Third, any types of cancer were not included. Additional studies are therefore still required to assess the prognostic value of KIFC1 in other types of malignant tumors. Furthermore, most studies were based on Asian patients, and are, therefore, not representative of the entire world population. Finally, some survival data were extracted from survival curves rather than direct data, which may have led to errors.
This meta-analysis confirms the prognostic role of KIFC1 in various malignancies. In addition, a number of studies suggest that KIFC1 might be a novel therapeutic target. Recently, the 3 small-molecule KIFC1 inhibitors AZ82, CW069, and PJ34, have been identified.[24–26] The ATP hydrolysis sites and microtubule-binding site in the motor domain of KIFC1 are common inhibitory sites.[9,27] Inhibition of KIFC1 expression induces multipolar spindle formation, and ultimately decreases cancer cells proliferation.[28] Therefore, KIFC1 may be an effective chemotherapy target in centrosome-amplified cancers.
5. Conclusion
In conclusion, increased KIFC1 expression is significantly associated with worse OS, RFS, positive lymph node metastasis, and advanced TNM stage in malignant tumors. KIFC1 can, therefore, be considered a prognostic biomarker for patients with various cancers.
Author contributions
Conceptualization: Zhenhong Zou.
Data curation: Yuting Sun, Yi Zhang, Zhiquan Lang.
Formal analysis: Yuting Sun, Yi Zhang, Zhiquan Lang, Junfu Huang.
Funding acquisition: Zhenhong Zou.
Resources: Yuting Sun, Junfu Huang.
Software: Yuting Sun, Yi Zhang.
Validation: Zhenhong Zou.
Writing – original draft: Yuting Sun, Yi Zhang, Zhiquan Lang.
Writing – review & editing: Junfu Huang, Zhenhong Zou.
Footnotes
Abbreviations: CI = confidence interval, HR = hazard ratio, KIFC1 = kinesin family member C1, OS = overall survival, RFS = relapse-free survival, RR = relative risk.
How to cite this article: Sun Y, Zhang Y, Lang Z, Huang J, Zou Z. Prognostic and clinicopathological significance of kinesin family member C1 in various cancers. Medicine. 2019;98:40(e17346).
YS, YZ, ZL, and JH are co-first authors.
This research was supported by the National Natural Science Foundation of China (No. 81860420 and No. 81760438), the Youth Foundation project of the Jiangxi provincial science and Technology Department (No. 20171BAB215038).
The authors have no conflicts of interest to disclose.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [2].DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 2014;64:252–71. [DOI] [PubMed] [Google Scholar]
- [3].DeLuca JG, Newton CN, Himes RH, et al. Purification and characterization of native conventional kinesin, HSET, and CENP-E from mitotic hela cells. J Biol Chem 2001;276:28014–21. [DOI] [PubMed] [Google Scholar]
- [4].Ando AK YY, Kawata H, Okamoto N, et al. Cloning of a new kinesin-related gene located at the centromeric end of the human MHC region. Immunogenetics 1994;39:194–200. [DOI] [PubMed] [Google Scholar]
- [5].Mountain V, Simerly C, Howard L, et al. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol 1999;147:351–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nath S, Bananis E, Sarkar S, et al. Kif5B and Kifc1 interact and are required for motility and fission of early endocytic vesicles in mouse liver. Mol Biol Cell 2007;18:1839–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Maurizio E, Wisniewski JR, Ciani Y, et al. Translating proteomic into functional data: an high mobility group A1 (HMGA1) proteomic signature has prognostic value in breast cancer. Mol Cell Proteomics 2016;15:109–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Levine MS, Bakker B, Boeckx B, et al. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Developmental Cell 2017;40:313–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xiao Y-X, Yang W-X. KIFC1: a promising chemotherapy target for cancer treatment? Oncotarget 2016;7:48656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pannu V, Rida PC, Ogden A, et al. HSET overexpression fuels tumor progression via centrosome clustering-independent mechanisms in breast cancer patients. Oncotarget 2015;6:6076–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pawar S, Donthamsetty S, Pannu V, et al. KIFCI, a novel putative prognostic biomarker for ovarian adenocarcinomas: delineating protein interaction networks and signaling circuitries. J Ovarian Res 2014;7:53–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu Y, Zhan P, Zhou Z, et al. The overexpression of KIFC1 was associated with the proliferation and prognosis of non-small cell lung cancer. J Thorac Dis 2016;8:2911–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fu X, Zhu Y, Zheng B, et al. KIFC1, a novel potential prognostic factor and therapeutic target in hepatocellular carcinoma. Int J Oncol 2018;52:1912–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].De S, Cipriano R, Jackson MW, et al. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res 2009;69:8035–42. [DOI] [PubMed] [Google Scholar]
- [15].Grinberg-Rashi H, Ofek E, Perelman M, et al. The expression of three genes in primary non–small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res 2009;15:1755–61. [DOI] [PubMed] [Google Scholar]
- [16].Chen J, Li S, Zhou S, et al. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther 2017;13:651–9. [DOI] [PubMed] [Google Scholar]
- [17].Imai T, Oue N, Yamamoto Y, et al. Overexpression of KIFC1 and its association with spheroid formation in esophageal squamous cell carcinoma. Pathol Res Pract 2017;213:1388–93. [DOI] [PubMed] [Google Scholar]
- [18].Li G, Chong T, Yang J, et al. Kinesin motor protein KIFC1 is a target protein of miR-338-3p and associated with poor prognosis and progression of renal cell carcinoma. Oncol Res 2018;27:125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mittal K, Choi DH, Klimov S, et al. A centrosome clustering protein, KIFC1, predicts aggressive disease course in serous ovarian adenocarcinomas. J Ovarian Res 2016;9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Oue N, Mukai S, Imai T, et al. Induction of KIFC1 expression in gastric cancer spheroids. Oncol Rep 2016;36:349–55. [DOI] [PubMed] [Google Scholar]
- [21].Sekino Y, Oue N, Shigematsu Y, et al. KIFC1 induces resistance to docetaxel and is associated with survival of patients with prostate cancer. Urol Oncol 2017;35:31.e13–20. [DOI] [PubMed] [Google Scholar]
- [22].Ogden A, Garlapati C, Li XB, et al. Multi-institutional study of nuclear KIFC1 as a biomarker of poor prognosis in African American women with triple-negative breast cancer. Sci Rep 2017;7:42289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kim N, Song K. KIFC1 is essential for bipolar spindle formation and genomic stability in the primary human fibroblast IMR-90 cell. Cell Struct Funct 2013;38:21–30. [DOI] [PubMed] [Google Scholar]
- [24].Li Y, Lu W, Chen D, et al. KIFC1 is a novel potential therapeutic target for breast cancer. Cancer Biol Ther 2015;16:1316–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wu J, Mikule K, Wang W, et al. Discovery and mechanistic study of a small molecule inhibitor for motor protein KIFC1. ACS Chem Biol 2013;8:2201–8. [DOI] [PubMed] [Google Scholar]
- [26].Watts CA, Richards FM, Bender A, et al. Design, synthesis, and biological evaluation of an allosteric inhibitor of HSET that targets cancer cells with supernumerary centrosomes. Chem Biol 2013;20:1399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang W, Cao L, Wang C, et al. Kinesin, 30 years later: recent insights from structural studies. Protein Sci 2015;24:1047–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xiao Y-X, Shen H-Q, She Z-Y, et al. C-terminal kinesin motor KIFC1 participates in facilitating proper cell division of human seminoma. Oncotarget 2017;8:61373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]


