Supplemental Digital Content is available in the text
Keywords: gene polymorphism, interleukin-10, meta-analysis, periodontitis
Abstract
Background:
Periodontitis is a common disease with an unclear pathological mechanism. No precise consensus has been reached to evaluate the association between the IL-10 rs1800872 (- 592, -590, -597 C>A) polymorphism and periodontal disease. Thus, we performed this meta-analysis to collect more evidence-based information.
Methods:
Four online databases, PubMed, Embase, Web of Science, and China Biology Medicine disc (CBM), were searched in August 2018. An odds ratio (OR) with a 95% confidence interval (CI) was applied to evaluate the association of the rs1800872 with periodontitis susceptibility.
Results:
Twenty three case–control studies with 2714 patients and 2373 healthy controls were evaluated. The overall analyses verified that the IL-10 rs1800872 polymorphism was significantly associated with an increased risk of periodontitis in the allelic model, homozygote model, dominant model, and recessive model (A vs C: OR = 1.28, 95%CI = 1.11–1.49, P = .00, I2 = 56.87%; AA vs CC: OR = 2.06, 95%CI = 1.32–3.23, P = .00, I2 = 73.3%; AA + AC vs CC: OR = 1.42, 95%CI = 1.03–1.96, P = .03, I2 = 76.2%; AA vs AC + CC: OR = 1.78, 95%CI = 1.26–2.56, P = .00, I2 = 76.7%). Moreover, the subgroup analysis based on ethnicity, periodontitis type, and smoking status showed significant differences.
Conclusions:
The results of our meta-analysis demonstrate that rs1800872 is associated with periodontitis susceptibility in Caucasians and Asians. Moreover, A allele, AA genotype, CC genotype may be closely associated with chronic periodontitis (CP), while A allele, AA genotype may be closely associated with aggressive periodontitis (AgP).
1. Introduction
Periodontal disease is a group of inflammatory disorders, primarily initiated by a chronic bacterial infection and related to the host response.[1] Interestingly, approximately 5000 years ago, a description of periodontal diseases that is now named periodontitis had been found in ancient Egyptian and Chinese writings.[2] Since then, the concept of periodontitis has prevailed and developed more comprehensively. Currently, periodontal diseases are the most common inflammatory conditions of humans worldwide and affect approximately 50% of adults and 60% of people over 65 years old.[3] Chronic bacterial infection and persistent inflammation lead to connective tissue breakdown and alveolar bone destruction. Inflammatory mediators and periodontal tissue breakdown products have already been detected in gingival tissues, gingival crevicular fluid, saliva, and even plasma.[4–6] Periodontitis can be classified into 2 main types: chronic and aggressive. Periodontitis is a progressive infectious disease of periodontal tissue and can lead to tooth loss and impaired functioning of dentition.[7,8]
Interleukin-10 (IL-10), a highly pleiotropic cytokine[9] produced by various cell types, including macrophages and T cells, plays a critical role in interconnected cellular and humoral host responses.[10] Interestingly, IL-10 is thought to be an anti-inflammatory cytokine that suppresses immune and inflammatory responses.[11,12] Some research has been conducted to demonstrate the changes in various kinds of cytokines secreted in normal periodontal tissues and pathological conditions,[13] but the expression profile of IL-10 in periodontal conditions has yet to be elucidated.
Polymorphism associated with IL-10, which could regulate gene expression and protein function, has been widely performed to detect the underlying pathophysiology of periodontitis. Many single nucleotide polymorphisms (SNPs) have been reported to play important roles in regulating IL-10 promoter activity, one of which is situated at position -592 (rs1800872) and is related to the translational start site.[14] Several studies have been performed to investigate the relationship between IL-10 rs1800872 (- 592, -590, -597 C>A) and periodontal diseases but yielded inconsistent results. Toker et al indicated that the IL-10 rs1800872 polymorphisms in Caucasians were associated with the susceptibility of the CP and AgP[18]; however, Gonzales insisted that IL-10 rs1800872 promoter polymorphisms investigated in Caucasians did not reach statistical significance for an association with AgP or CP.[17] what's more, Hu et al revealed that rs1800872 polymorphisms were associated with a lower risk for developing CP and AgP in Asians,[23] but Atanasovska-Stojanovska et al and Luo et al found that there were no significant differences between rs1800872 polymorphisms with periodontitis susceptibility in CP or AgP in Asians.[25,45] To date, plenty of epidemiological studies have focused on the association of IL-10 rs1800872 polymorphisms with CP or AgP susceptibility. However, the available studies remain ambiguous and controversial, so further studies need to be performed to resolve the problems whether there are significant differences between rs1800872 polymorphisms and CP or AgP susceptibility in different ethnic groups.
Therefore, we conducted this meta-analysis to determine whether there is an association between rs1800872 and periodontitis susceptibility, and we anticipate obtaining more substantive evidence of the pathogenesis and progression of periodontitis. Comprehensive understanding of the role of rs1800872 in periodontitis will promote the therapeutics of periodontitis in the future.
2. Methods
2.1. Literature search
Four electronic databases, namely, PubMed, Embase, Web of Science, and China Biology Medicine disc (CBM) databases, were searched in August 2018 by 2 independent reviewers (WZ and LYF). The language of the published articles was restricted to English or Chinese. The following search items were used: (“interleukin 10” OR “interleukin-10” or “IL-10” or “IL 10” or “IL-10–592” or “rs1800872”) and (“periodontal disease” or “periodontitis”) and (“polymorphism” or “polymorphisms” or “SNP” or “SNPs” or “gene”). Furthermore, references in related studies or reviews were also reviewed by hand searching to identify additional eligible studies. Ethical approval and informed consent were not required, as this study was based on previously published studies and had no patient contact or direct influences on patient treatment.
2.2. Inclusion and exclusion criteria
Two reviewers (WZ and LYF) independently evaluated all of the search results, and the following criteria were designed and used for including the identified studies in this meta-analysis: clinical case-control studies investigating the association between rs1800872 and periodontitis; the frequencies of alleles or genotypes in case (periodontitis patients) and control (periodontitis-free subjects) groups can be extracted; periodontal patients and control subjects are clearly described and confirmed; and the studies use validated genotyping methods to calculate the value of the odds ratios (ORs), 95% confidence intervals (95%CIs), and the Hardy–Weinberg equilibrium (HWE). The exclusion criteria were animal studies or in vitro studies; reviews, letters, case reports or comments; and studies that did not provide sufficient information about genotype or allelic frequency that could be extracted.
According to both criteria above, the search results were evaluated by 2 reviewers (WZ and LYF), and any dispute was resolved through discussion with a third reviewer (ZYH).
2.3. Data extraction
Two reviewers (WZ and LYF) independently extracted the following data from the included studies: the first author, the year of publication, ethnicity (Asians, Caucasians, and Brazilian), periodontitis type (CP, AgP), smoking status (nonsmokers, mixed), the study design, and genotyping type, genotyping method, frequency of alleles or genotypes in cases and controls. Any disagreements were resolved by a third reviewer (ZYH).
This meta-analysis collected data that are presented in the tables. In our study, allele model (A vs C), heterozygote model (AC vs CC), homozygote model (AA vs CC), dominant model (AA + AC vs CC), and recessive model (AA vs AC + CC) were established to explore the relationship between IL-10–592A/C gene polymorphism and periodontal disease.
2.4. Quality score assessment and Hardy-Weinberg equilibrium
The Newcastle–Ottawa scale was used to assess the quality of included studies by 2 researchers (QYQ and LYF). In accordance with the scale, study selection, comparability, and outcome were used to assess the methodological quality of the included studies with a maximum of 9 points. The included studies were categorized into poor (scored 0–3), fair (scored 3–6), and good (scored 7–9) study quality groups.
The deviation from HWE was tested by comparing genotype distributions in control groups by Chi-Squared statistic, and P < .05 was considered a significant departure from HWE.
2.5. Statistical analysis
Statistical analyses were performed by using Stata software (Version 12.0; Stata Corp., College Station, TX). The odds ratio (OR) value and the 95% confidence interval (CI) of each study were calculated to assess the strength of the association between rs1800872 and periodontitis.
The statistical heterogeneity was tested by I2 statistics, and values of 25%, 50%, and 75% indicated low, moderate, and high heterogeneity, respectively. A fixed-effect model was pooled to estimate the OR and 95%CI when heterogeneity was low (I2 ≤ 50%), while the random effect model was used when heterogeneity was high (I2 > 50%). Sensitivity analysis (Fig. S1) was conducted to analyze the stability of the pooled results. Publication bias in each model was evaluated by the Egger test. Subgroup analysis was performed to reveal the association among characteristics of the studies and the value of the overall OR and 95%CI, and P < .05 was considered statistically significant.
3. Results
3.1. Study selection and characteristics
A total of 420 published articles were identified from 4 databases, and 2 articles were found by hand research. The flow diagram of the search process is shown in Figure 1. Of the 422 articles, 79 articles were excluded as a result of duplication, and 294 articles were excluded because they were not relevant to our study. After reading the full text and assessing the eligibility, 23 articles that met all of the inclusion criteria were pooled in the meta-analysis, 20 of which were in English[18–35] and 3[36–38] in Chinese. In total, 23 studies containing 2714 patients and 2373 controls investigating the association between rs1800872 and periodontitis were included in the meta-analysis.
Figure 1.
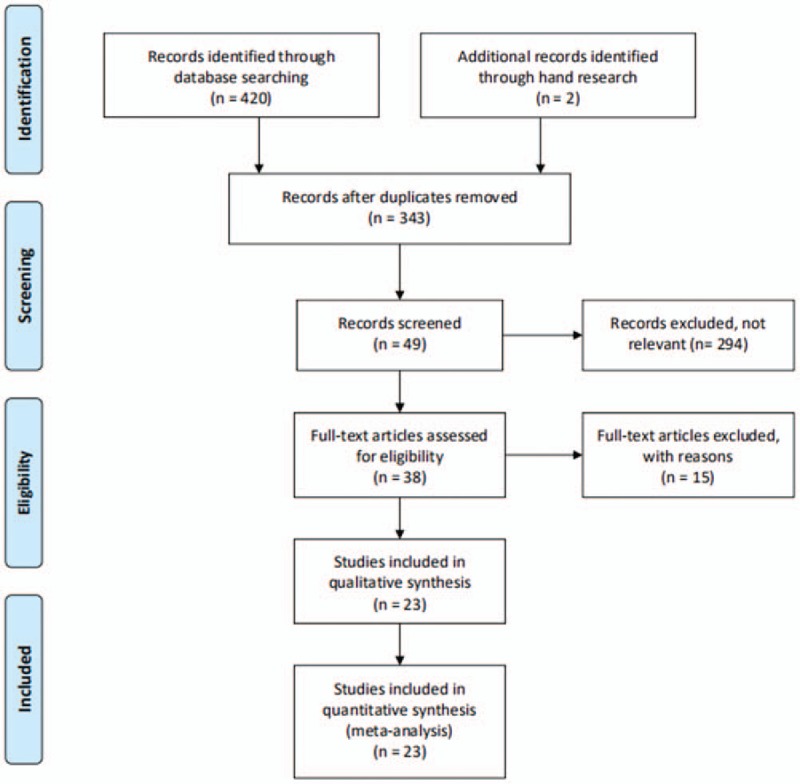
Flow diagram of the identification of eligible studies.
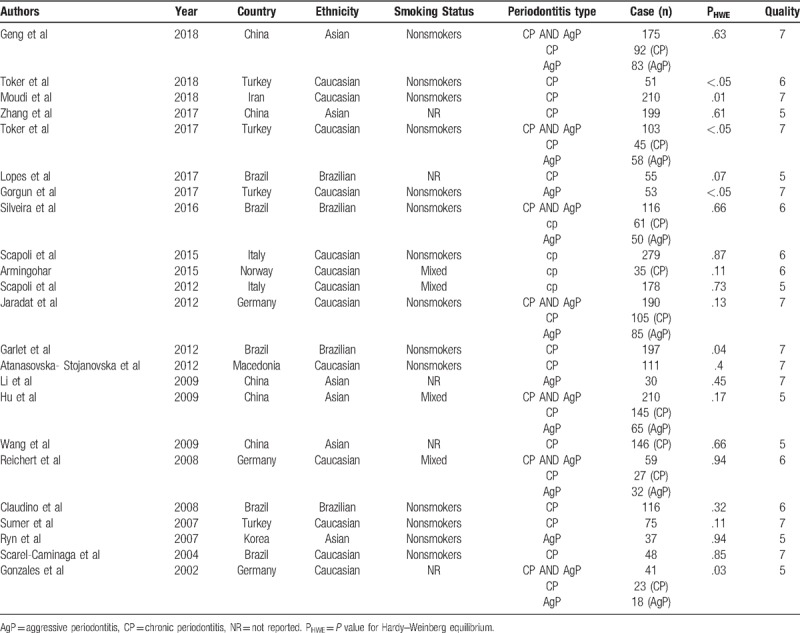
The publication dates of the 23 included studies ranged from 2002 to 2018; 2714 periodontitis patients and 2373 periodontitis-free control subjects from 9 different countries were studied. Six of the included studies reported on Asians,[23,29,31,36–38] 13 studies reported on Caucasians,[15,17,18,19,21,25–28,31,33–35] and the last 4 studies reported on Brazilians.[20,22,24,32] In terms of the type of periodontitis, 13 studies focused on chronic periodontitis (CP),[18,20–22,24,26,27,31,33–35,37,38] 3 studies focused on aggressive periodontitis (AgP),[28,31,36] and 7 studies focused on both CP and AgP.[15,17,19,23,25,29,32] The subjects of 14 studies were nonsmoker,[15,18,19,21,22,24,26–30,31–33] 4 studies included both smokers and nonsmoker (mixed group),[23,25,34,35] and 5 studies did not provide this information.[17,20,36–38] In the 23 included studies, 17 studies were in accordance with HWE for the genotype distribution,[20,21,23–27,36] whereas 6 represented a significant departure from HWE.[17–19,22,28,30] The characteristics of the included studies and patients are summarized in Table 1.
Table 1.
Characteristics of studies included in the meta-analysis.
3.2. Study quality assessment
Twenty three included studies had a quality score ≥5 (moderate–high quality), of which 9 studies were considered high quality and 14 studies were considered moderate quality, as shown in Table 1 and Table S2. Discrepancies between the 2 investigators (QYQ and LYF) were resolved by discussion to reach a consensus.
3.3. Meta-analysis results
3.3.1. Overall OR and 95% CI
The search yielded a total of 23 studies to assess the association between rs1800872 and the risk of periodontitis. The pooled results revealed significant differences in the relationship between the rs1800872 and periodontitis susceptibility, as shown in Table 2 and Figures 2 to 5 (A vs C: OR = 1.28, 95%CI = 1.11–1.49, P = .00, I2 = 56.87%; AA vs CC: OR = 2.06, 95%CI = 1.32–3.23, P = .00, I2 = 73.3%; AA + AC vs CC: OR = 1.42, 95%CI = 1.03–1.96, P = .03, I2 = 76.2%; AA vs AC + CC: OR = 1.78, 95%CI = 1.26–2.56, P = .00, I2 = 76.7%; Table 2), while only the heterozygote model did not show statistical significance (AC vs CC: OR = 1.29, 95%CI = .93–1.77, P = .13, I2 = 73.6%; Table 2). Meta-analyses showed that rs1800872 was associated with increased risks of periodontitis.
Table 2.
The association between rs1800872 and periodontitis.
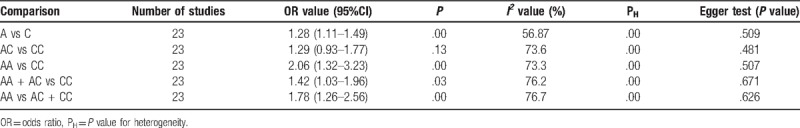
Figure 2.
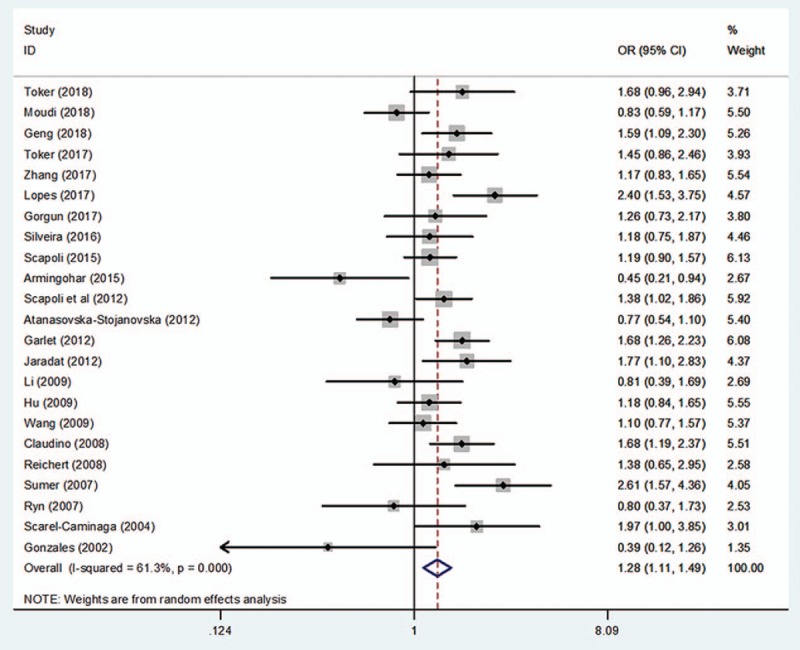
Forest plot of rs1800872 and periodontitis susceptibility in the allelic model (A vs C). CI = confidence interval, OR = odds ratio.
Figure 5.
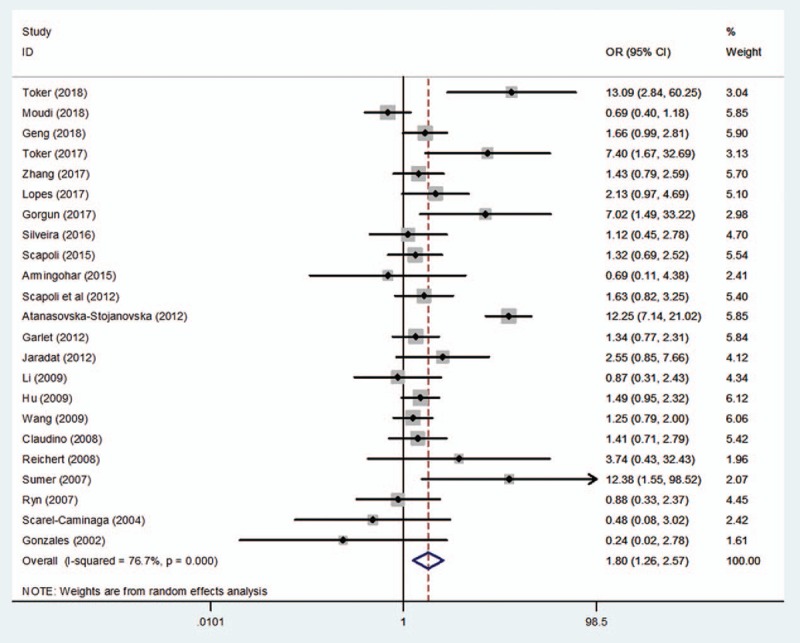
Forest plot of rs1800872 and periodontitis susceptibility in the recessive model (AA vs AC + CC). CI = confidence interval, OR = odds ratio.
Figure 3.
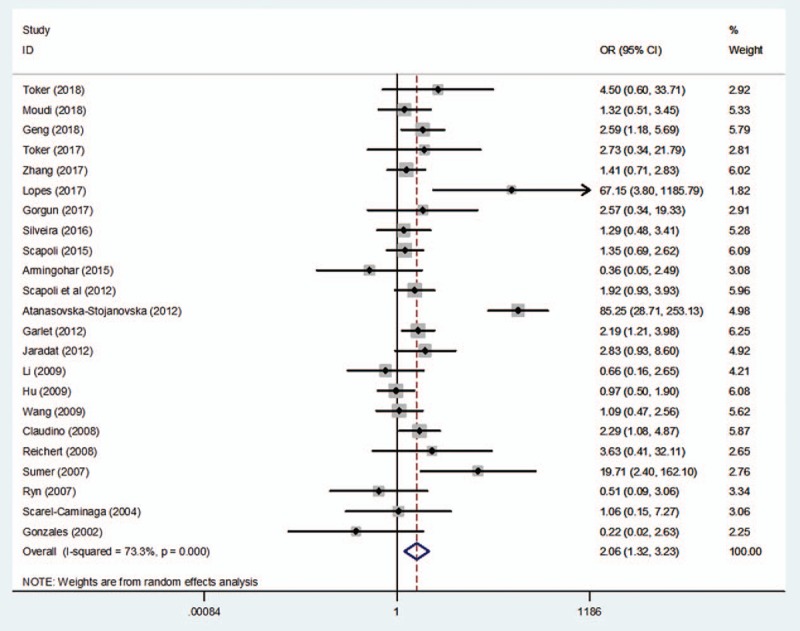
Forest plot of rs1800872 and periodontitis susceptibility in the homozygote model (AA vs CC). CI = confidence interval, OR = odds ratio.
Figure 4.
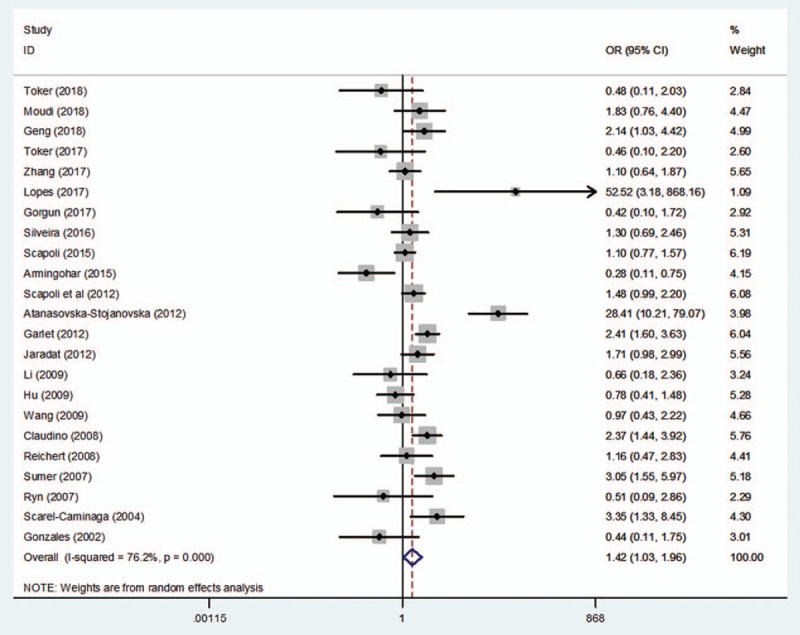
Forest plot of rs1800872 and periodontitis susceptibility in the dominant model (AA + AC vs CC). CI = confidence interval, OR = odds ratio.
3.3.2. Publication bias and sensitivity analysis
Moreover, no significant publication bias was identified in all comparisons by performing the Egger test. In the sensitivity analysis, the influence of each individual study on the pooled OR was assessed by sequentially removing one study each time in each genetic model. The results revealed that statistical heterogeneity and the overall OR did not change significantly.
3.3.3. Subgroup analysis
The meta-analysis results are summarized in Table 3. To assess the potential effect of ethnicity, and type of periodontitis on the genotype and allelic distributions, 3 subgroups analyses were conducted for further evaluating the overall OR value and P value. In the subgroup analysis based on ethnicity, significant associations between the rs1800872 and periodontitis susceptibility can be observed in Caucasians in the homozygote model and recessive model (AA vs CC: OR = 2.65, 95%CI = 1.14–6.17, P = .02; AA vs AC + CC: OR = 2.54, 95%CI = 1.19–5.44, P = .02; Table 3) and in the allelic model and recessive model in Asians (A vs C: OR = 1.18, 95%CI = 1.00–1.40, P < .05; AA vs AC + CC: OR = 1.37, 95%CI = 1.08–1.73, P = .01). Each genetic model in the Brazilian subgroup was statistically significant (A vs C: OR = 1.69, 95%CI = 1.41–2.02, P = .00; AC vs CC: OR = 2.30, 95%CI = 1.31–4.06, P = .00; AA vs CC: OR = 2.36, 95%CI = 1.13–4.92, P = .02; AA + AC vs CC: OR = 2.26, 95%CI = 1.29–3.94, P = .00; AA vs AC + CC: OR = 1.44, 95% = 1.01–2.03, P = .04; Table 3).
Table 3.
Statistics for subgroup analysis.
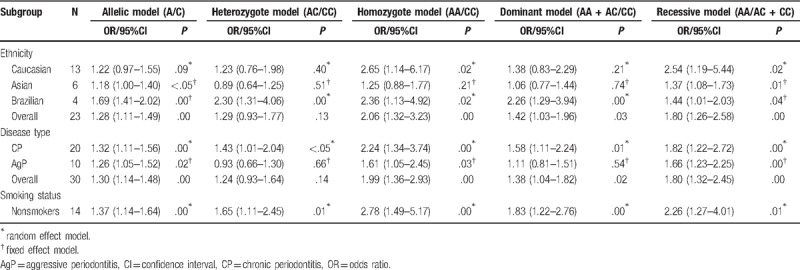
In the subgroup analyses categorized by the type of periodontitis, the pooled results presented significant differences in the allelic model, homozygote model, dominant model, and recessive model (A vs C: OR = 1.30, 95%CI = 1.14–1.48, P = .00; AA vs CC: OR = 1.99, 95%CI = 1.36–2.93, P = .00; AA + AC vs CC: OR = 1.38, 95%CI = 1.04–1.82, P = .02; AA vs AC + CC: OR = 1.80, 95%CI = 1.32–2.45, P = .00; Table 3) but not the heterozygote model (AC vs CC: OR = 1.24, 95%CI = 0.93–1.64, P = .14; Table 3). Interestingly, the data indicated that each model in the CP subgroup had significant differences (A vs C: OR = 1.32, 95%CI = 1.11–1.56, P = .00; AC vs CC: OR = 1.43, 95%CI = 1.01–2.04, P < .05; AA vs CC: OR = 2.24, 95%CI = 1.34–3.74, P = .00; AA + AC vs CC: OR = 1.58, 95%CI = 1.11–2.24, P = .01; AA vs AC + CC: OR = 1.82, 95%CI = 1.22–2.72, P = .00; Table 3). However, in the AgP subgroup, the allelic model, homozygote model and recessive model conveyed significant differences (A vs C: OR = 1.26, 95%CI = 1.05–1.52, P = .02; AA vs CC: OR = 1.61, 95%CI = 1.05–2.45, P = .03; AA vs AC + CC: OR = 1.66, 95%CI = 1.23–2.25, P = .00; Table 3).
In terms of smoking status, all genetic models showed significant differences in the nonsmoker subgroup between rs1800872 and periodontitis susceptibility (A vs C: OR = 1.37, 95%CI = 1.14–1.64, P = .00; AC vs CC: OR = 1.65, 95%CI = 1.11–2.45, P = .01; AA vs CC: OR = 2.78, 95%CI = 1.49–5.17, P = .00; AA+AC vs CC: OR = 1.83, 95%CI = 1.22–2.76, P = .00; AA vs AC+CC: OR = 2.26, 95%CI = 1.27–4.01, P = .01; Table 3). The AA genotype carriers more frequently suffered from periodontitis (AA vs CC: OR = 2.78, 95%CI = 1.49–5.17, P = .00; Table 3).
4. Discussion
Periodontitis develops as a result of imbalance in the oral microbiota, leading to an immune response of the host, thus carrying enormous influence on periodontium protection and support. IL-10, a factor produced by Th2 cells, inhibits the production of cytokines by Th1 cells. IL-10 can protect against bone resorption and attenuate the inflammatory reaction by affecting the bone protection factor system and can directly promote cytokine synthesis, such as IL-1, IL-6, TNF-α, gelatinase, TIMPs and OPG, and collagenase.[39–41] Therefore, individuals who are high producers of IL-10 are most likely protected against PD due to the anti-inflammatory cytokines that negatively regulate the immune response against periodontopathogenic bacteria. Additionally, some studies found that the interruption of IL-10 would result in accelerating alveolar bone absorption and decreasing bone formation.[42,43] More importantly, some studies demonstrated that polymorphism of IL-10 gene promoter were involved in the development of periodontal diseases. The specific genotype (-592AA) with low IL-10 expression might aggravate the inflammatory response.[44] Claudino et al[25] stated that the IL-10 promoter -592A/C single nucleotide polymorphism is associated with a decrease in IL-10 production. Hence, understanding the biological role of IL-10 and its gene polymorphism is necessary for further therapeutic approaches against periodontitis.
Discrepancy was also evident for rs1800872 in the included studies. A study by Summer et al[26] confirmed that a statistically significant difference in the frequencies of genotypes (AA vs CC + CA: OR = 12.37, 95%CI = 2.74–7.77, P < .05; CA + AA vs CC: OR = 3.05, 95%CI = 1.47–6.33, P < .05) and alleles (A vs C: OR = 2.61, 95%CI = 1.52–4.51, P < .05) at position -592C to A between CP patients and healthy controls. AA homozygosity was also found to be an increased risk for CP by others.[15,24,27] In contrast, Gonzales et al[17] reported that IL-10 loci were not associated with CP or AgP. Nevertheless, supporting Gonzales et al,[17] Reichert et al,[25] Atanasovska-Stojanovska et al[25] and Brett et al,[44] respectively, researched the German, Macedonian, and English populations, and they found no association between CP and IL-10–592A/C gene polymorphism. However, in this meta-analysis, the pooled data of the studies on periodontitis demonstrated significant relationships in 4 genetic models (A/C; AA/CC; AA +AC/CC; AA/AC+CC; Table 2; Figs. 2–5), except the heterozygote model (AC vs CC: OR = 1.29, 95%CI = 0.93–1.77, P = .13, I2 = 73.6%; Table 2). In addition, Jaradat et al[15] and Li et al[36] indicated that no significant differences were observed in the genotype frequencies between AgP patients and controls. However, as with Summer et al,[26] our investigation also found significant differences between AgP and rs1800872, and discovered that the AA carriers appeared to have the highest risk for AgP (AA vs AC+CC: OR = 1.66, 95%CI = 1.23–2.25, P = .00; Table 3).
Additionally, the sensitivity analysis performed by sequentially removing individual studies did not alter the high heterogeneity, which could be caused by many factors, such as race, sample sizes, smoking habits, and deviations of allele distributions from the HWE. Although we detected high heterogeneity in our meta-analysis, we decided not to eliminate more studies because our database had a limited number of included studies; if we excluded some of them, it would not be possible to establish a strong meta-analysis. We decided to generate subgroups by ethnicity, periodontitis type and smoking status to lower the statistical heterogeneity, and to discuss the results. No significant publication bias was identified using the Egger regression method.
To evaluate the effect of ethnicity, the included studies were stratified into Caucasian, Asian, and Brazilian groups that consisted of 13, 6 and 4 studies, respectively. The pooled results demonstrated that there was statistical significance in Caucasians in 2 genetic models, namely, the homozygote model and recessive model (AA vs CC: OR = 2.65, 95%CI = 1.14–6.17, P = .02; AA vs AC + CC: OR = 2.54, 95%CI = 1.19–5.44, P = .02; Table 3), but not in the allelic model, heterozygote model and dominant model (A vs C: OR = OR = 1.22, 95%CI = 0.97–1.55, P = .09; AC vs CC: OR = 1.23, 95%CI = 0.76–1.98, P = .40; AA + AC vs CC: OR = 1.38, 95%CI = 0.83–2.29, P = .21; Table 3). The Brazilian group consists of Europeans, Africans, Amerindians and unknown races; due to the complexity of their genetic backgrounds, we believe the evidence and pooled result in this group is controversial. More importantly, the homozygote model appeared to be the highest risk in Caucasians (AA vs CC: OR = 2.65, 95%CI = 1.14–6.17, P = .02; Table 3) and Brazilians (AA vs CC: OR = 2.36, 95%CI = 1.13–4.92, P = .02; Table 3). In comparison, the highest risk in Asians is the recessive model (AA vs AC + CC: OR = 1.37, 95%CI = 1.08–1.73, P = .01). Gorgun et al[46] reached a similar conclusion that the AA genotype carriers were more susceptible to aggressive periodontitis in Caucasians, and neither age nor gender had effects on genotype diversity, while Jaradat et al[15] reached the opposite conclusion in Caucasians. Nevertheless, the highest risk in Asians is the recessive model (AA vs AC + CC: OR = 1.37, 95%CI = 1.08–1.73, P = .01). Geng et al[29] also found that IL-10–592 AA occurred more frequently in patients with chronic periodontitis than in healthy controls, but no statistically significant difference in the distribution of interleukin-10 polymorphisms at -592 was found between generalized aggressive periodontitis and healthy control groups in Asians.[23,29,36]
We divided the included studies into CP and AgP subgroups that consisted of 20 and 10 studies, respectively. As is known to us, the CP and AgP are different from the rate and mechanism of the periodontal attachment loss and bone destruction, so it is necessary to conduct this subgroup analysis. There were obvious significant differences between the rs1800872 with CP in each model (A/C; AC/CC; AA/CC; AA + AC/CC; AA/AC + CC; Table 3). However, some studies were in disagreement with our results, which found no association between the -592 A/C allele and CP.[47] Furthermore, in this AgP subgroup, only the allelic model, homozygote model and recessive model (A/C; AA/CC; AA/AC+CC; Table 3) conveyed significant differences. The previous study performed by Toker et al[18] supported the conclusion that IL-10 -592 AA genotype appeared to be an increased risk factor for chronic periodontitis. Contrary to our results, Gonzales et al[17] insisted that no statistical significance was found in IL-10–592 homozygous positive individuals between controls and AgP. As with our finding, Hu et al[23] found no association between IL-10 AA genotype and AgP either. In summary, further in-depth studies should be conducted, and many more cases should be collected to explore these controversial problems comprehensively.
Some studies had demonstrated smoking was an important risk factor, which could increase the occurrence of periodontitis, directly, or indirectly.[48,49] In the smoking status subgroup, there was no significantly different association between rs1800872 and periodontitis susceptibility in the mixed group (smokers and nonsmokers), which might be related with the small sample size and selection bias leading to the lower power of the meta-analysis. Therefore, the study of periodontitis and rs1800872 should provide more reliable and robust evidence to demonstrate whether smoking was a critical risk factor. However, this finding should be interpreted with caution in the nonsmoker subgroup; all comparison models showed significant association between rs1800872 and periodontitis susceptibility, in which the homozygote model had the highest risk (AA vs CC: OR = 2.78, 95%CI = 1.49–5.17, P = .00; Table 3).
In some related studies, possibilities existed that some positive results might be hidden and some negative findings might be a consequence of low statistical power. It could be due to their small sample size or methodological shortcomings, such as the selection of an appropriate case group or control group. It is necessary to detect rs1800872 polymorphisms in our meta-analysis in well-defined subgroups of phenotypes with larger number of participants in order to reach more accurate conclusions regarding the genetic background for the development of CP and AgP.
To our knowledge, this is the first meta-analysis conducted to assess the association between rs1800872 and periodontitis susceptibility with 23 studies by subgroup analysis based on ethnicity, periodontitis type, and smoking status, independently. However, our meta-analysis has 4 limitations. First, heterogeneity among the included studies was high, both in the overall effect analysis and the subgroup analyses, which might affect the stability of the pooled results. Second, the search strategy was confined to English language and Chinese language studies, which might have caused a selection bias into this meta-analysis. Third, due to the limited information provided by the included studies, we did not explore other factors, such as the dental clinical parameters related with bleeding on probing, probing depth and clinical attachment loss that could distort the pooled results. Moreover, the environmental factors and others genetic variations might influence the results. Fourth, the number of eligible studies and patients was limited in the overall effect analysis and the subgroup analyses; therefore, more high-quality and well-designed studies are needed to further analyze the association between rs1800872 and periodontitis susceptibility, especially in the smoking population.
5. Conclusion
In summary, our meta-analysis identified a significant different association between rs1800872 and periodontitis susceptibility, which could help to understand properly the complex causal mechanisms of PD. In IL-10 rs1800872 polymorphism, A allele, AA genotype may confer an increased risk for the PD, while CC genotype may be a decreased risk for PD susceptibility. Moreover, AA genotype may be a high risk factor for PD in Caucasians, while A allele, AA genotype may be a relative risk factor for PD in Asians. Besides, A allele, AA genotype, CC genotype may be closely associated with CP, while A allele, AA genotype may be closely associated with AgP. Additionally, A allele, AA genotype, CC genotype may be an increased risk for the PD in nonsmokers. Hence, IL-10 rs1800872 polymorphism may be appropriate gene markers for gene screening.
Acknowledgments
The authors acknowledge that Xiaolong Guo and Quan Shi provided many useful suggestions for this article and thank them for their effort and help.
Author contributions
Data curation: Yafang Li.
Formal analysis: Yafang Li, Yanheng Zhou, Yiqiang Qiao.
Funding acquisition: Yiqiang Qiao.
Software: Zao Wang.
Supervision: Yanheng Zhou, Yiqiang Qiao.
Writing – original draft: Zao Wang.
Writing – review & editing: Yafang Li, Yanheng Zhou, Yiqiang Qiao.
Supplementary Material
Footnotes
Abbreviations: AgP = aggressive periodontitis, CI = confidence interval, CP = chronic periodontitis, HWE = Hardy–Weinberg equilibrium, OR = odds ratio, SNP = single nucleotide polymorphism.
How to cite this article: Wang Z, Li Y, Zhou Y, Qiao Y. Association between the IL-10 rs1800872 polymorphisms and periodontitis susceptibility. Medicine. 2019;98:40(e17113).
The datasets supporting the conclusions of this article are included within the article.
This study was supported in part by grants from the National Natural Science Foundation of China (U1704187).
The authors have no funding and conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Cavalla F, Biguetti CC, Colavite PM, et al. TBX21-1993T/C (rs4794067) polymorphism is associated with increased risk of chronic periodontitis and increased T-bet expression in periodontal lesions, but does not significantly impact the IFN-g transcriptional level or the pattern of periodontophatic bacterial infection. Virulence 2015;6:293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Highfield J. Diagnosis and classification of periodontal disease. Aust Dent J 2009;54Suppl 1:S11–26. [DOI] [PubMed] [Google Scholar]
- [3].Chapple ILC, Genco R. Diabetes and periodontal diseases: consensus report of the Joint EFP/AAP Workshop on Periodontitis and Systemic Diseases. J Periodontol 2013;84:S106–12. [DOI] [PubMed] [Google Scholar]
- [4].Miller CS, King CJ, Langub MC, et al. Salivary biomarkers of existing periodontal disease: a cross-sectional study. J Am Dent Assoc 2006;137:322–9. [DOI] [PubMed] [Google Scholar]
- [5].Gumus P, Nizam N, Nalbantsoy A, et al. Saliva and serum levels of pentraxin-3 and interleukin-1beta in generalized aggressive or chronic periodontitis. J Periodontol 2014;85:e40–6. [DOI] [PubMed] [Google Scholar]
- [6].Gumus P, Nizam N, Nalbantsoy A, et al. Saliva, serum levels of interleukin-21, -33 and prostaglandin E2 in patients with generalised aggressive or chronic periodontitis. Oral Health Prev Dent 2017;15:385–90. [DOI] [PubMed] [Google Scholar]
- [7].Duarte PM, Bastos MF, Fermiano D, et al. Do subjects with aggressive and chronic periodontitis exhibit a different cytokine/chemokine profile in the gingival crevicular fluid? A systematic review. J Periodontal Res 2015;50:18–27. [DOI] [PubMed] [Google Scholar]
- [8].Nibali L, Farias BC, Vajgel A, et al. Tooth loss in aggressive periodontitis: a systematic review. J Dent Res 2013;92:868–75. [DOI] [PubMed] [Google Scholar]
- [9].Mannino MH, Zhu Z, Xiao H, et al. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett 2015;367:103–7. [DOI] [PubMed] [Google Scholar]
- [10].Maynard CL, Weaver CT. Diversity in the contribution of interleukin-10 to T-cell-mediated immune regulation. Immunol Rev 2008;226:219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Al-Rasheed A, Scheerens H, Rennick DM, et al. Accelerated alveolar bone loss in mice lacking interleukin-10. J Dent Res 2003;82:632–5. [DOI] [PubMed] [Google Scholar]
- [12].Iyer SS, Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol 2012;32:23–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Javed F, Al-Askar M, Al-Hezaimi K. Cytokine profile in the gingival crevicular fluid of periodontitis patients with and without type 2 diabetes: a literature review. J Periodontol 2012;83:156–61. [DOI] [PubMed] [Google Scholar]
- [14].Turner DM, Williams DM, Sankaran D, et al. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 1997;24:1–8. [DOI] [PubMed] [Google Scholar]
- [15].Jaradat SM, Ababneh KT, Jaradat SA, et al. Association of interleukin-10 gene promoter polymorphisms with chronic and aggressive periodontitis. Oral Dis 2012;18:271–9. [DOI] [PubMed] [Google Scholar]
- [16].Babel N, Cherepnev G, Babel D, et al. Analysis of tumor necrosis factor-alpha, transforming growth factor-beta, interleukin-10, IL-6, and interferon-gamma gene polymorphisms in patients with chronic periodontitis. J Periodontol 2006;77:1978–83. [DOI] [PubMed] [Google Scholar]
- [17].Gonzales JR, Michel J, Diete A, et al. Analysis of genetic polymorphisms at the interleukin-10 loci in aggressive and chronic periodontitis. J Clin Periodontol 2002;29:816–22. [DOI] [PubMed] [Google Scholar]
- [18].Toker H, Gorgun EP, Korkmaz EM, et al. The effects of IL-10 gene polymorphism on serum, and gingival crevicular fluid levels of IL-6 and IL-10 in chronic periodontitis. J Appl Oral Sci 2018;26:e20170232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Toker H, Gorgun EP, Korkmaz EM. Analysis of IL-6, IL-10 and NF-kappaB gene polymorphisms in aggressive and chronic periodontitis. Cent Eur J Public Health 2017;25:157–62. [DOI] [PubMed] [Google Scholar]
- [20].Lopes CB, Barroso R, Burbano R, et al. Effect of ancestry on interleukin-10 haplotypes in chronic periodontitis. Front Biosci (Elite Ed) 2017;9:276–85. [DOI] [PubMed] [Google Scholar]
- [21].Atanasovska-Stojanovska A, Trajkov D, Popovska M, et al. IL10-1082, IL10-819 and IL10-592 polymorphisms are associated with chronic periodontitis in a Macedonian population. Hum Immunol 2012;73:753–8. [DOI] [PubMed] [Google Scholar]
- [22].Garlet GP, Trombone AP, Menezes R, et al. The use of chronic gingivitis as reference status increases the power and odds of periodontitis genetic studies: a proposal based in the exposure concept and clearer resistance and susceptibility phenotypes definition. J Clin Periodontol 2012;39:323–32. [DOI] [PubMed] [Google Scholar]
- [23].Hu KF, Huang KC, Ho YP, et al. Interleukin-10 (-592 C/A) and interleukin-12B (+16974 A/C) gene polymorphisms and the interleukin-10 ATA haplotype are associated with periodontitis in a Taiwanese population. J Periodontal Res 2009;44:378–85. [DOI] [PubMed] [Google Scholar]
- [24].Claudino M, Trombone AP, Cardoso CR, et al. The broad effects of the functional IL-10 promoter-592 polymorphism: modulation of IL-10, TIMP-3, and OPG expression and their association with periodontal disease outcome. J Leukoc Biol 2008;84:1565–73. [DOI] [PubMed] [Google Scholar]
- [25].Reichert S, Machulla HK, Klapproth J, et al. The interleukin-10 promoter haplotype ATA is a putative risk factor for aggressive periodontitis. J Periodontal Res 2008;43:40–7. [DOI] [PubMed] [Google Scholar]
- [26].Sumer AP, Kara N, Keles GC, et al. Association of interleukin-10 gene polymorphisms with severe generalized chronic periodontitis. J Periodontol 2007;78:493–7. [DOI] [PubMed] [Google Scholar]
- [27].Scarel-Caminaga RM, Trevilatto PC, Souza AP, et al. Interleukin 10 gene promoter polymorphisms are associated with chronic periodontitis. J Clin Periodontol 2004;31:443–8. [DOI] [PubMed] [Google Scholar]
- [28].Gorgun EP, Toker H, Korkmaz EM, et al. IL-6 and IL-10 gene polymorphisms in patients with aggressive periodontitis: effects on GCF, serum and clinic parameters. Braz Oral Res 2017;31:e12. [DOI] [PubMed] [Google Scholar]
- [29].Geng Y, Li L, Wang X, et al. Interleukin-10 polymorphisms affect the key periodontal pathogens in Chinese periodontitis patients. Sci Rep 2018;8:9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Moudi B, Heidari Z, Mahmoudzadeh-Sagheb H, et al. Analysis of interleukin-10 gene polymorphisms in patients with chronic periodontitis and healthy controls. Dent Res J (Isfahan) 2018;15:71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ji-Sun Ryn OK. Patients, IL- gene promoter polymorphisms IL-10 gene promoter polymorphisms in Korean gener_x0002_alized aggressive periodontitis patients. J Period Impl Sci 2007;37:563–73. [Google Scholar]
- [32].Silveira VR, Pigossi SC, Scarel-Caminaga RM, et al. Analysis of polymorphisms in Interleukin 10, NOS2A, and ESR2 genes in chronic and aggressive periodontitis. Braz Oral Res 2016;30:e105. [DOI] [PubMed] [Google Scholar]
- [33].Scapoli L, Girardi A, Palmieri A, et al. Interleukin-6 gene polymorphism modulates the risk of periodontal diseases. J Biol Regul Homeost Agents 2015;29:111–6. [PubMed] [Google Scholar]
- [34].Scapoli L, Girardi A, Palmieri A, et al. IL6 and IL10 are genetic susceptibility factors of periodontal disease. Dent Res J (Isfahan) 2012;9:S197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Armingohar Z, Jorgensen JJ, Kristoffersen AK, et al. Polymorphisms in the interleukin-10 gene and chronic periodontitis in patients with atherosclerotic and aortic aneurysmal vascular diseases. J Oral Microbiol 2015;7:26051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li Y, Zhao H. Interleukin-10 gene promoter polymorphism in chinese patients with generalized aggressive periodontitis. J Dental Prev Treat 2009;10:472–5. [Google Scholar]
- [37].Wang C. Investigations on the gene polymorphisms of vitamin D receptor and IL-10 as the risk factors for chronic periodontitis and type 2 diabetes mellitus. (Doctoral Dissertation). 2009. [Google Scholar]
- [38].Zhang Yuhui HPLJ. Correlation between interleukin-10 polymorphisms and susceptibility to chronic periodontitis among Uygur adults in the Moyu area. West Chin J Stomatol 2017;35:514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yu P, Hu Y, Liu Z, et al. Local Induction of B cell interleukin-10 competency alleviates inflammation and bone loss in ligature-induced experimental periodontitis in mice. Infect Immun 2017;85: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang Q, Chen B, Yan F, et al. Interleukin-10 inhibits bone resorption: a potential therapeutic strategy in periodontitis and other bone loss diseases. Biomed Res Int 2014;2014:284836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang X, Teng YT. Interleukin-10 inhibits gram-negative-microbe-specific human receptor activator of NF-kappaB ligand-positive CD4+-Th1-cell-associated alveolar bone loss in vivo. Infect Immun 2006;74:4927–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Claudino M, Garlet TP, Cardoso CR, et al. Down-regulation of expression of osteoblast and osteocyte markers in periodontal tissues associated with the spontaneous alveolar bone loss of interleukin-10 knockout mice. Eur J Oral Sci 2010;118:19–28. [DOI] [PubMed] [Google Scholar]
- [43].Al-Rasheed A, Scheerens H, Srivastava AK, et al. Accelerated alveolar bone loss in mice lacking interleukin-10: late onset. J Periodontal Res 2004;39:194–8. [DOI] [PubMed] [Google Scholar]
- [44].Luo Y, Gong Y, Yu Y. Interleukin-10 gene promoter polymorphisms are associated with cyclosporin A-induced gingival overgrowth in renal transplant patients. Arch Oral Biol 2013;58:1199–207. [DOI] [PubMed] [Google Scholar]
- [45].Brett PM, Zygogianni P, Griffiths GS, et al. Functional gene polymorphisms in aggressive and chronic periodontitis. J Dent Res 2005;84:1149–53. [DOI] [PubMed] [Google Scholar]
- [46].Pirim GE, Toker H, Korkmaz EM, et al. IL-6 and IL-10 gene polymorphisms in patients with aggressive periodontitis: effects on GCF, serum and clinic parameters. Braz Oral Res 2017;31:e12. [DOI] [PubMed] [Google Scholar]
- [47].Yamazaki K, Tabeta K, Nakajima T, et al. Interleukin-10 gene promoter polymorphism in Japanese patients with adult and early-onset periodontitis. J Clin Periodontol 2001;28:828–32. [DOI] [PubMed] [Google Scholar]
- [48].Ribeiro FV, Pino DS, Franck FC, et al. Resveratrol inhibits periodontitis-related bone loss in rats subjected to cigarette smoke inhalation. J Periodontol 2017;88:788–98. [DOI] [PubMed] [Google Scholar]
- [49].Fokkema SJ. Peripheral blood monocyte responses in periodontitis. Int J Dent Hyg 2012;10:229–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


