Abstract
The zinc-dependent UDP-3-O-acyl-N-acetylglucosamine deacetylase (LpxC) catalyzes the first committed step in the biosynthesis of lipid A, the hydrophobic anchor of lipopolysaccharide (LPS) that constitutes the outermost monolayer of Gram- negative bacteria. As LpxC is crucial for the survival of Gram-negative organisms and has no sequence homology to known mammalian deacetylases or amidases, it is an excellent target for the design of new antibiotics. The solution structure of LpxC from Aquifex aeolicus in complex with a substrate-analog inhibitor, TU-514, reveals a novel α/β fold, a unique zinc-binding motif and a hydrophobic passage that captures the acyl chain of the inhibitor. On the basis of biochemical and structural studies, we propose a catalytic mechanism for LpxC, suggest a model for substrate binding and provide evidence that mobility and dynamics in structural motifs close to the active site have key roles in the capture of the substrate.
Infection by Gram-negative bacteria remains an important public health concern throughout the world, especially with the growing prevalence of antibiotic-resistant Gram-negative strains. The development of new antibiotics against Gram-negative organisms is a major focus of current research, and the identification and characterization of molecular targets within bacteria is essential to this work. Gram-negative bacteria contain a unique structure in the outer monolayers of their outer membranes known as lipopolysaccharide (LPS), which is anchored into the membrane by the hydrophobic moiety lipid A. A minimal structure of lipid A and two 3-deoxy-D-manno-octulosonic acid (Kdo) sugars (Fig. 1) is essential for viability, providing the bacterial cell with crucial protection from external agents such as antibiotics and detergents1. Lipid A is an attractive target for the development of new antibiotics, both because it is required for bacterial survival and because it and its biosynthetic enzymes have no mammalian homologs.
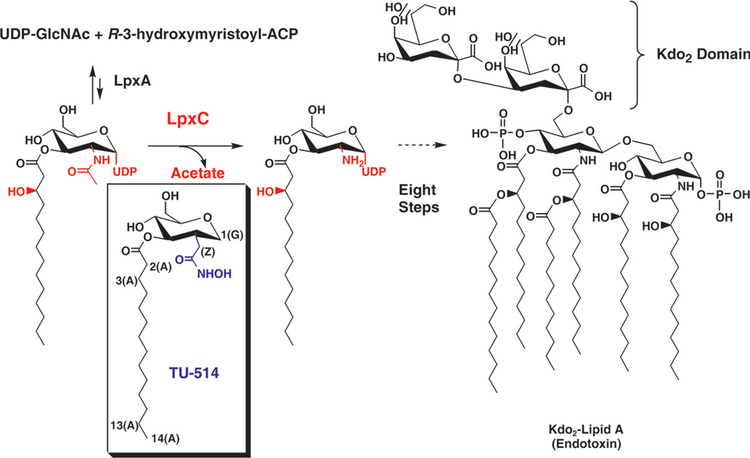
Figure 1.
LpxC reaction and structure of TU-514. Functional groups found on substrate but not on TU-514 are in red; those unique to TU-514 are in blue. Kdo, 3-deoxy-D-manno-octulosonic acid; ACP, acyl carrier protein. Carbons of TU-514 hexose ring are numbered as with an equivalent sugar, from 1 to 6. Protons at position 1 are distinguished between axial (1a) and equatorial (1e). The α-methylene of the hydroxamate group is labeled Z. Acyl chain is numbered from 1 (carbonyl carbon) to 14 (terminal methyl). Atom s in the acyl chain are labeled (A), those from the glucose-like ring (G) and those in the hydroxamate (H).
Lipid A, a hexa-acylated β(1′→6)-linked disaccharide of glucosamine, is synthesized through a nine-step enzymatic process in most Gram-negative bacteria1. The second and committed step in this pathway is catalyzed by the cytoplasmic protein UDP-3-O-acyl-N-acetylglucosamine deacetylase, also known as LpxC (Fig. 1)2–5. LpxC uses a single Zn2+ ion to catalyze deacetylation but is inhibited by excess zinc, suggesting another binding site for a second, inhibitory Zn2+ ion6. LpxC does not share sequence homology with other metalloamidases and seems to contain a unique zinc-binding motif, as shown by site-directed mutagenesis of conserved residues7.
LpxC has attracted attention as a target for the development of antibiotics against Gram-negative bacteria. The first LpxC inhibitors discovered were phenyloxazolines containing a hydroxamate functional group that binds Zn2+ and inhibits zinc-containing enzymes8. One of the most potent inhibitors identified to date, L-161,240, is bactericidal against Escherichia coli with an efficacy comparable to that of ampicillin. It was discovered early on, however, that phenyloxazolines do not inhibit all LpxC orthologs with equal potency. More recent efforts to develop broad-spectrum LpxC inhibitors with antibiotic capabilities have included substrate-analog inhibitors and additional variants on the oxazoline and isoxazoline moieties, but these efforts have not yet produced an antibiotic with potent activity against the complete spectrum of Gram-negative bacteria9,10. The structural characterization of the LpxC enzymes would therefore facilitate further progress toward the development of more effective LpxC inhibitors.
Proteins of the LpxC family share substantial sequence similarity. Among these orthologs, the 32 kDa, 282-residue LpxC enzyme from the hyperthermophilic bacterium A. aeolicus is particularly well suited for NMR structural studies because of its size and thermal stability. TU-514 (1,5-anhydro-2-C-(carboxymethyl-N-hydroxyamide)-2-deoxy-3-O-myristoyl-D-glucitol; Fig. 1) is the most potent inhibitor of the A. aeolicus enzyme reported to date, with an IC50 of 7.0 μM at 30 °C (estimated Ki of 3.9 μM)9,11. Because it is a substrate-analog inhibitor, the binding mode of TU-514 is likely to be shared across the LpxC family. Here we report the solution structure of A. aeolicus LpxC in complex with TU-514.
RESULTS
LpxC contains a novel α/β fold
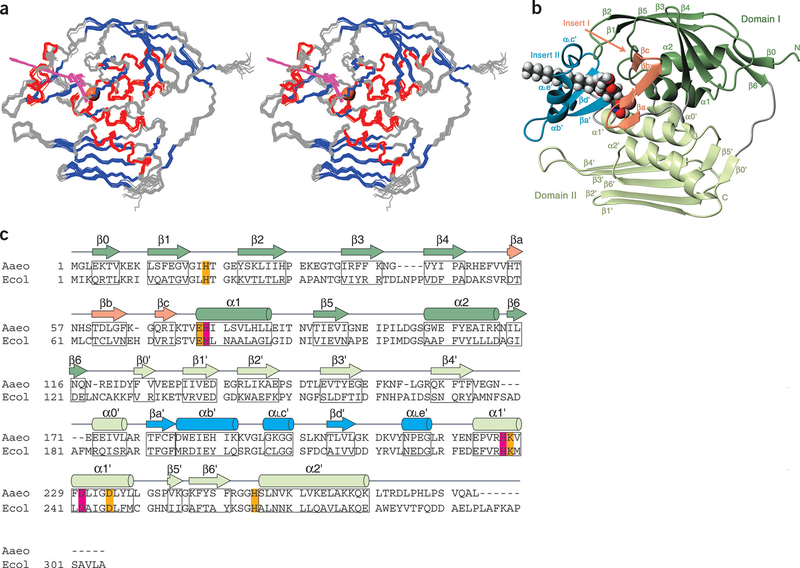
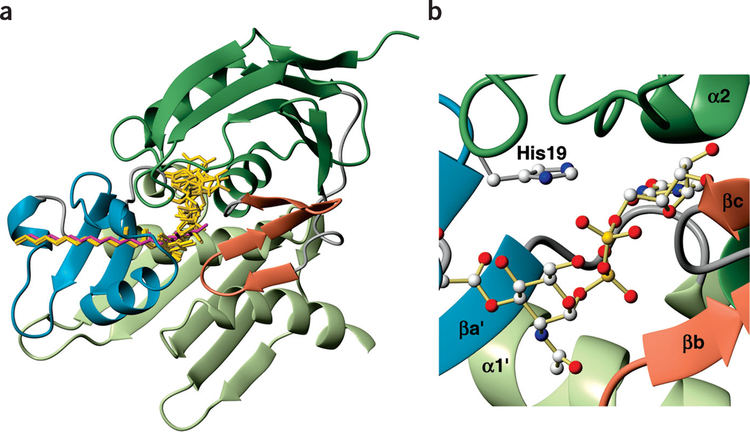
LpxC has two domains with similar folds: each contains two layers of secondary structural elements, with a layer of α-helices packing against a primary β-sheet (Fig. 2a–c). The main β-sheets are each composed of five strands of 4–6 residues, with mixed parallel and antiparallel orientation. The β-sheet of domain I is severely distorted, whereas that of domain II is essentially flat. Domain I features two helices (α1 and α2) with a hydrophobic pocket between them, whereas in domain II each of these helices (α1′ and α2′) crosses three β-strands at a similar angle (60–70°).
Figure 2.
Solution structure of LpxC in complex with TU-514. (a) Stereo view of backbone traces from the 15 final structures of the com plex, colored by secondary structure (α-helices, red; β-strands, blue; loop regions, gray). Residues 271–28 2 are disordered and are not shown. TU-514 is shown in magenta. Zinc ion locations from these structures are superimposed in a space-filling representation (coral). (b) Ribbon representation of structure colored by domain or domain insert. Linkers between domains and between domains and their inserts are colored gray. TU-514 is shown as a space-filling model with CPK coloring (carbon, black; hydrogen, white; oxygen, red; nitrogen, blue); zinc ion is shown as a space-filling model beside it. (c) Sequences of the LpxC enzymes from A. aeolicus and E. coli are aligned, with zinc-coordinating residues in magenta and conserved residues important for catalysis in orange. Secondary structure of A. aeolicus LpxC is indicated above the sequence, colored as in b, and the relevant residues are boxed in. Panels a and b were generated with MolMol45.
In addition to the distinct α-α-β-β-β-β-β structural motif, each domain contains an insert region that differs substantially between the domains. The insert region of domain I features a small, three-stranded antiparallel β-sheet (βa, βb and βc), Whereas the domain II insert features a β-α-αL-β motif. Both inserts approximately perpendicular to the main β-sheets.
The two domains pack against each other such that the β-sheets form the exterior layers of the protein and the α-helices form interior layers constituting the domain interface, with α1′ of domain II intersecting α1 of domain I at ~70°. The gap between the domains is sealed by a short 310-helix (α0′) from domain II, which is positioned next to α1′ and α2, forming a structure resembling a four-helix bundle (comprised of α1, α2, α0′ and α1′). The two insert regions are positioned on the same side of the molecule, together forming the active site, with access to the accessory pocket between the two helices of domain I; the linker between the domains is located on the opposite side of the molecule. Analysis of the LpxC structure using the Dali12 server did not find any known structure with a similar fold.
LpxC captures TU-514 with a hydrophobic passage
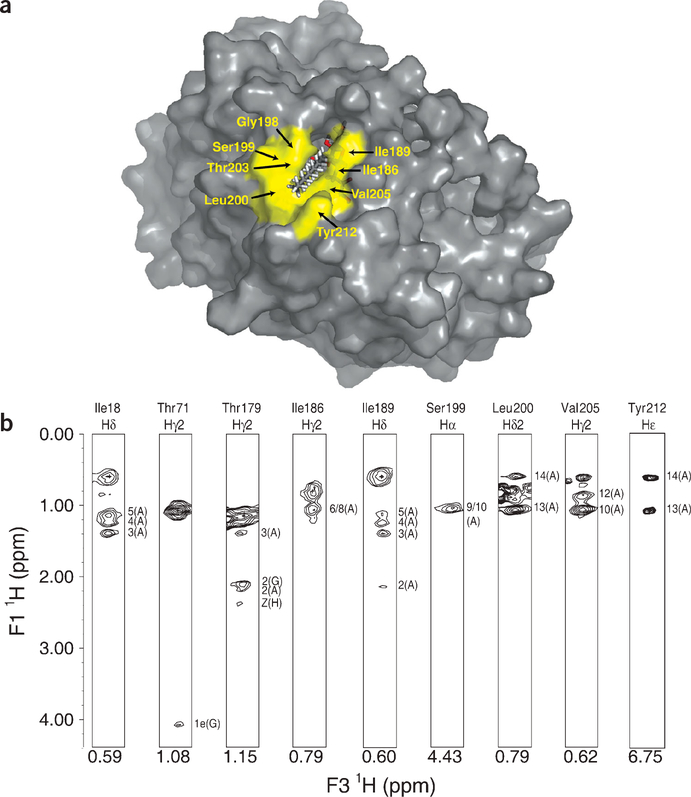
LpxC binds TU-514 via a hydrophobic passage formed by the β-α-αL-β motif of domain II, with the hexose ring buried in the active site cavity and the acyl chain extending out through the passage (Figs. 2b and 3a). The acyl chain is recognized by the hydro-phobic side chains of Thr179, Ile186, Ile189, Leu200, Thr203, Val205 and Tyr212 and by backbone atoms in Gly198 and Ser199. These strong interactions provide an excellent explanation for the ability of LpxC to capture its substrate with high affinity, as well as for its >5 × 106-fold lower activity with substrates lacking an acyl chain6. The positioning of the TU-514 acyl chain within the hydrophobic passage is well determined from numerous intermolecular NOE crosspeaks, some of which are shown in Figure 3b. Although the TU-514 spectrum shows high chemical-shift degeneracy among acyl-chain resonances (methylene groups 6–11), resonances from the terminal methyl, the penultimate methylene and the α-and β-methylenes are well dispersed. The aromatic ring of Tyr212 and a methyl group of Leu200 both form van der Waals contacts with the terminal methyl of the acyl chain of TU-514, whereas the a protons of the acyl chain are in close proximity to the methyl group of Thr179. The average distance between Thr179 and Tyr212 is ~ 15 Å, consistent with an extended conformation for the acyl chain of TU-514 from the α-methylene to the terminal methyl group. Although the binding of the acyl chain is important, as UDP-N-acetylglucosamine is a poor substrate, it is not clear whether the recognition of a full-length acyl chain is required, as LpxC does not distinguish between acyl chains of > 10 carbons (refs. 13, 14).
Figure 3.
The TU-514 acyl chain binds in a hydrophobic passage. (a) Surface of complex with TU-514 protruding. Residues that interact with the acyl chain of TU-514 are colored yellow and labeled, except for Thr179, which is at the bottom of the passage underneath the acyl chain. (b) Sample intermolecular NOE crosspeaks between TU-514 and LpxC. Strips corresponding to particular resonances of LpxC are shown, and the atoms on TU-514 correlated by each crosspeak are indicated. The chemical shift of each diagonal peak is labeled below that strip. TU-514 atom nomenclature is described in Figure 1 legend. Panel a was generated with PyMOL (DeLano Scientific).
The hexose ring of TU-514 adopts a chair conformation and sits in a largely enclosed pocket on the interior side of the β-α-αL-β motif and beneath the β-β-β motif, and binds through both polar and hydrophobic interactions. The backbone amide of Thr71 and the hydroxyl groups of Thr71 and Ser59 are positioned to form an intricate hydrogen bond network to recognize O5 and O6 of the hexose ring in TU-514. Thr179 has a pivotal role in supporting the hexose ring at its junction with the acyl chain, and exhibits NOE crosspeaks with H2 of the ring, the α and β protons of the acyl chain and the hydroxamate α-methylene.
Location of the active-size Zn2+ ion
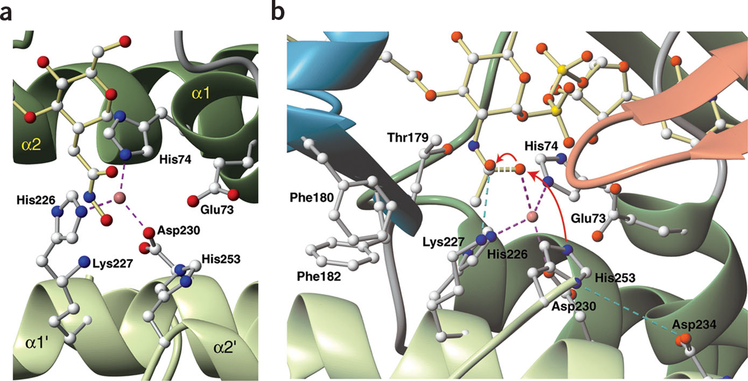
By analogy to other zinc metalloamidases, it has been assumed that the zinc in LpxC binds four ligands with tetrahedral geometry15–18. Previous mutagenesis studies of LpxC suggested that two of these ligands are His74 and His226, with either His253 or Asp234 as the third coordinating residue and a water molecule as the fourth ligand7 (Fig. 2c). In the solution structure of the LpxC-TU-514 complex, helix packing between helices α1 from domain I and α1′ from domain II positioned His74 and His226 directly beside each other, with the hydroxamate of TU-514 located adjacently, providing excellent geometry for a Zn2+ ion to be coordinated between them (Fig. 4a). Neither His253 nor Asp234 is positioned to coordinate Zn2+, as they are located 4.5 Å and 8.5 Å, respectively, away from the position estimated for Zn2+ (Fig. 4a,b). Asp230, however, is also completely conserved within the LpxC family (Fig. 2c) and is located perfectly to serve as a third zinc ligand (Fig. 4a,b). Mutation of Asp230 to alanine reduced the observed activity >1,000-fold (data not shown), consistent with a role of Asp230 as a zinc ligand. The hydroxamate oxygen serves as the fourth ligand in the LpxC-TU-514 structure, and is presumably replaced by a water molecule when the native substrate is bound (Fig. 4a,b). On the basis of analysis of the initial structures, a zinc ion was included during the final stages of structural refinement, coordinated between these residues (His74, His226 and Asp230) and the hydroxamate.
Figure 4.
Active-site structure and proposed catalytic mechanism. (a) Active site of LpxC with TU-514 bound. Coordination between His74, His226 and Asp230 and the zinc ion is shown with dashed lines (magenta). (b) Proposed mechanism of LpxC showing model of reaction transition state. Water activation by His253 and nucleophilic attack of the carbonyl carbon by the activated water molecule are shown with red arrows. Dashed lines indicate partial bonding between water and the substrate (yellow), coordination of ligands to zinc (magenta) and salt bridges between the oxyanion and Lys227 and between His253 and Asp234 (blue). Ribbons are colored by domain as in Figure 2. Generated with MolMol45.
Zinc metalloamidases discovered previously incorporate an HEXXH, HXXE or HXXEH zinc-binding motif, in which the first and last residues of the consensus sequence together with a downstream residue coordinate the zinc ion15,16,18. The LpxC family has a unique HKΦΦD zinc-binding motif (where Φ indicates a hydrophobic residue; Figs. 2c and 4a,b) in which His226 and Asp230, which are one turn apart on the same helix, and an upstream histidine serve as the zinc ligands. The conserved lysine helps stabilize the transition state (see below). These invariant residues are separated by two hydro- phobic residues, which are required to pack the helix to neighboring structural elements. The motif reinforces the designation of LpxC as a unique enzyme unrelated in both sequence and structural homology to other metalloamidases.
The catalytic mechanism of LpxC
Mechanisms of zinc metalloamidases have several key features: (i) a zinc-bound water molecule is activated by a general base through the abstraction of a proton; (ii) this activated water then attacks the carbonyl carbon of the substrate, with the Zn2+ polarizing the carbonyl oxygen, to produce a tetrahedral intermediate19,20; (iii) the oxygen of the former carbonyl group becomes an oxyanion, which is stabilized by positively charged or hydrogen bond-donating residues in the vicinity. With few exceptions21,22, all zinc metalloamidases discovered to date use a glutamate as the catalytic general base. Although LpxC has a glutamate in its active site (Glu73), previously published mutagenesis data7, which we have replicated and confirmed, show that an E73A mutant retains significant enzymatic activity (~10%), indicating that Glu73 is unlikely to be the catalytic general base. His253 is also positioned so that it might activate the water, either directly or through a ‘proton wire’ mediated by an additional water molecule20 (Fig. 4b). In contrast to Glu73, mutations of His253 (H253A and H253Q) inactivate the enzyme completely7, suggesting that His253 might serve as the catalytic base. A similar role for histidine has been proposed in a histone deacetylase homolog22. Mutagenesis studies have also suggested that Asp234 is important to catalysis, as D234A and D234S mutants are inactive and the activity of D234N mutants is greatly reduced7. Asp234 is located one helical turn away from the zinc-coordinating Asp230, with His253 in the vicinity (Fig. 4b). On the basis of the structure and biochemical data, we propose that Asp234 stabilizes the protonated His253 through electrostatic interaction, forming a potential charge relay system consisting of Asp–His–H20.
A variety of different residues have been observed to stabilize the oxyanion of the transition state19–21. In LpxC, Lys227 is well positioned to serve this purpose (Fig. 4b). Consistent with this proposition, a K227E mutant was completely inactive, confirming this residue’s importance to catalysis.
Because the E73A mutant shows greatly reduced but still significant activity (10%), we propose that this residue plays a secondary role in catalysis. Zinc enzymes have often featured conserved glutamate or aspartate residues that form hydrogen bonds to the direct zinc ligands, serving as an outer coordination shell18,23. Glu73 is positioned such that it could act in this manner, forming a hydrogen bond to the zinc- binding residue His74. Alternatively, Glu73 may assist with the stabilization of His253 or may help orient the catalytic water molecule.
Previous biochemical studies of LpxC have suggested that it can bind a second, inhibitory zinc ion in the active site6. Structures of other zinc metalloamidases that bind both a catalytic and an inhibitory Zn2+ show that the inhibitory zinc binds to the enzyme residue serving as the general base, and to the catalytic water molecule to form a bridged structure between the two zinc ions24–27. His253, the general base, and Glu73 are both positioned so that an inhibitory zinc ion bound to these residues could form a bridged structure with the catalytic water. The role of Glu73 in binding an inhibitory zinc is further supported by mutagenesis studies, which show that an E73A mutant cannot be inhibited by the addition of excess zinc7. Additional studies will be required to determine the fourth group that binds to this inhibitory zinc.
DISCUSSION
A model for native substrate binding
The binding of the native LpxC substrate is likely to mimic that of TU-514, raising the question of how LpxC accommodates the UDP moiety that is found on the native substrate but not present in the inhibitor. We noted an accessory hydrophobic pocket extending from the active site into the space between the two helices of domain I, which seems well positioned to bind UDP, taking into account the stereochemistry of the hexose ring and the corresponding restrictions on phosphate positioning. To determine whether this could be a binding pocket for the substrate, we conducted simulated annealing calculations using CNS with LpxC and the substrate UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine, together with a set of distance constraints for the acyl chain and hexose ring derived from the experimental TU-514 constraints28. The UDP moiety was allowed to rotate freely. When the five lowest-energy structures calculated were overlaid with the structure of TU-514 (Fig. 5a), the results indicated that UDP can be accommodated within this accessory pocket. The UDP moiety may be positioned in several different ways in this site, as a minor change in the orientation of the hexose ring may affect the position of the UDP moiety substantially. Resolving the details of this binding will require structural studies with this moiety present.
Figure 5.
Model for native substrate binding. (a) Overlay of the five lowest-energy calculated structures (gold). Position of TU-514 is shown in magenta. LpxC ribbon diagram is colored by domain as in Figure 2. (b) Model of His19 mediating the negative charges on the phosphates of the substrate. Generated with MolMol45.
The natural substrate also differs from TU-514 in that it has an acetyl group instead of a hydroxamate in the active site, which may be recognized by van der Waals contacts with Thr179 and Phe180 and potentially with Phe182 (Fig. 4b). The R-3-hydroxy group on the acyl chain of the native substrate could hydrogen bond to the hydroxyl group of Thr203.
Our model for substrate binding suggests an important role for His19 in mediating one or both of the negatively charged phosphate groups in the substrate (Fig. 5b). The E. coli strain envA1 incorporating the point mutation H19Y was first identified because of its unusual phenotype characterized by increased cell permeability and delayed cell separation, and it has long been known that His19 is important but not essential for LpxC function5. Subsequent in vitro mutagenesis studies have confirmed this assessment7. All of the calculated structures support this potential role for His19 in substrate capture and binding.
Protein dynamics and substrate binding
One of the most striking features of the LpxC-TU-514 complex is the hydrophobic passage that interacts with the acyl chain of TU-514. Inserting a long chain through a topologically closed loop and simultaneously inserting the UDP moiety into the binding pocket would undoubtedly entail a large entropic cost, even if it could be somewhat compensated for by enthalpy changes resulting from interactions with the hydrophobic passage and pocket.
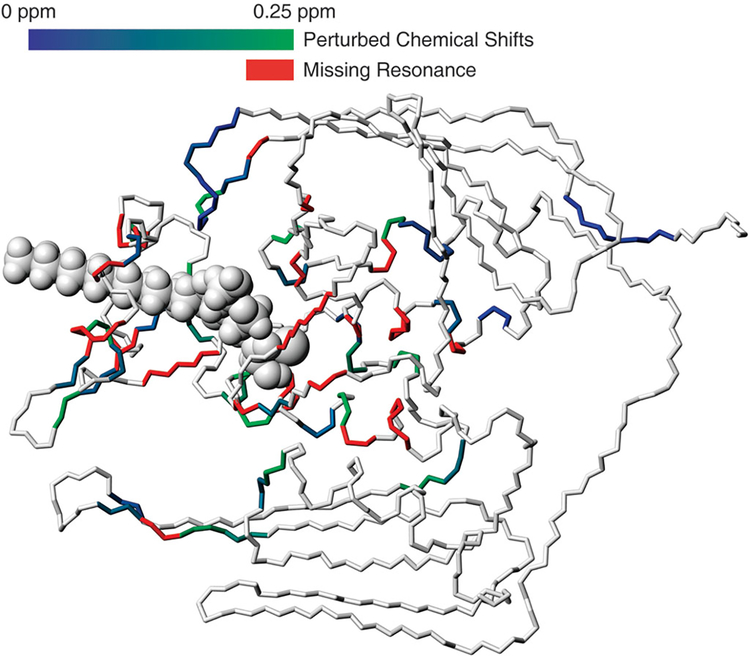
A comparison of the 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of the LpxC-TU-514 complex with that of free LpxC indicates that a number of resonances undergo chemical-shift perturbation, a reduction in line broadening, or both upon ligand binding. In the absence of the ligand, a number of residues do not exhibit resonance peaks; this may be caused by line broadening resulting from conformational exchange (Fig. 6). On the basis of this evidence, it seems unlikely that the rigid recognition motif exists in the free protein, but instead likely that a substantial portion of the enzyme either is in conformational exchange between several ordered states or is disordered. In particular, the residues in the β-α-αL-β motif, those in the direct vicinity of the active site and those forming the walls of the accessory pocket seem involved in motion in the absence of a substrate or inhibitor. In the absence of a ligand, multiple structural motifs, such as the β-α-αL-β motif and the loops close to the active site, may adopt different conformations or be disordered. The arrangement of α1 with α2 and the loop connecting β5 to α2 that form the proposed UDP pocket may also be perturbed. LpxC samples one or more ‘open’ conformations that provide convenient access to the active site; upon binding to a ligand, LpxC then locks into a single ‘closed’ conformation. Developing a complete understanding of the complex conformational changes that LpxC undergoes on ligand binding, which will be critical for rational inhibitor design, will require solving the structure of the free enzyme and completing dynamics studies. An assessment of the subtle variations in structure and dynamics between LpxC enzymes from different Gram-negative species will also be required to explain the observed specificity for phenyloxazoline-based inhibitors.
Figure 6.
Perturbations and missing resonances of free LpxC mapped to the structure of the LpxC-TU-514 complex. Chemical shift perturbations are calculated as (δH2 + 0.2δN2)1/2. Residues with perturbations are shown with colors on a gradient between deep blue (little change) and green (largest change); missing resonances in the absence of TU-514 are colored red. Generated with MolMol45.
METHODS
Expression and purification of the protein and preparation of samples.
A. aeolicus LpxC was overexpressed in BL21(DE3)STAR cells (Invitrogen) grown in M9 minimal medium from the pET21 vector pAaLpxC7. Isotope-enriched protein was prepared using [13C]glucose and 15NH4Cl (Cambridge Isotope Laboratory) as the sole carbon and nitrogen sources. Deuterium labeling was carried out by growing the cells in M9 with 100% D2O and deuterated [13C]glucose (Cambridge Isotope Laboratory). Samples were prepared and labeled with U-15N, U-13C/15N, U-2H/13C/15N, 50% 2H, U-13C/15N and 10% 13C labeling. A selective labeling protocol was used to prepare samples labeled only with 15N-Val, Ile, Leu, Lys, Phe or Tyr29,30. In addition, a uniformly 2H/13C/15N-labeled, Val-Ile-Leu (VIL)-methyl protonated sample was prepared according to a previously described protocol31. LpxC was purified using anion-exchange (Q-Sepharose Fast Flow, Amersham) and size-exclusion (Sephacryl S-200 HR, Amersham) chromatography. The LpxC-TU-514 complex was prepared by adding TU-514 to the purified LpxC protein in a roughly equimolar amount and allowing the complex to form at room temperature over 1 h. Samples were concentrated and buffer-exchanged into 25 mM sodium phosphate, pH 6.5, with 150 mM KCl, 4 mM dithiothreitol, 5% (v/v) D2O and 5% (v/v) deuterated dimethylsulfoxide (DMSO). An estimated 10–25% molar excess of TU-514 was then added to all samples of the complex, except for the samples used to record the F1/F2 15N/13C-filtered two-dimensional (2D) NOESY experiment, which were subjected to extensive dialysis to remove TU-514 that was not bound to LpxC. NMR samples contained 0.3–0.5 mM LpxC.
Preliminary studies.
The thermal stability of LpxC was assessed by circular dichroism (CD) spectroscopy. Melting curves were determined by monitoring the CD signal at 220 nm from 10 °C to 95 °C in 1 °C increments using an Aviv 202 CD spectrometer. A. aeolicus LpxC remained fully folded at temperatures as high as 95 °C, whereas denaturation of E. coli LpxC (prepared as described9) between 55 °C and 70 °C, with a midpoint of 62 °C, was observed by CD. Likewise, 1H-15N HSQC spectra of A. aeolicus LpxC collected in 5 °C increments from 25 °C to 70 °C did not show alterations, confirming that the protein remains folded at these temperatures.
Titration of 15N-labeled LpxC with increasing quantities of TU-514 was used to determine the timescale of ligand exchange and the ratio of binding. 1H-15N HSQC spectra were collected at TU-514/LpxC molar ratios of 0:1, 1:3, 2:3, 1:1, 2:1 and 3:1. To exclude the effects of DMSO interactions with LpxC, a control HSQC spectrum was recorded using free LpxC in the presence of 5% (v/v) DMSO. Ligand exchange was found to be slow on the NMR timescale, and the binding ratio was 1:1.
NMR spectroscopy.
All NMR experiments were conducted at 50 °C using Varian INOVA 600 MHz and 800 MHz spectrometers. Data were processed using FELIX (Accelrys) and analyzed with XEASY32. Sequential assignments were determined from HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB and HNCO experiments, along with 1H-15N HSQC of selectively labeled samples, using the PACES program, and confirmed by manual analysis33–35. Side chain resonances were assigned through 3D 15N-TOCSY-HSQC, HN(COCA)HA and 3D HCCH-TOCSY experiments36,37. Additional assignments were obtained through an H(CCO)NH-TOCSY experiment recorded on the 50% 2H, U-13C/15N-labeled sample38. Aromatic resonances were assigned based on 2D-homonuclear NOESY, 2D-homonuclear TOCSY and 13C-aromatic NOESY-HSQC experiments36. Stereospecific assignment of valine and leucine methyl groups was carried out using a 1H-13C HSQC spectrum of the sample labeled with 10% 13C (ref. 39). Resonances of TU-514 were assigned using F1/F2 15N/13C-filtered 2D NOESY experiments recorded with a perdeuterated, 15N/13C-labeled protein sample of the complex36,40.
Structure calculation.
Distance constraints for LpxC were derived from 3D 15N-NOESY-HSQC, 3D 13C-NOESY-HSQC and homonuclear 2D NOESY with mixing times of 60 ms, 80 ms and 50 ms, respectively36. Distance constraints obtained from these experiments were autocalibrated using the CALIBA module included in the DYANA package41. Loose constraints were also derived from methyl-methyl NOE experiments with a mixing time of 175 ms using a uniformly 2H/13C/15N-labeled and VIL-methyl-proton labeled sample42. Dihedral angle restraints were derived from TALOS analysis of the chemical shift information and from the HABAS routine based on the analysis of NOE patterns41,43. Two constraints per hydrogen bond (dHN-O ≤ 2.5 Å and dN-O ≤ 3.5 Å) were added for those amide protons protected from solvent exchange. Intermolecular NOEs between LpxC and TU-514 were detected through 3D F1-13C-filtered, F3-edited 13C-NOESYHSQC (100 ms mixing time), 3D F1-13C-filtered, F3-edited 13C-aromatic NOESY-HSQC (100 ms mixing time) using a 13C-labeled sample or 3D F1-13C-15N-filtered, F3-edited 15N-NOESY-HSQC (200 ms mixing time) experiments with a uniformly 2H/13C/15N-labeled complex36,40. Intermolecular NOEs were assigned to three classes—strong (≤3.0 Å), medium (≤4.0 Å) and weak (≤5.0 Å)—for conversion into distance constraints. Initial structures were generated by using DYANA41 starting from random conformations and refined using the simulated annealing protocol in CNS28. The 15 structures with no NOE violations >0.4 Å and no dihedral angle violations >4° were reported.
Site-directed mutagenesis and activity assays.
K227E and D230A mutants of LpxC were prepared using the QuikChange site-directed mutagenesis kit (Stratagene). The presence of appropriate mutants was confirmed by DNA sequencing. The preparation of the E73A mutant was described previously7. All mutants were prepared as pET21 constructs, which were overexpressed in BLR(DE3)pLysS (Novagen) grown in LB medium. Cell extracts were prepared and assayed at varying concentrations using the radiochemical assay described previously6,44.
Coordinates.
Atomic coordinates for the complex have been deposited with the Protein Data Bank (accession numbers 1NZT, RCSB018397). Chemical shifts for LpxC and TU-514 have been deposited with the BioMagResBank (accession number 5627).
Table 1.
Structural statistics for the LpxC-TU-514 complex (15 structures)a
| Protein (LpxC) | |
| NOE distance constraints | 3,614 |
| Intra-residue | 1,608 |
| Sequential (|i - j| = 1) | 780 |
| Medium-range (|i - j| ≤ 4) | 252 |
| i, i + 2 | 88 |
| i, i + 3 | 131 |
| i, i + 4 | 33 |
| Long-range (|i - j| ≥ 5) | 754 |
| Hydrogen bondsb | 220 |
| Dihedral angle constraintsc | 727 |
| Ramachandran plot (%)d | |
| Most-favored region | 87.8 |
| Additionally allowed region | 11.8 |
| Generously allowed region | 0.0 |
| Disallowed regione | 0.4 |
| Ligand (TU-514) | |
| NOE distance constraints | 29 |
| Dihedral angle constraintsf | 6 |
| Protein-ligand interface | |
| NOE distance constraints | 32 |
| Deviations from idealized geometryg | |
| Bonds(Å) | 0.0015 ± 0.0000 |
| Angles (°) | 0.338 ± 0.004 |
| Impropers (°) | 0.186 ± 0.008 |
| Average r.m.s. deviation to the mean structure (Å) | |
| Backbone (residue 4–268) | 0.63 |
| Heavy atoms (residue 4–268) | 1.19 |
None of these structures exhibit distance violations >0.4 Å or dihedral angle violations >4°.
Two constraints per hydrogen bond (dHN–O ≤ 2.5 Å and dN–O ≤ 3.5 Å) are implemented for amide protons protected from solvent exchange.
Dihedral angle constraints were generated by TALOS based on backbone-atom chemical shifts, and by the HABAS module in DYANA based on NOE constraints41,43.
PROCHECK-NMR46 was used to assess the quality of the structures.
γ-turn conformations are under-represented in the Ramachandran plot used by PROCHECK-NMR. This conformation is in the allowed region in an updated Ramachandran plot47.
Six dihedral angle constraints were used to maintain the chair conformation of the hexose ring of TU-514, which is consistent with the NOE patterns observed.
Average and standard deviation from idealized geometry for all 15 structures were calculated using CNS parameters (protein-allhdg.param).
ACKNOWLEDGMENTS
We thank J. Rudolph for helpful and stimulating discussions. This work was supported by grants from the US National Institutes of Health (NIH) and National Institute of General Medical Sciences (to C.R.H.R.), the NIH and National Institute of Allergy and Infectious Diseases (to P.Z.), the Natural Science and Engineering Research Council of Canada (to O.H.) and the Whitehead Institute (to P.Z.). X.L. is the recipient of a graduate scholarship in carbohydrate chemistry from the Alberta Research Council.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Raetz CRH & Whitfield C Lipopolysaccharide endotoxins. Annu. Rev. Biochem 71, 635–700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson MS, Bulawa CE & Raetz CRH The biosynthesis of Gram-negative endotoxin: formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J. Biol. Chem 260, 15536–15541 (1985). [PubMed] [Google Scholar]
- 3.Anderson MS, Robertson AD, Macher I & Raetz CRH Biosynthesis of lipid A in Escherichia coli: identification of UDP-3-O-(R-3-hydroxym yristoyl)-α-D-glucosamine as a precursor of UDP-N2,O3-bis(R-3-hydroxym yristoyl)-α-D-glucosamine. Biochemistry 27, 1908–1917 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Anderson MS et al. UDP-N-acetylglucosamine acyltransferase of Escherichia coli: the first step of endotoxin biosynthesis is thermodynamically unfavorable. J. Biol. Chem 268, 19858–19865 (1993). [PubMed] [Google Scholar]
- 5.Young K et al. The envA permeability/cell division gene of Escherichia c li encodes the second enzyme of lipid A biosynthesis. J. Biol. Chem 270, 30384–30391 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Jackman JE., Raetz CRH & Fierke CA UDP-3-O-(R-3-hydroxym yristoyl)-N- acetylglucosamine deacetylase of Escherichia coli is a zinc metalloamidase. Biochemistry 38, 1902–1911 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Jackman JE., Raetz CRH & Fierke CA Site-directed mutagenesis of the bacterial metalloamidase UDP-(3-O-acyl)-N -acetylglucosamine deacetylase (LpxC). Identification of the zinc binding site. Biochemistry 40, 514–523 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Onishi HR et al. Antibacterial agents that inhibit lipid A biosynthesis. Science 274, 980–982 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Jackman JE et al. Antibacteria agents that target lipid A biosynthesis in Gram-negative bacteria. J. Biol. Chem 275, 11002–11009 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Pirrung M et al. Inhibition of the antibacterial target UDP-(3-O-acyl)-N-acetyl-glucosamine deacetylase (LpxC): isoxazoline zinc amidase inhibitors bearing diverse metal binding groups. J. Med. Chem 45, 4359–4370 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Li X, Uchiyama T, Raetz CRH & Hindsgaul O Synthesis of a derived hydroxamic acid inhibitor of the bacterial enzyme (LpxC) involved in lipid A biosynthesis. Organic Letters 4, 539–541 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Holm L & Sander C Protein structure comparison by alignment of distance matrices. J. Mol. Biol 233, 123–138 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Williamson JM, Anderson MS & Raetz CRH Acyl-acyl carrier protein specificity of UDP-GlcNAc acyltransferases from Gram-negative bacteria: relationship to lipid A structure. J. Bacteriol 173, 3591–3596 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyland SA, Eveland SS & Anderson MS Cloning, expression, and purification of UDP-3-O-acyl-GlcNAc deacetylase from Pseudomonas aeruginosa: a metalloamidase of the lipid A biosynthesis pathway. J. Bacteriol 179, 2029–2037 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallee BL & Auld DS Active-site zinc ligands and activated H2 O of zinc enzymes. Proc. Natl. Acad. Sci. USA 87, 220–224 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallee BL & Auld DS Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 29, 5647–5659 (1990). [DOI] [PubMed] [Google Scholar]
- 17.Lipscomb WN & Sträter N Recent advances in zinc enzymology. Chem. Rev 96, 2375–2433 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Alberts IL, Nadassy K & Wodak SJ Analysis of zinc binding sites in protein crystal structures. Protein Sci. 7, 1700–1716 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christianson DW Carboxypeptidase A. Acc. Chem. Res 22, 62–69 (1989). [Google Scholar]
- 20.Christianson DW & Cox JD Catalysis by metal-activated hydroxide in zinc and manganese metalloenzymes. Annu. Rev. Biochem 68, 33–57 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Cheng X, Zhang X, Pflugrath JW & Studier FW The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 91, 4034–4038 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finnin MS et al. Structures of a histone deacetylase homologue bound to the T and SAHA inhibitors. Nature 401, 188–193 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Christianson DW & Alexander RS Carboxylate-histidine-zinc interactions in protein structure and function. J. Am. Chem. Soc 111, 6412–6419 (1989). [Google Scholar]
- 24.Larsen KS & Auld DS Carboxypeptidase A: mechanism of zinc Biochemistry 28, 9620–9625 (1989). [DOI] [PubMed] [Google Scholar]
- 25.Larsen KS & Auld DS Characterization of an inhibitory metal binding site in carboxypeptidase A. Biochemistry 30, 2613–2618 (1991). [DOI] [PubMed] [Google Scholar]
- 26.Holland DH, Hausrath AC, Juers D & Matthews BW Structural analysis of zinc substitutions in the active site of thermolysin. Protein Sci. 4, 1955–1965 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez-Ortiz M, Gomis-Rüth FX, Huber R & Aviles FX Inhibition of carboxypeptidase A by excess zinc: analysis of the structural determinants by X-ray crystallography. FEBS Lett. 400, 336–340 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT et al. Crystallography & NMR system: a new software suite for macro-molecular structure determination. Acta Crystallogr. D 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 29.LeMaster DM & Richards FM 1H-15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry 24, 7263–7268 (1985). [DOI] [PubMed] [Google Scholar]
- 30.LeMaster DM & Richards FM NMR sequential assignment of Escherichia coli thioredoxin utilizing random fractional deuteration. Biochemistry 27, 142–150 (1988). [DOI] [PubMed] [Google Scholar]
- 31.Goto NK, Gardner KH, Mueller GA, Willis RC & Kay LE A robust and cost-effective method for the production of Val, Leu, Ile (δ1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J. Biomol. NMR 13, 369–374 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Bartels C, Xia T-H, Billeter M, Güntert P & Wüthrich K The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 5, 1–10 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR & Kay LE A suite of triple resonance NMR experiments for the backbone assignment of 15N, 13C, 2H-labeled proteins with high sensitivity. J. Am. Chem. Soc 116, 11655–11666 (1994). [Google Scholar]
- 34.Yamazaki T et al. An HNCA pulse scheme for the backbone assignment of 2H-labeled proteins: application to a 37-kDa Trp repressor-DNA complex. J. Am. Chem. Soc 116, 6464–6465 (1994). [Google Scholar]
- 35.Coggins BE & Zhou P PACES: protein sequential assignment by computer-assisted exhaustive search. J. Biomol. N M R 26, 93–111 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Clore GM & Gronenborn AM Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 16, 22–34 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Olejniczak ET, Xu RX, Petros AM. & Fesik SW Optimized constant-time 4D HNCAHA and HN(CO)CAHA experiments: applications to the backbone assignments of the FKBP/ascomycin complex. J. Magn. Reson 100, 444–450 (1992). [Google Scholar]
- 38.Lin Y & Wagner G Efficient side-chain and backbone assignment in large proteins: application to tGCN5. J. Biomol. NMR 15, 227–239 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Szyperski T, Neri D, Leiting B, Otting G & Wüthrich K Support of 1H NMR assignments in proteins by biosynthetically directed fractional 13C-labeling. J. Biomol. NMR 2, 323–334 (1992). [DOI] [PubMed] [Google Scholar]
- 40.Breeze AL Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog. Nucl. Magn. Reson. Spectrosc 36, 323–372 (2000). [Google Scholar]
- 41.Güntert P, Mumenthaler C & Wüthrich K Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol 273, 283–298 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Zwahlen C et al. An NMR experiment for measuring methyl-methyl NOEs in 13C-labeled proteins with high resolution. J. Am. Chem. Soc 120, 7617–7625 (1998). [Google Scholar]
- 43.Cornilescu G, Delaglio F & Bax A Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Kelly TM, Stachula SA, Raetz CRH & Anderson MS The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase: the third step of endotoxin biosynthesis. J. Biol. Chem 268, 19866–19874 (1993). [PubMed] [Google Scholar]
- 45.Koradi R, Billeter M & Wuthrich K MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph 14, 51–55, 29–32 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R & Thornton JM AQ U A and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biol. Chem 8, 477–486 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Lovell SC et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 50, 437–450 (2003). [DOI] [PubMed] [Google Scholar]