Abstract
Using a mouse model, our group recently described an association between chronic paternal alcohol use prior to conception and deficits in offspring growth. Here, we sought to determine the impact of alcohol exposure on male reproductive physiology and the association of sperm-inherited noncoding RNAs with the transmission of the observed growth defects. Alcohol exposure did not appreciably alter male reproductive physiology or fertility. However, chronic alcohol use reproducibly induced late-term fetal growth restriction in the offspring, which correlated with a shift in the proportional ratio of transfer RNA-derived small RNAs to Piwi-interacting RNAs, as well as altered enrichment of microRNAs miR21, miR30, and miR142 in alcohol-exposed sperm. Although our dataset share similarities to prior works examining the impact of paternal stress on offspring phenotype, we were unable to identify any changes in plasma corticosterone, indicating alcohol may alter sperm-inherited noncoding RNAs through distinct mechanisms.
Keywords: paternal alcohol use, preconception exposure, epigenetic programming, growth restriction, sperm, noncoding RNAs, microRNAs, epigenetic
1. Introduction
Developmental plasticity refers to the dynamic ability of one genotype to produce multiple phenotypes in response to different environmental stimuli[1]. This phenomena enables the best chances of reproductive success but can also associate with the development of functional deficits and disease. Indeed, there are multiple instances where the predisposition of offspring to develop diseases later in life can be traced to a fetal compensation to an early life stressor[2]. For example, small-for-gestational-age babies have an increased risk of developing metabolic diseases, such as type two diabetes, and both cardiovascular morbidity and mortality as adults[3,4]. These observations helped found the Developmental Origins of Health and Disease hypothesis, and established the current recognition of the importance intrauterine development has in lifelong health.
Less well defined are the abilities of parental life history to influence organism phenotype. Although not as well characterized as intrauterine encounters, parental exposures prior to conception also exert a significant impact on offspring health and development[5]. Thus, in addition to uterine programming, the processes of germline programming that occur during gametogenesis also have the ability to impact offspring phenotype and influence the developmental plasticity of the next generation. Recently, preconception male exposures to a range of environmental factors have been linked to alterations in the developmental program of sperm and correlated with increased rates of structural and metabolic defects in the next generation[6–25]. These studies challenge the singular importance of maternal in utero exposures and implicate paternal exposure history as an additional and important mediator of both developmental defects and environmentally-induced disease.
The molecular mechanisms by which preconception stressors heritably influence cellular phenotypes are still very poorly understood. Molecular processes that allow the stable propagation of either chromatin-states or epigenetic information from one generation of cells to the next are hypothesized to play a role in transmitting the cellular memories of past exposures through gametogenesis to the offspring[26]. Specifically, mature sperm carries epigenetic information in patterns of DNA methylation, the region-specific retention of histones and DNA binding factors (like CTCF), as well as populations of small noncoding RNAs (ncRNAs). However, studies examining paternally-inherited abnormalities in growth and metabolic function have provided evidence to both support and refute the involvement of sperm-inherited changes in DNA methylation in the transmission of these phenotypes[7–9,16,17,23,25]. Similarly, although select regions of the sperm genome retain histones, it is unclear if this epigenetic information persists through the remodeling of the paternal genome during syngamy, or have the ability to transmit through the cell cycle[27,28]. The strongest candidate to date has been the transmission of paternally inherited ncRNAs. In these studies, the injection of either total sperm RNAs or a subset of sperm RNAs (for example, microRNAs or tRNA-derived small RNAs) into normal zygotes can recapitulate some paternal phenotypes induced by mental or metabolic stressors[29]. However, the mechanisms through which the effects of sperm-inherited ncRNAs persist into later life remain poorly defined[30].
In the United States, 70% of men drink and 40% drink heavily, with 8.3% reporting the routine consumption of more than two drinks per day[31–33]. Despite the nearly ubiquitous and constant exposure during reproductive ages, we currently have a very poor understanding of the effects chronic preconception alcohol use has on male reproductive physiology and the sperm-inherited developmental program. Using a mouse model of voluntary consumption, our lab recently described an association between chronic preconception paternal alcohol use and deficits in both placental function and offspring growth[23,34]. Importantly, these phenotypes did not associate with any measurable alterations in sperm-inherited patterns of DNA methylation[23]. Therefore, the question of how the memory of chronic alcohol use transmits to the offspring remains unresolved.
Mechanistic studies in rodents have revealed that alcohol impairs the endocrine-reproductive axis, indicating male alcohol use may impact foundational aspects of reproductive function[35,36]. In addition, multiple studies in humans and rodents have indicated that chronic alcohol use negatively impacts the integrity of the sperm nucleus[37–42]. Finally, using an inhalation model of exposure, Rompala and colleagues recently described alcohol-induced alterations in the profile of sperm derived ncRNAs[24]. However, whether facets of male reproductive function, sperm nuclear structure or the profile of sperm-inherited ncRNAs are altered in our model of voluntary alcohol consumption and associate with the development of the observed growth defects remains to be resolved.
2. Materials and Methods
2.1. Animal work
All experiments were conducted under AUP 2017–0308 and approved by the Texas A&M University IACUC. Individually caged C57BL/6J (RRID:IMSR_JAX:000664) postnatal day 90 adult males were obtained and housed in the Texas A&M Institute for Genomic Medicine, fed a standard diet (catalog# 2019, Teklad Diets, Madison, WI, USA) and maintained on a 12-hour light/dark cycle. In this study, we employed a voluntary model of alcohol exposure known as Drinking in the Dark. This model of exposure avoids the stress associated with forced feeding and mitigates the known impact of stress on the sperm-inherited epigenetic program[13]. Using methods described previously[23], males were provided limited access to ethanol during a four-hour window, beginning one hour after the initiation of the dark cycle. During this four-hour window, experimental males were provided access to a solution of 10% (w⁄v) ethanol (catalog# E7023, Millipore-Sigma, St. Louis, MO, USA) and 0.066% (w⁄v) Sweet’N Low (Cumberland Packing Corp, Brooklyn NY, USA). Control males received a solution of 0.066% (w⁄v) Sweet’N Low alone. After each session, the amount of fluid consumed by each mouse was recorded.
Once consistent patterns of drinking were established, males were maintained on this protocol for a period of 70 days. Subsequently, two naturally cycling females were placed in a new cage along with each exposed male. During these matings, males were not provided access to the alcohol/control preconception treatments. The next morning, matings were confirmed by the presence of a vaginal plug and both the male and female mice were returned to their original cages. Males were allowed a 24-hour rest period, during which the preconception exposure was resumed and then used in a subsequent mating. This procedure was repeated until each male had produced a minimum of two pregnancies. Subsequently, males were euthanized by CO2 asphyxiation and cervical dislocation, blood collected post-mortem and the male reproductive tract excised. Pregnant dams were maintained on a Breeder diet (catalog# 5058, LabDiet, St. Louis, MO, USA), subjected to minimal handling and euthanized by CO2 asphyxiation and cervical dislocation on gestational day 16.5. The female reproductive tract was excised, the gestational sac removed and fetal tissues weighed.
2.2. Measurement of Physiological Parameters
Male plasma alcohol concentrations were measured using the Ethanol Assay Kit (catalog# ECET100, BioAssay Systems, Hayward, CA, USA) according to the manufacturer’s protocol. Levels of serum testosterone were determined using an ELISA at the Ligand Assay and Analysis Core at the University of Virginia Center for Research in Reproduction. Serum levels of corticosterone were measured using the Corticosterone ELISA kit (catalog# EC3001–1, AssayPro, St. Charles, MO, USA) according to the recommended protocol.
2.3. Sperm and Tissue Collection & Histology
The male reproductive tract, liver, and spleen were excised, trimmed of fat and their weights recorded. To collect mature sperm, cauda epididymides and roughly 1 cm of vas deferens were placed in 500 μL of pre-warmed (37ºC) Human Tubal Fluid (HTF) medium (catalog# ZHTF-100, Zenith Biotech, Blue Bell, PA, USA) in 12 well plates. Four or five incisions were made to the cauda to allow sperm to swim out and the vas deferens were carefully squeezed using forceps to expel their contents. Plates were allowed to incubate at 37ºC for 10 minutes. Sperm counts were performed by diluting a 10µl aliquot 1:50 in diH2O and counting on a Neubauer chamber slide. Post-incubation, HTF media containing mature sperm were carefully layered over a pre-warmed (37ºC) gradient consisting of 350 μL each of 40% and 80% PureSperm (catalog#s PS40 and PS80, Spectrum Technologies, Healdsburg, CA, USA) in 1.5 mL microcentrifuge tubes and spun at 650g for 25 min. The pellet was then washed in 1mL of 1% BSA in PBS, and either immediately fixed for microscopic analyses or spun down, snap frozen in liquid Nitrogen and stored at −80ºC. To determine cell counts, 10 μL of caudal sperm from the last wash was used. Using microscopy, the purity of all samples was judged to be greater than 99%. After removal of encapsulating tunica, one testis from each animal was snap frozen in liquid nitrogen. The other testis was punctured with a 21 G needle and then fixed overnight in Bouin’s solution. Bouin’s fixed testes were paraffin embedded and stained using standard procedures for Hematoxylin and Eosin (H&E). H&E stained sections were imaged under bright field using the Leica DMi8 microscope (Leica microsystems, Germany). Cross-sectional area of tubules were measured using the area line tool in the Leica Application Suite X (LAS X) analysis software package. All tubules exhibiting longitudinal sectioning were excluded, while all tubules cut transversely into cross-sections were included in the analysis. Two non-sequential stained sections per animal were analyzed and a total of three animals per treatment were used in the comparison of calculated areas.
2.4. CMA3 staining
An equal volume of 3:1 methanol:acetic acid was added to PBS suspended sperm and samples incubated at 4ºC for 5 min. Samples were spread on glass slides and allowed to air dry at room temperature. Slides were treated for 20 minutes with 100 μL CMA3 solution: 0.25 mg/mL CMA (catalog# C2659, Millipore-Sigma, St. Louis, MO, USA) and 10mM MgCl2, McIlvain’s buffer (17 mM citric acid, 164 mM Na2HPO4, pH 7.0). Slides were rinsed with PBS and mounted with Prolong Gold Antifade Mountant (catalog# P36930, Thermo-Fisher, Waltham, MA, USA) and then kept at 4ºC overnight. Evaluation of fluorescence was done for a minimum of 200 spermatozoa on each slide.
2.5. Sperm TUNEL assay
The APO-DIRECT kit (catalog# 556381, BD Pharmingen, San Jose, CA, USA) was used for terminal deoxynucleotidyl transferase dUTP nick-end-labeling (TUNEL) to assess DNA damage. Approximately 3 million sperm per sample were resuspended in 4% PFA at room temperature for 1 hour. Samples were then spun down at 300xg for 5 minutes and washed twice with PBS. Pellets were suspended in ice-cold 70% ethanol and held at −20ºC for at least 12–18h. Following this incubation, preparation and staining of samples and appropriate controls were carried out as per manufacturer’s instructions. Stained samples were settled onto poly-L-lysine coated glass slides, mounted in Prolong Gold Antifade Mountant, and visualized on a Leica DMi8 microscope with SOLA SE 365 light source and Chroma filters for FITC detection. Total cell counts and sperm morphology of the PFA fixed sperm was visualized by differential Interference contrast microscopy at 63x. At least three fields of view and 200 sperm were scored for each sample to determine percent TUNEL positive.
2.6. Sperm RNA Isolation
Approximately 10 million mature sperm per sample were lysed in 1mL TRIzol Reagent (catalog# 15596018, Thermo-Fisher, Waltham, MA, USA) plus 10 µl β-ME (catalog# M3148, Millipore-Sigma, St. Louis, MO, USA) using homogenizing pestles (catalog# 6478820, Electron Microscopy Sciences, Hatfield, PA, USA). Subsequently, 200µl of 1-Bromo-3-chloropropane (catalog# B9673, Millipore-Sigma, St. Louis, MO, USA) was added to separate the aqueous phase, which was collected and precipitated using an equal volume of isopropanol. After two ice-cold, 70% ethanol washes, RNA samples were reconstituted in RNase free water and stored at −80ºC.
2.7. Sperm RNA Sequencing
Illumina single-end cDNA libraries were synthesized from size-selected RNAs (<50 bases) derived from 100 ng of sperm total RNA using the TruSeq Stranded mRNA kit. Four biological replicates per treatment group were multiplexed and sequenced on a single HiSeq2000 lane (Illumina) within the sequencing core of the Whitehead Institute. Using Bowtie2 (RRID:SCR_016368) and Tophat (RRID:SCR_013035), small RNA reads were aligned to the Mus musculus (UCSC version mm10) reference genome[43]. Small RNA reads with a single allowable mismatch were selected for further analysis. Small RNA annotation was performed by separately aligning reads to the microRNA database(http://www.mirbase.org/), tRNA database(http://gtrnadb.ucsc.edu/), and piRNA database(http://pirnabank.ibab.ac.in/index.shtml), as described previously[44]. Differentially expressed microRNAs were quantified using the miRDEEP2 ver2.0.0.7 pipeline[45]. The referenced tRNA database holds sequences for both tRNA anticodons and tRNA fragments (tRFs), which are selected as reads partially matching the tRNA anticodon but are less than 34 nucleotides in length[19]. To quantify differentially expressed tRFs, piRNAs, and tRNAs, total counts of mapped reads were calculated using the featureCounts pipeline[46] and then normalized to the total mapped reads of each class of small RNA species, as described previously[19]. Generated volcano plots contrast the differential enrichment of ncRNAs by raw p-value and not by FDR selection.
2.8. Data Handling and Statistical Analysis
For all experiments, measures were input into the statistical analysis program GraphPad (RRID:SCR_002798; GraphPad Software, Inc., La Jolla, CA, USA) and statistical significance was set at alpha = 0.05. For all datasets, normality was first verified using the Brown-Forsythe test. In this study, the effect of treatment was assessed using either an unpaired Student’s t-test or two-way analysis of variance test (ANOVA), with differences among the means evaluated using Sidak’s posthoc test of contrast. In all instances, we have marked statistically significant differences with an asterisk. For the comparisons of testicular, epididymal and seminal vesicle weights expressed as a percentage of total paternal body weight, as well as the percentage of TUNEL and CMA3 positive sperm, data were arcsine transformed and an unpaired t-test with Welch’s correction applied. To calculate the tRFs:piRNA ratio, the percentage of mapped tRFs per sample was divided by the percentage of mapped piRNAs and differences compared between treatments using an unpaired student’s t-test.
3. Results
3.1. Daily ethanol exposures induce pharmacologically meaningful blood alcohol concentrations but do not impact paternal body weight.
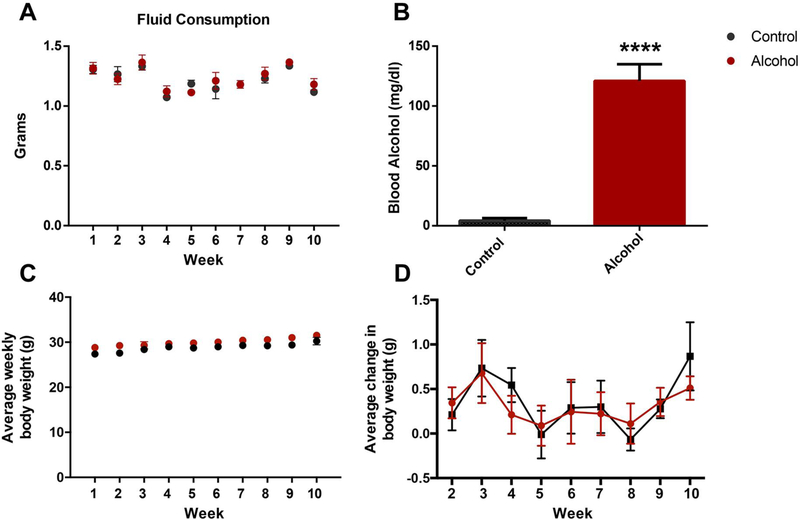
To define the long-term impact alcohol exposure has on both reproductive function and the male-inherited developmental program, we returned to our established mouse model of chronic alcohol exposure[23, 34]. Here, postnatal day 90 adult males were provided limited access to either the ethanol or control preconception treatments during a four-hour window, that began one hour into the night cycle. Once consistent patterns of alcohol consumption were established, males were maintained on this protocol for a period of 70 days, which corresponds to the length of two complete spermatogenic cycles and ensures that sperm formed prior to alcohol treatment are not able to confound the resulting phenotypes. For each male, the amount of fluid consumed per day was recorded. No differences in fluid consumption between the two preconception treatment groups were observed (Fig. 1A).
Figure 1.
Chronic alcohol exposure using a limited access model induces physiologically relevant plasma alcohol levels but does not alter paternal weight. A) Average daily fluid consumption compared between the preconception treatment groups (n=11). B) Average plasma alcohol levels between preconception treatment groups, as measured one month after the beginning of treatment (n=9). C) Average weight of males in each treatment group over the experimental course (n=11). Average weekly weight gain of males in each preconception treatment group (n=11). Data were analyzed using an unpaired t-test, error bars represent the standard error of the mean (**** p < 0.0001).
The rodent model we employed (Drinking in the Dark) promotes the daily, voluntary consumption of ethanol in sufficient quantities to achieve pharmacologically meaningful blood alcohol concentrations, typically around 125 mg/dL or 1.5x the legal limit (Fig. 1B). In the United States, 16% of men report engaging in high-risk drinking, which is defined as exceeding five or more standard drinks on any day within a given week and importantly, 8.3% of men routinely consume more than two drinks per day[33,47]. Therefore, the blood alcohol levels observed in our model of preconception male alcohol exposure are physiologically relevant. Further, chronic, daily alcohol use among males is both prevalent and a significant health concern. During the treatment course, no differences in body weight or changes in the rate of weight gain were observed between the preconception treatment groups (Fig 1C–D).
3.2. Chronic paternal alcohol exposure induces late-term fetal growth restriction and reductions in placental efficiency within the offspring.
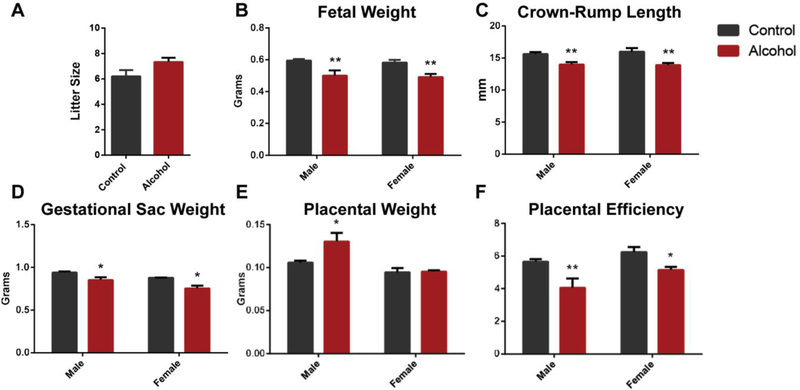
In our previous studies examining the offspring of alcohol-exposed males, we identified fetal growth restriction in only the female offspring at gestational day 14.5[23], while at birth, both male and female offspring exhibited significant growth restriction[34]. Therefore, we examined fetal growth at day 16.5 of gestation, which corresponds to the period when the mouse fetus experiences a dramatic increase in growth rate[48]. After 70 days of exposure, males undergoing the described preconception treatments were mated to unexposed females, and at gestational day 16.5, dams were sacrificed and offspring evaluated for growth. No differences in litter size were observed between the preconception treatment groups (Fig. 2A). At this developmental stage, the male and female offspring of alcohol-exposed sires displayed a ~15% reduction in fetal weight (p < 0.01), a ~10% reduction in crown-rump length (p < 0.01) and a respective 9% and 14% reduction in the weight of the gestational sac (p < 0.05) (Fig. 2B–D). These reductions in fetal growth were accompanied by an 18% increase in the placental weight of the male offspring of alcohol-exposed sires, while placental weights of the female offspring were identical to the controls (Fig. 2D). Collectively, a respective 28% and 17% reduction in placental efficiency (grams of fetus produced per gram of placenta[49]) was observed for the male and female offspring of the alcohol-exposed sires (Fig. 2E). These observations indicate that the growth restriction associated with paternal alcohol use predominantly manifest during the later phases of pregnancy and correlate with reductions in placental efficiency.
Figure 2.
Chronic preconception paternal alcohol exposure induces fetal growth restriction and decreased placental efficiency in the offspring at gestation day 16.5. A) Comparison of litter size between matings sired by control and ethanol-exposed males (n=5 control 6 alcohol). Comparisons of B) fetal weight, C) crown-rump length, D) gestational sac weight and E) placental weights between male and female offspring sired by control and ethanol-exposed males (n=10 male and 12 female offspring). F) Placental efficiencies (gram of fetus produced per gram of placenta) compared between the male and female offspring of ethanol-exposed sires (n=10 male and 12 female offspring). Data were analyzed using either an unpaired t-test or a two-way ANOVA followed by Sidak’s post hoc analysis. Error bars represent the standard error of the mean (* p<0.05, ** p < 0.01).
3.3. Chronic paternal alcohol use does not overtly impact male reproductive physiology
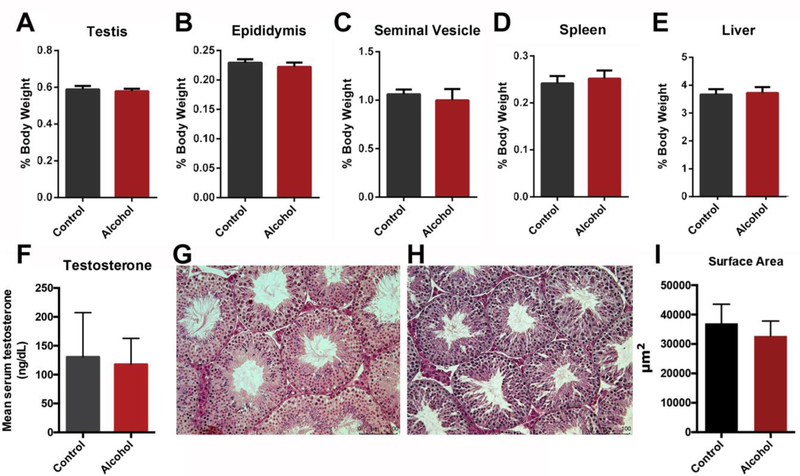
To better understand how the memory of chronic paternal alcohol use transmits to the offspring, we began by examining large-scale measures of male reproductive function. No differences in the absolute or proportional weights of the testis, epididymis or seminal vesicles, as well as the spleen or liver could be identified (Fig. 3A–E). Mean values and variations in serum testosterone levels we similar between the treatment groups and were consistent with established values[50] (Fig. 3F). Histological examinations of testicular sections could not identify any overt morphological differences and no changes in the mean surface area of the seminiferous tubules could be identified between the preconception treatment groups (Fig 3G–I). From these results, we conclude that the dose and duration of chronic alcohol exposure employed here do not overtly impact male reproductive physiology.
Figure 3.
Chronic alcohol exposure does not impact macro measures of male reproductive physiology. Comparisons of proportional A) testicular, B) epididymal, C) seminal vesicle, D) spleen and E) liver weights between preconception treatment groups (n=11). F) Levels of plasma testosterone were compared between preconception treatment groups (n=6). Representative hematoxylin and eosin stained sections of G) control and H) alcohol-exposed testes (n=3, with two, non-consecutive sections examined for each testis). I) Surface area of sectioned seminiferous tubules was determined using the LASX software package and compared between preconception treatment groups (n=3). Percent organ weights were arcsine transformed and an unpaired t-test with Welch’s correction used to compare treatments. For all other comparisons, an unpaired t-test was applied. Error bars represent the standard error of the mean.
3.4. Chronic paternal alcohol use does not impact sperm production, morphology, viability or large-scale measures of chromatin structure
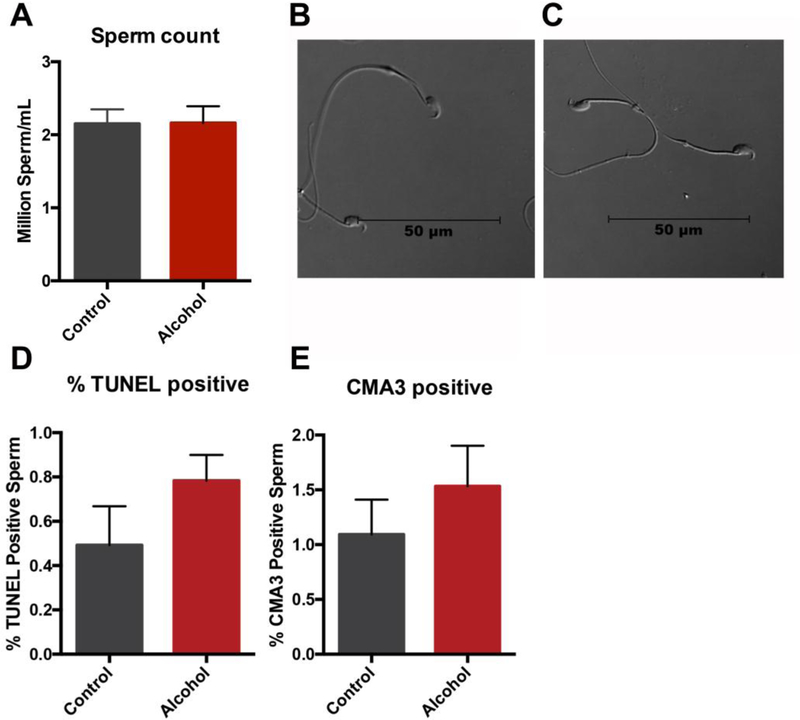
We next assayed the impact of chronic male alcohol use on sperm production. No differences in total sperm counts or changes in sperm morphology could be identified between treatment groups (Fig. 4A–B). Using the TUNEL assay, we were unable to identify any increases in sperm DNA fragmentation (Fig. 4C). Chromomycin A3 (CMA3) is a fluorochrome, which has been shown to compete with protamines for binding to the minor groove of DNA and is, therefore, indicative of compromised nuclear packaging. Using this stain, we could not detect any increases in the sperm of males chronically exposed to alcohol (Fig. 4D). Thus, we were unable to identify any large-scale changes in sperm production, DNA fragmentation (TUNEL assay) or nuclear packaging (CMA3 staining).
Figure 4.
Chronic paternal alcohol exposures do not measurably impede sperm production or alter either sperm DNA fragmentation or nuclear packaging. A) Total sperm counts were compared between preconception treatment groups (n=11). Representative light micrographs comparing B) control and C) alcohol-exposed sperm (n=11). The percentage of D) TUNEL positive and E) CMA3 positive sperm were compared between preconception treatment groups (n=4). Data comparing the percentage of stained sperm were arcsine transformed and an unpaired t-test with Welch’s correction applied. All other comparisons were conducted using an unpaired t-test. Error bars represent the standard error of the mean.
3.5. Chronic paternal alcohol alters the profile of sperm-inherited non-coding RNAs.
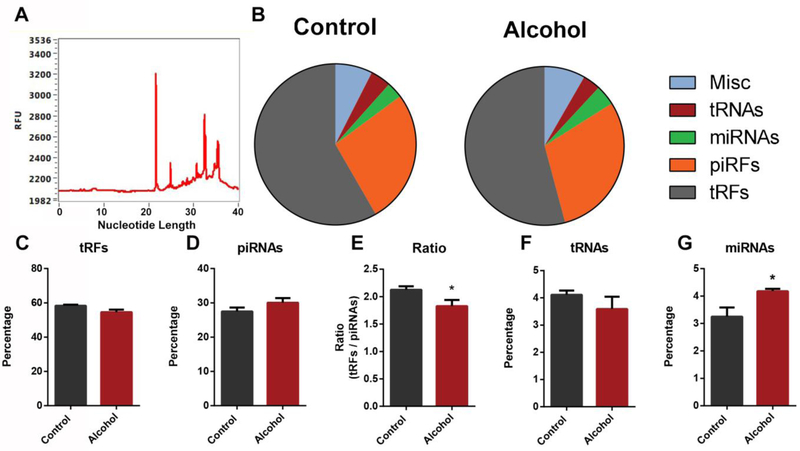
Using an inhalation model of exposure, Rompala and colleagues recently described alterations in sperm derived ncRNAs induced by a 5-week exposure to alcohol[24]. These studies achieved similar blood alcohol levels (125–175mg/dL) to those observed in our model. However, whether these separate models of exposure and different durations induce similar or distinct impacts on the profile of sperm-inherited ncRNAs is unknown. To examine this further, mature sperm were collected from the cauda epididymides and vas deferens of the control and ethanol-exposed males used to sire the offspring analyzed in Figure 2 (see materials and methods). The purity of sperm was judged to be greater than 99% as evaluated by microscopy. Similar to previous reports[51], isolated RNAs predominantly ranged from 20 to 40 nucleotides in length (Figure 5A). Small RNAs from control and ethanol-exposed males were subjected to deep sequencing analysis, with an average of 25 million mappable reads obtained per sample (n=4). Similar to previous studies describing the small RNA profiles of mouse sperm[19,24,51], we found that the majority of small RNA reads mapped to transfer RNA-derived small RNAs (~60% tRFs) and Piwi-interacting RNAs (~30% piRNAs) (Fig. 5B). The remaining small RNAs predominantly mapped to transfer RNAs (~5% tRNAs) and microRNAs (~5% miRNAs) (Fig 5B). A recent study by Sharma et al., described proportional changes in the ratio of tRFs and piRNAs as sperm undergo maturation in the epididymis[44]. Here, a progressive increase in tRFs and a loss of piRNAs were observed as sperm mature. Although sperm derived from alcohol-exposed males tended to have proportionally fewer mappable tRFs and a greater abundance of piRNAs, individually, these trends did not reach statistical significance (p=0.0552 and p=0.1086) (Fig. 5C–D). However, a ratio comparing tRFs:piRNAs revealed a significant shift (p<0.05) between the two preconception treatment groups (Fig. 5E). Further, while populations of tRNAs were similar between treatments, we observed a significant (p=0.03), ~30% increase in the abundance of miRNAs in sperm derived from alcohol-exposed males (Fig 5F–G). These observations reveal that chronic alcohol consumption shifts the profile of sperm-inherited non-coding RNAs, with miRNAs exhibiting the greatest change.
Figure 5.
Alcohol-induced alterations to the profile of sperm-inherited non-coding RNAs. A) Representative graph depicting the size distribution of RNAs isolated from sperm. B) The proportional abundance of transfer RNA-derived small RNAs (~60% tRFs) Piwi-interacting RNAs (~30% piRNAs), transfer RNAs (~5% tRNAs) and microRNAs (~5% miRNAs) between sperm derived from the two preconception treatment groups (n=4). Individual comparison of the percentage of C) tRFs and D) piRNAs mapped between preconception treatment groups. E) The ratio of tRFs:piRNAs in sperm derived from control and alcohol-exposed males. Individual comparison of the percentage of F) tRNAs and G) miRNAs mapped between preconception treatment groups. For comparison of percentages mapped, data were arcsine transformed and an unpaired t-test with Welch’s correction applied. All other comparisons were conducted using an unpaired t-test. Error bars represent the standard error of the mean (* p<0.05).
3.6. Alterations in the abundance of miR21, miR30, and miR142 in alcohol-exposed sperm.
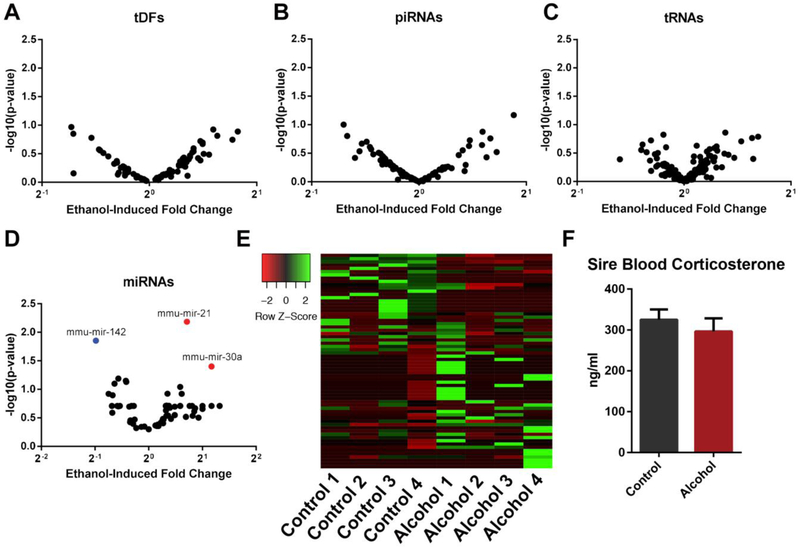
Using the Bowtie2 and miRDEEP2 pipelines, we compared the abundance of individual candidate small non-coding RNAs between control and alcohol-exposed sperm. No differentially enriched tRFs, piRNAs or tRNAs could be identified between treatment groups (Fig. 6A–C). In contrast, three differentially enriched miRNAs could be identified between treatments (miR21, miR30, and miR142) (Fig. 6D). Of these, miR21 and miR142 were abundantly enriched in both treatment groups (miR21 500 (C) and 900 (A) fpkm, miR142 600 (C) and 300 (A) fpkm). However, differences in the abundance of these two candidates offset each other, and therefore, do not explain the 30% increase in miRNA enrichment observed in alcohol-exposed sperm (Fig. 5F). In contrast to these candidates, the remaining miRNAs identified displayed large variations both across and within treatment groups (Fig. 6E). Collectively, these observations indicate that the 30% increase in miRNA abundance represents a general increase and is not linked to any specific candidate. Recently, alterations in the profile of sperm-inherited miRNAs induced by chronic stress have been directly linked to increased circulating levels of corticosterone[52]. We, therefore, assayed the levels of this hormone in our model. No differences in corticosterone could be identified between preconception treatment groups (Fig. 6F). Therefore, the 30% increase in miRNA enrichment and differences in miR21, miR30 and miR142 cannot be linked to alcohol-induced changes in the profile of circulating corticosterone.
Figure 6.
Alcohol-induced changes in the abundance of sperm-inherited miR21, miR30, and miR142. Volcano plots comparing the differential enrichment of candidate A) tRFs, B) piRNAs, C) tRNAs and D) miRNAs of sperm derived from the two preconception treatment groups. E) Heatmap comparing the variation of sperm derived miRNAs between treatment groups. F) Comparison of circulating levels of corticosterone between preconception treatment groups. An unpaired t-test was applied to compare the levels of corticosterone. Error bars represent the standard error of the mean, miRNAs identified with either blue or red dots were differentially enriched (p<0.05).
4. Discussion
Using a mouse model of voluntary alcohol consumption, our group recently described an association between chronic preconception paternal alcohol use and deficits in both placental function and fetal growth within the offspring[23]. Subsequent studies revealed that these alcohol-induced growth phenotypes were accompanied by a prolonged period of fetal gestation and sex-specific patterns of postnatal growth restriction[34]. These deficits in growth are similar to phenotypes described in long-term clinical studies of children with fetal alcohol spectrum disorders[53,54] and join a growing body of literature indicating preconception paternal alcohol use is a significant, yet under-recognized contributor to alcohol-induced growth defects (reviewed here[55–57]). However, the question of how the memory of chronic alcohol use transmits from father to offspring remains unresolved.
The literature examining the impacts of chronic alcohol use on male reproductive physiology is varied and highly inconsistent[58,59]. To this point, of three published studies using similar rodent models of exposure, two identified systemic decreases in testosterone concentrations[60,61], while the third was unable to identify any reproducible changes[62]. Similarly, the human literature is equally varied, with studies describing both alcohol-associated decreases and increases in testosterone levels[63–65]. However, combined with the negative correlations observed between alcohol use and successful outcomes in human in vitro fertilization[66,67], the prevailing feeling is that this teratogen exerts a negative effect on male reproductive function[58,59].
In this study, we returned to our voluntary model of alcohol consumption to determine the impact chronic ethanol use has on male reproductive physiology and the association of sperm-inherited noncoding RNAs with the transmission of alcohol-induced growth defects. Here, we first confirmed that chronic preconception paternal alcohol exposure induced fetal growth restriction in the offspring and extended our previous findings by demonstrating that these deficits in growth primarily manifest during the later phase of pregnancy (Fig. 2). We then assayed large-scale measures of male reproductive health, including testicular, epididymal and seminal vesicle weights, as well as testicular morphology and testosterone levels. However, we were unable to identify changes in any of these criteria. In addition, no differences in total sperm counts, sperm DNA fragmentation or sperm nuclear packaging were observed between the preconception treatment groups. Combined with the observed similarities in litter size between preconception treatments, we conclude that the dose and duration of alcohol exposure employed in our model do not impact macro-measures of male reproductive physiology.
As no observable changes in either sperm DNA methylation[23] or macro-measures of nuclear structure (Fig. 4) could be observed, we examined alcohol-induced alterations in the profile of sperm-inherited noncoding RNAs. Using an inhalation model of exposure, Rompala and colleagues recently identified alcohol-induced changes in the profile of tRFs and miRNAs, as well as select mitochondrial small mRNAs in sperm[24]. In contrast to these observations, we were only able to identify differences in select miRNAs, none of which were consistent with this published study. Although both models result in sex-specific patterns of postnatal growth restriction[68], these two separate models of exposure may induce distinct epigenetic changes. However, we did observe some similarities between our dataset and previous works examining the impact of paternal stress on offspring phenotype. Specifically, miR-30 and miR-21 were both up-regulated in the present study as well as in that of three previous reports examining stress-induced changes in sperm noncoding RNAs[13,52,69]. Although the levels of plasma corticosterone observed in our mice were higher than those reported by Rodgers et al.[69], we presume this difference is due to the techniques used to measure corticosterone levels and note that our results are consistent with those of two other publications employing ELISA-based measurements[52,70]. Regardless, we could not identify any changes in the levels of plasma corticosterone between treatment groups, indicating alcohol may alter sperm-inherited noncoding RNAs through distinct mechanisms. Importantly, the three differentially enriched candidate miRNAs identified in this study are all known to be modulated by alcohol exposure[71,72].
Recent studies indicate that dynamic changes in the levels of tRFs, piRNAs, and miRNAs are a core feature of sperm maturation[44]. Specifically, progressive increases in tRFs are observed during epididymal transit, while conversely, piRNAs become reduced. These changes are hypothesized to be integral to sperm maturation and the reproductive success of the conceptus[44]. In this study, we observed a shift in the ratio of tRFs and piRNAs indicating that the sperm of alcohol-exposed males has proportionally fewer tRFs and more piRNAs. This may indicate that the complement of non-coding RNAs in alcohol-exposed sperm are less mature than the controls. Of note, miR21, which is comparatively rare in testicular sperm populations, is nearly absent in the in the epididymal soma but is highly abundant in epididymosomes of the caput region of the epididymis[73–75]. This suggests that the alcohol-induced shifts in noncoding RNAs may primarily be mediated by effects on epididymal trafficking. However, further studies are needed to confirm this hypothesis.
At this point, we do not know if the modest changes in noncoding RNA abundance identified in this study are directly linked to the alcohol-induced growth restriction phenotypes observed in the offspring. The comparative contribution of sperm-inherited RNAs to the vast repertoire found in the early conceptus is negligible[76] and further, no mechanisms have been identified by which this small contribution could stably alter the gene expression profile of the early embryo and the long-term health of the offspring. In light of this, it is challenging to see how modest changes (50% decrease in miR142, 1.5 fold increases in miR21 and miR30) in the identified candidates could induce fetal growth restriction with the consistency observed in our model, as well as mediate the long-term impacts on offspring metabolic health. One possibility is that the paternally-inherited RNAs are post-transcriptionally modified to confer dramatically enhanced stability, which may potentiate their impacts beyond preimplantation development and influence the processes of lineage specification[29]. It may also be that unrelated alterations in sperm histone retention and chromatin looping mediate the observed effects or a combinatorial interaction between multiple epigenetic mediators. Finally, we have not ruled out the possibility that these phenotypes may be due to alcohol-induced changes in the seminal plasma. In previous studies, ablating the seminal vesicle gland induced placental hypertrophy in late gestation and sex-specific effects on the long-term growth and metabolic health of the offspring[77]. Although the effects on offspring growth were opposite to those observed in our model, the impact on placental growth is compelling. Future studies using IVF will be necessary to determine the impact of male seminal plasma in mediating the effects of paternal alcohol use on the offspring.
Supplementary Material
Highlights:
Chronic preconception paternal alcohol exposure associates with late-term fetal growth restriction and a loss of placental efficiency in the offspring.
The model of chronic alcohol consumption employed in this study did not impact macro-measures of male reproductive physiology or negatively affect fertility.
Chronic alcohol use alters the ratio of transfer RNA-derived small RNAs to Piwi-interacting RNAs, which has recently been identified as a core facet of the epigenetic maturation of sperm.
Chronic alcohol exposure induces a 30% increase in the abundance of sperminherited miRNAs, with miR21, miR30 and miR142 exhibiting the greatest changes.
Although our data share some similarities to recent work examining stressinduced changes in paternally-inherited miRNAs, we did not observe any differences in the levels of plasma corticosterone, indicating a novel mechanism underlies the observed changes.
Acknowledgements
This work was supported by the National Institutes of Health Piolot Grant P30 ES023512 (MCG), the Texas A&M Triads for Transformation multidisciplinary seed-grant (MCG) and Eunice Kennedy Shriver NICHD/NIH Grant R00-HD081204 (TMC). The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core is supported by the Eunice Kennedy Shriver NICHD/NIH (NCTRI) Grant P50-HD28934.
Abbreviations
- ncRNAs
noncoding RNAs
- tRFs
transfer RNA-derived small RNAs
- piRNAs
Piwi-interacting RNAs
- tRNAs
transfer RNAs
- miRNAs
microRNAs
- CMA3
Chromomycin A3
- fpkm
fragments per kilobase of transcript per million mapped reads
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hochberg Z, Feil R, Constancia M, Fraga M, Junien C, Carel JC, Boileau P, Le Bouc Y, Deal CL, Lillycrop K, Scharfmann R, Sheppard A, Skinner M, Szyf M, Waterland RA, Waxman DJ, Whitelaw E, Ong K, and Albertsson-Wikland K, Child health, developmental plasticity, and epigenetic programming, Endocr Rev 32 (2011) 159–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Feil R, and Fraga MF, Epigenetics and the environment: emerging patterns and implications, Nat Rev Genet 13 (2011) 97–109. [DOI] [PubMed] [Google Scholar]
- [3].Gluckman PD, Hanson MA, Buklijas T, Low FM, and Beedle AS, Epigenetic mechanisms that underpin metabolic and cardiovascular diseases, Nat Rev Endocrinol 5 (2009) 401–8. [DOI] [PubMed] [Google Scholar]
- [4].Padmanabhan V, Cardoso RC, and Puttabyatappa M, Developmental Programming, a Pathway to Disease, Endocrinology 157 (2016) 1328–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lane M, Robker RL, and Robertson SA, Parenting from before conception, Science 345 (2014) 756–60. [DOI] [PubMed] [Google Scholar]
- [6].Anway MD, Cupp AS, Uzumcu M, and Skinner MK, Epigenetic transgenerational actions of endocrine disruptors and male fertility, Science 308 (2005) 1466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, and Rando OJ, Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals, Cell 143 (2010) 1084–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, and Morris MJ, Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring, Nature 467 (2010) 963–6. [DOI] [PubMed] [Google Scholar]
- [9].Radford EJ, Isganaitis E, Jimenez-Chillaron J, Schroeder J, Molla M, Andrews S, Didier N, Charalambous M, McEwen K, Marazzi G, Sassoon D, Patti ME, and Ferguson-Smith AC, An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming, PLoS Genet 8 (2012) e1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zeybel M, Hardy T, Wong YK, Mathers JC, Fox CR, Gackowska A, Oakley F, Burt AD, Wilson CL, Anstee QM, Barter MJ, Masson S, Elsharkawy AM, Mann DA, and Mann J, Multigenerational epigenetic adaptation of the hepatic wound-healing response, Nat Med 18 (2012) 1369–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fullston T, Ohlsson Teague EM, Palmer NO, DeBlasio MJ, Mitchell M, Corbett M, Print CG, Owens JA, and Lane M, Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content, FASEB J 27 (2013) 4226–43. [DOI] [PubMed] [Google Scholar]
- [12].Lambrot R, Xu C, Saint-Phar S, Chountalos G, Cohen T, Paquet M, Suderman M, Hallett M, and Kimmins S, Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes, Nat Commun 4 (2013) 2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, and Mansuy IM, Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice, Nat Neurosci 17 (2014) 667–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dias BG, and Ressler KJ, Parental olfactory experience influences behavior and neural structure in subsequent generations, Nat Neurosci 17 (2014) 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, Seisenberger S, Hore TA, Reik W, Erkek S, Peters AH, Patti ME, and Ferguson-Smith AC, In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism, Science 345 (2014) 1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, and Sun QY, Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals, Proc Natl Acad Sci U S A 111 (2014) 1873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shea JM, Serra RW, Carone BR, Shulha HP, Kucukural A, Ziller MJ, Vallaster MP, Gu H, Tapper AR, Gardner PD, Meissner A, Garber M, and Rando OJ, Genetic and Epigenetic Variation, but Not Diet, Shape the Sperm Methylome, Dev Cell 35 (2015) 750–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Terashima M, Barbour S, Ren J, Yu W, Han Y, and Muegge K, Effect of high fat diet on paternal sperm histone distribution and male offspring liver gene expression, Epigenetics 10 (2015) 861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, and Rando OJ, Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals, Science 351 (2016) 391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, and Zhou Q, Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder, Science 351 (2016) 397–400. [DOI] [PubMed] [Google Scholar]
- [21].Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L, Mortensen B, Appel EV, Jørgensen N, Kristiansen VB, Hansen T, Workman CT, Zierath JR, and Barrès R, Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans, Cell Metab 23 (2016) 369–78. [DOI] [PubMed] [Google Scholar]
- [22].Esakky P, Hansen DA, Drury AM, Felder P, Cusumano A, and Moley KH, Paternal exposure to cigarette smoke condensate leads to reproductive sequelae and developmental abnormalities in the offspring of mice, Reprod Toxicol 65 (2016) 283–294. [DOI] [PubMed] [Google Scholar]
- [23].Chang RC, Skiles WM, Chronister SS, Wang H, Sutton GI, Bedi YS, Snyder M, Long CR, and Golding MC, DNA methylation-independent growth restriction and altered developmental programming in a mouse model of preconception male alcohol exposure, Epigenetics 12 (2017) 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rompala GR, Mounier A, Wolfe CM, Lin Q, Lefterov I, and Homanics GE, Heavy Chronic Intermittent Ethanol Exposure Alters Small Noncoding RNAs in Mouse Sperm and Epididymosomes, Front Genet 9 (2018) 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun W, Dong H, Becker AS, Dapito DH, Modica S, Grandl G, Opitz L, Efthymiou V, Straub LG, Sarker G, Balaz M, Balazova L, Perdikari A, Kiehlmann E, Bacanovic S, Zellweger C, Peleg-Raibstein D, Pelczar P, Reik W, Burger IA, von Meyenn F, and Wolfrum C, Cold-induced epigenetic programming of the sperm enhances brown adipose tissue activity in the offspring, Nat Med 24 (2018) 1372–1383. [DOI] [PubMed] [Google Scholar]
- [26].Rando OJ, and Simmons RA, I’m eating for two: parental dietary effects on offspring metabolism, Cell 161 (2015) 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Henikoff S, and Shilatifard A, Histone modification: cause or cog? Trends Genet 27 (2011) 389–96. [DOI] [PubMed] [Google Scholar]
- [28].O’Doherty AM, and McGettigan PA, Epigenetic processes in the male germline, Reprod Fertil Dev 27 (2015) 725–38. [DOI] [PubMed] [Google Scholar]
- [29].Chen Q, Yan W, and Duan E, Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications, Nat Rev Genet 17 (2016) 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Conine CC, Sun F, Song L, Rivera-Pérez JA, and Rando OJ, Small RNAs Gained during Epididymal Transit of Sperm Are Essential for Embryonic Development in Mice, Dev Cell 46 (2018) 470–480.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Naimi TS, Brewer RD, Mokdad A, Denny C, Serdula MK, and Marks JS, Binge drinking among US adults, JAMA 289 (2003) 70–5. [DOI] [PubMed] [Google Scholar]
- [32].White AM, Kraus CL, and Swartzwelder H, Many college freshmen drink at levels far beyond the binge threshold, Alcohol Clin Exp Res 30 (2006) 1006–10. [DOI] [PubMed] [Google Scholar]
- [33].Bose J, Hedden SL, Lipari RN, and Park-Lee E, Key substance use and mental health indicators in the United States: Results from the 2017 National Survey on Drug Use and Health, Center for Behavioral Health Statistics and Quality, Substance Abuse and Mental Health Services Administration: Rockville, MD, USA: SMA 18–5068 (2018). [Google Scholar]
- [34].Chang RC, Wang H, Bedi Y, and Golding MC, Preconception paternal alcohol exposure exerts sex-specific effects on offspring growth and long-term metabolic programming, Epigenetics & Chromatin 12 (2019) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Canteros G, Rettori V, Franchi A, Genaro A, Cebral E, Faletti A, Gimeno M, and McCann SM, Ethanol inhibits luteinizing hormone-releasing hormone (LHRH) secretion by blocking the response of LHRH neuronal terminals to nitric oxide, Proc Natl Acad Sci U S A 92 (1995) 3416–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lee HY, Naseer MI, Lee SY, and Kim MO, Time-dependent effect of ethanol on GnRH and GnRH receptor mRNA expression in hypothalamus and testis of adult and pubertal rats, Neurosci Lett 471 (2010) 25–9. [DOI] [PubMed] [Google Scholar]
- [37].Joo KJ, Kwon YW, Myung SC, and Kim TH, The effects of smoking and alcohol intake on sperm quality: light and transmission electron microscopy findings, J Int Med Res 40 (2012) 2327–35. [DOI] [PubMed] [Google Scholar]
- [38].Anifandis G, Bounartzi T, Messini CI, Dafopoulos K, Sotiriou S, and Messinis IE, The impact of cigarette smoking and alcohol consumption on sperm parameters and sperm DNA fragmentation (SDF) measured by Halosperm(®), Arch Gynecol Obstet 290 (2014) 777–82. [DOI] [PubMed] [Google Scholar]
- [39].Komiya A, Kato T, Kawauchi Y, Watanabe A, and Fuse H, Clinical factors associated with sperm DNA fragmentation in male patients with infertility, ScientificWorldJournal 2014 (2014) 868303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Talebi AR, Sarcheshmeh AA, Khalili MA, and Tabibnejad N, Effects of ethanol consumption on chromatin condensation and DNA integrity of epididymal spermatozoa in rat, Alcohol 45 (2011) 403–9. [DOI] [PubMed] [Google Scholar]
- [41].Rahimipour M, Talebi AR, Anvari M, Sarcheshmeh AA, and Omidi M, Effects of different doses of ethanol on sperm parameters, chromatin structure and apoptosis in adult mice, Eur J Obstet Gynecol Reprod Biol 170 (2013) 423–8. [DOI] [PubMed] [Google Scholar]
- [42].Sánchez MC, Fontana VA, Galotto C, Cambiasso MY, Sobarzo CMA, Calvo L, Calvo JC, and Cebral E, Murine sperm capacitation, oocyte penetration and decondensation following moderate alcohol intake, Reproduction 155 (2018) 529–541. [DOI] [PubMed] [Google Scholar]
- [43].Langmead B, and Salzberg SL, Fast gapped-read alignment with Bowtie 2, Nat Methods 9 (2012) 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, and Rando OJ, Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm, Dev Cell 46 (2018) 481–494.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Friedländer MR, Mackowiak SD, Li N, Chen W, and Rajewsky N, miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades, Nucleic Acids Res 40 (2012) 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liao Y, Smyth GK, and Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics 30 (2014) 923–30. [DOI] [PubMed] [Google Scholar]
- [47].Grant BF, Chou SP, Saha TD, Pickering RP, Kerridge BT, Ruan WJ, Huang B, Jung J, Zhang H, Fan A, and Hasin DS, Prevalence of 12-Month Alcohol Use, High-Risk Drinking, and DSM-IV Alcohol Use Disorder in the United States, 2001–2002 to 2012–2013: Results From the National Epidemiologic Survey on Alcohol and Related Conditions, JAMA Psychiatry 74 (2017) 911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].MCLAREN A, GENETIC AND ENVIRONMENTAL EFFECTS ON FOETAL AND PLACENTAL GROWTH IN MICE, J Reprod Fertil 9 (1965) 79–98. [DOI] [PubMed] [Google Scholar]
- [49].Wilson ME, and Ford SP, Comparative aspects of placental efficiency, Reprod Suppl 58 (2001) 223–32. [PubMed] [Google Scholar]
- [50].Bartke A, Steele RE, Musto N, and Caldwell BV, Fluctuations in plasma testosterone levels in adult male rats and mice, Endocrinology 92 (1973) 1223–8. [DOI] [PubMed] [Google Scholar]
- [51].Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, Zhou Q, Chen Q, and Duan E, A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm, Cell Res 22 (2012) 1609–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Short AK, Fennell KA, Perreau VM, Fox A, O’Bryan MK, Kim JH, Bredy TW, Pang TY, and Hannan AJ, Elevated paternal glucocorticoid exposure alters the small noncoding RNA profile in sperm and modifies anxiety and depressive phenotypes in the offspring, Transl Psychiatry 6 (2016) e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Carter RC, Jacobson JL, Sokol RJ, Avison MJ, and Jacobson SW, Fetal alcoholrelated growth restriction from birth through young adulthood and moderating effects of maternal prepregnancy weight, Alcohol Clin Exp Res 37 (2013) 452–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Moore EM, and Riley EP, What Happens When Children with Fetal Alcohol Spectrum Disorders Become Adults? Curr Dev Disord Rep 2 (2015) 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Finegersh A, Rompala GR, Martin DI, and Homanics GE, Drinking beyond a lifetime: New and emerging insights into paternal alcohol exposure on subsequent generations, Alcohol 49 (2015) 461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Day J, Savani S, Krempley BD, Nguyen M, and Kitlinska JB, Influence of paternal preconception exposures on their offspring: through epigenetics to phenotype, Am J Stem Cells 5 (2016) 11–8. [PMC free article] [PubMed] [Google Scholar]
- [57].Rompala GR, and Homanics GE, Intergenerational effects of alcohol: a review of paternal preconception ethanol exposure studies and epigenetic mechanisms in the male germline, Alcohol Clin Exp Res (2019). [DOI] [PMC free article] [PubMed]
- [58].Condorelli RA, Calogero AE, Vicari E, and La Vignera S, Chronic consumption of alcohol and sperm parameters: our experience and the main evidences, Andrologia 47 (2015) 368–79. [DOI] [PubMed] [Google Scholar]
- [59].Van Heertum K, and Rossi B, Alcohol and fertility: how much is too much? Fertil Res Pract 3 (2017) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Anderson RA, Willis BR, Oswald C, Reddy JM, Beyler SA, and Zaneveld LJ, Hormonal imbalance and alterations in testicular morphology induced by chronic ingestion of ethanol, Biochem Pharmacol 29 (1980) 1409–19. [DOI] [PubMed] [Google Scholar]
- [61].Emanuele NV, LaPaglia N, Benefield J, and Emanuele MA, Ethanol-induced hypogonadism is not dependent on activation of the hypothalamic-pituitary-adrenal axis, Endocr Res 27 (2001) 465–72. [DOI] [PubMed] [Google Scholar]
- [62].Salonen I, Pakarinen P, and Huhtaniemi I, Effect of chronic ethanol diet on expression of gonadotropin genes in the male rat, J Pharmacol Exp Ther 260 (1992) 463–7. [PubMed] [Google Scholar]
- [63].Välimäki M, Tuominen JA, Huhtaniemi I, and Ylikahri R, The pulsatile secretion of gonadotropins and growth hormone, and the biological activity of luteinizing hormone in men acutely intoxicated with ethanol, Alcohol Clin Exp Res 14 (1990) 928–31. [DOI] [PubMed] [Google Scholar]
- [64].Muthusami KR, and Chinnaswamy P, Effect of chronic alcoholism on male fertility hormones and semen quality, Fertil Steril 84 (2005) 919–24. [DOI] [PubMed] [Google Scholar]
- [65].Jensen TK, Swan S, Jørgensen N, Toppari J, Redmon B, Punab M, Drobnis EZ, Haugen TB, Zilaitiene B, Sparks AE, Irvine DS, Wang C, Jouannet P, Brazil C, Paasch U, Salzbrunn A, Skakkebæk NE, and Andersson AM, Alcohol and male reproductive health: a cross-sectional study of 8344 healthy men from Europe and the USA, Hum Reprod 29 (2014) 1801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Klonoff-Cohen H, Lam-Kruglick P, and Gonzalez C, Effects of maternal and paternal alcohol consumption on the success rates of in vitro fertilization and gamete intrafallopian transfer, Fertil Steril 79 (2003) 330–9. [DOI] [PubMed] [Google Scholar]
- [67].Rossi BV, Berry KF, Hornstein MD, Cramer DW, Ehrlich S, and Missmer SA, Effect of alcohol consumption on in vitro fertilization, Obstet Gynecol 117 (2011) 136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rompala GR, Finegersh A, Slater M, and Homanics GE, Paternal preconception alcohol exposure imparts intergenerational alcohol-related behaviors to male offspring on a pure C57BL/6J background, Alcohol 60 (2017) 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rodgers AB, Morgan CP, Bronson SL, Revello S, and Bale TL, Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation, J Neurosci 33 (2013) 9003–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kim JG, Jung HS, Kim KJ, Min SS, and Yoon BJ, Basal blood corticosterone level is correlated with susceptibility to chronic restraint stress in mice, Neurosci Lett 555 (2013) 137–42. [DOI] [PubMed] [Google Scholar]
- [71].Saad MA, Kuo SZ, Rahimy E, Zou AE, Korrapati A, Rahimy M, Kim E, Zheng H, Yu MA, Wang-Rodriguez J, and Ongkeko WM, Alcohol-dysregulated miR-30a and miR-934 in head and neck squamous cell carcinoma, Mol Cancer 14 (2015) 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Osterndorff-Kahanek EA, Tiwari GR, Lopez MF, Becker HC, Harris RA, and Mayfield RD, Long-term ethanol exposure: Temporal pattern of microRNA expression and associated mRNA gene networks in mouse brain, PLoS One 13 (2018) e0190841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Belleannée C, Calvo É, Caballero J, and Sullivan R, Epididymosomes convey different repertoires of microRNAs throughout the bovine epididymis, Biol Reprod 89 (2013) 30. [DOI] [PubMed] [Google Scholar]
- [74].Nixon B, Stanger SJ, Mihalas BP, Reilly JN, Anderson AL, Tyagi S, Holt JE, and McLaughlin EA, The microRNA signature of mouse spermatozoa is substantially modified during epididymal maturation, Biol Reprod 93 (2015) 91. [DOI] [PubMed] [Google Scholar]
- [75].Reilly JN, McLaughlin EA, Stanger SJ, Anderson AL, Hutcheon K, Church K, Mihalas BP, Tyagi S, Holt JE, Eamens AL, and Nixon B, Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome, Sci Rep 6 (2016) 31794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yang Q, Lin J, Liu M, Li R, Tian B, Zhang X, Xu B, Liu M, Zhang X, Li Y, Shi H, and Wu L, Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos, Sci Adv 2 (2016) e1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bromfield JJ, Schjenken JE, Chin PY, Care AS, Jasper MJ, and Robertson SA, Maternal tract factors contribute to paternal seminal fluid impact on metabolic phenotype in offspring, Proc Natl Acad Sci U S A 111 (2014) 2200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.