Summary
In addition to the conventional release of free, individual virions, virus dispersal can involve multi-virion assemblies that collectively infect cells. However, the implications of collective infection for viral fitness remain largely unexplored. Using vesicular stomatitis virus, here we compare the fitness of free versus saliva-aggregated viral particles. We find that aggregation has a positive effect on early progeny production, conferring a fitness advantage relative to equal numbers of free particles in most cell types. The advantage of aggregation resides, at least partially, in increasing the cellular multiplicity of infection. In mouse embryonic fibroblasts, the per-capita, short-term viral progeny production peaked for a dose of ca. three infectious particles per cell. This reveals an Allee effect restricting early viral proliferation at the cellular level, which should select for dispersal in groups. We find that genetic complementation between deleterious mutants is probably not the mechanism underlying the fitness advantage of collective infection. Instead, this advantage is cell-type dependent and correlates with cellular permissivity to the virus, as well as with the ability of host cells to mount an antiviral innate immune response.
Keywords: collective infectious unit, virus aggregation, dispersal, virus transmission, Allee effect, infectivity, innate immunity, genetic complementation, vesicular stomatitis virus
Introduction
In contrast to the traditional view that viruses propagate among cells as individual virions, recent work has described viral spread in groups [1, 2]. For instance, in vesicular stomatitis virus (VSV) [3] and poliovirus [4], virion aggregates can infect cells collectively. In enteroviruses, lipid vesicles containing multiple virions are released from infected cells before lysis and serve as intercellular vehicles for viral spread [5]. Similarly, multiple marseillevirus particles are jointly wrapped in large extracellular vesicles derived from the amoebal endoplasmic reticulum [6]. In baculoviruses, inter-host transmission takes place in the form of pools of virions encased in a protein matrix, called occlusion bodies [7]. Use of cell-cell contacts for the delivery of large number of viral particles has also been shown in many viruses. For instance, human T-cell leukaemia virions accumulate in extracellular matrix components called “viral biofilms”, which are used for cell-cell virion transfer [8]. Other viruses including HIV-1, measles virus, vaccinia virus, and herpes virus subvert existing cell-cell contacts or induce new ones, using cells as vehicles for the joint transfer of multiple particles [9, 10]. In fecal-oral transmitted viruses such as poliovirus, gut bacteria act as attractors of viral particles, also promoting their joint spread [11]. Finally, inter-host transmission in a cell-associated manner is also possible but remains understudied, except for HIV-1 where it has been shown to be an important transmission route [12].
Despite evidence for these various modes of collective spread in viruses, little is known about their implications for viral fitness. All else being equal, multi-virion propagules should be disfavored because they reduce dispersal capacity and, hence, the efficiency of intra-host spread and inter-host transmission. To illustrate this, consider infected cells (or hosts) producing N groups each containing S particles. The infection will reach at most N cells (hosts) in the next viral generation, whereas an equal amount of non-aggregated progeny would reach NS cells (hosts). Therefore, for collective spread to be selectively advantageous, there should be benefits offsetting this cost, yet these benefits remain unclear. Specifically, spread in groups will increase viral fitness if the average progeny yield of such groups exceeds NS, which means that cellular resources should be exploited more efficiently, compared to individual virions. Dispersal in groups can be advantageous if invasion of new spots (here, cells) is subject to an Allee effect, defined as a positive relationship between fitness and population size or density. The Allee effect is believed to be an important factor limiting population establishment because it slows down proliferation or even leads to extinction of small founder populations [13]. Allee effects have been documented in many animal and plant species, including invasive and/or parasitic species [14] such as, for instance, entomopathogenic nematodes [15]. Interestingly, the Allee effect has been demonstrated recently in Vibrio bacteria [16]. However, there is no previous evidence for Allee effects in viruses.
One way to examine the fitness effects of dispersal in groups is to compare equal numbers of viral particles in monodisperse versus aggregated form. VSV offers an excellent system to achieve this goal, because virions can be aggregated experimentally [3]. Here we show that VSV aggregates produce progeny more rapidly than equal numbers of non-aggregated viral particles during the first cellular infection cycle, and that this confers a competitive advantage to populations founded by aggregates compared to those founded by monodisperse particles. By measuring viral progeny production in cells inoculated with different doses of the virus, we show that the advantage of aggregation may reside in increasing the number of founder particles per cell. Specifically, we found that the per capita progeny production depends on the multiplicity of infection in a manner consistent with an Allee effect at the cellular level. The fitness advantage of aggregation is cell-type-dependent and correlates with cellular permissivity to infection. We suggest that invading cells with multiple viral particles reduces the risk of early stochastic loss and/or provides the virus a head start relative to innate immune responses, increasing the chances of establishing a successful infection locally.
Results
Aggregation of founder particles increases viral fitness in mouse embryonic fibroblasts (MEFs)
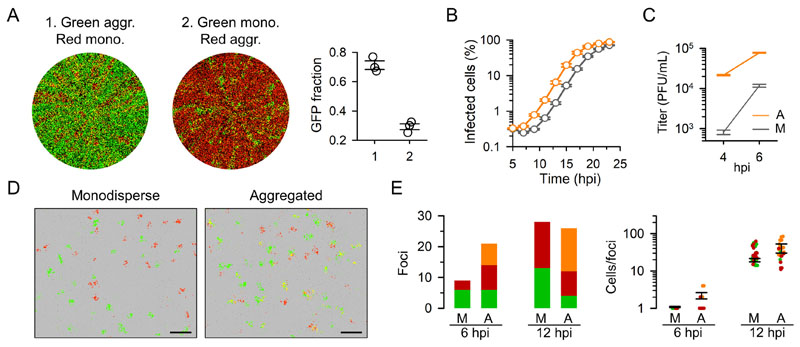
In previous work, we showed that VSV particles aggregate in the presence of saliva from some donors [3]. To test how initiating the infection with such aggregates determines viral fitness, we competed two recombinant viruses encoding different fluorescent reporters (GFP versus mCherry). For this, we aggregated VSV-GFP with human saliva and kept VSV-mCherry untreated (i.e. monodisperse), then mixed the two viruses at a 1:1 particle ratio (as determined by titration before aggregation), and inoculated MEFs at low initial viral density. The infection progressed until invading the entire cell culture at approximately 24 hours post inoculation (hpi). At this time point, 71.3 ± 3.0 % of the total fluorescent area corresponded to GFP, as determined by whole-well fluorescence microscopy. In contrast, when we competed monodisperse VSV-GFP against aggregated VSV-mCherry, GFP accounted for only 29.3 ± 2.0 % of total fluorescence at 24 hpi (t-test: P < 0.001; Figure 1A). Therefore, for an equal input of viral particles, initiating the infection in an aggregated manner increased viral fitness in competition assays. Real-time fluorescence microscopy revealed that the growth curves of viral populations founded by aggregates were left-shifted compared to those founded by monodisperse particles, with estimated half-times of 16.7 ± 0.2 h and 19.1 ± 0.1 h, respectively (Figure 1B). To confirm this short-term advantage, we titrated supernatants by the plaque assay at time points within or at the boundary of the first infection cycle (4 and 6 hpi). The 4 hpi titer in cells inoculated with monodisperse particles was below the inoculum size, hence showing no evidence of progeny production (eclipse phase). In contrast, aggregates produced a 27-fold higher titer at this time point, exceeding 104 plaque forming units (PFU) per mL (t-test, P < 0.001; Figure 1C). At 6 hpi, cells inoculated with aggregates still produced 6.5 times more progeny (t-test, P < 0.001). Therefore, viral aggregation accelerated viral proliferation during the first infection cycle, providing a fitness advantage relative to non-aggregated founders.
Figure 1. Aggregation increases viral fitness by promoting early growth in MEFs.
A. Whole-well fluorescence microscopy of MEFs inoculated with aggregated VSV-GFP and monodisperse VSV-mCherry particles (1), and with monodisperse VSV-GFP and aggregated VSV-mCherry particles (2). Images were taken at 24 hpi and correspond to one of the three replicate assays performed. The fraction of total fluorescent area occupied by GFP-positive cells is shown in the right graph for the three replicates. Error bars: SEM. B. Growth curve of VSV in MEFs obtained by real-time whole-well fluorescence microscopy. The percentage of fluorescent cells in the well is shown. Orange: aggregated inoculum. Grey: monodisperse inoculum. Each data point is the average of three replicates. Error bars: SEM. C. Viral titers in cultures inoculated with aggregates (A) versus monodisperse (M) particles (inoculum: 5000 PFU). Three replicates were performed. Error bars: SEM. D. Foci produced in MEFs inoculated with equal numbers of monodisperse versus aggregated particles (14 hpi). VSV-GFP and VSV-mCherry were mixed prior to aggregation. Cells expressing both VSV-GFP and VSV-mCherry appear in yellow. The cell monolayer is shown in phase contrast. Scale bar: 1 mm. E. Analysis of individual infection foci at different time points in MEFs inoculated with monodisperse or aggregated particles. Left: bars indicate the number of foci positive for GFP (green), mCherry (red), or both (orange). Right: number of cells within infection foci (same color legend). Notice that foci containing both VSV-Cherry and VSV-GFP tend to be bigger than those containing a single type. Error bars: SEM. See Figure S1 for results obtained in BHK-21 cells.
Aggregation does not reduce dispersal capacity in MEFs
In principle, because each aggregate contains multiple particles, aggregation should reduce the number of infectious units and, hence, reduce dispersal capacity, as argued above. To test whether aggregates infected fewer cells than an equal number of monodisperse particles, we co-incubated VSV-GFP and VSV-mCherry in saliva and counted the number of infection foci produced in MEFs, compared with the foci produced by this same mix without the saliva treatment. A large fraction of the foci produced by saliva-treated viruses were positive for both GFP and mCherry, confirming that aggregates deliver multiple viral genomes to target cells [3] (Figure 1D). Despite this, we did not observe a reduction in the number of foci in cells inoculated with the saliva-aggregated virus compared to those inoculated with untreated viruses. We explored this further by counting infected cells at early time points in an entire well using a minimal VSV-GFP/VSV-Cherry inoculum to ensure sufficient separation of infection foci. At 6 hpi, monodisperse particles produced only 9 individual infected cells, whereas aggregates produced 21 foci of 1.4 ± 0.2 cells on average, infecting 30 total cells (Figure 1E). At 12 hpi, we found similar numbers of foci in cultures infected with monodisperse and aggregated particles (28 and 26, respectively), and foci were slightly but not significantly larger in the latter (31.5 ± 2.8 and 38.7 ± 4.0 cells, respectively; t-test: P = 0.232). Therefore, aggregation increased the per-particle probability of successfully initiating infection, as determined by the production of infection foci.
The per-capita viral progeny production exhibits an Allee effect at the cellular level
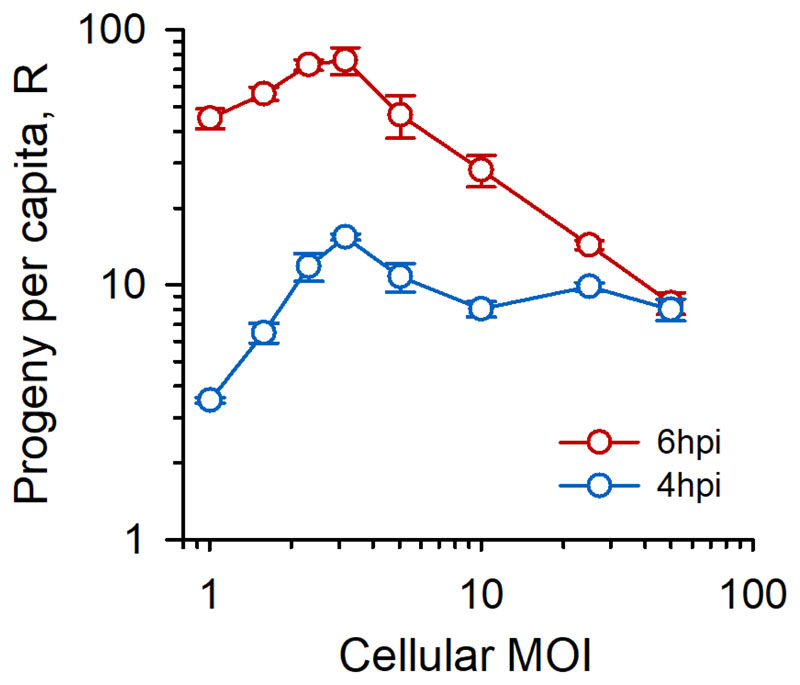
We speculated that viral aggregation was advantageous because it increased the cellular multiplicity of infection (cMOI, in PFU/cell units), defined as the average number of particles that initiate the infection of a cell. However, other processes were in principle possible, such as, for instance, more efficient cell binding and/or entry of aggregates compared to individual viral particles. To analyze how the cMOI determined progeny production, we inoculated MEFs with doses of monodisperse particles at densities ranging from D = 0.01 to D = 50 PFU/cell. The cMOI was calculated based on a Poisson model, as follows: where P(0) = e−D is the Poisson distribution null class (notice that cMOI ≥ 1). We determined viral titers at 4-6 hpi to obtain the per-capita progeny production during the first cell infection cycle, defined as the final/input titer ratio (R). In the absence of an Allee effect, R should decrease with the cMOI as a result of competition for limited resources, or remain constant if cellular resources were not limiting. In contrast, at 4 hpi R was always higher for cMOI > 1 than for cMOI = 1, that is, it always paid off for the virus to invade cells with multiple particles, and R peaked at approximately cMOI = 3 (Figure 2). For instance, doubling the cMOI from 1.58 to 3.16 increased the titer at 4 hpi by sevenfold. At 6 hpi, R peaked at a similar cMOI value but then dropped, probably because of cellular resource exhaustion. At this time point, it still paid off for the virus to invade N cells with S particles each, rather than NS cells with one particle each, provided that S ≤ 5 PFU/cell. These data show that a demographic Allee effect operates at the cellular level, supporting the possibility that the fitness advantage of aggregation resides at least partially in the collective infection of cells.
Figure 2. Early viral progeny production exhibits a demographic Allee effect.
Per capita progeny production (R, defined as final/initial titer) in MEFs for different cellular MOI values (defined as the average number of initial PFU per infected cell). Each data point is the average of three replicates. Error bars: SEM. See Figure S2 for results obtained in BHK-21 cells.
The fitness benefit of aggregation does not correlate with mutational load
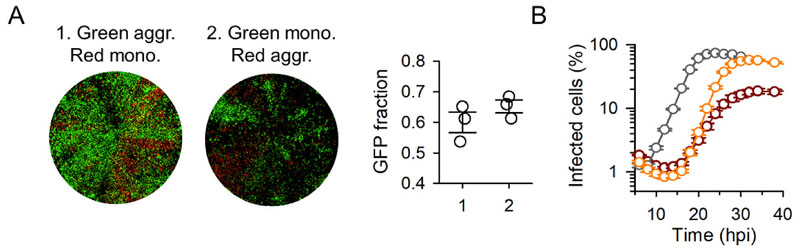
A possible mechanism explaining the advantage of collective infection is genetic complementation (sometimes referred to as multiplicity reactivation), whereby viral genomes carrying lethal or deleterious mutations regain infectivity/fitness in cells coinfected by other genomes that do not contain the same genetic defects [17, 18]. According to this hypothesis, a fraction of lethal mutations would be rescued in aggregates, increasing the number of viral genomes that effectively initiate the infection. Because this process should correlate with the population mutational load, to explore the role of genetic complementation we subjected the virus to chemical mutagenesis by performing three serial transfers in the presence the base analogue 5-fluorouracil (5-FU), as detailed previously [19]. VSV-GFP and VSV-mCherry were mutagenized separately and then used to perform competition assays, as above. The fluorescence signal was weaker and grew more slowly in 5-FU-treated viruses than in untreated viruses, consistent with an increased abundance of deleterious mutations. Yet, aggregation had no significant effect on fitness, as determined by the fraction of GFP to total fluorescence at endpoint (34 hpi; 60.0 ± 3.4% in competitions with aggregated VSV-GFP versus monodisperse VSV-mCherry particles, 65.2 ± 2.0% using monodisperse VSV-GFP versus aggregated VSV-mCherry; t-test: P = 0.261; Figure 3A). Furthermore, in mutagenized viruses, aggregation slowed down viral spread, reducing both the exponential growth rate (0.553 ± 0.001 versus 0.315 ± 0.012; t-test: P < 0.001) and the maximal fluorescent area (58.1 ± 2.0 % versus 20.0 ± 1.6%; t-test: P < 0.001; Figure 3B). Therefore, increasing the mutational load reversed the fitness benefits of aggregation, contradicting the genetic complementation hypothesis.
Figure 3. The fitness benefit of aggregation is lost in mutagenized populations.
A. Whole-well fluorescence microscopy of MEFs inoculated with aggregated VSV-GFP and monodisperse VSV-mCherry particles (1), and with monodisperse VSV-GFP and aggregated VSV-mCherry particles (2). VSV-GFP and VSV-mCherry were mutagenized prior to competition assays using 5-FU. Images were taken at 34 hpi and correspond to one of the three replicate assays. The fraction of total fluorescent area occupied by GFP-positive cells is shown in the right graph for the three replicates. Error bars: SEM. B. Growth curve of VSV in MEFs by real-time whole-well fluorescence microscopy. The percentage of fluorescent cells in the well is shown. Grey: non-mutagenized, monodisperse inoculum. Orange: mutagenized, monodisperse inoculum. Red: mutagenized, aggregated inoculum. Each data point is the average of three replicates. Error bars: SEM.
The benefits of aggregation correlate with cell permissivity to infection
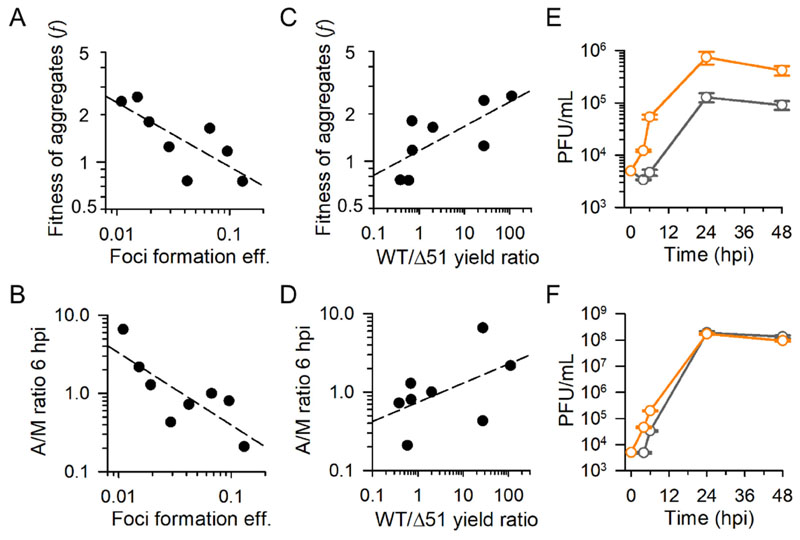
We set out to test whether our results were reproduced in baby hamster kidney fibroblasts (BHK-21), which are used routinely for VSV studies in part due to their high permissivity to VSV but which, as opposed to MEFs, are highly abnormal, tumoral cells. Competition assays, titrations at 4-6 hpi, and analysis of foci production showed that initiating the infection with saliva-aggregated particles provided a fitness advantage, although weaker than in MEFs (Figure S1). Titration of supernatants from BHK-21 cells inoculated at increasing cMOI values were also compatible with an Allee effect at the cellular level, but less markedly than in MEFs (Figure S2). To examine such cell dependence in more detail, we used 4T1 mouse mammary gland tumor cells (epithelial), CT26 mouse colon cancer cells (fibroblasts), Neuro2a mouse neuroblastoma cells (neural stem-like cells), A549 human lung adenocarcinoma cells (alveolar epithelium), MRC5 human lung fibroblasts, and African green monkey Vero kidney epithelial cells. For this extended analysis, we focused only on competition assays with GFP/mCherry reporters and on titrations at 4-6 hpi. Initiating the infection with aggregates was selectively beneficial in MRC5, CT26, A549, and 4T1 in addition to MEFs and BHK-21, whereas it was detrimental in Vero and Neuro2a cells, as determined by the ratio of GFP to total fluorescence at endpoint (Figure S3; t-tests: P < 0.001 in all cases). Hence, aggregation was most highly advantageous in the two non-tumoral cells lines tested (MEFs and MRC5). To measure cellular permissivity to VSV, we calculated the foci formation efficiency in each cell type, defined as the ratio of foci to viral particles present in a given inoculum (also termed specific infectivity), were viral particle concentration was determined by nanoparticle tracking analysis (Figure S4). The fitness benefit of aggregation, as indicated by competition assays, correlated inversely with foci formation efficiency among cells (Pearson correlation in log-scale: r = − 0.770; P = 0.025; Figure 4A), indicating that aggregation tended to be more beneficial in less permissive cells. Titration assays showed that in most cells the viral progeny derived from monodisperse inocula was not detectable at 4 hpi, whereas aggregates tended to be slightly more productive, except in Neuro2a cells (Table S1). The effects of aggregation on viral titers were more easily quantified at 6 hpi and showed a significant increase in MEFs and MRC5 cells (t-tests: P < 0.05) and a reduction in Neuro 2a, Vero, and 4T1 cells (P < 0.05; Table S1). Overall, aggregation accelerated progeny production at 6 hpi in less productive cells, as determined by the ratio of titers produced by aggregated and monodisperse inocula (r = − 0.795; P = 0.018; Figure 4B; Figure S5). Hence, again, aggregation tended to be more beneficial in difficult-to-infect cells.
Figure 4. The fitness effects of aggregation depend on cell type and innate immunity.
A-D. Analysis of MEFs, MRC5, CT26, A549, 4T1, BHK-21, Vero, and Neuro2a cells. Each data points corresponds to one cell type (average of three measurements). Dashed lines indicate least-squares regressions. A, B. The foci formation efficiency (ratio of foci to viral particles in the inoculum) was used as an indicator of cell permissivity to infection. The particle concentration was determined by nanoparticle tracking analysis (Figure S4). A, C. The fitness of aggregates was calculated as f = PG/PR, where PG is the fraction of GFP/total fluorescence in competitions between aggregated VSV-GFP and monodisperse VSV-Cherry founders, and PR the GFP/total fluorescence in competitions between monodisperse VSV-GFP and aggregated VSV-Cherry founders. Whole-well images used for inferring f are shown in Figure S3. B, D. The viral titers produced at 6 hpi by aggregates (A) and monodisperse (M) inocula were determined by the plaque assay. The ratio of these two titers is shown. Titers for each M and A inocula are shown in Figure S5. C, D. The ratio of maximal titers reached by the wild type (WT) and Δ51 variants was used as an indicator of the ability of a given cell type to mount an antiviral innate immune response. E, F. Growth curves produced by monodisperse (grey) and aggregated (orange) founders in MEFs primed with cytokines from a previous Δ51 infection (E), and in non-primed MEFs (F). Each data point is the average of three replicates. Error bars: SEM. Data are provided in Table S1.
Aggregation is more advantageous in cells displaying stronger innate immune antiviral signaling
The ability of VSV to infect a given cell type depends, among other factors, on whether cells can mount a robust antiviral response. To explore the association between the fitness advantage of aggregation and innate immunity, we used a VSV variant that carries a deletion in methionine 51 of the matrix protein M (Δ51) [20]. Whereas the wild-type protein M inhibits host gene expression, this function is severely impaired in the Δ51 variant, allowing cells to express antiviral genes more strongly [20–22]. As a result, VSV-Δ51 is attenuated in normal cells, but not in cells with innate immunity defects, such as most tumor cells [20]. Therefore, by comparing the titer yield of wild-type VSV and VSV-Δ51, we can obtain information about the ability of a given cell type to signal an innate immunity response. We found that the wild-type/Δ51 titer ratio at 24 hpi correlated positively with the fitness benefit of aggregation, suggesting that initiating the infection in an aggregated manner increases the chances of the virus to overcome innate immunity (r = 0.722; P = 0.043; Figure 4C, 4D). To explore this further, we tested whether aggregation afforded a fitness benefit in cytokine-stimulated cells. For this, we first infected MEFs with VSV-Δ51 and filtered the supernatant through 50 nm size-exclusion columns to remove the virus and collect interferon and other cytokines. Then, we primed MEFs with this conditioned medium for 1 h before adding the virus. In primed MEFs, viral spread was highly inefficient, resulting in a final titer reduction of more than two orders of magnitude compared to non-primed cells (<106 versus > 108 PFU/mL; Figure 4E, 4F). Under these conditions, fast initial growth strongly determined viral yield, and infections founded by aggregates achieved an endpoint titer fivefold higher than those founded with monodisperse particles (t-test: P = 0.006; Figure 4E), whereas in non-primed cells final yields were similar (t-test: P = 0.097; Figure 4F). Therefore, the fitness benefit of aggregation was exacerbated in cells displaying a strong antiviral response.
The benefits of aggregation precede cytokine-mediated innate immunity in non-primed cells
The fitness benefits of aggregation occurred during the first infection cycle, but cytokine-mediated antiviral responses tend to be deployed later [23]. We therefore reasoned that the advantage of aggregation did not reside in blocking innate immunity, but in producing a burst of progeny that could spread to other cells before the onset of antiviral responses. To test this, we measured expression levels of the interferon-stimulated gene Mx2 by RT-qPCR at 6 hpi in MEFs inoculated with aggregated versus monodisperse particles. In both cases, Mx2 expression was more than a thousand fold lower than that of the house keeping gene actin, consistent with the benefits of aggregation occurring before the innate immune response. Furthermore, Mx2 expression levels were higher in cells infected with aggregates than in those infected with monodisperse particles (log10 ratio of Mx2/actin mRNA levels: −3.926 ± 0.039 versus −4.316 ± 0.114; t-test: P = 0.032). Hence, interferon-signaled immunity was not blocked in cells infected with viral aggregates but, rather, appeared to simply follow virus infection.
Faster growth of aggregates also occurs in the absence of saliva
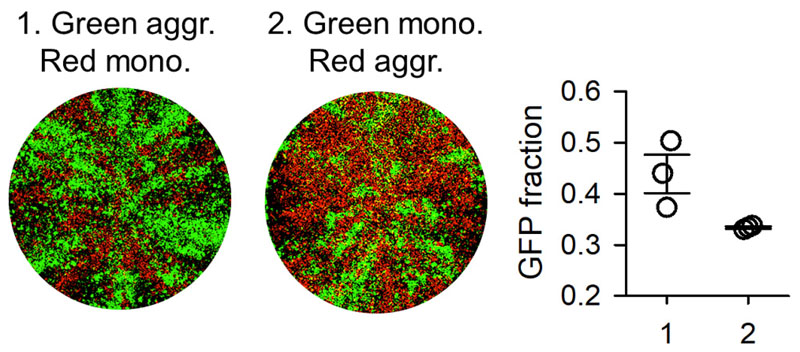
Finally, we set out to explore whether the fitness advantage of aggregation could be driven by some component of the saliva promoting infectivity. In principle, this was unlikely because saliva-treated particles were diluted strongly prior to inoculation (>1000 fold). Furthermore, this would not explain why aggregates outcompeted monodisperse particles in the same culture dish, except if the hypothetical proviral component was attached to virus aggregates and increased the permissivity of cells infected by aggregates, but not of other cells. To test whether aggregation increased fitness in the absence of saliva, we took advantage of the fact that aggregation occurs spontaneously at 37°C in high-titer preparations of gradient-purified virions [3]. Because this process is accompanied by significant virion degradation, we quantified the effect of degradation to establish our comparison for an equal infectious particle input. In competition assays between aggregated VSV-GFP and monodisperse VSV-mCherry, 43.9 ± 3.8 % of the total fluorescence signal corresponded to GFP at 24 hpi, whereas this fraction dropped to 33.4 ± 0.2 % when we competed monodisperse VSV-GFP against aggregated VSV-mCherry (t-test: P = 0.049; Figure 5). Hence, initiating the infection with particles aggregated by this method also increased viral fitness, albeit the effect was clearly less marked than with saliva. We also performed titrations at 4-6hpi. Aggregation produced a 4.2-fold increase in titer at 4 hpi (t-test: P < 0.001) and an 8.6-fold increase at 6 hpi (P < 0.001) after correcting for particle degradation (Table 1). Hence, the effect of aggregation on early viral proliferation also occurred in the absence of saliva. We note, however, that the benefits of aggregation with and without saliva are difficult to compare because of particle degradation and because the sizes of the aggregates might differ. At present, hence, we cannot formally discard the possibility that saliva might contain a proviral component that associates with viral aggregates.
Figure 5. Fitness effects of spontaneous viral aggregation in the absence of saliva.
Whole-well fluorescence microscopy images of cultures inoculated with aggregated VSV-GFP and monodisperse VSV-mCherry particles (1), and with monodisperse VSV-GFP and aggregated VSV-mCherry particles (2). Images were taken at 24 hpi and correspond to one of the three replicate assays performed. The fraction of total fluorescent area occupied by GFP-positive cells is shown on the right graph for the three replicates. Error bars: SEM.
Table 1. Early viral progeny production as determined by titration assays (PFU/mL) in MEFs inoculated with purified, monodisperse versus aggregated virions.
| Monodisperse | Aggregated (uncorrected) | Aggregated (corrected for degradation) | |
|---|---|---|---|
| 4 hpi | (7.6 ± 0.5) × 103 | (8.2 ± 0.7) × 103 | (3.1 ± 0.3) × 104 |
| 6 hpi | (2.6 ± 0.1) × 105 | (5.8 ± 0.3) × 105 | (2.2 ± 0.1) × 106 |
Discussion
Under the common perception that viruses propagate as individual virions, high cellular MOI values occur once intra-host viral densities have reached values that are sufficiently high to produce frequent coinfections by independent particles. Thus, under this conception it is difficult to envisage how a virus could overcome Allee effects operating at the cellular level during the early stages of infection. In contrast, if viruses can propagate as collective infectious units, high cMOI values can be reached even in the very first infected cells, and even if infections are initiated by a very small number of viral particles. As such, collective spread may help overcome Allee effects taking place during inter-host transmission, as well as during intra-host dissemination to new tissues/organs. The oral cavity is a preferred site of VSV replication and shedding in infected animals [24–26] and, hence, the aggregating effect of saliva could have implications for the efficacy of virus transmission in nature.
A critical or strong Allee takes place when populations go extinct or fail to establish below a certain density threshold, whereas a weak or non-critical Allee effect takes place when fitness drops at low densities but is not low enough to trigger extinction [14]. Addressing this question in viruses is complicated by the fact that individuals (i.e. potentially infectious particles) are not counted directly. Although it is possible to use physical methods to enumerate viral particles, such as electron microscopy, nanoparticle tracking analysis or tunable resistive pulse sensing, among others [27], a highly variable and often large fraction of particles are believed to be intrinsically non-infectious due to structural and/or genetic defects [28]. For this reason, viruses are typically enumerated using infectivity assays such as the plaque/foci assay and/or related methods. However, this precludes detection of critical Allee effects, because population sizes are estimated based on methods that count units capable of establishing a productive infection. In VSV infecting BHK-21, it has been suggested that the PFU/particle ratio is close to 1.0 [29], meaning that most individual particles are capable of infecting cells efficiently and, hence, that no critical Allee effect could be operating. In contrast, here we have estimated that this this ratio is on the order of 0.1 for VSV in BHK-21 cells, and as low as 0.01 in MEFs. We hence argue that the majority of viral particles fail to initiate infection in MEFs and other cell types despite containing no intrinsic structural/genetic defects, allowing for critical Allee effects. In vivo, viruses typically require a minimal dose for establishing infections, also suggesting critical Allee effects, but whether this barrier to infection operates at the cellular level or at other levels (organs, entire host) remains to be clarified.
In principle, genetic complementation offers a plausible mechanism for critical Allee effects in viruses, particularly in RNA viruses, which exhibit high mutation rates and hence high mutational loads [30]. However, the competitive advantage of aggregation was lost in our mutagenized viruses. Furthermore, aggregation reduced the overall growth of mutagenized populations. A possible explanation for these results is that genetic complementation allowed deleterious/lethal mutants present in aggregates to initiate infection, but that these mutants interfered with fitter variants at one or several steps of the infection cycle, reducing their fitness. A possible way of producing this interference is by triggering host antiviral responses. This could contribute to explaining why populations founded by aggregates grew more poorly than those founded by monodisperse particles since, in the latter, these mutants would fail to replicate and hence may not trigger innate immunity efficiently.
The fitness benefit of initiating the infection with aggregates was cell-type-dependent. In the two non-tumoral cells examined (MEFs and MRC5), aggregation provided a clear fitness benefit as judged by plaque assays, whereas in tumoral cells the effects were weaker and variable. Undoubtedly, the results obtained with MEFs and MRC5 cells are more likely to capture processes occurring in natural infections than those obtained with tumoral cells. In addition, the observed differences between tumoral and normal cells suggests candidate mechanisms for the Allee effect. Most tumoral cells exhibit innate immunity defects, rendering them more susceptible to infection than normal cells [20]. Even if the advantage of initiating cellular infection collectively preceded the onset of cytokine-mediated innate immunity, rapid diffusion of interferon in the medium might limit viral spread in neighbor cells [23, 31]. Concentrating the infection in fewer cells each receiving a higher dose of particles may allow the virus to stay ahead of such cellular innate immunity responses. It is also possible that the benefits of collective infection are related to other, earlier barriers to infection. These could include interferon-independent antiviral mechanisms triggered by pathogen-associated molecular patterns in the first infected cells. Alternatively, collective infection might reduce the risk of stochastic loss of essential viral components during the very first stages of the cellular infection cycle. All VSV proteins are essential for infection, but are expressed at different levels. Failure to produce sufficient levels of rate-limiting proteins, such as for instance the viral polymerase, could result in abortive infection, particularly in less active cells (here, non-tumoral cells). Entry of multiple particles to the same cell would reduce the risk of abortive infection because viral proteins produced by different infecting genomes would be shared, and hence early expression defects could be compensated.
Although Allee effects have not been examined directly in viruses previously, there is some evidence suggesting that collective infectious units are fitter than individual particles. For example, in poliovirus, cells infected with vesicle-encapsulated virions exhibited higher viral RNA production than free virions [5]. Also, in infectious bursal disease virus, polyploid capsids showed greater infectivity than haploid capsids [32]. Interestingly, inoculation of individual cells with vaccinia virus particles using microfluidics revealed that the probability of establishing a successful infection increased logistically with the number of particles placed per cell, revealing cooperative interactions during the early stages of infection suggestive of an Allee effect [33]. However, since in these studies per-capita viral proliferation rates were not determined, it was not possible to assess the net effect of collective infection on viral fitness. In future work, it would be interesting to evaluate whether our results apply to other viruses.
Star Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by Rafael Sanjuán (rafael.sanjuan@uv.es).
Experimental Model and Subject Details
Cell lines and culture
MEFs derived from C57BL/6 mice embryos were isolated in previous work by Dr. Carmen Rivas (Universidad de Santiago de Compostela, Spain) following standard procedures [35]. No information about the gender or exact age of the source animals was available. BHK-21 (CCL-10), A549 (CCL-185), 4T1 (CRL-2539), CT26 (CRL-2639), MRC5 (CCL-171), Neuro2a (CCL-131) and Vero (CCL-81) cells were obtained from the American Type Culture Collection (ATCC, reference number indicated in parentheses) and cultivated from low-passage stocks (typically below passage 10) for no more than 20 additional cell doublings. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in a 5% CO2 humidified incubator and tested mycoplasma-negative by PCR.
Virus
VSV was recovered from a cDNA clone originally created by Lawson et al. [34] and kindly provided by Dr. Valery Z. Grdzelishvili (University of North Carolina). Two variants of this clone were used with the GFP or mCherry gene cloned at the intergenic region between the G and L genes.
Saliva collection
Human saliva was provided with informed consent after seeking approval from the Ethics Committee of the Universitat de València. Saliva was filtered through a 0.45 μm cellulose filter to remove bacteria and debris, aliquoted, and stored at −70°C until use.
Methods Details
Viral titration
Confluent BHK-21 monolayers were used for virus titrations. The monolayers were inoculated with 200 μL of diluted viral suspensions for 45 min under standard culturing conditions (37°C, 5% CO2) and then overlaid with DMEM supplemented with 2% FBS and 0.5% or 0.6% agar. After 20-24 h, cells were fixed with 10% formaldehyde, stained with 2% crystal violet in 10% formaldehyde, and plaques were counted.
Viral aggregation in the presence of saliva
The efficacy of saliva to produce viral aggregates varies among donors [3]. All human saliva used in this work was provided by a unique female volunteer who showed the strongest aggregating effects in our previous work [3]. A virus stock (ca. 109 PFU/mL) was diluted 1:10 in saliva, incubated at 37°C for 1 h, and diluted conveniently to infect cells (ca. 1000-fold).
Gradient purification of virions
Purified VSV-GFP and VSV-mCherry stocks were prepared by inoculating eight BHK-21 confluent T175 flasks at an MOI of 0.1 PFU/cell and collecting supernatants upon appearance of the first obvious cytopathic effects (ca. 14 hpi). Large cellular debris was removed by spinning (780 g, 5 min) and filtering (0.22 μm) supernatants. Virions were then pelleted at 30,000 g, 4°C, and 1.5h in a Sorvall LYNX 6000 high-speed centrifuge with an A27-8 x 50 fixed-angle rotor. The virus pellet was resuspended in 2 mL 100mM NaCl, 0.5 mM EDTA.Na2·2H2O, 50mM Tris-HCl buffer (pH 7.4), laid on a iodixanol (Optiprep, Sigma) gradient in Nalgene polycarbonate oak ridge tubes, and centrifuged at 80,000 g, 4°C, 4.5 h. Approximately 700 μL of a whitish band generated at mid-gradient were collected, aliquoted and stored at −70°C.
Aggregation of gradient-purified virions
Gradient-purified stocks undergo spontaneous aggregation upon incubation at 37°C, but not at room temperature [3]. In high-titer purified stocks, a 30 min incubation produced frequent dual GFP/mCherry infection foci. However, this was accompanied by virion degradation. To correct for the degradation effect, we performed a calibration experiment in which we diluted the purified virions to 105 PFU/mL in the same buffer used for gradient purification. Under these conditions, aggregation became negligible because virion-virion contacts were highly unlikely, as shown by the fact that we saw no mixed GFP/mCherry foci. Hence, any change in titer was due to degradation. In this calibration experiment, after 30 min at 37°C the viral titer decayed by 3.8 ± 0.3 fold. Hence, we used this ratio as a correction factor to account for degradation, allowing us to compare the fitness of monodisperse and aggregated viruses for an equal infectious particle input.
Determination of foci formation efficiency
Gradient-purified virions were used for nanoparticle tracking analysis, which is based on video tracking of the light scattering produced by individual nanoparticles to measure their Brownian motion, obtain their diffusion coefficient, and infer their hydrodynamic diameter using Stokes-Einstein equation. Purified virions were loaded into a NanoSight NS300 equipment (Malvern) with the aid of a syringe pump to increase particle flow through the chamber, and analyzed by built-in software using default parameters. The size and concentration of virions was inferred by counting particles in five video captures of 10 s each. Then, we performed plaque assays by inoculating monolayers of each cell line with known numbers of particles and counted the number of foci produced to determine the foci formation efficiency (foci to particle ratio).
Quantitative analysis of foci
Confluent cell monolayers were inoculated with a mix of VSV-GFP/VSV-mCherry containing aggregated or monodisperse viral particles for microscopic examination of short-term viral spread. Cells were fixed at different post-inoculation times with 4% paraformaldehyde, stored overnight at 4°C, rinsed with PBS 1X, stained with DAPI (Roche) and preserved in phosphate buffer with 0.05 % azide until image acquisition. Multichannel fluorescence imaging of representative areas of the infected monolayers was performed on an IN Cell Analyzer 2000 (GE Healthcare). Filter channels used for imaging were FITC 490/20 nm excitation, 525/36 nm emission for GFP fluorescence; Texas Red 579/34 nm excitation, 624/40 nm emission for mCherry fluorescence, and DAPI 350/50 nm excitation, 455/50 nm emission for DAPI staining fluorescence with an exposure time of 400, 800 and 100 ms, respectively. Acquired images were visualized with Fiji (ImageJ) and the number of cells per infection foci was determined by manual counting, since high cell confluence extremely complicate an automated cell discrimination, especially in BHK-21 cells.
Automated real-time fluorescence microscopy
Imaging was performed in an IncuCyte S3 Live-Cell Analysis System (Essen BioScience) housed inside a humidified tissue culture incubator at 37°C and 5% CO2. Images were acquired using phase contrast, green (300-ms exposure) and red (400-ms exposure) channels in the IncuCyte S3 platform with a 4X objective in triplicate. Representative images of various time points and experimental conditions were selected and used as a reference to define image analysis masks for each acquisition channel. Images were segmented by defining a fluorescence intensity threshold after applying a background correction using the Top-Hat method. When needed, masks were fine-tuned by setting maximum and minimum object areas and eccentricity values. To infer growth parameters we used a logistic growth model of the form where t is the infection time, Pt is the percent of virus-positive cells (% confluence), PM is the maximum Pt value, r is the exponential growth rate, and c sets the initial conditions. Half times were calculated as t1/2 = c/r. The model was fit to the data by non-linear least-squares regression. In competition, assays the fraction of the total fluorescent area, i.e. GFP/(GFP+mCherry), was determined at the time point yielding maximal total fluorescence (i.e. at the growth curve plateau). The fitness of populations derived from aggregated relative to monodisperse founders was calculated as f = PG/PR, where PG is the fraction of GFP/total fluorescence in competitions between aggregated VSV-GFP and monodisperse VSV-Cherry founders, and PR the GFP/total fluorescence in competitions between monodisperse VSV-GFP and aggregated VSV-Cherry founders.
Chemical mutagenesis
VSV-GFP and VSV-mCherry were subjected separately to three serial transfers in BHK-21 cells in the presence of 5-FU 40 ug/mL. For each passage, confluent cell monolayers were pre-treated with 5-FU for 6 h, inoculated with the virus (D = 0.1 PFU/cell) and incubated for 24 h in standard infection medium supplemented with 5-FU. After each transfer, supernatants were collected and titrated by the plaque assay. 5-FU was cleared from transfer-3 supernatants by pelleting the virus at 30,000 g for 1.5 h and resuspending the pellet in medium not containing 5-FU. Next, a one-step amplification was carried out to increase the viral titer, since saliva-driven aggregation is titer-dependent. This amplification step was done by inoculating BHK-21 monolayers at high density (D = 10 PFU/cell) and collecting the supernatant at 8 hpi. A high viral density inoculum was used to favour the maintenance of deleterious mutants produced during the mutagenesis transfers. To reach a titer similar to those of non-mutagenized stocks (109 PFU/mL), we finally concentrated the viruses tenfold by centrifugation at 30,000 g for 1.5 h. Saliva-driven aggregation was verified by mixing VSV-GFP and VSV-mCherry mutagenized stocks, incubating the mix in saliva (37°C, 1 h), and enumerating doubly fluorescent foci in standard plaque assays.
Purification of cytokine-conditioned medium
Cytokine-containing conditioned medium was obtained by infecting a confluent MEF monolayer with VSV-Δ51 mutant at (D = 10 PFU/cell) and collecting the infection medium approximately at 24 hpi. Cell debris was removed from the crude infection medium by centrifugation at 5000 g for 10 min, and the supernatant was cleared of viruses and other small particles using a 0.05 μm cellulose filter (MF-Millipore; VMWP02500). The resulting purified medium was then aliquoted, stored at –70°C, and checked for the absence of viruses by plaque assays of undiluted aliquots in BHK-21 cells.
RT-qPCR
Confluent MEF monolayers cultured in 6-well plates and inoculated (D = 0.1 PFU/cell) with equal-virion, aggregated/non-aggregated VSV-GFP suspensions were used for total RNA isolation using the acid guanidinium-thiocyanate-phenol-chloroform method (TRI Reagent Solution, Invitrogen), following manufacturer’s instructions. Infected monolayers were overlaid with DMEM supplemented with 10% FBS after 45 min of virus incubation under standard culture conditions (37°C, 5% CO2). At 6 hpi, the culture medium was removed and RNA was isolated. Output concentrations for all RNA samples were adjusted to 150 ng/μL, and 3 μL were added to reverse transcription (RT) reactions carried out with gene-specific primers for either mouse Mx2 mRNA (5´tggagtcggattgacatctctg) or the β-actin mRNA (5´cagaggcatacagggacagc), and SuperScript IV Reverse Transcriptase (Invitrogen). RT reactions were performed at 55°C, following manufacturer’s instructions. The linear range of detection for the RT reaction was determined by serial dilutions of extracted mRNAs. The qPCR was performed with primers for mouse Mx2 mRNA (5´acacggtcactgaaattgtacg, 5´tcatcttttcacggttggctt) or actin mRNA (5´ctggcaccacaccttctaca, 5´tcatcttttcacggttggctt) using the 2X Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent) on an AriaMx machine (Agilent). The absence of contaminating DNA, primer-dimers and multiple amplicons in the reactions were tested by melting curve analysis and including no-RT and no-template controls. The following thermal profile was used for amplification: 95°C for 3 min, and 40 cycles of 95°C for 15 s and 60°C for 20 s.
Quantification and Statistical Analysis
All virus infections were conducted in triplicates and all measurements are reported as mean ± SEM. Statistical analyses were performed using SPSS software (IBM Analytics), and are indicated in the main text and SI figure legends. Model fitting explained in Methods Details was carried out by non-linear least squares regression using the nls function implemented in R package (www.r-project.org) and model parameters were calculated for each replicate.
Supplementary Material
Saliva-driven virion aggregation tends to increase short-term viral fitness
Infecting cells with multiple virions accelerates the per-virus release of progeny
The fitness effect of aggregation depends on cellular permissivity to infection
The benefits of aggregation correlate with the strength of cellular innate immunity
Acknowledgments
We thank María Durán-Moreno for technical assistance. This work was funded by ERC Consolidator Grant 724519 (Vis-a-Vis). I.A-M. was funded by a PhD fellowship from the Spanish Ministerio de Educación.
Footnotes
Author Contributions
I.A-M. conducted the experiments, acquired data, contributed data analysis, and contributed manuscript preparation. R.S. designed research, contributed data analysis, and wrote the article.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Sanjuán R. Collective infectious units in viruses. Trends Microbiol. 2017;22:402–412. doi: 10.1016/j.tim.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altan-Bonnet N. Extracellular vesicles are the Trojan horses of viral infection. Curr Opin Microbiol. 2016;32:77–81. doi: 10.1016/j.mib.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuevas JM, Durán-Moreno M, Sanjuán R. Multi-virion infectious units arise from free viral particles in an enveloped virus. Nat Microbiol. 2017;2:17078. doi: 10.1038/nmicrobiol.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilera ER, Erickson AK, Jesudhasan PR, Robinson CM, Pfeiffer JK. Plaques formed by mutagenized viral populations have elevated coinfection frequencies. MBio. 2017;8:e02020–02016. doi: 10.1128/mBio.02020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arantes TS, Rodrigues RA, Dos Santos Silva LK, Oliveira GP, de Souza HL, Khalil JY, de Oliveira DB, Torres AA, da Silva LL, Colson P, et al. The large Marseillevirus explores different entry pathways by forming giant infectious vesicles. J Virol. 2016;90:5246–5255. doi: 10.1128/JVI.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slack J, Arif BM. The baculoviruses occlusion-derived virus: virion structure and function. Adv Virus Res. 2007;69:99–165. doi: 10.1016/S0065-3527(06)69003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pais-Correia AM, Sachse M, Guadagnini S, Robbiati V, Lasserre R, Gessain A, Gout O, Alcover A, Thoulouze MI. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat Med. 2010;16:83–89. doi: 10.1038/nm.2065. [DOI] [PubMed] [Google Scholar]
- 9.Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, Tager AM, Luster AD, Mempel TR. HIV-infected T cells are migratory vehicles for viral dissemination. Nature. 2012;490:283–287. doi: 10.1038/nature11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mothes W, Sherer NM, Jin J, Zhong P. Virus cell-to-cell transmission. J Virol. 2010;84:8360–8368. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK. Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell Host Microbe. 2018;23:77–88 e75. doi: 10.1016/j.chom.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson DJ, Le Grand R. Cell-associated HIV mucosal transmission: the neglected pathway. J Infect Dis. 2014;210(Suppl 3):S606–608. doi: 10.1093/infdis/jiu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor CM, H A. Allee effects in biological invasions. Ecol Lett. 2005;8:14. doi: 10.1111/j.1461-0248.2011.01614.x. [DOI] [PubMed] [Google Scholar]
- 14.Kramer AM, D B, Liebhold AM, Drake JM. The evidence for Allee effects. Popul Ecol. 2009;51:14. [Google Scholar]
- 15.Shapiro-Ilan DI, Lewis EE, Schliekelman P. Aggregative group behavior in insect parasitic nematode dispersal. Int J Parasitol. 2014;44:49–54. doi: 10.1016/j.ijpara.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Kaul RB, Kramer AM, Dobbs FC, Drake JM. Experimental demonstration of an Allee effect in microbial populations. Biol Lett. 2016;12 doi: 10.1098/rsbl.2016.0070. 20160070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanjuán R. Collective properties of viral infectivity. Curr Opin Virol. 2018 doi: 10.1016/j.coviro.2018.06.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andino R, Domingo E. Viral quasispecies. Virology. 2015;479–480C:46–51. doi: 10.1016/j.virol.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanjuán R, Cuevas JM, Furio V, Holmes EC, Moya A. Selection for robustness in mutagenized RNA viruses. PLoS Genet. 2007;3:e93. doi: 10.1371/journal.pgen.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 21.Rajani KR, Pettit Kneller EL, McKenzie MO, Horita DA, Chou JW, Lyles DS. Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription. PLoS Pathog. 2012;8:e1002929. doi: 10.1371/journal.ppat.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quan B, Seo HS, Blobel G, Ren Y. Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 * Nup98) Proc Natl Acad Sci USA. 2014;111:9127–9132. doi: 10.1073/pnas.1409076111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voigt EA, Swick A, Yin J. Rapid induction and persistence of paracrine-induced cellular antiviral states arrest viral infection spread in A549 cells. Virology. 2016;496:59–66. doi: 10.1016/j.virol.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scherer CF, O'Donnell V, Golde WT, Gregg D, Estes DM, Rodriguez LL. Vesicular stomatitis New Jersey virus (VSNJV) infects keratinocytes and is restricted to lesion sites and local lymph nodes in the bovine, a natural host. Vet Res. 2007;38:375–390. doi: 10.1051/vetres:2007001. [DOI] [PubMed] [Google Scholar]
- 25.Smith PF, Howerth EW, Carter D, Gray EW, Noblet R, Berghaus RD, Stallknecht DE, Mead DG. Host predilection and transmissibility of vesicular stomatitis New Jersey virus strains in domestic cattle (Bos taurus) and swine (Sus scrofa) BMC Vet Res. 2012;8:183–188. doi: 10.1186/1746-6148-8-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith PF, Howerth EW, Carter D, Gray EW, Noblet R, Mead DG. Mechanical transmission of vesicular stomatitis New Jersey virus by Simulium vittatum (Diptera: Simuliidae) to domestic swine (Sus scrofa) J Med Entomol. 2009;46:1537–1540. doi: 10.1603/033.046.0643. [DOI] [PubMed] [Google Scholar]
- 27.Heider S, Metzner C. Quantitative real-time single particle analysis of virions. Virology. 2014;462–463:199–206. doi: 10.1016/j.virol.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klasse PJ. Molecular determinants of the ratio of inert to infectious virus particles. Prog Mol Biol Transl Sci. 2015;129:285–326. doi: 10.1016/bs.pmbts.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akpinar F, Yin J. Characterization of vesicular stomatitis virus populations by tunable resistive pulse sensing. J Virol Methods. 2015;218:71–76. doi: 10.1016/j.jviromet.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanjuan R, Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howat TJ, Barreca C, O'Hare P, Gog JR, Grenfell BT. Modelling dynamics of the type I interferon response to in vitro viral infection. J R Soc Interface. 2006;3:699–709. doi: 10.1098/rsif.2006.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luque D, Rivas G, Alfonso C, Carrascosa JL, Rodriguez JF, Caston JR. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc Natl Acad Sci USA. 2009;106:2148–2152. doi: 10.1073/pnas.0808498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stiefel P, Schmidt FI, Dorig P, Behr P, Zambelli T, Vorholt JA, Mercer J. Cooperative vaccinia infection demonstrated at the single-cell level using FluidFM. Nano Lett. 2012;12:4219–4227. doi: 10.1021/nl3018109. [DOI] [PubMed] [Google Scholar]
- 34.Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmero I, Serrano M. Induction of senescence by oncogenic Ras. Methods Enzymol. 2001;333:247–256. doi: 10.1016/s0076-6879(01)33060-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.