Summary
Benthic foraminifera are unicellular eukaryotes inhabiting sediments of aquatic environments. Several species were shown to store and use nitrate for complete denitrification, a unique energy metabolism among eukaryotes. The population of benthic foraminifera reaches high densities in oxygen-depleted marine habitats where they play a key role in the marine nitrogen cycle. However, the mechanisms of denitrification in foraminifera are still unknown, and the possibility of a contribution of associated bacteria is debated. Here, we present evidence for a novel eukaryotic denitrification pathway that is encoded in foraminiferal genomes. Large-scale genome and transcriptomes analyses reveal the presence of a denitrification pathway in foraminifera species of the genus Globobulimina. This includes the enzymes nitrite reductase (NirK) and nitric oxide reductase (Nor) as well as a wide range of nitrite/nitrate transporters (Nrt). A phylogenetic reconstruction of the enzymes’ evolutionary history uncovers evidence for an ancient acquisition of the foraminiferal denitrification pathway from prokaryotes. We propose a model for denitrification in foraminifera where a common electron transport chain is used for anaerobic and aerobic respiration. The evolution of hybrid respiration in foraminifera likely contributed to their ecological success, which is well documented in palaeontological records since the Cambrian period.
Keywords: Protists, Foraminifera, Globobulimina, Energy metabolism, Denitrification, Eukaryotic evolution, Phylogenetics, Transcriptomics, Metagenomics
Introduction
Production of biologically inaccessible dinitrogen (N2) is attributed to anaerobic oxidation of ammonium (NH4+) and to the anaerobic respiration of nitrate (NO3-) to N2, named denitrification[1]. These processes in the oceans are considered of major importance in the global nitrogen cycle[2]. Indeed, oxygen-depleted environments that are densely populated by denitrifying organisms constitute major sinks for bioavailable nitrogen-species [1]. While the production of N2 by denitrification is widespread among prokaryotes[3, 4], in eukaryotes it has been reported only in foraminifera[5].
Foraminifera are known to colonise a wide range of marine habitats where abiotic factors, especially fluctuation or depletion of oxygen availability, are key to species diversity and success[6]. Several benthic species of the order Rotaliida show denitrification activity[5, 7]. Recent studies predicted that denitrifying foraminifera contribute up to 100 % of total benthic denitrification in the Peruvian oxygen minimum zone[8], where foraminifera reach abundances of >500 individuals per cm2 [8, 9]. Furthermore, foraminifera were shown to have a NO3- storage that is suggested to accumulate in intracellular vacuoles[10] and sustains denitrification activity for months[11].
Several foraminifera species (e.g., Buliminella tenuata, and Virgulinella fragilis) were shown to harbour intracellular bacteria[12–15], and it has been suggested that these bacteria perform the denitrification reported in those species[14, 16]. In contrast, the denitrifying foraminifera Globobulimina turgida[17] (previously determined as G. pseudospinescens[18]) harbours intracellular bacteria in a low abundance, such that the denitrification rates measured for this species cannot be accounted for a substantial bacterial contribution[5]. A eukaryotic denitrification pathway has previously been described in fungi[19, 20]. However, the fungal denitrification is incomplete, where the end product is nitrous oxide (N2O) rather than N2. So far, only prokaryotes are known to encode the genetic repertoire to perform complete denitrification.
The commonly known denitrification pathway includes four enzymatic steps that catalyse the reactions NO3- → NO2- → NO → N2O → N2. Dissimilatory reduction of NO3- to NO2- is facilitated by periplasmic or membrane-bound nitrate reductase (NapA or NarG, respectively). The second denitrification step is catalysed by cd1-containing (NirS) or copper-containing (NirK) nitrite reductase. While NirS is exclusively found in prokaryotes, NirK homologs are found in a few protists and fungi[21]. Further reduction of NO to the greenhouse gas N2O is catalysed in prokaryotes by nitric oxide reductase (Nor), while an alternative enzyme (P450Nor) is documented in fungi. The last step of denitrification in prokaryotes is catalysed by nitrous oxide reductase (NosZ). Therefore, all denitrifying organisms share a similar gene repertoire of the denitrification pathway. Nonetheless, the denitrification gene set in foraminifera remains unknown. Here we investigate the genetic repertoire of the denitrification pathway in foraminifera. We analysed the whole genomes and transcriptomes of two Globobulimina species from the Gullmar Fjord (Sweden). We find eukaryotic genes encoding for the denitrification enzymes in foraminifera and investigate their evolutionary origin.
Results
Homologs of denitrification enzymes in Globobulimina spp
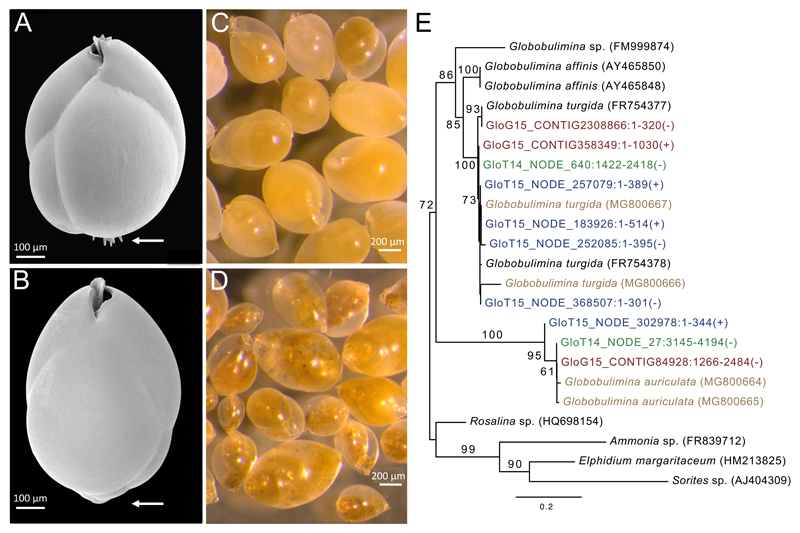
Foraminifera were obtained from sediment sampled in the Gullmar Fjord, Sweden. A total of 3,360 viable individuals of Globobulimina turgida and Globobulimina auriculata[17] were manually picked based on their morphological characteristics (Figure 1). Denitrification rates for both species were calculated from N2O production measurements following protocols previously established for foraminifera[5, 7, 22]. The denitrification rate measured for G. turgida ranges between 28 and 1712 pmol individual-1 day-1 (Table S1); and are within the range of previously measured rates for this species [5, 11]. The rate calculated for G. auriculata ranges between 29 and 124 pmol individual-1 day-1.
Figure 1. Globobulimina turgida and G. auriculata.
See also Tables S1 and S3. A) Scanning electron micrograph of a G. turgida test. This species is characterised by spines at the proloculus (arrow) and a serrated toothplate margin. B) Scanning electron micrograph of a G. auriculata test demonstrating the species’ lack of spines (arrow). C, D) Stereo micrographs showing numerous living G. turgida (C) and G. auriculata (D) individuals highlighting the differences in cytoplasm colour and test transparency. E) A species phylogenetic tree based on 18S rRNA gene sequences. GloG15 and GloT14/GloT15 prefixes correspond to genomics and transcriptomics data from our study, respectively (in red and green/blue). 18S rRNA gene sequences from specific individuals are shown in brown. Identifiers of sequences derived from assemblies of the current study include the contig identifier, sequence position and strand orientation. The tree was rooted with 18S rRNA gene sequences from other foraminifera species as outgroup. Bootstrap support values (> 50) are given at the branches, and the scale bar refers to substitutions per site. The tree supports the taxonomic classification of the species in our study and demonstrates a clear divergence of the two species into distinct clades.
The genome and the eukaryotic transcriptome were sequenced from batches of pooled individuals of both species. Genomic sequences were sorted into genomic bins based on coverage and tetra-nucleotide frequencies. Six bins containing substantial genomic information of Globobulimina, as validated by the transcriptome information, were classified as Globobulimina draft genome. The draft genome includes 48,370 contigs covering a total length of ~70 megabases. The number of duplicates per gene shows that the Globobulimina draft genome is enriched for a single species hence the species heterogeneity is low (see STAR★METHODS). Additional 26 bins were classified as bacterial taxa, and their coverage was sufficient to assemble draft genomes, while the remaining contigs (~2,260,000 contigs) were classified as unassigned bins. The presence of genes involved in the denitrification pathway in Globobulimina was tested by sequence similarity search using prokaryotic and eukaryotic query sequences. This revealed Globobulimina homologs of nitrate/nitrite transporters (Nrt) and two enzymes in the denitrification pathway: copper-containing nitrite reductase (NirK) and nitric oxide reductase (Nor).
Evidence for functional conservation of Globobulimina spp. Nrt, NirK & Nor protein sequences
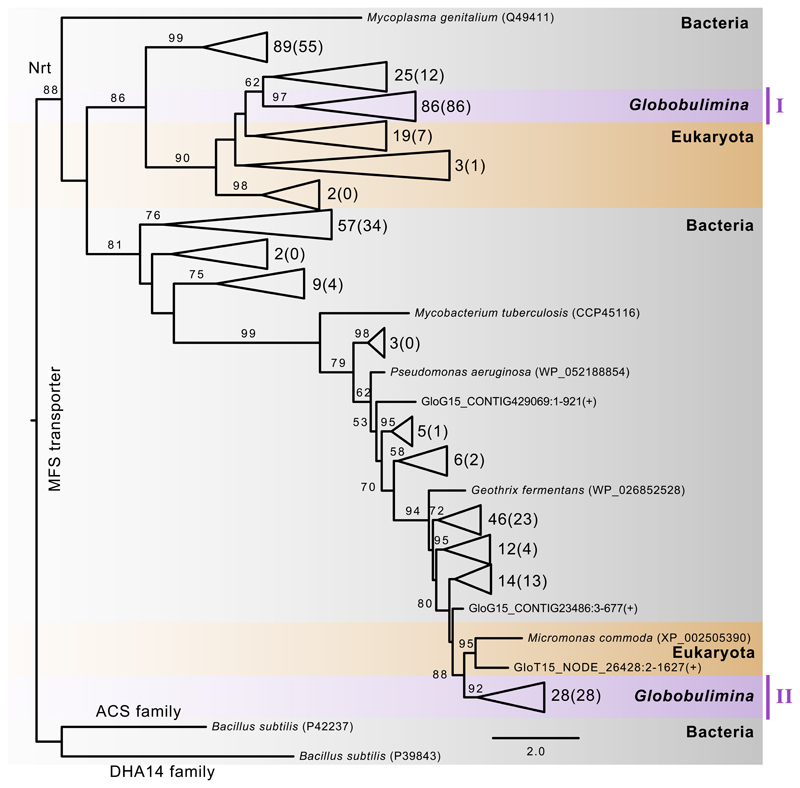
The utilization of NO3- in denitrification suggests that foraminifera harbour mechanisms to transport NO3- or NO2- The search for Nrt homologs in the Globobulimina draft genome revealed multiple protein coding sequences related to the NarK protein superfamily (Data S1A). A phylogenetic analysis of Nrt protein sequences and identified homologs show two distinct Globobulimina clades that likely correspond to two diverged protein families (Clades I and II; Figure 2). Within each clade we observe a multitude of diversified homologs (Data S2A). Clade I clusters with bacterial encoded Nrt, which would suggest an origin of the Globobulimina Nrt that is independent from other eukaryotes. To validate the bacterial neighbourhood we tested an alternative topology where we constrained a monophyletic clade including the eukaryotic homologs and Globobulimina clade I. This revealed that the likelihood of eukaryotic monophyly is not significantly different from the bacterial neighbourhood position (p-value 0.352, using AU-test). The second Globobulimina clade (clade II) has a highly supported eukaryotic nearest neighbour (Micromonas commoda); from this we conclude that the Nrt is of eukaryotic ancestry. This and a massive diversification of homologs in clade I and II indicate that NO3-/NO2- transport is an important property of Globobulimina.
Figure 2. Phylogeny of the Globobulimina nitrate/nitrite transporter (Nrt).
See also Tables S2, S4, S5 and Data S1, S2. Colour coding indicates taxonomic classification where Globobulimina clades are highlighted in purple. The tree was rooted with homologs of the DHA14 and ACS protein family as outgroup. The number of sequences included in collapsed branches is shown next to the corresponding triangles (the number of sequences derived from the current study is indicated in brackets). Additional sequences are from the genome (GloG15) and transcriptome assembly (GloT15). Bootstrap support values using 100 replicates (> 50) are denoted at the branches, and the scale bar refers to substitutions per site (see complete phylogeny in Data S2A).
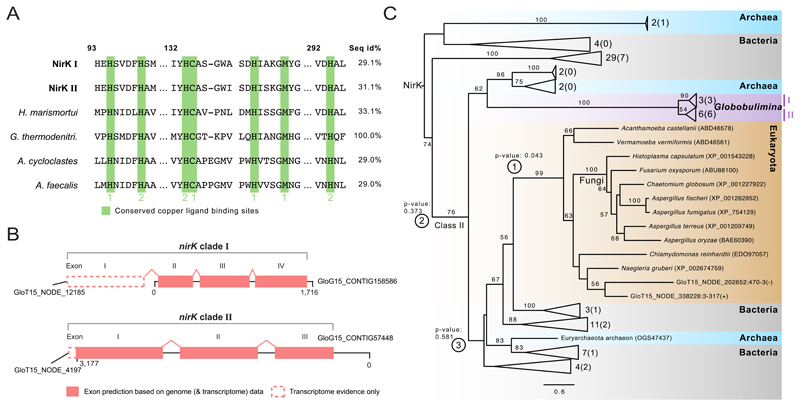
Our analysis identified homologs of NirK in Globobulimina, which contain cupredoxin domains and conserved copper binding sites that are typical for that enzyme, supporting their functional annotation as a nitrite reductase (Figure 3A; Data S1B). Notably, the Globobulimina homologs contain introns that are flanked by the canonical eukaryotic splice sites[23](Figure 3B; Data S1B). A phylogenetic reconstruction of NirK homologs reveals two highly supported sister clades (Figure 3C) including homologs having two (clade I) or three (clade II) introns (Figure 3B). Exon sequences of the NirK-encoding gene from both clades were further validated experimentally (Figure S1A-B). The phylogenetic tree further shows that the Globobulimina NirK homologs are neighbours to a prokaryotic clade (Figure 3C). To further validate the prokaryotic neighbourhood of the Globobulimina NirK, we tested the likelihood of alternative tree topologies. A eukaryotic neighbourhood of the Globobulimina clade could be rejected, albeit at a marginal confidence level (p-value 0.043, using AU-test). This shows that the Globobulimina clade is deeply branching in the tree, rather than grouping with a specific NirK sub-clade. Consequently, we conclude that the NirK origin in Globobulimina is ancient. Indeed, multiple topologies of deeper branching positions could not be statistically rejected (Figure 3C). Further search for the alternative nitrite reductase NirS reveals the presence of homologs, which are found in genomes of associated bacteria in our dataset but not in the Globobulimina transcriptome or draft genome (Data S2C).
Figure 3. Sequence and phylogeny of the Globobulimina nitrite reductase (NirK).
See also Figure S1, Tables S2–S5 and Data S1, S2. A) Sequence conservation of copper ligand binding sites in Globobulimina NirK. Copper ligand binding sites are highlighted in green. Type 1 or 2 copper binding sites are indicated. Column numbers refer to the corresponding sequence positions in the Geobacillus thermodenitrificans sequence. ‘Seq id%’ refers to the local alignment sequence identity compared to Geobacillus thermodenitrificans (100 %). For details and the complete alignment see Data S1B. B) Gene structure in Globobulimina nirK clades based on genomic contigs. Dashed lines indicate regions that are supported by transcriptome assembly but were not covered by genomic contigs. Although the automatic annotation of both genomic representatives of Globobulimina NirK was predicted to encode complete N-termini, homology to predicted transcriptome contigs indicate an incomplete gene start. C) Collapsed phylogenetic tree of Globobulimina NirK. Colour coding indicates taxonomic classification where two Globobulimina clades (clade I and II) are highlighted and form a monophyletic clade. The number of sequences represented by collapsed branches is shown next to the corresponding triangles. (The number of sequences derived from the current study is indicated in brackets.) Alternative tested topologies of the Globobulimina clades are indicated by circled numbers (① Grouping of the Globobulimina clade with other eukaryotes in contrast to all remaining sequences in the tree. ② The Globobulimina clade groups independently from NirK class II clade. ③ Grouping of the Globobulimina clade with a eukaryote-containing subclade of NirK class II). For more details and the complete phylogeny see Data S2B. The tree was rooted with the MAD method. Bootstrap support values using 100 replicates (>50) are denoted at the branches. The scale bar refers to substitutions per site.
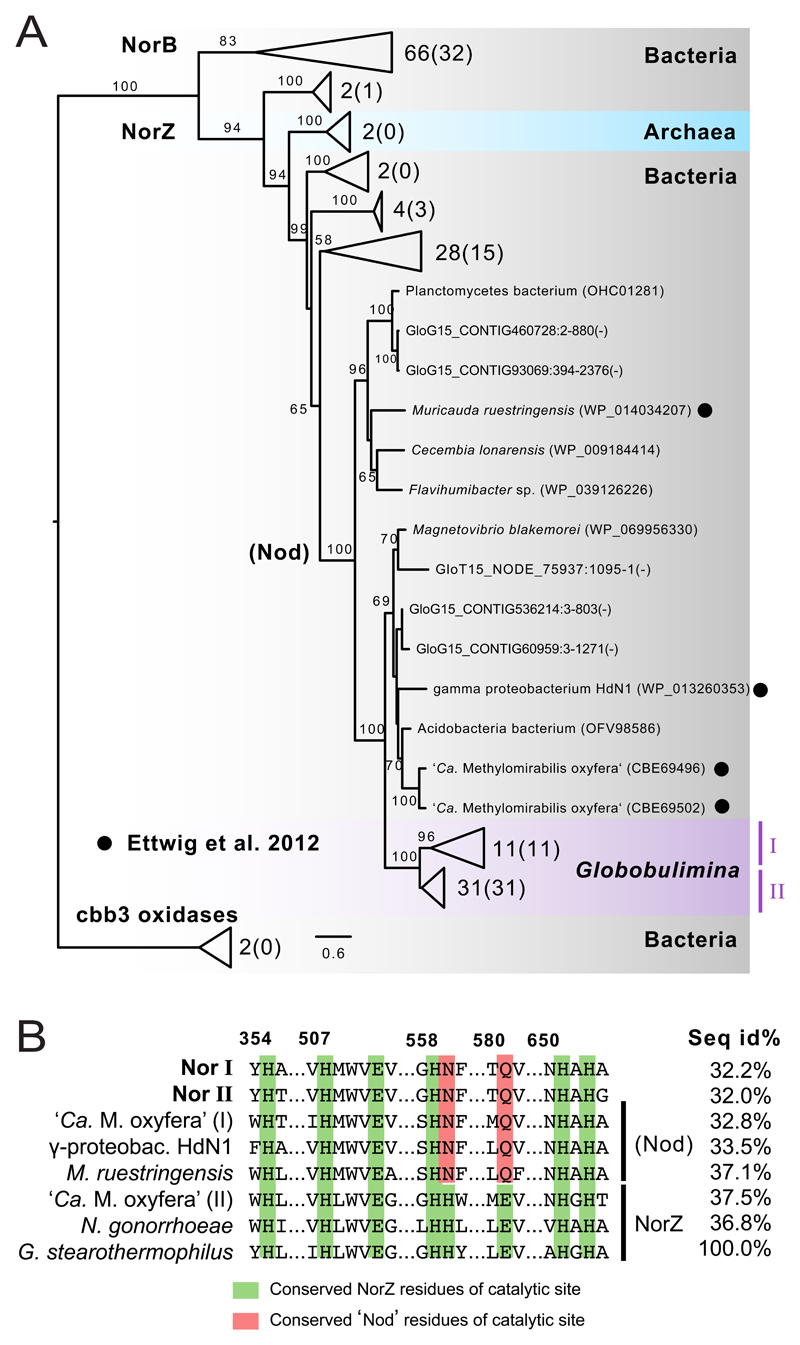
A search for Nor homologs revealed that the Globobulimina genome encodes the prokaryotic Nor whose functional annotation is supported by the presence of a conserved cytochrome oxidase I domain and a conserved catalytic site (Data S1C). One gene encoding a Nor homolog is located adjacent to a eukaryote-specific gene, and the Nor-encoding gene sequence could be validated experimentally (Figure S1C). These findings provide further support for the conclusion that Nor is indeed encoded in the Globobulimina genome. A phylogenetic reconstruction shows that the Globobulimina Nor homologs are monophyletic and nested within the bacterial NorZ clade (Figure 4A). Notably, the Globobulimina clade is neighbour to a clade containing two ‘Candidatus Methylomirabilis oxyfera’ Nor homologs. Denitrification in ‘Ca. M. oxyfera’ was reported to lack the intermediary N2O where N2 and O2 are released[24]. Consequently, it has been proposed that the ‘Ca. M. oxyfera’ Nor homolog is, in fact, a nitric oxide dismutase (Nod) that catalyses the enzymatic reaction 2NO → N2 + O2[25]. Amino acid residues of the Globobulimina Nor enzyme catalytic site are similar to Nod where two of the eight sites are identical (Figure 4B). However, since we measured N2O production in both foraminifera species in our study we can conclude that the Globobulimina Nor function is not Nod-like.
Figure 4. Phylogeny and conserved amino acid residues of the Globobulimina nitric oxide reductase (Nor).
See also Figure S1, Tables S2–S5 and Data S1, S2. A) Colour-coding indicates for taxonomic classification. Two Globobulimina clades (clade I and II) are monophyletic and highlighted in purple colour. The number of sequences represented by collapsed branches is shown next to the corresponding triangles (The number of sequences derived from the current study is indicated in brackets). The tree was rooted with two cbb3 oxidases as outgroup. Nod candidates as in Ettwig et al. 2012[24, 25] are highlighted by black circles. Uncollapsed sequences represent protein predictions from the genome (GloG15) and transcriptome assembly (GloT15). Bootstrap support values using 100 replicates are denoted at the branches (>50). The scale bar refers to substitutions per site. For more details and the complete phylogeny see Data S2D. B) Sequence conservation of catalytic sites. Amino acid residues of the catalytic site are highlighted in green and red. Two different sequences found in ‘Candidatus Methylomirabilis oxyfera’ are indicated by ‘(I)’ and ‘(II)’. ‘Seq id%’ refers to the local alignment sequence identity compared to Geobacillus thermodenitrificans (100 %), and column numbers refer to the corresponding sequence positions in G. thermodenitrificans. Details and the complete alignment are shown in Data S1C.
Missing enzymatic reactions and bacterial contribution
Our search for known denitrification genes did not detect Globobulimina homologs to enzymes catalysing the first and last steps of denitrification. Homologs of the dissimilatory nitrate reductases (NapA and NarG) as well as NosZ were found in the associated bacteria draft genomes (Figure S2; Data S2E-G). However, the possibility of bacterial contribution to the Globobulimina denitrification has been previously discarded due to their low abundance [5]. Considering the presence of nitrate storage in Globobulimina [5] it is likely that nitrate reduction occurs within the cell. Therefore, if Globobulimina encodes a dissimilatory nitrate reductase, it has to be a yet uncharacterised gene. Notably, we found homologs of the eukaryotic assimilatory nitrate reductase (Nr) in the Globobulimina draft genome. This enzyme has been shown to catalyse denitrification in the fungus Cylindrocarpon tonkinense under specific conditions[26]. The phylogenetic reconstruction and assessment of functional domains reveal that the Globobulimina Nr homologs are more closely related to sulfite oxidases than to Nr; therefore, they are better described as sulfite oxidases (Data S2H; Figure S3). Previous studies of sulfite oxidases in animals showed that these enzymes can acquire nitrate reducing activity following a replacement of only two amino acid residues (e.g., in human and chicken[27]). Thus, it is possible that the Globobulimina Nr homolog has an assimilatory nitrate reductase function.
To further validate the presence of denitrification enzymes in foraminifera, we searched the public databases for homologs to the Globobulimina enzymes. Foraminifera sequencing data is scarce in public databases yet we could find several representatives (Table S2). Only one highly fragmented draft genome of the distant species Reticulomyxa filosa [28] was found in databases, and no genomes or transcriptomes of species reported to denitrify have been published. Homologs to NirK were found in the rotaliids Brizalina sp. and Rosalina sp., while Nor homologs were found in Brizalina only. Homologs of Nrt were found in the intertidal rotaliids Elphidium margaritaceum and Ammonia spp., the miliolid foraminifera Sorites sp. as well as Rosalina sp., Nonionellina sp. and Bulimina marginata. Most of those homologous sequences are short gene fragments; nonetheless, their phylogenetic reconstruction shows a robust clustering with Globobulimina (Data S2I-K).
Discussion
Our results demonstrate that the main denitrification enzymes are encoded in the genome of Globobulimina, revealing a so far undescribed eukaryotic denitrification pathway. The pathway origin is independent of known eukaryotic enzymes and has a prokaryotic ancestry. Most of the eukaryotic genes of prokaryotic ancestry were shown to have been acquired by endosymbiotic gene transfer (EGT) from the mitochondrion ancestor at the origins of eukaryotes[29, 30]. This process is still ongoing[31], and the mechanisms involved are beginning to be unravelled[32]. Eukaryotic gene acquisition from prokaryotic donors by lateral gene transfer (LGT, as opposed to EGT) is frequently reported; however, these are often hampered by signatures of bacterial contamination, and, therefore, their interpretation is often complicated[33, 34]. We note that in our study we controlled for possible prokaryotic contamination by analysing the genome and transcriptome of Globobulimina in parallel. Furthermore, gene origin in our pipeline was determined by genomic binning approach, genomic context and phylogenetics.
The phylogeny of Nrt supports an ancient origin of the nitrate metabolism in foraminifera. The foraminiferal NirK phylogeny and the absence of eukaryotic homologs to the Nor found in foraminifera further indicate that the origin of the foraminiferal denitrification pathway is independent from the known fungal pathway. The fungal NirK gene origin is likely an EGT[21], and indeed, it is clustering with other eukaryotic taxa in our phylogenetic analysis as expected for genes of endosymbiotic origin (Figure 3C; Data S2B). The Globobulimina NirK and Nor are deep branching in all phylogenies, and the enzymes’ monophyly in foraminifera is supported by the presence of homologs in other rotaliids (Data S2I-K). These indicate an ancient origin of denitrification enzymes in foraminifera by LGT from a prokaryotic donor.
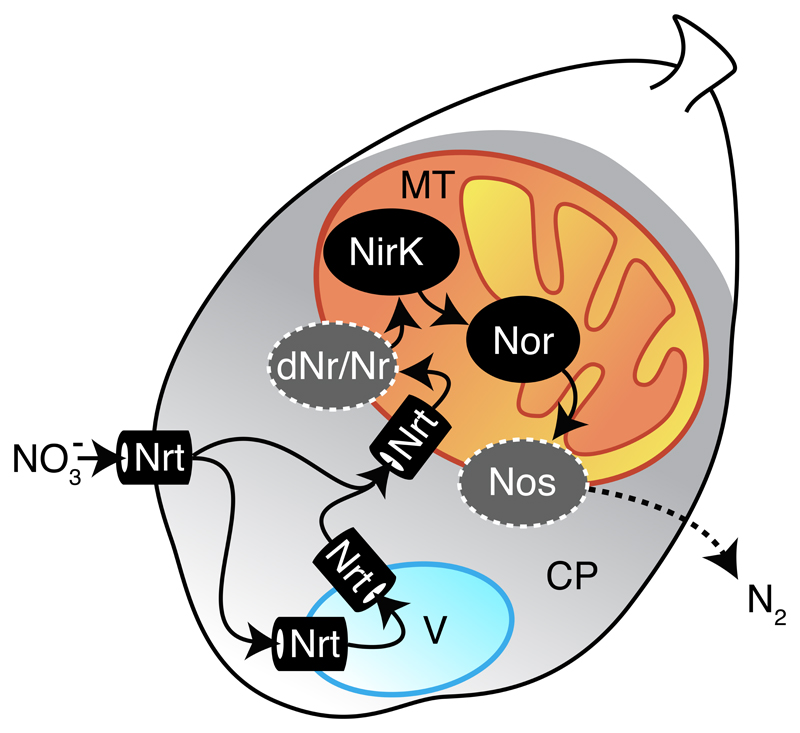
Our findings demonstrate that denitrification is performed by foraminifera rather than associated bacteria. Based on our results, we propose a denitrification model for foraminifera that draws upon known denitrification properties of fungi (Figure 5). Nitrate transport is a necessary step preceding denitrification that can be facilitated by the Nrt. The Nrt function must not be limited to transport into the cell, it can also be integrated into NO3- storage vacuoles and NO3- transport into the mitochondria. In certain foraminifera species (family Bolivinidae), mitochondria were observed to cluster near the tests’ pores in oxygen-depleted environments[35, 36]. Consequently, denitrification can be performed inside the mitochondrion, as previously reported for fungi[37] and suggested for the foraminifera Bolivina spissa[38]. We hypothesise that enzymatic components of foraminifera localise similarly to their fungal and bacterial homologs. Therefore, NirK can be localised in the mitochondrial intermembrane space and Nor in the mitochondrial inner membrane[37, 39]. Our study is lacking evidence for foraminiferal genes performing the first and last denitrification reactions. The Globobulimina sulfite oxidases homologous to Nr (Figure S3) could, hypothetically, catalyse the nitrate reduction step. However, it is also possible that so far uncharacterised or unrecognised proteins are catalysing the missing denitrification reactions. Considering a tight association of denitrifying enzymes with the respiratory chain, the putative dissimilatory nitrate reductase (dNr) and nitrous oxide reductase (Nos) enzymes are likely localised inside the mitochondrion.
Figure 5. A model for denitrification in foraminifera.
Related to Figures S2 and S3. A schematic representation of a foraminiferal cell containing cytoplasm, a vacuole and a mitochondrion is shown. Arrows indicate the suggested flow of nitrogen compounds in foraminiferal denitrification from NO3- to N2. A dashed arrow indicates for the diffusion of gaseous N2 to the outside of the cell. Hypothesised enzymatic reactions for the first and the last step of denitrification are highlighted by dark grey colour and surrounded by a dashed line. Putative bacterial contribution is shown as well. Abbreviations: MT: mitochondrion; CP: cytoplasm; Nrt: nitrate/nitrite transporter; dNr: dissimilatory nitrate reductase; Nr: eukaryotic nitrate reductase; NirK: copper-containing nitrite reductase; Nor: nitric oxide reductase; Nos: nitrous oxide reductase.
The proposed foraminiferal model suggests sharing of a common electron transport chain between aerobic respiration and denitrification, permitting the use of both electron acceptors in parallel without the need of assembling new protein complexes as reported in the fungus Fusarium oxysporum[37]. We speculate that this ability lends foraminifera a substantial ecological advantage when exposed to hypoxia or in response to fluctuating oxygen levels, and explains their success in populating a wide range of marine habitats.
Star★Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christian Woehle (cwoehle@ifam.uni-kiel.de).
Experimental Model and Subject Details
Sites description and sampling
Living foraminifera (Globobulimina turgida and Globobulimina auriculata) were sampled during two consecutive expeditions (2014 & 2015) to the Gullmar Fjord, Sweden. Sediment samples were obtained from the Alsbäck Deep (58°19.38’N, 11°32.74’E) at a depth of ca. 117 m using a 4-tube sediment interface corer (Mini Muc K/MT410). The top 3 cm of the sediments were sampled and wet sieved directly upon return to the station. Foraminifera were individually picked from the 125 to 2000 µm size fraction and cleaned in sterile artificial seawater (ASW) with 30 psu and a nitrate concentration of 20 µmol l-1 (see below, modified from Kester et al.[40]). Batches of foraminifera were frozen in liquid nitrogen (ambient condition) or incubated in culturing vessels filled with sterile ASW (see next paragraph) and flash frozen after ca. 45 h (Figure S4). The culturing vessels were incorporated to one of two culturing systems assembled to reproduce different Alsbäck Deep environmental conditions. One system was dedicated to natural oxygen concentration of the Alsbäck Deep (≈125 μmol/l) during summer whereas the second system was completely drawn down of oxygen (<10 μmol/l). Each system was filled with sterile ASW and sparged to oxygen concentration of interests with N2, CO2 and O2 pre-mixed gas (AGA Gas A; gas entries indicated by red arrows) before and during the length of the experiments. Frozen samples were stored at - 80 °C.
The sterile ASW used for rinsing and incubations was based on the natural salinity and nitrate concentration based on CTD and nutrients measurements. The water was obtained by dissolving the following salts in 3/5 of the end volume. Here we present weights and molecular weight (MW) for a 1 liter end volume ASW. A total of 23.38 g of NaCL (MW = 58.44 g/mol), 4.93 g MGSO4·7H2O (MW = 246.48 g/mol), 1.11 g CaCl2·2H2O (MW = 147.02 g/mol), 0.2 g KBr (MW = 119.01 g/mol), 0.75 g KCL (MW = 74.56 g/mol), 4.06 g MgCL2·6H20 (MW = 203.3 g/mol), and 1 ml H3BO3 (of 61.83 g/mol) were added to double-distilled-water. After the addition of the previous salts 0.023 ml of NaNO3 was added, the pH adjusted to 8 and the media autoclaved. It was important to dissolve each salt individually before the addition of a new one to decrease chance of salt precipitation. The nutrients necessary to produce the adequate ASW were autoclave separately in 10 ml glass vials. In our case 1 ml of NaH2PO4·H2O, 1 ml Na2SiO3·9H2O, and 1 ml of trace metal mix (see below) containing 100 μl selenium (H2SeO3, MW= 128,97) were added to the medium after sterilisation. The carbonate system was also added after sterilisation of the medium by sterile filtering (0.22 um) 0.17 g of NaHCO3 dissolved in 30 ml double-distilled-water and 500 μl of the vitamins directly to the medium.
The trace metal mix used for the ASW consists of 11,65 μM FeCl3·6H2O (MW = 270.3 g/mol), 11.71 μM of Na2EDTA·2H2O (MW=372.24 g/mol), 0.039 μM of CuSO4·5H2O (MW = 249.7 g/mol), 0.026 μM of Na2MoO4·2H2O (MW = 241.9 g/mol), 0.077 μM of ZnSO4·7H2O (MW = 287.54 g/mol), 0.042 μM of CoCl2·6H2O (MW = 237.93 g/mol), and 0.91 μM of MnCl2·4H2O (MW = 197.9 g/mol). Vitamins were made from 0.002 μM of Biotin (MW = 244.32 g/mol), 0.0004 μM of B12 (MW=1355.38 g/mol) and 0.30 μM of Thiamine-HCL (MW = 337.28 g/mol). The vitamins mix needed to be sterile filtered and kept in the dark.
Method Details
Nucleic acid isolation and sequencing
Foraminifera lysis and disruption (including lysozyme buffer) were done via immersion in liquid nitrogen followed by pestle-crashing. Genomic DNA and total RNA were simultaneously isolated using the AllPrep DNA/RNA Micro Kit (Qiagen) or innuPREP DNA/RNA Mini Kit (AnalytikJena). Transcriptome libraries of two consecutives years (2 independent biological replicates) were synthesised from cDNA and enrichment for eukaryotic transcripts. Libraries were prepared with TotalScript™ RNA-Seq kit (Epicentre®) with Oligo (dT) primers or NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB) with mRNA isolation performed with poly-A mRNA beads. Transcriptomics paired end libraries (2x 100 bp and 2x 125 bp) were sequenced on an Illumina HiSeq 2000 platform. Genomic libraries were prepared with the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® and were directly used for whole genome shotgun sequencing on an Illumina HiSeq 2000 platform (2x 100 bp). All samples were quantified and qualified using a Qubit® fluorometer (Invitrogen by Life Technologies™) and a Bioanalyzer & TapeStation (Agilent technology). Sequence data were deposited in NCBI (BioSamples SAMN07821823, SAMN07821824).
Validation of gene sequences
The gene sequence of the genes encoding NirK and Nor was confirmed by amplifying exon fragments using RT-PCR with cDNA synthesised with Oligo (dT) primers (NEB) as template. The reactions were performed with iTaq Universal SYBR® on the Bio-Rad CFX connect Real-Time System (Bio-Rad Laboratories). The reaction was composed of 10 μl iTaq Universal SYBR® master mix (Bio-Rad), 0.5 μl of 10 μM forward and reverse primers each (Table S3), 8 μl PCR-H2O and 1 μl of poly-A cDNA template. RT-PCR cycling conditions were 3 min at 95 °C (once), 39 cycles of 0:10 min at 95 °C, 0:30 min at 60 °C, followed by a melt curve from 58 °C to 98 °C (increment 0.2 °C 0:50). Duplicates of one biological sample per gene and no template control (NTC) were run for each primer pair.
Taxonomy
Six foraminifera-specific hypervariability 18S rRNA gene loci were amplified as described by Pawlowski and Lecroq 2010[41] and used to separate closely related species. The foraminiferal 18S rRNA gene sequence was amplified from 25–44 individuals using Phusion polymerase (NEB) and the primers 14F1 and B (Table S3). This yielded a fragment of about 1200 bp for the Globobulimina species. The PCR products were evaluated by electrophoresis, and positive samples were purified (GeneJet Gel extraction and DNA Cleanup Micro Kit; Thermo Scientific) for Sanger sequencing. Duplicates are shown in the 18S phylogenetic tree (Figure 1E). Additional 18S rRNA gene homologs were detected in NCBI public databases and the genomes and transcriptomes presented in the current study by BLAST[42]. All 18S rRNA gene sequences were aligned using MAFFT[43] 7.123b (‘linsi’ option), and a phylogeny was reconstructed using PhyML[44] ver. 20131022 (‘-b 100 -m HKY85’; Figure 1E).
Foraminiferal denitrification rate
The foraminiferal denitrification rates were calculated from linear steady state gradients of N2O in microtubes with 3–5 individuals (at least quadruplicates measurements per species). Nitrous oxide reductase was inhibited with acetylene, thereby altering the complete denitrification by making N2O the final product[45]. The steady state diffusion fluxes in the tubes corresponding to the respiration rates were calculated by Fick's first law of diffusion: Eq1: J = -D.dC/dx where J = flux; dC/dx = the measured concentration gradient; D = free diffusion coefficient of N2O. All measurements were performed in a cooling room at a constant temperature of 9 °C. However, since the temperature could not be continuously recorded inside the microchamber medium, sporadic temperature measurements were taken within a water bath next to the incubation chamber (average temperature 9.2 ± 0.1 °C; N = 19). The production of N2O was measured with a N2O microsensor[46] (Table S1), following methods previously established for foraminifera[5, 7, 11, 22]. N2O production microprofiles of ambient foraminifera were performed with freshly picked foraminifera directly after wet sieving and rinsing twice with nitrate free ASW (red sea salt) prior to the transfer to a microchamber.
Microscopic visualizations of tests and living individuals of Globobulimina spp
Specimens of the two Globobulimina species were removed from sediment samples, washed with clean seawater, dehydrated in a graded ethanol series (70 %, 80 %, 90 %, 96 % and two times 100 %; 15 min each), air-dried for 12 h in a desiccator and mounted on aluminium stubs (PLANO GmbH) using conductive and adhesive carbon pads (PLANO GmbH). Subsequently, the preparations were sputter-coated with a 10-nm-thick gold-palladium (80/20) layer using a high vacuum sputter coater Leica EM SCD500 (Leica Microsystems GmbH) and visualized with a Hitachi S-4800 field emission scanning electron microscope (Hitachi High-Technologies Corporation) at an acceleration voltage of 3 kV and an emission current of 10 mA applying a combination of the upper detector and the lower detector.
Processing of sequencing data
Sequencing resulted in total in 1.3 billion paired-end reads. This includes transcriptome datasets of two consecutive years (SRA accessions: SRR6202056, SRR6202059–SRR6202078) as well as genome data of Globobulimina (SRA accessions: SRR6202052–SRR6202055, SRR6202057–SRR6202058). Reads were quality-checked by FastQC ver. 0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc; Aug 2016). Filtering and trimming of the reads was performed using Trimmomatic[47] ver. 0.36 (Parameters: ILLUMINACLIP:primers.fa:2:30:10 LEADING:5 TRAILING:5 SLIDINGWINDOW:4:5 MINLEN:21; the file ‘primers.fa’ contains adaptor and contaminant sequences provided by Trimmomatic and FastQC). Processed paired-end reads of the transcriptome samples were assembled into contigs using SPAdes[48] ver. 3.9.1 (‘--rna’ option), which yielded 161,222 (termed ‘GloT14’, NCBI accession: GGCE00000000) and 906,588 (termed ‘GloT15’, NCBI accession: GGCD00000000) transcripts. The contigs shorter than 200 nucleotides and contamination identified by NCBI Transcriptome Shotgun Assembly (TSA) submission pipeline were excluded. Protein sequences were translated from transcripts as the longest open reading frame (ORF) using TransDecoder[49] ver. 3.0.1 (‘-m 30’ option). Transcript abundances of individual transcriptome datasets refer to Transcripts Per Million (TPM) determined by the Trinity pipeline[49] 2.4.0 (Trinity script ‘align_and_estimate_abundance.pl’) via RSEM[50] ver. 1.2.30 and Bowtie[51] 2.1.0 using paired-end reads. Paired-end reads of Globobulimina spp. genome datasets were assembled using IDBA-UD[52] ver. 1.1.1 (parameters: --pre_correction --mink 20 --maxk 120). Resulting contig sequences (termed ‘GloG15’) were classified into 241 genomic bins by MaxBin[53] ver. 2.2 (parameter: -min_contig_length 500) providing reads of individual sequencing samples separately. We assessed the quality of prokaryotic genomic bins, their completeness, coverage and first prediction of protein sequences on binned contigs with checkM[54] ver. 1.0.5. Thresholds of completeness ≥ 80 % and contamination ≤ 15 % were applied to classify bacterial bins as draft genomes (26 in total; Figure S2). The protein sequences obtained by checkM were combined with those predictions for unclassified contigs using MetaProdigal[55] ver. 2.6.2, and additional protein sequences were predicted based on genome mapping of transcriptome paired-end reads using HISAT2[56] ver. 2.0.5 (‘--non-deterministic’ option) and BRAKER[57] ver. 1.9. Similar sequences were clustered with CD-HIT[58] ver. 4.6 (‘-c 0.98’ option) in order to reduce redundancy of the complete protein catalogue. The final protein names consist of the contig IDs followed by the sequence positions covered by the CDS and an indicator for the forwards (+) or reverse (-) strand. In case of multiple exons, individual regions are separated by comma. For a high resolution taxonomic classification of prokaryotic genomic bins, DIAMOND[59] tool 0.7.11.60 (‘-k 10’ option) similarity searches of proteins predicted via checkM were applied. The first best hit (by score) per protein sequence to the NCBI non-redundant database (NR; January 2017; e-value ≤ 1e-10) was obtained, and the corresponding NCBI taxonomy assignment was further used. Identical protein taxonomy assignments per genomic bin were counted and sorted accordingly. Beginning with the most abundant taxonomy, the lowest taxonomic rank was searched that was supported by > 50 % of bin protein hits and accepted as taxonomic classification of the corresponding draft genome. Taxonomic assignments containing ‘environmental samples’ and the rank ‘Cellular organisms’ were not considered. Contigs that were not classified as bacterial or Globobulimina draft genome represent the unclassified metagenome associated with Globobulimina (NCBI accession: PJEL00000000). Six genomic bins (No. 179, 190–194) were classified as the Globobulimina draft genome (NCBI accession: PIVH00000000). All of them were annotated as eukaryotic, based on the previously described approach. Additionally, each of these bins covers more than 5 % of transcriptome read pairs mapped to binned contigs, and their coverage over different samples is highly correlated (Pairwise Pearson correlations ≥ 0.99, FDR adjusted p-values < 10-7). Overall, 93.3 % of transcriptome read pairs mapped to the Globobulimina draft genome. The corresponding assembly consists of 48,370 contigs with a GC-content of 34.25 % and a N50 of 1,797 nucleotides. The assessment of genome completeness by Benchmarking Universal Single-Copy Orthologs[60] (BUSCO v3; lineage ‘eukaryota’) method was done using all 132,080 protein predictions for Globobulimina genome contigs and recovered 63.7 % of BUSCOs completely, which shows that more than half of the genome is covered. We analysed pooled samples of two species of the genus Globobulimina (G. turgida & G. auriculata) and therefore expected our sequencing results to contain sequences derived from both genomes. As BUSCOs represent universal single-copy genes, the presence of multiple BUSCOs of the same type indicates multiple organisms in the analysis (i.e., species heterogeneity within the dataset). In the described BUSCO analysis, 44.1 % of recovered BUSCOs were inferred as multi-copy orthologs. However, this still includes duplicated predictions for the same gene locus that resulted by differing predictions based on prodigal or BRAKER. Focussing only on the 27,238 protein sequences predicted via BRAKER reduced the species heterogeneity such that only 7.12 % of complete BUSCOs were found multiple times. From this, we conclude that the Globobulimina draft genome is enriched for a single species, either G. turgida or G. auriculata. The BUSCO analysis of the transcriptome assemblies revealed a higher level of heterogeneity where 37.5 % (GloT14) or 86 % (GloT15) of the complete orthologs represented multiple copies.
Gene identification and phylogenies
To identify homologs of enzymes in the denitrification pathway, we collected query protein sequences of known enzymes from Swiss-Prot[61] and the literature (Table S4). The search for denitrification enzymes homologs in our (and external) datasets was performed with BLASTP[42] applying an e-value ≤ 1e-5 threshold. Protein sequences of hits with query coverage ≥ 40 % and sequence identity ≥ 20 % were extracted to obtain a first set of homologs. With this preliminary set, we reiterated the search in NR and RefSeq 79 database[62] using GHOSTZ[63] 1.0.0 applying a threshold of e-value ≤ 1e-5. Hit sequences were obtained and clustered with CD-HIT 4.6 (option: ‘-c 0.98’) to reduce sequence redundancy. Protein sequences were aligned with MAFFT (‘linsi’ option), and phylogenetic trees were reconstructed with FastTree[64] 2.1.7. Putative paralogs were excluded as following: i) the trees were rooted using outgroups (See Table S4), if available, and/or the Minimal Ancestor Deviation (MAD) method[65]; ii) the resulting root separated the tree into two main clades. Only one of these clades, containing orthologs, was retained, which was defined by sequence annotation and sequence representation of proteins from literature (Table S4). Further sequences that exhibit long branches or increase redundancy of the phylogenetic information were as well excluded. For a complete list of homologous sequences that were excluded from the final phylogenies see Table S5. The refined sequence set was used for the reconstruction of phylogenetic trees using PhyML (‘-m LG -b 100’ options). The resulting trees were rooted using outgroups or MAD. Clades of protein sequences encoded by the Globobulimina spp. were defined by representation in the Globobulimina genome data and transcriptome assemblies. Statistical support of alternative topologies was estimated using CONSEL[66] ver. 1.20. Homologs for comparison of functional domains and conserved amino acid residues of copper binding and catalytic sites were chosen based on available literature[25, 67, 68] (Table S4). For each enzyme, sequences were aligned using MAFFT (‘linsi’ option). The protein domains (shown in Data S1 & Figure S3) were determined using the CD-Search webserver at NCBI[69] (Aug–Nov 2017), and regions spanned by hits to specific protein domains were extracted. Pairwise local sequence identities were obtained using ‘water’ tool in the EMBOSS[70] package.
In the search for homologs of Globobulimina NirK, Nor and Nrt in further foraminifera species and related taxa, Globobulimina protein sequences were used as queries for BLAST (with a threshold of e-value ≤ 1e-5). Datasets in use include NCBI genome data (Reticulomyxa filosa, GCA_000512085.1; Astrammina rara, GCA_000211355.2), NCBI transcriptomic reads (Nonionellina sp., SRR2003403; Bulimina marginata, SRR2003397; Brizalina sp., SRR2003388; Ammonia sp., SRR2003283) and MMETSP[71] transcriptomes (Rosalina sp., MMETSP0190; Sorites sp., MMETSP0191; Ammonia sp., MMETSP1384; Elphidium margaritaceum, MMETSP1385). Nrt homologs with high sequence identity (≥ 50 %) were identified in protein sequences of MMETSP datasets and in sequencing reads of NCBI transcriptomes. Protein sequences of hits were used without modification, and the obtained sequencing reads were assembled into contigs using CAP3[72]. Final Nrt protein sequences were determined by predicting longest ORF using TransDecoder on contigs and remaining unassembled reads. Hits for Globobulimina NirK and Nor with high sequence identity (≥ 50 %) in the same dataset were only observed for Brizalina sp. sequencing reads. Further 24 reads of Rosalina were covered by hits to NirK using the same cut-off. Read sequences of individual hits to NirK and Nor were obtained and individually assembled using CAP3. Rosalina sp. associated reads were short, and the parameters recommended for assembly of short sequences (‘-i 30 -j 31 -o 18 -s 300’ options) were applied. Resulting contigs were translated into the reading frame that exhibited most sequence similarity to corresponding proteins. The sequence similarity of Brizalina sp. sequences to NirK was limited to a short region on one read (accession: SRR2003388.68811). The homologous region was extracted and translated. If represented, stop codons were replaced by ‘X’s. For protein sequences of gene fragments obtained see Table S2. Predicted protein sequences that did not show sequence similarity to corresponding query protein sequences were discarded. Protein sequences obtained in this way were added to the alignments of the denitrification proteins described before (MAFFT; ‘--addfragments’ option) for subsequent phylogenetic reconstruction via PhyML (Data S2I–K).
Quantification and Statistical Analysis
The standard deviation (σsd), standard of the mean (σSEM) and the number of individuals (N) for the denitrification measurements are presented in Table S1. An Approximate Unbiased (AU) test was used to compare optimal phylogenetic trees to alternative topologies without application of further testing procedure. Alternative topologies were inferred by constraining individual clades for tree reconstruction and their tree likelihoods were compared to the unconstrained phylogeny using AU-test via CONSEL (See Figure 2, eukaryotic monophyly of clade I: p-value 0.352; See Figure 3, Grouping of the Globobulimina clade with other eukaryotes: p-value 0.043, The Globobulimina clade groups independently from NirK class II clade: p-value 0.373, Grouping of the Globobulimina clade with a eukaryote-containing subclade of NirK class II: p-value 0.581). An α of 0.05 was applied to determine a significant difference of the alternative topology in contrast to the optimal tree.
Data and Software Availability
Data availability
Sequencing reads are available from the single read archive (SRA) accessions SRR6202052 to SRR6202078. Transcriptome assemblies were deposited at the transcriptome sequencing archive (TSA) accessions GGCE00000000 and GGCD00000000. The genome sequencing assembly is available at NCBI with the accessions PIVH00000000 to PIWH00000000 representing draft genomes and PJEL00000000 the unassigned contigs. Individually amplified 18S rRNA gene sequences of G. turgida and G. auriculata were submitted to GenBank (accessions: MG800664 to MG800667). All other information on accessing data analysed in this study is included in the manuscript or in the supplemental information. All additional datasets generated during the current study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank Giddy Landan, Chuan Ku and Nils Hülter for critical comments on the manuscript. The authors thank the Sven Lovén Centre for Marine Sciences, Kristineberg, Sweden, for their support during the sampling expeditions. We thank David Bogumil for his assistance during the 2014 expedition. We are grateful to the Kiel Marine Organism Culture Centre (KIMOCC) funded by the cluster of excellence ‘The Future Ocean’ at Kiel University who provided financial and technical support for the culturing systems. The technicians of the Biology Department of the Kiel University were also of great help in developing and producing miscellaneous components for the incubation systems. N.G. would like to thank Niels Peter Revsbech and Signe Høgslund for the kind introduction into the microprofiling techniques to measure foraminiferal denitrification rates. Furthermore, A.-S.R. would like to thank Magali Schweizer and Maria Holzmann for the help with the barcoding of the two species. The study was supported by the European Research Council (Grant No. 281357 to T.D.), the cluster of excellence ‘The Future Ocean’ and by the Deutsche Forschungsgemeinschaft (DFG) via the SFB 754 on Climate-Biogeochemistry Interactions in the Tropical Ocean and a Royal Swedish Academy of Sciences fund from the University of Gothenburg (to A.-S.R.).
Footnotes
Author contributions
A.-S.R., J.W., T.W., C.W., J.S., N.G. & T.D. designed the research strategy and performed the sampling. A.-S.R. carried out the experimental lab work. C.W. performed the bioinformatics analysis. C.H. built the incubation systems. N.G. performed the denitrification rate measurements. J.M. and A.-S.R. created the scanning electron and stereo micrographs. P.R. sequenced the genomes and transcriptomes. All authors interpreted the results and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Thamdrup B. New Pathways and Processes in the Global Nitrogen Cycle. Annu Rev Ecol Evol Syst. 2012;43:407–428. [Google Scholar]
- 2.Canfield DE, Glazer AN, Falkowski PG. The Evolution and Future of Earth’s Nitrogen Cycle. Science. 2010;330:192–196. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 3.Philippot L. Denitrifying genes in bacterial and Archaeal genomes. Biochim Biophys Acta. 2002;1577:355–376. doi: 10.1016/s0167-4781(02)00420-7. [DOI] [PubMed] [Google Scholar]
- 4.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risgaard-Petersen N, Langezaal AM, Ingvardsen S, Schmid MC, Jetten MSM, Op den Camp HJM, Derksen JWM, Piña-Ochoa E, Eriksson SP, Nielsen LP, et al. Evidence for complete denitrification in a benthic foraminifer. Nature. 2006;443:93–96. doi: 10.1038/nature05070. [DOI] [PubMed] [Google Scholar]
- 6.Murray JW. Ecology and applications of benthic foraminifera. Cambridge University Press; 2006. [Google Scholar]
- 7.Piña-Ochoa E, Høgslund S, Geslin E, Cedhagen T, Revsbech NP, Nielsen LP, Schweizer M, Jorissen F, Rysgaard S, Risgaard-Petersen N. Widespread occurrence of nitrate storage and denitrification among Foraminifera and Gromiida. Proc Natl Acad Sci USA. 2010;107:1148–1153. doi: 10.1073/pnas.0908440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glock N, Schönfeld J, Eisenhauer A, Hensen C, Mallon J, Sommer S. The role of benthic foraminifera in the benthic nitrogen cycle of the Peruvian oxygen minimum zone. Biogeosciences. 2013;10:4767–4783. [Google Scholar]
- 9.Gooday AJ, Bernhard JM, Levin LA, Suhr SB. Foraminifera in the Arabian Sea oxygen minimum zone and other oxygen-deficient settings: taxonomic composition, diversity, and relation to metazoan faunas. Deep Sea Research Part II: Topical Studies in Oceanography. 2000;47:25–54. [Google Scholar]
- 10.Bernhard JM, Edgcomb VP, Casciotti KL, McIlvin MR, Beaudoin DJ. Denitrification likely catalyzed by endobionts in an allogromiid foraminifer. ISME J. 2012;6:951–960. doi: 10.1038/ismej.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piña-Ochoa E, Koho K, Geslin E. Survival and life strategy of the foraminiferan Globobulimina turgida through nitrate storage and denitrification. Mar Ecol Prog Ser. 2010;417:39–49. [Google Scholar]
- 12.Nomaki H, Chikaraishi Y, Tsuchiya M, Toyofuku T, Ohkouchi N, Uematsu K, Tame A, Kitazato H. Nitrate uptake by foraminifera and use in conjunction with endobionts under anoxic conditions. Limnology and Oceanography. 2014;59:1879–1888. [Google Scholar]
- 13.Bernhard JM, Martin JB, Rathburn AE. Combined carbonate carbon isotopic and cellular ultrastructural studies of individual benthic foraminifera: 2. Toward an understanding of apparent disequilibrium in hydrocarbon seeps. Paleoceanography. 2010;25:PA4206. [Google Scholar]
- 14.Bernhard JM, Habura A, Bowser SS. An endobiont-bearing allogromiid from the Santa Barbara Basin: Implications for the early diversification of foraminifera. J Geophys Res. 2006;111:399. [Google Scholar]
- 15.Bernhard JM. Potential symbionts in bathyal foraminifera. Science. 2003;299:861–861. doi: 10.1126/science.1077314. [DOI] [PubMed] [Google Scholar]
- 16.Bernhard JM, Casciotti KL, McIlvin MR, Beaudoin DJ, Visscher PT, Edgcomb VP. Potential importance of physiologically diverse benthic foraminifera in sedimentary nitrate storage and respiration. J Geophys Res. 2012;117:G03002. [Google Scholar]
- 17.Bailey JW. Microscopical Examination of Soundings, Made by the U. S. Coast Survey Off the Atlantic Coast of the U. S. Smithsonian Contributions to Knowledge. 1851;2:1–15. [Google Scholar]
- 18.Emiliani C. Studio micropaleontologico di una serie calabriana. Riv Ital Paleont e Strat. 1949;54:75–77. [Google Scholar]
- 19.Shoun H, Fushinobu S, Jiang L, Kim S-W, Wakagi T. Fungal denitrification and nitric oxide reductase cytochrome P450nor. Philos Trans R Soc Lond B Biol Sci. 2012;367:1186–1194. doi: 10.1098/rstb.2011.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoun H, Kim DH, Uchiyama H, Sugiyama J. Denitrification by fungi. FEMS Microbiology Letters. 1992;73:277–281. doi: 10.1016/0378-1097(92)90643-3. [DOI] [PubMed] [Google Scholar]
- 21.Kim S-W, Fushinobu S, Zhou S, Wakagi T, Shoun H. Eukaryotic nirK genes encoding copper-containing nitrite reductase: originating from the protomitochondrion? Appl Environ Microbiol. 2009;75:2652–2658. doi: 10.1128/AEM.02536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Høgslund S, Revsbech NP, Cedhagen T, Nielsen LP, Gallardo VA. Denitrification, nitrate turnover, and aerobic respiration by benthic foraminiferans in the oxygen minimum zone off Chile. Journal of Experimental Marine Biology and Ecology. 2008;359:85–91. [Google Scholar]
- 23.Thanaraj TA, Clark F. Human GC-AG alternative intron isoforms with weak donor sites show enhanced consensus at acceptor exon positions. Nucleic Acids Res. 2001;29:2581–2593. doi: 10.1093/nar/29.12.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 25.Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MSM, Keltjens JT. Bacterial oxygen production in the dark. Front Microbiol. 2012;3:273. doi: 10.3389/fmicb.2012.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watsuji T-O, Takaya N, Nakamura A, Shoun H. Denitrification of nitrate by the fungus Cylindrocarpon tonkinense. Biosci Biotechnol Biochem. 2003;67:1115–1120. doi: 10.1271/bbb.67.1115. [DOI] [PubMed] [Google Scholar]
- 27.Qiu JA, Wilson HL, Rajagopalan KV. Structure-based alteration of substrate specificity and catalytic activity of sulfite oxidase from sulfite oxidation to nitrate reduction. Biochemistry. 2012;51:1134–1147. doi: 10.1021/bi201206v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glöckner G, Hülsmann N, Schleicher M, Noegel AA, Eichinger L, Gallinger C, Pawlowski J, Sierra R, Euteneuer U, Pillet L, et al. The genome of the foraminiferan Reticulomyxa filosa. Curr Biol. 2014;24:11–18. doi: 10.1016/j.cub.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 29.Ku C, Nelson-Sathi S, Roettger M, Sousa FL, Lockhart PJ, Bryant D, Hazkani-Covo E, McInerney JO, Landan G, Martin WF. Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524:427–432. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
- 30.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 31.Ju YS, Tubio JMC, Mifsud W, Fu B, Davies HR, Ramakrishna M, Li Y, Yates L, Gundem G, Tarpey PS, et al. Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome Res. 2015;25:814–824. doi: 10.1101/gr.190470.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diner RE, Noddings CM, Lian NC, Kang AK, McQuaid JB, Jablanovic J, Espinoza JL, Nguyen NA, Anzelmatti MA, Jansson J, et al. Diatom centromeres suggest a mechanism for nuclear DNA acquisition. Proc Natl Acad Sci USA. 2017;114:E6015–E6024. doi: 10.1073/pnas.1700764114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ku C, Martin WF. A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70 % rule. BMC Biol. 2016;14:89. doi: 10.1186/s12915-016-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koutsovoulos G, Kumar S, Laetsch DR, Stevens L, Daub J, Conlon C, Maroon H, Thomas F, Aboobaker AA, Blaxter M. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc Natl Acad Sci USA. 2016;113:5053–5058. doi: 10.1073/pnas.1600338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leutenegger S, Hansen HJ. Ultrastructural and radiotracer studies of pore function in Foraminifera. Marine Biology. 1979;54:11–16. [Google Scholar]
- 36.Bernhard JM, Goldstein ST, Bowser SS. An ectobiont-bearing foraminiferan, Bolivina pacifica, that inhabits microxic pore waters: cell-biological and paleoceanographic insights. Environ Microbiol. 2010;12:2107–2119. doi: 10.1111/j.1462-2920.2009.02073.x. [DOI] [PubMed] [Google Scholar]
- 37.Takaya N, Kuwazaki S, Adachi Y, Suzuki S, Kikuchi T, Nakamura H, Shiro Y, Shoun H. Hybrid respiration in the denitrifying mitochondria of Fusarium oxysporum. J Biochem. 2003;133:461–465. doi: 10.1093/jb/mvg060. [DOI] [PubMed] [Google Scholar]
- 38.Glock N, Eisenhauer A, Milker Y, Liebetrau V, Schönfeld J, Mallon J, Sommer S, Hensen C. Environmental influences on the pore-density in tests of Bolivina spissa. Journal of Foraminiferal Research. 2011;41:22–32. [Google Scholar]
- 39.Berks BC, Ferguson SJ, Moir JW, Richardson DJ. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 40.Kester DR, Duedall IW, Connors DN, Pytkowicz RM. Preparation of artificial seawater. Limnology and Oceanography. 1967;12:176–179. [Google Scholar]
- 41.Pawlowski J, Lecroq B. Short rDNA barcodes for species identification in foraminifera. J Eukaryotic Microbiology. 2010;57:197–205. doi: 10.1111/j.1550-7408.2009.00468.x. [DOI] [PubMed] [Google Scholar]
- 42.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 45.Smith MS, Firestone MK, Tiedje JM. The Acetylene Inhibition Method for Short-term Measurement of Soil Denitrification and its Evaluation Using Nitrogen-131. Soil Sci Soc Am J. 1978;42:611–5. [Google Scholar]
- 46.Andersen K, Kjær T, Revsbech NP. An oxygen insensitive microsensor for nitrous oxide. Sensors and Actuators. 2001;81B:42–48. [Google Scholar]
- 47.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 53.Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 54.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics. 2012;28:2223–2230. doi: 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]
- 56.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2016;32:767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 59.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014 doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 60.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 61.Boeckmann B, Bairoch A, Apweiler R, Blatter M-C, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki S, Kakuta M, Ishida T, Akiyama Y. Faster sequence homology searches by clustering subsequences. Bioinformatics. 2015;31:1183–1190. doi: 10.1093/bioinformatics/btu780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tria FDK, Landan G, Dagan T. Phylogenetic rooting using minimal ancestor deviation. Nat Ecol Evol. 2017;1 doi: 10.1038/s41559-017-0193. 0193. [DOI] [PubMed] [Google Scholar]
- 66.Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 67.Matsumoto Y, Tosha T, Pisliakov AV, Hino T, Sugimoto H, Nagano S, Sugita Y, Shiro Y. Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus. Nat Struct Mol Biol. 2012;19:238–245. doi: 10.1038/nsmb.2213. [DOI] [PubMed] [Google Scholar]
- 68.Adman ET, Godden JW, Turley S. The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted. J Biol Chem. 1995;270:27458–27474. doi: 10.1074/jbc.270.46.27458. [DOI] [PubMed] [Google Scholar]
- 69.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–31. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 71.Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data availability
Sequencing reads are available from the single read archive (SRA) accessions SRR6202052 to SRR6202078. Transcriptome assemblies were deposited at the transcriptome sequencing archive (TSA) accessions GGCE00000000 and GGCD00000000. The genome sequencing assembly is available at NCBI with the accessions PIVH00000000 to PIWH00000000 representing draft genomes and PJEL00000000 the unassigned contigs. Individually amplified 18S rRNA gene sequences of G. turgida and G. auriculata were submitted to GenBank (accessions: MG800664 to MG800667). All other information on accessing data analysed in this study is included in the manuscript or in the supplemental information. All additional datasets generated during the current study are available from the corresponding authors upon request.
Sequencing reads are available from the single read archive (SRA) accessions SRR6202052 to SRR6202078. Transcriptome assemblies were deposited at the transcriptome sequencing archive (TSA) accessions GGCE00000000 and GGCD00000000. The genome sequencing assembly is available at NCBI with the accessions PIVH00000000 to PIWH00000000 representing draft genomes and PJEL00000000 the unassigned contigs. Individually amplified 18S rRNA gene sequences of G. turgida and G. auriculata were submitted to GenBank (accessions: MG800664 to MG800667). All other information on accessing data analysed in this study is included in the manuscript or in the supplemental information. All additional datasets generated during the current study are available from the corresponding authors upon request.