Abstract
The risk of APOE for Alzheimer’s Disease (AD) is modified by age. Beyond APOE, the polygenic architecture may also be heterogeneous across age. We aim to investigate age-related genetic heterogeneity of AD and identify genomic loci with differential effects across age. Stratified gene-based genome-wide association studies (GWAS) and polygenic variation analyses were performed in the younger (60–79 years, N = 14,895) and older (≥ 80 years, N = 6,559) age-at-onset groups using Alzheimer’s Disease Genetics Consortium data. We showed a moderate genetic correlation (rg = 0.64) between the two age groups, supporting genetic heterogeneity. Heritability explained by variants on chromosome 19 (harboring APOE) was significantly larger in younger than in older onset group (P < 0.05). APOE region, BIN1, OR2S2, MS4A4E and PICALM were identified at the gene-based genome-wide significance (P < 2.73×10−6) with larger effects at younger age (except MS4A4E). For the novel gene OR2S2, we further performed leave-one-out analyses, which showed consistent effects across subsamples. Our results suggest using genetically more homogeneous individuals may help detect additional susceptible loci.
Keywords: Alzheimer’s disease, genetic heterogeneity, genetic correlation, stratified GWAS, gene-based analysis
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder. It is the most common form of dementia and is characterized by progressive memory loss and cognitive impairment(Winblad et al., 2016). The population with dementia worldwide, estimated to be 46.8 million in 2015, is expected to double every 20 years(Prince et al., 2015). AD is becoming one of the leading causes of death in the United States. In 2013 it was the sixth-leading cause of death and deaths attributed to AD increased 71%, while other major causes decreased between 2000 and 2013(Alzheimer’s, 2016).
Age is the most important risk factor of AD. Although this may result from a considerable accumulation of environmental exposures, genetic components also play a substantial role in AD, with heritability estimates of approximately 60% based on the twin study design(Gatz et al., 2006) and 24–53% based on GWAS(Lee et al., 2013; Ridge et al., 2016; Ridge et al., 2013). For late onset sporadic AD, APOE is the most hazardous susceptibility gene with moderate risk allele (ε4 allele) frequency (~15% in the US(Raber et al., 2004)). People with one or two copies of the ε4 allele were found to have, respectively, 3 or 12 times higher risk(Bertram et al., 2007) and earlier ages of AD onset(Raber et al., 2004) compared to non-carriers of the ε4 allele(Bertram et al., 2007). The lifetime risk of AD by age 85 was estimated to be 18–35% for one-copy-ε4 carriers and 51–68% for two-copy-ε4 carriers, relative to the estimated 4–12% risk for non-ε4 carriers in the European ancestry population(Genin et al., 2011). However, APOE alone only accounts for 6% of the phenotypic variations(Ridge et al., 2013). Several international collaborative consortia have conducted genome-wide association studies (GWAS) and identified at least 20 susceptibility loci with common allele frequencies but smaller effects (odds ratio < 2) on AD than APOE ε4(Harold et al., 2009; Hollingworth et al., 2011; Lambert et al., 2009; Lambert et al., 2013; Naj et al., 2011; Seshadri et al., 2010). These genetic loci associated with AD reaching GWAS significance accounted for only 2% of the phenotypic variations(Ridge et al., 2013). In addition, these susceptibility genes are largely involved in cholesterol and lipid metabolism, immune response and endosomal vesicle cycling pathways. Some are also associated with putative related etiology of AD including clearance of amyloid β and tau toxicity(Van Cauwenberghe et al., 2016). Beyond APOE and known susceptibility loci of AD, 25% of the phenotypic variations remains to be attributable to genetic variations(Ridge et al., 2013), suggesting that many risk loci are still to be discovered.
One potential reason for the relatively few identified risk genes is that the population of AD is genetically heterogeneous. APOE effects on AD have been shown to be heterogeneous across age in both retrospective(Farrer et al., 1997; Genin et al., 2011) and prospective(Bonham et al., 2016) studies, and in particular, the ε4 allele was associated with greater increase in risk among those aged 60–75 years than in older adults(Bonham et al., 2016; Farrer et al., 1997; Genin et al., 2011). The association between age at onset of AD cases and APOE has been observed in genome-wide linkage studies(Choi et al., 2011; Li et al., 2002) and recently, confirmed by two GWAS(Kamboh et al., 2012; Naj et al., 2014) which reported that one additional copy of the ε4 allele decreased age at onset in patients by 2.45 years(Naj et al., 2014). In addition, genome-wide studies also found that multiple chromosomal regions and loci other than APOE were involved in age at onset(Choi et al., 2011; Dickson et al., 2008; Holmans et al., 2005; Kamboh et al., 2012; Li et al., 2002; Naj et al., 2014), suggesting that the effect of these loci on AD may interact with age in a similar way as APOE. Such genetic heterogeneity across age has not been investigated in genome-wide studies.
Here, we investigated the genetic heterogeneity of AD by performing stratified analyses for two age-at-onset groups (60–79 years and ≥ 80 years, details in Materials and Methods) in terms of single nucleotide polymorphism (SNP) heritability estimates and their genetic correlation using a Genome-wide Complex Trait Analysis (GCTA) tool based on a multi-cohort sample from Alzheimer’s Disease Genetics Consortium (ADGC). We hypothesized that stratified analyses would reveal genetic heterogeneity between younger and older age at onset of AD, and enable identification of loci with differential effects on the two age groups.
2. Materials and methods
2.1. ADGC sample
Briefly, phase 1 of ADGC enrolled 15 cohorts from 1989 to 2011 based on case-control data, including 18,844 individuals with European ancestry aged ≥ 60 years and known covariates such as age, sex, and top 10 principal components for correcting population stratification. Phase 2 enrolled 15 cohorts, including 5,342 European ancestry individuals with covariates as phase 1 above, in which all participants were aged ≥ 60 years (except one AD case with age at onset of 58 years). The details of each cohort in phase 1(Jun et al., 2010; Naj et al., 2011; Naj et al., 2014) and phase 2(Jun et al., 2016) have been described elsewhere. The sample quality control included genotyping call rate, X-chromosome analysis for sex, identity by descent for relatedness and sample duplication(Naj et al., 2011; Naj et al., 2014). Genotyped SNPs were excluded due to low minor allele frequencies (< 0.02 for Affymetrix chips or < 0.01 for Illumina chips) or violation of Hardy-Weinberg equilibrium (P value < 10−6). Genome-wide SNP imputation was performed in each cohort using 1000 Genomes reference panel and imputed SNPs were removed if imputation quality (R2) < 0.5(Naj et al., 2014).
In our study, age of participants was defined as age at onset for AD cases and age at last visit for unaffected individuals(Desikan et al., 2017). We stratified participants into two age-at-onset groups in the stratification analyses of heritability and GWAS; 60–79 years (including one case with age 58 years) and ≥ 80 years. We chose an age cutoff of ≥ 80 years based on prior findings that ε4 effects are reduced in this age group relative to younger ages(Bonham et al., 2016). Additionally, our preliminary analyses revealed a smaller genetic correlation (rg), indicating greater genetic heterogeneity, between the older and younger groups based on the age ≥ 80 (rg = 0.64) cut-off than when age ≥ 75 years (rg = 0.75) was selected as the cut-off.
2.2. Statistical analysis
2.2.1. Whole-genome heritability and genetic correlation estimation
The heritability of AD was estimated by calculating the proportion of phenotypic variance explained by SNPs from the whole genome, which is implemented by Genome-wide Complex Trait Analysis (GCTA)(Yang et al., 2011a). GCTA fits effects of all SNPs simultaneously as random effects and effects of other covariates as fixed effects in a mixed linear model. In the regression model, the variance explained by SNPs can be estimated by the restricted maximum likelihood (REML) approach using the genetic relationship matrix (GRM), which reflects the genetic correlations across all SNPs between individuals(Yang et al., 2010). In our analysis, SNPs with minor allele frequencies > 0.01 were retained to estimate genetic relationship matrix (GRM) and we excluded related individuals using an individual-pairwise GRM threshold of < 0.025. The overall and stratified heritability estimates in the two age groups (60–79 years and ≥ 80 years) were calculated based on GRMs (random effects) in the mixed-model regression analyses with covariates (fixed effects) such as age, sex, cohort indicators, and 10 principal components. The heritability estimates were also partitioned into chromosome 19 and other chromosomes by using two GRMs, one generated from chromosome 19 and the other generated from the other 21 autosomal chromosomes, in the mixed model(Yang et al., 2011b). After removing individuals with GRM > 0.025, we combined ADGC phase 1 and 2 samples, including 12,698 and 5,198 individuals in age groups of 60–79 years and ≥ 80 years, respectively, for estimations of heritability and genetic correlation(Lee et al., 2012). The genetic correlation between the two age groups was estimated using the bivariate REML method(Lee et al., 2012), and we determined whether the resulting correlation significantly differed from 1, which implies genetic heterogeneity between the two groups. In the case-control study design, the prevalence of AD in the population should be used to correct ascertainment due to oversampled cases in the case-control ADGC sample(Lee et al., 2011). In our study, we assigned the prevalence of AD for the population aged ≥ 60 years at 0.0613, aged 60–79 years at 0.0259, and aged ≥ 80 years at 0.2217. Detailed information of the prevalence of AD source and calculation is shown in the section of “Prevalence of AD.”
2.2.2. Estimation of genetic effects of APOE ε4 alleles on AD
In whole-genome heritability estimation, heritability can be partitioned by chromosome. We estimated heritability for chromosome 19 and the other 21 chromosomes simultaneously in a mixed linear model including two GRMs and covariates (such as age, sex, cohort indicators, and 10 principal components). To estimate heritability of the APOE ε4 alleles, in the mixed linear model, we only included one GRM generated from the other 21 autosomal chromosomes and covariates and then calculated the best linear unbiased prediction (BLUP), which is the total genetic effect, and residual effect, which is the difference between BLUP and the phenotypic value for each individual(Yang et al., 2011a). We regressed residuals generated from BLUP estimation on the number of APOE ε4 alleles and obtained R2, which is the proportion of the variance of residuals that can be explained by APOE ε4 alleles. Therefore, the heritability of APOE ε4 alleles was denoted as R2 in combined ADGC phase 1 and 2 samples (N = 17,046) and across the two age groups (60–79 years, N = 12,064, and ≥ 80 years, N = 4,982). We also estimated effects of ε4 alleles in terms of odds ratios using logistic regression in the younger and older groups. We simultaneously calculated effects of one ε4 allele and two ε4 alleles for AD in the same model. The sample sizes were slightly smaller than those for whole-genome estimation due to missing APOE ε4 status for some individuals.
2.2.3. Prevalence of AD
As AD accounts for the majority of dementia cases, we used age-specific dementia prevalence (Supplementary Table 1) in the United State from a systemic meta-analysis, which included 5-year prevalence for those over 60 years of age, to estimate the prevalence of AD(Prince et al., 2013). We recalculated average prevalences for those aged 60–79 years and ≥ 80 years (Supplementary Table 1) weighted by annual estimates of the resident population by single year of age of the United States in 2015 from United States Census Bureau (Supplementary Table 1, https://www2.census.gov/programs-surveys/popest/datasets/2010-2015/national/asrh/nc-est2015-agesex-res.csv).
2.2.4. Stratified SNP-based GWAS in age 60–79 and ≥ 80 years
Genome-wide association analyses for 38,043,082 SNPs were performed in the two age-at-onset groups using logistic regressions implemented in PLINK 1.9(Chang et al., 2015). Age, sex, cohort indicators and the top 10 principal components for population structure correction were included as covariates. Individual-pairwise GRM > 0.1 were excluded from analyses to ensure sample independence. In the age 60–79 years group, 11,358 individuals (5,703 cases and 5,655 controls) in phase 1 and 3,537 individuals (1,613 cases and 1,924 controls) in phase 2 were included; in the age ≥ 80 years group, 4,801 individuals (1,942 cases and 2,859 controls) in phase 1 and 1,758 individuals (457 cases and 1,301 controls) in phase 2 were included (see details in Supplementary Table 2). We combined phase 1 and 2 samples for association analyses in the two age groups (N = 14,895 and 6,559 for the age 60–79 and ≥ 80 years groups), respectively, and obtained significant SNPs at the genome-wide significance level of P value = 5×10−8. Linkage disequilibrium (LD)-independent SNPs were identified within the significant loci after removing correlated SNPs at LD r2 > 0.1 that are within 250 kb of the top SNP base on empirical LD from European reference panel of 1000 Genomes Project phase 3 (released in May 2013)(Genomes Project et al., 2015) using PLINK 1.9. These remaining SNPs were LD-independent and the most significant in the LD block.
2.2.5. Stratified gene-based analyses in age 60–79 and ≥ 80 years
To reduce the number of tests that were conducted in SNP-based GWAS and aggregate the weak effect of each SNP within a gene, we then performed gene-based analyses using MAGMA(de Leeuw et al., 2015) implemented in FUMA(Watanabe et al., 2017). We used stratified whole GWAS results in the two age groups. The gene-based P value was calculated based on the mean of the summary statistic (χ2 statistic) of GWAS for the SNPs in a gene(de Leeuw et al., 2015; Watanabe et al., 2017). SNPs with minor allele frequencies ≥ 0.01 in the European reference panel of 1000 Genomes Project were included. The distance between two LD blocks < 250 kb were merged into a locus. In our analyses, SNPs within the genes were mapped to 18,334 loci (genes). The P value significance threshold was corrected by Bonferroni method, which is 2.73×10−6, i.e., 0.05 divided by the number of genes (18,334) and, in addition, the suggestive threshold was set to be 10−5. The stratified gene-based analyses in the age 60–79 and ≥ 80 years using summary statistics generated by stratified GWAS were performed to obtain significant genes for the two age groups, respectively. In addition, we showed the most significant SNP within each gene for the two age groups and performed Cochran’s Q-test implemented in METAL(Willer et al., 2010) for heterogeneity of the SNP effects between the younger and older groups.
3. Results
3.1. Heritability estimates and genetic correlations
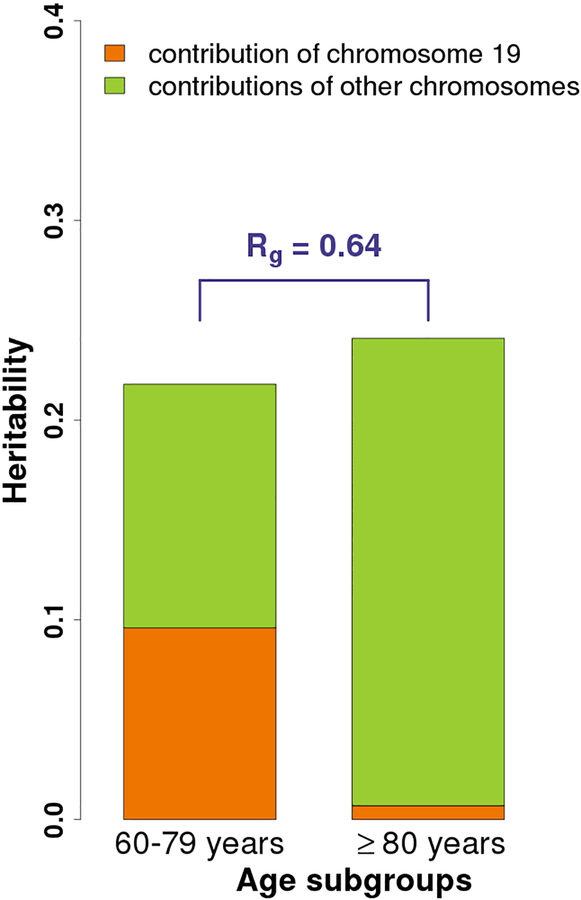
The heritability estimate of AD in combined phase 1 and 2 samples (N = 17,896) was 18.8% (95% CI 15.0% to 22.6%) for the full sample using GCTA. The heritability estimates in the two age groups and chromosomal partitioned estimates are shown in Fig. 1. The contributions of chromosome 19 were considerably different between the two age groups, with chromosome 19 having a larger impact on the younger population of AD. The genetic correlation (rg) between the two age groups was 0.64 (95% CI 0.30–0.97, P value for H0: rg = 1 was 0.043), suggesting divergent genetic components in the two age groups.
Fig. 1.
The overall and partitioned heritability estimates in combined phase 1 and 2 samples across two age groups (age 60–79 and ≥ 80 years). The heritability estimates are 16.9% (95% CI 12.9 to 20.9%) and 24.1% (95% CI 5.6 to 42.6%) for the younger (N = 12,698) and older (N = 5,198) age-at-onset groups, respectively. In the younger group, heritability estimates of chromosome 19 and others are 9.6% (95% CI 8.1 to 11.1%) and 12.2% (8.5 to 15.9%), whereas in the older group, they are 0.7% (0 to 3.8%) and 23.4% (5.1 to 41.8%), respectively. In addition, the genetic correlation (rg) between the two age groups is 0.64 (95% CI 0.30–0.97) which significantly differs from 1 (P value for H0: rg = 1 is 0.043).
3.2. Heritability of APOE and APOE effects stratified by age
The heritability of APOE ε4 was estimated to be 9.56% in the combined phase 1 and 2 samples (N = 17,046) and, 12.49% and 4.30% in the younger (N = 12,064) and older (N = 4,982) age groups, respectively. In terms of APOE effects (odds ratio) on AD, one APOE ε4 allele was estimated to increase risk of AD by 4.59 (95% CI 4.17 to 5.04) fold and two alleles 14.98 (95% CI 12.22 to 18.52) fold in the younger group. Corresponding values in the older group were 2.83 (95% CI 2.46 to 3.27) and 3.62 (95% CI 2.16 to 6.20) fold. The results suggest differential contribution of APOE ε4 alleles to risk of AD in the two age groups, with ε4 being a more important risk factor in the younger population of AD.
3.3. Stratified SNP-based GWAS: younger (age 60–79 years) and older (age ≥ 80) group
To characterize diverse genetic impacts between the younger and older age groups we performed SNP-based and gene-based genome-wide association analyses in each group.
In the younger group (N = 11,358), we identified 28 significant LD-independent SNPs on four different chromosomes (Supplementary Table 3) at P value < 5×10−8. Among those SNPs, 24 SNPs were located on chromosome 19 and four SNPs on chromosomes 1, 2 and 11. In addition to chromosome 19, three SNPs on chromosomes 2 and 11 were significant in the younger group but not (P value > 0.05) in the older group and their genetic effects for AD were stronger in the younger than the older group (Supplementary Table 3). The loci on chromosomes 1, 2 and 11 where significant SNPs were located have been reported in previous GWAS(Harold et al., 2009; Lambert et al., 2009; Lambert et al., 2013). In the older group (N = 4,801), we identified two significant LD-independent SNPs on chromosomes 19 (Supplementary Table 3), which were also identified in the younger group within APOE (Supplementary Table 3).
3.4. Stratified gene-based GWAS: younger (age 60–79 years) and older (age ≥ 80) group
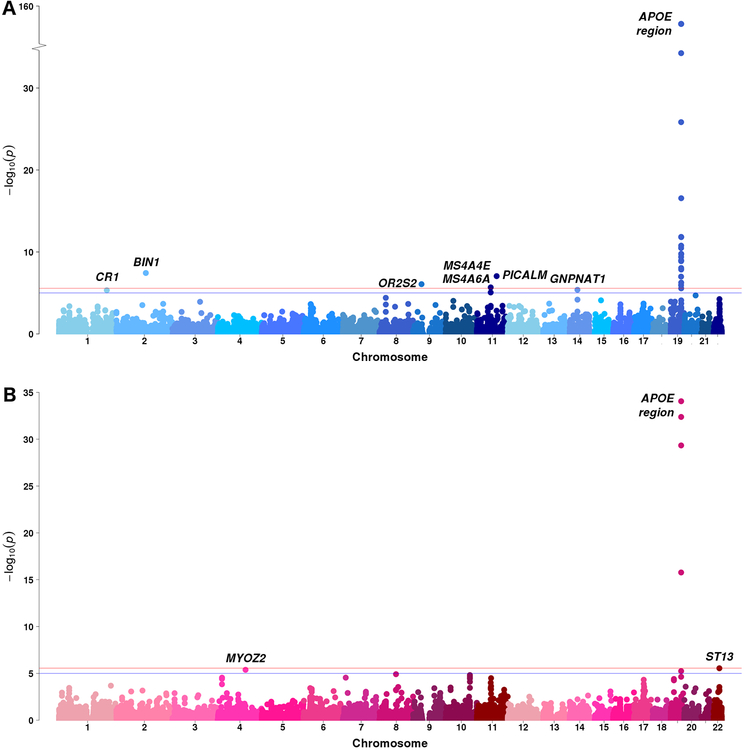
To detect novel associated loci for AD, stratified gene-based analyses were then performed in the two age groups. In the younger group (N = 14,895), in addition to genes on chromosome 19 surrounding APOE region, we identified four significant genes (BIN1, OR2S2, MS4A4E and PICALM) on chromosomes 2, 9 and 11 at P value < 2.73×10−6, and three genes (CR1, MS4A6A and GNPNAT1) on chromosomes 1, 11 and 14 are suggestive at P value < 10−5 (Table 1 and Fig. 2A). Two novel genes, OR2S2 (at the significance level) and GNPNAT1(at the suggestive level), were not reported in previous GWAS of AD. It is notable that effects of BIN1, OR2S2, PICALM and GNPNAT1 were suggestively significant (P value < 10−5) in the younger group but not (P value > 0.05) in older group in terms of the SNPs with smallest P values within genes (Table 1). The confidence intervals of odds ratios of four SNPs (rs6431219, rs1237868, rs639012 and rs73298734) in the younger and older groups were largely non-overlapping and their P values for testing heterogeneity (Cochran’s Q-test) between the two age groups were significant, except rs1237868 which was borderline significant. The results indicated their genetic effects for AD were distinct in the younger and older groups and, furthermore, they were stronger in the younger than the older group. (Table 1). In the older group (N = 6,559), other than chromosome 19, we identified two suggestive genes, MYOZ2 and ST13, on chromosomes 4 and 22 (Table 1 and Fig. 2B), which are novel for AD. Similarly, their effects had lower P values (P value ≈ 10−5) in the older group but not (P value > 0.05) in the younger with non-overlapping confidence intervals (significant for heterogeneity from the Cochran’s Q-test) and their genetic effects for AD were stronger in the older group than in the younger group (Table 1).
Table 1.
Significant (P value < 2.73×10−6) and suggestive (P value < 10−5) genes identified by stratified gene-based genome-wide analyses in ages 60–79 and ≥ 80 years using ADGC combined phase 1 and 2 samples. The top SNPs with smallest P values within genes are shown.
| P value | Top SNP in gene | A1/A2 | Age 60–79 years (younger) | Age ≥ 80 years (older) | Heterogeneity between the younger & older age groups | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Chr | Frq | N | OR (95%CI) | P value | Frq | N | OR (95%CI) | P value | P value | |||
| Top genes in age 60–79 years | |||||||||||||
| CR1 | 1 | 4.70×10−6 | rs3818361 | A/G | 0.204 | 14867 | 1.21 (1.14, 1.29) | 5.41×10−9 | 0.195 | 6554 | 1.12 (1.01, 1.23) | 0.027 | 0.159 |
| BIN1 | 2 | 3.67×10−8 | rs6431219 | T/C | 0.419 | 14225 | 1.23 (1.17, 1.30) | 6.06×10−14 | 0.408 | 6341 | 1.04 (0.96, 1.13) | 0.340 | 6.86×10−4 |
| OR2S2 | 9 | 8.45×10−7 | rs12378268 | T/C | 0.320 | 14185 | 1.14 (1.08, 1.21) | 5.43×10−6 | 0.320 | 6339 | 1.04 (0.96, 1.13) | 0.363 | 0.0695 |
| MS4A6A | 11 | 8.68×10−6 | rs1834550 | C/T | 0.389 | 14677 | 0.89 (0.84, 0.94) | 1.08×10−5 | 0.402 | 6473 | 0.86 (0.80, 0.94) | 3.38×10−4 | 0.587 |
| MS4A4E | 11 | 2.10×10−6 | rs184909761 | C/T | 0.353 | 12169 | 0.86 (0.81, 0.92) | 1.45×10−6 | 0.360 | 5340 | 0.85 (0.78, 0.93) | 5.53×10−4 | 0.863 |
| PICALM | 11 | 8.90×10−8 | rs639012 | A/G | 0.309 | 14880 | 0.86 (0.82, 0.91) | 1.86×10−7 | 0.322 | 6552 | 0.98 (0.90, 1.06) | 0.601 | 0.0132 |
| GNPNAT1 | 14 | 4.16×10−6 | rs73298734 | G/A | 0.108 | 14297 | 1.23 (1.13, 1.35) | 1.69×10−6 | 0.096 | 6311 | 0.95 (0.83, 1.09) | 0.455 | 1.29×10−3 |
| APOE* | 19 | 1.49×10−154 | rs429358 | C/T | 0.248 | 13641 | 3.93 (3.65, 4.23) | 4.90×10−286 | 0.126 | 6212 | 2.39 (2.11, 2.70) | 3.90×10−43 | 1.41×10−11 |
| Top genes in age ≥ 80 years | |||||||||||||
| MYOZ2 | 4 | 4.13×10−6 | rs4277762 | T/C | 0.299 | 14704 | 0.98 (0.93, 1.04) | 0.476 | 0.306 | 6466 | 1.20 (1.10, 1.30) | 2.05×10−5 | 8.32×10−5 |
| APOE* | 19 | 4.57×10−30 | rs429358 | C/T | 0.248 | 13641 | 3.93 (3.65, 4.23) | 4.90×10−286 | 0.126 | 6212 | 2.39 (2.11, 2.70) | 3.90×10−43 | 1.41×10−11 |
| ST13 | 22 | 2.82×10−6 | rs6002167 | T/A | 0.052 | 14844 | 1.04 (0.93, 1.17) | 0.502 | 0.051 | 6543 | 0.67 (0.55, 0.80) | 2.27×10−5 | 8.01×10−5 |
Chr: chromosome; A1: effect allele; A2: non-effect allele; Frq: allele frequency of A1; N: sample size; OR: odds ratio; 95% CI: 95% confidence interval
APOE is shown as representatives of significant genes on chromosome 19.
The genes in bold face are shown distinct genetic effects in the younger and older groups for AD in addition to genes on chromosome 19. Their effects are significant in the younger or older group but not (P value > 0.05) in the other group. The confidence intervals of those effects in the two age groups are largely non-overlapping and significantly different based on Cochran’s Q-test for heterogeneity.
Fig. 2.
Manhattan plots of gene-based genome-wide association analyses in ADGC combined phase 1 and phase 2 samples of (A) the younger age (age 60–79 years, N = 11,358) and (B) the older age (age ≥ 80 years, N = 4,801). The red line denotes the gene-based genome-wide significance level of P value = 2.73×10−6 and blue line denotes the suggestive level of P value = 2.73×10−6. The gene symbols are shown here if their P values calculated by gene-based analyses are less than the suggestive level. APOE region is shown as representatives of significant genes on chromosome 19.
For the significant locus, OR2S2, that we identified by gene-based GWAS in the younger group, we verified that the differential effects across age were observed in the phase 1 and phase 2 datasets (phase 1: Z value: 4.07, P value: 2.37 × 10−5 for the younger group, and Z value: −1.24, P value > 0.05 for the older group; phase 2: Z value: 1.59, P value = 0.056 for the younger group, and P value > 0.05 for the older group). Because the phase 2 data are under power, we were only able to observe the trend. Furthermore, to evaluate the consistency of associations within the individual cohorts in Phase 1 and Phase 2, we performed stratified gene-based analyses in the subsets of the whole sample (ADGC combined phase 1 and 2 samples) using a leave-out one cohort approach. All analytic procedures and covariates for adjustment followed the stratified gene-based GWAS. This leave-one-out method verified that P values of OR2S2 in the all subsets (Supplementary Table 4) were consistent with the P value calculated from the whole ADGC sample (Table 1).
4. Discussion
The partitioned heritability results showed that chromosome 19 explained approximately a half of heritability in the younger age-at-onset group compared to a very small proportion in the older age-at-onset group, suggesting different genetic architectures between these two age groups (Fig. 1). Using age-stratified gene-based GWAS, in addition to APOE region on chromosome 19, we identified one significant novel locus, OR2S2, which were not reported in previous GWAS using AD cases across all age groups, in the younger group (age 60–79 years). The APOE region, BIN1, PICALM and OR2S2 that we identified had stronger effects in the younger than the older group. We further performed leave-one-out analyses, which showed consistent effects of OR2S2 across subsamples. Our findings suggested that analysis in more restricted age groups with genetically homogeneous AD cases may help detect potential susceptibility loci.
An interaction of APOE with age in risk of developing AD has been shown in candidate-gene approach(Bonham et al., 2016; Farrer et al., 1997; Genin et al., 2011; Sando et al., 2008), genome-wide linkage analysis(Choi et al., 2011; Li et al., 2002), and GWAS, using age at onset as the outcome variable(Kamboh et al., 2012; Naj et al., 2014). In particular, a recent longitudinal study observed that the risk of AD for APOE ε4 carriers showed an inverted U-shaped function with peak risk between ages 70 and 75(Bonham et al., 2016). Our results based on genome-wide analyses from the large ADGC sample are consistent with this finding. We found that APOE ε4 was more deleterious for the younger cases of AD (age 60–79 vs. ≥ 80 years). In addition to AD, APOE ε4 has been reported to increase the risk of other age-related phenotypes(Ang et al., 2008), such as cognitive decline(Ihle et al., 2012; Izaks et al., 2011; Schiepers et al., 2012), cardiovascular disease(Eichner et al., 2002; Wilson et al., 1994), and mortality(Rosvall et al., 2009). Collectively, these findings support the idea that younger carriers of APOE ε4 are more vulnerable to these adverse outcomes than older carriers. Once carriers age successfully, the risks associated with APOE ε4 appear to be reduced, suggesting the potential presence of counteracting effects(Bonham et al., 2016), such as other protective genes and/or behaviors, e.g., physical activity which may reduce age-related neuroinflammation(Soto et al., 2015).
In addition to several significant hits on chromosome 19, four significant genes (BIN1, OR2S2, MS4A4E and PICALM) and three suggestive genes (CR1, MS4A6A and GNPNAT1) on chromosomes 2, 9 and 11 were identified in age 60–79 years (Table 1). We also found that APOE, BIN1, OR2S2 and PICALM have larger effects for AD in younger age group. Our findings supported associations between age at onset and know susceptibility loci of AD (BIN1, PICALM and APOE) which has been reported in a GWAS using age at onset as the outcome variable(Naj et al., 2014). The novel gene, OR2S2 (olfactory receptor family 2 subfamily S member 2), is a member of olfactory receptors involved in a neuronal response that triggers the perception of a smell and has been reported to be associated with urate levels(Huffman et al., 2015) in previous GWAS.
Chromosome 19 contributes the partition of heritability by 9.6% and 0.7% for the younger and older age groups, respectively (Fig. 1). Similarly, APOE ε4 explained 12.79% and 4.45% of the phenotypic variation in the younger and older groups, respectively. The chromosomal partition heritability was estimated using GCTA based on the restricted maximum likelihood which fits all SNPs jointly in a random-effect model so that each SNP effect is fitted conditioning on the joint effects of all the other SNPs (i.e., it accounts for LD between the SNPs)(Yang et al., 2016), and therefore, in this case, APOE ε4 explains higher variations in the regression model(Yang et al., 2016). Our result highly supported that AD is polygenic, particularly in older AD cases with higher heritability which may be contributed from other genes in addition to APOE.
In our study, heritability of AD was estimated to be 18.8% (N = 17,896, including ADGC phase 1 and 2) compared with 53.24% (N = 9,699, ADGC phase 1) in a recent study(Ridge et al., 2016), that also used subsets of the ADGC sample. The discrepancy of heritability estimates between the two studies might be due to differences in covariate adjustments, prevalences of AD, quality control criteria for including subjects, and methods to generate principal components. First, we included covariates, such as age, sex, source cohorts of individuals recruited (cohort indicators) and 10 principal components, but cohort indicators were not included in the study of Ridge et al.. In order to compare with the previous estimations we then excluded cohort indicators from the model of the current study, and thereafter heritability was estimated to be 31.9% (95% CI 27.7% to 36.2%), which is comparable to 33% from a previous study of Ridge et al. in 2013 using a subset of ADGC phase 1 sample(Ridge et al., 2013). As a result, adjustment for cohort indicators showed high impact on heritability reduction in our study. Second, for case-control study design with ascertainment bias of proportion of cases, heritability estimate is required to adjust for the prevalence of AD in the population(Lee et al., 2011). Based on 2015 age-specific population from United States Census Bureau and age-specific dementia prevalence from a systemic meta-analysis(Prince et al., 2013), we estimated the prevalence of AD to be 0.0613 among population over age 60 years (Supplementary Table 1), in contrast with 0.13 among population over age 65 years(Alzheimer’s, 2012) applied in the study of Ridge et al.(Ridge et al., 2016). Third, all individuals in our heritability analysis were non-missing for AD status, age, sex and principal components as well as their GRM < 0.025, whereas Ridge et al. removed individuals with more closely related than third cousins, and those with missing data for AD status, age, sex, principal components, APOE genotype or genotypes of the 21 known susceptibility loci of AD(Ridge et al., 2016). Finally, 10 principal components were calculated within each cohort in our study, whereas Ridge et al. estimated principal components using whole ADGC sample(Ridge et al., 2016).
In our stratified analyses to investigate heterogeneity of AD based on a continuous variable, age, we were confronted with three challenges: constant diagnostic accuracy across age, dichotomization of age and reduction of sample size in each age group. First, the previous study showed that the positive predictive value of AD based on the clinical diagnosis was 83%(Beach et al., 2012), which indicates that 17% of AD cases might be misdiagnosed. If the positive predictive value decreases with age at onset, we will include more non-cases in the older group than in the younger, and such misclassification will dilute effect sizes of susceptibility genes of AD and lead to being undetected. Second, the detrimental effect of APOE ε4 allele is higher among those aged 60–75 years and gradually diminishes in older adults(Bonham et al., 2016; Farrer et al., 1997; Genin et al., 2011). Moreover, our results from polygenic analyses showed contributions from other genetic risks become more evident to support genetic heterogeneity between AD cases below and above 80. Third, in general for stratified analysis that splits the sample into two subgroups sample size is halved at most in one group and GWAS are vulnerable to insufficient sample size, although GWAS power may increase in genetically more homogeneous groups. Future studies that include more older cases with AD (age ≥ 80 years), for which APOE have a moderate impact, may aid in identifying potential genes that may function to neutralize or postpone the effect of APOE.
Our analytical strategy for stratified analyses first used polygenic modeling to detect genetic heterogeneity of AD in terms of age at onset (60–79 vs. ≥ 80 years) and second used GWAS to uncover susceptibility genes (i.e., APOE, BIN1, OR2S2 and PICALM) with different effects in younger and older cases with AD. This strategy may help identify divergent biological mechanisms for AD cases with distinct features and/or subtypes of AD cases and provide insights for pharmaceutical development in personalized medicine.
Supplementary Material
Highlight.
Genetic heterogeneity of Alzheimer’s disease between the younger and older patients.
Heritability explained by chromosome 19 was markedly larger at younger than older age.
A novel gene, OR2S2, was found in the younger individuals using gene-based genome-wide association studies.
APOE, BIN1, OR2S2 and PICALM had stronger effects in the younger than the older age.
Our strategy may help identify divergent biological mechanisms and Alzheimer’s disease subtypes.
Acknowledgments
This study was supported by National Institute of Health R01MH100351 and R56AG061163. The Alzheimer’s Disease Genetics Consortium (ADGC) were supported by a grant from the National Institute on Aging/National Institutes of Health UO1AG032984 and complete acknowledgments for ADGC are detailed in the ADGC website (http://www.adgenetics.org/content/acknowledgements).
Footnotes
Disclosure statement
Min-Tzu Lo is employed by and receives salaries from Ambry Genetics after the first submission of this manuscript. Other authors report no actual or potential conflict of interest.
Reference
- Alzheimer’s, A., 2012. 2012 Alzheimer’s disease facts and figures. Alzheimers Dement 8(2), 131–168. [DOI] [PubMed] [Google Scholar]
- Alzheimer’s, A., 2016. 2016 Alzheimer’s disease facts and figures. Alzheimers Dement 12(4), 459–509. [DOI] [PubMed] [Google Scholar]
- Ang LS, Cruz RP, Hendel A, Granville DJ, 2008. Apolipoprotein E, an important player in longevity and age-related diseases. Exp Gerontol 43(7), 615–622. [DOI] [PubMed] [Google Scholar]
- Beach TG, Monsell SE, Phillips LE, Kukull W, 2012. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005–2010. J Neuropathol Exp Neurol 71(4), 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE, 2007. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet 39(1), 17–23. [DOI] [PubMed] [Google Scholar]
- Bonham LW, Geier EG, Fan CC, Leong JK, Besser L, Kukull WA, Kornak J, Andreassen OA, Schellenberg GD, Rosen HJ, Dillon WP, Hess CP, Miller BL, Dale AM, Desikan RS, Yokoyama JS, 2016. Age-dependent effects of APOE epsilon4 in preclinical Alzheimer’s disease. Ann Clin Transl Neurol 3(9), 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ, 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Marchani EE, Bird TD, Steinbart EJ, Blacker D, Wijsman EM, 2011. Genome scan of age-at-onset in the NIMH Alzheimer disease sample uncovers multiple loci, along with evidence of both genetic and sample heterogeneity. Am J Med Genet B Neuropsychiatr Genet 156B(7), 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D, 2015. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11(4), e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples LA, Thompson WK, Besser L, Kukull WA, Holland D, Chen CH, Brewer JB, Karow DS, Kauppi K, Witoelar A, Karch CM, Bonham LW, Yokoyama JS, Rosen HJ, Miller BL, Dillon WP, Wilson DM, Hess CP, Pericak-Vance M, Haines JL, Farrer LA, Mayeux R, Hardy J, Goate AM, Hyman BT, Schellenberg GD, McEvoy LK, Andreassen OA, Dale AM, 2017. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med 14(3), e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson MR, Li J, Wiener HW, Perry RT, Blacker D, Bassett SS, Go RC, 2008. A genomic scan for age at onset of Alzheimer’s disease in 437 families from the NIMH Genetic Initiative. Am J Med Genet B Neuropsychiatr Genet 147B(6), 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC, 2002. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am J Epidemiol 155(6), 487–495. [DOI] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM, 1997. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278(16), 1349–1356. [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL, 2006. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 63(2), 168–174. [DOI] [PubMed] [Google Scholar]
- Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, Bullido MJ, Engelborghs S, De Deyn P, Berr C, Pasquier F, Dubois B, Tognoni G, Fievet N, Brouwers N, Bettens K, Arosio B, Coto E, Del Zompo M, Mateo I, Epelbaum J, Frank-Garcia A, Helisalmi S, Porcellini E, Pilotto A, Forti P, Ferri R, Scarpini E, Siciliano G, Solfrizzi V, Sorbi S, Spalletta G, Valdivieso F, Vepsalainen S, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Hanon O, Piccardi P, Annoni G, Seripa D, Galimberti D, Licastro F, Soininen H, Dartigues JF, Kamboh MI, Van Broeckhoven C, Lambert JC, Amouyel P, Campion D, 2011. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry 16(9), 903–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C., Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR, 2015. A global reference for human genetic variation. Nature 526(7571), 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J, 2009. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41(10), 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, Alzheimer’s Disease Neuroimaging I., van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, consortium C., Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, consortium E., Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J, 2011. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43(5), 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Hamshere M, Hollingworth P, Rice F, Tunstall N, Jones S, Moore P, Wavrant DeVrieze F, Myers A, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, O’Donovan M, Jones L, Hardy J, Goate A, Lovestone S, Owen M, Williams J, 2005. Genome screen for loci influencing age at onset and rate of decline in late onset Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet 135B(1), 24–32. [DOI] [PubMed] [Google Scholar]
- Huffman JE, Albrecht E, Teumer A, Mangino M, Kapur K, Johnson T, Kutalik Z, Pirastu N, Pistis G, Lopez LM, Haller T, Salo P, Goel A, Li M, Tanaka T, Dehghan A, Ruggiero D, Malerba G, Smith AV, Nolte IM, Portas L, Phipps-Green A, Boteva L, Navarro P, Johansson A, Hicks AA, Polasek O, Esko T, Peden JF, Harris SE, Murgia F, Wild SH, Tenesa A, Tin A, Mihailov E, Grotevendt A, Gislason GK, Coresh J, D’Adamo P, Ulivi S, Vollenweider P, Waeber G, Campbell S, Kolcic I, Fisher K, Viigimaa M, Metter JE, Masciullo C, Trabetti E, Bombieri C, Sorice R, Doring A, Reischl E, Strauch K, Hofman A, Uitterlinden AG, Waldenberger M, Wichmann HE, Davies G, Gow AJ, Dalbeth N, Stamp L, Smit JH, Kirin M, Nagaraja R, Nauck M, Schurmann C, Budde K, Farrington SM, Theodoratou E, Jula A, Salomaa V, Sala C, Hengstenberg C, Burnier M, Magi R, Klopp N, Kloiber S, Schipf S, Ripatti S, Cabras S, Soranzo N, Homuth G, Nutile T, Munroe PB, Hastie N, Campbell H, Rudan I, Cabrera C, Haley C, Franco OH, Merriman TR, Gudnason V, Pirastu M, Penninx BW, Snieder H, Metspalu A, Ciullo M, Pramstaller PP, van Duijn CM, Ferrucci L, Gambaro G, Deary IJ, Dunlop MG, Wilson JF, Gasparini P, Gyllensten U, Spector TD, Wright AF, Hayward C, Watkins H, Perola M, Bochud M, Kao WH, Caulfield M, Toniolo D, Volzke H, Gieger C, Kottgen A, Vitart V, 2015. Modulation of genetic associations with serum urate levels by body-mass-index in humans. PLoS One 10(3), e0119752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle A, Bunce D, Kliegel M, 2012. APOE epsilon4 and cognitive function in early life: a meta-analysis. Neuropsychology 26(3), 267–277. [DOI] [PubMed] [Google Scholar]
- Izaks GJ, Gansevoort RT, van der Knaap AM, Navis G, Dullaart RP, Slaets JP, 2011. The association of APOE genotype with cognitive function in persons aged 35 years or older. PLoS One 6(11), e27415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC, Kunkle BW, Wang LS, Bis JC, Bellenguez C, Harold D, Lunetta KL, Destefano AL, Grenier-Boley B, Sims R, Beecham GW, Smith AV, Chouraki V, Hamilton-Nelson KL, Ikram MA, Fievet N, Denning N, Martin ER, Schmidt H, Kamatani Y, Dunstan ML, Valladares O, Laza AR, Zelenika D, Ramirez A, Foroud TM, Choi SH, Boland A, Becker T, Kukull WA, van der Lee SJ, Pasquier F, Cruchaga C, Beekly D, Fitzpatrick AL, Hanon O, Gill M, Barber R, Gudnason V, Campion D, Love S, Bennett DA, Amin N, Berr C, Tsolaki M, Buxbaum JD, Lopez OL, Deramecourt V, Fox NC, Cantwell LB, Tarraga L, Dufouil C, Hardy J, Crane PK, Eiriksdottir G, Hannequin D, Clarke R, Evans D, Mosley TH Jr., Letenneur L, Brayne C, Maier W, De Jager P, Emilsson V, Dartigues JF, Hampel H, Kamboh MI, de Bruijn RF, Tzourio C, Pastor P, Larson EB, Rotter JI, O’Donovan MC, Montine TJ, Nalls MA, Mead S, Reiman EM, Jonsson PV, Holmes C, St George-Hyslop PH, Boada M, Passmore P, Wendland JR, Schmidt R, Morgan K, Winslow AR, Powell JF, Carasquillo M, Younkin SG, Jakobsdottir J, Kauwe JS, Wilhelmsen KC, Rujescu D, Nothen MM, Hofman A, Jones L, Consortium I., Haines JL, Psaty BM, Van Broeckhoven C, Holmans P, Launer LJ, Mayeux R, Lathrop M, Goate AM, Escott-Price V, Seshadri S, Pericak-Vance MA, Amouyel P, Williams J, van Duijn CM, Schellenberg GD, Farrer LA, 2016. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry 21(1), 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, Buxbaum JD, Ertekin-Taner N, Fallin MD, Friedland R, Inzelberg R, Kramer P, Rogaeva E, St George-Hyslop P, Alzheimer’s Disease Genetics C., Cantwell LB, Dombroski BA, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Lunetta KL, Martin ER, Montine TJ, Goate AM, Blacker D, Tsuang DW, Beekly D, Cupples LA, Hakonarson H, Kukull W, Foroud TM, Haines J, Mayeux R, Farrer LA, Pericak-Vance MA, Schellenberg GD, 2010. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol 67(12), 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboh MI, Barmada MM, Demirci FY, Minster RL, Carrasquillo MM, Pankratz VS, Younkin SG, Saykin AJ, Alzheimer’s Disease Neuroimaging I., Sweet RA, Feingold E, DeKosky ST, Lopez OL, 2012. Genome-wide association analysis of age-at-onset in Alzheimer’s disease. Mol Psychiatry 17(12), 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, European Alzheimer’s Disease Initiative, I., de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P, 2009. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41(10), 1094–1099. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer's Disease, I., Genetic, Environmental Risk in Alzheimer's, D., Alzheimer's Disease Genetic, C., Cohorts for, H., Aging Research in Genomic, E., Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH Jr., Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P, 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45(12), 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Harold D, Nyholt DR, Consortium A.N., International Endogene C., Genetic, Environmental Risk for Alzheimer’s disease, C., Goddard ME, Zondervan KT, Williams J, Montgomery GW, Wray NR, Visscher PM, 2013. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Hum Mol Genet 22(4), 832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM, 2011. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet 88(3), 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR, 2012. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics 28(19), 2540–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YJ, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, Watts RL, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner BC, Jankovic J, Allen FA Jr., Goetz CG, Mastaglia F, Stajich JM, Gibson RA, Middleton LT, Saunders AM, Scott BL, Small GW, Nicodemus KK, Reed AD, Schmechel DE, Welsh-Bohmer KA, Conneally PM, Roses AD, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA, 2002. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet 70(4), 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, 2011. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43(5), 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Reitz C, Kunkle BW, Perry W, Park YS, Beecham GW, Rajbhandary RA, Hamilton-Nelson KL, Wang LS, Kauwe JS, Huentelman MJ, Myers AJ, Bird TD, Boeve BF, Baldwin CT, Jarvik GP, Crane PK, Rogaeva E, Barmada MM, Demirci FY, Cruchaga C, Kramer PL, Ertekin-Taner N, Hardy J, Graff-Radford NR, Green RC, Larson EB, St George-Hyslop PH, Buxbaum JD, Evans DA, Schneider JA, Lunetta KL, Kamboh MI, Saykin AJ, Reiman EM, De Jager PL, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Martin ER, Haines JL, Mayeux RP, Farrer LA, Schellenberg GD, Pericak-Vance MA, Alzheimer Disease Genetics C, Albert MS, Albin RL, Apostolova LG, Arnold SE, Barber R, Barnes LL, Beach TG, Becker JT, Beekly D, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Cantwell LB, Cao C, Carlson CS, Carney RM, Carrasquillo MM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cribbs DH, Crocco EA, DeCarli C, DeKosky ST, Dick M, Dickson DW, Duara R, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Kramer JH, LaFerla FM, Lah JJ, Leverenz JB, Levey AI, Li G, Lieberman AP, Lin CF, Lopez OL, Lyketsos CG, Mack WJ, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Murrell JR, Olichney JM, Pankratz VS, Parisi JE, Paulson HL, Peskind E, Petersen RC, Pierce A, Poon WW, Potter H, Quinn JF, Raj A, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosen HJ, Rosenberg RN, Sano M, Schneider LS, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Tanzi RE, Thornton-Wells TA, Trojanowski JQ, Troncoso JC, Valladares O, Van Deerlin VM, Van Eldik LJ, Vardarajan BN, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Wishnek S, Woltjer RL, Wright CB, Younkin SG, Yu CE, Yu L, 2014. Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol 71(11), 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince M, Bryce R, Albanese E, Wimo A, Ribeiro W, Ferri CP, 2013. The global prevalence of dementia: a systematic review and metaanalysis. Alzheimers Dement 9(1), 63–75 e62. [DOI] [PubMed] [Google Scholar]
- Prince M, Wimo A, Guerchet M, Ali GC, Wu Y, Prina AM, 2015. World Alzheimer Report 2015: The global impact of dementia An analysis of prevalence, incidence, costs and trends. London: Alzheimer’s Disease International. [Google Scholar]
- Raber J, Huang Y, Ashford JW, 2004. ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiol Aging 25(5), 641–650. [DOI] [PubMed] [Google Scholar]
- Ridge PG, Hoyt KB, Boehme K, Mukherjee S, Crane PK, Haines JL, Mayeux R, Farrer LA, Pericak-Vance MA, Schellenberg GD, Kauwe JS, Alzheimer’s Disease Genetics C., 2016. Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol Aging 41, 200 e213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge PG, Mukherjee S, Crane PK, Kauwe JS, Alzheimer’s Disease Genetics C., 2013. Alzheimer’s disease: analyzing the missing heritability. PLoS One 8(11), e79771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall L, Rizzuto D, Wang HX, Winblad B, Graff C, Fratiglioni L, 2009. APOE-related mortality: effect of dementia, cardiovascular disease and gender. Neurobiol Aging 30(10), 1545–1551. [DOI] [PubMed] [Google Scholar]
- Sando SB, Melquist S, Cannon A, Hutton ML, Sletvold O, Saltvedt I, White LR, Lydersen S, Aasly JO, 2008. APOE epsilon 4 lowers age at onset and is a high risk factor for Alzheimer’s disease; a case control study from central Norway. BMC Neurol 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiepers OJ, Harris SE, Gow AJ, Pattie A, Brett CE, Starr JM, Deary IJ, 2012. APOE E4 status predicts age-related cognitive decline in the ninth decade: longitudinal follow-up of the Lothian Birth Cohort 1921. Mol Psychiatry 17(3), 315–324. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT Jr., Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM, Consortium C., Consortium G., Consortium E., 2010. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303(18), 1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto I, Graham LC, Richter HJ, Simeone SN, Radell JE, Grabowska W, Funkhouser WK, Howell MC, Howell GR, 2015. APOE Stabilization by Exercise Prevents Aging Neurovascular Dysfunction and Complement Induction. PLoS Biol 13(10), e1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauwenberghe C, Van Broeckhoven C, Sleegers K, 2016. The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet Med 18(5), 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D, 2017. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8(1), 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR, 2010. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26(17), 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PW, Myers RH, Larson MG, Ordovas JM, Wolf PA, Schaefer EJ, 1994. Apolipoprotein E alleles, dyslipidemia, and coronary heart disease. The Framingham Offspring Study. JAMA 272(21), 1666–1671. [PubMed] [Google Scholar]
- Winblad B, Amouyel P, Andrieu S, Ballard C, Brayne C, Brodaty H, Cedazo-Minguez A, Dubois B, Edvardsson D, Feldman H, Fratiglioni L, Frisoni GB, Gauthier S, Georges J, Graff C, Iqbal K, Jessen F, Johansson G, Jonsson L, Kivipelto M, Knapp M, Mangialasche F, Melis R, Nordberg A, Rikkert MO, Qiu C, Sakmar TP, Scheltens P, Schneider LS, Sperling R, Tjernberg LO, Waldemar G, Wimo A, Zetterberg H, 2016. Defeating Alzheimer’s disease and other dementias: a priority for European science and society. Lancet Neurol 15(5), 455–532. [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM, 2010. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42(7), 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM, 2011a. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88(1), 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Wray NR, Goddard ME, Visscher PM, 2016. GCTA-GREML accounts for linkage disequilibrium when estimating genetic variance from genome-wide SNPs. Proc Natl Acad Sci U S A 113(32), E4579–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM, 2011b. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet 43(6), 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.