Abstract
Viral infections in human are leading cause of mortality and morbidity across the globe. Several viruses (including HIV and Herpesvirus), have evolved ingenious strategies to evade host-immune system and persist life-long. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) is an ancient antiviral system recently discovered in bacteria that has shown tremendous potential as a precise, invariant genome editing tool. Using CRISPR-Cas based system to activate host defenses or genetic modification of viral genome can provide novel, exciting and successful antiviral mechanisms and treatment modalities. In this review, we will provide progress on the CRISPR-Cas based antiviral approaches that facilitate clearance of virus-infected cells and/or prohibit virus infection or replication. We will discuss on the possibilities of CRIPSR-Cas as prophylaxis and therapy in viral infections and review the challenges of this potent gene editing technology.
Keywords: CRISPR-Cas, Viruses, Antiviral immunity, Gene editing, Restriction factors
1. Introduction
1.1. Genome Engineering
Genetic engineering is a powerful technology that has resulted in the advancements of many fields such as medicine, industrial biotechnology and agriculture [1,2]. Specifically, the ability to alter genetic makeup of an organism has revolutionized medical research and expanded our ability to understand disease development and prevention. The application of various genome engineering tools such as zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALENs) and, most recently, CRISPR-Cas systems have advanced biological research and allowed for novel discoveries to be made.
ZFN and TALEN are both synthetic proteins with nucleus activity that achieve specific DNA binding via protein-DNA interactions. However, retargeting the specific activity of these nucleases is achieved through protein engineering or cloning which limits its versatility, feasibility and application. Recently, harnessing the CRISPR-Cas system, initially observed in prokaryotes, as a gene editing tool in eukaryotic cells has revolutionized the field as a robust and efficient tool for precise genome editing [3-5]. CRISPR/Cas system is a prokaryotic adaptive immune system that confers resistance to foreign genetic elements and has been repurposed by researchers as an invariant genome editing tool that can be more easily redirected to a new target [6].
The use of genome editing technology, primarily CRISPR, is reaching exciting stages and surpassing milestones in basic science research with applications extending beyond genome editing. More recently, CRISPR is being used as an antiviral by targeting genes in both the human genome as well as the invading viral genome. Both in vitro and in vivo studies have shown effective targeting of key viral genes important for the virus life cycle as well as host genes involved in viral entry, replication and persistence [7].
1.2. CRISPR/Cas9: Precise genome editing
CRISPR is an abbreviation for clustered regularly interspaced short palindromic repeats, which were initially observed in Escherichia coli, and found to be separated by non-repeated DNA sequences called spacers. These spacers, known as CRISPR arrays, where later identified as acquired copies of previously encountered foreign DNA and provides the organism with memory of the plasmid and phage genomes [8-10]. Upon encountering the foreign genetic element again, the bacteria produce RNA segments from the CRISPR arrays to target the pathogen directly and employ an adaptive immune response. Adjacent to CRISPR arrays are well conserved CRISPR-associated (Cas) genes that have been grouped into families and subtypes based on sequence similarity of encoded proteins [11]. There are six types of CRISPR/Cas system that have been divided into two classes: class 1 CRISPR-Cas systems (types I, III, and IV) employ multi-Cas protein complexes for interference, whereas in class 2 systems (types II, V, and VI), interference is accomplished by a single effector protein [12].
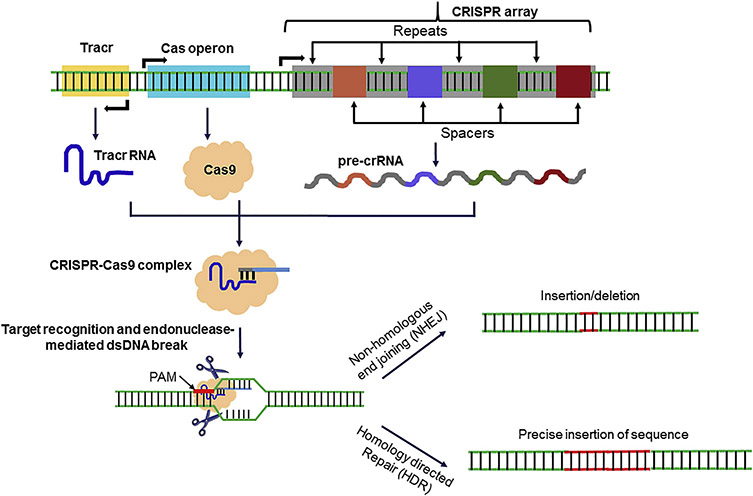
Type II CRISPR/Cas9 system of Streptococcus pyogenes (Sp) is the most studied and used system. Figure 1 describes generalized biogenesis and function of CRISPR/Cas9 system. The CRISPR array, which contains unique protospacer sequences that have homology to foreign DNA, is transcribed to make long precursor CRISPR RNA (pre-crRNA). Pre-crRNA are then processed to individual crRNA which guide the interference machinery to cleave complementary sequences or protospacers and ultimately eliminate the foreign DNA [13]. An invariant trans-activating CRISPR RNA (tracrRNA) is required to bind crRNA in a sequence complementary manner and recruit RNAse III and CRISPR-associated 9 (Cas9) enzymes [14]. Together, tracrRNA, RNAse III and Cas9 break up and form a complex with each individual unique crRNAs [15]. Protospacer adjacent motifs (PAMs), located directly after where the crRNA would bind, are critical to this system and contribute to the CRISPR targeting specificity [16]. Cas protein motifs have helicase and nuclease activity that are guided by crRNA resulting in DNA cleavage upon target binding, which stimulates non-homologous end joining (NHEJ) or homology-directed repair (HDR)-mediated genome editing, resulting in the elimination of invading foreign DNA [17-19].
Figure 1.
Overview of guide RNA biogenesis and genomic targeting by CRISPR-Cas9. Key components of Streptococcus pyogenes CRISPR-Cas locus are shown. Tracr, Cas operon and CRISPR locus (containing clustered repeats and spacers) are transcribed to tracr RNA with stem loop structure, Cas9 endonuclease and pre-crRNA, respectively. RNase III then generate mature crRNA which is eventually incorporated into Cas9 along with tracr RNA. Guide RNA (gRNA) along with RNA-protein complex binds to the specific genomic locus with complementary sequence. Binding of guide RNA and target sequence is facilitated by protospacer adjacent motif (PAM). DNA is cleaved by Cas9 and other accompanying endonucleases thus generating double strand DNA breaks which are eventually repaired by non-homologous end joining (NHEJ) or by homology directed repair (HDR).
Cas9 effector endonuclease have been repurposed by researchers as a tool for precise gene editing to eliminate or integrate target genes in the human genome [6,20]. Specifically, the tracrRNA:crRNA complex have been engineered into a single guide RNA (sgRNA), guiding CRISPR-Cas9 to desired targets in the genome [21]. The use of this system and its gene editing potential in eukaryotic cells proved to be successful and the application of CRIPSR/Cas9 is now widely adapted in multiple research areas [5,6,20,22].
1.3. The need for a new antiviral: CRISPR/Cas9
Multiple viruses still persist and infect a large number of the human population while still without a cure [23-26]. Current antiviral treatments are able to block active viral replication or subside symptoms of infection; however, they lack the ability to completely abolish the virus from the host, primarily due to escape mutants or establishment of latent infection [27-29]. CRISPR/Cas system has been adapted from its natural function as an immune response in prokaryotes against phage infection to studying its gene editing potential in eukaryotes. This application has extended to the development of a novel antiviral; where CRISPR/Cas systems are directed to target viral genome or host genes that aide in the persistence of the virus [30,31]. In this review we will discuss these approaches that have been studied as well as other potential applications against viruses including Herpesvirus, Human immunodeficiency virus (HIV), Hepatitis B Virus (HBV) and Influenza A Virus (IAV). Different CRISPR/Cas based antiviral strategies targeting host and viral genomes are illustrated in Figure 2.
Figure 2. Schematic illustration of different antiviral strategies targeting host and viral genomes using CRISPR/Cas.
(A) Viral entry into host cell is generally mediated through direct interaction of viral and host surface proteins. Targeting host entry receptors that are required for virus to gain access to host cellular machinery can hamper viral spread and also interfere with the viral tropism. Editing host entry receptor motifs required for virus-host interaction or utilizing naturally selected mutations for genomic modification using CRISPR can prevent entry of virus into host cells.
Viral replication and virion packaging requires numerous host factors. Targeting pro-viral and anti-viral host factors can restrict viral spread. (B) Dependency on host factors is a key characteristic of viruses. Some host proteins facilitate virus survival and spread. Targeting such pro-virus host genes may prevent viral replication and render virus more susceptible to host antiviral responses. CRISPR/Cas mediated functional knock-down of such genes will inhibit viral replication or virion production. (C) Induced expression of host restriction factors. Transcriptional activation of host factors that can block viral replication is achieved by coupling gRNA to a catalytically inactive Cas9(dCas9). Upon targeting, host restriction factor expression is activated leading to reduced viral replication and/or viral RNA transcription.
(D) Targeting viruses that integrate their DNA in the host’s genome. Integrated viral genome can be inactivated either by disruption of the essential viral genes or multiple sgRNAs can be targeted to the termini of the integrated viral DNA. Both strategies will lead to the reduced viral replication. Similar strategy can also be used to disrupt viral genes or fragmentation of viral genome of dsDNA genome of DNA viruses or RNA viruses with a non-integrated DNA intermediate.
Of the viruses listed, herpesvirus has been recognized as the most adapted human pathogen resulting in lifelong infections and the development of a wide range of innocuous diseases [32-34]. The Herpesviridae family is divided into three subfamilies of double stranded DNA viruses: Alpha-, Beta- and Gammaherpesvirinae [35]. Herpes simplex virus-1 and 2 (HSV-1 and HSV-2) are neurotropic viruses belonging to the Alphaherpesvirinae family and are capable of establishing lifelong latency in the central nervous system. HSV-2 is responsible for genital herpes resulting in painful ulcers while HSV-1 primarily infects the corneal epithelium resulting in lesions and the development herpes simplex keratitis as well as other less frequent diseases such as encephalitis [36-39]. Human Cytomegalovirus (HCMV), belongs to the Betaherpesvirinae family, is capable of infecting multiple cell types including epithelial, endothelial, myeloid, neuronal and fibroblast cells [40]. Epstein–Barr virus (EBV) belongs to the Gammaherpesvirinae family, primarily infecting B cells and epithelial cells causing infectious mononucleosis [41]. Acyclovir and Ganciclovir, nucleoside analogs that inhibit viral replication upon incorporation into viral DNA, are the current drug choice for treatment of HSV and CMV disease respectively [42,43]. While these drugs are highly effective in most cases, drug resistant viral strains have evolved [44-46]. On the other hand, EBV is reported to infect 95% of the population and no current treatment is available [47].
Another prominent virus, Hepatitis B Virus (HBV), remains a global health threat as infection results in life-threatening complications including elevated risks for liver damage and liver cancer [48]. HBV belongs to the family of Hepadnaviridae and replicates in infected hepatocytes in the liver via reverse transcription. HBV virions contain a partial dsDNA genome which is converted to double-stranded covalently closed circular (ccc)DNA in the nucleus shortly after infection [49]. The cccDNA is maintained as a ‘minichromosome’ and serves as a template for transcription of viral pregenomic RNA and mRNAs [50]. Current antivirals against HBV include IFN-α, which bolsters the host antiviral immune response, and nucleoside/nucleotide analogues to inhibit reverse transcriptase activity. However, due to persistence of cccDNA, long-term treatment is required is not a feasible option for some patients and has resulted in drug resistance among some strains [51].
Human Immunodeficiency virus (HIV) is also among the viruses that are highly prevalent worldwide and still remain without a cure. HIV is a single-stranded enveloped RNA virus that predominately infects human T cells and results in the development of acquired immune deficiency syndrome (AIDS). An estimated number of 35 million individuals suffer from HIV infection worldwide and require life-long treatment, which has resulted in many side effects and the emergence of escape mutants [52,53]. Antiretroviral therapy is the current mode of treatment and while it does efficiently limit virus replication to undetectable levels in HIV patients, it suffers from serious drawbacks [54].
Influenza A virus (IAV) is considered the most serious influenza capable of causing widespread outbreaks and disease [55]. IAV infection attacks the respiratory system and is able to rapidly evolve, resulting in seasonal epidemics and the constant emergence of variant- and drug-resistant strains [56]. The genome of IAV, which consists of multiple segments of single-stranded negative-sense RNA, contributes to its susceptibility to mutations which allow for evasion of the immune system and resistance to antiviral strategies [57]. Current vaccinations are limited in their efficacy and therapies only alleviate the symptoms and manage disease development to prevent hospitalization and death in patients [58].
The conventional antiviral drugs pose many concerning disadvantages. First of all, resistance among infected patients for all current FDA approved antiviral drugs, especially in the case of IAV, have been repeatedly reported [27,58-60]. Also, while current antivirals are effective in clearing productive viral replication in most cases, they are unable to eradicate the virus from latently infected cells [61,62]. This causes the infection to persist throughout the lifetime of the host with the potential of reactivation and recurrence. For these cases, life-long treatments are prescribed to patients in order to suppress viral reactivation, which results in side effects and escape mutants in many cases.
The viruses mentioned here remain a global health threat and continued efforts are aimed towards developing an effective antiviral or vaccination that eliminates viral genomes from the host. Most efforts to develop antivirals in the past have been directed towards targeting the viral genome in order to dismantle the virus’ ability to infect. However, with the emergence of escape mutants and drug resistance, efforts are now also being directed towards targeting the host cell machinery [31,63]. To prevent viral reactivation, drug resistance and protein expression, new antiviral strategies are needed that provide a durable and feasible antiviral effect. Ideally, eradication of the integrated proviral DNA in all infected cells, including the latent reservoir is required to effectively combat viral infection.
2. Targeting Host Factors Using CRISPR/Cas9
While targeting viral genomes with CRISPR/Cas9 may be subjected to the same disadvantages as current therapeutics, targeting host factors provides a more promising solution to abolish the virus from the host. Clinical advantages of host-based interventions using CRISPR/Cas9 include lower chances of drug resistances due to genetic stability of host factors, higher efficacy due to larger therapeutic timeframe and reduced side effects due to minimal dosing requirements.
Upon infection, the virus exploits host factors to complete its life cycle [64]. While the host’s innate immune system works to clear viral infection, the virus hijacks cell signaling pathways to escape immune surveillance and persist. Exploiting the dynamic virus-host interactions is a strategy used to block virus infection and make the virus less susceptible to survival in the cellular environment [65]. CRISPR/Cas systems have been used in multiple studies for genome editing of dispensable target host genes involved in viral replication and persistence. Recent advancements in CRISPR/Cas9 technology has also allowed for genome-wide CRISPR knockout (GeCKO) screening, providing a powerful tool to discover host factors and to highlight biological pathways essential for the replication of intracellular pathogens [66].
2.1. Host factors involved in viral entry
While current drug targets include viral proteins required for cell entry, most of the developed drugs have been regarded as ineffective over long term control of infection. In an effort to produce antivirals with long-lasting effects, reduced risks of developed resistance and toxicity, new mechanisms of action need to be developed. With the recent advancements made in gene editing, CRISPR/Cas9 and other tools have been used to target both the viral and host genome for factors involved in viral entry (Table 1). Recent studies have focused on inhibiting viral entry by targeting host factors such as cell surface receptors. Results from these studies have shown been high antiviral efficacy against HIV and IAV.
Table 1:
List of host genes targeted by CRISPR(/Cas9) strategy and their impact on viral replication and persistence.
| Virus | Viral infection model |
Target Gene(s) |
Functional impact |
Reference |
|---|---|---|---|---|
| HIV | Various cell lines and primary CD4+ T cells | CCR5 | Restriction of HIV-1 infection | [79,80,82] |
| Mice transplanted with human CD34+ HSPCs with CCR5-ablation treatment | ||||
| Human CD4+ T cells | CXCR4 | Restriction of HIV-1 infection | [83] | |
| HeLa and 293T cells | APOBEC3B and 3G | Increase expression of antiviral effectors | [88] | |
| IAV | human lung carcinoma cell line A549; H1N1 (A/Puerto Rico/8/1934; PR8) and H3N2 (A/Hong Kong/1/1968; HK68) | SLC35A1 | resistance against IAV infection due to inefficient binding of IAV particles to cell surface | [86] |
| CIC, JAK2, PIAS2 | CIC-deficient cells demonstrated upregulated antiviral gene expression and decreased replication of multiple viruses. | [86] | ||
| 293T and human lung carcinoma cell line A549; human IAV strain (A/Puerto Rico/8/1934 H1N1) | B4GALNT2 | Prevented infection by multiple avian IAV strain | [87] | |
| human lung carcinoma cell line A549; human IAV strain (A/Puerto Rico/8/1934 H1N1) | p53 | significantly reduced viral propagation | [92] |
2.1.1. Targeting HIV cell-surface entry receptors
CC chemokine receptor 5 (CCR5) and C-X-C chemokine receptor type 4 (CXCR4) are cell surface receptors involved in viral entry during primary infection [67]. Individuals homozygous for a naturally occurring deletion of 32 nt in the CCR5 gene prevents the expression of CCR5 on the cell surface and deems them highly resistant to HIV infection [68]. This discovery proposed a new antiviral approach to combat HIV: targeting cell surface entry receptors in order to disrupt membrane fusion between the virus and cell [69]. A competitive inhibitor of CCR5, maraviroc, was clinically approved and showed affective anti-HIV activity. However, development of resistance by selection for escape mutants was later observed by viral adaptations to the drug [70]. While CCR5 is the predominant receptor involved in HIV entry and primary infection, viral tropism towards CXCR4 usage occurs at later stages of infection and maraviroc was also dispensable in the presence of CXCR4-tropic HIV strains [71]. To combat CCR5-tropic HIV strains, gene therapy approaches to block CCR5 expression were evaluated and found to be promising both in vitro and in vivo [72]. Perez and colleagues published a study using ZFN-guided genomic editing of CCR5 in primary human CD4+ T lymphocytes showing high specificity, tolerance and robust protection against HIV-1 and soon underwent clinical trials targeting mature CD4+ T cells [73,74].
More interest has been developed in the past decade following a report that an acute myeloid leukemia patient who received a hematopoietic stem/progenitor cell (HSC) transplant from a CCR5Δ32 donor was also cured from HIV-1 [75,76]. Subsequent studies in mice transplanted with CCR5-disrupted HSCs showed significant reduction in viral load when challenged with CCR5-tropic HIV-1 strain, as well as protection of human T cell populations in key tissues that HIV-1 infects. Long-term control of HIV-1 replication was observed for the first time using this technique [77]. This study suggested that modification of autologous HSCs by CCR5-ZFNs could provide a permanent supply of HIV-resistant progeny, leading to immune reconstitution and long-term control of viral replication [78]. While ZFN and TALEN are effective in enabling mutagenesis in a variety of cells, they both exhibit challenges including variability in mutation frequencies, lack of specificity, feasibility and off-target effects. The most important off-target sites associated with CCR5-specific ZFNs and TALENs reside in CCR2, a close homolog of CCR5, located 15kpb upstream of CCR5.
In order to increase specificity and reduce undesired off-target effects, CRISPR/Cas9 has been employed to induce a site-specific genome modification in human cells and in vivo using mouse model of infection [79-82]. Mulitiple groups successfully transduced primary CD4+ T-lymphocytes and disrupted CCR5 by gene editing using adenovirus-delivered CRISPR/Cas9 targeting the open reading frames (ORF) of CCR5. This resulted in inhibition of HIV-1 infection with no significant off-target effects [81]. Targeting CCR5 in both HSC and CD4+ T cells has shown promise as a gene-editing strategy for persistent resistance to HIV. However, its ineffectiveness towards CXCR4-tropic HIV strains and therefore lacks conclusive evidence for its use as a cure. Studies have also directed Cas9-mediated gene editing of CXCR4 with high precision and efficacy, showing negligible off-target effects and resistance to HIV infection [79,83]. Targeting both coreceptors CCR5 and CXCR4 may provide more breadth in protection and use in clinical settings.
2.1.2. Targeting host factors involved in IAV entry
CRISPR/Cas9 has also been used to limit IAV infection by targeting host factors involved in viral entry. IAV enters host cells after attachment of its hemagglutinin (HA) to the cell surface receptor sialic acid. Entry inhibitors have been developed in the case of IAV that either bind to the HA globular head to prevent interaction with the receptor or by acting on the sialic acid receptor itself [84,85]. However, currently available drugs are ineffective against newly emerging viruses with developed resistance. Recent studies using a genome-wide CRISPR knockout (GeCKO) screen demonstrated that a CMP-sialic acid transporter (SLC35A1) is an essential host factor for IAV entry [86]. Validation studies revealed that loss of SLC35A1 resulted in resistance against IAV infection due to the absence of cell-surface sialic acid. Clonal knockouts infected with H5N1, H1N1 and H3N2 observed a significant reduction in viral titers, confirming the role of SLC35A1 in replication of multiple IAV strains. SLC35A1 knockout cell lines also showed inability to support recombinant HA binding and therefore resulting in a robust reduction of IAV entry.
Genome wide overexpression screen using CRISPR/dCas9 was recently used as a tool to uncover IAV restriction factors that, when overexpressed, are sufficient to prevent infection. A glycosyltransferase (B4GALNT2) that modifies sialic acid-containing glycans, and thus prevents IAV infection, was identified as a top hit, leading to greater than 100-fold reduction in IAV infection. The inhibitory activity of B4GALNT2 was restricted to viruses with avian a2,3-linked receptor which include avian influenza viruses H7N2, H9N2, H5N9, H10N4, some of which infect humans as well [87].
2.2. Antiviral Host Effectors (Cell-intrinsic immunity)
Upon viral infection, the host triggers a robust innate immune response as a defense mechanism to clear the pathogen [64]. The dynamic virus-host interaction that occurs at early stages of infection determines whether the cell will overcome the viral infection or succumb to it. While the host cells have evolved to express restriction factors that block virus infection, the virus in turn exploits host factors and uses specific strategies to escape immune surveillance by manipulating its environment. Studies of this dynamic interaction has allowed for the identification of host effectors that aid or suppress the cell-intrinsic immunity. These host effectors are now key targets for developing novel antiviral strategies through the use of CRISPR/Cas9.
In regard to HIV, APOBEC3G (A3G) and APOBEC3B (A3B) are restriction factors in human cells that have recently been targeted. A3G is expressed in human T cells and macrophages, which are usually the target cells of HIV-1, and is capable of restricting the virus. However, HIV-1 Vif protein is able to target and degrade this host effector. A3B on the other hand is restricted to expression in pluripotent cell types and is not targeted for degradation by Vif. In a recent study, a gain-of-function approach using a cleavage defective Cas9 (dCas9) allowed for specific and effective induction of A3G and A3B genes in cell culture [88]. Potent inhibition of HIV-1ΔVif infectivity was achieved by induced endogenous A3B and A3G in 293 T cells, which typically lack both proteins, and a decrease in HIV-1WT infectivity upon A3B expression. This study shows that CRISPR/Cas gene editing technology can be used as a novel platform to combat infection by efficiently targeting human genes that regulate virus replication.
Using a genome-wide CRISPR/Cas9 screen, key regulators of cell-intrinsic immunity upon IAV infection have been identified and serve as targets for further investigation [86]. In this study, capicua (CIC) was shown to play a role in suppressing antiviral gene expression by its transcriptional repressor activity. Confirming this, knocking down CIC using CRISPR/Cas9 resulted in decreased replication of both IAV and VSV by the observed heightened antiviral responses. Similarly, JAK2 was also identified as a host effector involved in negatively regulating cell-intrinsic immunity by lowering antiviral gene expression. While more studies are required to elucidate the exact role JAK2 plays at different stages of IAV infection, using CRISPR/Cas9 this group was able to identify key factors that contribute to virus survival against the innate immune system.
Similarly, a genome-wide CRISPR/Cas9 loss-of-function screen was conducted to identify host effectors critical for EBV induced B cell-transformation leading to B cell malignancies and how oncoproteins evade tumor suppressor responses [89]. The screen identified BATF, IRF4, IRF2, CDK6 and cyclin D2 as lymphoblastoid cell lines (LCLs) dependency factors that are highly upregulated in EBV infection of primary human B cells. Another gene, cFLIP, was highlighted as a potential EBV therapeutic target for its role as a survival factor in LCLs by blocking TNFα-mediated apoptosis and necroptosis.
Other host factors recently identified as having an integral role in viral infection are interferon-induced transmembrane proteins (IFITM) which restrict the entry of diverse enveloped viruses through unknown mechanism [90,91]. In the case of IAV, initial infection upregulates p53, a protein involved in key cellular stress mechanisms, which in turn downregulates IFITMs and therefore promotes virus survival in the host. Since IFITMs were detected as downstream targets that are negatively regulated by p53, CRISPR/Cas9 was used to generate p53null cells to investigate its role in influenza infectivity. While p53 was not shown to affect initial IAV entry, overexpression of IFITMs reduced cell susceptibility to IAV infection by blocking viral entry in A549 cells [92]. The role of IFITM on restricting HIV-1 infection was also investigated using CRISPR/Cas9 by directly targeting IFITMs in U87 CD4+ CCR5+ cells. While similar findings as those regarding IAV were concluded from the study, the clear mechanism of IFITM upon viral infection is still poorly understood and lacks in vivo relevance [93].
2.3. Host Factors involved in viral transcription and replication
Infectious particles take over various cellular pathways and processes in order to ensure its survival and persistence in the host. Certain cellular genes are upregulated in the presence of viral particles and aid in viral transcription and replication. For example, genes known to support HBV replication include sodium-taurocholate cotransporting polypeptide (NTCP), heat stress cognate 70 (Hsc70), heat shock protein Hsp90 and tyrosyl-DNA-phosphodiesterase 2 (TDP2) [31]. While studies have not shown the antiviral effect of knocking down these genes using CRISPR/Cas9, these are still potential targets to be considered.
Various host factors identified as restricting HIV replication include APOBEC3C, TRIM5α, SAMHD1, and tetherin, and also serve as potential CRISPR/Cas targets in developing therapeutics [31,94,95]. Many of these are involved in eliciting robust immune responses upon detection of HIV, and ultimately acting as restriction factors to terminate HIV replication by blocking viral entry or release from the host cell. TRIM5α is associated with antiviral activity by recognizing retroviral capsid proteins and, in turn, transducing diverse cellular signaling pathways, including triggering innate immunity [95]. Studies showed efficient antiviral activity using TRIM-cyclophilin fusion protein both in vitro and in vivo with robust inhibition of HIV-1 replication [96]. Similarly, SAMHD1 is a restriction factor that prevents HIV replication. SAMHD1 is a deoxynucleotide triphosphate (dNTP) hydrolase that is activated by the binding of GTP to its allosteric site which consequently reduces the intracellular dNTP pool below levels that support HIV-1 reverse transcription [97]. This results in the blockage of HIV-1 replication and prevents the induction of IFN responses. However, transducing the activity of SAMHD1 is counteracted by Viral Protein X (VpX) which targets and degrades the protein, leading to the restoration of dNTP levels and allowing for HIV-1 replication [98,99]. Tetherin is another restriction factor of HIV induced by interferon and prevents viral replication by restricting viral particle release [100]. While HIV-1 Viral Protein U (Vpu) counteracts tetherin activity, a single amino acid substitution in tetherin results in Vpu-resistance [101]. These restriction factors serve as promising targets for the use of CRIPSR/Cas9 in either activating these restriction factors or inducing the mutations that make the proteins resistant to inhibition by viral proteins.
Similarly, various studies have investigated the host-pathogen interaction that occurs upon IAV infection in order to identify key host genes involved. A genome-wide RNAi study identified hundreds of host genes involved in restricting or promoting cellular infection by RNA viruses, including IAV [102]. Of the genes identified, COPI-protein families, fibroblast growth factor receptor (FGFR) proteins, glycogen synthase kinase 3 (GSK3)-beta and calcium/calmodulin-dependent protein kinase (CaM kinase) II beta (CAMK2B) were significantly enriched and found to be involved in early steps of influenza virus replication. This study also showed that inhibition of these identified targets using small molecules resulted in attenuated viral replication; further proving that these may be suitable targets using CRISPR/Cas9.
A study conducting a genetic screening using CRISPR/Cas9 identified host factor dependencies of flavivirus replication including proteins involved in endocytosis, heparin sulfation and endoplasmic reticulum membrane complex (EMC) [103]. This study serves as a resource and paves the way for drug targeting and development. More recently, CIRPSR/Cas9 knock-out of viperin, an interferon stimulated gene (ISG) involved in innate immunity, further elucidated its role in Zika virus (ZIKV) infection [104]. ZIKV was originally isolated from a sentinel monkey in Uganda and belongs to the Flaviviridae family of positive-sense RNA viruses that includes dengue (DENV), West Nile (WNV) and Japanese encephalitis (JEV) [105]. Vanwalscappel and colleagues identified a TLR agonists with potent inhibition of ZIKV infection and identified enrichment of viperin upon treatment with the TLR agonist. Gene editing tools such as CRISPR/Cas9 may be used to target and induce antiviral host machinery to combat the infection.
2.4. Boosting Host Immunity with CRISPR/Cas9
Dysregulated immune responses are central to pathogenesis of various diseases and autoimmune disorders. Dysregulation could be a result of genetic mutation, pathogen-mediated suppression of effector immune responses or altered phenotype of cells in diseased microenvironment. Devising CRISPR/Cas system to correct these genetic errors or enhancement of immune cell function by targeted modification can benefit humans against numerous diseases. Multiple studies have shown positive outcomes with CRISPR/Cas-mediated improvement of T cells that are central to adaptive immune response. While specific role in boosting antiviral T cell immunity has not been studied, evidences support CRISPR/Cas-based enhancement of T cell-mediated immunity in cancer immunotherapy.
To test CRISPR/Cas system-mediated enhancement of T cell function, Rupp et al. examined disruption of programmed cell death protein 1 (PD-1) in chimeric antigen receptor (CAR) T cells which are modified to detect specific tumor antigens [145]. Recognition of tumor cells by CAR T cells alone is not sufficient to clear cancerous cells due to the immunosuppressive tumor microenvironment and expression of inhibitory ligands, such as PD-L1, on both tumor cells and surrounding tissues. PD-L1 can interact with its ligand PD-1 on T cells and trigger immunosuppressive signals leading to T cell exhaustion and hypo-function. This scenario is frequently observed in chronic viral infections and tumor infiltrating lymphocytes in patients with advanced malignancy. Using a Cas9 RNP gene editing (targeting PDCD-1) and lentiviral transduction (encoding the anti-CD19 CAR) of primary human T cells, PDCD-1 gene was ablated while CD19 CAR expression was simultaneously achieved in primary human T cells. The genetically modified T cells (PD-1−: CD19 CAR+) when cultured with K562 tumor cells (CD19+; PD-L1+) displayed efficient at killing PD-L1+ tumor cells. The efficacy of PD-1−: CD19 CAR+ T cells was assessed in vivo in NOD scid gamma (NSG) mice. Mice with adoptively transferred PD-1−: CD19 CAR+ T cells, following subcutaneous injection of CD19+ PD-L1+ K562 cells and establishment of tumors, showed reduced tumor growth, increased survival and accelerated tumor clearance compared with controls. Overall, CRISPR-mediated targeted disruption of the Pdcd1 locus in human CAR T cells strongly enhance in vivo anti-tumor efficacy.
Eyquem et al. employed a different approach of targeted delivery of CD19-specific CAR to the T-cell receptor α constant (TRAC) locus using CRISPR/Cas system [146]. They demonstrated that CAR expression is uniform in T cells and these genetically modified cells are more potent as in their antitumor activity and outperform CAR T cell generated using conventional approach in a mouse model of acute lymphoblastic leukaemia (AML). For instance, markers of T cell exhaustion such as PD1, LAG3 and TIM3 were expressed at higher levels in conventional (retrovirally delivered) CAR compared to TRAC-CAR T cells and the former showed accelerated terminal differentiation further correlating with higher antitumor activity of TRAC-CAR T cells. It can be noted that in vitro functional studies did not reveal any notable differences between TRAC-encoded and randomly integrated 1928z, in terms of both cytotoxicity and T-cell proliferation in response to weekly stimulation with CD19+ antigen-presenting cells strongly suggesting that readouts of in vitro and in vivo studies may marked vary thereby signifying the need to perform in vivo experiments. These findings clearly show that targeted delivery of CAR using CRISPR/Cas can reduces tonic signalling, averts accelerated T-cell differentiation and exhaustion, and increases the therapeutic potency of engineered T cells in vivo.
In another study, a different strategy was employed to evaluate CRISPR/Cas-mediated genomic editing to reprogram human T cells by correcting a gene locus in a genetic disease as well as selective targeting of cancer cell [147]. An electroporation was used to deliver crRNA and tracrRNA into T cells in order to generate genetically modified, highly viable, therapeutic grade T cells with no significant evidence of off-target integrations. The efficacy and targeted editing of the approach was tested on individuals (three siblings) with monogenic primary immune deficiency, an autoimmune disease caused by recessive loss-of-function mutations in the gene encoding the IL-2 alpha receptor (IL2RA), required for healthy regulatory T cells [148-149]. Two different mutations were identified on the ILRA of the subjects (1) c.530A>G, creates a premature stop codon and (2) c.800delA, causes a frameshift in the reading frame. Following restoration of genetic mutation, T cells were functionally assessed and active IL2 signaling was demonstrated by increased STAT5 phosphorylation upon IL-2 treatment, a hallmark of productive signaling. Furthermore, this CRISPR/Cas strategy was employed to assess in vivo anti-tumor function of T cells. A pair of T cell receptor (TCR) genes viz., TCR-β and TCR-α which recognizes the NY-ESO-1 tumor antigen was incorporated into the TRAC locus of polyclonal T cells isolated from healthy human donors [150]. Interestingly, it was observed that NY-ESO-1 TCR knock-in T cells preferentially localized to, persisted at, and proliferated in the tumor rather than the spleen. Adoptive transfer of sorted NY-ESO-1 TCR T cells also reduced the tumor burden in treated animals. These findings clearly support successful use of non-viral templates to harness potency of CRISPR/Cas genome editing capability in human health and disease. Most importantly, this methodology is cost-effective, absence of viral templates poses low risk of adverse impact on human health and is easy to adapt good manufacturing practices (GMP) for clinical use. However, a key limitation of using non-viral templates is low efficiency of genome editing. All of these studies strongly support that CRISPR/Cas can be utilized to boost T cell immunity and selectively targeting of cancer cells by T cells in vivo. Further investigations on long-term impact on the functions of genetically-modified T cells and how their phenotype is impacted in different individuals should be thoroughly examined. It will be important to assess CRISPR/Cas technology in boosting antiviral immunity with specific focus on CD8+ T cell and natural killer (NK) cell which are central to antiviral immunity and actively participate in clearing virus infected cells.
3. Targeting Viral Genomes
CRISPR/Cas9 has also been used to directly target and edit viral genome to impair its activity and eradicate it from the host (Table 2). While drugs targeting the virus, such as inhibitors that neutralize viral particles and nucleoside analogues that terminate viral replication have been developed, most have resulted in development of escape mutants from repeated use or ineffective against some viral strains. Using CRISPR/Cas9 to target one or multiple parts of the viral genome may be more effective in abolishing the virus from the host.
Table 2:
List of viral genes targeted by CRISPR/Cas9 strategy and their impact on viral replication and persistence.
| Virus | Viral infection model |
Target Gene(s) |
Functional impact | Reference |
|---|---|---|---|---|
| HSV-1 | Vero cells (4 gRNAs per target site); HSV-1-eGFP | essential viral genes (UL15; UL27; UL29; UL30; UL36; UL37; UL42; UL5; UL52; UL8; UL54; UL9) | Impaired HSV-1 replication | [106] |
| nonessential viral genes (US3; US8) | Impaired HSV-1 replication to a lesser extent | |||
| Vero cells, HEK293T cells, and intranasal infection of BALB/b mice | UL7 | Impaired HSV-1 replication | [109] | |
| HEK293 T and U-2 OS Cells; HSV-1 KOS and GFP strain | ICP0 | Modification of ICP0 gene with no off target effects | [107] | |
| HCMV | MRC5 cells (4 gRNAs per target site); | essential viral genes (UL54; UL44; UL57; UL70; UL105; UL86; UL84) | Impaired HCMV replication | [106] |
| HCMV strain TB40/E or AD169 | nonessential viral genes (US6; US7; US11) | No impact on HCMV replication | ||
| MRC5 and U-251 MG cells; HCMV strains: TB40GFP, Toledo and VR1814 | UL122/123 | Impaired HCMV replication | [110] | |
| EBV | Latently infected SNU-719 tumor cells | miRNA BART5, BART6 or BART 16 | Enhanced expression of miRNA target reporter | [106] |
| Akata-Bx1 cells carrying recombinant EBV expressing eGFP (2 gRNAs per target site) | EBNA1 | ~50% loss in GFP expressing cells with single gRNA and ~90% loss with two gRNAs targeting different sites on the viral genome | ||
| OriP (multiple sites) | ||||
| EBNA1+ OriP | ||||
| Raji cells: Burkitt’s lymphoma cell line with latent EBV infection | EBNA1 | EBV genome content was absent or reduced, cell proliferation arrest and apoptosis | [108] | |
| OriP LMP1 | ||||
| HBV | NTCP expressing HepG2 cells | Targeting cccDNA via: ENII/CP and Pre-C ORFs | Inhibition of HBV infection by 8-fold | [112] |
| Huh-7 cells | HBV-specific gRNAs P1, S1, XCp, PS2, | reduced HBV protein production | [113] | |
| HBV hydrodynamics-mouse model | HBV- specific gRNAs P1 and XCp | reduced serum levels of HBsAg | ||
| Huh-7 and HepG2.2.15 cells | X/L and X ORFs | Reduced HBsAg and HBeAg level in cell culture medium and intracellular cccDNA Reduced levels of HBsAg and HBeAg level in serum and intrahepatic cccDNA |
[121,141] | |
| Hydrodynamic-mouse model | ||||
| Huh-7, HepAD38, HepaRG, HepG2 cells | P, S, X and C ORFs | Reduced levels of HBV antigens | [114-117,119,141] | |
| Hydrodynamic-mouse model | Reduced serum levels of HBsAg and viral DNA | [117,119,141] | ||
| HIV | Primary CD4+ T cells, Jurkat cells, SupT1 T cells, HEK 293T cells | long terminal repeat (LTR) promoter region | Disruption and excision of the latent provirus from human cells | [122,125,142] |
| JLat10.6 HIV-latent cell line, HEK293T cells, HeLa cells | LTR, pol gene and second exon of tat/rev | Tat/rev target gene mutated with high frequency and no off target effects | [124,126] | |
| VACV | N1L and A46R genes | Efficient targeting of viral genes | [128] | |
| JCPyV | SVG-A cells; JCPyV-MAD1 virus | non-coding control region (NCCR), Capsid proteins VP1 and VP2 | Inhibition of viral replication | [129] |
| HPV | Cervical HPV-16-positive cell line SiHa | E6, E7 genes, promoter of E6/E7 | Reduction in proliferation of cervical cancer cells in vitro and inhibition of tumorigenicity in xenograft studies | (130) |
The use of CRISPR/Cas9 to directly mutate viral genomes and impair viral replication was demonstrated by multiple groups that targeted viral genetic elements important for both productive and latent herpesvirus infection [106-108]. Diemen el al. designed gRNAs targeting viral microRNAs BART5, BART6, and BART15 in latently infected gastric carcinoma cell line NSU-719 and showed direct editing of the EBV genome [106]. This study, along with others, showed clearance of latent viral infection in cell culture by effectively targeting EBNA1, which is crucial for EBV functions and latent genome replication, with CRISPR/Cas9. Direct editing of viral genome was proved to be possible, allowing for continued efforts to be made in this direction.
Along the same lines, multiple gRNAs targeting essential genes of HSV-1 and HCMV also showed high efficacy in abrogating virus replication in vitro. Specifically, the use of double gRNAs showed complete blocking of HSV-1 replication and was suggested to be more effective in preventing the outgrowth of resistant strains. Other studies have shown efficient use of CRISPR/Cas9 to edit both essential and nonessential genes in the HSV genome and this in turn has accelerated our understanding of HSV [106,109]. gRNAs targeting UL7 gene, a tegument protein of HSV, were designed by Xu et al. in 2016 using CRISPR/Cas9 and found that the replication capacity of this mutant strain was 10-fold lower than wild-type HSV-1 [109]. Similarly, a different group showed high efficacy in directly targeting HSV-1 genome by modifying ICP0, an immediate early gene involved in HSV-1 gene expression and replication, with no off-target modifications [107]. In 2018, a group designed a multiplex antiviral CRISPR/Cas9 system targeting UL122/123 gene in the HCMV genome which are important for regulating lytic replication and reactivation from latency. This study showed effective targeting of the viral genome and abrogation of viral replication in primary fibroblasts and U-251 MG cells [110]. The authors also suspect reduction in viral reactivation from latency following the destruction of the IL122/123 HCMV gene.
Multiple studies have shown effective targeting of cccDNA genome of HBV by Cas9-induced double-stranded DNA break [111,112]. Seeger et al. designed several sgRNAs spanning the ENII/CP region and the PC of the HBV genome and showed effective CRISPR/Cas9 targeting of HBV cccDNA in HEPG2 cells expressing HBV receptor sodium taurocholate cotransporting polypeptide (NTCP). A different group showed the eradication of persistent HBV infection both in vitro, using Huh-7 cells transfected with an HBV-expression vector, and in vivo, using a hydrodynamics-HBV persistence mouse model, using CRISPR/Cas9 [113]. In vitro, reduced production of HBV core and surface proteins was observed using two out of the eight designed HBV-specific gRNAs. Reduced serum surface antigen levels were observed in vivo, further suggesting a robust role of CRISPR/Cas9 -mediated disruption of HBV infection. Soon after, multiple studies showed the complete eradication of HBV infection and elimination of persistent HBV genome, including full-length integrated HBV DNA and HBV cccDNA [113-121].
CRISPR/Cas9 was also used to directly target HIV-1 proviral genome and showed elimination from latently infected human T-cells, and therefore inhibition of HIV-1 reactivation, without off-target effects [122]. Whole genome sequencing verified CRISPR/Cas9 mediated-excision of provirus of two copies of HIV-1 by designing sgRNAs with specific DNA sequences positioned within the HIV-1 promoter spanning the 5’ long terminal sequence (LTR). In another approach to target latent HIV-1 provirus, viral reactivation was achieved using Cas9 -synergistic activation mediators (dCas9-SAM) targeting the enhancer of the HIV-1 LTR promoter. This was done in an effort to develop an additional “shock and kill” therapeutic strategy where following viral reaction, the clearing of the viral genomes is achieved by mechanisms of viral cytotoxicity or host immune defense. While this approach suffers from limitations and requires more investigation, it proposes a unique therapeutic strategy with clinical significance [123].
In a different study, 10 sgRNAs were designed targeting different regions of the HIV-1 genome: LTR, pol gene, and the second exon of tat/rev [124]. The site in the second exon of Rev exhibited the highest level of mutation by Cas9 and as a result showed diminished levels of HIV-1 gene expression and virus production. Combined use of different gRNAs, which is predicted to diminish the change of HIV-1 escape mutants arising, targeting the viral genome at once showed reduced viral yields by more than 24-fold. Similarly, Wang and collogues further elucidate the impact of using combinatorial CRISPR/Cas9 as an antiviral to extinguish HIV-1 DNA without consequences such as escape mutants and resistance [125]. Most recently, Ophinni et al. demonstrated CRISPR/Cas9 transduction in 293T and HeLa cells using tat and rev-targeting regions of HIV-1 genome [126]. This study confirms no off-target effects as well as no effects on cell viability, further showing the therapeutic potential of CRISPR/Cas9 as a single-time intervention to clear viral genomic persistence and thus become a new cure for HIV-1.
Systemic removal of HIV from various cellular reservoirs is important for complete cure, however, during latency, viruses may reside in more than one cell or tissue. CRISPR/Cas technology has also been employed, both in vivo and ex vivo, to remove latent viruses (EBV, HSV-1, HCMV, etc.) including viruses that incorporate into host genome like HIV-1 [108]. While various studies have demonstrated successful use of CRISPR/Cas system to inhibit virus replication by targeting critical virus-encoded, efficient delivery of CRISPR/Cas components may vary in vivo and ex vivo/in vitro and this may impact the excision of desired pathogenic sequences hidden inside host cells. Kaminski et al. demonstrated CRISPR/Cas-mediated excision of HIV sequence in mice and rats infected with HIV-1 [143]. They utilized a recombinant adeno-associated virus (AAV) to express modified version of Staphylococcusaureus Cas9 (saCas9) and guide RNAs, targeting viral DNA sequences within the 5’-LTR and the Gag gene, for systemic delivery using tail vein (mice) or retroorbital inoculation (rats) in HIV infected animals. Deletion of target viral DNA region was noticed as well as reduced expression of viral transcripts indicating efficient systemic knockout of integrated viral genomes. The same research group further examined the feasibility and efficiency of HIV-1 excision using previously tested recombinant AAV-mediated delivery of saCas9 and a multiplex of sgRNA targeting various regions of HIV- genome editing in humanized bone marrow/liver/thymus (BLT) mice inoculated with HIV-1 and in mice during an acute infection of EcoHIV [144]. While targeting of LTR was sufficient to induce HIV genome excision, a combination of LTR and gal or pol targeting induces potent excision of viral genome suggesting multiple targeting is more efficient. However, further studies are required on possible off-target effects of CRISPR/Cas system in vivo and how this system can be effectively used in patients with other systemic illnesses. Additionally, the functional impact of CRISPR/Cas9 knockout on host cells should be thoroughly investigated. Whether the effector functions of these cells are similar to normal cells also demand a comprehensive analysis.
CRISPR/Cas9 has also successfully been used to edit the viral genomes of other viruses such as Vaccinia virus (VACV). JC polyomavirus (JCPyV) and human papillomavirus (HPV). In VACV, CRISPR/Cas9 has been used to generate efficient site-specific mutations in the genome of the virus that can be employed as vaccine vectors against infectious diseases. Specifically, sgRNAs targeting N1L and A46R genes were constructed due to their role in virulence and regulating host immune response to VACV [127, 128]. Homologous recombination was efficiently induced (62.5% and 85% respectively) by the gRNA-guided Cas9, showing direct mutation of the viral genome. Another study utilized CRISPR/Cas9 to inhibit virus replication and viral protein expression of JCPyV, a member of the Polyomaviridae family, by targeting the noncoding control region and the late gene open reading frame of the JCPyV genome [129]. Similarly, a study using CRISPR/Cas9 to eliminate the oncogenic function of HPV constructed gRNAs targeting E6 and E7 genes which are essential for malignant phenotype of cervical cancer by inactivating tumor suppressor proteins p53 and pRb [130]. CRISPR/Cas9 has been applied to a variety of viruses and shown to be both versatile and effective in most cases.
4. Current limitations to the use of CRISPR/Cas9 as an antiviral
While the use of CRISPR/Cas9 as an antiviral has shown to be both effective and precise in various studies, limitations to its use still exist. Most of the published studies discussed in this review have been performed in cell-culture-based systems and, while showing high antiviral efficacy, lack convincing in vivo data from animal model studies. Although these studies give an indication regarding the application of the system, they lack various considerations and therefore fail to elucidate its therapeutic potential in humans. This includes ensuring safe and efficient delivery of Cas9 and gRNA.
The time of addition, dosage and delivery vehicle of the CRISPR materials are all critical to ensure proper use and exert the desired therapeutic benefits [131, 132]. Although various studies suggest the activity of Cas9 is highly precise and specific, studies have indicated unintended mutations resulting from the repair process of DNA breaks generated by CRISPR/Cas [133, 134]. Additionally, the development of CRISPR/Cas9-induced virus escape mutants and, as a result, the development of more pathogenic virus strains has been reported in recent studies. This has been resolved by the use of multiple gRNAs directed towards various sites of the genome but still presents itself as an issue in some cases [110, 136].
The consideration of viral vectors used for therapeutic delivery of Cas9:gRNA is also a key component to ensure success and efficiency [30, 137]. The large size of Cas9 proteins and delivery to specific cell types and tissues limits the vehicles that can be used. Lentiviruses induce long-term expression by integration into the target cell genome, exhibit high infection efficacy and have demonstrated safe delivery in vivo [138]. However; lentiviruses yield low virus titers and makes them ineffective in cases where large numbers of virally infected cells need to be targeted. Additionally, a high potential for insertional mutagenesis due to long-term and systematic expression has been observed [139]. In contrast, adeno-associated virus (AAV) is another common option that does not integrate into the genome, yields high viral titers and is capable of infecting various cell types. AAV is suitable in many cases, however, the relatively small viral genome size limits the DNA cargo that can be delivered to target cells [140]. The development of a safe and efficient delivery system is crucial for the success of CRIPSR/Cas9 in clinics and targeting tissues or cells in human body still remains to be a challenge.
As we inch towards personalized gene editing and medicine, it is of utmost importance to identify and evaluate adverse effect of antiviral sgRNAs on host physiology and absolutely assuring that they do not impair genome integrity as well as transcriptome profiles. With these challenges resolved, CRISPR/Cas will be an attractive tool to eradicate numerous viral infections most of which remain without cure.
5. Conclusion
The recent advancements regarding gene editing technology has achieved broad application and progressed our knowledge in multiple research areas, including microbiology and immunology. Achieving precise genome editing in eukaryotic cells has long been studied with programmable endonucleases, such as ZFNs and TALENs, to interfere with certain genes. However, the feasibility and precision of these tools has been at question. CRISPR/Cas9 presents a more powerful platform with wide application, easier retargeting ability and reduced off-target events.
CRISPR/Cas9 has been widely used to target human genomic DNA and recently has been directed towards viral DNA in hopes of finding a cure for viruses such as herpesvirus, HBV, HIV and IAV. This review summarizes the application of CRISPR/Cas9 in abolishing these viruses from the host by directly targeting viral genomes or particular host genes that aid in viral replication and persistence. First and foremost, the goal is to identify host and virus genes that are indispensable for virus infection. Once these targets are ascertained, a major challenge will be to develop sgRNA that can specifically target gene-of-interest with no off-target effect. Another challenge is to thoroughly characterize sgRNA impact on cellular physiology both in vitro and in vivo. Using CRISPR/Cas9 in human as antiviral may be a near future reality, however, researchers must proceed with extreme caution regarding potential biological and ethical consequences.
Acknowledgments
Funding
Part of this work was funded by the NIH/NIDCR R21 DE026259-01A1 and R01 DE027980 to ARN and 2R01 EY024710-04 to DS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- [1].Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biology 2014;12[6]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346[6213]:1258096. [DOI] [PubMed] [Google Scholar]
- [3].Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem 2016;85:227–264. [DOI] [PubMed] [Google Scholar]
- [4].Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys 2017;46:505–529. [DOI] [PubMed] [Google Scholar]
- [5].Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014;157[6]:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell 2016;164[1-2]:29–44. [DOI] [PubMed] [Google Scholar]
- [7].Jin D, Berkhout B. CRISPR-Cas Antiviral Strategies. Virus Research 2018;244:285. [DOI] [PubMed] [Google Scholar]
- [8].Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 2005;60[2]:174–182. [DOI] [PubMed] [Google Scholar]
- [9].Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007;315[5819]: 1709–1712. [DOI] [PubMed] [Google Scholar]
- [10].Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature 2015;526[7571]:55–61. [DOI] [PubMed] [Google Scholar]
- [11].Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nature Reviews Microbiology 2011;9[6]:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, et al. An updated evolutionary classification of CRISPR-Cas systems. Nature Reviews Microbiology 2015;13[11]:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008;321[5891]:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011;471[7340]:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA biology 2013;10[5]:841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009;155[Pt 3]:733–740. [DOI] [PubMed] [Google Scholar]
- [17].Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32[4]:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014;507[7490]:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 2011;30[7]:1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 2016;17[1]:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012;337[6096]:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol 2016;34[9]:933–941. [DOI] [PubMed] [Google Scholar]
- [23].Kane M, Golovkina T. Common Threads in Persistent Viral Infections. J Virol 2010;84[9]:4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jaishankar D, Shukla D. Genital Herpes: Insights into Sexually Transmitted Infectious Disease. Microb Cell 2016;3[9]:438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].International AIDS Society Scientific Working Group on HIV Cure, Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 2012;12[8]:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Testoni B, Levrero M, Zoulim F. Challenges to a Cure for HBV Infection. Semin Liver Dis 2017;37[3]:231–242. [DOI] [PubMed] [Google Scholar]
- [27].Richman DD. Antiviral drug resistance. Chichester [u.a.]: Wiley; 1996. [Google Scholar]
- [28].Endy D, Yin J. Toward Antiviral Strategies That Resist Viral Escape. Antimicrob Agents Chemother 2000;44[4]:1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Domingo E, Menendez-Arias L, Quinones-Mateu ME, Holguin A, Gutierrez-Rivas M, Martinez MA, et al. Viral quasispecies and the problem of vaccine-escape and drug-resistant mutants. Prog Drug Res 1997;48:99–128. [DOI] [PubMed] [Google Scholar]
- [30].Soppe JA, Lebbink RJ. Antiviral Goes Viral: Harnessing CRISPR/Cas9 to Combat Viruses in Humans. Trends Microbiol 2017;25[10]:833–850. [DOI] [PubMed] [Google Scholar]
- [31].Chen S, Yu X, Guo D. CRISPR-Cas Targeting of Host Genes as an Antiviral Strategy. Viruses 2018; 10[1]: 10.3390/v10010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sehrawat S, Kumar D, Rouse BT. Herpesviruses: Harmonious Pathogens but Relevant Cofactors in Other Diseases? Frontiers in cellular and infection microbiology 2018;8:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Grinde B Herpesviruses: latency and reactivation - viral strategies and host response. Journal of oral microbiology 2013;5: 10.3402/jom.v5i0.22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell 2009;138[1]:30–50. [DOI] [PubMed] [Google Scholar]
- [35].Whitley RJ. Herpesviruses In: 4th, Baron S, editors. Medical Microbiology Galveston [TX]: The University of Texas Medical Branch at Galveston; 1996. [PubMed] [Google Scholar]
- [36].Lobo AM, Agelidis AM, Shukla D. Pathogenesis of herpes simplex keratitis: The host cell response and ocular surface sequelae to infection and inflammation. Ocul Surf 2019;17[1]:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tsatsos M, MacGregor C, Athanasiadis I, Moschos MM, Hossain P, Anderson D. Herpes simplex virus keratitis: an update of the pathogenesis and current treatment with oral and topical antiviral agents. Clin Exp Ophthalmol 2016;44[9]:824–837. [DOI] [PubMed] [Google Scholar]
- [38].Biswas PS, Rouse BT. Early events in HSV keratitis - Setting the stage for a blinding disease. Microbes Infect 2005;7[4]:799–810. [DOI] [PubMed] [Google Scholar]
- [39].Jaggi U, Wang S, Tormanen K, Matundan H, Ljubimov AV, Ghiasi H. Role of Herpes Simplex Virus Type 1 (HSV-1) Glycoprotein K (gK) Pathogenic CD8+ T Cells in Exacerbation of Eye Disease. Frontiers in Immunology 2018;9:2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Vanarsdall AL, Johnson DC. Human cytomegalovirus entry into cells. Current opinion in virology 2012;2[1]:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kuppers R B cells under influence: transformation of B cells by Epstein-Barr virus. Nat Rev Immunol 2003;3[10]:801–812. [DOI] [PubMed] [Google Scholar]
- [42].Elion GB. Mechanism of action and selectivity of acyclovir. Am J Med 1982;73[1]:7–13. [DOI] [PubMed] [Google Scholar]
- [43].Elion GB. Acyclovir: discovery, mechanism of action, and selectivity. J Med Virol 1993;Suppl 1:2–6. [DOI] [PubMed] [Google Scholar]
- [44].Field AK, Biron KK. "The end of innocence" revisited: resistance of herpesviruses to antiviral drugs. Clin Microbiol Rev 1994;7[1]:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Englund JA, Zimmerman ME, Swierkosz EM, Goodman JL, Scholl DR, Balfour HH Jr. Herpes simplex virus resistant to acyclovir. A study in a tertiary care center. Ann Intern Med 1990;112[6]:416–422. [DOI] [PubMed] [Google Scholar]
- [46].Piret J, Boivin G. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob Agents Chemother 2011;55[2]:459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cohen JI. Epstein-barr virus vaccines. Clinical & translational immunology 2015;4[1]:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li X, Gou C, Yao L, Lei Z, Gu T, Ren F, et al. Patients with HBV-related acute-on-chronic liver failure have increased concentrations of extracellular histones aggravating cellular damage and systemic inflammation. J Viral Hepat 2017;24[1]:59–67. [DOI] [PubMed] [Google Scholar]
- [49].Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol 2009;51[3]:581–592. [DOI] [PubMed] [Google Scholar]
- [50].Allweiss L, Dandri M. The Role of cccDNA in HBV Maintenance. Viruses 2017;9[6]: 10.3390/v9060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lok AS, McMahon BJ, Brown RS Jr, Wong JB, Ahmed AT, Farah W, et al. Antiviral therapy for chronic hepatitis B viral infection in adults: A systematic review and meta-analysis. Hepatology 2016;63[1]:284–306. [DOI] [PubMed] [Google Scholar]
- [52].Maartens G, Celum C, Lewin SR. HIV infection: epidemiology, pathogenesis, treatment, and prevention. Lancet 2014;384[9939]:258–271. [DOI] [PubMed] [Google Scholar]
- [53].Deeks SG, Overbaugh J, Phillips A, Buchbinder S. HIV infection. Nat Rev Dis Primers 2015;1:15035. [DOI] [PubMed] [Google Scholar]
- [54].Palmisano L, Vella S. A brief history of antiretroviral therapy of HIV infection: success and challenges. Ann Ist Super Sanita 2011;47[1]:44–48. [DOI] [PubMed] [Google Scholar]
- [55].Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host & microbe 2010;7[6]:440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Taubenberger JK, Morens DM. Pandemic influenza--including a risk assessment of H5N1. Rev Sci Tech 2009;28[1]:187–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hale BG, Albrecht RA, García-Sastre A. Innate immune evasion strategies of influenza viruses. Future Microbiology 2010;5[1]:23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rajão D,S, Pérez D,R Universal Vaccines and Vaccine Platforms to Protect against Influenza Viruses in Humans and Agriculture. Frontiers Microbiology 2018;9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Irwin KK, Renzette N, Kowalik TF, Jensen JD. Antiviral drug resistance as an adaptive process. Virus evolution 2016;2[1]:vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hussain M, Galvin HD, Haw TY, Nutsford AN, Husain M. Drug resistance in influenza A virus: the epidemiology and management. Infection & Drug Resistance 2017;10:121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lieberman PM. Epigenetics and Genetics of Viral Latency. Cell Host & microbe 2016;19[5]:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Strasfeld L, Chou S. Antiviral drug resistance: mechanisms and clinical implications. Infect Dis Clin North Am 2010;24[2]:413–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Targeting host genes for therapy. Nature 2016;540:487. [DOI] [PubMed] [Google Scholar]
- [64].Takeuchi o, Akira S. Innate immunity to virus infection. Immunol Rev 2009;227[1]:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].de Chassey B, Meyniel-Schicklin L, Vonderscher J, André P, Lotteau V. Virus-host interactomics: new insights and opportunities for antiviral drug discovery. Genome Medicine 2014;6[11]:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Shalem o, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014;343[6166]:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wilen CB, Tilton JC, Doms RW. HIV: cell binding and entry. Cold Spring Harbor Perspectives in Medicine 2012;2[8]:a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA 2014;111[26]:9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lopalco L CCR5: From Natural Resistance to a New Anti-HIV Strategy. Viruses 2010;2[2]:574–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].MacArthur RD, Novak RM. Maraviroc: The First of a New Class of Antiretroviral Agents. Clinical Infectious Diseases 2008;47[2]:236–241. [DOI] [PubMed] [Google Scholar]
- [71].Fatkenheuer G, Pozniak AL, Johnson MA, Plettenberg A, Staszewski S, Hoepelman AI, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med 2005. November;11[11]:1170–1172. [DOI] [PubMed] [Google Scholar]
- [72].Hütter G, Bodor J, Ledger S, Boyd M, Millington M, Tsie M, et al. CCR5 Targeted Cell Therapy for HIV and Prevention of Viral Escape. Viruses 2015;7[8]:4186–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene Editing of CCR5 in Autologous CD4 T Cells of Persons Infected with HIV. N Engl J Med 2014;370[10]:901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 2008;26[7]:808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, et al. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood 2011;117[10]:2791–2799. [DOI] [PubMed] [Google Scholar]
- [76].Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 2009;360[7]:692–698. [DOI] [PubMed] [Google Scholar]
- [77].Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 2010;28[8]:839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Li L, Krymskaya L, Wang J, Henley J, Rao A, Cao L, et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Molecular therapy : the journal of the American Society of Gene Therapy 2013;21[6]:1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Liu Z, Chen S, Jin X, Wang Q, Yang K, Li C, et al. Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4[+] T cells from HIV-1 infection. Cell & bioscience 2017;7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Xu L, Yang H, Gao Y, Chen Z, Xie L, Liu Y, et al. CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Mol Ther 2017;25[8]:1782–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Li C, Guan X, Du T, Jin W, Wu B, Liu Y, et al. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J Gen Virol 2015;96[8]:2381–2393. [DOI] [PubMed] [Google Scholar]
- [82].Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014;24[1]:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hou P, Chen S, Wang S, Yu X, Chen Y, Jiang M, et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci Rep 2015;5:15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Yang J, Li M, Shen X, Liu S. Influenza A virus entry inhibitors targeting the hemagglutinin. Viruses 2013;5[1]:352–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Edinger TO, Pohl MO, Stertz S. Entry of influenza A virus: host factors and antiviral targets. J Gen Virol 2014;95[Pt 2]:263–277. [DOI] [PubMed] [Google Scholar]
- [86].Han J, Perez JT, Chen C, Li Y, Benitez A, Kandasamy M, et al. Genome-wide CRISPR/Cas9 Screen Identifies Host Factors Essential for Influenza Virus Replication. Cell Rep 2018;23[2]:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Heaton BE, Kennedy EM, Dumm RE, Harding AT, Sacco MT, Sachs D, et al. A CRISPR Activation Screen Identifies a Pan-avian Influenza Virus Inhibitory Host Factor. Cell Rep 2017;20[7]:1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bogerd HP, Kornepati AVR, Marshall JB, Kennedy EM, Cullen BR. Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proc Natl Acad Sci U S A 2015;112[52]:E7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ma Y, Walsh MJ, Bernhardt K, Ashbaugh CW, Trudeau SJ, Ashbaugh IY, et al. CRISPR/Cas9 Screens Reveal Epstein-Barr Virus-Transformed B Cell Host Dependency Factors. Cell Host Microbe 2017;21[5]:591.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Yu J, Li M, Wilkins J, Ding S, Swartz TH, Esposito AM, et al. IFITM Proteins Restrict HIV-1 Infection by Antagonizing the Envelope Glycoprotein. Cell Rep 2015;13[1]:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog 2014;10[4]:e1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Wang B, Lam TH, Soh MK, Ye Z, Chen J, Ren EC. Influenza A Virus Facilitates Its Infectivity by Activating p53 to Inhibit the Expression of Interferon-Induced Transmembrane Proteins. Front Immunol 2018;9:1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Foster TL, Wilson H, Iyer SS, Coss K, Doores K, Smith S, et al. Resistance of Transmitted Founder HIV-1 to IFITM-Mediated Restriction. Cell Host Microbe 2016;20[4]:429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Santa-Marta M, de Brito PM, Godinho-Santos A, Goncalves J. Host Factors and HIV-1 Replication: Clinical Evidence and Potential Therapeutic Approaches. Frontiers in immunology 2013;4:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Grütter M G, Luban J TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Current opinion in virology 2012;2[2]:142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Neagu MR, Ziegler P, Pertel T, Strambio-De-Castillia C, Grutter C, Martinetti G, et al. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J Clin Invest 2009;119[10]:3035–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011;480[7377]:379–382. [DOI] [PubMed] [Google Scholar]
- [98].Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 2011;474[7353]:658–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011. May 25;474[7353]:654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 2009;139[3]:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Neil SJD, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 2008;451:425. [DOI] [PubMed] [Google Scholar]
- [102].König R, Stertz S, Zhou Y, Inoue A, Hoffmann H, Bhattacharyya S, et al. Human host factors required for influenza virus replication. Nature 2010;463[7282]:813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, et al. Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Reports 2016;16[1]:232–246. [DOI] [PubMed] [Google Scholar]
- [104].Vanwalscappel B, Tada T, Landau NR. Toll-like receptor agonist R848 blocks Zika virus replication by inducing the antiviral protein viperin. Virology 2018;522:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].DICK GW, KITCHEN SF, HADDOW AJ. Zika virus I Isolations and serological specificity. Trans R Soc Trop Med Hyg 1952;46[5]:509–520. [DOI] [PubMed] [Google Scholar]
- [106].van Diemen FR, Kruse EM, Hooykaas MJ, Bruggeling CE, Schurch AC, van Ham PM, et al. CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections. PLoS Pathog 2016;12[6]:e1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Lin C, Li H, Hao M, Xiong D, Luo Y, Huang C, et al. Increasing the Efficiency of CRISPR/Cas9-mediated Precise Genome Editing of HSV-1 Virus in Human Cells. Scientific Reports 2016;6:34531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci U S A 2014;111[36]:13157–13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Xu X, Fan S, Zhou J, Zhang Y, Che Y, Cai H, et al. The mutated tegument protein UL7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of alpha-4 gene transcription. Virol J 2016;13:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gergen J, Coulon F, Creneguy A, Elain-Duret N, Gutierrez A, Pinkenburg O, et al. Multiplex CRISPR/Cas9 system impairs HCMV replication by excising an essential viral gene. PLoS One 2018;13[2]:e0192602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Seeger C, Sohn JA. Complete Spectrum of CRISPR/Cas9-induced Mutations on HBV cccDNA. Mol Ther 2016;24[7]:1258–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Mol Ther Nucleic Acids 2014;3:e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids 2014;3:e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Karimova M, Beschorner N, Dammermann W, Chemnitz J, Indenbirken D, Bockmann JH, et al. CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Sci Rep 2015;5:13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Wang J, Xu ZW, Liu S, Zhang RY, Ding SL, Xie XM, et al. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J Gastroenterol 2015;21[32]:9554–9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kennedy EM, Bassit LC, Mueller H, Kornepati AVR, Bogerd HP, Nie T, et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology 2015;476:196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, et al. Harnessing the clustered regularly interspaced short palindromic repeat [CRISPR]/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther 2015;22[5]:404–412. [DOI] [PubMed] [Google Scholar]
- [118].Lin G, Zhang K, Li J. Application of CRISPR/Cas9 Technology to HBV. International journal of molecular sciences 2015;16[11]:26077–26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Liu X, Hao R, Chen S, Guo D, Chen Y. Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J Gen Virol 2015;96[8]:2252–2261. [DOI] [PubMed] [Google Scholar]
- [120].Li H, Sheng C, Wang S, Yang L, Liang Y, Huang Y, et al. Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9. Frontiers Cellular & Infection Microbiology 2017;7:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Dong C, Qu L, Wang H, Wei L, Dong Y, Xiong S. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral Res 2015;118:110–117. [DOI] [PubMed] [Google Scholar]
- [122].Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, et al. Elimination of HIV-1 Genomes from Human T-lymphoid Cells by CRISPR/Cas9 Gene Editing. Scientific Reports 2016;6:22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zhang Y, Yin C, Zhang T, Li F, Yang W, Kaminski R, et al. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Scientific reports 2015;5:16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Zhu W, Lei R, Le Duff Y, Li J, Guo F, Wainberg MA, et al. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 2015;12:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Wang G, Zhao N, Berkhout B, Das AT. A Combinatorial CRISPR-Cas9 Attack on HIV-1 DNA Extinguishes All Infectious Provirus in Infected T Cell Cultures. Cell Reports 2016;17[11]:2819–2826. [DOI] [PubMed] [Google Scholar]
- [126].Ophinni Y, Inoue M, Kotaki T, Kameoka M. CRISPR/Cas9 system targeting regulatory genes of HIV-1 inhibits viral replication in infected T-cell cultures. Scientific Reports 2018;8(1):7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Yuan M, Gao X, Chard LS, Ali Z, Ahmed J, Li Y, et al. A marker-free system for highly efficient construction of vaccinia virus vectors using CRISPR Cas9. Mol Ther Methods Clin Dev 2015. September 16;2:15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yuan M, Zhang W, Wang J, Al Yaghchi C, Ahmed J, Chard L, et al. Efficiently editing the vaccinia virus genome by using the CRISPR-Cas9 system. J Virol 2015. May;89(9):5176–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Chou YY, Krupp A, Kaynor C, Gaudin R, Ma M, Cahir-McFarland E, et al. Inhibition of JCPyV infection mediated by targeted viral genome editing using CRISPR/Cas9. Sci Rep 2016. November 14;6:36921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun 2014. August 8;450(4):1422–1426. [DOI] [PubMed] [Google Scholar]
- [131].Glass Z, Lee M, Li Y, Xu Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends in Biotechnology 2018. February;36(2):173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release 2017. November 28;266:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Ran FA, Hsu PD, Lin C, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013;154(6):1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013. September;31(9):822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Lei L, Chen H, Xue W, Yang B, Hu B, Wei J, et al. APOBEC3 induces mutations during repair of CRISPR–Cas9-generated DNA breaks. Nature Structural & Molecular Biology 2018;25(1):45–52. [DOI] [PubMed] [Google Scholar]
- [136].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013;339(6121):819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release 2017. November 28;266:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 1998. December;72(12):9873–9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013. September;31(9):822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther 2010. January;18(1):80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Ramanan V, Shlomai A, Cox DBT, Schwartz RE, Michailidis E, Bhatta A, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Scientific Reports 2015;5:10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Lebbink RJ, de Jong DC, Wolters F, Kruse EM, van Ham PM, Wiertz EJ, et al. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci Rep 2017. February 8;7:41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Kaminski R, Bella R, Yin C, Otte J, Ferrante P, Gendelman HE, Li H, Booze R, Gordon J, Hu W, Khalili K. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 2016. August;23(8-9):690–5. doi: 10.1038/gt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Yin C, Zhang T, Qu X, Zhang Y, Putatunda R, Xiao X, Li F, Xiao W, Zhao H, Dai S, Qin X, Mo X, Young WB, Khalili K, Hu W. In Vivo Excision of HIV-1 Provirus by saCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol Ther. 2017. May 3;25(5):1168–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Rupp LJ, Schumann K, Roybal KT, Gate RE, Ye CJ, Lim WA, Marson A. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptorT cells. Sci Rep. 2017. April 7;7(1):737. doi: 10.1038/s41598-017-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gönen M, Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017. March 2;543(7643):113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Krystofinski P, Li H, Tobin V, Nguyen DN, Lee MR, Putnam AL, Ferris AL, Marson A. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 2018. July;559(7714):405–409. doi: 10.1038/s41586-018-03265.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Sharfe N, Dadi HK, Shahar M & Roifman CM. Human immune disorder arising from mutation of the alpha chain of the interleukin-2 receptor. Proc. Natl. Acad. Sci. U. S. A. 94, 3168–3171 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Sakaguchi S, Sakaguchi N, Asano M, Itoh M & Toda M. Immunologic Self-Tolerance Maintained by Activated T Cells Expressing 11-2 Receptor a-Chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol 155, 1152–1164(1995). [PubMed] [Google Scholar]
- [150].Robbins PF et al. Single and Dual Amino Acid Substitutions in TCR CDRs Can Enhance Antigen-Specific T Cell Functions. J. Immunol. 180, 6116–6131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]