Summary
Arp2/3 complex nucleates branched actin filaments important for cellular motility, endocytosis, meiosis, and cellular differentiation [1–4]. WASP proteins, the prototypical Arp2/3 complex activators, activate Arp2/3 complex only once it is bound to the side of an actin filament [5,6]. This ensures WASP-activated Arp2/3 complex only nucleates branched actin filaments but means branched actin networks must be seeded with an initial preformed filament. Dip1 and other WISH/DIP/SPIN family proteins activate Arp2/3 complex without preformed filaments [7], creating seed filaments that activate WASP-bound Arp2/3 complex [8]. Importantly, Dip1-mediated activation of Arp2/3 complex creates linear filaments instead of branches [7]. Cells may therefore need to limit Dip1 activity relative to WASP to preserve the dendritic nature of actin networks, although it is unclear whether such regulatory mechanisms exist. Here, we use TIRF microscopy to show that Dip1 causes actin assembled with WASP and Arp2/3 complex to form disconnected networks with many linear filaments rather than highly branched arrays. We discover a key biochemical difference between Dip1 and WASP that may limit linear filament nucleation in cells; while WASP must be released for nucleation, Dip1 stays associated with Arp2/3 complex on the pointed ends of nucleated actin filaments, so Dip1 is consumed in the reaction. Using live cell imaging of fission yeast we provide evidence that Dip1 is a single turnover activator of Arp2/3 complex in vivo, revealing a mechanism by which Dip1 can initiate branched actin networks at endocytic sites without disrupting their branched architectures.
eTOC Blurb
Balzer et al. show that Dip1 activates Arp2/3 complex through a single turnover mechanism that provides a potential regulatory mechanism to control the balance of linear versus branched actin filaments at endocytic sites.
Results
Dip1 induces disconnected linear actin network architectures, even in the presence of Wsp1
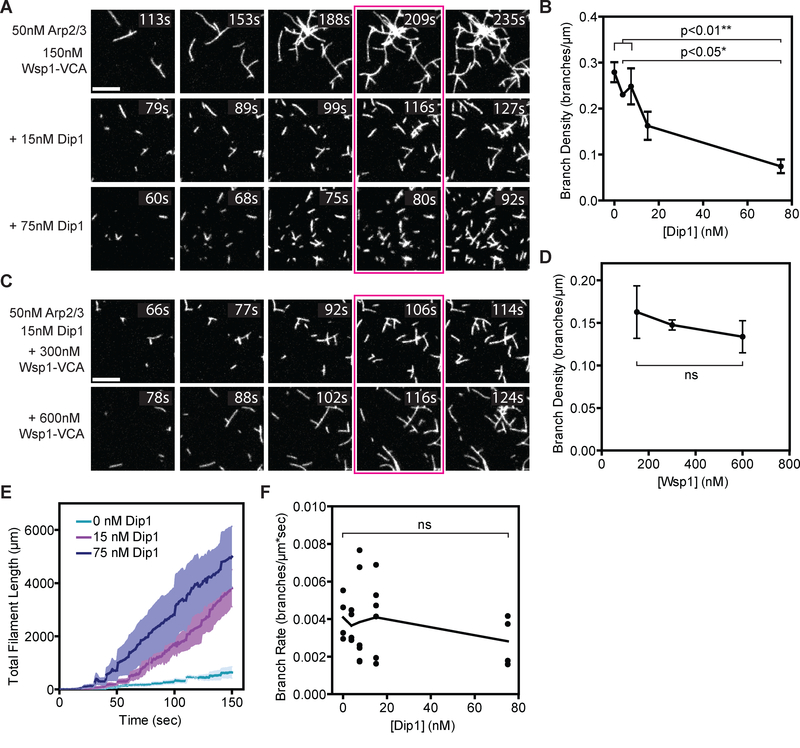
The branching nucleation activity of Arp2/3 complex allows it to assemble networks of short, crosslinked actin filaments that are thought to be optimal for pushing against broad surfaces [9, 10]. However, unlike Wsp1, the S. pombe Wiskott-Aldrich Syndrome (WASP) family protein, the S. pombe WISH/DIP/SPIN90 family protein, Dip1, activates Arp2/3 complex to nucleate linear actin filaments instead of branches [7,11]. Both Dip1 and Wsp1 are present at sites of endocytosis [11,12], so we wondered how their simultaneous activity on Arp2/3 complex influences actin network assembly. To address this, we used TIRF microscopy to visualize in vitro actin assembly mediated by Wsp1 and Arp2/3 complex with increasing concentrations of Dip1 (Figure 1A). In reactions lacking Dip1, many branches grew from a few spontaneously nucleated filaments, generating networks with high branching densities (Figure 1B). Adding Dip1 to the reactions caused network architectures to become less dendritic and more disconnected when comparing networks with the same amount of total polymer (Figure 1A). At 75 nM Dip1, the branch density decreased ~ 4-fold compared to reactions without Dip1, even though 150 nM Wsp1-VCA was present (Figure 1B). These data show that Dip1 induces Arp2/3 complex to produce poorly connected filament networks with greater proportions of linear filaments, even in the presence of Wsp1. For technical reasons (see below, and Figure S1), we could not measure the branch density at equivalent timepoints across the range of Dip1 concentrations we tested. However, the influence of Dip1 on actin network architectures is not due to the fact that we compared actin networks with equal total polymer length rather than at equivalent reaction time points, since reactions containing low (3.75 nM) versus high (75 nM) Dip1 showed that adding Dip1 decreased the branching density and the branch to linear filament ratio at a single timepoint (80 s) (Figure S2).
Figure 1: Arp2/3 complex preferentially nucleates linear actin filaments in the presence of both Dip1 and Wsp1.
A. TIRF microscopy images of actin polymerization assays containing 150 nM GST-Wsp1-VCA, 1.5 μM 33% Oregon Green-labeled actin, 50 nM SpArp2/3 complex and the indicated concentrations of Dip1. The reaction time is indicated in the upper right corner of each panel. Panels are aligned by total filament length in the full uncropped frame from which each panel is derived. The corresponding total filament length for each panel from left to right is 250, 500, 1000, 1500, and 2000 μm. Scale bar: 5 μm. B. Plot of the branch density versus Dip1 concentration for reactions as shown in Panel A. Branches were counted when the total filament length was 1500 μm (panels boxed in magenta in A). Error bars: SE from 4 to 6 regions of interest containing at least 130 filaments from 2–3 separate TIRF reactions. C. TIRF microscopy images of actin polymerization assays containing 15 nM Dip1, 1.5 μM 33% Oregon Green-labeled actin, 50 nM SpArp2/3 complex and indicated concentrations of GST-Wsp1-VCA. The reaction time is indicated in the upper right corner of each panel. Panels are aligned by total filament length as described in panel (A). Scale Bar: 5 μm. D. Plot of the branch density versus the concentration of GST-Wsp1-VCA as shown in panel C. Branches were counted when the total filament length was 1500 μm (panels boxed in magenta in C). Error Bars: SE from 4 to 6 regions of interest containing at least 340 total filaments from 2–3 separate TIRF reactions. E. Plot of total actin filament length in the field of view versus time for reactions described in panel (A) with the indicated concentration of Dip1. The standard error is calculated from the measurements of the full field of view from 2 or 3 reactions and is shown in a lighter shade around the means at each time point (dark lines). The increased polymer accumulation rate is due to faster nucleation in the Dip1-containing reactions, since Dip1 does not influence filament barbed end elongation rates [7,8]. F. Plot of the branch rate versus the concentration of Dip1 added to reactions as shown in panel (A). Measurements were made when the reaction reached a total filament length of 1500 μm. The time component of the rate is based on an extrapolation of the lifetime of the filament determined from filament lengths and the barbed elongation rate (see methods). Each point represents the average branching rate within a region of interest containing between 19 and 230 filaments. See also Figures S1–S2.
The influence of Dip1 on actin network assembly by Arp2/3 complex and Wsp1 initially led us to hypothesize that Dip1 competes with Wsp1 for activation of Arp2/3 complex. While structural and biochemical studies demonstrate that WASP and WDS family proteins bind to distinct sites on Arp2/3 complex [13–20], eliminating the possibility of direct competition, the two NPFs could potentially allosterically reduce each other’s binding, causing negative cooperativity. Therefore, we asked if increasing the concentration of Wsp1 would counteract the effect of Dip1, increasing the proportion of branched filaments, presumably by decreasing the amount of Dip1-bound Arp2/3 complex. Unexpectedly, increasing the Wsp1 concentration from l50 nM to 600 nM in the presence of l5 nM Dip1 did not increase the branching density when comparing networks with the same amount of total polymer (Figures 1C and 1D). These data argue against a simple negative cooperativity model in which allosteric competition between WASP and Dip1 for binding Arp2/3 complex controls flux though linear versus branched filament pathways.
We previously showed that Dip1 activates Arp2/3 complex with a reduced lag phase because it can trigger nucleation by Arp2/3 complex without waiting for slow spontaneous nucleation of actin filaments, unlike WASP [7]. Therefore, we wondered if the disconnected architectures observed in the presence of both NPFs result from rapid activation of Arp2/3 complex by Dip1 and accumulation of many linear filaments before a significant fraction of WASP-bound complexes can be activated by pre-existing filaments. Consistent with this hypothesis, we found that actin polymer accumulates more rapidly in the presence of Dip1, with linear filaments appearing much more rapidly than branches (Figures 1A and 1E). However, because previous data showed that WDS proteins compete with actin filaments for binding to Arp2/3 complex [16], it is also possible that linear filament generation by Dip1 is faster than branching because Dip1 blocks Arp2/3 complex from binding pre-existing filaments, thereby inhibiting WASP-mediated activation [16]. Importantly, the branching rate was not decreased by adding up to 75 nM Dip1, indicating that this concentration of Dip1 does not effectively prevent Wsp1-bound Arp2/3 complex from binding to actin filaments (Figures 1F and S2).
Dip1 remains bound to the pointed ends of actin filaments for hundreds of seconds after activating Arp2/3 complex
Programmed release of WASP allows it to catalyze multiple rounds of Arp2/3 complex activation at the attachment zone between a polymerizing network and a membrane [21,22]. This multi-turnover capability allows prolonged motility of WASP-coated beads in reconstituted motility assays and may also be important for continuous assembly of actin networks in cells [23,24]. Whether release of Dip1 from Arp2/3 complex is programmed into its activation mechanism, like WASP, or if it instead stays bound to the complex after triggering nucleation is unclear. This distinction dictates how fast a Dip1 molecule can be turned over to catalyze additional rounds of nucleation, so is critical in understanding the balance between linear and branched filament generating NPFs at endocytic sites.
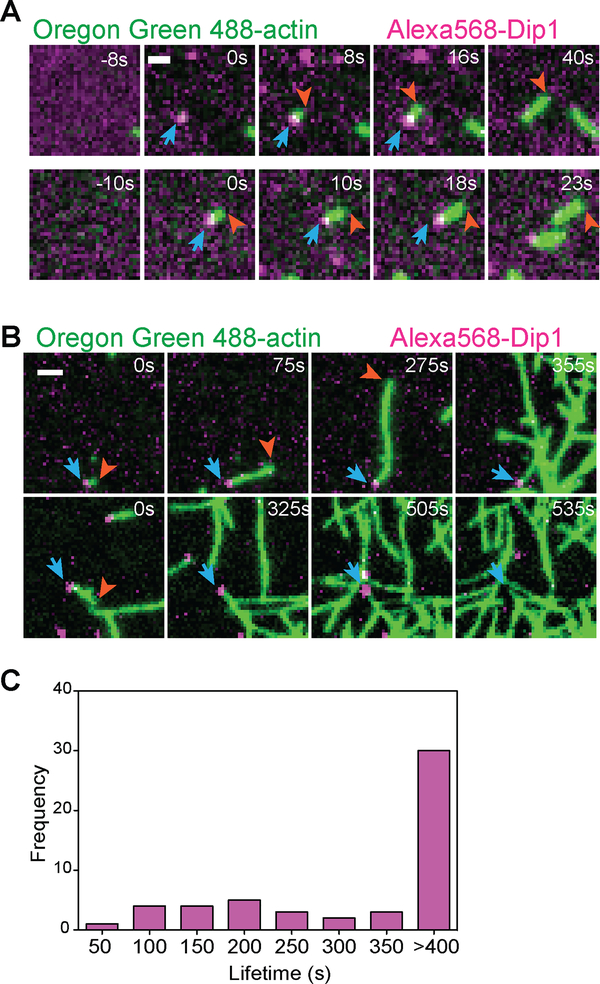
To determine if “programmed” Dip1 release is a feature of its activation mechanism, we labeled it with Alexa568 (Alexa568-Dip1) and visualized its action on Arp2/3 complex in reactions containing Oregon Green-labeled actin using TIRF microcopy. As we reported previously, these reactions produced multiple events in which a new linear actin filament grew from an Alexa568-Dip1 molecule non-specifically adsorbed to the surface [8](Figure 2A). Dip1 does not nucleate filaments on its own and does not bind actin filaments in the absence of Arp2/3 complex, so we interpret these events as Dip1 and Arp2/3 complex nucleating a linear filament [7,8]. Importantly, Dip1 did not release from filament ends upon nucleation, and the Alexa568 signal was visible at the filament end for many seconds as the filament elongated (Figure 2A). To determine how long Dip1 stays bound to Arp2/3 complex after nucleation, we measured the average length of Dip1 binding events. Under our initial imaging conditions, the average lifetime of the Alexa568-Dip1 signal on filament ends was ~l95 seconds. To determine if this lifetime was controlled by photobleaching or dissociation, we decreased the laser exposure by increasing the time interval between frames during the acquisition (Figure 2B). At the lowest exposure levels we tested, the majority of filament-bound Dip1 molecules failed to release (or photobleach) before the end of the experiment (Figures 2B, 2C and Video S1), so we could not determine the rate constant for dissociation of Dip1 from Arp2/3 complex on filament ends. However, these measurements set a lower limit on the average Dip1 lifetime on the filament end at 325 seconds (Figure 2C). Together, these data show that Dip1-mediated activation of the complex does not require Dip1 release, and that Dip1 stays bound to Arp2/3 complex after nucleation and throughout elongation of the filament.
Figure 2: Dip1 remains bound to actin filaments for hundreds of seconds after nucleation.
A. TIRF microscopy images of the polymerization of 1.5 μM 33% Oregon Green-labeled actin in the presence of 6 nM Alexa568-Dip1, 250 nM GST-Wsp1-VCA, and 500 nM SpArp2/3 complex collected with 50 ms exposure times at 200 ms intervals. Top row shows an event in which Dip1 non-specifically adsorbed to the surface activates Arp2/3 complex to nucleate a linear filament. Bottom row shows an event in which a Dip1-bound actin filament landed on the imaging surface. Blue arrows indicate the position of Dip1 and orange arrows mark the barbed end of the growing filament. B. Same as A, except that the interval for data collection was changed to 5s. The laser power was kept constant. The times indicated in the upper right corner of panels (A) and (B) correspond to the time since the appearance of the Alexa568-Dip1 molecule. C. Histogram of single molecule lifetimes on actin filament ends for videos collected as described in (B). See also Video S1.
Dip1 binds to treadmilling actin networks in S. pombe while most Wsp1 remains cortical
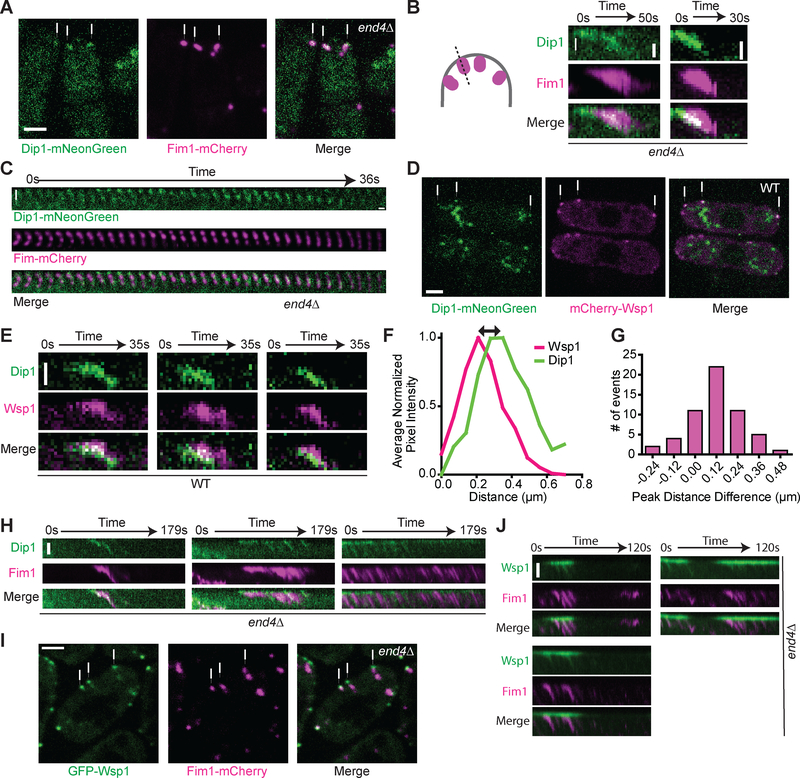
Our in vitro data show that Dip1 stays bound to actin filament ends for hundreds of seconds after nucleation of the linear filament, demonstrating that - unlike WASP - release of Dip1 is not programmed into the nucleation mechanism. We wondered if Dip1 also remains bound to Arp2/3 complex after it activates nucleation of actin filaments at endocytic sites in S. pombe. Arp2/3 complex becomes incorporated into the actin networks it nucleates [24–27], so if Dip1 fails to release from the complex after triggering nucleation, it will also become incorporated into the network. In wild type S. pombe, it can be difficult to distinguish between proteins that stay bound to the cortex or incorporate into the actin network because of the small size of endocytic actin patches (~200 nm) [28,29]. To circumvent this issue, we labeled Dip1 and the S. pombe actin patch marker, Fim1, in the context of an end4Δ strain and imaged the equatorial plane of the cells using spinning disk confocal microscopy (Figure 3A). END4 deletion converts punctate and transient endocytic actin networks into semi-continuously polymerizing actin comet tails that treadmill away from endocytic adaptors on the cortex and into the cytoplasm [12,30] (Figure 3A).
Figure 3: Localization of Dip1 and Wsp1 at wild type endocytic actin networks and treadmilling actin networks induced by deletion of END4.
A. Spinning disk confocal images of Dip1-mNeonGreen (green) and Fim1-mCherry (magenta) in end4Δ S. pombe cells. The white arrows indicate representative treadmilling actin networks. Scale bar: 2 μm. B. Kymographs showing Dip1-mNeonGreen (green) and Fim1-mCherry (magenta) signal in treadmilling actin networks in end4Δ cells. The kymographs were made from a one pixel-wide line drawn from the exterior of the cell into the cytoplasm as depicted in the cartoon. The top of each panel is the cortex and the white arrow shows the direction of internalization into the cytoplasm. Scale bar: 0.5 μm. C. Montage of Dip1-mNeonGreen (green) and Fim1-mCherry (magenta) signals in a treadmilling actin network in end4Δ cells. The interval between each tile is l second. Scale bar: 0.5 μm. D. Spinning disk confocal images of Dip1-mNeonGreen (green) and mCherry-Wsp1 (magenta) in wildtype S. pombe cells. The white arrows indicate representative endocytic patch sites. The large structures in the Dip1-mNeonGreen channel are autofluorescence (Figure S4). Scale bar: 2 μm. E. Kymographs of Dip1-mNeonGreen (green) and mCherry-Wsp1 (magenta) dynamics in wild type cells. The kymographs are generated as described in panel C. Scale Bar: 0.5 μm. F. Plot showing the average normalized pixel intensity of mCherry-Wsp1 and Dip1-mNeonGreen along a line drawn through an endocytic patch from the exterior of the cell into the cytoplasm as depicted in the cartoon in panel B. Measurements were made when the total Wsp1 was at its peak intensity. The peak pixel intensity was aligned to 0.2l μm, the average distance of the peak pixel intensity of Wsp1 from the cortex at this timepoint. Traces represent the average of l5 endocytic patches selected proportionally from the histogram in panel G. G. Histogram of the distance between the peak signals of mCherry-Wsp1 and Dip1-mNeonGreen, as depicted by the black arrow in panel F. Positive numbers correspond to patches in which Dip1 is further from the cortex than Wsp1. On average, Dip1-mNeonGreen peak signal is 120 nm further from the cortex than mCherry-Wsp1 peak signal. See Materials and Methods for additional details (n = 56 patches, one sample t-test p < 0.0001). H. Kymographs of Dip1-mNeonGreen (green) and Fim1-mCherry (magenta) at cortical sites with multiple actin treadmilling events in end4Δ cells. Scale Bar: l μm. I. Spinning disk confocal images of GFP-Wsp1 (green) and Fim1-mCherry (magenta) in end4Δ S. pombe cells. The white arrows indicate representative treadmills. Scale bar: 2 μm. J. Kymographs of GFP-Wsp1 (green) and Fim1-mCherry (magenta) at repetitively treadmilling comet tails in end4Δ cells. Scale bar: l μm. See also Figures S3–S4 and Videos S2–S4.
In an end4 deleted strain expressing Dip1-mNeonGreen and Fim1-mCherry, Dip1 colocalized with Fim1-marked actin networks (Figure 3A), consistent with a previous report showing that slightly overexpressed Dip1 shows a comet tail-like localization pattern in an end4Δ strain [12]. Monitoring the dynamics of Dip1 at comet tail sites revealed that Dip1 treadmills inward from the cortex at the same rate as Fim1-marked actin (Figure 3B). We previously showed that Dip1 does not bind actin filaments directly, and only binds strongly to Arp2/3 complex on an actin filament pointed end if it has cooperated with the complex to nucleate that filament [8]. Therefore, our observation that Dip1 treadmills is consistent with a model in which it remains bound to Arp2/3 complex after triggering nucleation. Interestingly, Dip1 tended to incorporate in punctate patterns rather than spreading evenly throughout the entire treadmill, suggesting that bursts of Dip1 activity may occur at the cortex (Figure 3C and Video S2).
We next examined Dip1 localization in endocytic actin patches from wild type cells. During endocytosis, Wsp1 remains cortical as the actin network assembles, and then moves inward slightly when the vesicle internalizes [11,29]. To determine whether Dip1 localization is distinct from Wsp1 in a wild type background, we labeled Wsp1 with mCherry and Dip1 with mNeonGreen. In most endocytic patches, Dip1 and Wsp1 partially colocalized, but Dip1 was further from the cortex than Wsp1 (Figures 3D, 3E and Video S3). Analysis of endocytic events from several cells revealed that the peak Dip1 signal is on average ~120 nm further from the cortex than the Wsp1 peak signal when the Wsp1 intensity is maximal (Figures 3F and 3G).
To more clearly visualize differences between Wsp1 and Dip1, we compared the behavior of each NPF in the context of an end4 deletion strain. Actin comet tails that formed in the end4 deletion strain treadmilled for variable lengths of time before releasing from the membrane and moving into the cytoplasm (VideoS2 and S4). In many instances, multiple comet tails grew from a single site at the cortex, with the actin network reinitiating after each instance of comet tail release (VideoS2 and S4). The peak Dip1 signal frequently moved inward with the treadmilling actin network, in some cases leaving little or no signal behind at the cortex (Figures 3H, S3, Video S2). In contrast, most Wsp1 remained at the cortex, with only a small portion of GFP-Wsp1 fluorescence evenly distributed within the comet tail (Figures 3I, 3J and Video S4). This observation is consistent with a model in which cortical Wsp1 molecules can catalyze multiple rounds of nucleation by Arp2/3 complex. This mechanism may help cells control the balance between branching and linear filament nucleation at endocytic sites (see discussion).
Discussion
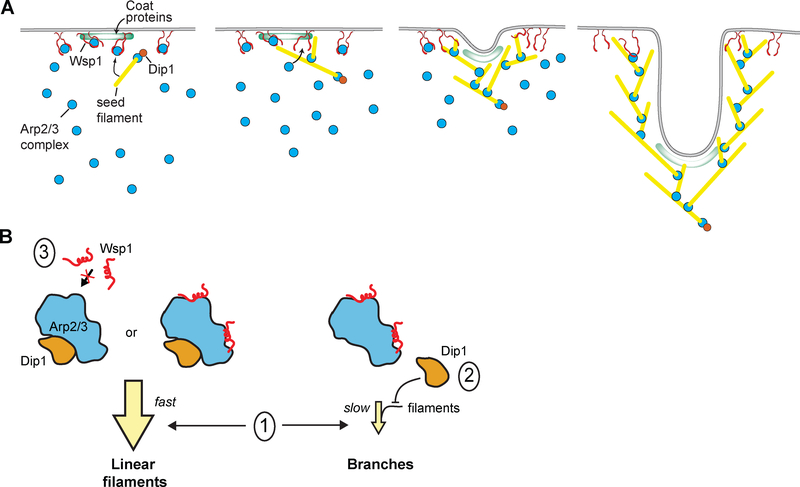
Our data reveal a critical mechanistic difference between WASP and WDS family NPFs. While WASP release is programmed into the nucleation mechanism, Dip1 release is not. Instead, Dip1 stays bound to Arp2/3 complex on the pointed ends of actin filaments for an average of over 5 minutes, more than 15 times longer than the average lifetime of an endocytic actin patch [31]. This suggests that Dip1 is a single turnover activator of Arp2/3 complex at endocytic sites. Our in vivo data support this model, showing that Dip1 incorporates into treadmilling actin networks, presumably because it is bound to Arp2/3 complex and thus unable to stimulate additional rounds of nucleation. The single turnover mechanism of Dip1-mediated activation of Arp2/3 complex may be important in limiting the linear filament nucleation activity of Dip1, since the catalytic activity of an NPF is proportional to its turnover rate. Endocytic actin patches contain at peak ~20 Dip1 molecules, ~150–230 Wsp1 molecules and ~320 Arp2/3 complexes [12,31]. Because Dip1 is consumed in the reaction, 20 likely represents the upper limit on the number of Dip1-Arp2/3 nucleated linear filaments per patch (Figure 4A).
Figure 4: Proposed models of Dip1 activity and dynamics in vitro and at endocytic patches.
A. Cartoon model showing activity of Dip1 (orange circle), Wsp1 (paired red lines), Arp2/3 complex (cyan circles), coat proteins (green zone) and actin (yellow lines) during membrane invagination of an endocytic patch. The number of molecules depicted is roughly proportional to the measured concentrations of these proteins at endocytic sites [12,31]. Dip1 activates Arp2/3 complex to create a linear filament that initiates assembly of the actin network. Dip1 remains bound to Arp2/3 complex so it can only activate a single Arp2/3 complex during assembly of the network. Most Wsp1 molecules remain bound to the cortex allowing them to catalyze multiple rounds of branching. The Dip1-bound linear filament that primed network assembly moves inward as Wsp1-mediated activation of the complex nucleates branches at the cortex. B. Cartoon model showing potential mechanisms of coordinate Arp2/3 complex regulation by Dip1 and Wsp1. The three Arp2/3 complexes represent the different possible binding states of the complex in the presence of both NPFs. Given that WDS proteins have been shown to block actin filament binding, we assume that Arp2/3 complex bound to both activators would create a linear actin filament [16]. Model l: faster kinetics of activation of the Arp2/3 complex by Dip1 compared to Wsp1-bound Arp2/3 complex. Model l explains the dominance of Dip1 over Wsp1 in vitro. Model 2: competition between Dip1 and actin filaments for binding to Arp2/3 complex. Our data indicate that model 2 does not contribute to the dominance of Dip1 over Wsp1 in vitro, though this competition may be important in vivo. Model 3: competition between Dip1 and Wsp1 for binding to Arp2/3 complex. Our data argue against a role for model 3 either in vitro or in vivo.
In contrast to Dip1, our data here and several other studies have shown that most WASP remains cortically localized rather than incorporating into treadmilling actin networks [24,30]. Therefore, it is possible that single WASP molecules catalyze multiple rounds of branching nucleation in treadmilling networks in vivo to allow continued branched actin assembly at the cortex, as occurs in reconstituted systems in which WASP is attached to polystyrene beads or glass surfaces [23,32,33]. While it is still unclear how actin filaments are organized to generate force at endocytic sites [34,35], some experiments support a model in which WASP nucleates a network that assembles at the base of the endocytic invagination, pushing inward on a disk of adaptor proteins attached to the tip of the invagination [36,37]. This model is consistent with our observation that Dip1 is more cytoplasmic than Wsp1 in wild type yeast, as Dip1 molecules bound to the initial filaments that seeded the network would move inward as the actin network grows away from the cortex (Figure 4A).
Here we show that in vitro, linear filament nucleation by Dip1-activated Arp2/3 complex is dominant over Wsp1-mediated branching. This suggests that cells must employ mechanisms to limit linear filament generation by Dip1-activated Arp2/3 complex. Under the conditions we tested, the dominance of Dip1 is due to the fast kinetics of Arp2/3 complex activation by Dip1 relative to activation by Wsp1 (Figure 4B, model 1). Given that the mammalian WDS protein, SPIN90, blocks Arp2/3 complex from interacting with actin filaments [16], we expected competition between Dip1 and actin filaments for Arp2/3 complex binding would provide another mechanism (Figure 4B, model 2) by which linear filament generation could dominate in vitro. However, our data indicate that this mechanism contributes little to the dominance of Dip1 under the conditions here, since increasing the concentration of Dip1 up to 75 nM did not decrease branching rates. We suspect that the inability of Dip1 to block branching at these concentrations is due to its relatively weak affinity for the isolated Arp2/3 complex [7]; it is likely that only a small percentage of the complex binds Dip1 at this concentration. Our data also indicate that Dip1 and Wsp1 do not compete for binding and activation of the complex, either directly or allosterically (Figure 4B, model 3). This observation indicates that increasing the local concentration of activated Wsp1 is not an effective way to limit the linear filament nucleation activity of Dip1. Therefore, the ~8 fold higher concentration of Wsp1 at endocytic sites likely does little to drive endocytic networks toward highly branched topologies [12,31]. However, because actin filaments trigger Wsp1-mediated activation of the complex but likely block Dip1 from activating, the relative flux between the linear verses branched filament nucleation pathways likely depends on the local concentration of actin filaments (Figure 4B, model 1). We speculate that when a branched network assembles on a membrane, the high local concertation of actin filaments may allow WASP proteins to better compete with WDS family proteins, providing a mechanism by which Dip1 activity could be limited to the initiation phase of branched actin network assembly. This effect of actin filaments, combined with the consumption of Dip1 after a single round of Arp2/3 complex activation, may sufficiently limit linear filament generation at endocytic sites. We note that it was not possible under the conditions of our TIRF assays to deplete the pool of Dip1, so Dip1 activity dominated over Wsp1 despite the single turnover nature of the activity of Dip1.
An important future direction will be to investigate the coordinate regulation of Arp2/3 complex by Dip1 and full length Wsp1. While we use only the Arp2/3 complex-activating region (VCA) of Wsp1 in our studies here, the N-terminal region of Wsp1 harbors a WH1 domain that likely binds verprolin (an actin monomer binding protein [11]), and polyproline segments that have a variety of potential binding partners [38]. Understanding how these potential interaction partners influence network propagation will be important for understanding how Wsp1 controls actin network assembly in the presence of Dip1 in cells.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Brad Nolen (bnolen@uoregon.edu).
Schizosaccharomyces pombe strains generated in this study are available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
This study uses Schizosaccharomyces pombe cells which were grown at 30°C in YE5S media with shaking at 180 rpm unless otherwise noted. S. pombe cells were maintained in exponential phase for 24–48 hours before harvesting for imaging. A list of all strains used in this manuscript can be found in the Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Antibodies | ||
| Bacterial and Virus Strains | ||
| BL21-CodonPlus(DE3)-RIL | Agilent | Cat. #230245 |
| Biological Samples | ||
| Rabbit Muscle Acetone Powder | Pel-Freeze Biologicals | Cat. #41995–2 |
| S. pombe Arp2/3 complex | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Alexa Fluor 568 C5 maleimide | ThermoFisher Scientific | Cat. # A20341 |
| Oregon Green 488 maleimide | ThermoFisher Scientific | Cat. # O6034 |
| (3-Aminopropyl)triethoxysilane (APTES) | MilliporeSigma | Cat. # 440140 |
| EZ-Link-NHS-PEG12-biotin | ThermoFisher Scientific | Cat. # 21312 |
| Methoxy PEG succinimidyl succinate, MW5000 | JenKem | Cat. # A3011–1 |
| Biotin-PEG NHS ester, MW5000 | JenKem | Cat. # A5027–1 |
| NeutrAvidin | ThermoFisher Scientific | Cat. # 31000 |
| Catalase | Sigma | Cat. # C3515 |
| Glucose Oxidase | MP Biomedicals | Cat. # 195196 |
| Myosin II, rabbit skeletal muscle | Cytoskeleton | Cat. # MYO2 |
| Albumin, from bovine serum | Sigma | Cat. # A2153 |
| cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail tablets | Sigma | Cat. # 11836170001 |
| Zymolyase-100T | Nacalai tesque | Cat. # 07665–55 |
| Salmon Sperm DNA, sheared (10 mg/mL) | Invitrogen | CAS # 7732–18–5 |
| S. pombe Dip1 | This study | N/A |
| S. pombe Dip1(6CysAla) | This study | N/A |
| S. pombe Wsp1-VCA | This study | N/A |
| Critical Commercial Assays | ||
| In-Fusion HD Cloning Plus CE | Takara | Cat. # 638916 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| S. pombe TP150 (protease deficient, h- leu1) | Vladimir Sirotkin | SpBN110 |
| S. pombe VS872 (h- end4::ura4+ ade6-M216, his3-D1, leu1–32, ura4-D18) | Vladimir Sirotkin | SpBN109 |
| S. pombe VS1124a (h+ kanMX6-Pwsp1-mGFP-wsp1, fim1-mCherry-natMX6, ade6-M210, his3-D1, leu1–32, ura4-D18) | Vladimir Sirotkin | SpBN282 |
| S. pombe VS1025–7 (h- kanMX6-Pwsp1-mCherry-wsp1, ade6-M216, his3-D1, leu1–32, ura4-D18) | Vladimir Sirotkin | SpBN259 |
| S. pombe SpBN157–6 (end4::ura4+, kanMX6-Pwsp1-mGFP-wsp1, fim1-mCherry-natMX6, his3-D1, leu1–32, ura4-D18) | This study | SpBN157–6 |
| S. pombe SpBN165–1 (end4::ura4+, dip1-mGFP-kanMX6, fim1-mCherry-natMX6, his3-D1, leu1–32, ura4-D18) | This study | SpBN165–1 |
| S. pombe SpBN278–3 (end4::ura4+, dip1-mNeonGreen-hphMX6, fim1-mCherry-natMX6, his3-D1, leu1–32, ura4-D18) | This study | SpBN278–3 |
| S. pombe SpBN280–1 (h- kanMX6-Pwsp1-mCherry-wsp1, dip1-mNeonGreen-hphMX6, ade6-M216, his3-D1, leu1–32, ura4-D18) | This study | SpBN280–1 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| pGV67_SpDip1 | [7] | Plasmid #174 |
| pGV67_SpDip1(6cysAla) | [8] | Plasmid #180 |
| pGV67_SpWsp1(VCA)-497 | [44] | Plasmid #4 |
| pGEX6_NWASP-WA | [45] | Plasmid #222 |
| pRK1043 | [46] | N/A |
| pJK148_SpDip1-mNeonGreen-hphMX6 | This study | Pombe vector #181 |
| Software and Algorithms | ||
| Fiji/ImageJ | [41] | https://fiji.sc/ |
| MATLAB | MathWorks | N/A |
| ImageJ filament tracking plugin | Gift from Jeff Kuhn | N/A |
| ImageJ StackProfileData macro | Michael Schmid | https://imagej.nih.gov/ij/macros/StackProfileData.txt |
| Other | ||
| Amicon Ultra-4 Centrifugal Filters 10,000 MWCO | MilliporeSigma | Cat. # UFC9010 |
| Vivaspin Turbo 15 30,000 MWCO | Sartorius | Cat. # VS15T21 |
| Vivaspin Turbo 15 3,500 MWCO | Sartorius | Cat. # VS15T91 |
| Richard-Allan Scientific Slip-Rite Cover Glass (24 × 60 #1.5 coverslips) | ThermoFisher Scientific | Cat. # 152460 |
METHOD DETAILS
Protein Expression, Purification, and Fluorescent Labeling
To generate a Dip1 construct for site specific labeling with a cysteine-reactive fluorescent dye, the six endogenous cysteines were mutated to alanine by amplifying pGV67-Dip1 (described previously [8]) with non-overlapping 5’phosphoryated primers encoding the mutation. The final expressed protein after TEV cleavage included the full coding sequence for Dip1 along with a short N-terminal polypeptide sequence left after TEV cleavage (GSMEFELRRQACGR). The cysteine in this N-terminal polypeptide was exploited for tagging with the fluorescent dye. For expression and labeling, BL21(DE3)RIL E. coli transformed with the pGV67 Dip1 expression vector was grown to an O.D.595 of 0.6–0.7, induced with 0.4 mM isopropyl 1-thio-β-D-galactopyranoside, and grown overnight at 22 °C. Cells were lysed by sonication in lysis buffer: 20 mM Tris pH 8.0, 140 mM NaCl, 2 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and protease inhibitor tablets (Roche). The lysate was clarified by centrifugation, and the soluble fraction was loaded on a glutathione sepharose column and eluted with 20 mM Tris pH 8.0, 140 mM NaCl and 50 mM glutathione. Peak fractions were pooled and a 25:1 ratio (by mass) of TEV protease to recombinant proteins was added. The reaction mix was dialyzed overnight against 20 mM Tris pH 8.0, 50 mM NaCl and 1 mM dithiothreitol at 4°C. The sample was loaded onto a 6ml Resource Q column at pH 8.0 and eluted with a gradient of 50 mM to 500 mM NaCl. Protein was then concentrated in an Amicon-Ultra concentration device and loaded onto a Superdex 200 HiLoad 16/60 gel filtration column equilibrated in 20 mM HEPES pH 7.0, 50 mM NaCl. Peak fractions were pooled and concentrated to ~40 μM for labeling. A 10 mM solution of Alexa568 C5 Maleimide (Thermo Fisher) was prepared by dissolving in water according to the manufacturer’s protocol. Protein was labeled by the dropwise addition of a 10–40 molar ratio of dye:protein while stirring at 4°C. The reaction was quenched after 12–16 hours by dialyzing against 20 mM Tris pH 8.0, 50 mM NaCl, and 1 mM dithiothreitol for 24 hours with buffer exchanges after 4 and 8 hours. Labeled protein sample was loaded on a 5mL Hi-Trap desalting column and peak fractions were pooled and flash frozen in liquid nitrogen. The concentration of Alexa568 dye was determined by measuring the absorbance at 575 nm and dividing this by the dye extinction coefficient of 92,009 M−1cm−1. The concentration of 568-Dip1 was determined by subtracting the absorbance at 575 nm multiplied by the correction factor of 0.403 from the absorbance at 280 nm and then dividing this number by the Dip1 extinction coefficient of 36,330 M−1cm−1.
To purify S. pombe Arp2/3 complex, 10 mL of a turbid culture of S. pombe (TP150 strain) was added to 1 L of YE5S media in a 2.8 L Fernbach flask. The cultures we grown at 30°C for ~12 hours with shaking at 180 rpm. Additional YE5S media (35 g/L) was added and the cells were grown for an additional 4 hours. Following growth, all steps were carried out at 4°C. Cultures were brought to 2 mM EDTA and 0.5 mM PMSF before harvesting the cells by centrifugation. The cell pellet was resuspended in 2 mL of lysis buffer (20mM Tris pH 8.0, 50 mM NaCl, 1 mM EDTA, 1mM DTT) per gram of wet cell pellet, plus 6 protease inhibitor tablets per liter of lysis buffer. Lysis was carried out in a microfluidizer (Microfluidics Model M-110EH-30 Microfluidizer Processor) by passing cells through the interaction chamber 5 to 6 times at 25 kPSI. The lysate was spun down at 9000 rpm for 25 minutes in a JA-10 (Beckman) rotor after adding 0.5 mM PMSF. The supernatant was decanted into prechilled 70 mL polycarbonate centrifuge tubes (Beckman Coulter #355655) and spun in a Fiberlite F37L rotor (Thermo-Scientific) at 34,000 rpm for 75 minutes. The supernatant was filtered through cheesecloth into a prechilled graduated cylinder. Ammonium sulfate was added to the supernatant (0.243 g/mL) over ~30 minutes with heavy stirring. The solution was allowed to stir for an additional 30 minutes before it was pelleted at 34,000 rpm for 90 minutes in a Fiberlite F37L rotor. The pellet was resuspended in 50 mL PKME (25 mM PIPES, 50 mM KCl, 1 mM EGTA, 3 mM MgCl2, 1 mM DTT and 0.1 mM ATP), added to 50,000 MWCO dialysis tubing and dialyzed overnight against 8 L of PKME. The dialysate was centrifuged in the Fiberlite F37L rotor for 90 minutes at 34,000 rpm. The supernatant was added to a GST-VCA affinity column. The column contained 10 mL of GS4B beads pre-equilibrated in GST-binding buffer (20 mM Tris pH 8.0, 140 mM NaCl, 1 mM EDTA, and 1 mM DTT) and then charged with 15 mg of GST-N-WASP-VCA. Prior to addition of the supernatant, GST-binding buffer was added to the column until no protein was detectable by Bradford assay and the column was then equilibrated in PKME pH 7.0. The supernatant was loaded at 1 mL per minute and washed with ~45 mL of additional PKME. The column was then washed with ~30 mL of PKME + 150 mM KCl at which point no protein was detected in the flow through by Bradford assay. Protein was eluted from the column with PKME + 1 M NaCI into approximately 2 mL fractions until no further protein was detected by Bradford assay (~30 mL). All fractions containing Arp2/3 complex were pooled, added to 50,000 MWCO dialysis tubing and dialyzed against 2 L of QA buffer (10 mM PIPES, 25 mM NaCl, 0.25 mM EGTA, 0.25 mM MgCl2, pH 6.8 w/ KOH) overnight. The complex was then loaded onto a 1 mL MonoQ column attached to an FPLC and was eluted using a linear gradient of QA buffer to 100% QB buffer (10 mM PIPES, 500 nM NaCl, 0.25 mM EGTA, 0.25 mM MgCl2, pH 6.8 w/ KOH) over 40 column volumes at a flow rate of 0.5 mL/minute. Any fractions with Arp2/3 complex were pooled, added to 50,000 MWCO dialysis tubing and dialyzed against Tris pH 8.0, 50 mM NaCl and 1 mM DTT overnight. The dialysate was concentrated to ~1.5 mL in a 30,000 MWCO concentrator tube (Sartorius Vivaspin Turbo 15 #VS15T21) in a Fiberlite F13B rotor spinning at 2,500 rpm for 5 to 10 minute cycles. Between each cycle, the solution was gently pipetted to mix. The concentrated Arp2/3 complex was loaded on a Superdex 200 size exclusion column pre-equilibrated in Tris pH 8.0, 50 mM NaCl and 1 mM DTT. The complex was eluted into 1.25 mL fractions with the same buffer at 1 mL/minute. Fractions containing Arp2/3 complex were pooled and concentrated as described above before determining the final concentration. The final concentration was determined by measuring the absorbance at 290 nm (E290 = 139,030 M−1cm−1) and then the protein was aliquoted and flash frozen.
To express GST-Wsp1-VCA, BL21(DE3)-RIL cells were transformed with a pGV67-GST-Wsp1-VCA plasmid. A single colony was picked and used to inoculate 5 mL of LB plus 100 μg/mL ampicillin and 35 μg/mL chloramphenicol which was grown overnight with shaking at 37°C. A milliliter of this overnight culture was added to 50 mL of LB plus ampicillin and chloramphenicol and grown at 37°C with shaking at 180 rpm until the culture was turbid. From this starter culture, 10 mL was used to inoculate each liter of LB plus ampicillin and chloramphenicol. The 1 L cultures were grown at 37°C to an OD600 of between 0.4 and 0.6 and induced by adding 400 μL of 1 M IPTG per liter. The cultures were grown for 12 to 14 hours at 22°C before adding 2 mM EDTA and 0.5 mM PMSF. Cultures were centrifuged at 4°C for 20 minutes at 4000 rpm in the Fiberlite F8B rotor to pellet cells. The pellet was resuspended in 100 mL of lysis buffer (20 mM Tris pH 8.0, 140 mM NaCl, 1 mM DTT, 0.5 mM PMSF) plus 2 protease inhibitor pellets per 4 L of starting culture. Lysis was carried out by sonication on ice with intermittent pulses to ensure the temperature of the lysate remained below 10°C. The lysate was then centrifuged at 4°C for 45 minutes at 18,000 rpm in a JA-20 rotor (Beckman). The supernatant was loaded onto a column containing 10 mL of GS4B beads pre-equilibrated in GST-binding buffer (20 mM Tris pH 8.0, 140 mM NaCl, 2 mM EDTA, 1 mM DTT). The column was washed with ~70 mL of GST-binding buffer and protein was eluted with 30 mL of GST-elution buffer pH 8.0 (20 mM Tris pH 8.0, 100 mM NaCl, 1 mM DTT, 50 mM reduced L-glutathione). The elution was loaded into 3500 MWCO dialysis tubing and dialyzed overnight in 2 liters of 20 mM Tris pH 8.0, 50 mM NaCl and 1 mM DTT at 4°C. The dialysate was loaded onto a Source30Q column pre-equilibrated in QA buffer (20 mM Tris pH 8.0, 100 mM NaCl, 1 mM DTT) and GST-Wsp1-VCA was eluted from the column over a 20 column volume gradient to 100% QB buffer (20 mM Tris pH 8.0, 500 mM NaCl, 1 mM DTT). All fractions containing GST-Wsp1-VCA were pooled and concentrated to ~1.5 mL in a 3500 MWCO spin concentrator tube (Sartorius Vivaspin Turbo 15 #VS15T91) in the Fiberlite F13B rotor at 2,500 rpm for 5 to 10 minute cycles at 4°C. The GST-Wsp1-VCA was loaded onto a Superdex 75 size exclusion column pre-equilibrated in 20 mM Tris pH 8.0, 150 mM NaCl and 1 mM DTT and eluted from the column at 1 mL/minute into 1.25 mL fractions. Fractions containing pure GST-Wsp1-VCA were concentrated as described above before determining the final concentration. The final concentration was determined by measuring the absorbance at 280 nm (E280 =5,500M−1cm−1) and then the protein was aliquoted and flash frozen.
Fission Yeast Strains and Molecular Biology
The Key Resources Table lists all Schizosaccharomyces pombe strains used in this study. To make S. pombe strain SpBN157–6, we crossed two strains provided by Volodia Sirotkin, VS1124a (kanMX6-Pwsp1-GFP-wsp1, fim1-mCherry-clonNAT) and VS872 (end44::ura4+). Crossing and random spore analysis were carried out as previously described with some modifications [39]. A droplet of sterile water was pipetted onto an SPA5 mating plate and sterile wooden sticks were used to mix equal amounts of each strain being crossed into the droplet. The plate was incubated at 25°C for 2 to 4 days and tetrad formation was confirmed using a microscope. Tetrads were treated in Zymolyase solution (to 5 mL of sterile water add 0.5 mL of 1 mg/mL Zymolyase (20T) in Zymolyase buffer (1 M sorbitol, 50 mM potassium phosphate pH 7.5, and 5 mM EDTA pH 8.0)). A pinhead of mated cells were added into the 15 mL conical tube containing the Zymolyase solution and vortexed briefly to mix. The tetrads were incubated at 30 °C for 12 to 16 hours on a Labquake tube rotator. The spore concentration was measured with a hemocytometer and ~500 spores were plated on YE5S plates to germinate at 25°C for 3 to 5 days. Single colonies were isolated and checked for the appropriate genotype using a combination of growth on antibiotic plates, microscopy and genomic PCR and sequencing.
Using InFusion cloning (Takara), we constructed a plasmid containing 351 bp of the genomic DIP1 5′ UTR followed by the DIP1 ORF, mNeonGreen, the ADH1 terminator, the hphMX6 resistance cassette and 610 bp of the genomic DIP1 3′ UTR. To construct S. pombe strain SpBN278–3, this region of the plasmid was linearized and transformed into S. pombe cells with an end4 deletion and Fim1-mCherry background. To make S. pombe strain SpBN280–1, the region of the plasmid described above was transformed into another strain provided by Volodia Sirotkin, VS1025–7 (kanMX6-Pwsp1-mCherry-Wsp1). Transformations were carried out as previously described with some modifications [40]. Briefly, 20 to 50 mL of S. pombe cells were grown to a density of around 1 × 107 cells/mL before pelleting by centrifugation at 1300 × g for 5 minutes. The cells were washed by resuspending in 5 mL of sterile deionized water and pelleted by centrifugation as described above. Cells were again resuspended in 1 mL of sterile deionized water and transferred to a sterile microcentrifuge tube before pelleting at 16,000 × g for 1 min. The cells were then resuspended in 100 μL of sterile TE/LiAc solution (10 mM Tris-HCl, 1 mM EDTA, and 0.1 M lithium acetate, pH 7.5) and mixed with 2 μL of 10 mg/mL salmon sperm (carrier) DNA (Invitrogen) and 2 to 5 μg of the expression plasmid. This mixture was incubated at room temperature for 10 minutes, after which 260 μL of freshly prepared 40% TE/LiAc/PEG solution (40% w/v PEG4000 in sterile TE/LiAc) was added and allowed to incubate for an additional 1 hour at 30°C. After incubation, 43 μL of DMSO was added to the solution and mixed by gentle inversion before heat shock for 5 minutes at 42°C. The cells were then pelleted by centrifugation at 6000 × g for 1 minute, resuspended in 1 mL of sterile water, pelleted again, and then resuspended in 500 μL of sterile water. Finally, 250 μL of cells were spread onto EMM(4S)-uracil plates to select for successful transformants. Transformants typically appeared within 3 days to 1 week after growth at 30°C.
Preparation of S. pombe for Imaging
S. pombe cells were grown in YE5S media at 30°C with shaking at 180 rpm. Cells were back diluted to an optical density (600 nm) of ~0.2 in EMM(5S) and maintained in the exponential phase for 24 to 48 hours before imaging. Just before imaging, cells were collected by centrifuging at 900 × g for 3 minutes, washed once with EMM5S, and finally resuspended in 20–100mL EMM5S. Cells were mounted on 0.25% gelatin pads containing 10 mM propyl gallate and imaged within 30 minutes.
Confocal Microscopy Imaging of Fission Yeast
S. pombe cells on gelatin pads were imaged on a Nikon TE2000-U inverted microscope equipped with a 100×/1.49 numerical aperture TIRF objective, a 1× - 1.5 × intermediate magnification module, a Coherent OBIS 488 nm LS 60 mW laser, a Coherent Sapphire 50 mW 561 nm continuous wave solid state laser, an acousto-optic tunable filter (AOTF), a filter wheel (Applied Scientific Instrumentation) containing ET525/50, ET605/52 and ZET488/561m-TRF 25 mm Dia Mounted filters and a Yokogawa spinning disk scan head (CSU10). Images were collected on an EMCCD camera (iXon Ultra-897, Andor). For imaging of the mCherry-Wsp1 and Fim1-mCherry signal, images were typically taken using 200 to 300 ms exposures of 15 to 20 mW 561 nm laser at 1 second intervals with the ET605/52 filter. Dip1-mNeonGreen and GFP-Wsp1 were typically imaged using 300 to 400 ms exposures of 25 mW 488 nm laser at 1 second intervals with the ET525/50 filter. All images were collected at a single focal plane taken at the center of the cell.
Analysis of S. pombe Confocal Microscopy Images
Images were processed and analyzed using the FIJI distribution of Image J [41]. For spinning disk confocal images, the brightness and contrast were adjusted and then images were cropped. Videos were made using the Multi Stack Montage (BIOP) FIJI plugin and arrows were added with the Image, Stack and Timelapse Arrow Labelling Tool for ImageJ. Montages were made using the Make Montage plugin. Kymographs were created by drawing a single pixel wide line through an endocytic patch from the outside of the cell into the cytoplasm and then using the Reslice tool.
To determine if Dip1 has a different localization pattern in endocytic patches than Wsp1, the relative position of their peak intensities was measured. Any patch that was present in the cell between 20 and 40 seconds after the start of the video was measured. A straight line was drawn through the center of each measured patch from outside of the cell towards the cell center following the direction of the internalizing patch. The pixel intensity along this line was measured using the Plot Profile tool to determine the intensity of both mCherry-Wsp1 and Dip1-mNeonGreen at each position along the line. The distance between the peak signals of Wsp1 and Dip1 were measured when the average Wsp1 signal across the line was at its maximum. A total of 56 patches across 8 cells were measured. To determine if the position of Dip1 and Wsp1 in these patches was statistically different, a one-sample t test with a hypothetical mean of 0 was used. The result showed a p-value of < 0.0001 with a 95% confidence interval of 0.076 to 0.153 and a t-value of 5.98.
In Figure 3, panel F, the Dip1-mNeonGreen and mCherry-Wsp1 signals across 15 patches were normalized, averaged and plotted. The average pixel intensities were normalized to a range of 0 to 1 by first subtracting the lowest value across the line from each measurement and then dividing by the peak intensity. The 15 patches were selected proportionally from each bin of the histogram in panel G. The peak signal of Wsp1 was set to 0.21 μm and the signal of Dip1 was plotted based on its relative position to Wsp1 in that endocytic patch.
To determine the position of the peak signal of Dip1-mNeonGreen or GFP-Wsp1 in actin comet tails the ImageJ macro, StackProfileData (Michael Schmid), was used to plot the intensity of each pixel along a single pixel wide line through the center of the comet tail in each frame of the video. The peak signals of Dip1 and Wsp1 were analyzed from the initial appearance of Fim1-mCherry until the actin comet tail released from the membrane. The highest intensity pixel along the line in each frame was determined using the profile plot. If a higher intensity pixel was present within 2 pixels of either side of the profile line, this value was used as the peak intensity pixel. The position of the cortex was set by the location of the Dip1 or Wsp1 peak signal in the first analyzed frame for that comet tail. If the position of the peak signal was within 2 pixels ( ~0.32 μm) of the cortex, it was considered to be cortically localized.
Total Internal Reflection Fluorescence (TIRF) Microscopy Imaging of Actin Polymerization
TIRF flow chambers were constructed and reactions setup as previously described with slight modifications [42]. The coverslips were cleaned by sonicating for 25 minutes in acetone, rinsed with water, and sonicated for 25 minutes in 1 M KOH in Coplin jars. The coverslips were then rinsed twice with methanol before being aminosilanized by incubating in a 1% APTES (Sigma) and 5% acetic acid in methanol solution for l0 minutes, sonicating for 5 minutes, and then incubating for an additional 15 minutes at room temperature. Coverslips were then rinsed twice with methanol followed by deionized water and left to air dry. TIRF chambers were created by sandwiching double-sided tape (Scotch) between a glass microscope slide and a clean and dry coverslip (24 × 60 #1.5) to create a ~14 μL, 0.5 cm wide chamber. TIRF chambers were passivated by incubating for 4–5 hours in 300 mg/mL methoxy PEG succinimidyl succinate, MW5000 (JenKem) containing l-3% biotin-PEG NHS ester, MW5000 (JenKem) dissolved in 0.l M NaHCO3 pH 8.3. After incubation, 0.l M NaHCO3 pH 8.3 was flowed into chambers to wash away excess PEG. TIRF chambers were stored in deionized water at 4°C for no more than l week. Just before use, TIRF chambers were incubated for 8 minutes with l μM NeutrAvidin (ThermoFisher) followed by l00 nM biotin inactivated myosin (Cytoskeleton, Inc.), both prepared in high-salt (HS) TBS (50 mM Tris pH 7.5, 600 mM NaCl). Chambers were then washed 2 times with 20 mg/mL BSA in HS-TBS followed by 2 washes with 20 mg/mL BSA in low-salt (LS) TBS (50 mM Tris pH 7.5, l50 mM NaCl). Finally, TIRF chambers were pre-incubated with TIRF buffer (10 mM Imidazole pH 7.0, 1 mM MgCl2, l mM EGTA, 50 mM KCl, l00 mM DTT, 0.2 mM ATP, 25 mM Glucose, 0.5% Methylcellulose (400 cP at 2%), 0.02 mg/mL Catalase (Sigma) and 0.l mg/mL Glucose Oxidase (MP Biomedicals).
TIRF Microscopy Actin Polymerization Reactions
To initiate the reaction, 1 μL of 2.5 mM MgCl2 and 10 mM EGTA was mixed with 5 μL of 9 μM 33% Oregon Green-labeled actin and incubated for 2 min before adding 4 μL of this actin solution to 16 μL of 1.25 × TIRF buffer and any other proteins. Reactions were imaged on a Nikon TE2000-E inverted microscope equipped with a 100× 1.49 NA TIRF objective, 50 mW 488 nm and 561 nm Sapphire continuous wave solid state lase lines (Coherent), an acousto-optic tunable filter (AOTF), a dual band TIRF (zt488/561rpc) filter cube (Chroma C143315), a filter wheel (Finger Lakes Instrumentation) containing ET525/50 and ET605/52 filters, and a 1× to 1.5× intermediate magnification module. Images were taken using a 512×512 pixel EM-CCD camera (iXon3, Andor). For single color reactions, 50 ms exposures with the 488 nm laser at 10 mW were taken at 1 second intervals, and a typical polymerization reaction was imaged for 10–15 minutes. For two color reactions, typical imaging conditions were 50 ms exposures with the 488 nm laser at 10 mW and 50 ms exposures with the 561 nm laser at 35 mW with 200 ms intervals. For low exposure imaging conditions, the exposure times and laser powers remained the same; however, a 5 second interval between frames was used to decrease the overall exposure time of Alexa568-Dip1. The concentration of Alexa568-Dip1 was kept at 6 nM to minimize the background signal in the 561 channel from non-specifically adsorbed Alexa568-Dip1.
TIRF Microscopy Image Analysis
Images were prepared using the FIJI distribution of Image J [41]. For actin polymerization experiments, the backgrounds of image sequences were subtracted using a 10-pixel rolling ball radius. The total actin polymer length, as shown in Figure 1E, was calculated using a custom image processing script run in Matlab (Mathworks), described as follows. For each frame, pixels corresponding to filament fluorescence were identified using image segmentation followed by morphological area opening to remove non-filament small fluorescent objects. The final pixel number value was converted to micrometers (1px = 106.7 nm) to yield the total length of actin filaments in the image frame. To calculate the reported branch densities (Figures 1B,D), the number of branches were counted manually and divided by the length of the seed filament when the total polymer length in each video was approximately 1500 μm. The branching rate in Figure 1F was calculated as the number of primary branches divided by one half the seed filament length multiplied by the lifetime of the seed. The lifetime of the seed filament was calculated as the length of the seed multiplied by 370 actin monomers/μm divided by the barbed end elongation rate of 11.8 actin monomers/second [43]. To determine the branch to linear filament ratio, in each region of interest, the total number of branches was divided by the total number of seeds. To determine how long Dip1 stayed bound to Arp2/3 complex after nucleation, we measured the average length of Dip1 binding events from both filaments that grew from a Dip1 puncta already on the surface and also from filaments that landed on the surface after nucleation, extrapolating the birth time of the Dip1-Arp2/3 nucleated filaments in the latter based on the filament length and elongation rate of the filaments.
QUANTIFICATION AND STATISTICAL ANALYSIS
The numbers of replicates and description of error bars can be found in the figure legends. To determine if there were significant differences in the data in Figures 1B, D and F, a one-way analysis of variance (ANOVA) with a Tukey’s Multiple Comparison test was used. Data points with significant differences are indicated on each plot with the respective p-values. All other data points were not significantly different from one another. For Supplemental Figures, two-tailed t-tests assuming unequal variances were used to determine if the average values were significantly different. The respective p-values are reported in the figure panels above data points that were determined to be statistically different.
| P-value | 0.002 | |||
| Number of Groups | 5 | |||
| F | 6.52 | |||
| R-squared | 0.58 | |||
| Tukey’s Multiple Comparison Test | Mean Difference | q | Significance | 95% CI of difference |
| 0 nM vs 3.75 nM Dip1 | 0.049 | 1.48 | ns | −0.092 to 0.19 |
| 0 nM vs 7.5 nM Dip1 | 0.031 | 1.03 | ns | −0.097 to 0.16 |
| 0 nM vs 15 nM Dip1 | 0.12 | 3.86 | ns | −0.012 to 0.25 |
| 0 nM vs 75 nM Dip1 | 0.20 | 6.19 | ** | 0.064 to 0.35 |
| 3.75 nM vs 7.5 nM Dip1 | −0.018 | 0.59 | ns | −0.15 to 0.11 |
| 3.75 nM vs 15 nM Dip1 | 0.068 | 2.24 | ns | −0.061 to 0.20 |
| 3.75 nM vs 75 nM Dip1 | 0.16 | 4.71 | * | 0.015 to 0.30 |
| 7.5 nM vs 15 nM Dip1 | 0.086 | 3.17 | ns | −0.029 to 0.20 |
| 7.5 nM vs 75 nM Dip1 | 0.17 | 5.75 | ** | 0.045 to 0.30 |
| 15 nM vs 75 nM Dip1 | 0.088 | 2.92 | ns | −0.040 to 0.22 |
One-way ANOVA and Tukey’s Multiple Comparison test results for the data in Figure 1 panel B. For significance, “ns”, “*” and “**” corresponds to a p-values of > 0.05, ≤ 0.05 and ≤ 0.01, respectively.
| P-value | 0.719 | |||
| Number of Groups | 3 | |||
| F | 0.34 | |||
| R-squared | 0.06 | |||
| Tukey’s Multiple Comparison Test | Mean Difference | q | Significance | 95% CI of difference |
| 150 nM vs 300 nM Wsp1 | 0.015 | 0.60 | ns | −0.081 to 0.11 |
| 150 nM vs 600 nM Wsp1 | 0.029 | 1.16 | ns | −0.067 to 0.12 |
| 300 nM vs 600 nM Wsp1 | 0.014 | 0.50 | ns | −0.091 to 0.12 |
One-way ANOVA and Tukey’s Multiple Comparison test results for the data in Figure 1 panel D.
| P-value | 0.851 | |||
| Number of Groups | 5 | |||
| F | 0.33 | |||
| R-squared | 0.07 | |||
| Tukey’s Multiple Comparison Test | Mean Difference | q | Significance | 95% CI of difference |
| 0 nM vs 3.75 nM Dip1 | 0.00043 | 0.46 | ns | −0.0036 to 0.0044 |
| 0 nM vs 7.5 nM Dip1 | 0.00025 | 0.29 | ns | −0.0034 to 0.0039 |
| 0 nM vs 15 nM Dip1 | −0.0000077 | 0.01 | ns | −0.0037 to 0.0036 |
| 0 nM vs 75 nM Dip1 | 0.0013 | 1.35 | ns | −0.0027 to 0.0053 |
| 3.75 nM vs 7.5 nM Dip1 | −0.00018 | 0.21 | ns | −0.0038 to 0.0035 |
| 3.75 nM vs 15 nM Dip1 | −0.00044 | 0.51 | ns | −0.0041 to 0.0032 |
| 3.75 nM vs 75 nM Dip1 | 0.00084 | 0.89 | ns | −0.0032 to 0.0048 |
| 7.5 nM vs 15 nM Dip1 | −0.00026 | 0.34 | ns | −0.0035 to 0.0030 |
| 7.5 nM vs 75 nM Dip1 | 0.0010 | 1.19 | ns | −0.0026 to 0.0047 |
| 15 nM vs 75 nM Dip1 | 0.0013 | 1.49 | ns | −0.0024 to 0.0049 |
One-way ANOVA and Tukey’s Multiple Comparison test results for the data in Figure 1 panel F.
DATA AND CODE AVAILABILITY
This study did not generate any datasets or code.
Supplementary Material
Video S1. TIRF microscopy video taken under low exposure conditions showing signal for Alexa568-Dip1 (magenta) bound to the end of an elongating Oregon Green-labeled actin filament (green) for 8 minutes. Related to Figure 2. Conditions: 1.5 μM 33% Oregon Green-labeled actin in the presence of 6 nM Alexa568-Dip1, 250 nM GST-Wsp1-VCA and 500 nM SpArp2/3 complex collected with 50 ms exposure times at 5 second intervals.
Video S2. Spinning disk microscopy video showing end4Δ S. pombe cells expressing Dip1-mNeonGreen (green) and Fim1-mCherry (magenta). Related to Figure 3. White arrows show examples of Dip1 treadmilling with the Fim1-marked actin network. Scale bar: 2 μm.
Video S3. Spinning disk microscopy video showing wildtype S. pombe cells expressing Dip1-mNeonGreen (green) and mCherry-Wsp1 (magenta). Related to Figure 3. White arrows show an example of an endocytic site at which Dip1 is further into the cytoplasm than Wsp1. Scale bar: 1 μm.
Video S4. Spinning disk microscopy video showing end4Δ S. pombe cells expressing GFP-Wsp1 (green) and Fim1-mCherry (magenta). Related to Figure 3. White arrows show an example of Wsp1 signal remaining predominately at the cortex with only a small portion treadmilling with the Fim1-marked actin network. Scale bar: 2 μm.
Highlights.
Dip1 stimulates assembly of disconnected linear actin filament networks
Dip1 is a single turnover activator of Arp2/3 complex
Dip1 incorporates into actin filament networks at endocytic sites
Single turnover activation of nucleation by Dip1 may limit linear filament creation
Acknowledgements
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01 GM092917 and R01 GM127440 (B.J.N), T32 GM007759 (to C.J.B, A.R.W and L.A.H.) and by the American Heart Association, grants #18PRE33960110 (C.J.B) and #15PRE25900011 (A.R.W). We thank Vladamir Sirotkin for sharing S. pombe strains and for advice on strain construction.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mooren OL, Galletta BJ, and Cooper JA (2012). Roles for Actin Assembly in Endocytosis. Annual Review of Biochemistry 81, 661–686. [DOI] [PubMed] [Google Scholar]
- 2.Yi K, Unruh JR, Deng M, Slaughter BD, Rubinstein B, and Li R (2011). Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat. Cell Biol 13, 1252–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P-S, Chou F-S, Ramachandran S, Xia S, Chen H-Y, Guo F, Suraneni P, Maher BJ, and Li R (2016). Crucial roles of the Arp2/3 complex during mammalian corticogenesis. Development 143, 2741–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, and Bear JE (2012). Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148, 973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achard V, Martiel J-L, Michelot A, Guérin C, Reymann A-C, Blanchoin L, and Boujemaa-Paterski R (2010). A “Primer”-Based Mechanism Underlies Branched Actin Filament Network Formation and Motility. Current Biology 20, 423–428. [DOI] [PubMed] [Google Scholar]
- 6.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, and Pollard TD (1999). Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A 96, 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner AR, Luan Q, Liu S-L, and Nolen BJ (2013). WISH/DIP/SPIN90 proteins form a class of Arp2/3 complex activators that function without preformed actin filaments. Curr Biol 23, 1990–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balzer CJ, Wagner AR, Helgeson LA, and Nolen BJ (2018). Dip1 Co-opts Features of Branching Nucleation to Create Linear Actin Filaments that Activate WASP-Bound Arp2/3 Complex. Current Biology 28, 3886–3891.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollard TD, and Borisy GG (2003). Cellular Motility Driven by Assembly and Disassembly of Actin Filaments. Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- 10.Berro J, Michelot A, Blanchoin L, Kovar DR, and Martiel J-L (2007). Attachment Conditions Control Actin Filament Buckling and the Production of Forces. Biophys J 92, 2546–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sirotkin V, Beltzner CC, Marchand J-B, and Pollard TD (2005). Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. The Journal of Cell Biology 170, 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu R, and Chang F (2011). Characterization of Dip1p reveals a switch in Arp2/3-dependent actin assembly for fission yeast endocytosis. Curr Biol 21, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodnick-Smith M, Luan Q, Liu S-L, and Nolen BJ (2016). Role and structural mechanism of WASP-triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. PNAS 113, E3834–E3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padrick SB, Doolittle LK, Brautigam CA, King DS, and Rosen MK (2011). Arp2/3 complex is bound and activated by two WASP proteins. PNAS 108, E472–E479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ti S-C, Jurgenson CT, Nolen BJ, and Pollard TD (2011). Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci U S A 108, E463–E471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luan Q, Liu S-L, Helgeson LA, and Nolen BJ (2018). Structure of the nucleation-promoting factor SPIN90 bound to the actin filament nucleator Arp2/3 complex. The EMBO Journal 37, e100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luan Q, Zelter A, MacCoss MJ, Davis TN, and Nolen BJ (2018). Identification of Wiskott-Aldrich syndrome protein (WASP) binding sites on the branched actin filament nucleator Arp2/3 complex. PNAS 115, E1409–E1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boczkowska M, Rebowski G, Kast DJ, and Dominguez R (2014). Structural analysis of the transitional state of Arp2/3 complex activation by two actin-bound WCAs. Nat Commun 5, 3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurgenson CT, and Pollard TD (2015). Crystals of the Arp2/3 complex in two new space groups with structural information about actin-related protein 2 and potential WASP binding sites. Acta Crystallogr F Struct Biol Commun 71, 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodnick-Smith M, Liu S-L, Balzer CJ, Luan Q, and Nolen BJ (2016). Identification of an ATP-controlled allosteric switch that controls actin filament nucleation by Arp2/3 complex. Nat Commun 7, 12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin AC, Welch MD, and Drubin DG (2006). Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nature Cell Biology 8, 826–833. [DOI] [PubMed] [Google Scholar]
- 22.Smith BA, Padrick SB, Doolittle LK, Daugherty-Clarke K, Corrêa IR Jr, Xu M-Q, Goode BL, Rosen MK, and Gelles J (2013). Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife 2, e01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akin O, and Mullins RD (2008). Capping Protein Increases the Rate of Actin-based Motility by Promoting Filament Nucleation by the Arp2/3 Complex. Cell 133, 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millius A, Watanabe N, and Weiner OD (2012). Diffusion, capture and recycling of SCAR/WAVE and Arp2/3 complexes observed in cells by single-molecule imaging. J Cell Sci 125, 1165–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwasa JH, and Mullins RD (2007). Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol 17, 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith BA, Daugherty-Clarke K, Goode BL, and Gelles J (2013). Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc. Natl. Acad. Sci. U.S.A 110, 1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullins RD, Heuser JA, and Pollard TD (1998). The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U.S.A 95, 6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Idrissi F-Z, Blasco A, Espinal A, and Geli MI (2012). Ultrastructural dynamics of proteins involved in endocytic budding. Proc. Natl. Acad. Sci. U.S.A 109, E2587–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arasada R, Sayyad WA, Berro J, and Pollard TD (2018). High-speed superresolution imaging of the proteins in fission yeast clathrin-mediated endocytic actin patches. Mol. Biol. Cell 29, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaksonen M, Sun Y, and Drubin DG (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475–487. [DOI] [PubMed] [Google Scholar]
- 31.Sirotkin V, Berro J, Macmillan K, Zhao L, and Pollard TD (2010). Quantitative Analysis of the Mechanism of Endocytic Actin Patch Assembly and Disassembly in Fission Yeast. Mol Biol Cell 21, 2894–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reymann A-C, Martiel J-L, Cambier T, Blanchoin L, Boujemaa-Paterski R, and Théry M (2010). Nucleation geometry governs ordered actin networks structures. Nature Materials 9, 827–832. [DOI] [PubMed] [Google Scholar]
- 33.Bieling P, Hansen SD, Akin O, Li T-D, Hayden CC, Fletcher DA, and Mullins RD (2018). WH2 and proline-rich domains of WASP-family proteins collaborate to accelerate actin filament elongation. EMBO J. 37, 102–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaksonen M, Toret CP, and Drubin DG (2006). Harnessing actin dynamics for clathrin-mediated endocytosis. Nature Reviews Molecular Cell Biology 7, 404–414. [DOI] [PubMed] [Google Scholar]
- 35.Lacy MM, Ma R, Ravindra NG, and Berro J (2018). Molecular mechanisms of force production in clathrin-mediated endocytosis. FEBS Lett. 592, 3586–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mund M, van der Beek JA, Deschamps J, Dmitrieff S, Hoess P, Monster JL, Picco A, Nédélec F, Kaksonen M, and Ries J (2018). Systematic Nanoscale Analysis of Endocytosis Links Efficient Vesicle Formation to Patterned Actin Nucleation. Cell 174, 884–896.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picco A, Mund M, Ries J, Nédélec F, and Kaksonen M (2015). Visualizing the functional architecture of the endocytic machinery. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campellone KG, and Welch MD (2010). A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol 11, 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forsburg SL, and Rhind N (2006). Basic methods for fission yeast. Yeast 23, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagan Iain, Carr Antony M., and Nurse Paul (2016). Fission Yeast: A Laboratory Manual (Cold Spring Harbor Laboratory Press; ) Available at: https://cshlpress.com/default.tpl?cart=1558030115142488353&fromlink=T&linkaction=full&linksortby=oop_title&--eqSKUdatarq=1106 [Accessed May 16, 2019]. [Google Scholar]
- 41.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhn JR, and Pollard TD (2005). Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J 88, 1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollard TD (1986). Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol 103, 2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nolen BJ, and Pollard TD (2008). Structure and biochemical properties of fission yeast Arp2/3 complex lacking the Arp2 subunit. J. Biol. Chem 283, 26490–26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S-L, May JR, Helgeson LA, and Nolen BJ (2013). Insertions within the actin core of actin-related protein 3 (Arp3) modulate branching nucleation by Arp2/3 complex. J. Biol. Chem 288, 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, and Waugh DS (2001). Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. TIRF microscopy video taken under low exposure conditions showing signal for Alexa568-Dip1 (magenta) bound to the end of an elongating Oregon Green-labeled actin filament (green) for 8 minutes. Related to Figure 2. Conditions: 1.5 μM 33% Oregon Green-labeled actin in the presence of 6 nM Alexa568-Dip1, 250 nM GST-Wsp1-VCA and 500 nM SpArp2/3 complex collected with 50 ms exposure times at 5 second intervals.
Video S2. Spinning disk microscopy video showing end4Δ S. pombe cells expressing Dip1-mNeonGreen (green) and Fim1-mCherry (magenta). Related to Figure 3. White arrows show examples of Dip1 treadmilling with the Fim1-marked actin network. Scale bar: 2 μm.
Video S3. Spinning disk microscopy video showing wildtype S. pombe cells expressing Dip1-mNeonGreen (green) and mCherry-Wsp1 (magenta). Related to Figure 3. White arrows show an example of an endocytic site at which Dip1 is further into the cytoplasm than Wsp1. Scale bar: 1 μm.
Video S4. Spinning disk microscopy video showing end4Δ S. pombe cells expressing GFP-Wsp1 (green) and Fim1-mCherry (magenta). Related to Figure 3. White arrows show an example of Wsp1 signal remaining predominately at the cortex with only a small portion treadmilling with the Fim1-marked actin network. Scale bar: 2 μm.
Data Availability Statement
This study did not generate any datasets or code.