Abstract
Objective
Discovering subphenotypes of complex diseases can help characterize disease cohorts for investigative studies aimed at developing better diagnoses and treatments. Recent advances in unsupervised machine learning on electronic health record (EHR) data have enabled researchers to discover phenotypes without input from domain experts. However, most existing studies have ignored time and modeled diseases as discrete events. Uncovering the evolution of phenotypes – how they emerge, evolve and contribute to health outcomes – is essential to define more precise phenotypes and refine the understanding of disease progression. Our objective was to assess the benefits of an unsupervised approach that incorporates time to model diseases as dynamic processes in phenotype discovery.
Methods
In this study, we applied a constrained non-negative tensor-factorization approach to characterize the complexity of cardiovascular disease (CVD) patient cohort based on longitudinal EHR data. Through tensor-factorization, we identified a set of phenotypic topics (i.e., subphenotypes) that these patients established over the 10 years prior to the diagnosis of CVD, and showed the progress pattern. For each identified subphenotype, we examined its association with the risk for adverse cardiovascular outcomes estimated by the American College of Cardiology/American Heart Association Pooled Cohort Risk Equations, a conventional CVD-risk assessment tool frequently used in clinical practice. Furthermore, we compared the subsequent myocardial infarction (MI) rates among the six most prevalent subphenotypes using survival analysis.
Results
From a cohort of 12,380 adult CVD individuals with 1,068 unique PheCodes, we successfully identified 14 subphenotypes. Through the association analysis with estimated CVD risk for each subtype, we found some phenotypic topics such as Vitamin D deficiency and depression, Urinary infections cannot be explained by the conventional risk factors. Through a survival analysis, we found markedly different risks of subsequent MI following the diagnosis of CVD among the six most prevalent topics (p < 0.0001), indicating these topics may capture clinically meaningful subphenotypes of CVD.
Conclusion
This study demonstrates the potential benefits of using tensor-decomposition to model diseases as dynamic processes from longitudinal EHR data. Our results suggest that this data-driven approach may potentially help researchers identify complex and chronic disease subphenotypes in precision medicine research.
Keywords: deep phenotyping, computational phenotyping, tensor decomposition
Graphical Abstract
Introduction
Chronic diseases such as diabetes, Alzheimer’s disease, and cardiovascular disease (CVD) affect more than 50% of the population, drive up to 90% of healthcare spending in the United States, and are the leading causes of mortality and disability globally.1–5 Chronic diseases commonly have complex causal mechanisms involving the interplay between genetic, environmental, and lifestyle factors.6 Chronic diseases may present with numerous signs and symptoms involving multiple physiologic systems and typically co-occur with other conditions.4 For example, CVD patients often present with multiple comorbidities including hyperlipidemia, diabetes, and hypertension.7 However, known risk factors (e.g. Framingham study7) for CVD may only explain ~75% of major CVD events.8 A more precise characterization of complex chronic disease based on different patterns of comorbidities may help identify subpopulations to facilitate prevention, early detection and precise treatment.
Deep phenotyping is a method for identifying disease cohorts that incorporates multiple categories of EHR data. 9 Traditional phenotyping algorithms typically require feature engineering and multiple cycles of review from experts to specify inclusion and exclusion criteria on a data set. This does not scale well to large patient cohorts. 10–12 To address this problem, researchers have developed data-driven approaches to facilitate automated phenotyping. Using a training set including labeled cases and controls (i.e. gold standard), some methods use feature selection techniques 11 or machine learning classifiers 11,13 to build a phenotyping model.14 Other methods start with a set of core concepts and use natural language processing to automatically find the occurrence of matched terms in clinical notes, which can be used as input features to build a classifier to predict the target phenotype.15,16 Most approaches achieve success on a broad range of algorithms through a collaboration among informaticians and clinical experts 17 However, such supervised methods still need to specify an outcome variable (e.g. a target phenotype) and labeled dataset (e.g. cases or controls). Due to the complexity of the contributing factors of a complex disease and their intricate interactions, such supervised approaches are unsuited to the scenario in which we have limited knowledge to label the phenotypes, and we wish to discover them from the data.18
Recent advances in unsupervised computational phenotyping have allowed researchers to discover phenotypes from EHR with minimal human guidance.1,16,18,19 Rather than relying on the pre-defined outcomes, these unsupervised approaches can identify patterns in the entire data source.18 One such method is topic modeling approach including non-negative matrix factorization (NMF) and Latent Dirichlet Allocation (LDA).19–21 Researchers have applied these approaches to EHR to identify phenotype candidates –clusters of patients that met with clinical conditions– with no pre-defined phenotype definitions. 11,20–24 For example, we have previously used NMF to find disease topics (described by clusters of co-occurring symptoms) and study their associations with genetic variants.25 Aside from replicating known signals, we also reported a new correlation between a lipoprotein(a) (LPA) variant (rs10455872) and a topic enriched for lung cancer which was not previously identified via phenome-wide scanning. 25
Nevertheless, these studies typically overlook the valuable information conveyed by a temporal pattern of phenotypes input. The comorbidities and effects of complex chronic diseases like CVD typically evolve and progress over a long time. Thus, incorporating a time effect into phenotyping may help yield more precise phenotypes. Current unsupervised techniques such as LDA and NMF have difficulty capturing the temporal changes of topics26. Others have proposed training separate topic models for each time slice, but accurately measuring the connections of topics from different time slices remains a challenge.27–29
In this paper, we applied a constrained non-negative tensor-factorization approach to extract phenotypic topics across time scales. This approach models an individual’s medical data as a third-order tensor (i.e., a three-way data array, a cube instead of the matrix) with each order representing phenotypes, time and individuals respectively. By fitting a tensor-factorization model, we can identify a set of topics (i.e., subphenotypes), describe their progress, and obtain patients’ representations in such topics at the same time. Identifying subphenotypes enables us to generate testable hypotheses of how complex diseases differ across patients, such as in temporal disease progression. Furthermore, it may provide insight into differences in the temporal progression of the disease. To demonstrate the feasibility and utility of the approach, we apply this method to EHR data of a large cohort of CVD patients. We identify CVD topics that represent clinically meaningful subphenotypes, some of which are not captured by traditional clinical assessment tools for CVD risk. This approach may improve understanding of disease progression over time, and identify novel subphenotypes that may be used for enhancing precision treatment.
Background and related work
Tensor decomposition (or factorization) can be treated as an extended NMF on high-dimensional data.
Non-negative Matrix Factorization (NMF) for Topic Modeling
NMF assumes that given an n· m sparse data matrix X with n samples (e.g. documents) and m dimensions can be approximated by a small set of k basis vectors – an inner-product of two non-negative matrices W and H such that
| (1) |
is achieved subject to W ≥ 0, H ≥ 0 where || · ||2F is the Frobenius norm to measure the error between the original data and the approximation. In the context of topic modeling, H is a k × m (k ≤ m) topic – word matrix, indicating m words’ weights corresponding to k topics, and W is an n × k document-topic factor matrix, indicating n documents’ relevance score to k topics. From H, we can order each word’s weights in a particular topic and use the top words to describe topics. Furthermore, W can be used to represent documents for regression and clustering tasks. However, NMF fails to capture the temporal changes of topics.
Tensor decomposition
Tensor decomposition (or factorization), analogous to NMF, can learn latent factors from high dimensional data. After being introduced in 1927 30, tensor factorization regained popularity in the computer science community in the late 20th century 31,32, and until recently, proliferated into many other fields 33–35. Sun et al. have used PARAFAC2 36 on automated phenotyping for medically complex children who need intensive medical care due to multisystem dysfunction, technology dependence, or complex medication needs 37. Their extract disease topics, however, may not be independent because they did not impose any orthogonality restraints. Bahargam et al. have utilized tensor factorization in text mining to find dynamic topics from forums. 38 They introduced a method that can add orthogonality or sparsity constraints on each mode. In this study, we used this method to discover the phenotypic topic in longitudinal EHR.
Materials and Method
Data preparation
Study cohort
We used the data from Synthetic Derivative, a database that contains a de-identified copy of the EHR for every patient in the Vanderbilt University Medical Center (VUMC) system (>3 million patients). Our cohort included individuals: 1) phenotype codes (PheCodes) 39 of 411.* or 433.*, i.e., International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) 346.6*(Persistent migraine aura with cerebral infarction), 410–414 (Ischemic Heart Disease), 433–438 (Cerebrovascular Disease, excluded hemorrhage), 996.03 (Mechanical complication due to coronary bypass graft ), V12.54 (Personal history of transient ischemic attack [TIA], V45.81(Aortocoronary bypass status), V45.82 (Percutaneous transluminal coronary angioplasty status), and 2) had ≥ 10 years of EHR data prior to the first diagnosis. For each individual, we defined the baseline date (time 0) as 3 months before their first CVD event date. We extracted the diagnosis codes within the 10-year observation window before their baseline date.
Preprocessing data
From each individual’s records, all distinct ICD-9-CM billing codes within the observation window are captured and grouped into 1821 distinct PheCodes 39. This mapping scheme was developed and validated manually by our group to facilitate high-throughtput phenotypic analysis. 40 The phenotype codes have been used in several systematic analyses to successfully replicate previously known gene-disease associations and discover potentially novel pleiotropic associations. 39
We filtered out rare PheCodes (i.e. prevalence <=0.5%) to avoid overfitting and achieved 1068 unique PheCodes. We divided the 10-year observation period into one-year slice windows. For each PheCode, we used a binary value (i.e. 1 or 0) to indicate whether or not a patient had a particular diagnosis each year.
Constructing a tensor from the dataset
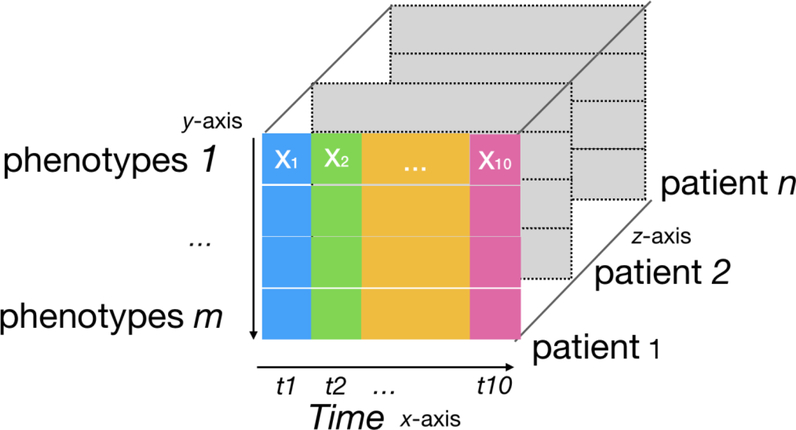
We introduce the following notation for the construction of the data tensors. First, for each patient i ∈ {1,...,n} we constructed a matrix, Xi, of dimensions m × t where m denotes the size of PheCodes, and t is the number of time windows; this matrix X can be treated as a vertical slice of the full tensor T. Then we concatenated each matrix Xi to construct a tensor T of three modes (i.e. axes) corresponding to phenotypes, time windows, and patients. Figure 1. illustrates this construction graphically. Each tensor cell contains a binary value (1 or 0) indicating whether each patient i-th has the PheCode m in t-th window. By slicing the T among different axis, we achieved three modes indicating different views of the data:
Figure 1.
Input data representation for tensor factorization. Each slice along the z-axis is a patient record. In such a slice, each row represents a phenotype (PheCode), and each colored column represents a one-year time interval.
Phenotype mode (x-z axis): a matrix of a phenotype assigned to all patients across time windows
Time mode (y-z axis): a matrix of phenotypes from all patients in the specific time window
Patient mode (x-y axis): a matrix of a patient’s phenotypes across all time windows
Detecting evolving-topics via tensor factorization
Problem formulation
Now that we have described the process of structuring our data in terms of three-dimensional tensors, we describe how to detect time-evolving topics. Given a tensor T, we factorize T into:
| (2) |
where af, bf, and cf correspond to the f-th columns of three-factor matrices: A, B, and C. Specifically, the interpretation for A, B, and C is:
The factor matrix A corresponds a phenotype – topic matrix of size m × k, where each row denotes phenotypes, and each column represents a topic. Each element aif represent a weight of i-th phenotype on the f-th topic.
The factor matrix B corresponds to a time – topic matrix of size t × k, where each row represents a time window and each column represents a topic. The element bjf represents the f-th topic’s weights or strength in j-th time window.
The factor matrix C is a patient- topic matrix of size n × k, where each row indicates a patient, and each column represents a topic. The element czf represents the z-th patient’s weight on f-th topic.
We normalize each column in the factor matrix (making the length of the column to 1).
The e indicates the error between the original input data T and approximation of the outer product of three factors. We can find A, B, and C that best approximate the input tensor by minimizing the e. Thus, equation (2) can be written as an optimization problem:
| (3) |
Each factor column af, bf, and cf corresponds to a disease topic (component) represented in an object in each mode.
To yield clinically interpretable results, we imposed the following constraints:
Non-negativity: For better interpretability, we restrict the factor matrices A, B, and C to be non-negative: A ≥ 0, B ≥ 0, C ≥ 0.
Orthogonality: To generate distinct (less overlapped) and independent topics, we imposed orthogonality constraints on the factor matrix (i.e., columns in a factor matrix should be close to orthogonal):
We assumed disease topics to be distinct (less overlapped) and independent. Thus, we imposed strict orthogonality on factor matrix a (i.e., word-topic model) with ∈A 0.05 (columns in a factor matrix should be close to orthogonal). We relaxed the constraints on B (time-topic) and C (patient-topic) factor matrices by setting ∈B = 1 and ∈c= 0.8, because certain topics may have similar time trend, and patients may be inherently similar to others.
Sparsity: Comparing to the large size of phenotypes, comorbidity or disease may present only a few of them. Similarly, a patient may only have a few comorbidities. Thus, we imposed L1-norm41 on each column in the factor matrix to fortify the sparsity:
To yield interpretable topics, we used sparsity constraints in this study γ = [0.8,1,1]. Small γ would yield more sparse weights (i.e. a lot of zero weights). We expected PheCode weights in the topic to be sparse for better interpretation. We did not limit sparsity on time and patient factor.
As the equation (2) is not a convex function, we utilized a PARAFAC decomposition with alternating least squares (ALS) to solve it 42,43. PARAFAC is one of several decomposition methods for multi-way data. Compared with other competitors, such as Tucker3 and unfolding of the multi-way array to a matrix and then performing standard two-way PCA, PARAFAC had fewer parameters and resulted in the most simple and restricted model. 43 Solving PARAFAC required a significant amount of time to run, but fortunately, ALS increased computational efficiency resulting in decreased required clock-time. Also, by iteratively fixing two factors, the equation (2) becomes convex for each intermediate step. We implemented the approach based on Matlab CMTF42 and tensor toolbox. The convergence criteria was 10e-5. 44
Choosing the number of disease topics
To decide the number of topics (k), we plotted the decay of eigenvalues of the unfolded-input data. The point of inflection can suggest a range of candidate k. To decide the specific k, we considered two metrics: the coherence within a topic (i.e., whether a topic can represent a single theme or similar concept) and the similarity between topics. We first measured topic coherence by using UMass,45 which calculates the co-concurrency of the topic descriptors in the original EHR data. Topics with higher UMass are easier to interpret. The similarity between topics can be measured with mean pairwise Jaccard similarity (mean_Jaccard),46 or cosine similarity 47, reflecting the overlapping degree between topics. We expected the topics should have less similarity. To select the K, we standardized the UMass and the 1/mean_Jaccard and summed these two as the topic quality score. We calculated the topic score for k from 5 to 15. We ranked the topic quality scores in descending order and selected the top 10 highest quality scores. Then we picked the largest K (within the suggestion range), because a larger number of topics may explain more of the input data variation. We involved three reviewers with clinical backgrounds to review the results 48. We designed two surveys at the topic and patient levels (Appendix B).
To visualize the topic results, we employed word clouds to present top-ranked phenotypes in a topic. Patients were represented in combined weights of topics (patient-topic factor matrix). We assign a patient to the topic group with the maximum weight. We used t-Distributed Stochastic Neighbor Embedding (t-SNE) to project the patient-topic on a 2-dimensional (2D) map to visualize the similarity between topic groups.
Statistical analysis
Correlation between topics and conventional assessments of CVD risk
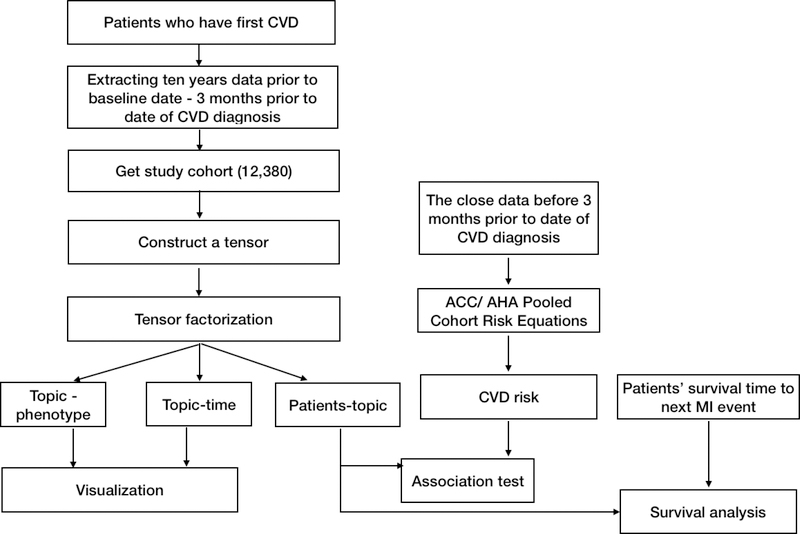
For each topic, we examined its association with the risk estimated by the American College of Cardiology/American Heart Association (ACC/AHA) Pooled Cohort Risk Equations 2 (ACC/AHA Pooled Equations), which is a widely used clinical tool to estimate 10-year risk of atherosclerotic cardiovascular disease (ASCVD) events - a composite of MI, stroke, or cardiovascular death - based on conventional risk factors, e.g., age, gender, hypertension and smoking status. The goal of the association analysis was to examine the correlation between each topic and the conventional assessed risk of significant cardiovascular outcomes. We hypothesized that our approach would identify subphenotypes where the CVD risk was not adequately captured by a conventional risk assessment tool.
To apply ACC/AHA Pooled Equations to the CVD patients (Figure 2), we collected demographics, hypertension drug usage, smoking status, the most recent physical measurements and lab values (i.e. SBP/DBP and high-density lipoprotein [HDL]- cholesterol level) before the baseline date. We replaced the missing physical or laboratory measures using the median values from the same age, gender and race group. We calculated the ASCVD risk for each individual based on the ACC/AHA Risk Equations.
Figure 2.
Study Design
For each topic, we examined its association with the 10-year CVD risk estimated by ACC/AHA equations using the Pearson correlation coefficient (PCC). The study reported the coefficient and p-value. The level of significance was set at p < 0.05 / number of topics (Bonferroni correction). We further stratified patients into high - (risk probability≥20%) and low - risk (risk probability<7.5%) category 50, and plotted the distribution of high/low -risk patients in the top six prevalent topics (Figure A.8). We aimed to find if high or low-risk patients would present any difference in such phenotypic topics.
Risk of subsequent MI among subphenotypes
To demonstrate the clinical importance of the topics, we evaluated the risk of subsequent MI event for topics through survival analysis. We hypothesized that topics would vary in the survival time from their initial diagnosis of CVD to subsequent MI. We first examined Kaplan-Meier plots stratified by topic. The start date of the time outcome (time period 0) was the date of the first CVD diagnosis. The observation time was between time period 0 and the subsequent MI. For patients who had no subsequent MI, we right-censored such patients at the last date they were observed at VUMC. We also applied a Cox proportional hazards regression model using patient’s weights of topics, age, and gender as covariates. We calculated hazard ratios for each topic along with their corresponding confidence intervals and p-values.
Results
Time-evolving disease topics
We identified 12,380 adult CVD patients and extracted 1068 unique PheCodes among them. We applied an unsupervised tensor-factorization on the dataset. In determining the number of topics (k), we tried k from 5 to 20 and set the number of topics as 14 in the study according to the measuring process (Figure A.1–A.3). Three reviewers with clinical practice or medical training background validated the results to ensure clinical relevance (Appendix B). The results showed that at the topic level, the average rating was 1.45 (an ordinal scale of 0–2 [i.e.0-not clinically meaningful;1- possibly clinically meaningful; 2 - clinically meaningful], with a Fleiss’ Kappa 51 score of 0.41 (moderate agreement). At the patient level, the mean of the three reviewers rating score was 1.16 (an ordinal scale of 0–2 [i.e.0-not relevant;1- relevant; 2- highly relevant], with a Fleiss’ Kappa score of 0.45, suggesting that the topics assigned to each patient were relevant to the patient’s history.
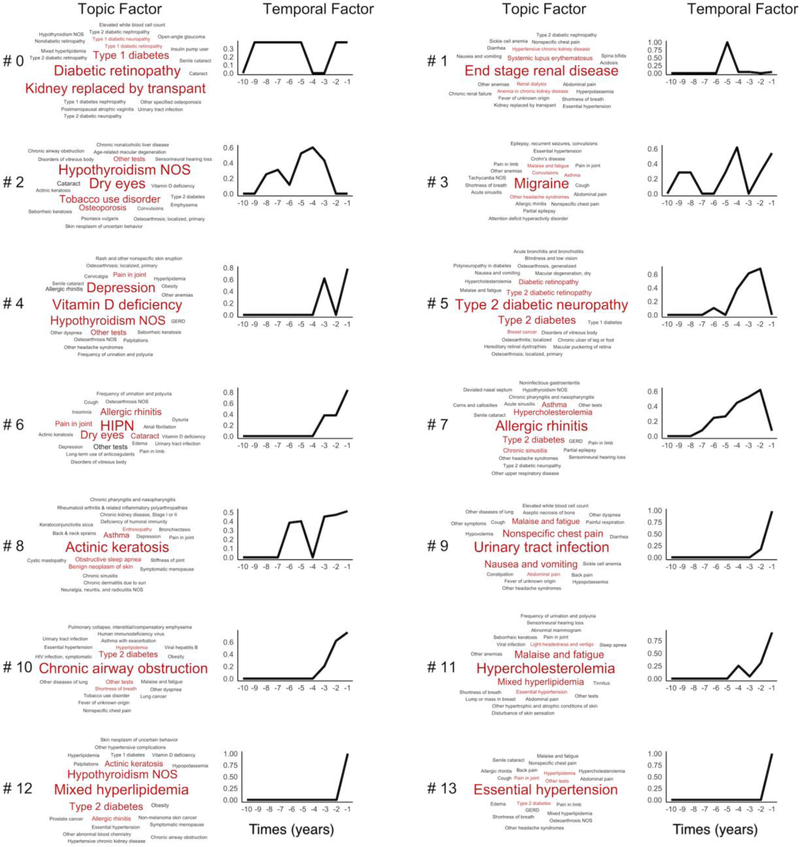
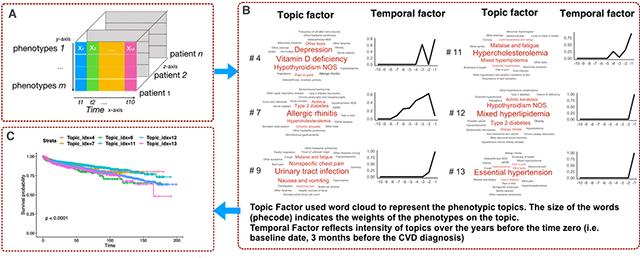
Figure 3 showed the learned 14 topics and the progression pattern over the 10 years prior to the first diagnosis of CVD. Topic #0 and Topic #1 were enriched for serious kidney disease, with more relevance with Type 1 Diabetes in Topic #0 than Topic #1. Topic #4 was enriched for Vitamin D deficiency, Depression, and Hypothyroidism NOS. Topic #5 clustered around complications of Type 2 diabetes, such as Type 2 diabetic neuropathy and Type 2 diabetic retinopathy. Topics #11 and #12 both had lipid disorders as their top diagnoses (Hypercholesterolemia and Mixed Hyperlipidemia, respectively). Yet, they differed substantially in associated diagnoses: Malaise and fatigue, Mixed hyperlipidemia, and Essential hypertension were enriched in Topic #11; while Hypothyroidism NOS and Type 2 diabetes were enriched in Topic #12.
Figure 3.
Tensor factorization on 10 year-diagnoses prior to CVD diagnosis. Each topic contains the topic factor, represented by a word cloud and a time factor showing the time trend. The word size of a PheCode is proportional to its weight (i.e. influence) for the topic (from factor A), e.g. Essential hypertension has a larger influence than GERD within Topic #13. The x-axis on the line charts represent the years prior to the diagnosis of CVD, increasing from 10 years to 1 year prior to CVD. The y-axis represents the mathematical weights (from time factor B) for the respective topics for each year prior to the diagnosis of CVD. HIPN in Topic #6: Hereditary and idiopathic peripheral neuropathy.
The time trend in Figure 3 shows the trajectory of each topic before the first diagnosis of CVD. Specifically, it reflects the changes in the combined intensity of the phenotypes (in the topic) in patients assigned to the topic group from the last 10 years to the first CVD diagnosis. For example, when close to the first CVD, patients in Topic #13 have a sharp increase, which means more Essential hypertension appear in the topic group. Patients in Topic #7 (Allergic rhinitis) increased the events slowly but have a small drop when close to the first CVD.
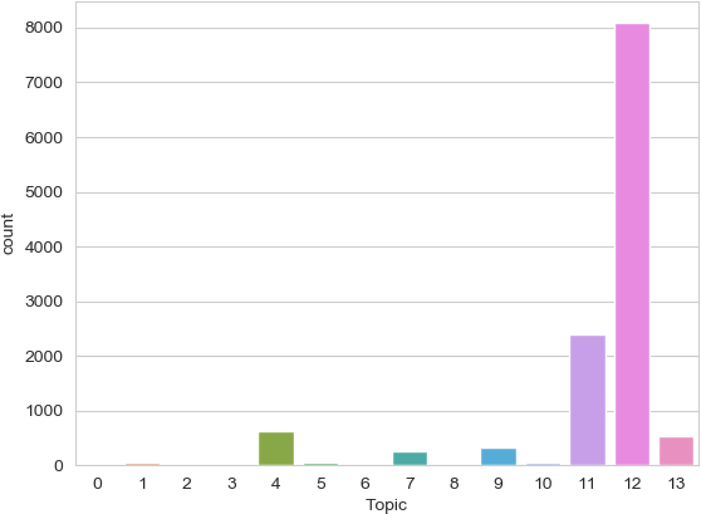
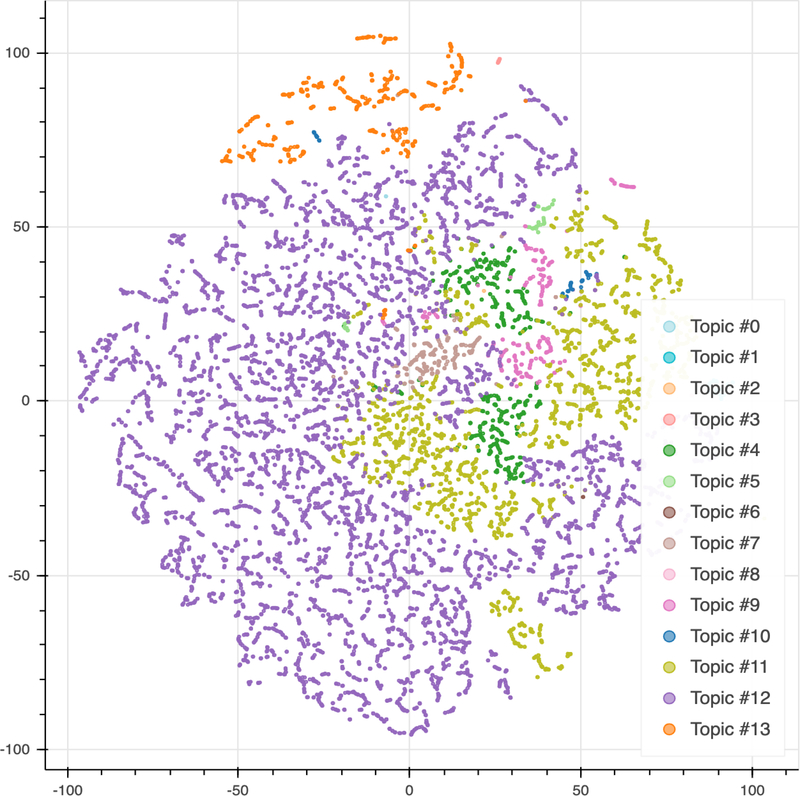
We assigned the patients into their best-fitting topic according to their maximum weights (Fig 4). Topic #12 (Mixed Hyperlipidemia) was the most prevalent topic in the cohort with 8078 patients. Topic #11 (Hypercholesterolemia), Topic #4 (Vitamin D deficiency) and Topic #13 (Essential hypertension) were other prevalent topics in the cohort with 2386, 627, and 541 patients, respectively. Figure 5 showed the visualization of patients-topics in a 2D map using t-Distributed Stochastic Neighbor Embedding (t-SNE). Topic #13 (Essential hypertension) formed a distinct group separate from other groups on the top of the figure.
Figure 4. Topic distribution in the cohort.
To visualize the frequency of each topic in the cohort, we assigned an individual to the topic with the maximum score. The vertical axis represents the number of patients. Topics are plotted on the horizontal axis.
Figure 5. t-SNE visualization of topic clusters.
Each data point represents an individual and the color of each data point represents the topic with the maximum score for that individual. We used principal component analysis (PCA) for t-SNE embedding initialization.
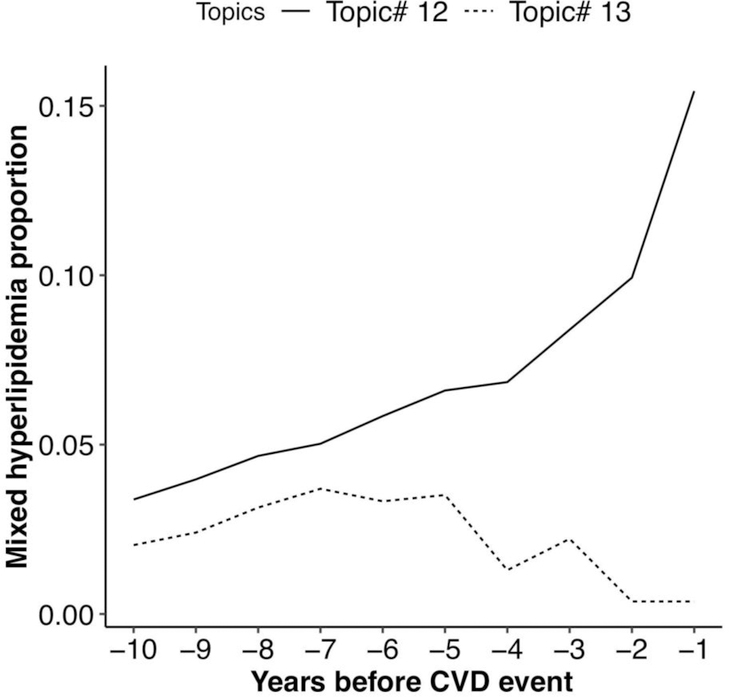
To illustrate the differences in mixed hyperlipidemia between topics, we compared the prevalence of mixed hyperlipidemia between the patients who were assigned with Topic #12 and Topic #13 (Figure 6). Overall, the proportion of patients with Mixed Hyperlipidemia in Topic #13 was smaller than Topic #12. Although the proportion of patients with Mixed Hyperlipidemia was initially similar, the proportions diverged starting at approximately 5 years prior to the CVD event. At that point, the proportion of patients with Mixed Hyperlipidemia in Topic #12 increased substantially, whereas Mixed Hyperlipidemia declined among patients in Topic #13.
Figure 6.
Proportion of patients with mixed hyperlipidemia in patients assigned with Topic #12 (Mixed hyperlipidemia) and Topic #13 (Essential hypertension).
Analysis of correlation between topics and conventional assessments of CVD risk
We applied ACC/AHA Pooled Cohort Risk Equations to calculate the 10-year ASCVD risk for each patient (Table A.1). Table 1 summarizes the association result between topics and the ASCVD risk estimated by ACC/AHA equations. Topics such as #11 (Hypercholesterolemia), Topic #12 (Mixed hyperlipidemia, Type 2 diabetes), and Topic #13 (Essential hypertension) significantly correlated with an increased estimated risk of ASCVD. Among the most prevalent topics, Topic #4 (Vitamin D deficiency, Depression), Topic #7 (Allergic rhinitis), and Topic #9 (Urinary tract infection) have no significant associations between the estimated risk of ASCVD. Figure A.8 further presents the proportion of stratified risk in the top prevalent topics. Topic#11, Topic #12 and Topic #13 contained a higher proportion of high-risk patients than low-risk patients. Both high and low-risk patients present in Topic #9 and Topic #4, with a similar proportion, suggesting that the conventional risk assessment tool did not capture the true risk of CVD for patients in these subphenotypes.
Table 1.
Pearson correlation coefficient testing between topics and estimated ASCVD risk. Significance level after Bonferroni correction is p<0.0036 (0.05/14).
| Topic | Top phenotypes | Co-efficient | p-value |
|---|---|---|---|
| #0 | Diabetic retinopathy, Kidney replaced by transplant, Type 1 diabetes, Type 1 diabetic retinopathy | −0.004 | 0.683 |
| #1 | End stage renal disease, Systemic lupus erythematosus, Renal dialysis, Anemia in chronic kidney disease | −0.002 | 0.817 |
| #2 | Dry eyes, Hypothyroidism NOS, Tobacco use disorder, Osteoporosis | 0.019 | 0.032 |
| #3 | Migraine, Convulsions, Other headache syndromes, Asthma | −0.020 | 0.025 |
| #4 | Vitamin D deficiency, Depression, Hypothyroidism NOS, Other tests | −0.001 | 0.016 |
| #5 | Type 2 diabetic neuropathy, Type 2 diabetes, Type 2 diabetic retinopathy, Diabetic retinopathy | 0.016 | 0.071 |
| #6 | Hereditary and idiopathic peripheral neuropathy, Dry eyes, Allergic rhinitis, Cataract | 0.016 | 0.06 |
| #7 | Allergic rhinitis, Type 2 diabetes, Hypercholesterolemia, Asthma | 0.010 | 0.267 |
| #8 | Actinic keratosis, Asthma, Obstructive sleep apnea, Benign neoplasm of skin | −0.006 | 0.528 |
| #9 | Urinary tract infection, Nonspecific chest pain, Nausea and vomiting, Malaise and fatigue | −0.011 | 0.197 |
| #10 | Chronic airway obstruction, Type 2 diabetes, Other tests, Hyperlipidemia | 0.043 | 1.932e-06 |
| #11 | Hypercholesterolemia, Malaise and fatigue, Mixed hyperlipidemia, Essential hypertension | 0.035 | 1.20e-04 |
| #12 | Mixed hyperlipidemia, Hypothyroidism NOS, Type 2 diabetes, Actinic keratosis | 0.061 | 1.063e-11 |
| #13 | Essential hypertension, Other tests, Type 2 diabetes, Pain in joint | 0.054 | 1.292e-09 |
Survival analysis
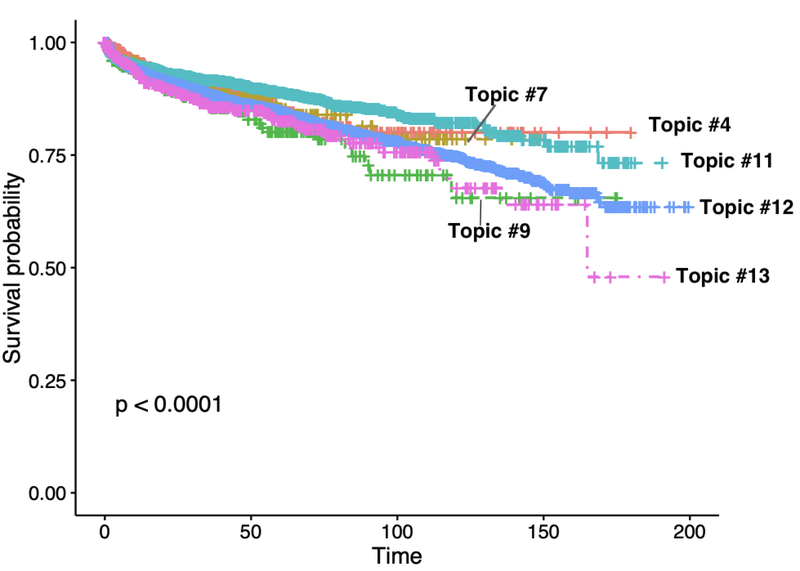
We assessed the risk of subsequent MI following the diagnosis of CVD by using a Kaplan-Meier plot. We stratified patients by the maximum observed weight for each individual among the six most prevalent topics: Topic #4 (Vitamin D deficiency, Depression), Topic #7 (Allergic rhinitis), Topic #9 (Urinary tract infection), Topic #11 (Hypercholesterolemia), Topic #12 (Mixed hyperlipidemia) and Topic #13 (Essential hypertension) We observed a significant difference among these groups (p<0.0001, Figure 7), implying these subgroups may have the differing risk of subsequent MI.
Figure 7.
Associate topic groups (top six prevalent) with the next myocardial infarction (MI) event by fitting a Kaplan-Meier model. The x-axis represents the months between the first diagnosis of CVD and the next MI event. The y-axis represents the probability of an individual surviving past time t with respect to second MI.
We then fit the Cox model with the topic weights as covariates using the lifelines Python package (0.20.0).52 Table 2 summarizes the topics that have a significant association with the subsequent MI. We observed that patients who have higher weights in topics #5 (Type 2 diabetic neuropathy, Type 2 diabetes), and Topic #13 (Essential hypertension) had increased the risk for subsequent MI. Patients with Topic #11 (Hypercholesterolemia) had decreased risk for a subsequent MI.
Table 2.
Cox regression models to examine the association between phenotypic topics and the subsequent MI event. We report topics significantly associated with subsequent MI event (statistically significant at the 0.05 level) and provide their corresponding hazard ratio, p-value and 95% confidence interval.
| Topic # | Top phenotypes | Hazard ratio (HR) | P | CI |
|---|---|---|---|---|
| 5 | Type 2 diabetic neuropathy, Type 2 diabetes, Type 2 diabetic retinopathy, Diabetic retinopathy | 29.66 | <0.005 | (7.6, 127) |
| 11 | Hypercholesterolemia, Malaise and fatigue, Mixed hyperlipidemia, Essential hypertension | 1.10e-14 | 0.01 | (1.589e-25, 1.132e-03) |
| 13 | Essential hypertension, Other tests, Type 2 diabetes, Pain in joint | 2.12 | 0.02 | (1.15, 3.86) |
Table 2 Cox regression models to examine the association between phenotypic topics and the subsequent MI event. We report topics significantly associated with subsequent MI event (statistically significant at the 0.05 level) and provide their corresponding hazard ratio, p-value and 95% confidence interval.
Discussion
In this study, we leveraged a constrained non-negative tensor-decomposition method to detect time-evolving phenotypes in CVD patients. Compared to previous studies,1 our approach captures both subphenotypes and their dynamics over time. We examined the association between the phenotypic topics and the estimated ASCVD risk calculated using conventional clinical tools. Results from this analysis showed that some phenotypic topics were not correlated with conventional CVD risk factors, suggesting that individuals with these subphenotypes may be affected by sub phenotypic diseases with different pathophysiologic causes. Furthermore, we observed significant differences in subsequent MI rates between the six most prevalent subphenotypes.
Comparison with traditional NMF
We compared our method with traditional NMF on each time slice of patients’ diagnosis (Appendix F). We measure the stability score (range [0, 1]) between topics of each time slice using Jaccard similarity and Hungarian method.49 From the stability plot (Figure A.10), we observed that the similarity between topics in the last first year and last 3 – 10 years were less than 50%, which means the topics changed over 10 years. It would be difficult to read, interpret and trace how topics change using traditional NMF. Further, it lacked a persuasive methodology to combine the weights of topics from each time slice to represent a single patient. Compared to the traditional NMF, our approach provides a solution for data exploration to 1) better show the dynamic changes of the subphenotypes; 2) generate the representations and topic membership for patients to facilitate the analysis.
Choosing the number of factors
Topic modeling and tensor decompositions and related unsupervised learning approaches require pre-specification of topic numbers, k. Choosing the optimal number of topics remains challenging. General approaches include: looking at residuals (e.g. construction errors), plotting the decay of singular values or the percentage of explained variance such as scree-plots and core consistency, and evaluating the interpretability of the factors. Selecting the approaches should consider the characteristics of input data (e.g. data size, sparsity, binary), the goal of the task, and the computing time. In this study, we first plotted the decay of singular values to get suggested candidates of k. As we aimed to yield distinct and interpretable subphenotypes and such metrics, we maximized the topic quality metric (coherence UMass and similarity) to determine the final k. Such approach has been widely used in evaluating the topic model.24,25,49 We also involved clinicians to validate whether topics were clinically meaningful. Larger k may allow the discovery of deep subphenotypes, but also increases the risk of fracturing meaningful phenotype clusters. Sun et. al applied Core Consistency Diagnostic CONCORDIA), 37 a method that can be used for the PARAFAC model to automatically discover the number of phenotypes. However, through the experiment, we found if we constrained the sparsity and normalized all factors, the core consistency is not accurate. We plotted the core consistency by removing sparsity constraints and only normalizing two factors (Figure A.3). The result showed k=14 achieved the highest core consistency, which matches the number of topics calculated using topic quality metric.
Adding orthogonal and sparsity constraints are important in generating an interpretable phenotype. We showed the topics using PARAFAC model without adding orthogonal and sparsity constraints (Figure A.5–A.6). From Figure A.5–A.6, we observed that it is highly possible to yield overlapped phenotypes.
Clinical relevance
This approach enabled us to obtain meaningful CVD subphenotypes and visualize their progression pattern. Topic #0 and Topic #1 are particularly enriched in individuals with kidney failure but may represent patients with different causes, e.g. uncontrolled Type 1 diabetes 53, and Systemic lupus erythematosus 54, respectively. Among the top six most prevalent topics, Topic #11(Hypercholesterolemia, Malaise, and fatigue), Topic #12 (Mixed hyperlipidemia and Type 2 diabetes), and Topic #13 (Essential hypertension) are well-described common comorbidities of CVD. Each of these topics had a time-progression pattern with a sharply increased intensity in the years leading up to the CVD event. Such results confirmed our previous findings in using the longitudinal EHR to predict 10-year CVD, that the most recent years’ value contributes more to the prediction.55 In contrast, some prevalent topics demonstrated different time-progressions. For example Topic #4 (Vitamin D Deficiency, Depression)56 had a wave change morphology with increased intensity at 3 to 4 years prior to the first diagnosis of CVD, followed by a drop-off, then a final increase in intensity at a year before the event.
As expected, we found statistically significant correlations between ASCVD risk and topics heavily enriched with traditional CVD comorbidities including Topic #11 (Hypercholesterolemia), Topic #12 (Mixed hyperlipidemia and Type 2 diabetes), and Topic #13 (Essential hypertension). Such results were in line with the Framingham study (Table A.1). The main phenotype Chronic airway obstruction in Topic #10, was not included in ACC/AHA ASCVD risk score, but evidence has shown that CVD is more common among patients with Chronic Obstructive Pulmonary Disease (COPD), a major cause of Chronic airway obstruction.57 COPD patients also have a higher risk of mortality from complications of CVD.58–60 Topic #10 had a positive correlation with an estimated risk of ASCVD, mainly because some risk factors of COPD such as age (mean age 69.5 [8.9]), smoking status, weight loss were also known risk factors of CVD. Therefore, ACC/AHA risk equation may still capture many of the important risk factors for CVD in this subphenotype.
We also observed topics such as Topic #4 (Vitamin D deficiency, Depression), and Topic #9 (Urinary tract infection, Nonspecific chest pain, Nausea and vomiting, Malaise and fatigue) had a negative correlation with the traditionally estimated risk. Topic #4 and Topic #9 contained a similar or larger proportion of low-risk patients than high-risk patients. Recent evidence has shown a strong correlation between Urinary tract infections (Topic #9) and cardiovascular events 61,62 There is also support for moderate to severe Vitamin D deficiency (Topic #4) as a risk factor for developing CVD.63 This suggests that conventional risk assessment tools could not accurately model risk of CVD for patients with these topics.
Topic #5 was not significantly correlated with the estimated risk of ASCVD by the ACC/AHA assessment tool, but it had a high hazard ratio for increased risk of subsequent MI (Table 2). Topic #5 appears to capture a subphenotype of patients with advanced Type 2 diabetes with microvascular complications including diabetic neuropathy and diabetic retinopathy. The presence of microvascular complications of diabetes such as Type 2 diabetic neuropathy portend an increased risk of CVD.64 Diabetic autonomic neuropathy is also an independent risk for recurrent CVD events in diabetic patients with a prior history of CVD.65 Furthermore, whereas intensive glucose control in patients with newly diagnosed or less advanced Type 2 diabetes may decrease risk of subsequent CVD 66; intensive glucose control in patients with long-standing Type 2 diabetes may not reduce the risk of developing CVD.67 Therefore, Topic #5 may identify a subphenotype of patients with advanced diabetes as their predominant risk for CVD, where conventional treatment aimed at controlling the risk comorbidity (diabetes) does not mitigate the risk of ultimately developing CVD and subsequent complications like MI.
Essential hypertension (Topic #13) and lipid disorders (Topic #11, Topic #12) are two well-known risk factors of CVD, however; they formed into separate distinct topics (Figure 5), suggesting they may represent distinct subphenotypes. Interestingly, Topic #13 represents a unique group enriched for Essential hypertension but with few hyperlipidemia events across the years (Figure 6). Notably, Topic #13 (Essential hypertension) had significantly increased risk of subsequent MI while high Topic #11 score (Hypercholesterolemia) had decreased risk. This may reflect an increased propensity for Topic #11 patients to receive cholesterol-lowering statin medications, which have a well-understood role in primary prevention of CVD events.68,69 Although both Topic #11 and Topic #12 contained mixed hyperlipidemia, Topic #12 also had Type 2 diabetes as the key descriptor and had a different evolving trend than Topic #11, indicating two different subphenotypes. The combination of Type 2 diabetes, Mixed Hyperlipidemia, Obesity, and Essential hypertension in the descriptors of Topic #12 capture many features of the metabolic syndrome, a constellation of comorbidities that result from abdominal obesity and insulin resistance.70 This suggests that although studies have traditionally treated Hyperlipidemia, Hypertension, and Type 2 diabetes as CVD risk factors, these phenotypes can be treated as different phenotype cohorts, with each having different optimal treatment regimens.
Limitations
This study has several limitations. First, we used binary values to indicate whether an individual received a diagnosis code in each year. A method accounting for disease severity (e.g., using counts of diagnosis codes) could be used in future studies and may improve the identification of subphenotypes. Second, we did not incorporate medications and laboratory tests in this study, which may also improve the identification of meaningful phenotype clusters. Third, although PheCodes are aggregates of ICD codes, there is still overlap among PheCodes, i.e., some distinct PheCodes defined closely related disease (e.g.Type 2 diabetic neuropathy [250.24], Type 2 diabetes [250.2]). Word-embedding models such as Word2Vec71 may precisely quantify the similarity between related terms. Finally, our study cohort did not include any controls of CVD. If we want to find predictors for CVD, we need to include the negative controls in future work.
Conclusion
This study demonstrated the feasibility of tensor-decomposition for learning time-evolving topics from longitudinal EHRs. The methodology may help identify potentially useful subphenotypes of complex diseases for precision medicine studies.
Supplementary Material
Highlight.
Present a method using Tensor Factorization to find subphenotypes from longitudinal EHR.
We applied this approach to 12380 patients with the diagnosis of CVD.
We identified 14 subphenotypes that patients established in 10 years prior to CVD, and showed the progress pattern.
Through an association analysis with estimated CVD risk, we found some topics e.g. Vitamin D deficiency, Urinary infections cannot be explained by conventional risk factors.
We identified a distinct subphenotype enriched for “Hypertension” with few “hyperlipidemia” that increased the risk of the subsequent MI.
Acknowledgment
We thanked Dr. Zhijun Yin and Dr. You Chen from Department of Biomedical Informatics at VUMC for helpful suggestions on measuring the quality of the topics; Dr. Yuankai Huo from Department of Electrical Engineering and Computer Science at Vanderbilt University for insights on tensor decomposition approach.
Funding
The project was supported by NIH grant P50 GM115305, R01 HL133786, R01 GM120523, T32 GM007347 from the National Institute of General Medical Studies for the Vanderbilt Medical-Scientist Training Program, 18AMTG34280063 from American Heart Association and T15 LM007450 from the National Library of Medicine for the Vanderbilt Biomedical Informatics Training Program, and Vanderbilt Faculty Research Scholar Fund. The dataset used for the analyses described were obtained from Vanderbilt University Medical Center’s resources, BioVU and the Synthetic Derivative, which are supported by institutional funding and by the Vanderbilt National Center for Advancing Translational Science grant 2UL1 TR000445-06 from NCATS/NIH. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing Interest
The authors have no competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Li L, Cheng W-Y, Glicksberg BS, Gottesman O, Tamler R, Chen R, et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med. 2015. October 28;7(311):311ra174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goff DC Jr, Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, Gibbons R, et al. 2013. ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014 June 24;129(25 Suppl 2):S49–73. [DOI] [PubMed] [Google Scholar]
- 3.Alkadhi K, Eriksen J. The complex and multifactorial nature of Alzheimer’s disease. Curr Neuropharmacol. 2011. December;9(4):586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buttorff C, Ruder T, Bauman M. Multiple chronic conditions in the United States. 2017. RAND Corporation: Santa Monica, CA: 2018; [Google Scholar]
- 5.About Chronic Diseases | CDC [Internet]. 2019. [cited 2019 May 15]. 2"Available from: https://www.cdc.gov/chronicdisease/about/index.htm
- 6.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet 2005. April;6(4):287–98. [DOI] [PubMed] [Google Scholar]
- 7.D’Agostino RB Sr, Vasan RS, Pencina MJ, Wolf PA, Cobain M Massaro JM, et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation. 2008. February 12;117(6):743–53. [DOI] [PubMed] [Google Scholar]
- 8.Kannel WB, Vasan RS. Adverse consequences of the 50% misconception. Am J Cardiol. 2009. February 1;103(3):426–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delude CM. Deep phenotyping: The details of disease Nature. 2015. November 5;527(7576):S14–5. [DOI] [PubMed] [Google Scholar]
- 10.Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016. November;23(6):1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH. Advances in Electronic Phenotyping: From Rule-Based Definitions to Machine Learning Models. Annu Rev Biomed Data Sci. 2018. July 20;1(1):53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei W-Q, Denny JC Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015. April 30;7(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni Y, Alwell K, Moomaw CJ, Woo D, Adeoye O, Flaherty ML, et al. Towards phenotyping stroke: Leveraging data from a large-scale epidemiological study to detect stroke diagnosis. PLoS One. 2018. February 14;13(2):e0192586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll RJ Eyler AE, Denny JC. Naïve electronic health record phenotype identification for rheumatoid arthritis. In: AMIA annual symposium proceedings American Medical informatics Association; 2011. p. 189. [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W-Q, Tao C, Jiang G, Chute CG. A high throughput semantic concept frequency based approach for patient identification: a case study using type 2 diabetes mellitus clinical notes. AMIA Annu Symp Proc. 2010. November 13;2010:857–61. [PMC free article] [PubMed] [Google Scholar]
- 16.Yu S, Liao KP, Shaw SY, Gainer VS, Churchill SE, Szolovits P, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc. 2015. September;22(5):993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013. June;20(e1):e147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasko TA, Denny JC, Levy MA. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PLoS One. 2013. June 24;8(6):e66341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pivovarov R, Perotte AJ, Grave E, Angiolillo J, Wiggins CH, Elhadad N. Learning probabilistic phenotypes from heterogeneous EHR data. J Biomed Inform. 2015. December;58:156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z, Dong W, Duan H. A probabilistic topic model for clinical risk stratification from electronic health records. J Biomed Inform. 2015. December;58:28–36. [DOI] [PubMed] [Google Scholar]
- 21.Lu H-M, Wei C-P, Hsiao F-Y. Modeling healthcare data using multiple-channel latent Dirichlet allocation. J Biomed Inform. 2016. April;60:210–23. [DOI] [PubMed] [Google Scholar]
- 22.Chan KR, Lou X, Karaletsos T, Crosbie C, Gardos S, Artz D, et al. An Empirical Analysis of Topic Modeling for Mining Cancer Clinical Notes. In: 2013 IEEE 13th International Conference on Data Mining Workshops 2013. p. 56–63. [Google Scholar]
- 23.Liu L, Tang L, Dong W, Yao S, Zhou W. An overview of topic modeling and its current applications in bioinformatics. Springerplus. 2016. September 20;5(1):1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Ghosh J, Bejan CA, Gunter CA Gupta S, Kho A, et al. Building bridges across electronic health record systems through inferred phenotypic topics. J Biomed Inform. 2015. June;55:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J, Feng Q, Wu P, Warner JL, Denny JC, Wei W-Q. Using topic modeling via non-negative matrix factorization to identify relationships between genetic variants and disease phenotypes: A case study of Lipoprotein(a) (LPA). PLoS One. 2019. February 13;14(2):e0212112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao X, Li C, Yan J, Yao X, Risacher SL, Saykin AJ, et al. Identification of associations between genotypes and longitudinal phenotypes via temporally-constrained group sparse canonical correlation analysis. Bioinformatics. 2017. July 15;33(14):i341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen y, Zhang H, Wu J, Wang X, Liu R, Lin M. Modeling Emerging, Evolving and Fading Topics using Dynamic Soft Orthogonal NMF with Sparse Representation. In: 2015 IEEE International Conference on Data Mining 2015. p. 61–70. [Google Scholar]
- 28.Greene D, Cross JP. Exploring the Political Agenda of the European Parliament Using a Dynamic Topic Modeling Approach. Polit Anal. 2017. January;25(1):77–94. [Google Scholar]
- 29.Cohen MJ, Grossman AD, Morabito D, Knudson MM, Butte AJ, Manley GT. Identification of complex metabolic states in critically injured patients using bioinformatic cluster analysis. Crit Care. 2010. February 2;14(1):R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hitchcock FL. The expression of a tensor or a polyadic as a sum of products. J Math Phys. 1927;6(1–4):164–89. [Google Scholar]
- 31.Sidiropoulos ND, De Lathauwer L, Fu X, Huang K, Papalexakis EE, Faloutsos C. Tensor Decomposition for Signal Processing and Machine Learning. IEEE Trans Signal Process. 2017. July;65(13):3551–82. [Google Scholar]
- 32.Rabanser S, Shchur O, Günnemann S. Introduction to Tensor Decompositions and their Applications in Machine Learning [Internet]. arXiv [stat.ML]. 2017. Available from: http://arxiv.org/abs/1711.10781 [Google Scholar]
- 33.Ho JC, Ghosh J, Steinhubl SR, Stewart WF, Denny JC, Malin BA, et al. Limestone: high-throughput candidate phenotype generation via tensor factorization. J Biomed Inform. 2014. December;52:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Chen R, Ghosh J, Denny JC, Kho A, Chen Y, et al. Rubik: Knowledge Guided Tensor Factorization and Completion for Health Data Analytics. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining New York, NY, USA: ACM; 2015. p. 1265–74. (KDD ‘15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams AH, Kim TH, Wang F, Vyas S, Ryu SI, Shenoy KV, et al. Unsupervised Discovery of Demixed, Low-Dimensional Neural Dynamics across Multiple Timescales through Tensor Component Analysis. Neuron. 2018. June 27;98(6):1099–115.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harshman RA. PARAFAC2: Mathematical and technical notes. UCLA working papers in phonetics. 1972;22(3044):122215. [Google Scholar]
- 37.Perros I, Papalexakis EE, Vuduc R, Searles E, Sun J Temporal Phenotyping of Medically Complex Children via PARAFAC2 Tensor Factorization. J Biomed Inform. 2019. February 8;103125. [DOI] [PubMed] [Google Scholar]
- 38.Bahargam S, Papalexakis EE A Constrained Coupled Matrix-Tensor Factorization for Learning Time-evolving and Emerging Topics [Internet]. arXiv [cs.IR]. 2018. Available from: http://arxiv.org/abs/1807.00122 [Google Scholar]
- 39.Wei W-Q, Bastarache LA, Carroll RJ, Marlo JE, Osterman TJ, Gamazon ER, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record [Internet]. Vol. 12, PLOS ONE. 2017. p. e0175508 Available from: 10.1371/journal.pone.0175508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin PA, Thorburn MJ, McNeil Smith-Read EH. Chromosomal rearrangements in three generations of a Jamaican family. Cytogenet Genome Res. 1970;9(5):360–8. [DOI] [PubMed] [Google Scholar]
- 41.Horn RA, Johnson CR. Norms for vectors and matrices [Internet]. Matrix analysis. p. 257–342. Available from: 10.1017/cbo9780511810817.007 [DOI] [Google Scholar]
- 42.Bahargam S, Papalexakis EE. Constrained Coupled Matrix-Tensor Factorization and its Application in Pattern and Topic Detection. In: 2018 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM) 2018. p. 91–4. [Google Scholar]
- 43.PARAFAC Bro R.. Tutorial and applications. Chemometrics Intellig Lab Syst. 1997. October 1;38(2):149–71. [Google Scholar]
- 44.Bader BW, Kolda TG, Others. Matlab tensor toolbox version 2.5. Available online, January. 2012;7.
- 45.Stevens K, Kegelmeyer P, Andrzejewski D, Buttler D. Exploring Topic Coherence over Many Models and Many Topics. In: Proceedings of the 2012 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning Stroudsburg, PA, USA: Association for Computational Linguistics; 2012. p. 952–61. (EMNLP-CoNLL ‘12). [Google Scholar]
- 46.O’Callaghan D, Greene D, Carthy J, Cunningham P. An analysis of the coherence of descriptors in topic modeling. Expert Syst Appl. 2015. August 1;42(13):5645–57. [Google Scholar]
- 47.Han J, Kamber M, Pei J. Data mining concepts and techniques third edition. Morgan Kaufmann [Internet]. 2011; Available from: http://www.academia.edu/download/43034828/Data_Mining_Concepts_And_Techniques_3rd_Edition.pdf
- 48.Bhatia S, Lau JH, Baldwin T. An Automatic Approach for Document-level Topic Model Evaluation. In: Proceedings of the 21st Conference on Computational Natural Language Learning (CoNLL 2017) Stroudsburg, PA, USA: Association for Computational Linguistics; 2017. p. 206–15. [Google Scholar]
- 49.Greene D, O’Callaghan D, Cunningham P. How Many Topics? Stability Analysis for Topic Models In: Machine Learning and Knowledge Discovery in Databases. Springer; Berlin Heidelberg; 2014. p. 498–513. [Google Scholar]
- 50.Grundy SM, Stone NJ. 2018. American Heart Association/American College of Cardiology Multisociety Guideline on the Management of Blood Cholesterol [Internet]. Vol. 4, JAMA Cardiology. 2019 p. 488 Available from: 10.1001/jamacardio.2019.0777 [DOI] [PubMed] [Google Scholar]
- 51.Fleiss JL. Measuring nominal scale agreement among many raters. Psychol Bull. 1971. November;76(5):378–82. [Google Scholar]
- 52.Davidson-Pilon C, Kalderstam J, Zivich P, Kuhn B, Fiore-Gartland A, Moneda L, et al. CamDavidsonPilon/lifelines: v0.20.4 [Internet]. 2019. Available from: https://zenodo.org/record/2611708
- 53.Orchard TJ, Costacou T, Kretowski A, Nesto RW. Type 1 diabetes and coronary artery disease. Diabetes Care.2006. November;29(11):2528–38. [DOI] [PubMed] [Google Scholar]
- 54.Sinicato NA, da Silva Cardoso PA, Appenzeller S. Risk factors in cardiovascular disease in systemic lupus erythematosus. Curr Cardiol Rev. 2013. February 1;9(1):15–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao J, Feng Q, Wu P, Lupu RA, Wilke RA, Wells QS, et al. Learning from Longitudinal Data in Electronic Health Record and Genetic Data to Improve Cardiovascular Event Prediction. Sci Rep. 2019. January 24;9(1):717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkins CH, Sheline YI, Roe CM, Birge SJ, Morris JC. Vitamin D deficiency is associated with low mood and worse cognitive performance in older adults. Am J Geriatr Psychiatry. 2006. December;14(12):1032–40. [DOI] [PubMed] [Google Scholar]
- 57.Feary JR, Rodrigues LC, Smith CJ, Hubbard RB, Gibson JE. Prevalence of major comorbidities in subjects with COPD and incidence of myocardial infarction and stroke: a comprehensive analysis using data from primary care [Internet]. Vol. 65, Thorax. 2010. p. 956–62. Available from: 10.1136/thx.2009.128082 [DOI] [PubMed] [Google Scholar]
- 58.Wakabayashi K, Gonzalez MA, Delhaye C, Ben-Dor I, Maluenda G, Collins SD, et al. Impact of chronic obstructive pulmonary disease on acute-phase outcome of myocardial infarction. Am J Cardiol. 2010. August 1;106(3):305–9. [DOI] [PubMed] [Google Scholar]
- 59.Enriquez JR, Parikh SV, Selzer F, Jacobs AK, Marroquin O, Mulukutla S, et al. Increased adverse events after percutaneous coronary intervention in patients with COPD: insights from the National Heart, Lung, and Blood Institute dynamic registry. Chest. 2011. September;140(3):604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bafadhel M, Russell REK. Are COPD and cardiovascular disease fundamentally intertwined? Eur Respir J. 2016. May;47(5):1307–9. [DOI] [PubMed] [Google Scholar]
- 61.Santos-Gallego CG, García-Ropero Á, Badimon JJ. Spark That Lights the Fire: Infection Triggers Cardiovascular Events. J Am Heart Assoc. 2018. November 20;7(22):e011175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cowan LT, Lutsey PL, Pankow JS, Matsushita K, Ishigami J, Lakshminarayan K. Inpatient and Outpatient Infection as a Trigger of Cardiovascular Disease: The ARIC Study. J Am Heart Assoc. 2018. November 20;7(22):e009683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang TJ, Pencina MJ, Booth SL, Jacques PF, Ingelsson E, Lanier K, et al. Vitamin D deficiency and risk of cardiovascular disease. Circulation. 2008. January 29;117(4):503–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papanas N, Ziegler D. Risk Factors and Comorbidities in Diabetic Neuropathy: An Update 2015. Rev Diabet Stud. 2015. August 10;12(1–2):48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cha S-A, Yun J-S, Lim T-S, Min K, Song K-H Yoo K-D, et al. Diabetic Cardiovascular Autonomic Neuropathy Predicts Recurrent Cardiovascular Diseases in Patients with Type 2 Diabetes. PLoS One. 2016. October 14;11(10):e0164807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HAW. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med. 2008. October 9;359(15):1577–89. [DOI] [PubMed] [Google Scholar]
- 67.Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet. 1998. September 12;352(9131):837–53. [PubMed] [Google Scholar]
- 68.Alenghat FJ, Davis AM. Management of Blood Cholesterol [Internet]. Vol. 321, JAMA. 2019. p. 800 Available from: 10.1001/jama.2019.0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sacks FM Pfeffer MA, Moye LA, Rouleau JL, Rutherford JD, Cole TG, et al. The Effect of Pravastatin on Coronary Events after Myocardial Infarction in Patients with Average Cholesterol Levels [Internet]. Vol. 335, New England Journal of Medicine. 1996. p. 1001–9. Available from: 10.1056/nejm199610033351401 [DOI] [PubMed] [Google Scholar]
- 70.Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Forceon Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World HeartFederation; International Atherosclerosis Society; and International Association for the Study of Obesity [Internet]. Vol., Obesity and metabolism. 2010. p. 63 Available from: 10.14341/2071-8713-5281 [DOI] [PubMed] [Google Scholar]
- 71.Neelakantan A, Shankar J, Passos A, McCallum A. Efficient Non-parametric Estimation of Multiple Embeddings per Word in Vector Space [Internet] Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) 2014. Available from: 10.3115/v1/d14-1113 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.