Abstract
We attempted to establish a magnetic resonance imaging (MRI)-based radiomic model for stratifying prognostic subgroups of newly diagnosed glioblastoma (GBM) patients and predicting O (6)-methylguanine-DNA methyltransferase promotor methylation (pMGMT-met) status of the tumor. Preoperative MRI scans from 201 newly diagnosed GBM patients were included in this study. A total of 489 texture features including the first-order feature, second-order features from 162 datasets, and location data from 182 datasets were collected. Supervised principal component analysis was used for prognostication and predictive modeling for pMGMT-met status was performed based on least absolute shrinkage and selection operator regression. 22 radiomic features that were correlated with prognosis were used to successfully stratify patients into high-risk and low-risk groups (p = 0.004, Log-rank test). The radiomic high- and low-risk stratification and pMGMT status were independent prognostic factors. As a matter of fact, predictive accuracy of the pMGMT methylation status was 67% when modeled by two significant radiomic features. A significant survival difference was observed among the combined high-risk group, combined intermediate-risk group (this group consists of radiomic low risk and pMGMT-unmet or radiomic high risk and pMGMT-met), and combined low-risk group (p = 0.0003, Log-rank test). Radiomics can be used to build a prognostic score for stratifying high- and low-risk GBM, which was an independent prognostic factor from pMGMT methylation status. On the other hand, predictive accuracy of the pMGMT methylation status by radiomic analysis was insufficient for practical use.
Subject terms: Prognostic markers, Cancer imaging
Introduction
Glioblastoma (GBM) shows poor prognosis despite development of multimodal treatment including surgery, radiation therapy, and chemotherapy. O (6)-methylguanine-DNA methyltransferase (MGMT) promotor methylation (pMGMT-met) is a favorable prognostic factor in GBM patients, and patients with GBM and pMGMT-met benefit from temozolomide1. On the other hand, meta-analysis of previous clinical trials revealed that clinical features such as age, neurological status, and extent of tumor removal or residual tumor volume post-surgery are independent prognostic factors in GBM. These facts indicate that prognosis of this malignant disease is impacted by both the biological nature of the disease and the clinical manifestation of the patients. Accurate prognostication for each patient is necessary not only to determine the most appropriate individual treatment strategy but also to identify prognostic factors for patient stratification in clinical trials. Furthermore, in clinical practice, identifying patients who will potentially have a good outcome despite harboring this devastating disease is of great benefit for not only the patients and their families but also for primary physicians to provide hope while the patient goes through the demanding treatment procedures.
In this investigation, we retrieved as much information as possible from the initial magnetic resonance images (MRIs) for patients with GBM by use of radiomics2–4. The hypothesis that radiomics of GBM could build a radiologically derived prognostic score was tested with respect to the tumor’s pMGMT-met status.
Materials and Methods
Patient cohort and inclusion criteria
This study was performed in accordance with the principles of the Helsinki Declaration and was approved by the internal ethical review boards of Wakayama Medical University, Osaka International Cancer Institute, and all collaborative institutes, the list of which can be found in the acknowledge section. Written informed consent was obtained from all patients.
Inclusion criteria for the present study were as follows: new in-house diagnosis of GBM with fresh or frozen tissue available for genomic analysis, and preoperative MRI available including T1-weighted images (WI), T2WI, and gadolinium-enhanced (Gd) T1WI. Finally, 201 cases from 10 institutions that belong to the Kansai Molecular Diagnosis Network for central nervous system tumors were eligible for analysis. Frozen or fresh tumor samples were obtained at the time of surgery, and tumor genomic DNA was extracted from those tissues for genetic analysis.
Genetic analysis and integrated diagnosis
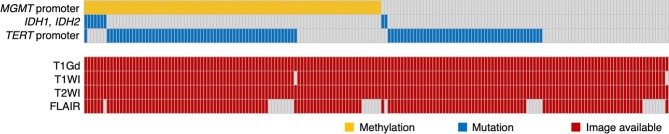
Genetic analyses were performed in the Osaka National Hospital according to the procedures previously described5. Briefly, the methylation status of the MGMT promoter was analyzed by quantitative methylation-specific PCR after bisulfite modification of genomic DNA, and we used a cut-off of ≥1% for MGMT promoter methylation5. The presence of hotspot mutations in IDH1 (R132) and IDH2 (R172)6 and the two mutation hotspots in the TERT promoter7 were analyzed by Sanger sequencing. The copy number status of 1p-19q was determined by multiplex ligation-dependent probe amplification (Oligodendroglioma 1p-19q probemix and EK1 reagent kit, MRC-Holland, Amsterdam, the Netherlands). Central pathology review was performed by a senior board-certified neuropathologist (Y.K.), and integrated diagnosis and WHO grading were made based on the 2016 WHO Classification of Tumors of the CNS (2016 WHO). Patient characteristics are listed in Fig. 1 and Supplementary Dataset.
Figure 1.
Overview of the analyzed cohort with landscape of genetic information. Genetic status is shown by color as indicated.
MRI data and clinical information
All MRI analyzed in the present study were preoperatively acquired using either 1.5- or 3.0-T MRI scanners according to the protocols in each institution. Magnetic field strength of the scanners used is detailed in Supplementary Dataset. Gd-T1WIs were available in 181 cases, T1WIs in 179 cases, and T2WIs in 181 cases. Overall survival was determined as the time from the date of the initial surgery for diagnosis to the date of death or the latest follow-up. Progression free survival was determined as the time from the date of the initial surgery for diagnosis to the date when first progression was identified or the latest follow-up. Visual inspection of each images was conducted in advance to sending these images for radiomics analysis in order to ensure that collected images are qualified for being used as GBM diagnosis. Clinical information such as age, sex, KPS at initial presentation and type of surgery performed was collected as well as details of post-surgical treatment including use of chemotherapy and/or radiation therapy. As for surgery, gross total removal was considered when more than 90% of the bulk of the tumor was surgically removed whereas partial removal was considered as significant amount of tumor being removed but not reaching more than 90% of it. Biopsy was defied as surgery aiming only histological confirmation of the tumor with no intention for cytoreduction of the tumor. Details can be found in Supplementary Dataset.
Radiomics
Radiomic analyses (radiomics) were conducted using image analyzing software developed in-house in combination with the Oxford Centre for Functional MRI of the Brain (FMRIB) Linear Image Registration Tool (FLIRT) provided by FMRIB Software Library (FSL)8–10. The in-house software was developed in Matlab (Mathworks, Natick, MA), and seamless data transfer was carried out between Matlab-based in-house software and FSL via FSL integration into Matlab. All Digital Imaging and Communications in Medicine format images were first converted to the Neuroimaging Informatics Technology Initiative format using MRIConvert (University of Oregon Lewis Center for Neuroimaging: http://lcni.uoregon.edu/~jolinda/MRIConvert/), followed by 256 gray-scale level conversion. Non-contrast T1WI and Gd-T1WI voxels that were in the top 0.1% in intensity were deleted as they were mainly high signal noise, and the remaining 99.9% were reallocated in 256 gray scale. For T2WI, 100% of the data range was reallocated in 256 gray scale. This procedure was necessary for intensity normalization across all images acquired by different MRI scanners. Previous study by the authors conducting radiomics in WHO grade 2 and 3 gliomas revealed that the above-mentioned intensity normalization was most suitable for further analysis11. Furthermore, T2Edge images were constructed by applying a Prewitt filter to T2WI. Gdzscore images were also constructed by performing a voxel-wise contrast enhancement calculation using non-contrast and Gd-T1WI. Tumors were delineated by manually tracing contrast enhancing lesions on Gd-T1WI and high-intensity lesions on T2WI in three dimensions by experienced surgical neuro-oncologists (T.S. and M.K.) to create two different voxels of interest (VOIs). A small subset of data was used for assessing interobserver agreement. VOIs of both Gd-T1WI and T2WI for case number 1, 2, 5, 6, 8, 9 were created by both of the neuro-oncologists and Dice index was calculated. Dice index for Gd-T1WI-VOI was 0.88 ± 0.05 and that for T2WI was 0.86 ± 0.03, indicating acceptable interobserver agreement (both in mean ± standard deviation).
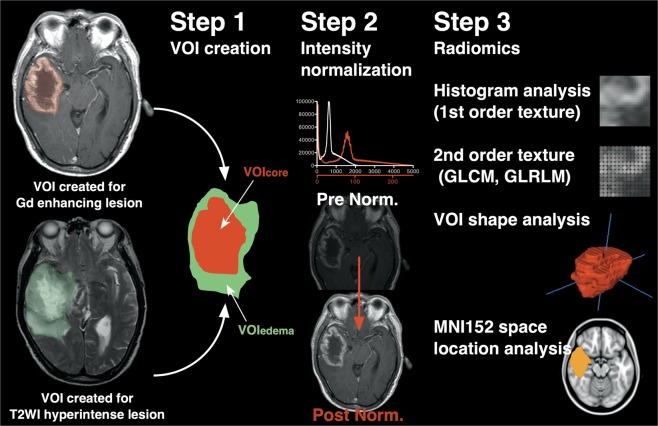
After VOIs were created, all different image sequences obtained from a single subject were co-registered to each other using a mutual information algorithm with 12 degrees of freedom transformation with FSL-FLIRT to obtain transformation matrices of different image sequences. Three-dimensional lesion VOIs modeled on Gd-T1WI and T2WI as mentioned above were deformed and resliced using the obtained transformation matrices via FSL-FLIRT for each specific image sequence. VOIs created on T2WI were subtracted from VOIs on Gd-T1WI to obtain VOIs for edema. VOIs created on Gd-T1WI were further denoted as “VOIcore,” and the VOIs on T2WI subtracted from VOIs on Gd-T1WI as “VOIedema”. Three different aspects of texture features of the two VOIs were measured on T1WI, T2WI, Gd-T1WI, T2Edge, and Gdzscore image series, i.e., histogram-based first-order texture, second-order texture, and shape characteristics of the VOIs (Fig. 2).
Figure 2.
Illustration showing the workflow for image analysis. Two types of VOIs were created based on Gd enhancement of the tumor and edema lesion identification on T2-weighted images. Both VOIs were co-registered, and VOIcore and VOIedema were generated. Subsequently, intensity normalization of all images was performed, and first-order and second-order texture analysis, VOI shape analysis, and location analysis were performed.
Furthermore, T2WIs and Gd-T1WIs were registered to a 1.0-mm isotropic, high-resolution T1-weighted brain atlas provided by MNI152 using a mutual information algorithm with 12 degrees of freedom transformation with FSL-FLIRT. VOIcore and VOIedema were then registered onto MNI152 by using the obtained transformation matrices. This procedure was necessary to perform lesion mapping of the VOIs on the standard MNI152 space. It should be noted that tissue classification i.e., delineation of VOIcore and VOIedema was performed prior to spatial normalization and spatial normalization of these VOIs were performed using transformation matrices calculated between the whole brain of the patient and MNI152. Thus, eliminating any contamination of spatial normalization procedure into the process of tissue classification except for the fact that non-linear correction of the distorted brain by the lesion was not performed. The workflow for radiomics is illustrated in Fig. 2 and the calculated radiomic values are listed in Supplementary Table.
Statistical analysis and predictive modeling
Statistical analysis was performed by M.K. using JMP Pro ver. 13 (SAS, Cary, NC). Survival analysis was performed with Wald test for continuous variables such as age and KPS and the Kaplan-Meier method for nominal variables. Multiple group comparison was performed by proportional hazard.
Supervised principal component analysis (SPCA) was used for prognostication using the referenced method2. SPCA was originally developed to identify subsets of patients with different survival outcome from a large set of available data such as gene expression profiles12 and has been successfully used for this purpose13–15. In this context, large data set such as gene expression profiles can be considered equivalent to radiomic data and although SPCA has some limitations such as limited number of subclass identification and ignoring potentially significant features12, SPCA has been shown to effectively perform in predicting overall survival in glioma radiomic study2. This analysis was performed on R using the Superpc package (https://cran.r-project.org/web/packages/superpc/superpc.pdf). The threshold for constructing a survival prediction was searched by 10-fold outer-loop-cross-validation using Superpc and a threshold parameter of 1.69 was achieved as the best tuned parameter for the Supervised Principal Component Predictor model. Importance score of each radiomic feature was calculated with the threshold hold of Supervised Principal Component Predictor model set as 1.69, enabling visualization of significant radiomic features predictive of patient survival. Finally, a binary radiomic risk classification was achieved using the default and parameters12 as suggested in the Superpc reference manual of the superpc.predict function. More specifically, n.components of 1 and prediction.type of discrete were used with the threshold set to 1.69 as mentioned above. The Superpc will in the end produce an “object”, which could be considered as a complex sum of algorithm that enables to classify each individual patient into either high- or low-radiomic risk groups according to the arguments which, in the current study, are the radiomic features. The most important concept that should be reminded is that this approach is designed to stratify high-risk (which also means short living) patients by comparing the survival dataset and the radiomics features.
Predictive modeling for pMGMT-met and long-term survivors, which is a different analysis from prognostication mentioned-above was performed based on the least absolute shrinkage and selection operator (LASSO) method to select features that were most significant to build predictive models for identifying patients that live longer than a given duration. λ, which is the tuning parameter for LASSO, was selected for the smallest cross-validation error (λ_min). The final predictive models were refit using the significant components chosen by LASSO and λ_min. Calculations were performed on R using the Glmnet package using 489 radiomic features and 162 datasets, which had T1WI, T2WI, and Gd-T1WIs available with 10-fold outer-loop-cross-validation, which was repeated five times. Each patient was assigned as long-term or shot-term survivor according to the defined cut-off of overall survival for predicting long-term survivors and this cut-off was continuously changed which enables predicting modeling of long-term survivors at various thresholds. Although the fundamental aim of this analysis is the same as to that of the above-mentioned SPCA, the basic philosophy of analysis is different, as LASSO analysis only takes into account whether the patient lived longer or shorter than a given threshold while SPCA includes the whole survival duration into analysis.
Results
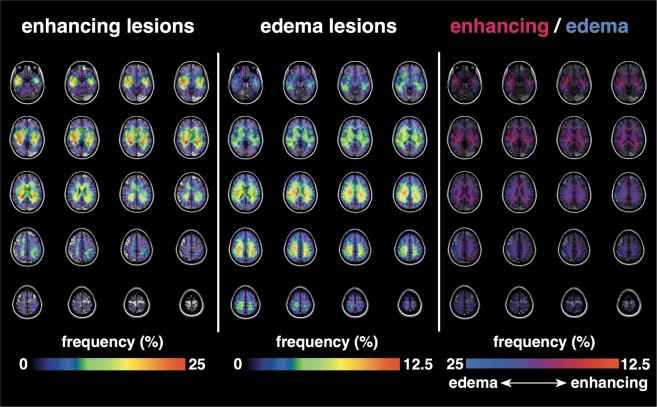
One hundred one cases were pMGMT-met, and 100 were MGMT promotor unmethylated (pMGMT-unmet). A total of 489 texture features including first-order texture features, second-order features (Gray level co-occurrence matrix and Grey level run length matrix), and shape characteristics of the VOIs were extracted from 162 datasets, and location data from 182 datasets were collected. Location analysis showed that VOIs of core and edema accumulated symmetrically around the periventricular white matter (Fig. 3).
Figure 3.
Lesion location mapping on the MNI152 standard brain atlas. Note that both enhancing and edema lesions were distributed symmetrically on both sides of the brain. Also note that enhancing lesions tended to occur in proximity to the ventricles compared with edema lesions.
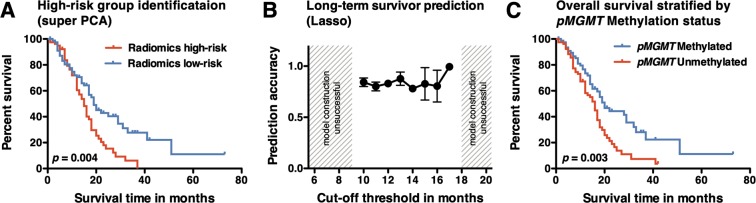
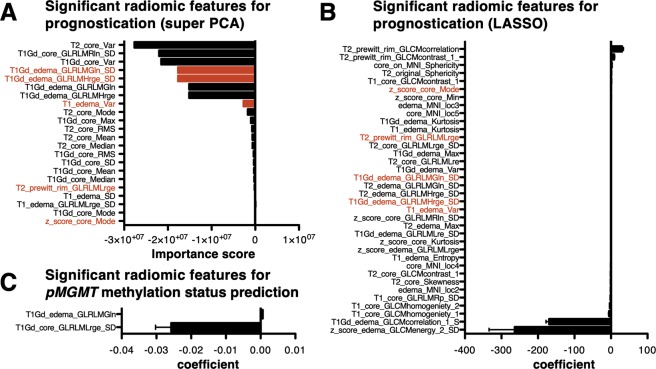
Next, prognostic modeling via radiomics was attempted by the aid of supervised principal component analysis, which was used to build a model capable of stratifying high- and low-risk GBM purely based on the initial MRI (Fig. 4A). The median overall survival time for radiomic high-risk GBM was 15 months, whereas that of radiomic low-risk GBM was 19 months (p = 0.004 Log-rank test). Furthermore, prediction of long-terms survivors at various overall survival cut-offs using LASSO also supported the finding by use of supervised principal component analysis. More specifically, predictions of patients surviving longer than 10, 11, 12, 13, 14, 15, 16 and 17 months were possible with high accuracy (Fig. 4B). These survival times are exactly when the Kaplan-Meier curves separates in Fig. 4A. As a reference, the median overall survival time of pMGMT-unmet GBM was 16 months, whereas that of pMGMT-met GBM was 20 months (p = 0.003 Log-rank test, Fig. 4C). The Supervised principal component analysis (SPCA) was able to identify 22 radiomic features that were significant for prognostication of GBM (Fig. 5A) and the trained prognostication model was able to label each subject into either a radiomic-high or -low risk class. Similarly, LASSO was able to identify 36 radiomic features that were significant for predicting patients surviving longer than 17 months (Fig. 5B). Five radiomic features were identified as prognostic features both by supervised principal component analysis and LASSO (Fig. 5A,B). Parametric survival analysis, however, using only these 5 radiomic features failed to converge to a meaningful predictive model for overall survival and univariate analysis revealed that only 3 among 5 features significantly correlated with survival duration of the patient (Supplementary Fig. 1). These results suggested that other listed radiomics factors were necessary for either SPCA or LASSO to construct their predictive model. On the other hand, attempt of predictive modeling of the pMGMT-met or unmet status via radiomics revealed to be unsatisfactory. Although radiomic feature selection predictive for pMGMT methylation status was pursued using LASSO regression (Fig. 5C), predictive accuracy of the pMGMT methylation status was as low as 67% on average measured by 10-fold cross-validation repeated five times (Table 1), suggesting limited use of this technique for pMGMT methylation status prediction. Univariate analysis revealed that age, pretreatment KPS, type of surgery, pMGMT methylation status and radiomic-risk stratification but not type of MRI used for analysis significantly correlated with overall survival (Fig. 4 and Supplementary Fig. 2). Cox regression analysis further revealed that age (p = 0.009), pMGMT methylation status (p = 0.001) and radiomic risk (p = 0.03) were independent prognostic factors. The hazard ratio of pMGMT-unmet was 2.04 (95% CI: 1.33–3.16, p = 0.001), and that of radiomic high-risk was 1.62 (95% CI: 1.04–2.52, p = 0.03). This result also supports the conclusion that radiomic risk score and pMGMT methylation status are independent prognostic factors (Table 2). Aside from radiomic analysis being able to stratify poor and favorable patients in terms of overall survival, this was not the case in progression free survival (Supplementary Fig. 3).
Figure 4.
Kaplan-Meier curve of the cohort stratified by radiomic risk score (A) and pMGMT methylation status (C). Both stratifications identified the poor-risk subgroup within the cohort. P values were calculated with the Log-rank test. LASSO was further used to predict long-term survivors using various cut-off in overall survival (B). In this analysis, predictive modeling was possible only when the cut-off was set within 10 to 17 months.
Figure 5.
(A) Significant radiomic features corresponding to the prognostic outcome within the cohort using supervised principal component analysis. Twenty-two features were identified by supervised principal component analysis. (B) Significant radiomic features corresponding to the prognostic outcome within the cohort using LASSO. 36 features were identified by supervised principal component analysis. Items colored in “red” are those both identified by supervised principal component analysis and LASSO. (C) Two radiomic features were identified to be predictive of pMGMT methylation status. Of note, higher T1Gd_core_GLRLMLrge_SD was indicative of pMGMT unmethylated glioblastomas.
Table 1.
Prediction accuracy of pMGMT methylation status with radiomics.
| Average of 5 repetitive measures | |
|---|---|
| Accuracy | 67% |
| Sensitivity | 67% |
| Specificity | 66% |
| Positive predictive value | 67% |
| Negative predictive value | 67% |
| Prevalence of pMGMT methylation | 50% |
Table 2.
Hazard ratio for overall survival of investigated factors.
| Factor | Hazard ratio (lower to upper 95% CI) | p value |
|---|---|---|
| Age | 1.02 (1.00–1.04)* | 0.009** |
| Pretreatment KPS | 0.99 (0.98–1.00)* | 0.114 |
| Type of Surgery | Partial to Total removal: 1.57 (0.97–2.51) | 0.138 |
| Biopsy to Partial removal: 0.94 (0.50–1.71) | ||
| Biopsy to Total removal: 1.47 (0.81–2.61) | ||
| pMGMT unmethylated | 2.04 (1.33–3.16) | 0.001** |
| Radiomic high-risk | 1.62 (1.04–2.52) | 0.031** |
CI; confidence interval, *per unit change in regressor, **considered statistically significant.
Based on these results, we established a combined risk stratification using both radiomic risk and pMGMT methylation status of GBM. The cohort was categorized into three groups: 1. those with pMGMT-unmet and radiomic high risk considered high risk, 2. those with pMGMT-met and radiomic low risk considered low risk, and 3. the others considered intermediate risk. The median overall survival times for these groups were 13, 20, and 18 months, respectively (p = 0.0003, Fig. 6). The survival difference was mainly attributable to the high- versus medium- or low-risk group and the survival difference between low- and medium-risk group was not statically significant (p = 0.21).
Figure 6.
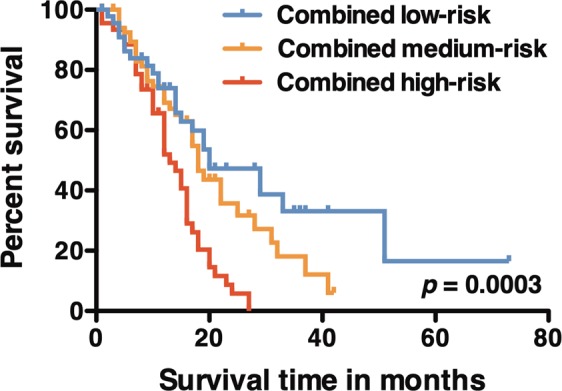
Kaplan-Meier curve of three different risk groups: low risk is composed of patients of radiomic low risk with pMGMT methylated status; medium risk is composed of patients of radiomic low risk with pMGMT unmethylated status or radiomic high risk with pMGMT methylated status; high risk is composed of patients of radiomic high risk with pMGMT unmethylated status. The P value was calculated with the Log-rank test.
Discussion
Development of objective and high-throughput analysis of lesion textures enabled acquisition of quantitative imaging features on various imaging modalities, which is now termed radiomics2–4,6,16. The main goal of radiomics in neuro-oncology is to identify imaging biomarker(s) to predict biological features of brain tumors such as genetic status or clinical outcomes. Although this approach is potentially powerful for identifying IDH mutation status in high- and low-grade gliomas6,11,17–19, whether this method can be successfully applied for predicting pMGMT methylation status of GBM is still controversial. The current study is an extension of the authors previous study focusing on radiomics and WHO grade 2 and 3 gliomas11 and attempted to predict pMGMT methylation status of GBM by building a radiomic model and further stratify a poor prognostic subgroup of GBM patients based on radiomic profiles. A revised analytical system except for constructing the classifier was used in this study as the previous system was not yet incorporating secondary texture analysis11.
In radiomic analysis, intensity normalization is one of the key steps to ensure correct quantitative measurements of qualitative images such as MRI. Although there are other image intensity normalization methods which in some cases takes into account the tissue types within the image2,20, we have adopted the simplest but yet the closest to clinical practice. Similar to diagnostic radiologist thresholding and narrowing the window of images when reading, the current method performed similar thresholding of images to achieve intensity normalization for further radiomic analysis. In the current research, radiomic analysis consisted of location and texture analysis. Location analysis was included in the analysis to quantitatively evaluate the distribution of tumor locations, as this information could harbor valuable information about the biological features of GBM as reported previously21,22. The current system is unique in that contrast enhancing tumor core (VOIcore) and the surrounding edematous lesions (VOIedema) were separately analyzed to retrieve as much radiomic information as possible.
Regarding the radiomic elements separating pMGMT-met from -unmet (Fig. 3), only two radiological features distinguished these two subgroups of GBM. Of note, second-order texture of the core lesion was heavily affected by the pMGMT methylation status. The sensitivity and selectivity, however, were not high enough to accurately predict pMGMT methylation status of GBM. Lesion location mapping also did not reveal any asymmetry in lesion occurrence between the two, a conclusion that is still controversial in the literature16,21,22. The present accuracy of 67% for predicting pMGMT methylation status is comparable to the accuracy described in previous investigations using different patient cohorts and analysis systems16,23. Those studies reported an accuracy ranging from 71 to 73%. Taking all these reports into consideration, the accuracy of radiomic prediction of pMGMT methylation status is at best around 70%. Considering that pMGMT methylation status is binary, this accuracy is far from satisfactory. Prediction of pMGMT methylation status may be limited by structural MRI alone.
On the other hand, when radiomics was compared with survival time of the cohort, radiomics was able to separate the cohort into poor versus good prognostic groups. As many as 68 among 162 cases (42%) were differently classified compared with the pMGMT methylation status. In fact, Cox regression analysis indeed identified that pMGMT methylation status and radiomic stratification were independent prognostic factors. It should further be noted that radiomic stratification was more prognostic than initial KPS or type of the surgery performed although residual tumor volume is now emerging as a key prognostic factor24–26. This finding is similar to that reported by Kickingereder et al. who used a different cohort and a different radiomic system and showed that radiomic risk scoring of GBM is independent of pMGMT methylation status27. It is also interesting that most of the radiomic features that the Supervised principal component analysis (SPCA) identified as prognostic derived from radiomic feature of the core lesions of GBM (14 out of 22). Although the reason in unclear, it is intuitively understandable that “enhancing core lesions” exhibit a more pronounce biological phenotype of the tumor than the surrounding edematous lesions. It is also of note that no location related information was identified as prognostic, which is different from our previous study focusing radiomic analysis and WHO grade 2 and 3 gliomas11. From a clinically practical point of view, as the SPCA model produces a “trained classifier object”, the actual numerical process of the predictive classification is not intuitive, which is difficult for clinician to incorporate this system into actual clinical practice. Furthermore, although the combination of pMGMT methylation status and radiomic risk score was able to identify an extremely high-risk group of patients (i.e. the combined high-risk group), the survival difference of combined low- and medium-risk was not statistically significant, requiring careful interpretation of the difference between these two groups.
Limitations of this study include the retrospective nature of the study and the absence of external cohort validation. It should also be pointed out that other machine learning methods for prognostication or pMGMT methylation status of GBM was not tested in this study and should be further pursued in future studies.
Conclusion
The current study revealed that radiomics can be used to build a prognostic score stratifying high- and low-risk GBM; this score was an independent prognostic factor from pMGMT methylation status. On the other hand, predictive accuracy of pMGMT methylation status by radiomic analysis was 67%, which remains insufficient for practical use. A combination of radiomic score and pMGMT methylation status effectively provided a more accurate stratification of clinical outcomes for newly diagnosed GBM patients.
Supplementary information
Acknowledgements
This research was supported by Japan Society for the Promotion of Science (JSPS grant numbers 16K10778 and 17H05308), Osaka Medical Research Foundation for Intractable Diseases, Uehara Memorial Foundation, Takeda Science Foundation and MSD Life Science Foundation. We would like to thank Ms. Ai Takada at the Institute for Clinical Research, Osaka National Hospital, National Hospital Organization for her excellent support in conducting this research. This research was approved by the local ethics committees/institutional review boards of the following institutions; Wakayama Rosai Hospital, Wakayama Medical University, Osaka International Cancer Institute, Osaka University Graduate School of Medicine, Osaka National Hospital, Kansai Medical University, Osaka City University Graduate School of Medicine, Kindai University Faculty of Medicine, Kansai Rosai Hospital, Osaka City General Hospital, Toyonaka Municipal Hospital, Tazuke Kofukai Foundation, Medical Research Institute, Kitano Hospital.
Author Contributions
Takahiro Sasaki: conceived and designed the study, acquired imaging data and performed annotations and preprocessing, wrote and edited the manuscript. Manabu Kinoshita: conceived and designed the study, acquired imaging data and performed annotations and preprocessing, provided the software for performing radiomics, performed statistical analysis, wrote and edited the manuscript. Koji Fujita: acquired imaging data and performed annotations and preprocessing. Junya Fukai: acquired imaging data and performed annotations and preprocessing. Nobuhide Hayashi: acquired imaging data and performed annotations and preprocessing. Yuji Uematsu: acquired imaging data and performed annotations and preprocessing. Yoshiko Okita: acquired imaging data and performed annotations and preprocessing. Masahiro Nonaka: acquired imaging data and performed annotations and preprocessing. Shusuke Moriuchi: acquired imaging data and performed annotations and preprocessing. Takehiro Uda: acquired imaging data and performed annotations and preprocessing. Naohiro Tsuyuguchi: acquired imaging data and performed annotations and preprocessing. Hideyuki Arita: conducted genetic analyses. Kanji Mori: acquired imaging data and performed annotations and preprocessing. Kenichi Ishibashi: acquired imaging data and performed annotations and preprocessing. Koji Takano: acquired imaging data and performed annotations and preprocessing. Namiko Nishida: acquired imaging data and performed annotations and preprocessing. Tomoko Shofuda: conducted genetic analyses. Ema Yoshioka: conducted genetic analyses. Daisuke Kanematsu: conducted genetic analyses. Yoshinori Kodama: conducted the central pathology review. Masayuki Mano: conducted the central pathology review. Naoyuki Nakao: acquired imaging data and performed annotations and preprocessing. Yonehiro Kanemura: conceived and designed the study, wrote and edited the manuscript. All authors read and approved the manuscript.
Data Availability
Raw analyzed data are available in Supplementary Dataset.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50849-y.
References
- 1.Chinot OL, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J. Clin. Oncol. 2007;25:1470–1475. doi: 10.1200/JCO.2006.07.4807. [DOI] [PubMed] [Google Scholar]
- 2.Kickingereder Philipp, Burth Sina, Wick Antje, Götz Michael, Eidel Oliver, Schlemmer Heinz-Peter, Maier-Hein Klaus H., Wick Wolfgang, Bendszus Martin, Radbruch Alexander, Bonekamp David. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology. 2016;280(3):880–889. doi: 10.1148/radiol.2016160845. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, et al. Relationship between Glioblastoma Heterogeneity and Survival Time: An MR Imaging Texture Analysis. American Journal of Neuroradiology. 2017;38:1–7. doi: 10.3174/ajnr.A5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smits M, van den Bent MJ. Imaging Correlates of Adult Glioma Genotypes. Radiology. 2017;284:316–331. doi: 10.1148/radiol.2017151930. [DOI] [PubMed] [Google Scholar]
- 5.Okita Y, et al. (11)C-methinine uptake correlates with MGMT promoter methylation in nonenhancing gliomas. Clin Neurol Neurosurg. 2014;125:212–216. doi: 10.1016/j.clineuro.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita M, et al. Introduction of High Throughput Magnetic Resonance T2-Weighted Image Texture Analysis for WHO Grade 2 and 3 Gliomas. PLoS ONE. 2016;11:e0164268. doi: 10.1371/journal.pone.0164268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arita H, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126:267–276. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 8.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1006/nimg.2002.1132. [DOI] [PubMed] [Google Scholar]
- 9.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–156. doi: 10.1016/S1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 10.Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage. 2009;48:63–72. doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arita H, et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-30273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bair E, Tibshirani R. Semi-Supervised Methods to Predict Patient Survival from Gene Expression Data. PLoS Biol. 2004;2:e108–12. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pellagatti A, et al. Identification of gene expression-based prognostic markers in the hematopoietic stem cells of patients with myelodysplastic syndromes. J. Clin. Oncol. 2013;31:3557–3564. doi: 10.1200/JCO.2012.45.5626. [DOI] [PubMed] [Google Scholar]
- 14.De Cecco L, Bossi P, Locati L, Canevari S, Licitra L. Comprehensive gene expression meta-analysis of head and neck squamous cell carcinoma microarray data defines a robust survival predictor. Annals of Oncology. 2014;25:1628–1635. doi: 10.1093/annonc/mdu173. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Wang L, Smith JD, Zhang B. Supervised principal component analysis for gene set enrichment of microarray data with continuous or survival outcomes. Bioinformatics. 2008;24:2474–2481. doi: 10.1093/bioinformatics/btn458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drabycz S, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. NeuroImage. 2010;49:1398–1405. doi: 10.1016/j.neuroimage.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro-Oncology. 2016;19:109–117. doi: 10.1093/neuonc/now121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou H, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro-Oncology. 2017;19:862–870. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C-F, et al. Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas. Clin. Cancer Res. 2018;24:4429–4436. doi: 10.1158/1078-0432.CCR-17-3445. [DOI] [PubMed] [Google Scholar]
- 20.Shinohara RT, et al. Statistical normalization techniques for magnetic resonance imaging. NeuroImage: Clinical. 2014;6:9–19. doi: 10.1016/j.nicl.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellingson B.M., Lai A., Harris R.J., Selfridge J.M., Yong W.H., Das K., Pope W.B., Nghiemphu P.L., Vinters H.V., Liau L.M., Mischel P.S., Cloughesy T.F. Probabilistic Radiographic Atlas of Glioblastoma Phenotypes. American Journal of Neuroradiology. 2012;34(3):533–540. doi: 10.3174/ajnr.A3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellingson BM, et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: A radiographic study in 358 de novo human glioblastomas. NeuroImage. 2012;59:908–916. doi: 10.1016/j.neuroimage.2011.09.076. [DOI] [PubMed] [Google Scholar]
- 23.Kanas VG, et al. Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput Methods Programs Biomed. 2017;140:249–257. doi: 10.1016/j.cmpb.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Ellingson BM, et al. Validation of postoperative residual contrast-enhancing tumor volume as an independent prognostic factor for overall survival in newly diagnosed glioblastoma. Neuro-Oncology. 2018;20:1240–1250. doi: 10.1093/neuonc/noy053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellingson Benjamin M, Aftab Dana T, Schwab Gisela M, Hessel Colin, Harris Robert J, Woodworth Davis C, Leu Kevin, Chakhoyan Ararat, Raymond Catalina, Drappatz Jan, de Groot John, Prados Michael D, Reardon David A, Schiff David, Chamberlain Marc, Mikkelsen Tom, Desjardins Annick, Holland Jaymes, Ping Jerry, Weitzman Ron, Wen Patrick Y, Cloughesy Timothy F. Volumetric response quantified using T1 subtraction predicts long-term survival benefit from cabozantinib monotherapy in recurrent glioblastoma. Neuro-Oncology. 2018;20(10):1411–1418. doi: 10.1093/neuonc/noy054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellingson BM, et al. Baseline pretreatment contrast enhancing tumor volume including central necrosis is a prognostic factor in recurrent glioblastoma: evidence from single and multicenter trials. Neuro-Oncology. 2016;19:89–98. doi: 10.1093/neuonc/now187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kickingereder Philipp, Neuberger Ulf, Bonekamp David, Piechotta Paula L, Götz Michael, Wick Antje, Sill Martin, Kratz Annekathrin, Shinohara Russell T, Jones David T W, Radbruch Alexander, Muschelli John, Unterberg Andreas, Debus Jürgen, Schlemmer Heinz-Peter, Herold-Mende Christel, Pfister Stefan, von Deimling Andreas, Wick Wolfgang, Capper David, Maier-Hein Klaus H, Bendszus Martin. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro-Oncology. 2017;20(6):848–857. doi: 10.1093/neuonc/nox188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw analyzed data are available in Supplementary Dataset.