Abstract
This study investigated the non-thermal effects of Wi-Fi radiofrequency radiation of 2.4 GHz on global gene expression in Escherichia coli K-12 DH5α. High-throughput RNA-sequencing of 2.4 GHz exposed and non-exposed bacteria revealed that 101 genes were differentially expressed (DEGs) at P ≤ 0.05. The up-regulated genes were 52 while the down-regulated ones were 49. QRT-PCR analysis of pgaD, fliC, cheY, malP, malZ, motB, alsC, alsK, appB and appX confirmed the RNA-seq results. About 7% of DEGs are involved in cellular component organization, 6% in response to stress stimulus, 6% in biological regulation, 6% in localization, 5% in locomotion and 3% in cell adhesion. Database for annotation, visualization and integrated discovery (DAVID) functional clustering revealed that DEGs with high enrichment score included genes for localization of cell, locomotion, chemotaxis, response to external stimulus and cell adhesion. Kyoto encyclopedia of genes and genomes (KEGG) pathways analysis showed that the pathways for flagellar assembly, chemotaxis and two-component system were affected. Go enrichment analysis indicated that the up-regulated DEGs are involved in metabolic pathways, transposition, response to stimuli, motility, chemotaxis and cell adhesion. The down-regulated DEGs are associated with metabolic pathways and localization of ions and organic molecules. Therefore, the exposure of E. coli DH5α to Wi-Fi radiofrequency radiation for 5 hours influenced several bacterial cellular and metabolic processes.
Subject terms: High-throughput screening, Transcriptomics, Bacterial genetics, Gene ontology, Functional clustering
Introduction
Electromagnetic Fields (EMF) effects on living organisms have been an important research topic for many years. Wireless fidelity (Wi-Fi) waves are part of the non-ionizing radiation of the electromagnetic spectrum. Several studies examined the non-thermal effects of high frequency electromagnetic fields of mobile phones and Wi-Fi on different strains of bacteria1–8. We have found previously that Wi-Fi exposure of Escherichia coli O157H7 increased antibiotic resistance, motility and ability to form biofilm9. Bacterial resistance is expanding to most commonly used antibiotics that has been considered as “global health crisis” by the World Health Organization (WHO)10. Flagella represent a critical virulence factor that permit bacterial motility and promote adhesion to the gastrointestinal mucins11. During environmental stress, bacteria produce a polysaccharide matrix and aggregate to form biofilms12. These virulence factors play key roles in infection initiation and development of diseases.
Gene expression in E. coli is influenced by environmental factors such as temperature, pH, and other stress factors13–16. Bacteria may activate strategies to adapt to various environmental stress. Here, we investigated the changes in global transcriptome in E. coli K12 DH5α after exposure to 2.4 GHz EMF emitted from a Wi-Fi router using high-throughput RNA sequencing. Escherichia coli K-12 DH5α strain was constructed by Douglas Hanahan and is a commonly used laboratory strain17,18. We used this strain as a model to understand the effects of Wi-Fi radiofrequency radiation on transcriptomes of E. coli bacteria.
The differences in the expression of selected genes were confirmed by RT-PCR assays.
Results
Transcriptome sequencing of exposed E. coli DH5α to Wi-Fi radiofrequency radiation
The whole transcriptome sequencing was performed to study the effects of Wi-Fi exposure on global gene expression in Escherichia coli DH5α. RNA-seq experiments were performed in triplicates of Wi-Fi radiation exposed and non-exposed bacteria. The trimmed reads were mapped to the reference genome using Bowtie software19. The reads per kilobase million (RPKM) value of the genes obtained through the RNA-seq were used as the original raw data. A number of 83 genes with zero RPKMs in the 6 samples were excluded leaving 4,461 genes. During data preprocessing, low quality transcripts were filtered resulting in 4,378 genes to be studied. The distribution of gene expression between the unexposed and exposed samples is represented by the volcano plot (Fig. 1). The genes located out of the borders of the line fold change FC ≥1.2 were considered to be differentially expressed genes (DEGs). The number of DEGs at FC ≥1.2 without P-value restraint was 468, which was reduced to 101 at P < 0.05. Genes situated from the left boundary are down-regulated while those at the right are up-regulated genes. In that data set, 52 genes were significantly up-regulated and 49 genes were down-regulated after the exposure to 2.4 GHz Wi-Fi radiation.
Figure 1.
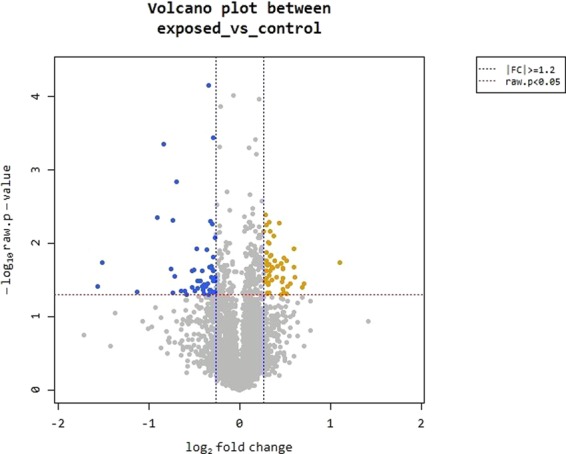
Volcano plot highlighting differentially expressed genes between unexposed and exposed bacteria. The genes are colored if they pass the thresholds for −log10 P value (P value = 0.05) and log fold change |FC| ≥1.2, yellow if they are up-regulated and blue if they are down-regulated.
Hierarchical clustering analysis of 101 DEGs was performed to group similar samples and genes. These results were graphically depicted using heat map and dendogram (Fig. 2). Heat map shows the results of hierarchical clustering analysis which clusters genes and samples by expression level from significant list.
Figure 2.
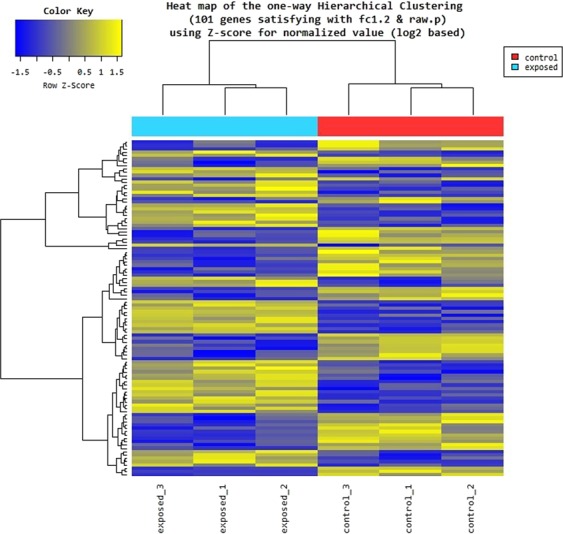
Heat map of DEGs in control and 2.4 GHz exposed bacteria (3 replicates each). Expression level is color patterned: yellow for up-regulated genes, gray for unchanged expression, and blue for down-regulated genes.
Functional classification of DEGs based on gene ontology
Gene ontology (GO) analysis classifies the differentially expressed genes into three groups: biological processes, molecular functions and cellular components. The 101 DEGs at FC ≥1.2 and P-value = 0.05 were classified into eight functional groups of the biological processes category (Fig. 3). About 36% of the DEGs are implicated in cellular process, 27% in metabolic processes, 7% in cellular component organization and 6% in biological regulation. In addition, 6% of the DEGs are involved in response to stimulus, 5% in localization, 5% in locomotion and 3% in cell adhesion.
Figure 3.
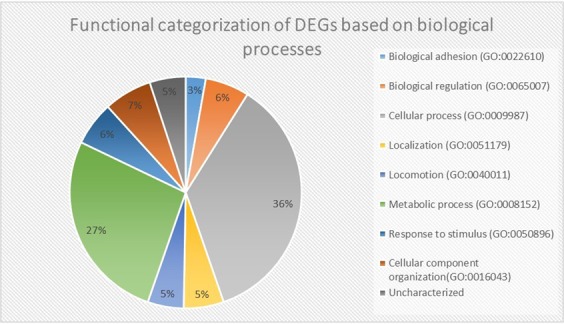
Gene ontology analysis of DEGs after exposure of E. coli DH5α to 2.4 GHz Wi-Fi.
Functional annotation clustering
Functional annotation clustering was done by DAVID tools version 6.8 to evaluate genes differentially expressed between the exposed and control bacterial cells20,21. Based on the enrichment score (ES), functional annotation clustering produced a total of 18 clusters from the 101 DEGs. The most enriched clusters were composed of genes involved in locomotion, localization of cell and bacterial-type flagellum-dependent cell motility (ES = 3.74) and chemotaxis and response to external stimuli (ES = 2.00). They also include genes associated with cell adhesion (ES = 1.08), cellular component organization (ES = 0.71), DNA repair and metabolism (ES = 0.63) and metabolic processes (ES = 0.53) (Table 1).
Table 1.
Summary of functional annotation clustering analysis by DAVID tools.
| Functional Annotation Cluster | Enrichment Score | Gene Count |
|---|---|---|
| Annotation Cluster 1 | 3.74 | |
| Locomotion | 9 | |
| Localization of cell | 8 | |
| Bacterial-type flagellum-dependent cell motility | 7 | |
| Annotation Cluster 2 | 2.00 | |
| chemotaxis | 5 | |
| Response to external stimulus and chemicals | 8 | |
| Annotation Cluster 3 | 1.08 | |
| Cell adhesion | 4 | |
| Annotation Cluster 4 | 0.71 | |
| Cellular component organization or biogenesis | 8 | |
| Annotation Cluster 5 | 0.63 | |
| DNA repair | 4 | |
| DNA metabolism | 6 | |
| Annotation Cluster 5 | 0.54 | |
| Nucleotide biosynthetic process | 5 | |
| Carbohydrate derivative metabolic process | 7 | |
| Organonitrogen compound metabolic process | 12 | |
Clusters involved in the same biological process were merged together. ES threshold was set to 0.5 and gene count ≥4.
GO enrichment analysis of DEGs
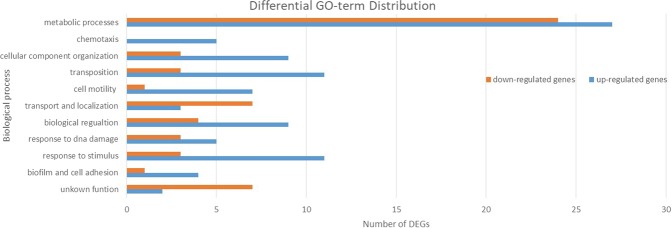
The different functional networks for down-regulated and up-regulated genes are shown in Fig. 4. Among the 52 up-regulated genes 50 were assigned to GO functions. The 49 down-regulated genes contained 42 with GO functions. The other 9 genes in the DEGs were coding for t-RNA or have uncharacterized functions. The entire list of the up-regulated and down-regulated genes can be found as Supplementary Tables S1 and S2. The higher percentage of DEGs are implicated in metabolic processes. The up-regulated DEGs are involved in cell motility, cell adhesion, chemotaxis, transposition, cellular component organization, response to stimulus and DNA damage. Most of the down-regulated DEGs are associated with transport and localization of nitrogen compounds, organic substances and ions (Fig. 4).
Figure 4.
GO term enrichment analysis of up- and down-regulated differentially expressed genes.
KEGG pathways analysis
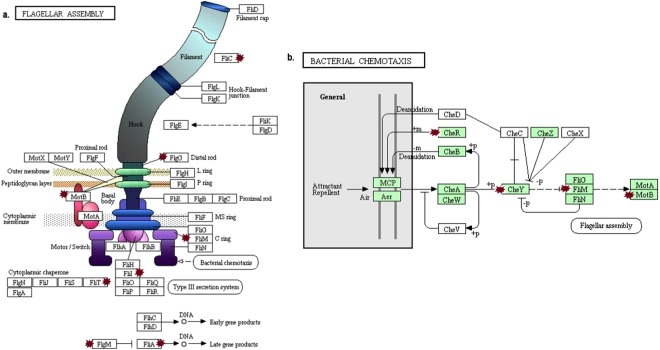
Kyoto encyclopedia of genes and genomes (KEGG) pathways were obtained within the analysis by DAVID22. KEGG Pathways affected by the exposure of 2.4 GHz Wi-Fi radiation are: flagellar assembly, bacterial chemotaxis and two-component system.
Figure 5 shows the genes involved in the synthesis, assembly of flagella and chemotaxis. The genes flgG, motB, fliM, fliL, fliT, fliC, fliA, flgM, cheR and cheY were up-regulated by the exposure to Wi-Fi waves. FLgG is a protein part of the flagellar basal-body which connects to components of the flagellar motor. MotB and FliM are elements of the flagellar motor required for the rotation of the flagellar23. FliL controls the rotational direction of flagella during chemotaxis. FliT acts as an export chaperone for the filament-capping protein FliD. The fliC gene code for the subunit protein flagellin which is polymerized to form the filaments of bacterial flagella. FliA (σ28) and FlgM (anti- σ28) control the expression of late flagella-related genes24. CheR is responsible for the methylation of the membrane-bound methyl-accepting chemotaxis proteins and CheY is implicated in the transduction of signals from the chemoreceptors to the flagellar motors23.
Figure 5.
Flagellar pathway assembly and bacterial chemotaxis from KEGG database. Red stars point to the up-regulated genes affected in (a) the flagelar assembly pathway (ko02040). (b) the bacterial chemotaxis pathway (ko02030). Some of the affected genes belong to the two-component system pathway (ko02020).
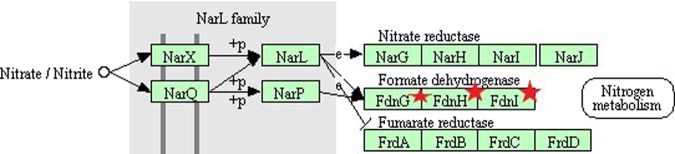
Figure 6 shows the genes involved in nitrogen metabolism. The genes fdnG, fdnI, and fdnH are down-regulated by the exposure to Wi-Fi radiation and are part of the two-component system pathway. These genes code for the subunits of the protein formate dehydrogenase25. During anaerobic respiration, E. coli utilize formate as electron donor and nitrate as electron acceptor through formate dehydrogenase26.
Figure 6.
Two-component system pathway from KEGG database (ko02020). Red stars point to the down-regulated genes affected in the nitrogen metabolism.
Validation of several DEGs by quantitative RT-PCR
We measured the expression of representative DEGs by qRT-PCR. These DEGs examined included pgaD, fliC, cheY, malP, malZ, motB, alsC, alsK, appB and appX. The qRT-PCR results were analyzed by REST 2009 software (Fig. 7). The housekeeping genes gyrA and frr were used for gene normalization. The results from qRT-PCR assays were found to be consistent with that obtained from RNA-sequencing; P < 0.05 (Table 2).
Figure 7.
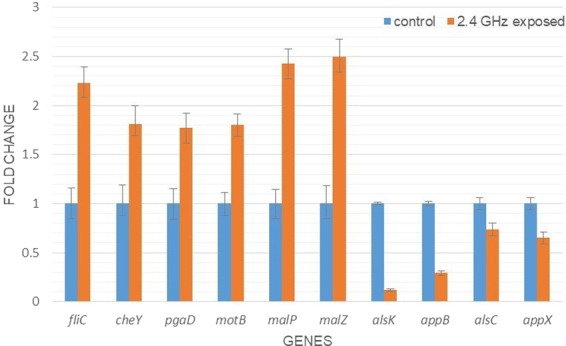
Quantitative real time PCR chart; relative expression level normalized with 2 housekeeping genes: gyrA and frr.
Table 2.
Comparison of fold change difference between RNA-seq and qRT-PCR.
| Gene | RNA-seq | qRT-PCR | ||
|---|---|---|---|---|
| *FC | p-value | **FC [std error] | p-value | |
| pgaD | 1.78 | 0.0329 | 1.775[1.612 to 1.924] | <0.001 |
| fliC | 2.12 | 0.0396 | 2.236[2.085 to 2.399] | <0.001 |
| cheY | 1.76 | 0.0348 | 1.813 [1.692 to 2.001] | <0.001 |
| malP | 2.4 | 0.0286 | 2.426[2.275 to 2.573] | 0.024 |
| malZ | 2.5 | 0.0382 | 2.495[2.339 to 2.674] | 0.039 |
| motB | 1.69 | 0.0355 | 1.804 [1.743 to 1.915] | 0.04 |
| alsC | −1.42 | 0.0388 | −1.35[−1.48 to −1.26] | <0.001 |
| alsK | −8.78 | 0.0459 | −8.33 [−9 to −7.63] | <0.001 |
| appB | −2.47 | 0.0473 | −3.38 [−3.69 to −3.15] | 0.036 |
| appX | −1.5 | 0.0183 | −1.53[−1.7 to −1.41] | 0.033 |
*FC: fold change.
**[std error]: standard error interval.
Discussion
EMF exposure induced different responses in bacteria depending on frequency, intensity, exposure time and the organism model27. Two hours of mobile phone exposure revealed no effects on the growth and susceptibility to different antibiotics of S. aureus, S. epidermis, and P. aeruginosa8. The exposure of S. aureus to mobile phone for one hour did not affect their ability to form biofilm3. Longer time of GSM and Wi-Fi exposure affected the growth, antibiotic susceptibility and biofilm formation of P. aeruginosa, E. coli, S. aureus, S. epidermis and L. monocytogenes6,7,9. The prolonged EMF exposure resulted in major effects in K. pneumonia before they reach an adaptation stage5. Some bacteria with resistance to multidrugs were detected in the vicinity of telecommunication stations28. Furthermore, the microbial growth of human skin microbiota was disrupted after exposure to radiofrequency EMF (RF-EMF)29.
The WHO recommended evaluation of the biological effects of existing EMF exposure in a population before providing permission for establishment of new EMF networks30,31. Understanding the mechanisms by which RF-EMF influences human health is also important to reduce the risk of incidence of diseases32.
In this study, most of the DEGs in DH5α cells exposed to Wi-Fi waves are involved in cellular and metabolic processes. The upregulated sulA, yjjQ, oxC and arsC genes are associated with stress response. The sulA, yadC, sbcB, recT and arsC genes are part of the defense system against DNA damage. SulA in E. coli is induced by the SOS response to DNA damage resulting in an inhibition of cell division by interaction with tubulin-like FTsZ. At the site of cell division, FtsZ assembles into a ring which is essential for bacterial cell division33. In parallel, a reduction of ZipA, coding for a stabilizer protein of FtsZ, was detected. This regulatory network elucidate morphological changes observed after exposure to extremely low frequency EMF (ELF-EMF) by affecting cell division34,35. Only 3 of the genes associated with response to DNA damage, udp, purN and yafO, were down-regulated.
The up-regulated ampE, yadC and ybhG genes are related to antibiotics response. AmpE is implicated in the expression of β-lactamase and plays a role in peptidoglycan murein recycling36,37. YadC is a fimbrial-like protein that is induced immediately after exposure to stress and provide antimicrobial protection38. YbhG is involved in the sensitivity control to chloramphenicol through the efflux pathway39. These results confirm the data of some previous studies indicated altered antibiotic resistance after exposure to RF-EMF5,6,9.
Our data showed that genes involved in chemotaxis and motility such as fliA, flgM, motB, fliC, cheY, cheR, fliM, fliL, flgG and fliT were up-regulated by the exposure to Wi-Fi waves. Overexpression of CheY in association with MotA and MotB increases motility40. These results confirm our prior finding that Wi-Fi radiation enhanced motility of E. coli O157 H7 by 28%9, which may be a strategy of the pathogen for survival after excessive exposure to radiation stress. E. coli O157H7 cells motility was increased under different stress conditions such as heat and acid41,42. Motility of E. coli is important for starting biofilm formation43. We have previously reported that Wi-Fi radiofrequency radiation triggers biofilm formation in E. coli9. Our data indicated that the exposure to Wi-Fi radiofrequency radiation induced the expression of pagD, yadM, yadC and fimI involved in cell adhesion and biofilm formation. The down-regulated genes included yadV which codes for a fimbrial chaperone involved in cell adhesion and yciG that is implicated in flagellum-dependent swarming motility. The changes in bacterial gene expression permit the survival in environments with rapidly changing conditions. Therefore, biofilm formation is a sort of bacterial defense against stressful conditions.
Gene ontology analysis revealed that the down-regulated DEGs are involved mostly in metabolic, cellular process and localization. For example, the ssuA, ppdB, xanP, gadC, ompL, sgcB, yacD and alsC genes are associated with localization and transport of different compounds. The ompL, alsC and sgcB genes have a role in carbohydrate transport. The ompL and gadC genes facilitate ions transport. The gadC, xanP and ppdB genes are related to nitrogen compound transport. In addition, the down-regulated pyrI, purE, pyrB and purN genes are involved in nucleotide biosynthetic pathway. We observed down-regulation of the appX and appB genes, which are part of cytochrome bd-II oxidase and are involved in aerobic electron transport chain44. Moreover, down-regulation of the ldrA, ldrB and ldrC genes type I toxin-antitoxin system that inhibits cell growth, were detected45. Many genes involved in anaerobic respiration were down-regulated. For instance, the down-regulated fdnI, fdnH and fdnG genes belongs to the two-component system pathway and plays a role in nitrogen metabolism25. The down-regulated hyaA, hyaB and hyaD genes are involved in fermentation. hya expression is affected by nutrients starvation and external pH and hence it is involved in response to stress46.
As consequence of regulatory network, we detected increased expression of yhdX and agaC genes that play a role in transport of amino acids and sugar, respectively. The up-regulated puuC, eutP, metC, kbaY genes are involved in metabolic pathways. In addition, we observed up-regulation of malP and malZ genes, part of the maltodextrin system, which are involved in the uptake of maltose by ABC transporter47.
We found that 11 of the up-regulated and 3 of the down-regulated genes are associated with bacterial transposition activity. Transposases increase genomic rearrangements by incorporation of adjacent genes during transpositions48. Nutritional and cellular stress as well as environmental adaptation trigger transposition in Escherichia coli49,50. It has been reported that extremely low frequency EMF affected transposition activity in E. coli51.
Microorganisms often switch between phases of growth and non-growth to adapt to environmental conditions and they can enter dormant state. In this viable but non-culturable state, bacteria can still preserve certain metabolic activity but are incapable to divide due to their minor adaptation capacity52. Alterations in bacterial gene expression allow bacteria to resist the stress conditions. Our data showed that both genes involved in aerobic and anaerobic respiration were down-regulated after exposure to Wi-Fi radiofrequency radiation. Decreased metabolic activity in bacteria has been linked with increased persistence. Part of the bacterial population enters into dormant state through metabolic inactivity and growth arrest in order to survive in the stress conditions53. Other persistence mechanisms are also found in E. coli such as SOS response, DNA repair, pump efflux and biofilm formation52,54,55. On the other hand, the exposure of bacteria to EMF can be valuable when interfering with these phenomena and may compromise therapeutic success. It has been shown that the exposure to ELF-EMF resulted in antibacterial effects as well as altered bacterial morphology and decreased the viability in biofilms suggesting a reduced persistence35,56,57. Further research is required to clarify the effects of EMF on their persistence and therapeutic success.
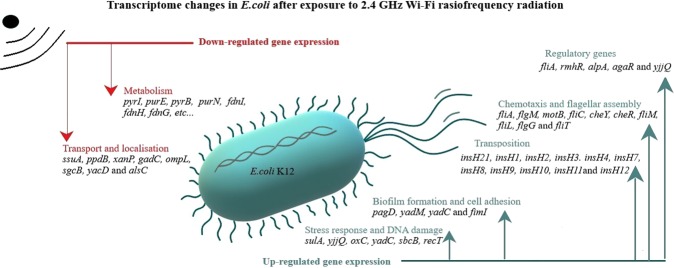
The changes in bacterial gene expression in response to exposure to Wi-Fi waves are summarized in Fig. 8. Our results showed alterations in the transcription of genes involved in metabolism, transport, flagellar assembly, response to DNA damage, transposition and biofilm formation. In addition, there was increased transcription of five regulatory genes agaR, yjjQ, alpA, rmhR and fliA after exposure to Wi-Fi waves. agaR is a repressor that regulate aga operon which is related to transportation and degradation of N-acetylgalactosamine58. The transcriptional regulator YjjQ is part of the LuxR family and play a role in detoxification of methylglyoxal59. AlpA is a transcriptional regulator of intA expression which results in excision of the cryptic prophage CP4-57 and play a role in biofilm formation60,61. RmhR is a transcriptional factor part of the rhm operon involved in L-rhamnonate utilization62. Moreover, sigma factor FliA (σ28) control the expression of late flagella-related genes24. Therefore, the exposure of E. coli to Wi-Fi radiofrequency radiation influenced the transcription of a network of regulatory genes that affected several biological processes.
Figure 8.
Proposal hypothesis for the mechanisms by which 2.4 GHz EMF influences the bacterial transciptome of E. coli K12.
Several studies investigated the effects of exposing human and rodent cells to Wi-Fi radiofrequency radiation on gene expression. Exposure of human cells HL-60 to 2.4 GHz waves for 2 h altered the expression of 221 genes. The up-regulated ones were associated with stress response and apoptosis while the down-regulated genes were involved in cell cycle, metabolism and transport63. Moreover, the exposure of rat hippocampus to Wi-Fi radiation for 12 hours influenced the expression of 41 genes related to heat shock proteins, metabolism and signal transduction64. Other studies showed that long term exposure to Wi-Fi waves changed microRNAs levels and gene expression in rat brain65,66.
In summary, the exposure of E. coli DH5α to Wi-Fi radiofrequency radiation for 5 hours up-regulated 52 genes and down-regulated 49 genes. The expression of detected genes was confirmed by qRT-PCR assays. Gene ontology analysis showed that the higher percentage of DEGs are implicated in cellular and metabolic processes. Most of up-regulated DEGs plays a role in transposition, response to stimuli, motility and chemotaxis while the down-regulated DEGs are mainly associated with transport and localization of nitrogen compounds, organic substances and ions. DAVID functional clustering indicated that DEGs with high enrichment score for localization of cell and bacterial-type flagellum-dependent cell motility was 3.74. The ES for chemotaxis and response to external stimulus was 2 and for cell adhesion was 1.08. KEGG pathways analysis revealed that the pathways for flagellar assembly, bacterial chemotaxis and two-component system were affected after the exposure to 2.4 GHz Wi-Fi radiation. Our results showed that the exposure to these waves influenced transcriptomes responsible for metabolic and cellular processes, localization, stress response, transposition, motility, chemotaxis and cell adhesion.
This is the first report investigating the alterations in the bacterial transcriptome profiling after exposure to Wi-Fi radiofrequency radiation. Detailed information of RNA-seq analysis following Wi-Fi radiation exposure in E. coli could be valuable to understand the effects of Wi-Fi radiation on pathogenic traits of bacteria, particularly antibiotic resistance, motility and biofilm formation. The results of this study open the door for further investigation of the mechanisms of effects of RF-EMF on pathogenic and non-pathogenic bacteria that may have influence in human health and disease. Further studies are required to explore deeply the mechanisms by which 2.4 GHz EMF influences the bacterial transcriptome. Undervaluing the problem of telecommunication exposure could cause further rise in infectious diseases or their complications.
Materials and Methods
Microwave exposure system
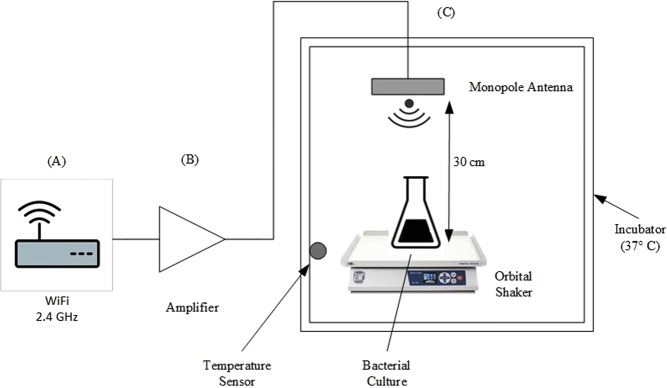
Wi-Fi radiofrequency radiation was generated by a wireless router extended range (TL-WR524G-Tp-Link -China) corresponding to 2.4 GHz frequency, connected to an amplifier and monopole antenna. The mounted system was placed in an incubator at 30 cm from the bacterial culture. E. coli cells were continuously exposed to Wi-Fi radiation for 5 hours (Fig. 9). The control bacteria were placed in Faraday bags to their exposure to limit any exterior radiation.
Figure 9.
Schematic flow chart of the Wi-Fi radiofrequency exposure model. (A) Wi-Fi router (B) amplifier (C) monopole antenna mounted in an incubator chamber with temperature control.
Bacterial growth conditions
E. coli K12 DH5α transformed with puc18 plasmid (Sigma-Aldrich-Germany) were cultured in Luria Bertani (LB) medium with 100 μg/ml ampicillin (Sigma-Aldrich-Germany) under aerobic conditions. Overnight cultures were inoculated at a dilution of 1:100 into a new LB medium with ampicillin and were incubated at 37 °C with shaking at 200 rpm until they reached mid-exponential phase (OD600 = 0.6). Bacteria protect reagent (Qiagen-Germany) was added and the bacterial cultures were centrifuged at 4000 rpm for 10 min. The bacterial pellets were collected for extraction of total RNA followed by next generation sequencing and quantitative real-time PCR (qRT-PCR) assays.
Extraction of total RNA and high throughput RNA sequencing
Total RNA was extracted from the Wi-Fi exposed and control bacteria by the bacteria protect-RNeasy kit (QIAGEN-Germany). Any remaining genomic DNA was digested by On-column RNase-Free DNase kit (Qiagen-Germany). The concentration of RNA was measured by a NanoDrop 2000 (Thermo Fisher Scientific- USA). Triplicates of RNA samples with RNA integrity number >7 were used for high throughput sequencing by Macrogen Next Generation Sequencing Division (Macrogen, Seoul, South Korea). Libraries were generated using Ribo-Zero rRNA Removal Kit (Bacteria) and TruSeq RNA Sample Prep (Illumina-USA). Sequencing was performed by NovaSeq. 6000 System (Illumina-USA) according to the user guide (Document #1000000019358). Bowtie aligner 1.1.2 was used to map sequence reads against the GCF_002848225.1_ASM284822v1 genome reference and read numbers were counted using the HTSeq version 0.10.019. Expression profile was calculated for each sample and gene as read count. Differentially expressed genes (DEGs) analysis of the 2.4 GHz exposed bacteria compared to the unexposed bacteria was performed on a comparison pair (exposed vs. control) using reads per kilobase million (RPKM). Genes with a fold change |FC| ≥1.2 and p-value < 0.05 between the two samples were considered as DEGs. The RNA-seq data obtained from this study has been deposited in the NCBI’s Gene Expression Omnibus and is accessible through GEO Series accession number GSE12658467.
Quantitative real-time PCR
cDNA was synthesized by the FIRE Script RT cDNA synthesis kit (Solis Biodyne-Estonia). qRT-PCR was performed using Rotor-Gene Q (QIAGEN-Germany). Each qRT-PCR reaction contained 1 uL cDNA, 5 μL QuantiTect SYBR Green PCR Master mix (Qiagen), and 0.5 μM specific primers. The reaction cycle was: denaturation at 95 °C for 5 min; 45 cycles of denaturation at 95 °C for 10 s, annealing temperature for 30 s and extension at 72 °C for 20 s. The annealing temperatures were experimentally determined using direct PCR. Primers are listed in Table S3 (Supplementary Information).
Data analysis of relative target gene expression
QRT-PCR efficiencies were measured from the slope of a linear regression model for each pair of primers where the reaction efficiency (E) = 10(−1/slope)68,69. The calibration curve was determined by the Ct with serial cDNA concentrations. The results were analyzed by the Rotor gene Q v. 2.3.1 software (QIAGEN-Germany). The relative expressions of pgaD, fliC, cheY, malP, malZ, motB, alsC, alsK, appB and appX were analyzed using Relative Expression Software Tool (REST 2009), taking in consideration the reaction efficiency and reference gene normalization using Pfaffl method68,69 (https://www.gene-quantification.de/rest-2009.html). The results were normalized to the levels of the two housekeeping genes (DNA gyrase subunit A (gyrA) and ribosome-recycling factor (frr)). Statistical significant difference was considered at P ≤ 0.05.
Gene ontology and clusters analysis
Database for annotation, visualization and integrated discovery (DAVID) online tool was used to find out the biological processes of gene ontology (GO) for the differentially expressed genes (http://david.abcc.ncifcrf.gov/)20,21. The functional annotation clustering and Kyoto encyclopedia of genes and genomes (KEGG) pathways were obtained within the analysis by DAVID22. Enrichment score (ES) threshold was fixed to 0.5, and clusters involved in the same biological process were combined. To analyze the co-expression network between the up-regulated and down-regulated DEGs, enrichment analysis was done using UniProt GO Annotation70. For clarity GO classifications by biological function were manually grouped into sub-categories.
Supplementary information
Acknowledgements
This study has been supported by a grant from the Lebanese University to F.A.J.
Author Contributions
We declare that this work was performed by the authors named in this article. All authors wrote, reviewed and approved the manuscript including figures and tables. F.A.J. and M.E.M. conceived the study. I.H.S. carried out the biological experiments, and performed the bioinformatics analysis. M.E.M., H.H.Y. and I.H.S. analyzed the data.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-51046-7.
References
- 1.Chang S-K, et al. Genotoxicity evaluation of electromagnetic fields generated by 835-MHz mobile phone frequency band. Eur. J. Cancer Prev. 2005;14:175–9. doi: 10.1097/00008469-200504000-00014. [DOI] [PubMed] [Google Scholar]
- 2.Cranfield C, Wieser HG, Al Madan J, Dobson J. Preliminary evaluation of nanoscale biogenic magnetite-based ferromagnetic transduction mechanisms for mobile phone bioeffects. IEEE Trans. Nanobioscience. 2003;2:40–3. doi: 10.1109/TNB.2003.810155. [DOI] [PubMed] [Google Scholar]
- 3.Mohd-zain Z, Buniyamin N. Effects of Mobile Phone Generated High Frequency Electromagnetic Field on the Viability and Biofilm Formation of Staphylococcus aureus. World Acad. Sci. Eng. Technol. 2012;6:221–224. [Google Scholar]
- 4.Nasri K, Daghfous D, Landoulsi A. Effects of microwave (2.45 GHz) irradiation on some biological characters of Salmonella typhimurium. Comptes Rendus - Biol. 2013;336:194–202. doi: 10.1016/j.crvi.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Taheri M, et al. Klebsiella pneumonia, a Microorganism that Approves the Non-linear Responses to Antibiotics and Window Theory after Exposure to Wi-Fi 2.4 GHz Electromagnetic Radiofrequency Radiation. J. Biomed. Phys. Eng. 2015;5:115–20. [PMC free article] [PubMed] [Google Scholar]
- 6.Taheri M, et al. Evaluation of the effect of radiofrequency radiation emitted from Wi-Fi router and mobile phone simulator on the antibacterial susceptibility of pathogenic bacteria listeria monocytogenes and Escherichia coli. Dose-Response. 2017;15:1–8. doi: 10.1177/1559325816688527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakouti I, Hobbs G, Teethaisong Y, Phipps D. A demonstration of athermal effects of continuous microwave irradiation on the growth and antibiotic sensitivity of Pseudomonas aeruginosa PAO1. Biotechnol. Prog. 2017;33:37–44. doi: 10.1002/btpr.2392. [DOI] [PubMed] [Google Scholar]
- 8.Salmen SH, Alharbi SA, Faden AA, Wainwright M. Evaluation of effect of high frequency electromagnetic field on growth and antibiotic sensitivity of bacteria. Saudi J. Biol. Sci. 2018;25:105–110. doi: 10.1016/j.sjbs.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salman, I. H. S., Jebaii, F. A., Yusef, H. H. & Moustafa, M. E. Evaluation of Wi-Fi radiation effects on antibiotic susceptibility, metabolic activity and biofilm formation by Escherichia coli O157H7, Staphylococcus aureus and Staphylococcus epidermis. J. Biomed. Phys. Eng. In press (2019). [DOI] [PMC free article] [PubMed]
- 10.High levels of antibiotic resistance found worldwide, new data shows. News release Available at, https://www.who.int/news-room/detail/29-01-2018-high-levels-of-antibiotic-resistance-found-worldwide-new-data-shows (2018).
- 11.Erdem L, Avelino F, Xicohtencatl-cortes J, Giro JA. Host Protein Binding and Adhesive Properties of H6 and H7 Flagella of Attaching and Effacing. Escherichia coli. J. Bacteriol. 2007;189:7426–7435. doi: 10.1128/JB.00464-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the Natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 13.Bury-Moné Stéphanie, Nomane Yanoura, Reymond Nancie, Barbet Romain, Jacquet Eric, Imbeaud Sandrine, Jacq Annick, Bouloc Philippe. Global Analysis of Extracytoplasmic Stress Signaling in Escherichia coli. PLoS Genetics. 2009;5(9):e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basak, S. & Jiang, R. Enhancing E. coli Tolerance towards Oxidative Stress via Engineering Its Global Regulator cAMP Receptor Protein (CRP). PLoS One7 (2012). [DOI] [PMC free article] [PubMed]
- 15.House B, et al. Acid-stress-induced changes in enterohaemorrhagic Escherichia coli O157: H7. virulence. 2009;155:2907–2918. doi: 10.1099/mic.0.025171-0. [DOI] [PubMed] [Google Scholar]
- 16.Guernec A, Robichaud-Rincon P, Saucier L. Whole-genome transcriptional analysis of Escherichia coli during heat inactivation processes related to industrial cooking. Appl. Environ. Microbiol. 2013;79:4940–4950. doi: 10.1128/AEM.00958-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–80. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 18.Taylor RG, Walker DC, Mclnnes RR. E. coli host strains significantly affect the quality of small scale plasmid DNA. Nucleic Acids Res. 1993;21:1677–1678. doi: 10.1093/nar/21.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP Binding Protein YcgR Controls Flagellar Motor Direction and Speed to Affect Chemotaxis by a “Backstop Brake” Mechanism. Mol. Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J. Bacteriol. 1986;168:1315–8. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg BL, Li J, Heider J, Stewart V. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. I. Nucleotide sequence of the fdnGHI operon and evidence that opal (UGA) encodes selenocysteine. J. Biol. Chem. 1991;266:22380–22385. [PubMed] [Google Scholar]
- 26.Jormakka M, Törnroth S, Byrne B, Iwata S. Molecular Basis of Proton Motive Force Generation: Structure of Formate Dehydrogenase-N. Science (80-.). 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 27.Salmen, S. H. Non-Thermal Biological Effects of Electromagnetic Field on Bacteria-A Review. American Journal of Research Communication4 (2016).
- 28.Adebayo E, Adeeyo A, Ayandele A, Omomowo I. Effect of Radiofrequency Radiation from Telecommunication Base Stations on Microbial Diversity and Antibiotic Resistance. J. Appl. Sci. Environ. Manag. 2015;18:669. [Google Scholar]
- 29.Crabtree DPE, Herrera BJ, Kang S. The response of human bacteria to static magnetic field and radiofrequency electromagnetic field. J. Microbiol. 2017;55:809–815. doi: 10.1007/s12275-017-7208-7. [DOI] [PubMed] [Google Scholar]
- 30.WHO | Framework for developing health-based EMF standards. WHO Available at, https://www.who.int/peh-emf/standards/framework/en/. (2016).
- 31.WHO | Standards and Guidelines. WHO Available at, https://www.who.int/peh-emf/standards/en/ (2016).
- 32.Prüss-Ustün A, et al. Diseases due to unhealthy environments: an updated estimate of the global burden of disease attributable to environmental determinants of health. J. Public Health (Bangkok). 2016;39:464–475. doi: 10.1093/pubmed/fdw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J. Bacteriol. 1998;180:3946–53. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inhan-Garip A, et al. Effect of extremely low frequency electromagnetic fields on growth rate and morphology of bacteria. Int. J. Radiat. Biol. 2011;87:1155–1161. doi: 10.3109/09553002.2011.560992. [DOI] [PubMed] [Google Scholar]
- 35.Cellini L, et al. Bacterial response to the exposure of 50 Hz electromagnetic fields. Bioelectromagnetics. 2008;29:302–311. doi: 10.1002/bem.20391. [DOI] [PubMed] [Google Scholar]
- 36.Lindquist S, Galleni M, Lindberg F, Normark S. Signalling proteins in enterobacterial AmpC beta-lactamase regulation. Mol. Microbiol. 1989;3:1091–102. doi: 10.1111/j.1365-2958.1989.tb00259.x. [DOI] [PubMed] [Google Scholar]
- 37.Park, J. T. & Uehara, T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72, 211–27, table of contents (2008). [DOI] [PMC free article] [PubMed]
- 38.Allen MJ, White GF, Morby AP. The response of Escherichia coli to exposure to the biocide polyhexamethylene biguanide. Microbiology. 2006;152:989–1000. doi: 10.1099/mic.0.28643-0. [DOI] [PubMed] [Google Scholar]
- 39.Yamanaka Y, Shimada T, Yamamoto K, Ishihama A. Transcription factor CecR (YbiH) regulates a set of genes affecting the sensitivity of Escherichia coli against cefoperazone and chloramphenicol. Microbiology. 2016;162:1253–1264. doi: 10.1099/mic.0.000292. [DOI] [PubMed] [Google Scholar]
- 40.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP Binding Protein YcgR Controls Flagellar Motor Direction and Speed to Affect Chemotaxis by a ‘Backstop Brake’ Mechanism. Mol. Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung HJ, Bang W, Drake MA. Stress Response of Escherichia coli. Compr. Rev. Food Sci. Food Saf. 2006;5:52–64. doi: 10.1111/j.1541-4337.2006.00002.x. [DOI] [Google Scholar]
- 42.House B, et al. Acid-stress-induced changes in enterohaemorrhagic Escherichia coli O157: H7 virulence. Microbiology. 2009;155:2907–2918. doi: 10.1099/mic.0.025171-0. [DOI] [PubMed] [Google Scholar]
- 43.Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013;37:849–71. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bekker M, de Vries S, Ter Beek A, Hellingwerf KJ, de Mattos MJT. Respiration of Escherichia coli Can Be Fully Uncoupled via the Nonelectrogenic Terminal Cytochrome bd-II Oxidase. J. Bacteriol. 2009;191:5510–5517. doi: 10.1128/JB.00562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi Y, Tokunaga N, Inouye M, Phadtare S. Characterization of LdrA (Long Direct Repeat A) Protein of Escherichia coli. J. Mol. Microbiol. Biotechnol. 2014;24:91–97. doi: 10.1159/000357949. [DOI] [PubMed] [Google Scholar]
- 46.King PW, Przybyla AE. Response of hya expression to external pH in. Escherichia coli. J. Bacteriol. 1999;181:5250–6. doi: 10.1128/jb.181.17.5250-5256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 1998;62:204–29. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Craig, N. L, Craigie, R & Gellert, M. L. A. Mobile DNA II. Am. Soc. Microbiol. DC 1204 (2002).
- 49.Casacuberta E, González J. The impact of transposable elements in environmental adaptation. Mol. Ecol. 2013;22:1503–1517. doi: 10.1111/mec.12170. [DOI] [PubMed] [Google Scholar]
- 50.Twiss E, Coros AM, Tavakoli NP, Derbyshire KM. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol. Microbiol. 2005;57:1593–1607. doi: 10.1111/j.1365-2958.2005.04794.x. [DOI] [PubMed] [Google Scholar]
- 51.Del RB, Bersani F, Agostini C, Mesirca P, Giorgi G. Various effects on transposition activity and survival of Escherichia coli cells due to different ELF-MF signals. Radiat. Environ. Biophys. 2004;43:265–270. doi: 10.1007/s00411-004-0260-9. [DOI] [PubMed] [Google Scholar]
- 52.Wood TK, Knabel SJ, Kwan BW. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013;79:7116–21. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dörr T, Vulić M, Lewis K. Ciprofloxacin Causes Persister Formation by Inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dörr T, Lewis K, Vulić M. SOS Response Induces Persistence to Fluoroquinolones in Escherichia coli. PLoS Genet. 2009;5:e1000760. doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui P, et al. Identification of Genes Involved in Bacteriostatic Antibiotic-Induced Persister Formation. Front. Microbiol. 2018;9:413. doi: 10.3389/fmicb.2018.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zohre R, Ali Y, Mostafa J, Samaneh R. Nondrug Antimicrobial Techniques: Electromagnetic Fields and Photodynamic Therapy. Biomed. Pharmacol. J. 2015;8:147–155. doi: 10.13005/bpj/571. [DOI] [Google Scholar]
- 57.Di Campli E, Di Bartolomeo S, Grande R, Di Giulio M, Cellini L. Effects of Extremely Low-Frequency Electromagnetic Fields on Helicobacter pylori Biofilm. Curr. Microbiol. 2010;60:412–418. doi: 10.1007/s00284-009-9558-9. [DOI] [PubMed] [Google Scholar]
- 58.Ray WK, Larson TJ. Application of AgaR repressor and dominant repressor variants for verification of a gene cluster involved in N-acetylgalactosamine metabolism in Escherichia coli K-12. Mol. Microbiol. 2003;51:813–826. doi: 10.1046/j.1365-2958.2003.03868.x. [DOI] [PubMed] [Google Scholar]
- 59.Kim I, Kim J, Min B, Lee C, Park C. Screening of genes related to methylglyoxal susceptibility. J. Microbiol. 2007;45:339–43. [PubMed] [Google Scholar]
- 60.Herzberg M, Kaye IK, Peti W, Wood TK. YdgG (TqsA) Controls Biofilm Formation in Escherichia coli K-12 through Autoinducer 2 Transport. J. Bacteriol. 2006;188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirby JE, Trempy JE, Gottesman S. Excision of a P4-like cryptic prophage leads to Alp protease expression in. Escherichia coli. J. Bacteriol. 1994;176:2068–2081. doi: 10.1128/jb.176.7.2068-2081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rakus JF, et al. Evolution of Enzymatic Activities in the Enolase Superfamily: l -Rhamnonate Dehydratase † ‡. Biochemistry. 2008;47:9944–9954. doi: 10.1021/bi800914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee S, et al. 2.45 GHz radiofrequency fields alter gene expression in cultured human cells. FEBS Lett. 2005;579:4829–4836. doi: 10.1016/j.febslet.2005.07.063. [DOI] [PubMed] [Google Scholar]
- 64.Yang X-S, et al. Exposure to 2.45GHz electromagnetic fields elicits an HSP-related stress response in rat hippocampus. Brain Res. Bull. 2012;88:371–378. doi: 10.1016/j.brainresbull.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 65.Dasdag S, et al. Eff ects of 2.4 GHz radiofrequency radiation emitted from Wi-Fi equipment on microRNA expression in brain tissue. Int. J. Radiat. Biol. 2015;91:555–561. doi: 10.3109/09553002.2015.1028599. [DOI] [PubMed] [Google Scholar]
- 66.Durna Dastan, S. et al. Hazardous Genomic Bioeffects of Home Wi-Fi Systems. NeuroQuantology16 (2018).
- 67.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.