Abstract
Gut microbiota acts as a barrier against intestinal pathogens, but species-specific protection of the host from infection remains relatively unexplored. Although lactobacilli and bifidobacteria produce beneficial lactic and short-chain fatty acids in the mammalian gut, the significance of intestinal Escherichia coli producing these acids is debatable. Taking a Koch’s postulates approach in reverse, we define Escherichia coli as health-promoting for naturally colonizing the gut of healthy mice and protecting them against intestinal colonization and concomitant mortality by Pseudomonas aeruginosa. Reintroduction of faecal bacteria and E. coli in antibiotic-treated mice establishes a high titre of E. coli in the host intestine and increases defence against P. aeruginosa colonization and mortality. Strikingly, high sugar concentration favours E. coli fermentation to lactic and acetic acid and inhibits P. aeruginosa growth and virulence in aerobic cultures and in a model of aerobic metabolism in flies, while dietary vegetable fats - not carbohydrates or proteins - favour E. coli fermentation and protect the host in the anaerobic mouse gut. Thus E. coli metabolic output is an important indicator of resistance to infection. Our work may also suggest that the lack of antimicrobial bacterial metabolites in mammalian lungs and wounds allows P. aeruginosa to be a formidable microbe at these sites.
Subject terms: Bacterial genetics, Pathogens
Introduction
Escherichia coli and streptococci are the first bacteria to colonize the gastrointestinal tract of humans upon birth, paving the way for the establishment of species of the Bifidobacterium, Bacteroides and other genera1. Bifidobacterium and Lactobacillus strains are considered efficient fermenters in the human gut2,3. E. coli on the other hand thrives aerobically, but may also ferment carbon sources anaerobically to produce short-chain fatty acids (SCFAs) such as acetic acid and related metabolic products such as lactic acid4,5. While it is an effective colonizer of the healthy anaerobic mammalian gut, commensal E. coli also has a fitness advantage upon gut inflammation and concomitant host-derived nitrate production5. Interestingly, the probiotic E. coli strain Nissle 1917 (EcN) is particularly beneficial to ulcerative colitis patients in maintaining disease remission6–8. EcN induces host immune defence against pathogens9,10, strengthens the intestinal barrier11,12, and directly inhibits pathogenic E. coli strains13,14. Yet the beneficial role of E. coli has so far only been demonstrated for EcN and is not linked to lactic acid and SCFA production, while lactic acid bacteria, such as lactobacilli and bifidobacteria, are considered the main probiotic fermenters in the mammalian gut.
Antibiotics can greatly reduce microbiota diversity and promote dysbiosis early in life15. In children and adults, opportunistic pathogens can take advantage of the antibiotic effect on commensal bacteria to infect the gut16. One such pathogen is the gram-negative human opportunistic bacterium Pseudomonas aeruginosa, which is frequently found in hospital-acquired infections17. While not a common clinical problem in the gut, P. aeruginosa colonizes the gastrointestinal tract of many hospitalized patients and to a lesser extent of healthy individuals18–21. P. aeruginosa can nevertheless cause frequent and severe wound and lung infections in immunocompromised individuals and the ears and eyes of seemingly healthy people22. It is responsible for more than 50,000 infections per year in the U.S. alone, causing acute, chronic and relapsing infections due to a wide variety of virulence factors. Many of its virulence genes are controlled by quorum sensing (QS), a bacterial communication system that promotes synchronized microbial behaviours such as the production of the oxidative agent pyocyanin by P. aeruginosa23.
Here we interrogate the contribution of E. coli in controlling P. aeruginosa intestinal colonization in a nutrient-dependent manner. We apply the Koch’s postulates in reverse to prove a causal role of commensal E. coli in fending off P. aeruginosa infection. We found that: (a) E. coli is detectable through culture-independent methods (16S sequencing) in the faeces of untreated mice but not of antibiotic-treated mice, which become susceptible to infection; (b) A candidate health-promoting commensal E. coli strain was isolated through culture-dependent microbiological analysis and archived as a pure culture in the laboratory; (c) This mouse E. coli strain and other E. coli strains ameliorate P. aeruginosa infection when introduced into antibiotic-treated mice; (d) The administered health-promoting E. coli strains can be identified in high titres in the faeces of mice in which resistance to infection was improved. Moreover, assessing three extremes and a conventional diet in mice we find that, while sugars are fermented by various E. coli strains to lactic and acetic acid in culture and in flies aerobically, in the anaerobic mouse gut a vegetable-fat-based rather than a carbohydrate- or protein-based diet boosts lactic acid production and helps E. coli to inhibit P. aeruginosa. Our findings support the notion that unbalanced diets or the use of antibiotics may eliminate not only lactic acid bacteria but also commensal E. coli, imposing a gut environment conducive to P. aeruginosa infection due to the depletion of lactic acid and SCFAs.
Methods
Bacterial strains
Pseudomonas aeruginosa strain UCCBP 14 (PA14) and isogenic gene deletion mutants Δmvfr, ΔphzS, ΔphzS and ΔrhlR/ΔlasR were previously described24,25. E. coli MGH is a human isolate obtained from Prof. Elizabeth Hohmann at Mass General Hospital (Boston, USA). Mouse E. coli (E. coli CD1) was isolated from the faeces of CD1 mice for this study and validated through colony PCR and biochemical analysis i.e. being positive for indole production and growth on selective chromogenic Tryptone Bile X-glucuronide (TBX) agar plates. Laboratory E. coli BW25113 and KEIO collection strains, including Δpgi, ΔadhE, ΔatpC, Δpta and ΔldhA, were previously described26. Laboratory E. coli BW25113 and Δtna, ΔsdiA, ΔluxS, strains were previously described27. Enteropathogenic (EPEC) E. coli O127:H6 E2348/69 was obtained from Prof. Tassos Economou and was previously described28.
Bacteria handling for in-culture experiments
E. coli and P. aeruginosa strains were grown at 37 °C overnight with shaking at 200 rpm in liquid LB from frozen LB-20% glycerol stocks. Cultures were then diluted to OD600nm 0.01 in fresh sterile LB to establish mono- or co-cultures. Sucrose or glucose was added to a final concentration of 4% w/v during growth assessments. Bacterial supernatants were produced by overnight bacterial cultures filter-sterilized and mixed in 1:1 volume ratio with fresh LB broth. Selective plates contained 50 μg/ml rifampicin for P. aeruginosa and 60 μg/ml kanamycin for E. coli Keio collection or TBX agar for wild-type E. coli.
Fly survival
For aerobic growth, strains were grown at 37 °C overnight with shaking at 200 rpm in liquid LB from frozen LB-20% glycerol stocks and then diluted to OD600nm 0.01 in fresh sterile LB grown over day to OD600nm 3. For anaerobic growth, strains were grown at 37 °C for 72 hours without shaking in liquid BHI from frozen BHI-20% glycerol stocks to OD600nm 1–2. Cultures were then pelleted and diluted to a final OD600nm 0.15 per strain in a 4% sugar (sucrose or glucose), 10% sterile LB infection medium. Wild-type Oregon R Drosophila melanogaster female flies 3–5 days old were starved for 6 hours prior to infection. 5 ml infection medium was added on a cotton ball at the bottom of a fly vial. Each vial contained 10 to 15 flies and observed twice a day for fly survival29.
Fly colonization
Germ-free flies were generated through dechorionation of collected eggs in 50% bleach. Adult Oregon R 3–5-day-old female flies were infected for 24 hours with a single bacterial culture or a mix of cultures grown as mentioned above, pelleted and diluted to a final OD600nm 0.02 per strain in a 4% sugar (sucrose or glucose) medium. Flies were then transferred to modified falcon tubes and maintained there with 200 μl 2% or 4% of sucrose or glucose as previously described24. At day 2 and day 5 flies were homogenized using the Qiagen Tissuelyser LT for 5 minutes at 50 Hz. Bacteria CFUs were enumerated on selective plates after overnight incubation at 37 °C.
KEIO E.coli gene deletion library screen
The Keio E. coli collection of gene knockouts was acquired from the Japanese National Institute of Genetics and contains 3884 E. coli mutants with unique gene deletions. Strains were grown overnight in sterile 96-well clear flat bottom plates containing 200 μl of sterile LB broth at 37 °C and 200 rpm shaking. P. aeruginosa was grown in glass tubes at standard overnight conditions. Over day co-cultures were incubated at 37 °C and 200 rpm in 96-well plates starting with 1:100 dilutions of P. aeruginosa and E. coli mutant overnight cultures in 200 μl LB broth supplemented with 4% glucose. At 24 hours pyocyanin production was observed visually using as positive controls PA14 monocultures and co-cultures of PA14 with E. coli mutants lacking inhibitory properties (e.g. Δpgi). Bacterial growth was measured at OD600nm on a plate reader. Bacterial co-cultures typically exhibit half the optical density of PA14 monocultures. Thus co-cultures with optical density equal to or higher than PA14 monocultures indicated antagonistic interactions.
Animal diets
Drosophila melanogaster Oregon R flies were reared in a cornmeal, yeast and sugar diet at 25 °C in a 12-hour day and night cycle. CD1 mice were reared 5–6 individuals per cage at 24 °C in a 12-hour day and night cycle. Standard chow diet was obtained from Mucedola s.r.l Italy (#4RF25 a complete balanced diet containing mainly starch 35.18%, sucrose 5.66%, crude protein 22%, crude oil 3.5%). Specialized diets based on either vegetable fats, carbohydrates or protein were manufactured by Mucedola s.r.l (#PF4550, PF4551 and PF4552) per Table 1 below30.
Table 1.
Composition of macronutrient diets (% by weight).
| Carbohydrate | Fat | Protein | |
|---|---|---|---|
| Corn starch | 58.11 | 0.00 | 0.00 |
| Powdered sugar | 29.06 | 0.00 | 0.00 |
| Casein | 0.00 | 0.00 | 87.17 |
| dl-Methionine | 0.11 | 0.20 | 0.11 |
| Vegetable shortening* | 0.00 | 75.12 | 0.00 |
| AIN-76A vitamin mix** | 0.77 | 1.49 | 0.77 |
| AIN-76A mineral mix** | 3.07 | 5.95 | 3.07 |
| Choline chloride | 0.18 | 0.34 | 0.18 |
| Cellulose (Alphacel) | 8.72 | 16.91 | 8.72 |
| Energy density, kcal/g | 3.53 | 6.85 | 3.53 |
*Crisco brand, a blend of soybean oil, fully hydrogenated palm oil, and partially hydrogenated palm and soybean oils. Contains 50% polyunsaturated fat, 20.8% monounsaturated fat, 0% trans fat and 25% saturated fat per weight.
**Vitamin (A and D3) and mineral (Fe, Mn, Zn, Cu, I, Se) mixes contain 97% and 12% sucrose, respectively.
Ethics statement
Animal protocols were approved by the Cyprus Veterinary Service inspectors under the license number CY/EXP/PR.L6/2018 for the Laboratory of Prof. Apidianakis at the University of Cyprus. The veterinary services act under the auspices of the Ministry of Agriculture in Cyprus and the project number is CY.EXP101. These national services abide by the National Law for Animal Welfare of 1994 and 2013 and the Law for Experiments with Animals of 2013 and 2017. All experiments were performed in accordance with these guidelines and regulations.
Mouse colonization assay
Female CD1 mice 7–8 weeks old were treated with an antibiotic cocktail of 0.1 mg/ml Rifampicin, 0.3 mg/ml Ampicillin and 2 mg/ml Streptomycin for 6 days to reduce endogenous gut bacteria. Subsequently, PA14 was provided daily for 7 days in the drinking water prepared from an over-day culture of OD600nm 3, centrifuged at 4610 RCF for 5 minutes to collect bacteria and diluted 1:10 to obtain ~3 × 108 bacteria/ml. Following infection (Day 0 of PA14 colonization) E. coli was provided for 1 day at the same concentration and CFUs for both bacteria were measured every other day from homogenized and plated mouse faeces.
16S Metagenomic
Mouse faecal samples were collected in Eppendorf tubes, weighed, snap frozen and stored at −80 °C. Bacterial DNA was extracted using the QIAamp DNA Stool Mini Kit (Qiagen). 16S Sequencing was performed using the Illumina metagenomics analyser. Kraken software was used to assign taxonomic sequence classification.
Mouse survival assay
Female CD1 mice 7–8 weeks old were given an antibiotic cocktail of 0.1 mg/ml Rifampicin, 0.3 mg/ml Ampicillin and 2 mg/ml Streptomycin in their drinking water for 6 days to reduce endogenous gut bacteria. Subsequently, E. coli strains were provided in drinking water for 24 hours prepared from an over-day culture of OD600nm 3 and/or anaerobic faecal culture grown to its maximum for 2 days, centrifuged at 4610 RCF for 5 minutes to collect bacteria and diluted 1:10 to obtain ~3 × 108 bacteria/ml. The next day P. aeruginosa (strain PA14) was provided daily for 7 days in the drinking water as for E. coli. Then mice were injected intraperitoneally with 150 mg/kg of body weight with cyclophosphamide (CP) and 3 days later with another dose of 100 mg/kg as previously described31. Survival was observed twice a day until all mice die or for up to 1 week.
Acid and sugar measurements
Lactic and acetic acid concentrations in culture supernatants and homogenized mouse faeces (produced via bead homogenization in water) were determined enzymatically using R-Biopharm kits No. 11112821035 and No. 10148261035 respectively, according to manufacturer’s instructions. Sugar concentrations in homogenized mouse faeces were determined using the Megazyme Sucrose/D-Fructose/D-Glucose Assay Kit (K-SUFRG) according to manufacturer’s instructions. Absorbance was measured using the NanoDrop 2000c Spectrophotometer.
Pyocyanin measurement
Overnight PA14 cultures were diluted to OD600nm 1, then 0.25 ml was used to inoculate 25 ml of LB. Cultures were grown at 37 °C, 200 rpm in 250 ml flasks. Supernatants were collected after centrifugation at 4800 RCF for 10 minutes. 4.5 ml of chloroform was added to 7.5 ml of supernatant and vortexed. Samples were then centrifuged at 4800 RCF for 10 minutes. 3 ml of the resulting blue layer at the bottom was transferred to a new tube. 1.5 ml of 0.2 M HCl was added to each tube and vortexed 2 times for 10 seconds. Samples were centrifuged for 3 minutes at 4800 RCF and 1 ml of the pink layer was transferred to cuvettes. Pyocyanin concentration (μg/ml) was calculated by multiplying the spectrophotometric measurements taken at OD520nm by 17.072, then multiplying them again by 1.5 due to the chloroform dilution.
Computational analysis
Pairwise comparisons of bacterial CFUs and other pairwise comparisons were evaluated using the two-sided Student’s t-test for samples of ≥10 and Mann–Whitney U-test or one-way analysis of variance (ANOVA) with post hoc Tukey’s multiple comparison test for samples <10. Survival curves of mice and flies were analysed with the Kaplan-Meier method and the log-rank test. All experiments were repeated at least twice with qualitatively similar results. Gene enrichment analysis was performed using the David’s functional annotation tool. Correlation coefficient (R) significance analyses of mouse faecal acid concentration vs. LT50 was done using Pearson correlation and an n = 6 (the average of six dietary conditions sampling 6 mice for each). The Acetic + Lactic acid Index for each of the 6 dietary conditions was computed by dividing each acid concentration of each dietary condition with the average concentration of that acid in all conditions and adding the normalized values of the two acids. For sucrose assimilation prediction we used BLASTN 2.8.1+ per Zhang et al. 200032 and found (a) an E. coli W sucrose hydrolase (98% identity), (b) a sucrose permease (98% identity), (c) a sucrose-specific IIBC component (100% identity) and (d) a sucrose-6-phosphate hydrolase (100% identity) present in E. coli O127:H6 str. E2348/69 (taxid:574521), but not in the genomes of E. coli BW25113 (taxid:679895) and E. coli DH5[alpha] (taxid:668369).
Results
Escherichia coli-secreted factors antagonize Pseudomonas aeruginosa growth in the presence of sugars
Screening for bacterial strains that may alleviate P. aeruginosa infection in Drosophila, we compared and combined the highly virulent P. aeruginosa strain, PA14, which kills all orally infected flies within 6 days29, with various E. coli strains (MGH, EPEC, BW25113 and DH5a), none of which was by itself significantly lethal to flies. The fly lethal time 50% (LT50%) extended beyond the 15 days for all E. coli strains, as exemplified with E. coli MGH and BW25113 shown in Fig. 1a and Suppl. Fig. 1a. Strikingly, P. aeruginosa-mediated fly lethality, fly colonization and P. aeruginosa growth in culture was dramatically inhibited by the human E. coli isolate MGH (Fig. 1a–d) and the laboratory E. coli strain BW25113 (Suppl. Fig. 1a–d) in the presence of 4% sucrose or 4% glucose, respectively. Noticeably, sucrose can be used by the E. coli strains MGH and EPEC to inhibit P. aeruginosa lethality and growth (fly LT50% > 10 days; Fig. 1a–d), because EPEC, for example, has 4 sucrose uptake and metabolism enzymes, namely, an E. coli W sucrose hydrolase, a sucrose permease, a sucrose-specific IIBC component and a sucrose-6-phosphate hydrolase. In contrast, the E. coli strains BW25113 and DH5a do not have these genes and were unable to utilize sucrose to inhibit P. aeruginosa in our experiments (fly LT50% < 7 days). As expected, when 4% glucose instead of sucrose was used in the infection mix, E. coli BW25113 gained the capacity to inhibit P. aeruginosa lethality, fly colonization and in culture growth (Suppl. Fig. 1a–d)33.
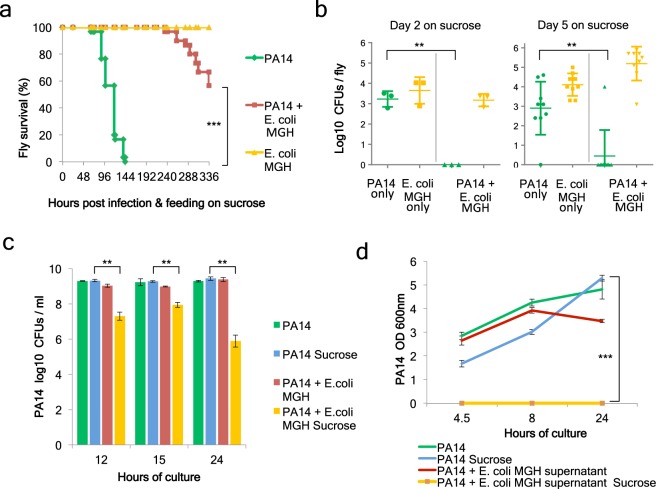
Figure 1.
E. coli MGH inhibits P. aeruginosa growth and virulence in the Drosophila gut and in culture in the presence of sucrose. (a) Survival of Drosophila melanogaster Oregon R flies infected with PA14, E. coli strain MGH or co-infected with E. coli MGH and P. aeruginosa PA14 [n = 30]. (b) Colonization levels measured in colony forming units (CFUs) at Day 2 [n = 3] and Day 5 [n = 9] post-PA14-infection only, E. coli MGH only, and upon co-infection (triangles for PA14, inverted triangles for MGH). (c) CFUs of PA14 growth in the presence or absence of 4% sucrose and E. coli MGH in LB cultures [n = 3]. (d) Optical density measurements at 600 nm of PA14 growth in half fresh LB, half liquid supernatant of E. coli LB cultures + /− 4% sucrose [n = 9]. **p < 0.005,***p < 0.0005. Error bars represent standard deviation of the mean.
Of note, E. coli antagonizes P. aeruginosa not only in fly survival but also in inhibiting its colonization. We assessed the bacterial loads of each bacterial strain independently in the fly and upon co-infection. Colony forming unit (CFU) measurements in selective media revealed that at 2 days and 5 days after infection with either P. aeruginosa or E. coli MGH flies harboured roughly 103 bacteria or more per fly (Fig. 1b). Upon co-infection with E. coli MGH, P. aeruginosa was almost eradicated, while E. coli MGH remained stable (Fig. 1b). Similarly, co-infection with the E. coli BW25113 strain and P. aeruginosa (PA14) resulted in lower PA14 CFUs and tentatively lower BW25113 CFUs at day 5 (Suppl. Fig. 1b), suggesting that mutual inhibition at the level of colonization is possible.
To assess if the inhibition between P. aeruginosa and E. coli is direct, we assessed bacterial growth in aerobic LB cultures. Interestingly, E. coli MGH did not inhibit P. aeruginosa growth in plain liquid LB (Fig. 1c). To assess if sucrose added in the fly infection media as a standard carbon source for the flies would make a difference in bacterial interactions in culture, we supplemented the LB media with 4% sucrose. Strikingly, in the presence of sucrose, P. aeruginosa CFUs were reduced by >1,000 fold when co-cultured with E. coli MGH, but no inhibition was noticed in the absence of sucrose (Fig. 1c). The monosaccharides glucose and fructose enable also the E. coli strain BW25113 to inhibit P. aeruginosa (Suppl. Fig. 1c). To assess whether secreted factors are responsible for P. aeruginosa growth inhibition we grew P. aeruginosa in a mix of 50% fresh LB and 50% filtered LB supernatant from an overnight E. coli culture that was supplemented or not with 4% sugar. The mix containing supernatant of E. coli MGH grown in sucrose and that of E. coli BW25113 grown in glucose was able to completely inhibit the growth of P. aeruginosa for at least 24 hours (Fig. 1d, Suppl. Fig. 1d).
E. coli inhibits P. aeruginosa intestinal colonization and lethality during mouse gut-derived sepsis
To model the antibiotic-induced dysbiosis of mammals we used a mouse assay of intestinal infection. We administered a regime of three broad-spectrum antibiotics in mice and assessed their gut microbiota at the genus level through 16S sequencing analysis. In the absence of antibiotics, the microbiota consisted primarily of Bacteroidetes, Firmicutes and Proteobacteria, including E. coli (Suppl. Fig. 2a). Using colony PCR sequencing we verified the presence of an endogenous E. coli strain (naming the respective cultured strain CD1) and further identified 7 easy-to-culture and potentially beneficial strains belonging to the Lactobaccillus, Bifidobacterium and Bacteroides genera in the faeces of mice (Suppl. Fig. 2b). Antibiotic treatment induced dysbiosis, which is exemplified by the eradication of E. coli, the reduction of all the prevalent phyla below the detection level (Suppl. Fig. 2c), and the eradication of all 8 cultured bacterial strains except for Bifidobacterium sp.2, which was reduced from 8.4 log10 to 7.2 log10 CFUs per gram of mouse faeces (Suppl. Fig. 2d).
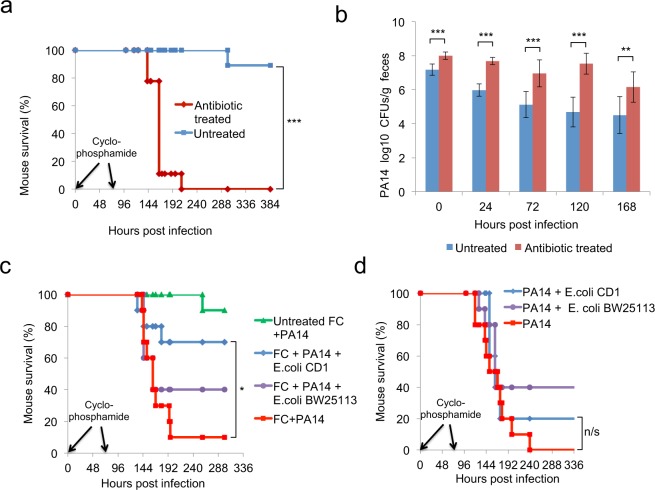
Antibiotic-treated mice subjected to immunosuppression via cyclophosphamide injections and infected with P. aeruginosa exhibit systemic spread of bacteria (Suppl. Fig. 3a–c) and die from sepsis as previously described31. Notably, all immunosuppressed dysbiotic mice died within 9 days of oral infection with P. aeruginosa, while 90% of the P. aeruginosa-infected immunocompromised mice that are not treated with antibiotics survived (Fig. 2a). Accordingly, P. aeruginosa load in the stools of infected mice bearing the healthy microbiota were significantly less at all time points than in mice treated with antibiotics, suggesting that commensal microbes inhibit colonisation by P. aeruginosa (Fig. 2b). To partly re-establish the mouse microbiome, we administered a faecal culture supplement (FC) prepared from a pelleted anaerobic stool culture. FC contained the endogenous Bacteroides, Bifidobacteria and Lactobacillus species and to a lesser extent the endogenous E. coli. The addition of FC in the drinking water had little to no effect by itself in protecting mice against lethality. However, FC fortified with the endogenous E. coli strain (E. coli CD1) rescued 70% of mice (Fig. 2c). On the other hand, the E. coli CD1 in the absence of FC did not protect mice against P. aeruginosa infection (Fig. 2d), suggesting a synergism between the endogenous E. coli CD1 and other members of the microbiota as a result of adaptation or co-evolution. Unlike the E. coli CD1 strain, the laboratory E. coli strain BW25113 showed only a trend in improving mouse survival due to P. aeruginosa infection, and this effect was not modifiable by FC (Fig. 2c,d). Despite the marginal effect on survival, E. coli BW25113 can stably colonize the mouse gut (Suppl. Fig. 3d) and reduces the P. aeruginosa burden significantly in the mouse gut within a week post-infection (Suppl. Fig. 3e).
Figure 2.
Commensal bacteria and E. coli protect antibiotic- and cyclophosphamide-treated mice from P. aeruginosa-induced lethality and colonization. (a) Survival to P. aeruginosa infection of immunocompromised mice pre-treated with antibiotics or untreated [n = 9]. (b) PA14 CFUs in faeces of immunocompromised mice pre-treated with antibiotics or untreated [n = 9]. (c) Survival to PA14 infection of immunocompromised mice feeding on a faecal culture (FC) fortified with E. coli BW25113 or mouse isolate E. coli CD1. Controls include antibiotic-treated and PA14-infected immunocompromised mice without E. coli, and mice without E. coli or antibiotic treatment [n = 10]. (d) Survival of antibiotic-treated PA14-infected immunocompromised mice feeding on the commensal mouse E. coli CD1 or the E. coli BW25113. Control immunocompromised mice were antibiotics-treated and infected with PA14, but no E. coli [n = 10]. n/s = p > 0.05, *p < 0.05, **p < 0.005, ***p < 0.0005. Error bars represent standard deviation of the mean.
Aerobic or anaerobic fermentation of glucose to lactic and acetic acid by E. coli is necessary for inhibiting P. aeruginosa growth
E. coli QS signalling and the production of the metabolite indole have been reported to inhibit P. aeruginosa growth27,34. To reveal E. coli factors that inhibit P. aeruginosa in our glucose-supplemented media, we assessed E. coli QS mutants and indole production genes previously implicated in bacterial competition27. We found that the E. coli QS genes luxS and sdiA are not necessary for P. aeruginosa inhibition in an LB culture supplemented with 4% glucose (Suppl. Fig. 4a). In addition, a deletion of the indole production enzyme tryptophanase (tna) essentially eliminated indole production (Suppl. Fig. 4b), but not the ability of E. coli BW25113 to inhibit P. aeruginosa (Suppl. Fig. 4c). Therefore we performed an unbiased screen of the KEIO collection of 3985 isogenic K-12 BW25113 gene mutants, identifying 45 genes that are necessary for the inhibition of P. aeruginosa in LB broth supplemented with 4% glucose. Gene enrichment analysis pinpointed glycolysis and the downstream pathways of oxidative phosphorylation and pentose phosphate as strongly enriched (Fig. 3a).
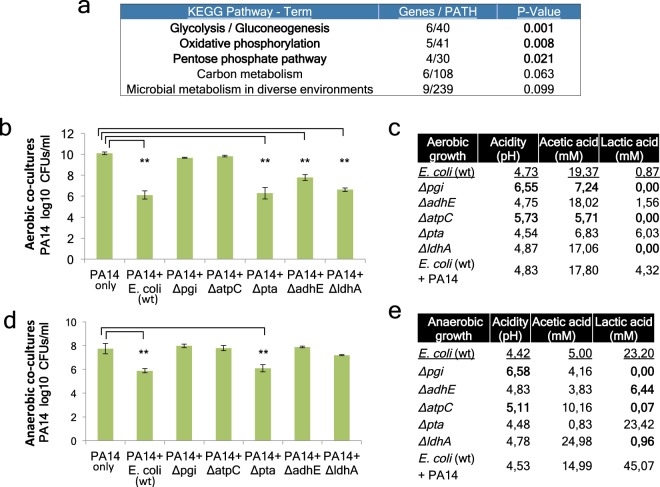
Figure 3.
E. coli mutants deficient in inhibiting P. aeruginosa and their acetic and lactic acid production profiles. (a) Enrichment analysis of E. coli genes identified as necessary for PA14 inhibition using EASE Score, a modified Fisher Exact P-Value. (b,d) PA14 CFUs in co-cultures with E. coli BW25113 (WT) and isogenic gene mutants at 5 hours of aerobic growth (b) or 24 hours of anaerobic growth (d) [n = 6]. (c,e) Liquid culture media pH and acid concentration (mM) at 5 hours of aerobic growth (c) or 24 hours of anaerobic growth (e) [n = 6]. Bold values indicate deviation from the wild type of mutant E. coli strains. **p < 0.005. Error bars represent standard deviation of the mean.
To assess the impact of E. coli glycolysis and oxidative phosphorylation on P. aeruginosa growth, we co-cultured P. aeruginosa with the core glycolysis and oxidative phosphorylation pathway mutants of E. coli, Δpgi and ΔatpC, respectively. In aerobic cultures using LB plus 4% glucose, wild-type E. coli BW25113 reduced P. aeruginosa CFUs by >1,000 fold, while Δpgi and ΔatpC mutants were unable to inhibit P. aeruginosa growth significantly (Fig. 3b). This is in line with the fermentation efficiency of the Δpgi and ΔatpC strains, which was severely compromised with no lactic acid and reduced acetic acid (>2 fold decrease) production and deficient acidification (pH > 5.5) of the liquid bacterial culture (Fig. 3c). In aerobic conditions lactic acid production is very low compared to acetic acid production, but none of the mixed acid fermentation mutants, Δpta, ΔadhE or ΔldhA, could abolish production of lactic acid and reduce acetic acid production at the same time (Fig. 3c). Accordingly, these mutants retained their ability to inhibit P. aeruginosa aerobically (Fig. 3b). On the other hand, the Δpgi and ΔatpC strains abolish lactic acid and reduce acetic acid production, and these mutants are the only ones unable to inhibit P. aeruginosa (Fig. 3b,c).
Because the environment in the mammalian gut is anaerobic and the fermentation process towards lactic acid production is much more efficient, we further tested this pathway anaerobically. As under aerobic conditions, the core metabolism E. coli mutants Δpgi and ΔatpC were unable to inhibit P. aeruginosa growth, acidify culture media and produce lactic acid in anaerobic cultures (Fig. 3d,e). Also E. coli ΔldhA and ΔadhE mutants exhibited significantly reduced lactic acid production [P < 0.001] (Fig. 3e) and an impaired ability to inhibit P. aeruginosa in an anaerobic culture (Fig. 3d). Thus, lactic acid production is crucial, while acetic acid production is helpful, in inhibiting P. aeruginosa growth either aerobically or anaerobically.
Lactic acid and acetic acid can inhibit P. aeruginosa growth and virulence
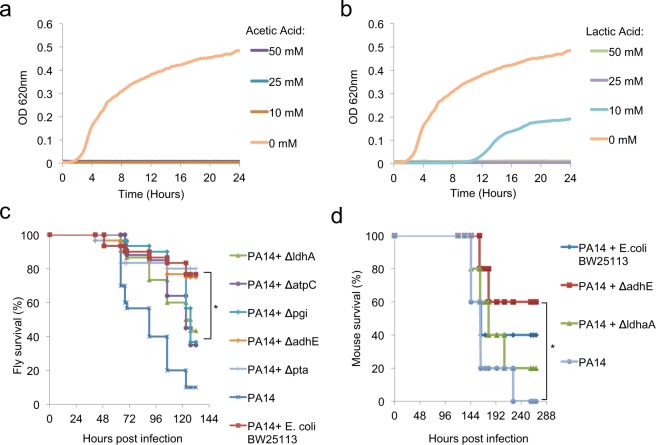
Supplementation of the E. coli mixed-acid fermentation products acetic acid and lactic acid have been reported to act as antimicrobials against P. aeruginosa35–37. We validated the role of these two metabolites in inhibiting P. aeruginosa growth at pH 5. Acidic pH of <5 is observed in an E. coli culture in the presence of sugars in either aerobic or anaerobic conditions (Fig. 3c,e). A concentration of 10 mM or more of acetic acid, which can be produced by E. coli in an aerobic liquid culture (Fig. 3c), abolished P. aeruginosa growth (Fig. 4a). Similarly, 10 mM or more of lactic acid, which can be produced by E. coli in an anaerobic culture (Fig. 3e), inhibited P. aeruginosa growth (Fig. 4b).
Figure 4.
Lactic and acetic acid capacity to inhibit P. aeruginosa growth in culture media and virulence in flies and mice. (a,b) PA14 growth in LB broth supplemented with 0, 10, 25 and 50 mM of acetic acid (a) or lactic acid (b) at pH 5 [n = 4]. (c) Drosophila survival curves upon co-infection with PA14 and wild-type (BW25113) or isogenic mutant E. coli strains [n = 30]. (d) Survival of PA14-infected immunocompromised mice complemented with a faecal culture (FC) and wild-type E. coli BW25113 or E. coli mutants, ΔldhA or ΔadhE, or no E. coli [n = 10]. *p < 0.05.
The inhibitory effect of E. coli lactic and acetic acid genes was further tested in animal models. In fly infection experiments, the ΔldhA E. coli mutant, specifically deficient in lactic acid production, and the core metabolism mutants Δpgi and ΔatpC, which are also unable to produce lactic acid aerobically or anaerobically, exhibited diminished ability to rescue flies infected with P. aeruginosa (Fig. 4c). In contrast, the Δpta and ΔadhE mutants, which cannot abolish lactic acid production in culture, rescued flies to the levels of the wild-type isogenic E. coli strain ΒW25113 (Fig. 4c). The same pattern was observed during co-infections in mice. We noticed that the E. coli mutant ΔadhE significantly rescued 60% of mice from lethality upon oral Pseudomonas infection in mice, comparable to the wild-type isogenic E. coli strain ΒW25113 that rescued 40% of mice, unlike the lactic-acid-defective strain ΔldhA that did not provide any significant rescue (Fig. 4d), suggesting that lactic acid production is the key for E. coli to inhibit P. aeruginosa in the host.
P. aeruginosa antagonizes E. coli strains unable to ferment sugars to lactic acid
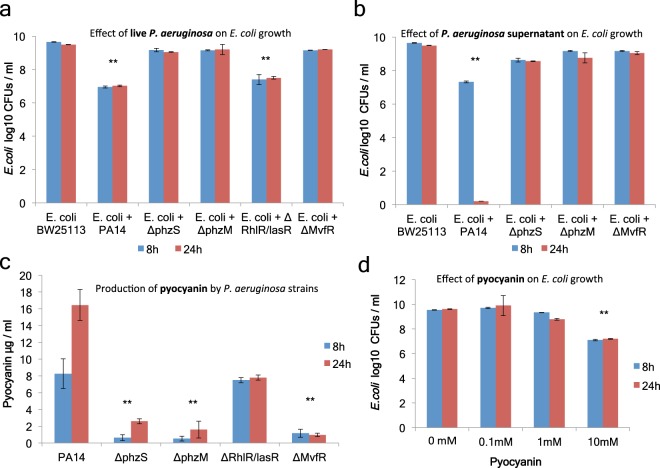
In the absence of added sugars in the culture media, P. aeruginosa inhibited E. coli growth rather than being inhibited by it (Fig. 5a,b). Screening for P. aeruginosa mutants implicated in this process, we identified the phenazine system and its known regulators (phzS, phzM and mvfR) as necessary for E. coli growth inhibition by >100 fold in culture (Fig. 5a,b). Pyocyanin, a redox-active secondary metabolite and a potent antibacterial, is produced and secreted by P. aeruginosa under the strict control of these quorum-sensing regulators. On the other hand the lasR/rhlR QS system regulators cannot fully control the expression of pyocyanin (Fig. 5c), and thus their mutation does not abolish the ability of P. aeruginosa to inhibit E. coli (Fig. 5a). Supplementation of 10 mM of pure pyocyanin was sufficient to inhibit E. coli in LB cultures to the same extent as in co-cultures with P. aeruginosa (Fig. 5a,d).
Figure 5.
P. aeruginosa toxin pyocyanin inhibits E. coli growth in culture. (a) CFUs of E. coli BW25113 in co-culture with wild-type PA14 vs. isogenic QS mutants at 8 and 24 hours [n = 6]. (b) CFUs of E. coli BW25113 in 1:1 LB to LB supernatant of PA14 vs. isogenic QS mutants at 8 and 24 hours [n = 6]. (c) Pyocyanin concentration in LB cultures of wild-type PA14 vs. isogenic QS mutants at 8 and 24 hours [n = 6]. (d) CFUs of BW25113 at 8 and 24 hours in LB with supplemented pure pyocyanin [n = 6]. **p < 0.005. Error bars represent standard deviation of the mean.
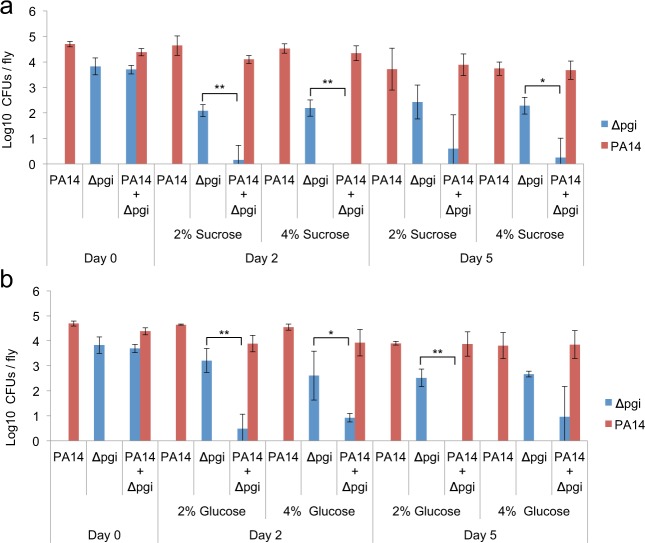
Moreover, the E. coli mutants, Δpgi and Δldha, which are deficient in glycolysis and lactic acid production respectively, were unable to inhibit P. aeruginosa growth in culture with and without supplementation of 4% glucose or sucrose (Suppl. Fig. 5a,b). Accordingly, we assessed the ability of P. aeruginosa to inhibit colonization by the E. coli mutants Δpgi and Δldha. Flies were inoculated with one or both species together and offered 2% or 4% glucose or sucrose as a necessary fly food nutrient. Co-inoculation of Drosophila with the E. coli glycolysis mutant Δpgi and P. aeruginosa showed reduced E. coli CFUs on day 2 and 5 compared to single inoculation with E. coli Δpgi (Fig. 6a,b). Moreover, flies mono-inoculated with P. aeruginosa or the E. coli lactic acid mutant Δldha could be colonized with an average of 4.1 (±0.5 SD) and 3.5 (±0.2 SD) log10 CFUs, respectively. Immediately after co-inoculation though E. coli Δldha CFUs were low, exhibiting an average of 1.9 (±0.2 SD) log10, while P. aeruginosa remained high at 3.8 (±1 SD) log10 CFUs. Thus with or without dietary sugars P. aeruginosa may inhibit gut colonization by E. coli mutants unable to ferment sugars into lactic acid.
Figure 6.
P. aeruginosa inhibits fermentation mutant E. coli in the Drosophila gut, despite dietary sucrose or glucose. (a,b) Drosophila intestine CFUs at 0, 2 and 5 days of flies orally infected with the E. coli BW25113 Δpgi core glycolysis mutant and/or PA14 [n = 6]. 2% or 4% sucrose (a) or glucose (b) was added in the infection medium and the fly food as a standard nutrient for the flies. *p < 0.05, **p < 0.005. Error bars represent standard deviation of the mean.
Lactic and acetic acid production rather than nutritional input defines the interaction between E. coli and P. aeruginosa in the mouse gut
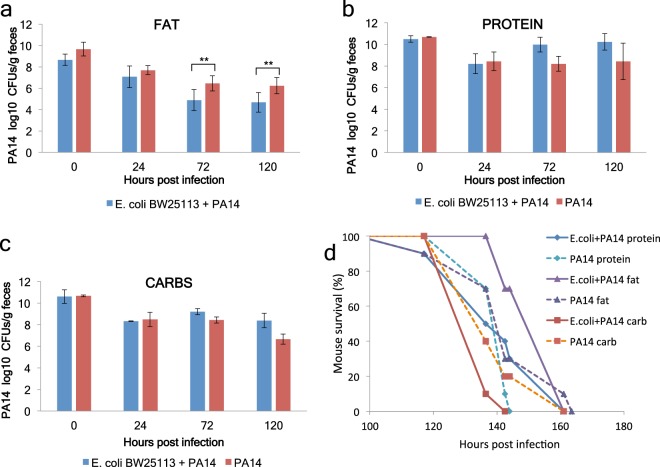
Diet is very important for the maintenance of a healthy microbiome and in shaping the intestinal immune response38–40. Our study shows that the interaction between E. coli and P. aeruginosa is shaped by the fermentation of sugars. Therefore we sought to investigate in mice the contribution of three nutritionally extreme diets: a protein-based, a vegetable fat-based and a carbohydrate-based diet. In mice on the carbohydrate-based diet the total sugar concentration (sucrose, glucose and fructose) in the faeces was 67.4 μg/ml, which was higher than any of the other diets, while it was only 18.7 μg/ml in the faeces of mice given E. coli orally (Suppl. Fig. 6a). This means that E. coli is consuming sugars in the mouse gut. Yet faecal lactic acid concentration was the highest in the fat-based diet group in the presence of E. coli (Suppl. Fig. 6b). Accordingly, P. aeruginosa CFUs were reduced by E. coli in mice fed with the vegetable fat-based diet (Fig. 7a), but not with the protein- or the carbohydrate-based diets (Fig. 7b,c). Similarly, mouse survival upon P. aeruginosa infection in immunosuppressed mice was the highest in mice fed the vegetable fat-based diet and co-inoculated with E. coli, as opposed to mice fed with the carbohydrate-based diet and co-inoculated with E. coli (Fig. 7d). However, the fat-based diet does not favour E. coli gut colonization, as the E. coli CFUs in the faeces are comparable between the carbohydrate- and the fat-based diets and lower than those of the protein-based diet (Fig. 8a).
Figure 7.
A fat-based diet, but not a carb- or protein-based diet enables E. coli to inhibit P. aeruginosa colonization and lethality in mice. (a) PA14 CFUs in faeces of immunocompromised mice fed on a fat- (a), protein- (b) or carbohydrate-based diet following inoculation with PA14 or co-inoculation with PA14 and E. coli BW25113 [n = 10]. **p < 0.005. Error bars represent standard deviation of the mean. (d) Survival of immunocompromised mice kept on a fat-, protein-, or carb-based diet and infected with PA14 or PA14 plus E. coli BW25113 [n = 10]. p = 0.04 for Fat PA14 + E. coli vs. Protein PA14 + E. coli and p = 0.0001 for Fat PA14 + E. coli vs. Carbs PA14 + E. coli.
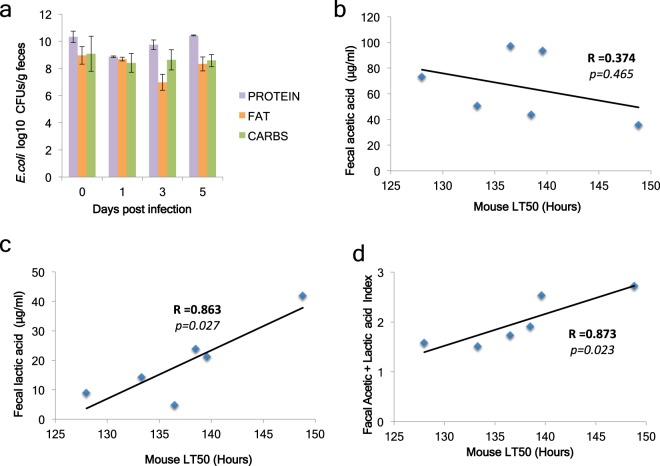
Figure 8.
Faecal concentration of lactic and acetic acid correlates with mouse survival upon P. aeruginosa regardless of faecal E. coli levels. (a) E. coli CFUs in faeces of immunocompromised mice infected with PA14 and E. coli BW25113 and kept on a protein-, fat- or carbohydrate-based diet [n = 10]. (b,c) Correlation plots and Pearson correlation coefficient (R) of faecal acetic acid (b) and lactic acid (c) concentration against the lethal time 50% (LT50) of the corresponding mice for each of the 6 conditions (3 mouse diets × 2 types of infection). (d) A combinatorial index of normalized mouse faecal acetic and lactic acid concentration correlated with LT50.
To assess if faecal acetic and lactic acid production may indicate protection against P. aeruginosa, we correlated acid concentration in the faeces with the lethal time 50% (LT50) of the corresponding sets of mice. We found that lactic but not acetic acid levels alone correlate significantly and positively with survival to infection (Fig. 8b,c), while an index of normalized concentration values for acetic and lactic acid combined gave also a clear correlation with survival (Fig. 8d). We conclude that the standard balanced diet (Fig. 4d; Suppl. Fig. 3e) and the vegetable fat-based diet (Fig. 7a), rather than the carbohydrate-based diet (Fig. 7c) facilitate the inhibitory effect of E. coli on P. aeruginosa; and we suggest that, given the complexity of the mammalian intestinal environment, the metabolic output in acetic and lactic acid production rather than the dietary input is indicative of susceptibility to intestinal P. aeruginosa infection.
Discussion
Despite primarily observations on bacterial antagonism dating more than 100 years ago41, there are not many cases supporting the model of one-pathogen-one-colonization-resistor, according to which specific bacterial strains protect the host against infection42. Known cases include the inhibitory effect of Clostridium scindens in resistance against C. difficile infection43, the non-toxigenic Bacteroides fragilis resistance against the enterotoxigenic Bacteroides fragilis44, the effect of E. coli O21:H+ against muscle atrophy due to infection, and the effect of E. coli EcN against intestinal pathogens42. Antagonistic interactions between a pathogenic and a non-pathogenic bacterial strain may include: (a) the direct inhibition of pathogen’s growth, colonization or virulence by the non-pathogenic strain, or (b) the indirect effect of the non-pathogenic strain in inducing or supporting the host defence to infection. To establish the mode of interaction between E. coli and P. aeruginosa we first examined whether Drosophila can be efficiently colonized with these species. Sugar-based diets allow stable bacterial colonization with either P. aeruginosa or E. coli strains. When flies are infected with both species, P. aeruginosa colonization and mortality is significantly reduced or eliminated. Antagonism between P. aeruginosa and other species is nevertheless specific. Human oropharyngeal bacteria are predominantly gram-positive, such as the Neisseria, Streptococcus, Staphylococcus and Actinomyces species, differ from those of the human intestine, and tend to induce rather than antagonize P. aeruginosa virulence45. Moreover, peptidoglycan, which is abundant in gram-positive bacteria, can directly induce the virulence of P. aeruginosa46. Thus E. coli, as opposed to many gram-positive bacteria, might serve as a safer inhibitor of P. aeruginosa by inhibiting its growth without inducing its virulence.
P. aeruginosa usually affects hospitalized and immunocompromised individuals. It causes life-threatening burn wound and lung infections, but humans often carry P. aeruginosa asymptomatically in their intestines47. During P. aeruginosa intestinal colonization, a healthy host primarily deploys innate immune responses recruiting macrophages and monocytes in the gut and then adaptive immune cells, such as B lymphocytes, through the induction of pro-inflammatory cytokines to control infection48. In immunocompromised patients, however, P. aeruginosa may disrupt the intestinal epithelial barrier and translocate extraluminally, leading to sepsis and death49. Moreover, virulent P. aeruginosa may facilitate this process by subverting the innate immune responses upon infection50. Another reason for the benignity of P. aeruginosa in the healthy human gut may be the action of intestinal microbiota, which are part of the host defence to intestinal infection51–53. Previous studies describe the use of antibiotic cocktails that favour P. aeruginosa intestinal colonization by compromising resistance by the intestinal microbiota54. Accordingly, we show that antibiotic use in mice diminishes all the prevalent phyla, eradicates E. coli, and induces dysbiosis. Using a Pseudomonas-induced gut-derived sepsis model to investigate infection in mice that exhibit neutropenia, lymphopenia, as well as mucosal damage55,56, we found that mice not given antibiotics mostly survived and were less colonized with P. aeruginosa, contrary to antibiotic-treated mice. In addition, reintroduction of the commensal microbes through a faecal culture of endogenous and potentially beneficial bacterial species was inefficient in improving mouse protection from lethality. Nevertheless, a high dose of the endogenous E. coli CD1 isolate in combination with faecal bacteria exhibited significant protection against P. aeruginosa. We postulate a symbiotic adaptation of the mouse-isolated E. coli strain with the mouse gut environment and its microbiota in protecting the host.
Contrary to a previous study27, we found that E. coli indole production had no effect on inhibiting the growth of P. aeruginosa in our experiments. This might be due to the inhibition of E. coli indole production by sugars added in our media or indole degradation via a higher induction of QS in the P. aeruginosa strain PA1427,57. Accordingly, we screened in an unbiased way and pinpointed E. coli glucose metabolism and fermentation mutants deficient in lactic and acetic acid production responsible for inhibiting P. aeruginosa. In antibiotic-treated mice, a similar trend was observed whereby the lactate-dehydrogenase-deficient E. coli mutant was unable to protect mice from P. aeruginosa infection and mortality. The anti-infective properties of lactic and acetic acid may be attributed to lowering the pH, but also to the permeabilization of the outer membrane of gram-negative bacteria37. On the other hand, P. aeruginosa produces many virulence factors regulated by QS, such as pyocyanin, which has bactericidal properties58. Accordingly, we notice that only strains of P. aeruginosa able to produce pyocyanin can inhibit E. coli unable to produce lactic or acetic acid due to the lack of sugars in the media. Interestingly, despite the fact that high sugar concentrations may inhibit P. aeruginosa QS59, P. aeruginosa grown in sugar-supplemented media can still inhibit E. coli strains with mutated fermentation pathway genes. Thus depending on the concentration of E. coli’s acetic and lactic acid or P. aeruginosa’s pyocyanin, the antagonistic growth may be shifted towards one or the other species.
The role of diet has been extensively studied in response to gut microbiota and host physiology60. Hence we explored three different diets, based either on carbohydrates (corn starch and sucrose), fat (vegetable shortening) or protein (casein). Mice feeding on these diets exhibited complex features: First, the carbohydrate-based diet did not improve the ability of E. coli to inhibit P. aeruginosa colonization and concomitant mortality. This might be because this carbohydrate-based diet does not deliver a significant amount of free sugars in the mouse colon. While sugars are higher in the faeces of mice fed with a carbohydrate-based diet, they may be too low to have the anticipated impact on E. coli. Second, the protein-based diet sustains more E. coli than the other diets, yet this didn’t translate into better inhibiting capacity against P. aeruginosa. This might be because casein inhibits or lacks the ability to fuel fermentation into lactic acid. Third, the vegetable fat-based diet, while not ideal for E. coli growth compared to the other diets, allows E. coli to produce more lactic acid that can inhibit P. aeruginosa growth. This is in line with evidence showing that unsaturated fat may benefit lactic acid bacteria in mice61.
Lactic acid in the mouse faeces is much lower than the lowest inhibitory concentration tested in culture. Nevertheless, E. coli mutants defective in lactic acid production are also defective in inhibiting P. aeruginosa in the fly and mouse gut. The ability of any chemical to inhibit bacterial growth depends on the environment and thus additional factors (e.g. additional antimicrobials or lactic acid metabolism products) in the fly and mouse gut may boost the ability of lactic acid to inhibit P. aeruginosa. In addition, high sugar levels may be difficult to achieve by a high carbohydrate diet because sugar is readily absorbed in the small intestine, and E. coli and other commensals may use dietary fat more efficiently towards lactic acid production. Thus the metabolic output in the colon rather than the dietary input might better dictate the balance between and among bacterial species. Metabolic output is nevertheless a result of diet, microbiota composition and the host physiology acting in concert. Accordingly, faecal metabolomics might prove very helpful in predicting the outcome of bacterial interactions in the human colon and the risk for an infection.
Supplementary information
Acknowledgements
We thank Christos Shammas from AVVA Pharmaceuticals for 16S sequencing; Robert J.C. McLean for E. coli strains and the NBRP (NIG, Japan) for the KEIO collection BW25113 library strains; Tassos Economou for the Enteropathogenic E. coli O127:H6 E2348/69 strain; and Laurence Rahme for critical reading and editing. We also thank Marie Curie CIG and Fondation Sante for funding to YA.
Author Contributions
T.C., S.P., I.D., C.M. performed experiments. T.C. provided novel insights and wrote the manuscript. Y.A. provided the concept, funding and editing of the manuscript.
Data Availability
All data will be available upon publication.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-51058-3.
References
- 1.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 2.Zoetendal EG, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macfarlane GT, Macfarlane S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011;45(Suppl):S120–7. doi: 10.1097/MCG.0b013e31822fecfe. [DOI] [PubMed] [Google Scholar]
- 4.Mitsuoka T. Intestinal flora and human health. Asia Pac. J. Clin. Nutr. 1996;5:2–9. [PubMed] [Google Scholar]
- 5.Winter SE, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruis W, et al. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 1997;11:853–858. doi: 10.1046/j.1365-2036.1997.00225.x. [DOI] [PubMed] [Google Scholar]
- 7.Kruis W, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53:1617–1623. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthes H, Krummenerl T, Giensch M, Wolff C, Schulze J. Clinical trial: probiotic treatment of acute distal ulcerative colitis with rectally administered Escherichia coli Nissle 1917 (EcN) BMC Complement. Altern. Med. 2010;10:13-6882–10-13. doi: 10.1186/1472-6882-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudeau J, Glasser AL, Julien S, Colombel JF, Darfeuille-Michaud A. Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn’s disease. Aliment. Pharmacol. Ther. 2003;18:45–56. doi: 10.1046/j.1365-2036.2003.01638.x. [DOI] [PubMed] [Google Scholar]
- 10.Schlee M, et al. Induction of human beta-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect. Immun. 2007;75:2399–2407. doi: 10.1128/IAI.01563-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ukena SN, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One. 2007;2:e1308. doi: 10.1371/journal.pone.0001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zyrek AA, et al. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell. Microbiol. 2007;9:804–816. doi: 10.1111/j.1462-5822.2006.00836.x. [DOI] [PubMed] [Google Scholar]
- 13.Reissbrodt R, et al. Inhibition of growth of Shiga toxin-producing Escherichia coli by nonpathogenic Escherichia coli. FEMS Microbiol. Lett. 2009;290:62–69. doi: 10.1111/j.1574-6968.2008.01405.x. [DOI] [PubMed] [Google Scholar]
- 14.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One. 2013;8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka S, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol. Med. Microbiol. 2009;56:80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 16.Chang JY, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 17.Markou P, Apidianakis Y. Pathogenesis of intestinal Pseudomonas aeruginosa infection in patients with cancer. Front. Cell. Infect. Microbiol. 2014;3:115. doi: 10.3389/fcimb.2013.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu K, et al. Altered gut flora and environment in patients with severe SIRS. J. Trauma. 2006;60:126–133. doi: 10.1097/01.ta.0000197374.99755.fe. [DOI] [PubMed] [Google Scholar]
- 19.Ohara T, Itoh K. Significance of Pseudomonas aeruginosa colonization of the gastrointestinal tract. Intern. Med. 2003;42:1072–1076. doi: 10.2169/internalmedicine.42.1072. [DOI] [PubMed] [Google Scholar]
- 20.Vincent JL, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 21.Chuang CH, et al. Shanghai fever: a distinct Pseudomonas aeruginosa enteric disease. Gut. 2014;63:736–743. doi: 10.1136/gutjnl-2013-304786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panayidou, S. & Apidianakis, Y. Pseudomonas aeruginosa. Laboratory Models for Foodborne Infections, 373–390 (2017).
- 23.Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 2004;72:4275–4278. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapsetaki SE, et al. The bacterial metabolite 2-aminoacetophenone promotes association of pathogenic bacteria with flies. Nat. Commun. 2014;5:4401. doi: 10.1038/ncomms5401. [DOI] [PubMed] [Google Scholar]
- 25.Rahme LG, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 26.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2(2006):0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu W, et al. Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl. Environ. Microbiol. 2012;78:411–419. doi: 10.1128/AEM.06396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine MM, et al. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;1:1119–1122. doi: 10.1016/S0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 29.Apidianakis Y, Pitsouli C, Perrimon N, Rahme L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc. Natl. Acad. Sci. USA. 2009;106:20883–20888. doi: 10.1073/pnas.0911797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith BK, York DA, Bray GA. Activation of hypothalamic serotonin receptors reduced intake of dietary fat and protein but not carbohydrate. Am. J. Physiol. 1999;277:R802–11. doi: 10.1152/ajpregu.1999.277.3.R802. [DOI] [PubMed] [Google Scholar]
- 31.Zuluaga AF, et al. Neutropenia induced in outbred mice by a simplified low-dose cyclophosphamide regimen: characterization and applicability to diverse experimental models of infectious diseases. BMC Infect. Dis. 2006;6:55-2334–6-55. doi: 10.1186/1471-2334-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 33.Jahreis K, et al. Adaptation of sucrose metabolism in the Escherichia coli wild-type strain EC3132. J. Bacteriol. 2002;184:5307–5316. doi: 10.1128/JB.184.19.5307-5316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, Attila C, Cirillo SL, Cirillo JD, Wood TK. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2009;2:75–90. doi: 10.1111/j.1751-7915.2008.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips I, Lobo AZ, Fernandes R, Gundara NS. Acetic acid in the treatment of superficial wounds infected by Pseudomonas aeruginosa. Lancet. 1968;1:11–14. doi: 10.1016/S0140-6736(68)90004-4. [DOI] [PubMed] [Google Scholar]
- 36.Levison ME. Effect of colon flora and short-chain fatty acids on growth in vitro of Pseudomonas aeruginsoa and Enterobacteriaceae. Infect. Immun. 1973;8:30–35. doi: 10.1128/iai.8.1.30-35.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alakomi HL, et al. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 2000;66:2001–2005. doi: 10.1128/AEM.66.5.2001-2005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4:1095–1119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tilg H, Moschen AR. Food, immunity, and the microbiome. Gastroenterology. 2015;148:1107–1119. doi: 10.1053/j.gastro.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 41.Rettger LF. The Antagonism of Bacteria and Their Products to Other Bacteria. J. Infect. Dis. 1905;2:562–568. doi: 10.1093/infdis/2.4.562. [DOI] [Google Scholar]
- 42.Byrd AL, Segre JA. Infectious disease. Adapting Koch’s postulates. Science. 2016;351:224–226. doi: 10.1126/science.aad6753. [DOI] [PubMed] [Google Scholar]
- 43.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner VE, et al. Effects of a gut pathobiont in a gnotobiotic mouse model of childhood undernutrition. Sci. Transl. Med. 2016;8:366ra164. doi: 10.1126/scitranslmed.aah4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sibley CD, et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. USA. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoodley BJ, Thom BT. Observations on the intestinal carriage of Pseudomonas aeruginosa. J. Med. Microbiol. 1970;3:367–375. doi: 10.1099/00222615-3-3-367. [DOI] [PubMed] [Google Scholar]
- 48.von Klitzing E, Ekmekciu I, Bereswill S, Heimesaat MM. Intestinal and Systemic Immune Responses upon Multi-drug Resistant Pseudomonas aeruginosa Colonization of Mice Harboring a Human Gut Microbiota. Front. Microbiol. 2017;8:2590. doi: 10.3389/fmicb.2017.02590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okuda J, et al. Translocation of Pseudomonas aeruginosa from the intestinal tract is mediated by the binding of ExoS to an Na,K-ATPase regulator, FXYD3. Infect. Immun. 2010;78:4511–4522. doi: 10.1128/IAI.00428-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Apidianakis Y, et al. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc. Natl. Acad. Sci. USA. 2005;102:2573–2578. doi: 10.1073/pnas.0409588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schubert AM, Sinani H, Schloss PD. Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile. MBio. 2015;6:e00974–15. doi: 10.1128/mBio.00974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ge X, et al. Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. J. Transl. Med. 2017;15:13-016–1105-4. doi: 10.1186/s12967-016-1105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hentges DJ, Stein AJ, Casey SW, Que JU. Protective role of intestinal flora against infection with Pseudomonas aeruginosa in mice: influence of antibiotics on colonization resistance. Infect. Immun. 1985;47:118–122. doi: 10.1128/iai.47.1.118-122.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koh AY, Priebe GP, Pier GB. Virulence of Pseudomonas aeruginosa in a murine model of gastrointestinal colonization and dissemination in neutropenia. Infect. Immun. 2005;73:2262–2272. doi: 10.1128/IAI.73.4.2262-2272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsumoto T, et al. Oral administration of Bifidobacterium longum prevents gut-derived Pseudomonas aeruginosa sepsis in mice. J. Appl. Microbiol. 2008;104:672–680. doi: 10.1111/j.1365-2672.2007.03593.x. [DOI] [PubMed] [Google Scholar]
- 57.Botsford JL, DeMoss RD. Catabolite repression of tryptophanase in Escherichia coli. J. Bacteriol. 1971;105:303–312. doi: 10.1128/jb.105.1.303-312.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baron SS, Rowe JJ. Antibiotic action of pyocyanin. Antimicrob. Agents Chemother. 1981;20:814–820. doi: 10.1128/AAC.20.6.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang R, Starkey M, Hazan R, Rahme LG. Honey’s Ability to Counter Bacterial Infections Arises from Both Bactericidal Compounds and QS Inhibition. Front. Microbiol. 2012;3:144. doi: 10.3389/fmicb.2012.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh RK, et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017;15:73-017–1175-y. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Backhed F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell. Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data will be available upon publication.