Abstract
Hereditary spastic paraplegia (HSP) comprises a heterogeneous group of neurodegenerative disorders, it share common symptom - of progressive lower spastic paraparesis. The most common autosomal dominant (AD) forms of HSP are SPG4 (SPAST gene) and SPG3 (ATL1 gene). In the current research we investigated for the first time the distribution of pathogenic mutations in SPAST and ATL1 genes within a large cohort of Russian HSP patients (122 probands; 69 famillial cases). We determined the frequencies of genetic abnormalities using Sanger sequencing, multiplex ligation-dependent probe amplification (MLPA), and Next Generation Sequencing (NGS) of targeted gene panels. As a result, SPG4 was diagnosed in 30.3% (37/122) of HSP cases, where the familial cases represented 37.7% (26/69) of SPG4. In total 31 pathogenic and likely pathogenic variants were detected in SPAST, with 14 new mutations. Among all detected SPAST variants, 29% were gross deletions and duplications. The proportion of SPG3 variants in Russian cohort was 8.2% (10/122) that were all familial cases. All 10 detected ATL1 mutations were missense substitutions, most of which were in the mutational hot spots of 4, 7, 8, 12 exons, with 2 novel mutations. This work will be helpful for the populational genetics of HSP understanding.
Subject terms: DNA sequencing, Next-generation sequencing, Motor neuron disease, Motor neuron disease, Genotype
Introduction
Hereditary spastic paraplegia (HSP) is genetically heterogeneous group of neurodegenerative disorders characterized by progressive pyramidal tract dysfunction due to retrograde degeneration of long axons of the corticospinal tracts. The most prevalent form of HSP is SPG4 (OMIM 182601). SPG4 is associated with mutations in the SPAST gene (protein: Spastin) and demonstrates autosomal dominant (AD) inheritance. It represents 12–25% of all HSP cases (including mutations with non-proven pathogenicity) and 40–60% within familial cases1–3. Spastin is a microtubule-severing protein that belongs to AAA (ATPase associated with various cellular activities) family of ATPases and regulates the number and mobility of microtubules. SPAST gene contains 17 exons where 723 mutations are mapped been scattered across the coding regions. Most of those mutations identified are missense, however deletions and duplications in SPAST also make a considerable contribution to the HSP pathogenesis4. So far, no mutational hotspot regions for SPAST gene were described and no frequent mutations were found. Therefore, screening of the complete coding sequence of SPAST is necessary for the detection of mutations. But if the new variant located in the region encoding the ATPase domain it is most likely to be pathogenic3.
SPG3 (SPG3A; OMIM 182600) form of HSP is caused by pathogenic variants in the ATL1 gene (protein: Atlastin). Alastin is a protein implicated in vesicle trafficking and neurite outgrowth. SPG3 is the second most common form of ADHSP and of HSP with other type of inheritance. Although its contribution is much less than that of SPG4, on average it accounts for 2–3% of total HSP cases and 8–10% of family AD HSP cases1,5,6. Forty-eight ATL1 mutations were identified so far and they are mainly missense changes. Large rearrangements make however a contribution to SPG3 pathogenesis. Four of 14 gene exons (4, 7, 8 exons, coding GBP/Ras-like GTPase domain, and 12th exon)4 are mutation hotspots in ATL1 whereas no frequent mutations have been described.
Many variants can cause different pathogenesis of HSP that requires different treatments. At the same time the mutational spectra of HSP varies strongly on the populational level. Thus the current study is intended to provide for the first time an overview of SPAST and ATL1 mutational landscape in a cohort of Russian HSP patients that would help diagnosis and treatment.
Methods
Patients
The current study analyzed DNA of 122 unrelated HSP patients (69 AD familial cases, 53 AD sporadic cases). Cohort included 68 men and 54 women aged between 4 and 68 years. The non-familial cases started to be selected after the next generation sequencing (NGS) panel being implemented.
Most patients were diagnosed at the Research and Counseling Department of the Research Centre for Medical Genetics (RCMG) including patients that were referred from the Genetic Counseling Department for the Moscow Region. While the others had been diagnosed by the Genetic Counseling Departments of Voronezh, Yekaterinburg, Khabarovsk and other regions.
The study was approved by the local ethics committee of the Federal State Budgetary Institution “Research Centre for Medical Genetics” (the approval number 2016-6/7) and all the patients gave written informed consent. All experiments were performed in accordance with the institutional guidelines.
Methods
Blood samples were collected, and DNA was extracted at the DNA-diagnostics Laboratory of RCMG. Molecular diagnostics of HSP patients have been performed using NGS, all detected variants were validated by Sanger sequencing. The DNA of HSP patients who were negative for sequencing mutations was analyzed using MLPA to quantify copy numbers.
Genomic DNA was extracted from whole venous blood by Wizard® Genomic DNA Purification Kit (Promega, USA) following the manufacturer’s protocol.
For the current research the Spastic Paraplegia Sequencing Panel of target genes was developed. It comprises the following HSP associated genes; GJC2, AP4B1, AMPD2, IBA57, ALDH18A1, ZFYVE27, NT5C2, ENTPD1, MTPAP, CAPN1, BSCL2, KLC2, KIF5A, C12orf65, MARS, VAMP1, B4GALNT1, SPG20, SACS, ATL1, ZFYVE26, DDHD1, TECPR2, AP4S1, NIPA1, SPG11, SPG21, AP4E1, USP8, SPG7, FA2H, ARL6IP1, KIF1C, AFG3L2, RTN2, PNPLA6, C19orf12, CPT1C, MAG, HSPD1, KIF1A, REEP1, PGAP1, MARS2, SPAST, SLC33A1, TFG, WDR48, CYP2U1, ARSI, ZFR, REEP2, AP5Z1, AP4M1, CYP7B1, KIAA0196, ERLIN2, VPS37A, DDHD2, GBA2, L1CAM, PLP1 and SLC16A2. Next generation sequencing of patient’s DNA was performed by Ion S5 next-generation sequencer (Thermo Fisher Scientific, USA) with an Ion AmpliSeq ™ Library Kit 2.0 according to the manufacturer’s protocol. Patient’s DNA samples were prepared using ultra rapid multiplex PCR technology combined with subsequent sequencing (AmpliSeq™).
Sequencing data was processed according to the standard bioinformatic algorithm from Thermo Fisher Scientific (Torrent Suite™) and Gene-Talk software (www.gene-talk.de/contact; Gene Talk GmbH, Germany). Sequenced fragments were visualized in Integrative Genomics Viewer (IGV) software (© 2013–2018 Broad Institute, and the Regents of the University of California, USA).
A beta release of the Genome Aggregation Database (gnomAD browser beta) was used to determine the frequencies of new variants.
MLPA method was used for the analysis of large deletions and duplications using a SALSA MLPA P-165-С2 HSP kit following the manufacturer’s protocol. MLPA data was analyzed with Coffalayser software (MRC-Holland).
Revealed SPAST and ATL1 modifications were designated in accordance with HGVS nomenclature (http://www.hgvs.org/mutnomen/) with reference sequences NM_199436.1 and NM_001127713.1, respectively (http://www.Ncbi.nlm.nih.gov/nuccore).
The following online prediction programs were used to determine pathogenicity in silico: Mutation Taster (http://www.mutationtaster.org/), UMD-predictor (http://umd-predictor.eu/); SIFT/Provean (http://provean.jcvi.org/index.php); PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml); and Human Splicing Finder (http://www.umd.be/HSF/).
Guidelines for interpretation of NGS data7,8 were used to define the clinical significance of newly discovered variants.
Results
In total 37 SPG4 cases were detected among 122 DNA samples of patients with HSP that represents 30.3% of cohort. Where SPAST mutations within AD HSP forms amounted for 37.7% (26/69) of cases and 20.7% (11/53) were among sporadic cases. A total of 31 pathogenic and likely pathogenic variants were detected in SPAST gene, encompassing 14 novel variants. Whereas 5 newly detected variants were repeated in 2 or more families. Twenty-two pathogenic and likely pathogenic variants in 27 out of total 37 unrelated SPG4 probands were found by sequencing techniques, namely NGS of targeted panel and Sanger sequencing. These were: 10 missense changes, 3 nonsense mutations, 7 micro-rearrangements and 2 splice site mutations. Most of the variants (19/22) were located in the AAA-domain of SPAST, while 2 were located in the promoter region and 1 in the microtubule interacting and trafficking (MIT) domain. Major limitation of the direct sequencing methods is that large deletions/duplications may not be detected. Therefore, 10 remaining patients out of the total 37 SPG4 patients that fail to reveal mutations by direct sequencing were examined using MPLA assay. As a result, 9 pathogenic variants in 10 unrelated probands, which amounts for 29.0% (9/31) of all pathogenic and likely pathogenic variants, were detected in SPAST.
The proportion of SPG3 in Russian patients comprised 8.2% (10/122), where the pathogenic variants were detected only in familial group and amounted to 14.5% (10/69). Seven pathogenic variants of ATL1 were detected in 10 unrelated probands. All detected mutations were missense substitutions. Large rearrangements were not detected in Russian cohort. The most frequent variants (6/8) in our study were located in the ATL1 gene mutational hotspots in exons 7, 8 and 12. Notably, the variants с.1041G > A (p.Met347Ile) and с.1213G > A (p.Val405Met) are described here for the first time. Three mutations were found in 2 or more families, whereas the other 5 were distinct in each family.
The data upon SPAST and ATL1gene mutational landscape is summarized in the Table 1. Altogether 34 pathogenic and likely pathogenic variants were detected in 42 patients. In total we found 16 novel mutations presented in the Table 2. The novel variants identified in this study were categorized according to the guidelines of the American College of Medical Genetics and Genomics (ACMG) (Table 2). The distribution of identified mutations according to the different HSP inheritance types, showed 36 probands (52.2%) among 69 of AD cases and 11 probands (20.7%) among 53 of sporadic cases. None of these new variants was registered before or were found with allele frequency higher than 0.01% in a control cohort annotated in the GnomAD project, 1000 Genomes Project, ESP6500 and Exome Aggregation Consortium.
Table 1.
Mutational spectrum of SPAST and ATL1 genes.
| Gene | Family # | Exon | Modification of nucleic acid sequence | Modification of protein sequence | HGMD reference number, references | Pathogenic variants |
|---|---|---|---|---|---|---|
| SPAST | 53о | 3 | c.551A > C | p.Asn184Thr | CM10358326, | Pathogenic |
| 27o | 7 | c.1070T > A | p.Ile357Asn | CM18898527, | Pathogenic | |
| 44o | 8 | c.1107A > G* | p.Thr369Thr* | Likely pathogenic | ||
| 116 | 8 | c.1116A > T* | p.Arg372Term* | Pathogenic | ||
| 32 | 8 | c.1139T > C | p.Leu380Pro | CM13162828, | Pathogenic | |
| 121 | 9 | c.1196C > T | p.Ser399Leu | CM02225029, | Pathogenic | |
| 73o, 73 | 9 | c.1216A > G | p.Ile406Val | CM06048519, | Pathogenic | |
| 204 | 10 | c.1252G > A* | p.Glu418Lys* | Likely pathogenic | ||
| 2o, 21o, 91 | 10 | c.1291C > T | p.Arg431Term | CM00043718, | Pathogenic | |
| 5 | 11 | с.1391A > G* | p.Glu464Gly* | Pathogenic | ||
| 35 | 13 | c.1507C > T | p.Arg503Trp | HM06005630, | Pathogenic | |
| 43o | 15 | c.1663G > T | p.Asp555Asn | CM10358226, | Pathogenic | |
| 172, 73 | 15 | c.1684C > T | p.Arg562Term | CM00044118, | Pathogenic | |
| 24o | 1 | c.284delC* | p.Ala95ArgfsTerm65* | Pathogenic | ||
| 44 | 1 | c.286delG | p.Ala96ArgfsTerm65 | CD00467931, | Pathogenic | |
| 120o | 8 | c.1162delA* | p.Lys388ArgfsTerm8* | Pathogenic | ||
| 35 | 10 | c.1271delG* | p.Ala425LeufsTerm13* | Pathogenic | ||
| 34 | 12 | c.1469delA* | p.Glu491SerfsTerm39* | Pathogenic | ||
| 19 | 17 | c.1750_1751delGAinsT* | p.Asp584SerfsTerm5* | Pathogenic | ||
| 126 | 17 | c.1840_1846del* | Pathogenic | |||
| 212 | 7 | c.1098 + 1G > A | CS06339032, | Pathogenic | ||
| 54, 183 | 9 | c.1245 + 1G > A | CS01184520, | Pathogenic | ||
| 33o, 107 | 1 | del.ex1 | 13, 21, 22 | Pathogenic | ||
| 88 | 1 | dup.ex1 | CN171006133, | Pathogenic | ||
| 42 | 1–17 | del.ex1-17 | CG07271622, | Pathogenic | ||
| 131 | 6 | del.ex6 | CG07272312, | Pathogenic | ||
| 26o | 6–16 | del.ex6-16* | Pathogenic | |||
| 247 | 8–16 | del. ex8-16* | Pathogenic | |||
| 59o | 10–12 | dup.ex10-12 | CN07714534, | Pathogenic | ||
| 20 | 10–13 | del.ex10-13* | Pathogenic | |||
| 84 | 15–16 | del.ex15-16* | Pathogenic | |||
| ATL1 | 13 | 7 | c.715C > T | p.Arg239Cys | CM01329035, | Pathogenic |
| 22, 121.1 | 8 | c.757G > A | p.Val253Ile | CM04358425, | Pathogenic | |
| 70 | 8 | c.773A > G | p.His258Arg | CM01329135, | Pathogenic | |
| 200 | 10 | c.1041G > A* | p.Met347Ile* | Likely pathogenic | ||
| 67o | 12 | c.1213G > A* | p.Val405Met* | Likely pathogenic | ||
| 46o,37o, 125 | 12 | c.1243C > T | p.Arg415Trp | CM04144424, | Pathogenic | |
| 51 | 12 | c.1483C > T | p.Arg495Trp | CM04358825, | Pathogenic |
*Novel mutations.
Table 2.
Pathogenicity of the novel variants.
| Gene | Variant | Pathogenicity | Criteria |
|---|---|---|---|
| SPAST | c.284delC (p.Ala95ArgfsTerm65*) | Pathogenic | PVS1, PM1, PM2, PM5, PP3 |
| c.1107A > G (p.Thr369Thr) | Likely pathogenic | PM1, PM5, PP1, PP3 | |
| c.1116A > T (p.Arg372Term) | Pathogenic | PVS1, PM1, PM2, PM5, PP3 | |
| c.1162delA (p.Lys388ArgfsTerm8) | Pathogenic | PVS1, PM1, PM2, PM5, PP1, PP3 | |
| c.1252G > A (p.Glu418Lys) | Likely pathogenic | PM1, PM2, PM5, PP2, PP3 | |
| c.1271delG (p.Ala425LeufsTerm13) | Pathogenic | PVS1, PM1, PM2, PM5, PP3 | |
| с.1391A > G (p.Glu464Gly) | Pathogenic | PS1, PM1,PM5, PP1, PP3 | |
| c.1469delA (p.Glu491SerfsTerm39) | Pathogenic | PVS1, PM1, PM2 PM5, PP3 | |
| c.1750_1751delGAinsT (p.Asp584Serfs*5) | Pathogenic | PVS1, PM1, PM2, PM5, PP3 | |
| c.1840_1846del | Pathogenic | PVS1, PM1, PM4, PP3 | |
| del.ex6-16 | Pathogenic | PVS1, PM1, PM4, PP3 | |
| del.ex8-16 | Pathogenic | PVS1, PM1, PM4, PP3 | |
| del.ex10-13 | Pathogenic | PVS1, PM1, PM4, PP3 | |
| del.ex15-16 | Pathogenic | PVS1, PM1, PM4, PP3 | |
| ATL1 | c.1041G > A (p.Met347Ile) | Likely pathogenic | PM1, PM2, PM5, PP3 |
| c.1213G > A (p.Val405Met) | Likely pathogenic | PM1, PM2, PP1, PP3 |
To determine pathogenicity of the novel variants, we used in silico prediction programs. At least 3 programs were used to confirm the pathogenic effect of each variant on the gene or gene products. In summary the most of discovered mutations were pathogenic and likely pathogenic with high probability.
Discussion
In the current study we analyzed for the first time the incidence rates of SPG4 and SPG3 forms of HSP in a large cohort of Russian patients. We determine that the frequencies of SPG4 and SPG3 were 30.3% (37/122) and 8.2% (10/122), respectively. In the other studies, however, these forms on average were found to be less frequent. Namely, percentages of pathogenic variants of SPAST range from 14.5% of cases in Spain to 28% of cases in Germany. While ATL1 forms impact range from 1.3% of HSP cases in Germany to 4.6% of HSP cases in Poland9–14; (Table 3). The larger proportions of SPG4 and SPG3 observed in the current study, might be because the majority of Russian patients consisted of AD family cases (69/122) in contrast to the other studies.
Table 3.
Comparison of the obtained results with published data.
| Country, reference | SPG4 in general cohort | SPG4 amongst AD cases | SPG3 in general cohort | SPG3 amongst AD cases |
|---|---|---|---|---|
| Russia, present study | 31.1% (38/122) | 39.1% (27/69) | 8.2% (10/122) | 15.9% (11/69) |
| Poland15, | 18.5% (40/2016) | 38.8% (33/85) | 4.6% (10/216) | 10.6% (9/85) |
| Hungary4, | 17% (10/58) | — | 1.7% (1/58) | — |
| Spain3, | 14.5% (54/370) | 31.2% (44/141) | 2.7% (10/370) | 11.3% (10/88) |
| Germany17, | 28.7% (149/519) | 61% (121/197) | 1.3% (7/519) | 3.1% (7/222) |
| Japan16, | 24.8% (32/129) | 55.1% (27/49) | 1.6% (2/129) | 2% (1/49) |
| China5, | 22.5% (27/120) | 44.4% (24/54) | 2.5% (3/120) | 3.7% (2/54) |
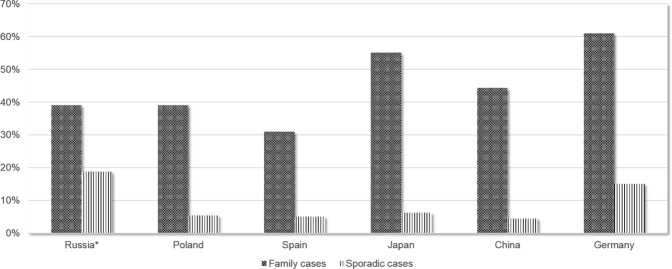
Here we show that the proportion of SPG4 among ADHSP cases is 37.7% (26/69), which is consistent with the results obtained in Spain, China and Poland9,11,12. However, it is significantly lower than the proportion of SPG4 among AD HSP unraveled by researchers from Germany (p = 0.002), e.g. 37.7% versus 61.0% (121/197)14. SPG4 fraction among AD HSP cases in Japan is also relatively high and amounts 55%13. This rates of Japan population however are not significantly higher compare to Russian cohort (p = 0.09). This might be explained by either real lack of differences, or by the smaller sample size that affects statistical significance (Figs 1, 2); (Table 3).
Figure 1.
Mutations rates among familial and sporadic cases for SPAST according to the data of researchers from different countries. * - present study.
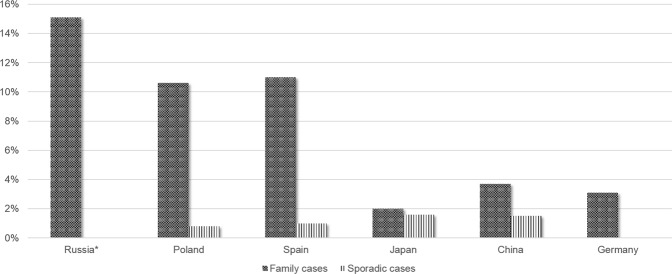
Figure 2.
Mutations rates among familial and sporadic cases for ATL1 according to the data obtained by researchers from different countries. * - the present study.
The proportion of SPG3 in Russian cohort of AD HSP is 15.9% (11/69), that is compatible to Spanish results showing 11.3% (p = 0.49) and Polish data showing 10.6% (p = 0.4)9,12. SPG3 incidence rates however are significantly higher in Russian cohort compare to Chinese data, showing 3.7% (p = 0.028), German data, showing 3.1% (p < 0.0001) and Japanese data, showing 2.0% (p = 0.014)11,13,14; (Fig. 2); (Table 3).
Mutations in SPAST and ATL1 may spread through the population and undergo fixation by random genetic drift. This can happen because SPG4 (that has mostly late onset) and SPG3 (which has mostly early onset), exhibit slow progression and a relatively mild symptoms, that don’t interfere with fertility and procreation. Also, particular HSP alleles may accumulate in different populations because of random genetic drift that depends on historical, religious, geographical and other reasons. In this way SPG4 is more common in Asian population (Chinese and Japanese), whereas SPG3, is more common among Europeans. Nevertheless there are exceptions, such as German population that differs from this trend since SPG4 demonstrate higher frequencies, whereas SPG3 is much less common compare to other European countries. In addition, proportions of SPG4 and SPG3 can be influenced by proportions of other forms of HSP in the particular cohorts. Because the research in the field of HSP genetics is very recent the complete mutational spectrum of this disease and its populational genetics in many countries has been described only partially or not at all. Consequently, new forms of HSP in certain populations, may lead to proportional decline of the other forms.
Mutations in SPAST gene identified here can be divided into 2 groups: large deletions/duplications detected by MLPA assay (29.1% of all SPAST mutations) and mutations detected via sequencing methods (70.9% of all SPAST mutations). The corresponding proportion of large deletions/duplications in SPAST gene in other cohorts was as follows: 37.5% in the Polish cohort12; 2.5% in the Spanish cohort9; 9.0% in The Republic of Bashkortostan cohort15; and 13.5% in the Australian cohort16. The proportion of large deletions/duplications detected in this study and their mutational spectrum, were similar to the results obtained by Polish researchers. This might be explained by the Slavic origin and long-standing historical relationship between these nations. In contrary, the percentage of large deletions/duplications in cohorts of Spain, The Republic of Bashkortostan, and Australia were significantly lower compare to Russian sample.
The current study did not reveal mutational hotspots or frequent mutations in SPAST gene. This is consistent with the most available data, except of the results from The Republic of Bashkortostan. Where the c.283delG (p.Ala95Profs*66) variant demonstrated high frequencies in families of Tatar ethnicity17. The c.283delG variant was not detected in our study. The pathogenic variant c.1291C > T (p.Arg431Term) described in 3 unrelated probands of our study was also identified in study from Fonknechten N. et al.18. However, there were no specific symptoms in clinical manifestation of HSP driven by this mutation.
Among repeated mutations, c.1216A > G (p.Ile406Val), c.1684C > T (p.Arg562Term), and c.1245 + 1G > A variants appeared twice in our cohort. Clinical features of patients carrying these variants were similar to those described in other studies18–20.
Other repeated variant that we found is an exon 1 deletion. It was observed in a couple of other studies with different genetic boundaries13,21,22. In the current studies boundaries of deletion were not identified.
Similar to Polish, Hungarian, Spanish and Chinese studies, our study did not unravel large deletions/duplications in ATL1 gene. Strikingly, the proportion of large deletions/duplications in the Bashkir study amounted to 1.8% (1/56) that distinguish this population15. Several mutational hotspots in exons 4, 7, 8 and 12, where pathogenic variants acquire more frequently compare to the rest of the coding sequence were described in ATL1. Most pathogenic ATL1mutations detected by our study were also located in the mutational hotspots in exons 7, 8 and 12, however we also found 1 mutation in exon 1023. Two variants c.1243C > T (p.Arg415Trp), and c.757G > A (p.Val253Ile) were reported repeatedly. Clinical profile of the patients carrying these mutations corresponds to pure HSP with age at onset (AAO) less than 10 years old24,25.
We estimated that new pathogenic variants compose 45.2% of all SPAST gene mutations identified in our study. This corresponds to the data from other countries and confirms that new mutations occur at a high rate in this gene9–13. Among 14 newly discovered variants, there were 2 missense variants, 1 non-sense variant, 6 micro-rearrangements and 4 large deletions (Table 2). Furthermore, we detected synonymous substitution in SPAST gene position c.1107A > G (p.Thr369Thr) that was predicted to be likely pathogenic in silico. Indeed, it segregates with the disease in a family.
Similar to our observations, The Human Gene Mutation Database (HGMD) describes a small number of repeated mutations in SPAST. In total only 9 variants out of 723 have been mentioned in 3 or more studies, with one mutational case per study.
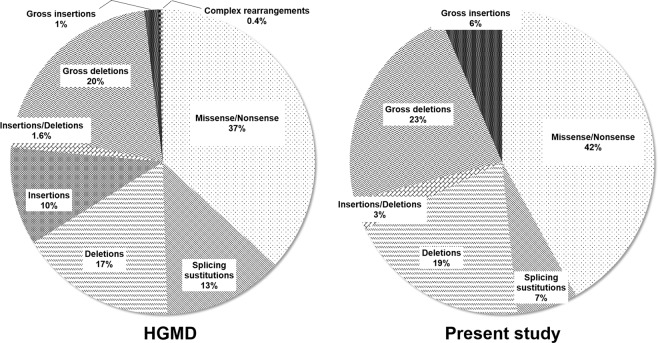
The comparison of SPAST gene mutational spectrum determined in the current study with the mutational spectrum presented in HGMD, unraveled that the proportion of large insertions in SPAST gene in Russian cohort (Fig. 3) was significantly higher than the worldwide average (p = 0.039). To another hand, splice-site modifications were less frequent in Russian population (p = 0.294). Insertions and complex rearrangements were not detected within our patients at all. The proportion of other types of mutations were similar in both samples. The observed differences may be due to the small sample size or because of regional specificity. More studies are needed to clarify this observation. Only 2 novel missense mutations were found in ATL1 gene in our study. Due to the small numbers of revealed variants the comparison with HGMD is impossible.
Figure 3.
Comparison between the mutational spectrum in SPAST gene of Russian population with the statistics of Human Gene Mutation Database.
In our and other studies, pathogenic mutations were easier to find in familial cases (52.2%)9–12. This may be because many sporadic cases misdiagnosed/confused with the other diseases.
Conclusions
HSP comprises a group of genetically heterogeneous neurodegenerative diseases that are hard to diagnose in clinics. Such difficulties are caused by the high number of clinical manifestations and clinical complications that can mask main symptoms. Also, differential diagnosis for HSPs is hampered by the existence of many phenocopies within the other non-hereditary neuropathologies. All the above factors emphasize the need of up-to-date molecular diagnostics for HSP.
The existence of frequent HSP forms and mutational hotspots in causative genes facilitates the development of a diagnostic algorithms. NGS techniques would perform in the most effective way because mutations can be found in individual genes and/or in many genes simultaneously by sequencing of gene panel, whole-exome or whole-genome. Due to the limitations of direct sequencing methods however large deletions/duplications can’t be detected by NGS. In this respect, it is necessary also to implement MLPA or other assays in the diagnostic algorithm to identify this type of mutations. The current study describes large deletions/duplications along with small DNA alterations in Russian cohort of HSP patients. Altogether that demonstrate necessity of comprehensive molecular examinations for accurate diagnose of HSP in Russian Federation.
In summary, we did not reveal any mutational hotspots or frequent mutations in SPAST gene. To another hand, in ATL1 gene the most of pathogenic and likely pathogenic variants were found in mutational hotspots of the gene (7, 8 and 12 exons). Altogether this re-confirms the global data.
In our study, we discovered new pathogenic and likely pathogenic variants of SPAST gene with 45.2% (14/31) of incidence. 20.0% of detected mutations in ATL1 were novel though the pathogenicity of the novel mutations has still to be confirmed. A comparison between obtained results and published data indicates that HSPs are extremely genetically pleomorphic and the proportions of different HSP forms can vary even among the cohorts of adjacent regions.
It was observed also that the incidence of pathogenic variants was remarkably higher among familial cases compare to sporadic cases (52.2% against 20.7%). However, patients without a family history should not be excluded from extensive genetic testing.
Acknowledgements
The research was funded by the government contract of Ministry of Science and Higher Education of Russian Federation. This work uses data provided by patients and collected by Research Centre For Medical Genetics (Moscow), Voronezh Regional Clinical Hospital №1 (Voronezh), Regional Clinical Hospital №2 (Khabarovsk). The research would not have been possible without access to patient’s data. We acknowledge all patients and their families, as well as medical doctors participated in our research for collaboration.
Author Contributions
O.P.R. and G.E.R. designed the study and planned the experiments. G.E.R. performed patients recruitment and diagnosis verification; I.G.S. and A.A.S. performed Sanger sequencing of DNA from some patients; V.A.K. performed NGS of targeted panel, MLPA-analysis and Sanger sequencing; V.A.K. analyzed data and made figures and tables; V.A.K. and O.P.R. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A. Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Exp. Neurol. 2014;261:518–539. doi: 10.1016/j.expneurol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Meszarosova AU, et al. Disease-Causing Variants in the ATL1 Gene Are a Rare Cause of Hereditary Spastic Paraplegia among Czech Patients. Ann. Hum. Genet. 2017;81:249–257. doi: 10.1111/ahg.12206. [DOI] [PubMed] [Google Scholar]
- 3.Klebe S, Stevanin G, Depienne C. Clinical and genetic heterogeneity in hereditary spastic paraplegias: from SPG1 to SPG72 and still counting. Rev. Neurol. 2015;171:505–530. doi: 10.1016/j.neurol.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Human Gene Mutation Database, Institute of Medical Genetics, Cardiff, UK. https://www.portal.biobase-international.com/hgmd/pro/start.php.
- 5.Kadnikova VA, Ryzhkova OP, Rudenskaya GE, Polyakov AV. Molecular Genetic Diversity and DNA Diagnostics of Hereditary Spastic Paraplegias. Advances in current biology. 2018;138:462–475. [Google Scholar]
- 6.Rudenskaya GE, Kadnikova VA, Ryzhkova OP. Hereditary spastic paraplegias in the era of next generation sequencing: genetic diversity. epidemiology, classification. Medical Genetics. 2018;17:3–12. [Google Scholar]
- 7.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryzhkova OP, et al. Guidelines for the interpretation of massive parallel sequencing variants (MPS) Medical Genetics. 2017;2017:4–17. [Google Scholar]
- 9.Alvarez V, et al. Mutational spectrum of the SPG4 (SPAST) and SPG3A (ATL1) genes in Spanish patients with hereditary spastic paraplegia. BMC Neurol. 2010;10:89–98. doi: 10.1186/1471-2377-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balicza P, et al. Genetic background of the hereditary spastic paraplegia phenotypes in Hungary — An analysis of 58 probands. J. Neurol. Sci. 2016;364:116–121. doi: 10.1016/j.jns.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Luo Y, et al. Mutation and clinical characteristics of autosomal-dominant hereditary spastic paraplegias in China. Neurodegener. Dis. 2014;14:176–183. doi: 10.1159/000365513. [DOI] [PubMed] [Google Scholar]
- 12.Elert-Dobkowska E, et al. Molecular spectrum of the SPAST, ATL1 and REEP1 gene mutations associated with the most common hereditary spastic paraplegias in a group of Polish patients. J. Neurol. Sci. 2015;359:35–39. doi: 10.1016/j.jns.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 13.Ishiura H, et al. Molecular epidemiology and clinical spectrum of hereditary spastic paraplegia in the Japanese population based on comprehensive mutational analyses. J. Hum. Genet. 2014;59:163–172. doi: 10.1038/jhg.2013.139. [DOI] [PubMed] [Google Scholar]
- 14.Schüle RWS, et al. Hereditary spastic paraplegia: Clinicogenetic lessons from 608 patients. Ann. Neurol. 2016;79:646–658. doi: 10.1002/ana.24611. [DOI] [PubMed] [Google Scholar]
- 15.Akhmetgaleeva AF, et al. Clinical genetic testing of the patients with hereditary spastic paraplegia in the Republic of Bashkortostan. MOLECULAR DIAGNOSTICS 2017, a collection of works of the IX All-Russian Scientific and Practical Conference with international participation. 2017;2017:47–48. [Google Scholar]
- 16.Vandebona H, Kerr NP, Liang C, Sue CM. SPAST mutations in Australian patients with hereditary spastic paraplegia. Intern. Med. J. 2012;42:1342–1347. doi: 10.1111/j.1445-5994.2012.02941.x. [DOI] [PubMed] [Google Scholar]
- 17.Khidiyatova IM, et al. Major mutation in the SPAST gene in patients with autosomal dominant spastic paraplegia from the Republic of Bashkortostan. Medical Genetics. 2019;55:229–233. [Google Scholar]
- 18.Fonknechten N, et al. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum. Mol. Genet. 2000;9:637–644. doi: 10.1093/hmg/9.4.637. [DOI] [PubMed] [Google Scholar]
- 19.Schickel J, et al. Unexpected pathogenic mechanism of a novel mutation in the coding sequence of SPG4 (spastin) Neurology. 2006;66:421–423. doi: 10.1212/01.wnl.0000196468.01815.55. [DOI] [PubMed] [Google Scholar]
- 20.Svenson IK, et al. Identification and expression analysis of spastin gene mutations in hereditary spastic paraplegia. Am. J. Hum. Genet. 2001;68:1077–1085. doi: 10.1086/320111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boone PM, et al. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am. J. Hum. Genet. 2014;95:43–61. doi: 10.1016/j.ajhg.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Depienne C, et al. Exon deletions of SPG4 are a frequent cause of hereditary spastic paraplegia. J. Med. Genet. 2007;44:281–284. doi: 10.1136/jmg.2006.046425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao GH, Liu XM. Clinical features and genotype-phenotype correlation analysis in patients with ATL1 mutations: A literature reanalysis. Transl. Neurodegener. 2017;6:9–15. doi: 10.1186/s40035-017-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Amico A, et al. Incomplete penetrance in an SPG3A-linked family with a new mutation in the atlastin gene. Neurology. 2004;62:2138–2139. doi: 10.1212/01.WNL.0000127698.88895.85. [DOI] [PubMed] [Google Scholar]
- 25.Durr A, et al. Atlastin1 mutations are frequent in young-onset autosomal dominant spastic paraplegia. Arch Neurol. 2004;61:1867–1872. doi: 10.1001/archneur.61.12.1867. [DOI] [PubMed] [Google Scholar]
- 26.Rudenskaia GE, et al. Hereditary spastic paraplegia type 4 (SPG4): clinical and molecular-genetic characteristics. Zh. Nevrol. Psikhiatr. Im. S.S. Korsakova. 2010;110:12–19. [PubMed] [Google Scholar]
- 27.Lu C, et al. Targeted next-generation sequencing improves diagnosis of hereditary spastic paraplegia in Chinese patients. J. Mol. Med. 2018;96:701–712. doi: 10.1007/s00109-018-1655-4. [DOI] [PubMed] [Google Scholar]
- 28.Loureiro J, et al. Autosomal dominant spastic paraplegias: a review of 89 families resulting from a portuguese survey. JAMA Neurol. 2013;70:481–487. doi: 10.1001/jamaneurol.2013.1956. [DOI] [PubMed] [Google Scholar]
- 29.Meijer IA, Hand CK, Cossette P, Figlewicz DA, Rouleau GA. Spectrum of SPG4 mutations in a large collection of North American families with hereditary spastic paraplegia. Arch. Neurol. 2002;59:281–286. doi: 10.1001/archneur.59.2.281. [DOI] [PubMed] [Google Scholar]
- 30.Depienne C, et al. Spastin mutations are frequent in sporadic spastic paraparesis and their spectrum is different from that observed in familial cases. J. Med. Genet. 2006;43:259–265. doi: 10.1136/jmg.2005.035311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsey JC, et al. Mutation analysis of the spastin gene (SPG4) in patients with hereditary spastic paraparesis. J. Med. Genet. 2000;37:759–765. doi: 10.1136/jmg.37.10.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDermott CJ, et al. Clinical features of hereditary spastic paraplegia due to spastin mutation. Neurology. 2006;67:45–51. doi: 10.1212/01.wnl.0000223315.62404.00. [DOI] [PubMed] [Google Scholar]
- 33.Chelban V, et al. Truncating mutations in SPAST patients are associated with a high rate of psychiatric comorbidities in hereditary spastic paraplegia. J. Neurol. Neurosurg. Psychiatry. 2017;88:681–687. doi: 10.1136/jnnp-2017-315796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitne-Neto M, et al. A multi-exonic SPG4 duplication underlies sex-dependent penetrance of hereditary spastic paraplegia in a large Brazilian pedigree. Eur. J. Hum. Genet. 2007;15:1276–1279. doi: 10.1038/sj.ejhg.5201924. [DOI] [PubMed] [Google Scholar]
- 35.Zhao X, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat. Genet. 2001;29:326–331. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]