Abstract
Enhancer RNA (eRNA) is a type of noncoding RNA transcribed from the enhancer. Although critical roles of eRNA in gene transcription control have been increasingly realized, the systemic landscape and potential function of eRNAs in cancer remains largely unexplored. Here, we report the integration of multi-omics and pharmacogenomics data across large-scale patient samples and cancer cell lines. We observe a cancer-/lineage-specificity of eRNAs, which may be largely driven by tissue-specific TFs. eRNAs are involved in multiple cancer signaling pathways through putatively regulating their target genes, including clinically actionable genes and immune checkpoints. They may also affect drug response by within-pathway or cross-pathway means. We characterize the oncogenic potential and therapeutic liability of one eRNA, NET1e, supporting the clinical feasibility of eRNA-targeted therapy. We identify a panel of clinically relevant eRNAs and developed a user-friendly data portal. Our study reveals the transcriptional landscape and clinical utility of eRNAs in cancer.
Subject terms: Cancer genomics, Non-coding RNAs
Enhancer RNA (eRNA) is a type of noncoding RNA transcribed from enhancer regions. Here, the authors investigate the transcriptional landscape of eRNA in cancer, incorporating pharmacogenomic data to identify potential target genes and therapeutic targets.
Introduction
Enhancer is a distal regulatory DNA that enhances the transcription of a target gene by interacting with target gene promoter1. Traditionally considered to be DNA elements that nucleate transcription factor (TF) binding, enhancers were recently found to also transcribe noncoding RNAs, which are referred to as enhancer RNAs (eRNAs)2. Tens of thousands of eRNAs have been identified in human cells, many of which were shown to play important roles in transcriptional circuitry to mediate the activation of target genes3.
In human cancers, activation of oncogenes or oncogenic signaling pathways often converges to enhancer activation and production of eRNAs. For example, the activation of ESR1 can globally increase eRNA transcription in breast cancer4. Oncogene-induced eRNAs can under certain circumstances directly promote tumorigenesis. For example, KLK3e, an androgen-induced eRNA regulating gene KLK3, can scaffold the androgen receptor (AR)-associated protein complex to control AR-dependent gene expression in prostate cancer5. Tumor suppressors can also induce eRNAs to contribute to tumor repression processes. For example, TP53-induced eRNAs were found to be involved in p53-dependent cell cycle arrest in multiple cancer cell lines6. Together these evidences reveal significant roles of eRNAs in tumorigenesis and suggest their clinical utility in eRNA-targeted therapy7.
The Encyclopedia of DNA Elements (ENCODE) project8, Functional Annotation of the Mammalian Genome (FANTOM) project9, and Roadmap Epigenomics project10 have annotated a large number of regulatory elements, including enhancers, while The Cancer Genome Atlas (TCGA) collected multi-omic data and clinical information in ~10,000 tumor samples11. In addition to patient samples, Cancer Cell Line Encyclopedia (CCLE)12 collected omics data in ~1000 cancer cell lines. Furthermore, Cancer Therapeutics Response Portal (CTRP)13, and Genomics of Drug Sensitivity in Cancer (GDSC)14 provided pharmacogenomics data from ~ 500 anticancer compounds across > 1000 cancer cell lines. These data resources provide unique opportunities to characterize the expression landscape, functions and drug response of eRNAs across different cancer types.
Results
Dynamic expression landscape of eRNAs in human cancers
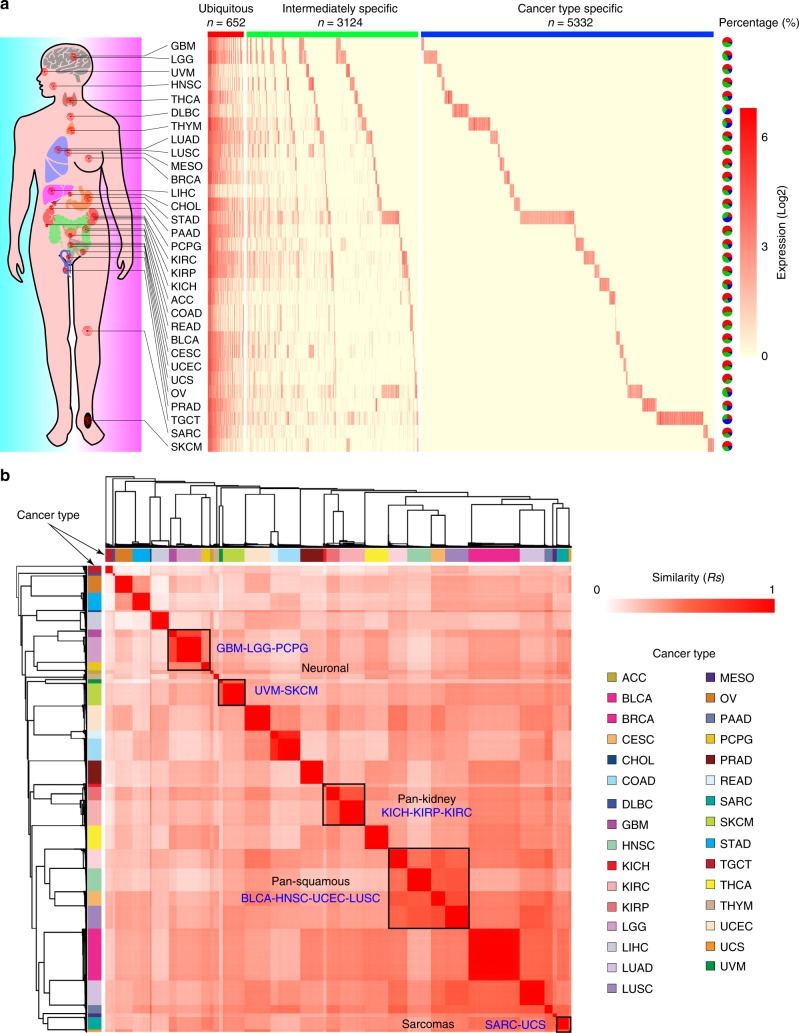
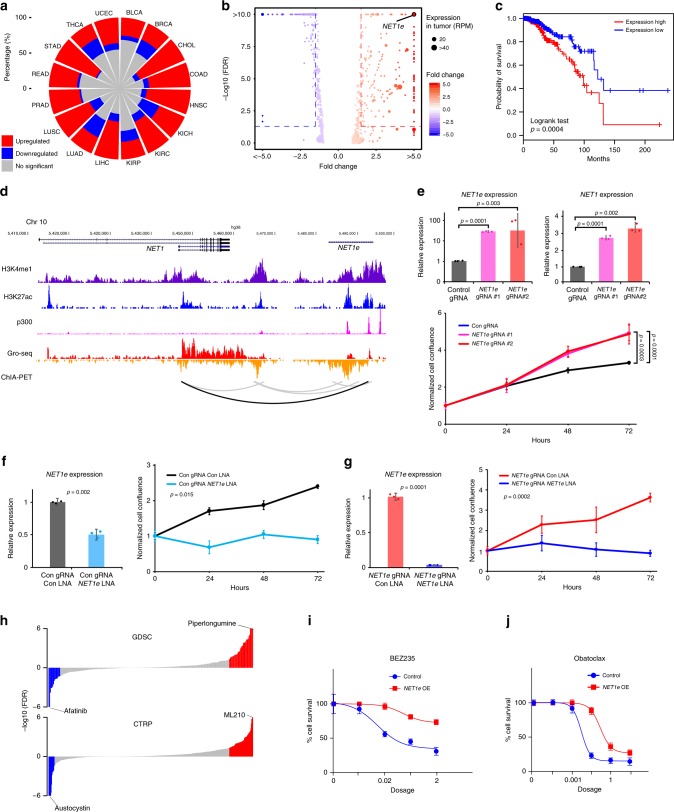
We obtained enhancer annotations from ENCODE, FANTOM, and Roadmap Epigenomics, and selected enhancers annotated in at least two datasets. Given the fact that eRNA transcription region could be wider than the enhancer ChIP-seq peaks15, we defined the ± 3 kb regions around the middle point of these annotated enhancers as potential eRNA-transcribing regions16. To avoid counting the transcriptional signal from known coding genes, we excluded eRNA regions that overlap with known coding genes and lncRNAs (with 1 kb extension from both transcription start site and transcription end site) (Supplementary Fig. 1A and Methods). To characterize the expression landscape of eRNAs across human cancers, we mapped TCGA RNA-seq reads to eRNA regions and defined those eRNAs with average expression value (reads per million, RPM) ≥1 as detectable eRNA for each cancer type (Supplementary Fig. 1A and Methods). This analysis identified a total of 9108 detectable eRNAs in human cancers (Fig. 1a and Supplementary Fig. 1B). The number of detectable eRNAs ranged from 457 in liver hepatocellular carcinoma (LIHC) to 2267 in stomach adenocarcinoma (STAD) (Supplementary Fig. 1B, Supplementary Data 1). We classified these detectable eRNAs into three groups: 652 ubiquitous eRNAs expressed in ≥10 cancer types, 3124 intermediately specific eRNAs that are expressed in 2–9 cancer types, and 5332 cancer-type-specific eRNAs that are expressed in only one cancer type (Fig. 1a). The ubiquitous eRNAs account for 20.0% of detectable eRNAs in STAD, but for 64.8% of eRNAs in uterine corpus endometrial carcinoma (UCEC). Interestingly, the ubiquitous eRNAs have higher expression levels than the intermediately specific eRNAs (Wilcoxon test p-value < 2.2 × 10–16) and the cancer-type-specific eRNAs (Wilcoxon test p-value < 2.2 × 10–16, Supplementary Fig. 1C). This phenomenon is reminiscent of features of protein-coding genes, among which the housekeeping genes are generally expressed at high levels as compared with tissue-specific genes17. Numbers of cancer-type-specific eRNAs showed a broad range, from 4 in colon adenocarcinoma (COAD) to 987 in STAD (Fig. 1a). We still observed cancer-type-specific pattern even with a much more stringent cutoff (RPM ≥ 5, Supplementary Fig. 1D), suggesting that the cancer-type-specific patterns of eRNA expression is not due to expression levels.
Fig. 1.
Expression landscape of eRNAs in human cancers. a Expression profile of eRNA in human cancers. Red, green and blue bars, respectively, denote ubiquitous, intermediately specific and cancer-type-specific eRNAs. Pie charts reflect the percentage of each category of eRNAs in each cancer type. Abbreviation of each cancer type is listed in Supplementary Data 1. Scale bar denotes the expression level of eRNAs (log2). Organ icons made by Freepik (https://www.freepik.com/). b Expression similarity among tumor samples. Color bar denotes each cancer type. Scale bar denotes the similarity of eRNA expression among TCGA samples
The expression similarity between any two tumor samples further showed a strong cancer-type-specific pattern, in that samples from the same cancer type clustered together (Fig. 1b). Furthermore, cancer types with similar histological features clustered at higher levels of hierarchy, such as pan-kidney cancers18 (kidney renal clear cell carcinoma [KIRC], kidney renal papillary cell carcinoma [KIRP] and kidney chromophobe [KICH]), pan-squamous cell carcinomas19 (bladder urothelial carcinoma [BLCA], head and neck squamous cell carcinoma [HNSC], cervical squamous cell carcinoma and endocervical adenocarcinoma [CESC] and lung squamous cell carcinoma [LUSC]), the sarcomas20 (sarcoma [SARC] and uterine carcinosarcoma [UCS]), and the neuronal cancers20 (glioblastoma multiforme [GBM], brain lower grade glioma [LGG], pheochromocytoma and paraganglioma [PCPG], skin cutaneous melanoma [SKCM], and uveal melanoma [UVM]). This cancer-type-specific pattern is further confirmed by t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis (Supplementary Fig. 1E), suggesting that eRNAs may be powerful biomarkers with clinical utility in specific cancer types7.
Analysis of transcription factor and eRNA relationship
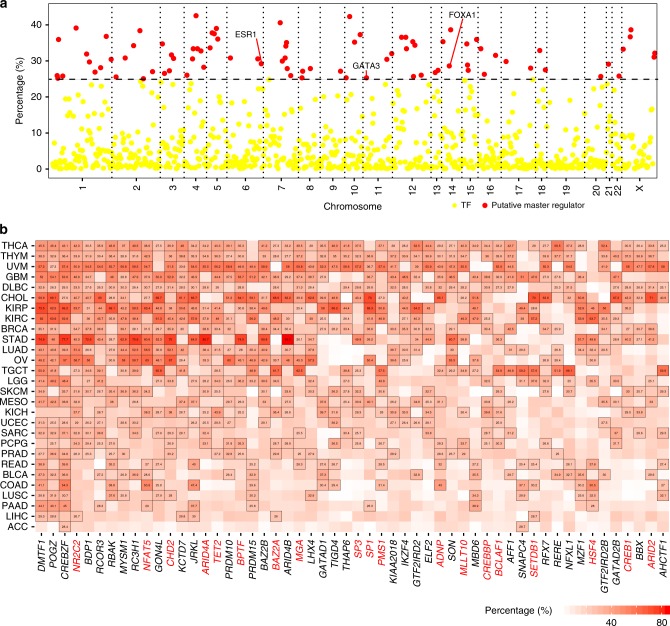
TFs have been shown to mediate the biogenesis of eRNAs2,8; however, the global regulation of eRNAs is still unclear. Here, we collected human transcription factors (TFs) from JASPAR21, DBD22, AnimalTFDB23, and TF2DNA24, and calculated Spearman’s correlation between individual TF expression and individual eRNA expression in each cancer type. We defined TFs that show significant correlation (Rs ≥ 0.3; false discovery rate [FDR] <0.05) with an individual eRNA in a specific cancer type as its putative regulators, and further defined those putative regulators significantly correlated to ≥ 25% of individual eRNAs in a specific cancer type as putative master regulators. Taking breast invasive carcinoma (BRCA) as an example, we identified 84 putative master regulators, including three well-known regulators, FOXA125 (highly correlated with 28.6% of all eRNAs in BRCA), ESR115 (29.2%), and GATA326 (25.3%, Fig. 2a and Supplementary Data 2). Applying this computational predictions, we have identified 845 putative master regulators across cancer types (Supplementary Fig. 2A and Supplementary Data 2). The majority of these putative master regulators (693/845, 82.0%) exhibits strong expression correlation with a large number of eRNAs in only one or a few cancer types (i.e., <5), suggesting that the TF-eRNA correlation is tissue-specific and may imply direct regulatory functions of these TFs in that cancer type (Supplementary Fig. 2B). For example, OLIG2 is a TF highly expressed in brain and highly correlate with the expression of 33.5% of eRNAs in LGG, suggesting its potential importance in enhancer/eRNA control therein (Supplementary Fig. 2C). Our global analysis of TF-eRNA correlation indicates that cancer- and/or lineage-specific patterns of eRNAs can be largely mediated by lineage-specific TFs.
Fig. 2.
Putative regulation of eRNA biogenesis in cancer. a Putative regulators of eRNAs in BRCA. Each dot represents one transcription factor (TF). Red dots denote putative master regulators significantly correlated to ≥25% of individual eRNAs. Three well-known TFs (ESR1, GATA3, and FOXA1) are highlighted. X-axis is chromosome; y-axis is percentage of eRNAs correlated to each TF. b General putative master regulators in human cancers. X-axis is symbol of putative master regulator and y-axis is cancer type. General putative master regulators involved in genomic instability are marked by red. Number in cells denotes the percentage of eRNAs regulated by each master regulator. Scale bar denotes percentage of eRNA correlated with TFs (x axis) across cancer types (y axis)
We further identified 54 general putative master regulators that play significant roles in ≥ 10 cancer types (Fig. 2b). We performed GO analyses and observed that these TFs are enriched in the functional categories related to transcriptional process (Supplementary Fig. 2D-2E). These general master regulators can be classified into 17 families base on Pfam annotation (https://pfam.xfam.org/), and they are significantly enriched in four families, including MDB, ARID, GTF2I, and MYB (FDR <0.05, Supplementary Fig. 2F). More importantly, we manually examined the functions of these TFs and found 35.2% (19/54) of them are associated with genomic instability (Fig. 2b). For example, NR2C2, which can mediate genomic rearrangements by a telomere-related pathway27, is highly correlated with eRNAs in 20 cancer types, ranging from 26.3% in PRAD to 53.7% in KIRP. NFAT5, which can induce genomic instability by regulating inflammation28,29, is highly correlated with eRNAs in 17 cancer types, ranging from 27.0% in READ to 63.4% in KIRP. These general master regulators are enriched in functions related to genomic instability, which provides a potential explanation to a previously observed positive correlation between somatic copy number alteration and enhancer activation in many cancer types30.
Putative effects of eRNAs on signaling pathways
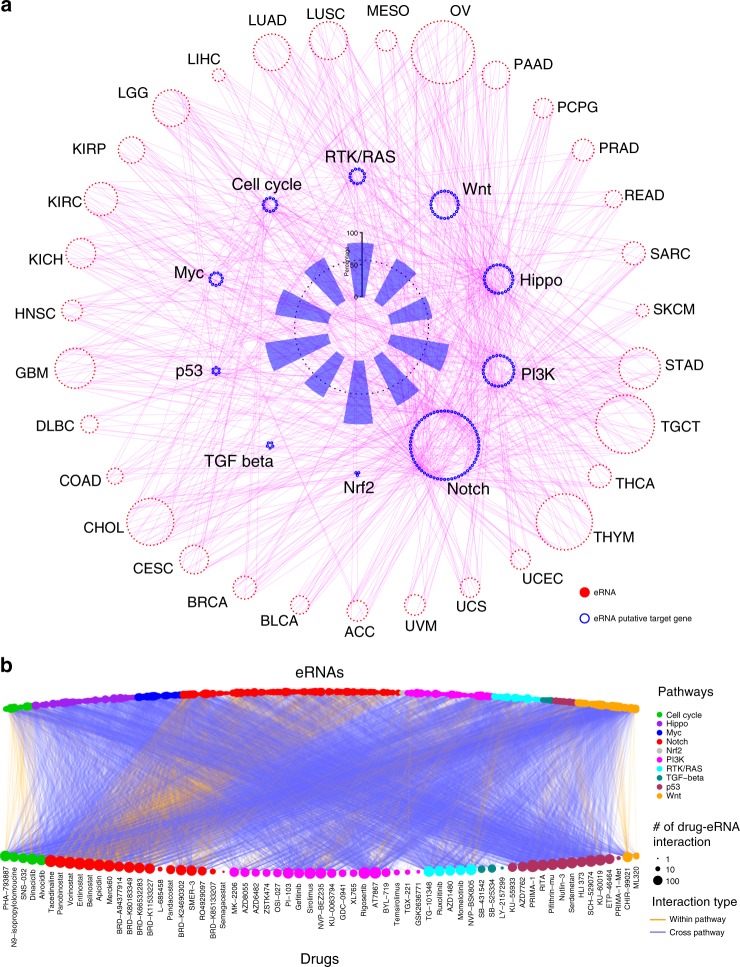
It remains a challenge to establish the direct interaction between eRNA and its target genes. We built a global eRNA-gene regulatory network across cancer types based on the physical distance (≤ 1MB) and co-expression between individual eRNAs and their putative target genes (Spearman’s correlation Rs ≥ 0.3, FDR < 0.05, Supplementary Fig. 3A)2. We identified 11,593 (56.5% of all protein-coding genes) putative target genes that are significantly correlated with 88.8% (8086/9108) of eRNAs in at least one cancer type. High-throughput chromosome conformation capture (Hi-C) data can reveal the interaction between an enhancer and its target gene, while active enhancer usually produce eRNA31–33. Therefore, we investigated Hi-C interaction for all putative eRNA-gene connections across 20 tissues, and observed that more than 80% eRNA-gene connections are supported by significant Hi-C interactions in at least one tissue (Supplementary Fig. 3B). The proportion of Hi-C supported eRNA-gene connection is significant higher than the background of random pairs (permutation test, bootstrap = 10,000, p < 0.0001, Supplementary Fig. 3B). To explore the regulatory roles of eRNAs in cancer, we collected 229 genes involved in 10 cancer signaling pathways, including Myc, PI3K, and p53 pathways34 (Supplementary Data 3). The majority (185/229, 80.8%) of these genes are correlated with eRNAs in at least one cancer type (Fig. 3a and Supplementary Data 3). For example, all six genes in the p53 pathway (i.e., TP53, MDM2, MDM4, ATM, CHEK2, and RPS6KA3) were found to correlate with eRNAs in at least one cancer type (Supplementary Fig. 3C and Supplementary Data 3). In support of this, most eRNA-gene associations in pathways (91.9%, 170/185) were found to form chromatin interaction by Hi-C (Supplementary Fig. 3D and 3E), including MAML2-associated eRNA (hereafter we will refer to eRNAs based on their associated, putative target gene, i.e., MAML2e, ENSR00000043746) and MAML2, CDK6-associated eRNA (CDK6e, ENSR00000215101) and CDK6, and TCF7L2-associated eRNA (TCF7L2e, ENSR00000033597) and TCF7L2 (Supplementary Fig. 3F). Our results suggested important roles played by eRNAs in regulating various cancer signaling pathways.
Fig. 3.
Putatively regulatory network and drug response of eRNAs on signaling pathways. a eRNA and putative target gene in 10 cancer signaling pathways. Red nodes in outer circle denote eRNAs that regulate genes in cancer signaling pathways across cancer types. Blue nodes in middle circle denote putative target gene across cancer signaling pathways. Blue bars in inner circle denote percentage of eRNA putative target genes across each pathway. Magenta links denote correlation between eRNAs and their putative target genes. b Association between eRNAs (top) and drugs (bottom) across different cancer signaling pathways. Orange and blue links, respectively, denote within- and cross-pathway relationships
To further understand the meaningful contributions of eRNAs in cancer signaling pathways on drug response, we calculated eRNA expression levels across ~1000 cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE), and then analyzed Spearman’s correlation between eRNA expression levels and drug sensitivity of these cells (Area Under Curve [AUC]), which is available from the Cancer Therapeutics Response Portal (CTRP). We identified 512 eRNAs in all 10 cancer signaling pathways, the expression of which displayed high correlation with 63 anticancer drugs (FDR < 0.0535, Fig. 3b and Supplementary Fig. 3G), suggesting significant roles of eRNAs in the response to anticancer drugs. For example, 217 eRNAs are highly correlated with belinostat, a drug that targets the Notch pathway. Among these, 32.7% (71/217) of their putative target genes are within the Notch pathway (Supplementary Fig. 3H), such as PSEN2-associated eRNA (PSEN2e, ENSR00000257043), RBX1-associated eRNA (PBX1e, ENSR00000257043), and NOTCH4 associated eRNA (NOTCH4e, ENSR00000320261). More interestingly, the putative target genes of the remaining eRNAs (146/217, 67.4%) are in cross-pathways, such as MDM2-associated eRNA (MDM2e, ENSR00000053727) in the p53 pathway, CDK6-associated eRNA (CDK6e, ENSR00000215101) in the cell cycle pathway, and RNF43-associated eRNA (RNF43e, ENSR00000096250) in the Wnt pathway (Supplementary Fig. 3H). We further confirmed this eRNA-drug connection using another drug database, Genomics of Drug Sensitivity in Cancer (GDSC), and observed some similar pattern (Supplementary Fig. 3I and 3J). Indeed, belinostat treatment could alter the expression of 46 eRNAs (35.7%) within the target pathway and 83 eRNAs (64.3%) in a cross-pathway in A549 cells (Supplementary Fig. 3K). Taken together, our results suggest a strong association between eRNAs and anticancer drugs, either within the target pathway or through a cross-pathway. It will an important future direction to examine the molecular basis of eRNA-gene-drug correlation, and potential roles eRNAs played in modulating cancer cell drug response.
Putatively regulation of eRNAs on CAGs and ICs
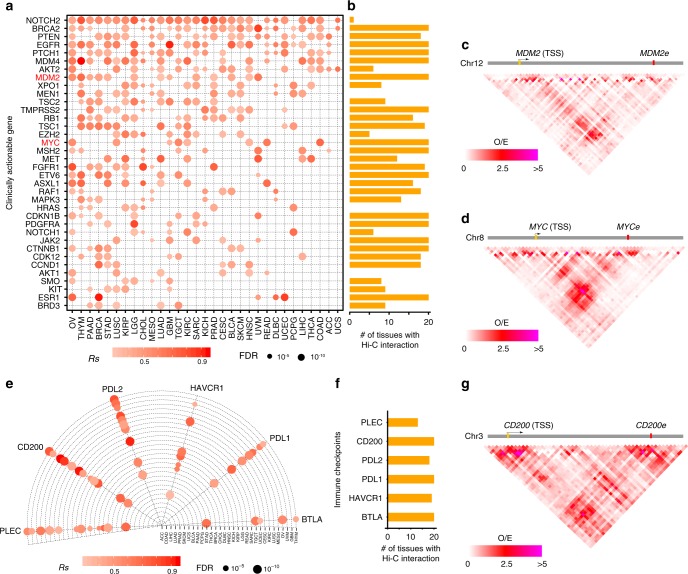
Based on the finding that eRNAs were tightly associated with cancer signaling pathways and drug-associated pathways, we further asked if eRNAs were directly linked to cancer therapy. We collected 135 clinically actionable genes (CAGs), and observed that 107 of them (79.3%) were correlated to eRNAs in at least one cancer type (distance ≤ 1MB, Spearman’s correlation Rs ≥ 0.3 and FDR < 0.05, Supplementary Data 4). Among these, 36 clinically actionable genes are correlated with eRNAs in at least five cancer types (Fig. 4a), suggesting that these genes are potentially regulated by eRNAs in multiple cancers. Increased numbers of samples enhance the ability to detect and analyze molecular data. In particular, the pan-cancer analysis will help to identify master events that play a critical functional role in different tumor contexts36–38. Furthermore, 91.7% of these correlations (33/36) could be supported by Hi-C interactions in at least one tissue, which further support the potential regulatory roles of eRNAs on clinically actionable genes (Fig. 4b and Supplementary Fig. 4A). For example, MDM2- and MDM2-associated eRNA (MDM2e, ENSR00000053727) are positively correlated in 12 cancer types, and Hi-C data supports their chromatin interaction in 20 tissues (Fig. 4c and Supplementary Fig. 4A). MYC- and MYC-associated eRNA (MYCe, ENSR00000333355) are positively correlated in six cancer types, and Hi-C data supports their interaction in all 20 tissues (Fig. 4d and Supplementary Fig. 4A).
Fig. 4.
Putatively regulatory effects of eRNAs on clinically actionable genes and immune checkpoints. a Correlation between eRNAs and clinically actionable genes in human cancers. Red dots denote significantly correlation. X-axis is cancer type and y-axis is symbol of gene. b Number of tissues with Hi-C interactions between clinically actionable genes and their eRNAs. c Hi-C interaction between MDM2 and MDM2e in blade. d Hi-C interaction between MYC and MYCe in lung. e Correlation between eRNA and cancer immune checkpoints in human cancers. f Number of tissues with remarkable Hi-C interaction between cancer immune checkpoints and their eRNA. g Hi-C interaction between CD200 and CD200e in ovary. Scale bars denote spearman correlation (Rs) in (a) and (e), and Hi-C O/E value in (b), (c), and (g), respectively
We further investigated the relationship between individual eRNAs and immune checkpoints (ICs, Supplementary Data 5), and observed six checkpoints were correlated with eRNAs in at least five cancer types (Fig. 4e). All these putative eRNA-checkpoints interactions again were supported by Hi-C data in at least one tissue (Fig. 4f and Supplementary Fig. 4B). For example, CD200- and CD200-associated eRNA (CD200e, ENSR00000156542) are positively correlated in 12 cancer types, and Hi-C data supports the interactions in all 20 tissues (Fig. 4g and Supplementary Fig. 4B). Taken together, our analysis showed putative interactions between eRNAs and clinically actionable genes and/or immune checkpoints, suggesting the potentially clinical utility of eRNAs in cancer therapy.
Characterizing the functional roles of eRNA in cancer
To further characterize the functional roles of eRNAs in cancer, we examined the differentially expressed eRNAs (|fold change| >1.5 and FDR <0.05) across 16 cancer types with ≥5 tumor-normal paired samples (Supplementary Data 6). Overall, there were more upregulated eRNAs in tumor samples, ranging from 22.0% in thyroid carcinoma (THCA) to 68.9% in cholangiocarcinoma (CHOL), with a median of 42.2%. The downregulated eRNAs ranged from 1.9% in STAD to 27.9% in KICH, with a median of 9.9% (Fig. 5a). Taking BRCA as an example, we identified 208 upregulated eRNAs and 166 downregulated eRNAs (Fig. 5b). Among these, one eRNA located ~ 90 kb downstream of the oncogene NET139, which we referred to as NET1-associated eRNA (NET1e, ENSR00000023843), showed the largest expression alteration between tumor and normal samples (fold change = 5.8, FDR = 3.7 × 10–13, Fig. 5b and Supplementary Fig. 5A). NET1e exhibited much higher expression levels in BRCA, including all subtypes (Supplementary Fig. 5B). High level of NET1e was associated with worse survival (log-rank test p-value = 0.0004, Fig. 5c). Of interest, NET1 gene itself is not associated with the breast cancer patient’s survival (Supplementary Fig. 5C), suggesting that NET1e may be a predictor irrelevant to NET1 in breast cancer patients. NET1e was highly correlated with NET1 across all BRCA subtypes (Fisher’s transformation, Rs’ = 0.45, p′ = 1.58 × 10–4), including the basal subtype (Spearman’s correlation, Rs = 0.53, p = 1.95 × 10–11, Supplementary Fig. 5D). We further examined NET1e signaling in MCF7, a breast cancer cell line (Fig. 5d). This region harbors classical enhancer features, such as the enrichment of histone H3K4me1 modification; it also exhibits strong enrichment of active enhancer markers such as histone H3K27ac modification and binding of transcription co-factor p300 (Fig. 5d). There are multiple p300 binding peaks densely distributed in the NET1e region, indicating it might be a potential super-enhancer in MCF7. NET1e transcription was also detected by GRO-seq data in MCF7 cells (Fig. 5d). Furthermore, we observed a chromatin interaction between NET1e and NET1 by RNA Pol II ChIA-PET21, suggesting a direct interaction for regulation.
Fig. 5.
Functional characterization of NET1e in cancer. a Expression alterations of eRNAs in human cancers. Red, blue and gray, respectively, denote percentage of upregulated, downregulated and no significant expression of eRNAs in each cancer type. b Differentially expressed eRNAs in BRCA. Scale bar denotes the fold change between tumor and normal samples. c Survival curves for NET1e in BRCA (top quarter in red vs. bottom quarter in blue). d Epigenetic features and expression of NET1e and NET1 region in MCF7. e Overexpression of NET1e in MCF7 and its functional consequences. Overexpression of NET1e by CRISPR/dCas9-SAM (top left) and NET1 (top right), and cell growth assay with overexpression of NET1e in MCF7 (bottom). f Knockdown NET1e by LNA (left) and cell growth assay (right) in MCF7. g Knockdown NET1e by LNA after activation (left) and cell growth assay (right) in MCF7. h NET1e-associated drugs in GDSC and CTRP database. i Response curve of compound BEZ235 in NET1e overexpressed cells and control. j Response curve of compound Obatoclax in NET1e overexpressed cells and control. Error bars in (e), (f), (g), (i), and (j) indicate mean ± SD
To further characterize NET1e, we applied CRISPR activation (CRISPR/dCas9-SAM)40 to epigenetically induce NET1e expression in MCF7 cells (Fig. 5e). We successfully achieved >30-fold NET1e upregulation by two different sgRNAs, which interestingly led to strong upregulation of NET1 mRNA (Fig. 5e). Consistent with the role of NET1 as a breast cancer oncogene39, CRISPR-SAM induction of NET1e increased cell proliferation significantly (Fig. 5e). To delineate a role of the eRNA per se, we designed three locked nucleic acid GapmeR (LNAs) to knockdown NET1e. With efficient reduction of NET1e expression, we found that cell proliferation was significantly reduced in both MCF7 cells (Fig. 5f) and MCF7 cells with CRISPR-SAM treatment (Fig. 5g). These data, together with their chromatin looping (Fig. 5d), suggested that NET1e contributes to breast cancer progression via upregulation of the important breast cancer oncogene NET1. In addition, knockdown of NET1e did not significantly impact cell proliferation in non-breast cancer cell lines, including MCF10A and Hela, in which NET1e shows low expression level (Supplementary Fig. 5E and 5F), suggesting a specific effect of NET1e in breast cancer growth. It supports a minimal off-target effects/toxicity of NET1e LNA and the potential to target cancer-specific eRNAs for effective treatment. More importantly, expression of NET1e is negatively correlated (sensitive, Spearman’s correlation, FDR <0.05) with 14 and 15 compounds response (AUC) while positively correlated (resistance, Spearman’s correlation, FDR < 0.05) with 56 and 31 compounds response (AUC) in CTRP and GDSC, respectively, which suggested that altered expression of NET1e could influence response to these drugs (Fig. 5h and Supplementary Data 7). Indeed, in situ overexpression of NET1e led to the resistance of MCF7 cells to a PI3K inhibitor, BEZ235 (Fig. 5i), and a BCL-2 Inhibitor, Obatoclax (Fig. 5j) in MCF7 cells. We also examined the effects for the other three drugs (CHIR-99021, BX-795, and (5Z)-7-Oxozeaenol) and observed a similar trend but not statistically significant. Cells showed strong growth inhibition when we knocked down NET1e in MCF7 (Fig. 5f, g), therefore we could not test drug response in NET1e KD cells. Of interest, NET1 is not significantly correlated with BEZ235 (FDR = 0.15) and obatoclax (FDR = 0.80). Taken together, these results revealed that NET1e is an oncogenic eRNA in BRCA and may be a promising target for eRNA therapy.
Identification of clinically relevant eRNAs
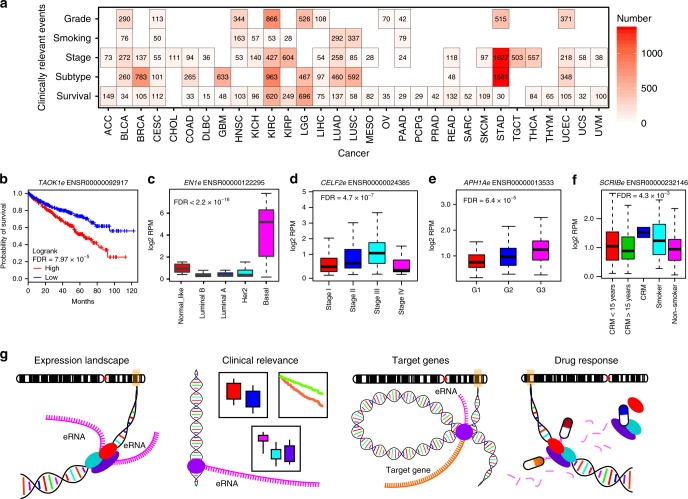
Clinical relevance is used to define cancer-related clinical features, including association with survival, differential expression among subtypes, stages, grade, and different groups of smoking history41–43. To further investigate the clinical utility of eRNAs, we identified 5715 clinically relevant eRNAs (i.e., associated with clinical relevance) that account for 62.7% (5715/9108) of the total detectable eRNAs in cancers (Fig. 6a and Supplementary Data 8). For example, TAOK1-associated eRNA (TAOK1e, ENSR00000092917), which putatively targets the Hippo signaling pathway gene TAOK144, is associated with overall survival in KIRC (Fig. 6b, log-rank test, FDR = 7.97 × 10–5); EN1-associated eRNA (EN1e, ENSR00000122295), which putatively targets the BRCA-basal marker gene EN145, is highly expressed in the BRCA-basal subtype (Fig. 6c, Analysis of variance [ANOVA], FDR < 2.2 × 10–16); CELF2-associated eRNA (CELF2e, ENSR00000024385), which putatively targets the tumor suppressor gene CELF246, is highly expressed in stage III STAD (Fig. 6d, ANOVA, FDR = 4.7 × 10–7); APH1A-associated eRNA (APH1Ae, ENSR00000013533), which putatively targets the oncogene APH1A47, is highly expressed in grade-3 LIHC (Fig. 6e, ANOVA, FDR = 6.4 × 10–5); and SCRIB-associated eRNA (SCRIBe, ENSR00000232146), which putatively targets the oncogene SCRIB48, is differentially expressed among patients with LUAD according to different categories of smoking history (Fig. 6f, ANOVA, FDR < 4.3 × 10–3). These results suggest that appreciable levels of eRNAs are clinically valuable.
Fig. 6.
Clinically relevant eRNAs and the eRNA data portal in cancer (eRic). a Number of clinically relevant eRNAs across different cancer types. For survival analyses, we use cox model to analyze the relationship between eRNA expression and survival time, and considered FDR < 0.05 as significant. For other clinical relevance, we used Student’s t test to test the difference between two groups and analysis of variance (ANOVA) to test the difference among multiple groups, and considered FDR < 0.05 as significant. Scale bar denotes number of clinically relevant eRNAs. b–f Examples of clinically relevant eRNAs for patient survival time (b), cancer subtype (c), tumor stage (d), tumor grade (e), and patient smoking history (f). The boxes in (c–f) show the median ± 1 quartile, with whiskers extending from the hinge to the smallest or largest value within 1.5 interquartile range from the box boundaries. g The four modules in eRic: expression landscape, clinical relevance, putative target genes, and drug response
A comprehensive data resource to explore eRNAs in cancer
We developed a user-friendly data portal, eRNA in cancer (eRic) (https://hanlab.uth.edu/eRic), to facilitate broad access to these data by the biomedical community. eRic includes four modules: expression, clinical relevance, target genes and drug response (Fig. 6g). In the eRNA-expression module, users can explore the expression of eRNA across TCGA cancer types and samples and the eRNA location by Ensembl ID or genomic location. The clinical relevance module aims to help users identify clinically relevant eRNAs, including those that showed differentially expressed patterns between tumor and normal samples among different groups of cancer subtypes, stages, and grades and different categories of patient smoking history, and in association with patient survival times. The target genes module allows users to identify eRNA target genes (Supplementary Fig. 6A). We also integrated the drug response data from GDSC and CTRP, which allows users to investigate whether an eRNA shows sensitivity or resistance to drugs (Supplementary Fig. 6B and 6C). In addition, eRic provides a download module, which allows users to download the expression, clinical relevance, targeted genes, and drug response data. This valuable resource will be of significant interest to the research community49.
Discussion
eRNA are increasingly realized to play important roles in the regulation of gene transcriptional circuitry in human cancers. We developed a computational pipeline to reveal the global expression landscape of eRNAs across multiple cancer types. By integrating multi-omics data from TCGA, CCLE, ENCODE, FANTOM, Roadmap Epigenomics, and 4D Nucleome projects, as well as pharmacogenomics datasets from GDSC and CTRP, we have revealed novel insights on the expression landscape and clinical utility of eRNAs in cancer (Supplementary Fig. 7 and Supplementary Table 1). We demonstrated a strong cancer-type-specific expression pattern of many eRNAs, suggesting that eRNAs may be powerful diagnostic and/or prognostic markers in cancer therapy. The cancer-type-specific pattern is aligned well with the previous studies characterizing the regulatory elements (e.g., enhancers) based on ATAC-seq50, as well as activated enhancers30. The sequencing depth may still cause some batch effects. For those cancer types with 75/76 bp pair-end reads, including OV, STAD, and GBM, only STAD showed the relative more eRNAs. In contrast, COAD, READ, and UCEC with 76 bp single-end reads have relative fewer eRNAs. The majority (25 out of 31) of cancer types with 48/50 bp pair-end reads showed vast difference in detectable eRNAs ranging from 457 in LIHC to 1790 in TGCT (Supplementary Data 1). The number of eRNAs is not correlated with the sequencing depth in these cancer types (Rs = 0.02, p = 0.93). These results suggested that the tissue-specific pattern of eRNAs is robust. We identified a series of transcription factors as the potential regulators for eRNA biogenesis, which greatly expanded our knowledge about eRNA biogenesis. Interestingly, we observed that the general putative master regulators of eRNAs, including NR2C2 and NFAT5, displayed an intriguing enrichment of functions in modulating genomic instability, suggesting a potential mechanistic link between eRNA expression/biogenesis and genome instability.
Integrative analysis showed that more than 80% genes in the canonical cancer signaling pathways are highly correlated with specific eRNAs in at least one cancer type, suggesting potentially important regulatory roles of eRNA in cancer. Due to the lack of Hi-C data in large number of tumor tissues, we can only confirm the eRNA-gene connections support by Hi-C interaction in at least one normal tissue. It will be more appropriate to use the Hi-C data in matched samples to confirm the regulatory roles of eRNAs. We also observed associations between eRNAs and anticancer drugs, either within the target pathway or through a cross-pathway. Furthermore, many clinically actionable genes and immune checkpoints were putatively regulated by eRNAs, emphasizing the clinical utility of eRNA in anticancer treatment. Nevertheless, our integrative analysis demonstrated the putative regulatory roles of eRNAs, and further experiments are necessary to confirm their regulatory roles.
We demonstrated the functional importance of an individual eRNA, NET1e, which is highly expressed in breast cancer. CRISPR activation of NET1e accelerated cell growth in MCF7, suggesting its oncogenic effect in cancer cell lines, while NET1e LNA specifically decreased cell proliferation in MCF7, and has shown limited or no off-target effects and toxicity. More importantly, in situ overexpression of NET1e will lead to drug resistance to BEZ235 and Obatoclax in MCF7 cells. To our knowledge, this is the first evidence showing that eRNA could affect drug response in cancer. Taken together, our results suggest the promising clinical importance of NET1e.
RNA-target drugs are now becoming a major new branch of pharmaceuticals. For example, US FDA has approved the first siRNA drug in 2018 (i.e., to use Patisiran infusion for the treatment of peripheral nerve disease like polyneuropathy), and there are extensive ongoing efforts to target disease-relevant RNAs in the pharmaceutical industry. We identified an appreciable number of clinically relevant eRNAs and further demonstrated their clinical utility in diagnostic and/or eRNA-targeted therapy. To facilitate utilization of the expression landscape and clinical relevance of eRNAs by the broad biomedical community, we have built a data portal, eRic, offering a comprehensive resource for further investigation of eRNA expression landscape, clinical relevance, target genes, functions in tumorigenesis or response to anticancer drugs. This is the first data portal in eRNA field and will be a valuable resource for further investigation of cancer therapy that targets eRNAs. In particular, the related data will help the researchers to identify key eRNAs in cancer patients, and to select the appropriate cancer cell lines for their functional investigations.
Methods
Data collection
We downloaded RNA-seq BAM files, clinical features and the mRNA expression matrix from TCGA data portal (https://portal.gdc.cancer.gov/)11. GRO-seq data and ChIP-seq for MCF7 were collected from our previous paper4. ChIA-PET data were obtained from WASHU EpiGenome Browser (https://epigenomegateway.wustl.edu/)21. RNA-seq data of cancer cell lines were downloaded from the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle/about)46. Drug sensitivity datasets were downloaded from GDSC14 and CTRP51. The Hi-C interactions across 20 human tissues were downloaded from http://promoter.bx.psu.edu/public/HiCPlus/matrix/52. Clinically actionable genes were collected from previous literatures37,38,53, and cancer immune checkpoints were collected from a previous literature54. RNA-seq data for A549 treated by belinostat (GSE96649) was downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), and processed by Hisat2 software55 and SAMtools toolkit56.
Quantification of eRNA expression
Annotation of enhancers were collected from Ensembl (https://useast.ensembl.org/)57, FANTOM (http://fantom.gsc.riken.jp/index.html)9, and Roadmap Epigenomics (http://www.roadmapepigenomics.org/)10. The annotation from ENCODE and Roadmap considered H3K4me1 and H3K27ac marks57, and annotation from FANTOM considered CAGE marks. We combined all three datasets and used those enhancers annotated in at least two datasets. Annotation of protein-coding genes was collected from GENECODE58 and UCSC Genome Browser59 (hg38). We used the ± 3 kb of the middle loci of enhancer to define eRNA region16. We also filtered out those eRNA regions that overlapped with known coding regions and lncRNAs (with 1 kb extension from both transcription start site and transcription end site). In particular, the ~ 500 bp (uaRNA) region was also excluded from our analysis. We also excluded all blacklist regions, including rRNA repeats.
We downloaded RNA-seq BAM files from TCGA data portal (https://portal.gdc.cancer.gov/)11. The RNA-seq raw data were processed by TCGA consortium as described on the official website (https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/Expression_mRNA_Pipeline/). We downloaded BAM files for our downstream analysis. We mapped RNA-seq data to these eRNA regions and calculated the expression level as RPM60 for each eRNA in each sample. We normalized eRNA expression by reads per million. We averaged all RPMs annotated to the eRNA from all samples in a cancer type, and defined those eRNAs with average expression level (RPM) ≥1 as a detectable eRNA. We converted different genomic versions of the human genome by liftover59. We present t-SNE analysis using R package Rtsne61. Our method may only detect a subset of polyadenylated eRNAs at their steady state since TCGA and CCLE only included the poly(A) RNA-seq. Our method may not distinguish functional eRNAs from those may just be the side effect of active enhancer.
Biogenesis of eRNAs
We collected TFs from multiple TF resources, including JASPAR (http://jaspar.genereg.net/)21, DBD (http://www.transcriptionfactor.org/)22, AnimalTFDB (http://bioinfo.life.hust.edu.cn/AnimalTFDB/)23, and TF2DNA (http://www.fiserlab.org/tf2dna_db/)24. We identified putative regulators of eRNAs based on the co-expression between individual eRNA and each TF in a given cancer type, and considered Spearman’s correlation Rs ≥ 0.3 and FDR < 0.05 as significant. For each cancer type, TFs that significantly correlated with more than 25% of the detectable eRNAs were defined as master regulators. Master regulators that exist in more than 10 cancer types were defined as general master regulators. The functional enrichment analyses of these general master regulator were performed by DAVID62 and GSEA63.
eRNA putative target genes and drugs
We identified eRNA putative target genes based on close distance (≤ 1MB) and co-expression (Spearman’s correlation Rs ≥ 0.3 and FDR < 0.05) between individual eRNAs and their putative target genes in each cancer type58. We filtered out eRNAs located in the intronic regions of target genes for correlation analysis. We collected 229 genes associated with 10 cancer signaling pathways34: p53, PI3K, Myc, RTK/RAS, cell cycle, Wnt, TGF beta, Nrf2, Notch, and Hippo. Due to the lack of Hi-C data in large number of tumor tissue, we used 20 Hi-C data from normal tissues52 to confirm the putative eRNA-gene connections. Hi-C interaction was evaluated by O/E value, which is calculated as observed value (estimated with normalized mapped reads) divided by expected value (estimated with a genome-wide model of interaction probability over the genomic distance)64. We also estimated the Hi-C interactions based on random eRNA-gene pairs throughout genome as background, and performed permutation test (bootstrap = 10,000) to compare with eRNA-gene pairs with the background of random pairs. For those eRNAs identified in TCGA, we examined their expression across ~1000 cancer cell lines in CCLE. GDSC and CTRP collected drug response data across >1000 cancer cell lines. We used matched cell lines to calculate the Spearman’s correlation between eRNA expression in CCLE and drug response value (AUC) of more than 500 anticancer drugs from CTRP and GDSC, and defined FDR < 0.05 as significant35.
Clinically relevant eRNAs
We used the Student’s t test to assess the statistical difference between tumor and paired normal samples and defined significantly aberrant expression as |fold change| > 1.5 and FDR < 0.05. We used Student’s t test for two groups and analysis of variance (ANOVA) for multiple groups to assess the statistical difference of patient smoking history and cancer subtype, stage, and grade (FDR <0.05). Only groups with ≥ 5 samples were included in these analyses. We used the univariate Cox model or log-rank test to assess whether eRNA expression was associated with the overall survival times of cancer patients and considered FDR < 0.05 as significant.
Lentivirus generation
A mixture of 3 μg of psPAX2, 1 μg of pMD2.G, and 4 μg of target sgRNA vector was transfected into 293T cells using Lipofectamine 2000 (Life Technologies). After 16 h, the media was changed, and the supernatants were collected at 48 and 72 h posttransfection for two independent infections. The collected supernatants were filtered using 0.45 μm syringe filter (Fisher) and used to infect MCF7 cells after being mixed with polybrene (final concentration of 8 μg ml−1, Sigma). Target cells were incubated in complete media with an equal amount of lentiviral particle-containing media for 24 h for each infection. After the second infection, the cells were selected over at least one week with selection markers to achieve a stable line.
Cell culture and transfection
We originally purchased MCF7 and MCF10A cells from American Type Culture Collection. We maintained the MCF7 and Hela cells in Dulbecco’s modified Eagle’s medium (DMEM) (Corning) media, supplemented with 10% fetal bovine solution (FBS) (GenDEPOT) and maintained the MCF10A cells in DMEM/F-12 (Corning) supplemented with 5% horse serum, 20 ng ml−1 EGF, 0.5 mg ml−1 hydrocortisone, 100 ng ml−1 cholera toxin, 10 μg ml−1 insulin in a 5% CO2 incubator at 37 °C4,65. Transfection of LNA GapmeRs (Qiagen) into the cells was carried out using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol and at a final concentration of 60 nM. For NET1e eRNA knockdown, a mixture of NET1e LNA 1, 2, and 3 was transfected into the cell. The sequence information for LNA is described in Supplementary Table 2.
CRISPR/dCas9-SAM
We followed the experimental procedures in Konermann et al.40. In brief, we generated the MCF7 stable cell line expressing dCAS9-VP64-Blast and Lenti MS2-p65-HSF1-Hygro using lentivirus. Infected cells were selected in DMEM supplemented with 10% FBS, 300 μg ml−1 Hygromycin, and 5 μg ml−1 Blasticidin. After 1 week of selection, we infected the stable cells with the lentiviral particle expressing sgRNA and selected the infected cells in DMEM with 10% FBS, 300 μg ml−1 Hygromycin, 5 μg ml−1 Blasticidin, and 300 μg ml−1 Zeocine for 1 additional week. All plasmids for CRISPR/dCAS9-SAM were purchased from Addgene (#61425, 61426, and 61427). The target gRNA sequence was chosen using http://crispr.mit.edu/. Target gRNA sequences were cloned into plasmid 61427 (Supplementary Table 2).
qRT-PCR for eRNA expression
RNA was extracted from cells using Quick RNA-miniprep (Zymo Research) and the RNA was reverse-transcribed using SuperScript® III Reverse Transcriptase with random hexamer (Invitrogen) or qScript XLT cDNA SuperMix (QuantaBio). We performed qRT-PCR in QuantStudio 3 qPCR systems (Applied Biosystems, Thermo Fisher) using 2X Ssoadvanced Universal Sybr Green Supermix (Bio-Rad). We used glyceraldehyde-3-phosphate dehydrogenase for normalization. We used a two-tailed Student’s t test to obtain the p-values. The sequences of qPCR primers are provided in Supplementary Table 2. All RT-qPCRs were performed with at least biological duplicates. Each biological replicate has three technical repeats.
Cell growth assay
Cells were trypsinized and plated at 3000 cells per well in a 96-well plate (Corning). Photos of each well were taken every 24 h using Incucyte Live Cell Imager (Essen Bioscience), and cell confluence was measured by Incucyte Software (Essen Bioscience) for 72 h. To normalize the confluences, the values for each time point were divided by the mean value at 0 h. In order to test the effects of NET1e to drug sensitivity in MCF7, we examined the half maximal inhibitory concentration (IC50) for PI3K inhibitor (BEZ235), BCL-2 Inhibitor (Obatoclax) in MCF7 Crispr/SAM control and NET1e using Incucyte live cell imager.
Data portal
We constructed the data portal based on Rscript and JavaScript. The expression profile, clinical relevance, putative target genes, Hi-C data, and drug responses of eRNAs are available on the data portal (https://hanlab.uth.edu/eRic/).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by the Cancer Prevention and Research Institute of Texas (grant no. RR150085 and RP190570) to CPRIT Scholar in Cancer Research (L.H.), Cancer Prevention and Research Institute of Texas (grant no. RR160083 and RP180734) to CPRIT Scholar in Cancer Research (W.L.). This work was also supported by funding from NIH/NCI (K22CA204468), NIH/NIGMS (R21GM132778), and Welch foundation (AU-2000-20190330) to W.L. We gratefully acknowledge contributions from TCGA Research Network. We thank LeeAnn Chastain for editorial assistance.
Author contributions
L.H. conceived and supervised the project. Z.Z, J-H.L., W.L. and L.H. designed and performed the research. Z.Z., H.R., Y.Y, Y.X., J.G. and L.D. performed the data analyses. Z.Z. and H.R. constructed the data portal. J-H.L., J.K., and Q.H. performed the experiments. Z.Z., B.Z., C.L., L.W., G.B.M., W.L. and L.H. interpreted the results. Z.Z., W.L., and L.H. wrote the paper with input from all the other authors.
Data availability
All accession codes, unique identifiers, or web links for publicly available datasets are described in the paper. All data supporting the findings of the current study are listed in Supplementary Data 1–8, Supplementary Fig. 7, and our online data portal (https://hanlab.uth.edu/eRic/).
Code availability
All codes are available upon reasonable request.
Competing interests
G.B.M. has sponsored research support from AstraZeneca, Critical Outcomes Technology, Karus, Illumina, Immunomet, Nanostring, Tarveda and Immunomet and is on the Scientific Advisory Board for AstraZeneca, Critical Outcomes Technology, ImmunoMet, Ionis, Nuevolution, Symphogen, and Tarveda. The other authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhao Zhang, Joo-Hyung Lee, Hang Ruan.
Contributor Information
Wenbo Li, Email: wenbo.li@uth.tmc.edu.
Leng Han, Email: leng.han@uth.tmc.edu.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-019-12543-5.
References
- 1.Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 2.Kim T-K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 4.Li W, et al. Condensin I and II complexes license full estrogen receptor α-dependent enhancer activation. Mol. Cell. 2015;59:188–202. doi: 10.1016/j.molcel.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh C-L, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl Acad. Sci. USA. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melo CA, et al. ERNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 7.Léveillé N, Melo CA, Agami R. Enhancer-associated RNAs as therapeutic targets. Expert Opin. Biol. Ther. 2015;15:723–734. doi: 10.1517/14712598.2015.1029452. [DOI] [PubMed] [Google Scholar]
- 8.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome . Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BE, et al. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghandi Mahmoud, Huang Franklin W., Jané-Valbuena Judit, Kryukov Gregory V., Lo Christopher C., McDonald E. Robert, Barretina Jordi, Gelfand Ellen T., Bielski Craig M., Li Haoxin, Hu Kevin, Andreev-Drakhlin Alexander Y., Kim Jaegil, Hess Julian M., Haas Brian J., Aguet François, Weir Barbara A., Rothberg Michael V., Paolella Brenton R., Lawrence Michael S., Akbani Rehan, Lu Yiling, Tiv Hong L., Gokhale Prafulla C., de Weck Antoine, Mansour Ali Amin, Oh Coyin, Shih Juliann, Hadi Kevin, Rosen Yanay, Bistline Jonathan, Venkatesan Kavitha, Reddy Anupama, Sonkin Dmitriy, Liu Manway, Lehar Joseph, Korn Joshua M., Porter Dale A., Jones Michael D., Golji Javad, Caponigro Giordano, Taylor Jordan E., Dunning Caitlin M., Creech Amanda L., Warren Allison C., McFarland James M., Zamanighomi Mahdi, Kauffmann Audrey, Stransky Nicolas, Imielinski Marcin, Maruvka Yosef E., Cherniack Andrew D., Tsherniak Aviad, Vazquez Francisca, Jaffe Jacob D., Lane Andrew A., Weinstock David M., Johannessen Cory M., Morrissey Michael P., Stegmeier Frank, Schlegel Robert, Hahn William C., Getz Gad, Mills Gordon B., Boehm Jesse S., Golub Todd R., Garraway Levi A., Sellers William R. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569(7757):503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rees MG, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016;12:109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Wanjuan, Soares Jorge, Greninger Patricia, Edelman Elena J., Lightfoot Howard, Forbes Simon, Bindal Nidhi, Beare Dave, Smith James A., Thompson I. Richard, Ramaswamy Sridhar, Futreal P. Andrew, Haber Daniel A., Stratton Michael R., Benes Cyril, McDermott Ultan, Garnett Mathew J. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Research. 2012;41(D1):D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hah N, Murakami S, Nagari A, Danko CG, Lee Kraus W. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorighi KM, et al. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol. Cell. 2017;66:568–576. doi: 10.1016/j.molcel.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, He F, Hu S, Yu J. On the nature of human housekeeping genes. Trends Genet. 2008;24:481–484. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Ricketts Christopher J., De Cubas Aguirre A., Fan Huihui, Smith Christof C., Lang Martin, Reznik Ed, Bowlby Reanne, Gibb Ewan A., Akbani Rehan, Beroukhim Rameen, Bottaro Donald P., Choueiri Toni K., Gibbs Richard A., Godwin Andrew K., Haake Scott, Hakimi A. Ari, Henske Elizabeth P., Hsieh James J., Ho Thai H., Kanchi Rupa S., Krishnan Bhavani, Kwiatkowski David J., Lui Wembin, Merino Maria J., Mills Gordon B., Myers Jerome, Nickerson Michael L., Reuter Victor E., Schmidt Laura S., Shelley C. Simon, Shen Hui, Shuch Brian, Signoretti Sabina, Srinivasan Ramaprasad, Tamboli Pheroze, Thomas George, Vincent Benjamin G., Vocke Cathy D., Wheeler David A., Yang Lixing, Kim William Y., Robertson A. Gordon, Spellman Paul T., Rathmell W. Kimryn, Linehan W. Marston, Caesar-Johnson Samantha J., Demchok John A., Felau Ina, Kasapi Melpomeni, Ferguson Martin L., Hutter Carolyn M., Sofia Heidi J., Tarnuzzer Roy, Wang Zhining, Yang Liming, Zenklusen Jean C., Zhang Jiashan (Julia), Chudamani Sudha, Liu Jia, Lolla Laxmi, Naresh Rashi, Pihl Todd, Sun Qiang, Wan Yunhu, Wu Ye, Cho Juok, DeFreitas Timothy, Frazer Scott, Gehlenborg Nils, Getz Gad, Heiman David I., Kim Jaegil, Lawrence Michael S., Lin Pei, Meier Sam, Noble Michael S., Saksena Gordon, Voet Doug, Zhang Hailei, Bernard Brady, Chambwe Nyasha, Dhankani Varsha, Knijnenburg Theo, Kramer Roger, Leinonen Kalle, Liu Yuexin, Miller Michael, Reynolds Sheila, Shmulevich Ilya, Thorsson Vesteinn, Zhang Wei, Akbani Rehan, Broom Bradley M., Hegde Apurva M., Ju Zhenlin, Kanchi Rupa S., Korkut Anil, Li Jun, Liang Han, Ling Shiyun, Liu Wenbin, Lu Yiling, Mills Gordon B., Ng Kwok-Shing, Rao Arvind, Ryan Michael, Wang Jing, Weinstein John N., Zhang Jiexin, Abeshouse Adam, Armenia Joshua, Chakravarty Debyani, Chatila Walid K., de Bruijn Ino, Gao Jianjiong, Gross Benjamin E., Heins Zachary J., Kundra Ritika, La Konnor, Ladanyi Marc, Luna Augustin, Nissan Moriah G., Ochoa Angelica, Phillips Sarah M., Reznik Ed, Sanchez-Vega Francisco, Sander Chris, Schultz Nikolaus, Sheridan Robert, Sumer S. Onur, Sun Yichao, Taylor Barry S., Wang Jioajiao, Zhang Hongxin, Anur Pavana, Peto Myron, Spellman Paul, Benz Christopher, Stuart Joshua M., Wong Christopher K., Yau Christina, Hayes D. Neil, Parker Joel S., Wilkerson Matthew D., Ally Adrian, Balasundaram Miruna, Bowlby Reanne, Brooks Denise, Carlsen Rebecca, Chuah Eric, Dhalla Noreen, Holt Robert, Jones Steven J.M., Kasaian Katayoon, Lee Darlene, Ma Yussanne, Marra Marco A., Mayo Michael, Moore Richard A., Mungall Andrew J., Mungall Karen, Robertson A. Gordon, Sadeghi Sara, Schein Jacqueline E., Sipahimalani Payal, Tam Angela, Thiessen Nina, Tse Kane, Wong Tina, Berger Ashton C., Beroukhim Rameen, Cherniack Andrew D., Cibulskis Carrie, Gabriel Stacey B., Gao Galen F., Ha Gavin, Meyerson Matthew, Schumacher Steven E., Shih Juliann, Kucherlapati Melanie H., Kucherlapati Raju S., Baylin Stephen, Cope Leslie, Danilova Ludmila, Bootwalla Moiz S., Lai Phillip H., Maglinte Dennis T., Van Den Berg David J., Weisenberger Daniel J., Auman J. Todd, Balu Saianand, Bodenheimer Tom, Fan Cheng, Hoadley Katherine A., Hoyle Alan P., Jefferys Stuart R., Jones Corbin D., Meng Shaowu, Mieczkowski Piotr A., Mose Lisle E., Perou Amy H., Perou Charles M., Roach Jeffrey, Shi Yan, Simons Janae V., Skelly Tara, Soloway Matthew G., Tan Donghui, Veluvolu Umadevi, Fan Huihui, Hinoue Toshinori, Laird Peter W., Shen Hui, Zhou Wanding, Bellair Michelle, Chang Kyle, Covington Kyle, Creighton Chad J., Dinh Huyen, Doddapaneni HarshaVardhan, Donehower Lawrence A., Drummond Jennifer, Gibbs Richard A., Glenn Robert, Hale Walker, Han Yi, Hu Jianhong, Korchina Viktoriya, Lee Sandra, Lewis Lora, Li Wei, Liu Xiuping, Morgan Margaret, Morton Donna, Muzny Donna, Santibanez Jireh, Sheth Margi, Shinbrot Eve, Wang Linghua, Wang Min, Wheeler David A., Xi Liu, Zhao Fengmei, Hess Julian, Appelbaum Elizabeth L., Bailey Matthew, Cordes Matthew G., Ding Li, Fronick Catrina C., Fulton Lucinda A., Fulton Robert S., Kandoth Cyriac, Mardis Elaine R., McLellan Michael D., Miller Christopher A., Schmidt Heather K., Wilson Richard K., Crain Daniel, Curley Erin, Gardner Johanna, Lau Kevin, Mallery David, Morris Scott, Paulauskis Joseph, Penny Robert, Shelton Candace, Shelton Troy, Sherman Mark, Thompson Eric, Yena Peggy, Bowen Jay, Gastier-Foster Julie M., Gerken Mark, Leraas Kristen M., Lichtenberg Tara M., Ramirez Nilsa C., Wise Lisa, Zmuda Erik, Corcoran Niall, Costello Tony, Hovens Christopher, Carvalho Andre L., de Carvalho Ana C., Fregnani José H., Longatto-Filho Adhemar, Reis Rui M., Scapulatempo-Neto Cristovam, Silveira Henrique C.S., Vidal Daniel O., Burnette Andrew, Eschbacher Jennifer, Hermes Beth, Noss Ardene, Singh Rosy, Anderson Matthew L., Castro Patricia D., Ittmann Michael, Huntsman David, Kohl Bernard, Le Xuan, Thorp Richard, Andry Chris, Duffy Elizabeth R., Lyadov Vladimir, Paklina Oxana, Setdikova Galiya, Shabunin Alexey, Tavobilov Mikhail, McPherson Christopher, Warnick Ronald, Berkowitz Ross, Cramer Daniel, Feltmate Colleen, Horowitz Neil, Kibel Adam, Muto Michael, Raut Chandrajit P., Malykh Andrei, Barnholtz-Sloan Jill S., Barrett Wendi, Devine Karen, Fulop Jordonna, Ostrom Quinn T., Shimmel Kristen, Wolinsky Yingli, Sloan Andrew E., De Rose Agostino, Giuliante Felice, Goodman Marc, Karlan Beth Y., Hagedorn Curt H., Eckman John, Harr Jodi, Myers Jerome, Tucker Kelinda, Zach Leigh Anne, Deyarmin Brenda, Hu Hai, Kvecher Leonid, Larson Caroline, Mural Richard J., Somiari Stella, Vicha Ales, Zelinka Tomas, Bennett Joseph, Iacocca Mary, Rabeno Brenda, Swanson Patricia, Latour Mathieu, Lacombe Louis, Têtu Bernard, Bergeron Alain, McGraw Mary, Staugaitis Susan M., Chabot John, Hibshoosh Hanina, Sepulveda Antonia, Su Tao, Wang Timothy, Potapova Olga, Voronina Olga, Desjardins Laurence, Mariani Odette, Roman-Roman Sergio, Sastre Xavier, Stern Marc-Henri, Cheng Feixiong, Signoretti Sabina, Berchuck Andrew, Bigner Darell, Lipp Eric, Marks Jeffrey, McCall Shannon, McLendon Roger, Secord Angeles, Sharp Alexis, Behera Madhusmita, Brat Daniel J., Chen Amy, Delman Keith, Force Seth, Khuri Fadlo, Magliocca Kelly, Maithel Shishir, Olson Jeffrey J., Owonikoko Taofeek, Pickens Alan, Ramalingam Suresh, Shin Dong M., Sica Gabriel, Van Meir Erwin G., Zhang Hongzheng, Eijckenboom Wil, Gillis Ad, Korpershoek Esther, Looijenga Leendert, Oosterhuis Wolter, Stoop Hans, van Kessel Kim E., Zwarthoff Ellen C., Calatozzolo Chiara, Cuppini Lucia, Cuzzubbo Stefania, DiMeco Francesco, Finocchiaro Gaetano, Mattei Luca, Perin Alessandro, Pollo Bianca, Chen Chu, Houck John, Lohavanichbutr Pawadee, Hartmann Arndt, Stoehr Christine, Stoehr Robert, Taubert Helge, Wach Sven, Wullich Bernd, Kycler Witold, Murawa Dawid, Wiznerowicz Maciej, Chung Ki, Edenfield W. Jeffrey, Martin Julie, Baudin Eric, Bubley Glenn, Bueno Raphael, De Rienzo Assunta, Richards William G., Kalkanis Steven, Mikkelsen Tom, Noushmehr Houtan, Scarpace Lisa, Girard Nicolas, Aymerich Marta, Campo Elias, Giné Eva, Guillermo Armando López, Van Bang Nguyen, Hanh Phan Thi, Phu Bui Duc, Tang Yufang, Colman Howard, Evason Kimberley, Dottino Peter R., Martignetti John A., Gabra Hani, Juhl Hartmut, Akeredolu Teniola, Stepa Serghei, Hoon Dave, Ahn Keunsoo, Kang Koo Jeong, Beuschlein Felix, Breggia Anne, Birrer Michael, Bell Debra, Borad Mitesh, Bryce Alan H., Castle Erik, Chandan Vishal, Cheville John, Copland John A., Farnell Michael, Flotte Thomas, Giama Nasra, Ho Thai, Kendrick Michael, Kocher Jean-Pierre, Kopp Karla, Moser Catherine, Nagorney David, O’Brien Daniel, O’Neill Brian Patrick, Patel Tushar, Petersen Gloria, Que Florencia, Rivera Michael, Roberts Lewis, Smallridge Robert, Smyrk Thomas, Stanton Melissa, Thompson R. Houston, Torbenson Michael, Yang Ju Dong, Zhang Lizhi, Brimo Fadi, Ajani Jaffer A., Gonzalez Ana Maria Angulo, Behrens Carmen, Bondaruk Jolanta, Broaddus Russell, Czerniak Bogdan, Esmaeli Bita, Fujimoto Junya, Gershenwald Jeffrey, Guo Charles, Lazar Alexander J., Logothetis Christopher, Meric-Bernstam Funda, Moran Cesar, Ramondetta Lois, Rice David, Sood Anil, Tamboli Pheroze, Thompson Timothy, Troncoso Patricia, Tsao Anne, Wistuba Ignacio, Carter Candace, Haydu Lauren, Hersey Peter, Jakrot Valerie, Kakavand Hojabr, Kefford Richard, Lee Kenneth, Long Georgina, Mann Graham, Quinn Michael, Saw Robyn, Scolyer Richard, Shannon Kerwin, Spillane Andrew, Stretch onathan, Synott Maria, Thompson John, Wilmott James, Al-Ahmadie Hikmat, Chan Timothy A., Ghossein Ronald, Gopalan Anuradha, Levine Douglas A., Reuter Victor, Singer Samuel, Singh Bhuvanesh, Tien Nguyen Viet, Broudy Thomas, Mirsaidi Cyrus, Nair Praveen, Drwiega Paul, Miller Judy, Smith Jennifer, Zaren Howard, Park Joong-Won, Hung Nguyen Phi, Kebebew Electron, Linehan W. Marston, Metwalli Adam R., Pacak Karel, Pinto Peter A., Schiffman Mark, Schmidt Laura S., Vocke Cathy D., Wentzensen Nicolas, Worrell Robert, Yang Hannah, Moncrieff Marc, Goparaju Chandra, Melamed Jonathan, Pass Harvey, Botnariuc Natalia, Caraman Irina, Cernat Mircea, Chemencedji Inga, Clipca Adrian, Doruc Serghei, Gorincioi Ghenadie, Mura Sergiu, Pirtac Maria, Stancul Irina, Tcaciuc Diana, Albert Monique, Alexopoulou Iakovina, Arnaout Angel, Bartlett John, Engel Jay, Gilbert Sebastien, Parfitt Jeremy, Sekhon Harman, Thomas George, Rassl Doris M., Rintoul Robert C., Bifulco Carlo, Tamakawa Raina, Urba Walter, Hayward Nicholas, Timmers Henri, Antenucci Anna, Facciolo Francesco, Grazi Gianluca, Marino Mirella, Merola Roberta, de Krijger Ronald, Gimenez-Roqueplo Anne-Paule, Piché Alain, Chevalier Simone, McKercher Ginette, Birsoy Kivanc, Barnett Gene, Brewer Cathy, Farver Carol, Naska Theresa, Pennell Nathan A., Raymond Daniel, Schilero Cathy, Smolenski Kathy, Williams Felicia, Morrison Carl, Borgia Jeffrey A., Liptay Michael J., Pool Mark, Seder Christopher W., Junker Kerstin, Omberg Larsson, Dinkin Mikhail, Manikhas George, Alvaro Domenico, Bragazzi Maria Consiglia, Cardinale Vincenzo, Carpino Guido, Gaudio Eugenio, Chesla David, Cottingham Sandra, Dubina Michael, Moiseenko Fedor, Dhanasekaran Renumathy, Becker Karl-Friedrich, Janssen Klaus-Peter, Slotta-Huspenina Julia, Abdel-Rahman Mohamed H., Aziz Dina, Bell Sue, Cebulla Colleen M., Davis Amy, Duell Rebecca, Elder J. Bradley, Hilty Joe, Kumar Bahavna, Lang James, Lehman Norman L., Mandt Randy, Nguyen Phuong, Pilarski Robert, Rai Karan, Schoenfield Lynn, Senecal Kelly, Wakely Paul, Hansen Paul, Lechan Ronald, Powers James, Tischler Arthur, Grizzle William E., Sexton Katherine C., Kastl Alison, Henderson Joel, Porten Sima, Waldmann Jens, Fassnacht Martin, Asa Sylvia L., Schadendorf Dirk, Couce Marta, Graefen Markus, Huland Hartwig, Sauter Guido, Schlomm Thorsten, Simon Ronald, Tennstedt Pierre, Olabode Oluwole, Nelson Mark, Bathe Oliver, Carroll Peter R., Chan June M., Disaia Philip, Glenn Pat, Kelley Robin K., Landen Charles N., Phillips Joanna, Prados Michael, Simko Jeffry, Smith-McCune Karen, VandenBerg Scott, Roggin Kevin, Fehrenbach Ashley, Kendler Ady, Sifri Suzanne, Steele Ruth, Jimeno Antonio, Carey Francis, Forgie Ian, Mannelli Massimo, Carney Michael, Hernandez Brenda, Campos Benito, Herold-Mende Christel, Jungk Christin, Unterberg Andreas, von Deimling Andreas, Bossler Aaron, Galbraith Joseph, Jacobus Laura, Knudson Michael, Knutson Tina, Ma Deqin, Milhem Mohammed, Sigmund Rita, Godwin Andrew K., Madan Rashna, Rosenthal Howard G., Adebamowo Clement, Adebamowo Sally N., Boussioutas Alex, Beer David, Giordano Thomas, Mes-Masson Anne-Marie, Saad Fred, Bocklage Therese, Landrum Lisa, Mannel Robert, Moore Kathleen, Moxley Katherine, Postier Russel, Walker Joan, Zuna Rosemary, Feldman Michael, Valdivieso Federico, Dhir Rajiv, Luketich James, Pinero Edna M. Mora, Quintero-Aguilo Mario, Carlotti Carlos Gilberto, Dos Santos Jose Sebastião, Kemp Rafael, Sankarankuty Ajith, Tirapelli Daniela, Catto James, Agnew Kathy, Swisher Elizabeth, Creaney Jenette, Robinson Bruce, Shelley Carl Simon, Godwin Eryn M., Kendall Sara, Shipman Cassaundra, Bradford Carol, Carey Thomas, Haddad Andrea, Moyer Jeffey, Peterson Lisa, Prince Mark, Rozek Laura, Wolf Gregory, Bowman Rayleen, Fong Kwun M., Yang Ian, Korst Robert, Rathmell W. Kimryn, Fantacone-Campbell J. Leigh, Hooke Jeffrey A., Kovatich Albert J., Shriver Craig D., DiPersio John, Drake Bettina, Govindan Ramaswamy, Heath Sharon, Ley Timothy, Van Tine Brian, Westervelt Peter, Rubin Mark A., Lee Jung Il, Aredes Natália D., Mariamidze Armaz. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Reports. 2018;23(1):313-326.e5. doi: 10.1016/j.celrep.2018.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell, J. D. et al. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas graphical. Cell Rep. 10.1016/j.celrep.2018.03.063194–212 (2018). [DOI] [PMC free article] [PubMed]
- 20.Malta TM, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–354.e15. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X, et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat. Methods. 2013;10:375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kummerfeld SK. DBD: a transcription factor prediction database. Nucleic Acids Res. 2006;34:D74–D81. doi: 10.1093/nar/gkj131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H.-M., Chen H., Liu W., Liu H., Gong J., Wang H., Guo A.-Y. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Research. 2011;40(D1):D144–D149. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 2011;30:546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernardo GM, et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene. 2013;32:554–563. doi: 10.1038/onc.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Zhijie, Merkurjev Daria, Yang Feng, Li Wenbo, Oh Soohwan, Friedman Meyer J., Song Xiaoyuan, Zhang Feng, Ma Qi, Ohgi Kenneth A., Krones Anna, Rosenfeld Michael G. Enhancer Activation Requires trans-Recruitment of a Mega Transcription Factor Complex. Cell. 2014;159(2):358–373. doi: 10.1016/j.cell.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marzec Paulina, Armenise Claudia, Pérot Gaëlle, Roumelioti Fani-Marlen, Basyuk Eugenia, Gagos Sarantis, Chibon Frédéric, Déjardin Jérôme. Nuclear-Receptor-Mediated Telomere Insertion Leads to Genome Instability in ALT Cancers. Cell. 2015;160(5):913–927. doi: 10.1016/j.cell.2015.01.044. [DOI] [PubMed] [Google Scholar]
- 28.Remo, A. et al. Systems biology analysis reveals NFAT5 as a novel biomarker and master regulator of inflammatory breast cancer. J. Transl. Med. 1–13 10.1186/s12967-015-0492-2 (2015). [DOI] [PMC free article] [PubMed]
- 29.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2018;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 30.Chen Han, Li Chunyan, Peng Xinxin, Zhou Zhicheng, Weinstein John N., Liang Han, Caesar-Johnson Samantha J., Demchok John A., Felau Ina, Kasapi Melpomeni, Ferguson Martin L., Hutter Carolyn M., Sofia Heidi J., Tarnuzzer Roy, Wang Zhining, Yang Liming, Zenklusen Jean C., Zhang Jiashan (Julia), Chudamani Sudha, Liu Jia, Lolla Laxmi, Naresh Rashi, Pihl Todd, Sun Qiang, Wan Yunhu, Wu Ye, Cho Juok, DeFreitas Timothy, Frazer Scott, Gehlenborg Nils, Getz Gad, Heiman David I., Kim Jaegil, Lawrence Michael S., Lin Pei, Meier Sam, Noble Michael S., Saksena Gordon, Voet Doug, Zhang Hailei, Bernard Brady, Chambwe Nyasha, Dhankani Varsha, Knijnenburg Theo, Kramer Roger, Leinonen Kalle, Liu Yuexin, Miller Michael, Reynolds Sheila, Shmulevich Ilya, Thorsson Vesteinn, Zhang Wei, Akbani Rehan, Broom Bradley M., Hegde Apurva M., Ju Zhenlin, Kanchi Rupa S., Korkut Anil, Li Jun, Liang Han, Ling Shiyun, Liu Wenbin, Lu Yiling, Mills Gordon B., Ng Kwok-Shing, Rao Arvind, Ryan Michael, Wang Jing, Weinstein John N., Zhang Jiexin, Abeshouse Adam, Armenia Joshua, Chakravarty Debyani, Chatila Walid K., de Bruijn Ino, Gao Jianjiong, Gross Benjamin E., Heins Zachary J., Kundra Ritika, La Konnor, Ladanyi Marc, Luna Augustin, Nissan Moriah G., Ochoa Angelica, Phillips Sarah M., Reznik Ed, Sanchez-Vega Francisco, Sander Chris, Schultz Nikolaus, Sheridan Robert, Sumer S. Onur, Sun Yichao, Taylor Barry S., Wang Jioajiao, Zhang Hongxin, Anur Pavana, Peto Myron, Spellman Paul, Benz Christopher, Stuart Joshua M., Wong Christopher K., Yau Christina, Hayes D. Neil, Parker Joel S., Wilkerson Matthew D., Ally Adrian, Balasundaram Miruna, Bowlby Reanne, Brooks Denise, Carlsen Rebecca, Chuah Eric, Dhalla Noreen, Holt Robert, Jones Steven J.M., Kasaian Katayoon, Lee Darlene, Ma Yussanne, Marra Marco A., Mayo Michael, Moore Richard A., Mungall Andrew J., Mungall Karen, Robertson A. Gordon, Sadeghi Sara, Schein Jacqueline E., Sipahimalani Payal, Tam Angela, Thiessen Nina, Tse Kane, Wong Tina, Berger Ashton C., Beroukhim Rameen, Cherniack Andrew D., Cibulskis Carrie, Gabriel Stacey B., Gao Galen F., Ha Gavin, Meyerson Matthew, Schumacher Steven E., Shih Juliann, Kucherlapati Melanie H., Kucherlapati Raju S., Baylin Stephen, Cope Leslie, Danilova Ludmila, Bootwalla Moiz S., Lai Phillip H., Maglinte Dennis T., Van Den Berg David J., Weisenberger Daniel J., Auman J. Todd, Balu Saianand, Bodenheimer Tom, Fan Cheng, Hoadley Katherine A., Hoyle Alan P., Jefferys Stuart R., Jones Corbin D., Meng Shaowu, Mieczkowski Piotr A., Mose Lisle E., Perou Amy H., Perou Charles M., Roach Jeffrey, Shi Yan, Simons Janae V., Skelly Tara, Soloway Matthew G., Tan Donghui, Veluvolu Umadevi, Fan Huihui, Hinoue Toshinori, Laird Peter W., Shen Hui, Zhou Wanding, Bellair Michelle, Chang Kyle, Covington Kyle, Creighton Chad J., Dinh Huyen, Doddapaneni HarshaVardhan, Donehower Lawrence A., Drummond Jennifer, Gibbs Richard A., Glenn Robert, Hale Walker, Han Yi, Hu Jianhong, Korchina Viktoriya, Lee Sandra, Lewis Lora, Li Wei, Liu Xiuping, Morgan Margaret, Morton Donna, Muzny Donna, Santibanez Jireh, Sheth Margi, Shinbrot Eve, Wang Linghua, Wang Min, Wheeler David A., Xi Liu, Zhao Fengmei, Hess Julian, Appelbaum Elizabeth L., Bailey Matthew, Cordes Matthew G., Ding Li, Fronick Catrina C., Fulton Lucinda A., Fulton Robert S., Kandoth Cyriac, Mardis Elaine R., McLellan Michael D., Miller Christopher A., Schmidt Heather K., Wilson Richard K., Crain Daniel, Curley Erin, Gardner Johanna, Lau Kevin, Mallery David, Morris Scott, Paulauskis Joseph, Penny Robert, Shelton Candace, Shelton Troy, Sherman Mark, Thompson Eric, Yena Peggy, Bowen Jay, Gastier-Foster Julie M., Gerken Mark, Leraas Kristen M., Lichtenberg Tara M., Ramirez Nilsa C., Wise Lisa, Zmuda Erik, Corcoran Niall, Costello Tony, Hovens Christopher, Carvalho Andre L., de Carvalho Ana C., Fregnani José H., Longatto-Filho Adhemar, Reis Rui M., Scapulatempo-Neto Cristovam, Silveira Henrique C.S., Vidal Daniel O., Burnette Andrew, Eschbacher Jennifer, Hermes Beth, Noss Ardene, Singh Rosy, Anderson Matthew L., Castro Patricia D., Ittmann Michael, Huntsman David, Kohl Bernard, Le Xuan, Thorp Richard, Andry Chris, Duffy Elizabeth R., Lyadov Vladimir, Paklina Oxana, Setdikova Galiya, Shabunin Alexey, Tavobilov Mikhail, McPherson Christopher, Warnick Ronald, Berkowitz Ross, Cramer Daniel, Feltmate Colleen, Horowitz Neil, Kibel Adam, Muto Michael, Raut Chandrajit P., Malykh Andrei, Barnholtz-Sloan Jill S., Barrett Wendi, Devine Karen, Fulop Jordonna, Ostrom Quinn T., Shimmel Kristen, Wolinsky Yingli, Sloan Andrew E., De Rose Agostino, Giuliante Felice, Goodman Marc, Karlan Beth Y., Hagedorn Curt H., Eckman John, Harr Jodi, Myers Jerome, Tucker Kelinda, Zach Leigh Anne, Deyarmin Brenda, Hu Hai, Kvecher Leonid, Larson Caroline, Mural Richard J., Somiari Stella, Vicha Ales, Zelinka Tomas, Bennett Joseph, Iacocca Mary, Rabeno Brenda, Swanson Patricia, Latour Mathieu, Lacombe Louis, Têtu Bernard, Bergeron Alain, McGraw Mary, Staugaitis Susan M., Chabot John, Hibshoosh Hanina, Sepulveda Antonia, Su Tao, Wang Timothy, Potapova Olga, Voronina Olga, Desjardins Laurence, Mariani Odette, Roman-Roman Sergio, Sastre Xavier, Stern Marc-Henri, Cheng Feixiong, Signoretti Sabina, Berchuck Andrew, Bigner Darell, Lipp Eric, Marks Jeffrey, McCall Shannon, McLendon Roger, Secord Angeles, Sharp Alexis, Behera Madhusmita, Brat Daniel J., Chen Amy, Delman Keith, Force Seth, Khuri Fadlo, Magliocca Kelly, Maithel Shishir, Olson Jeffrey J., Owonikoko Taofeek, Pickens Alan, Ramalingam Suresh, Shin Dong M., Sica Gabriel, Van Meir Erwin G., Zhang Hongzheng, Eijckenboom Wil, Gillis Ad, Korpershoek Esther, Looijenga Leendert, Oosterhuis Wolter, Stoop Hans, van Kessel Kim E., Zwarthoff Ellen C., Calatozzolo Chiara, Cuppini Lucia, Cuzzubbo Stefania, DiMeco Francesco, Finocchiaro Gaetano, Mattei Luca, Perin Alessandro, Pollo Bianca, Chen Chu, Houck John, Lohavanichbutr Pawadee, Hartmann Arndt, Stoehr Christine, Stoehr Robert, Taubert Helge, Wach Sven, Wullich Bernd, Kycler Witold, Murawa Dawid, Wiznerowicz Maciej, Chung Ki, Edenfield W. Jeffrey, Martin Julie, Baudin Eric, Bubley Glenn, Bueno Raphael, De Rienzo Assunta, Richards William G., Kalkanis Steven, Mikkelsen Tom, Noushmehr Houtan, Scarpace Lisa, Girard Nicolas, Aymerich Marta, Campo Elias, Giné Eva, Guillermo Armando López, Van Bang Nguyen, Hanh Phan Thi, Phu Bui Duc, Tang Yufang, Colman Howard, Evason Kimberley, Dottino Peter R., Martignetti John A., Gabra Hani, Juhl Hartmut, Akeredolu Teniola, Stepa Serghei, Hoon Dave, Ahn Keunsoo, Kang Koo Jeong, Beuschlein Felix, Breggia Anne, Birrer Michael, Bell Debra, Borad Mitesh, Bryce Alan H., Castle Erik, Chandan Vishal, Cheville John, Copland John A., Farnell Michael, Flotte Thomas, Giama Nasra, Ho Thai, Kendrick Michael, Kocher Jean-Pierre, Kopp Karla, Moser Catherine, Nagorney David, O’Brien Daniel, O’Neill Brian Patrick, Patel Tushar, Petersen Gloria, Que Florencia, Rivera Michael, Roberts Lewis, Smallridge Robert, Smyrk Thomas, Stanton Melissa, Thompson R. Houston, Torbenson Michael, Yang Ju Dong, Zhang Lizhi, Brimo Fadi, Ajani Jaffer A., Gonzalez Ana Maria Angulo, Behrens Carmen, Bondaruk Jolanta, Broaddus Russell, Czerniak Bogdan, Esmaeli Bita, Fujimoto Junya, Gershenwald Jeffrey, Guo Charles, Lazar Alexander J., Logothetis Christopher, Meric-Bernstam Funda, Moran Cesar, Ramondetta Lois, Rice David, Sood Anil, Tamboli Pheroze, Thompson Timothy, Troncoso Patricia, Tsao Anne, Wistuba Ignacio, Carter Candace, Haydu Lauren, Hersey Peter, Jakrot Valerie, Kakavand Hojabr, Kefford Richard, Lee Kenneth, Long Georgina, Mann Graham, Quinn Michael, Saw Robyn, Scolyer Richard, Shannon Kerwin, Spillane Andrew, Stretch Jonathan, Synott Maria, Thompson John, Wilmott James, Al-Ahmadie Hikmat, Chan Timothy A., Ghossein Ronald, Gopalan Anuradha, Levine Douglas A., Reuter Victor, Singer Samuel, Singh Bhuvanesh, Tien Nguyen Viet, Broudy Thomas, Mirsaidi Cyrus, Nair Praveen, Drwiega Paul, Miller Judy, Smith Jennifer, Zaren Howard, Park Joong-Won, Hung Nguyen Phi, Kebebew Electron, Linehan W. Marston, Metwalli Adam R., Pacak Karel, Pinto Peter A., Schiffman Mark, Schmidt Laura S., Vocke Cathy D., Wentzensen Nicolas, Worrell Robert, Yang Hannah, Moncrieff Marc, Goparaju Chandra, Melamed Jonathan, Pass Harvey, Botnariuc Natalia, Caraman Irina, Cernat Mircea, Chemencedji Inga, Clipca Adrian, Doruc Serghei, Gorincioi Ghenadie, Mura Sergiu, Pirtac Maria, Stancul Irina, Tcaciuc Diana, Albert Monique, Alexopoulou Iakovina, Arnaout Angel, Bartlett John, Engel Jay, Gilbert Sebastien, Parfitt Jeremy, Sekhon Harman, Thomas George, Rassl Doris M., Rintoul Robert C., Bifulco Carlo, Tamakawa Raina, Urba Walter, Hayward Nicholas, Timmers Henri, Antenucci Anna, Facciolo Francesco, Grazi Gianluca, Marino Mirella, Merola Roberta, de Krijger Ronald, Gimenez-Roqueplo Anne-Paule, Piché Alain, Chevalier Simone, McKercher Ginette, Birsoy Kivanc, Barnett Gene, Brewer Cathy, Farver Carol, Naska Theresa, Pennell Nathan A., Raymond Daniel, Schilero Cathy, Smolenski Kathy, Williams Felicia, Morrison Carl, Borgia Jeffrey A., Liptay Michael J., Pool Mark, Seder Christopher W., Junker Kerstin, Omberg Larsson, Dinkin Mikhail, Manikhas George, Alvaro Domenico, Bragazzi Maria Consiglia, Cardinale Vincenzo, Carpino Guido, Gaudio Eugenio, Chesla David, Cottingham Sandra, Dubina Michael, Moiseenko Fedor, Dhanasekaran Renumathy, Becker Karl-Friedrich, Janssen Klaus-Peter, Slotta-Huspenina Julia, Abdel-Rahman Mohamed H., Aziz Dina, Bell Sue, Cebulla Colleen M., Davis Amy, Duell Rebecca, Elder J. Bradley, Hilty Joe, Kumar Bahavna, Lang James, Lehman Norman L., Mandt Randy, Nguyen Phuong, Pilarski Robert, Rai Karan, Schoenfield Lynn, Senecal Kelly, Wakely Paul, Hansen Paul, Lechan Ronald, Powers James, Tischler Arthur, Grizzle William E., Sexton Katherine C., Kastl Alison, Henderson Joel, Porten Sima, Waldmann Jens, Fassnacht Martin, Asa Sylvia L., Schadendorf Dirk, Couce Marta, Graefen Markus, Huland Hartwig, Sauter Guido, Schlomm Thorsten, Simon Ronald, Tennstedt Pierre, Olabode Oluwole, Nelson Mark, Bathe Oliver, Carroll Peter R., Chan June M., Disaia Philip, Glenn Pat, Kelley Robin K., Landen Charles N., Phillips Joanna, Prados Michael, Simko Jeffry, Smith-McCune Karen, VandenBerg Scott, Roggin Kevin, Fehrenbach Ashley, Kendler Ady, Sifri Suzanne, Steele Ruth, Jimeno Antonio, Carey Francis, Forgie Ian, Mannelli Massimo, Carney Michael, Hernandez Brenda, Campos Benito, Herold-Mende Christel, Jungk Christin, Unterberg Andreas, von Deimling Andreas, Bossler Aaron, Galbraith Joseph, Jacobus Laura, Knudson Michael, Knutson Tina, Ma Deqin, Milhem Mohammed, Sigmund Rita, Godwin Andrew K., Madan Rashna, Rosenthal Howard G., Adebamowo Clement, Adebamowo Sally N., Boussioutas Alex, Beer David, Giordano Thomas, Mes-Masson Anne-Marie, Saad Fred, Bocklage Therese, Landrum Lisa, Mannel Robert, Moore Kathleen, Moxley Katherine, Postier Russel, Walker Joan, Zuna Rosemary, Feldman Michael, Valdivieso Federico, Dhir Rajiv, Luketich James, Pinero Edna M. Mora, Quintero-Aguilo Mario, Carlotti Carlos Gilberto, Dos Santos Jose Sebastião, Kemp Rafael, Sankarankuty Ajith, Tirapelli Daniela, Catto James, Agnew Kathy, Swisher Elizabeth, Creaney Jenette, Robinson Bruce, Shelley Carl Simon, Godwin Eryn M., Kendall Sara, Shipman Cassaundra, Bradford Carol, Carey Thomas, Haddad Andrea, Moyer Jeffey, Peterson Lisa, Prince Mark, Rozek Laura, Wolf Gregory, Bowman Rayleen, Fong Kwun M., Yang Ian, Korst Robert, Rathmell W. Kimryn, Fantacone-Campbell J. Leigh, Hooke Jeffrey A., Kovatich Albert J., Shriver Craig D., DiPersio John, Drake Bettina, Govindan Ramaswamy, Heath Sharon, Ley Timothy, Van Tine Brian, Westervelt Peter, Rubin Mark A., Lee Jung Il, Aredes Natália D., Mariamidze Armaz. A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell. 2018;173(2):386-399.e12. doi: 10.1016/j.cell.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren G, et al. CTCF-mediated enhancer-promoter interaction is a critical regulator of cell-to-cell variation of gene expression. Mol. Cell. 2017;67:1049–1058. doi: 10.1016/j.molcel.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ron, G., Globerson, Y., Moran, D. & Kaplan, T. Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat. Commun. 8, 2237 (2017). [DOI] [PMC free article] [PubMed]
- 33.Javierre BM, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Vega F, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, et al. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell. 2017;31:225–239. doi: 10.1016/j.ccell.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Z. et al. Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers. Commun. Biol. 1, 234 (2018). [DOI] [PMC free article] [PubMed]
- 37.Ye Y, et al. The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Syst. 2018;6:314–328. doi: 10.1016/j.cels.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang Y, et al. Comprehensive characterization of alternative polyadenylation in human cancer. J. Natl. Cancer Inst. 2018;110:379–389. doi: 10.1093/jnci/djx223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilcrease MZ, et al. Coexpression of α6β4 integrin and guanine nucleotide exchange factor Net1 identifies node-positive breast cancer patients at high risk for distant metastasis. Cancer Epidemiol. Biomark. Prev. 2009;18:80–87. doi: 10.1158/1055-9965.EPI-08-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Z. et al. tRic: a user-friendly data portal to explore the expression landscape of tRNAs in human cancers. RNA Biol. 1–6 10.1080/15476286.2019.1657744 (2019). [DOI] [PMC free article] [PubMed]
- 42.Ye Y, et al. Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat. Metab. 2019;1:431–444. doi: 10.1038/s42255-019-0045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong J, et al. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep. 2017;21:1968–1981. doi: 10.1016/j.celrep.2017.10.070. [DOI] [PubMed] [Google Scholar]
- 44.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 45.Beltran AS, Graves LM, Blancafort P. Novel role of Engrailed 1 as a prosurvival transcription factor in basal-like breast cancer and engineering of interference peptides block its oncogenic function. Oncogene. 2014;33:4767–4777. doi: 10.1038/onc.2013.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ran Y, et al. γ‐Secretase inhibitors in cancer clinical trials are pharmacologically and functionally distinct. EMBO Mol. Med. 2017;9:950–966. doi: 10.15252/emmm.201607265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elsum IA, et al. Scrib heterozygosity predisposes to lung cancer and cooperates with KRas hyperactivation to accelerate lung cancer progression in vivo. Oncogene. 2014;33:5523–5533. doi: 10.1038/onc.2013.498. [DOI] [PubMed] [Google Scholar]
- 49.Ruan H, et al. Comprehensive characterization of circular RNAs in ~1000 human cancer cell lines. Genome Med. 2019;11:55. doi: 10.1186/s13073-019-0663-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science362, eaav1898 (2018). [DOI] [PMC free article] [PubMed]
- 51.Iorio F, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, Y. et al. Enhancing Hi-C data resolution with deep convolutional neural network HiCPlus. Nat. Commun. 9, 750 (2018). [DOI] [PMC free article] [PubMed]
- 53.Van Allen EM, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Auslander Noam, Zhang Gao, Lee Joo Sang, Frederick Dennie T., Miao Benchun, Moll Tabea, Tian Tian, Wei Zhi, Madan Sanna, Sullivan Ryan J., Boland Genevieve, Flaherty Keith, Herlyn Meenhard, Ruppin Eytan. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nature Medicine. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zerbino, D. R., Wilder, S. P., Johnson, N., Juettemann, T. & Flicek, P. R. The ensembl regulatory build. Genome Biol. 16, 56 (2015). [DOI] [PMC free article] [PubMed]
- 58.Wright, J. C. et al. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat. Commun. 7, 11778 (2016). [DOI] [PMC free article] [PubMed]
- 59.Karolchik, D., Hinrichs, A. S. & Kent, W. J. The UCSC genome browser. Curr. Protoc. Hum. Genet. 10.1002/0471142905.hg1806s71 (2011). [DOI] [PMC free article] [PubMed]
- 60.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 61.der Maaten L, Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:85. [Google Scholar]
- 62.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 63.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All accession codes, unique identifiers, or web links for publicly available datasets are described in the paper. All data supporting the findings of the current study are listed in Supplementary Data 1–8, Supplementary Fig. 7, and our online data portal (https://hanlab.uth.edu/eRic/).
All codes are available upon reasonable request.