Abstract
Herein we present an assay allowing concurrent detection of oxidative DNA damage and photoproducts. We apply DNA repair enzymes specific for each lesion type to incorporate spectrally distinct fluorescent nucleotides, enableing simultaneous quantification of the lesions on individual DNA molecules. We follow the repair of both damage types in skin cells exposed to artificial sunlight.
Tens of thousands of DNA lesions are produced daily in living organisms from exogenous and endogenous exposures1. Genomic instability plays an initiating role in carcinogenesis2,3, is involved in aging4–6, and has been linked to Alzheimer’s7–9 and other medical conditions. DNA damage takes the form of double or single strand breaks and various chemical modifications to the bases themselves10,11. Given the importance of DNA damage and its role in genomic instability, there is a need for reliable experimental methods to measure and quantify DNA damage.
Single and double strand breaks are commonly measured indirectly through the use of strand break markers such like phospho-H2AX (γH2AX) or 53BP-1 and quantified by immunofluorescence or enzyme-linked immunosorbent assay (ELISA)12,13. Comet assay or Single cell Gel Electrophoresis allows more specific strand break detection in cells, eliminating the requirements for specific antibodies14. Detection of chemical modifications present an even more difficult challenge due to their variety. Two major types of damaged base modifications are oxidative15 and photoproducts16. Oxidative base lesions are the reaction products of reactive oxygen species (ROS) and nitrogenous bases17. More than 20 different oxidative base modifications have been identified with 8-oxo-2′deoxyguanosine (8-oxodG) as a notable example15. Photoproducts are crosslinks between adjacent pyrimidine bases, i.e., cyclobutane pyrimidine dimers (CPD) or (6–4) pyrimidine-pyrimidone structures, that result from direct exposure to ultraviolet (UV) radiation18,19. Chemical modifications can also occur in mixtures, with UV radiation producing ROS that creates oxidative base damage in addition to photoproducts20,21. Direct quantification of these damage types or their combinations is limited due to the need for antibodies specific to each lesion type.
Recent developments in single molecule approaches have demonstrated that various DNA base lesions could be characterized through analysis of individual DNA molecules22–25. Using a commercial cocktail of repair enzymes, we previously analysed the global amount of DNA damage and repair dynamics in human cell lines26. Although this approach showed very high sensitivity, it could not distinguish between different damage types due to the use of a mixture of repair enzymes recognizing various types of damage. To address the need for simultaneous detection of multiple specific damage types, we developed an assay for single-molecule multi-colour detection and relative quantification of photoproducts and oxidation products simultaneously.
To facilitate the distinction between damage types via labelling with different colours we first used two individual reactions for labelling oxidative damage and photoproducts. As a model for photoproducts, we used HEK cells exposed to either UVA, UVB or UVC radiation for increasing periods of time, resulting in an increasing dose of UV radiation. Oxidative damage was induced by exposing HEK cells to increasing concentrations of hydrogen peroxide. Labelling of photoproducts was performed with pyrimidine dimer glycosylase (T4 PDG), an enzyme that recognizes cis-syn-cyclobutane pyrimidine dimers caused by UV irradiation27. Oxidative damage was specifically labelled using human 8-oxoguanine DNA glycosylase (hOGG1), an enzyme that releases damaged purines from double stranded DNA28. For every experiment, a control DNA sample extracted from untreated cells served to evaluate the endogenous DNA damage baseline. We compared the results from our assays to the commercial cocktail of repair enzymes (PreCR mix) to verify their correlation. After labelling damage sites and without further cleaning, the backbone of the DNA was stained and then stretched on modified glass slides for imaging. Data were analysed using custom software that detects the stretched DNA molecules and counts the number of labels along the DNA contour, reporting on the number of damage sites as a function of overall DNA length measured29 (for error calculations see Fig. S1 and for experimental details see methods section, ESI†).
As expected, more DNA lesions were induced by longer irradiation time or higher hydrogen peroxide concentration in agreement with the results obtained with the PreCR mix (Fig. S2 and Fig. S3, ESI†). We also observed increased damage levels with shorter wavelength, higher energy UV. When treating the DNA extracted from irradiated cells with the hOGG1 mix, fluorescent labelling was observed, indicating that irradiation induced oxidative DNA damage in addition to photoproducts. Both damage types increased with UVC in a dose-dependent manner (see Fig. S2a, ESI†). UVB radiation showed only a slight increase in photoproducts with no increase in oxidative damage (see Fig. S2b, ESI†). The UVA radiation produced no detectable increase in any of the damage types measured over the dose range (see Fig. S2c, ESI†).
We found the results from our labelling assays to be more reproducible than those received with the PreCR mix (see Fig S4, ESI†). Several improvements were added to the optimized reactions. First, we found that hOGG1 performed better than the Formamidopyrimidine DNA glycosylase (Fpg) enzyme present in the commercial PreCR mix for repairing oxidative damage. Fpg produced a significant amount of non-specific labelling that prevented quantitative analysis (see Fig. S5, ESI†). In addition, in the optimized reactions we used Large Fragment Bst Polymerase instead of the Full Fragment Bst. Large Fragment Bst produced significantly less non-specific end labelling of DNA molecules and was thus better for quantitative damage assessment (see Fig. S6, ESI†). Importantly, we found that the reaction for photoproducts produced labelling in cells treated with hydrogen peroxide, where no photoproducts are expected (see Fig. S3, ESI†). This is due to the presence of Endonuclease IV, that is used for conditioning the gaps, but is also known to detect and excise oxidative damage and AP sites. However, eliminating the Endonuclease IV from this reaction drastically decreases the labelling efficiency of the assay, emphasizing its importance (see Fig. S7, ESI†).
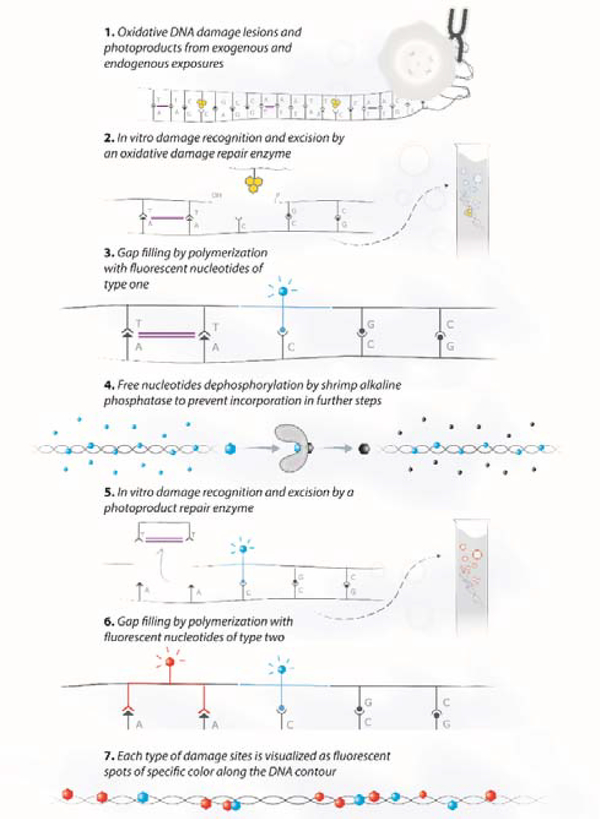
Given that UV radiation results in mixtures of base modifications on the same DNA molecules, quantifying both oxidative lesions and photoproducts simultaneously would provide new insight into DNA damage mechanisms and their repair dynamics. Spectrally distinct fluorescent labels offer the potential to assign different colours to different damage types within the same sample and on the same DNA molecule. To highlight multiple damage types on the same sample, we performed the two damage-specific reactions consecutively while enzymatically degrading the fluorescent nucleotides between labelling reactions (for experimental details see methods section, ESI†). We first labelled the oxidative damage with one colour using the hOGG1 mix and then the photoproducts with a second colour. For selective labelling of each damage type with a distinct fluorescent nucleotide, we used a heat sensitive phosphatase to cleave the phosphate groups from any residual fluorescent nucleotides from the first reaction, leaving a terminating hydroxyl group that prevents further incorporation by the DNA polymerase. The second enzyme mix is then applied along with spectrally distinct nucleotides that are inserted by the DNA polymerase during the second repair step. This way, the amount of fluorescent labelling at each colour reflects the amount of each DNA damage type. Scheme 1 represents the various steps of the multi-labelling reaction.
Scheme 1:
Schematic representation of the multi-labelling reaction that highlights multiple damage types on the same DNA sample. First, the oxidative DNA damage is repaired using hOGG1 and Endonuclease IV enzymes followed by ATTO550 dye labelling (represented with blue markers). Then the ATTO550 nucleotides are rendered inactive by an alkaline phosphatase that removes the phosphate groups from the unincorporated nucleotides, followed by heat inactivation. Finally, the photoproducts are repaired using the T4 PDG and Endonuclease IV enzymes followed by ATTO647N dye labelling (represented with red markers).
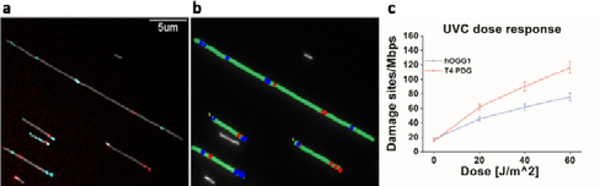
To validate our multi-labelling reaction, we used the previously analysed HEK cells exposed to UVC, as these samples showed an increase in both oxidative damage and photoproducts. Figure 1 shows some representative labelled DNA molecules from the imaged data and the image analysis results. Figure 1a was cropped from a typical field of view (see Fig. S8, ESI†) containing labelled DNA molecules from the imaged data. Each grey structure is a DNA molecule and the red dots represent repaired photoproduct sites while blue dots represent the repaired oxidative damage sites. Figure 1b shows the DNA molecules detected by the image analysis software (green) as well as the detected oxidative sites (blue) and photoproduct sites (red). Only linear molecules with a minimal length of 12 kbp that do not cross other molecules are analysed. Figure 1c shows an increase in both photoproducts and oxidative damage with increasing UVC dose. Results for both damage types are produced simultaneously from the same sample and correlate with the results received for the individual reactions (Figure S2c), although the absolute numbers vary due to different reaction conditions. We note that due to the ability of the reaction for photoproducts labelling to repair some oxidative lesions as well (see Fig. S3, ESI†), it is important to first apply the oxidation-damage specific hOGG1 mix and only then apply the photoproduct T4 PDG mix (see Fig. S9, ESI†).
Figure 1:
Simultaneous detection and quantification of photoproducts and oxidative DNA damage in HEK cells exposed to UVC radiation. (a) Fluorescence image of DNA molecules labelled with the multi-labelling reaction. Grey colour represents the backbone of the DNA molecules, red dots represent photoproduct sites, and blue dots represent oxidative damage sites. (b) Analysis results of the raw image using automatic software. Green – detected DNA molecules, red dots – detected photoproduct labels, blue dots –detected oxidative damage sites. (c) Dose response of HEK cells exposed to UVC radiation as detected by the multi-labelling reaction. Red represents the levels of detected photoproducts and blue the levels of oxidative damage.
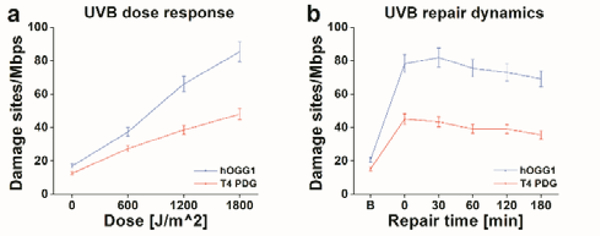
UVC shows the most dramatic induction of photoproducts and oxidative lesions; however, it is mostly absorbed by atmospheric ozone and is less relevant to human exposure. Ambient sunlight in the UV range is mostly UVA (90%–95%) and residual UVB (5%–10%). While the longer wavelength UVA penetrates deeply into the dermis, UVB is predominantly absorbed by the skin’s more superficial epidermal layers30. It is the main cause of sunburn and plays a key role in the development of skin cancer31,32. Therefore, to demonstrate the utility of our methods for studying exposure, we examined human skin cells exposed to “environmental UV” irradiation centred at 315–320nm, right at the interface between UVA and UVB light. We used immortalized human keratinocyte (HaCaT) cells and exposed them to this radiation with a power density of 1 W/m2, attempting to simulate ambient solar exposures at this wavelength33–35. We first examined the dose response of the HaCaT cells to the UVB, simultaneously following the increasing levels of both photoproducts and oxidative damage. Then, we tracked the repair process of these lesions by returning the cells to the incubator for given intervals of recovery time after irradiation and measuring the retention of photoproducts and oxidative lesions in the genomic DNA after each given repair time. As before, unexposed cells served as controls for basal damage levels in every experiment (for experimental details see methods section, ESI†). Figure 2 summarizes the damage measurements for the HaCaT cells experiments using our dual-colour reaction.
Figure 2:
“Environmental UV”-induced DNA damage and repair in the human skin model cell line HaCaT, as detected by the multi-labelling reaction. (a) HaCaT cells response to increasing dose of UVB radiation. (b) Repair dynamics over 180 minutes in HaCaT cells exposed to 1800 J/m2 of UVB radiation. In both graphs, red represents the levels of detected photoproducts and blue the levels of oxidative damage.
Figure 2a shows that increased irradiation time results in higher levels of photoproducts (red) and oxidative damage (blue) as expected. In Figure 2b, we show the DNA-damage repair dynamics of HaCaT cells exposed to 1800 J/m2 of “environmental UV.” A sharp increase in damage levels is observed immediately after irradiation and then a gradual decrease in damage levels is observed as a function of repair time, due to the natural repair process of the cells.
In summary, we demonstrated a single molecule approach for distinctive labelling of two major DNA damage types for simultaneous detection. In all experiments, only 50 ng of DNA was needed to produce efficient labelling, an important factor in cases of rare biological samples. The multi-labelling reaction does not require any cleaning or removal of free fluorescent nucleotides prior to imaging, improving yields and shortening preparation times. The automatic sample imaging and image analysis process greatly eased the overall procedure, making the entire assay robust and straightforward. Moreover, the new protocol for glass coverslip activation is much faster, reducing the assay time. Although this study focused on photoproducts and oxidative damage, this method is not limited to these damage types. This methodology could be used with any specific repair enzyme to label other damage types of interest and monitor repair dynamics. The approach can also be combined with epigenetic labelling to get insight into the relationship between DNA damage and epigenetic modulation36–39. Some of the most interesting questions in the field require knowledge of the location of specific damage lesions in the genome. The genomic distribution of DNA damage may potentially be mapped utilizing nano-channel technology and integration of this labelling approach with optical genome mapping as recently shown for DNA methylation40–43. Overall, this assay is highly adaptable for various biological studies and can easily be extended to reveal more information, while requiring minimal expertise.
Supplementary Material
Acknowledgments
This work was funded by the BeyondSeq consortium (EC program 634890), the European Research Council Proof of Concept grant (grant no. 767931), the European Research Council starter grant (grant No. 337830), the US National Institute of Health (grant R21ES028015-011) and the Joint Israeli German R&D nanotechnology (grant no. 61976).
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Jackson SP and Bartek J, Nature, 2009, 461, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loft S and Poulsen HE, J. Mol. Med, 1996, 74, 297–312. [DOI] [PubMed] [Google Scholar]
- 3.Wiseman H and Halliwell B, Biochem. J, 1996, 313 (Pt 1), 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ames BN, Mutat. Res. Mol. Mech. Mutagen, 1989, 214, 41–46. [DOI] [PubMed] [Google Scholar]
- 5.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC and Bohr VA, Carcinogenesis, 2008, 30, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoeijmakers JHJ, Engl N J. Med, 2009, 361, 1475–1485. [DOI] [PubMed] [Google Scholar]
- 7.Mecocci P, MacGarvey U and Beal MF, Ann. Neurol, 1994, 36, 747–751. [DOI] [PubMed] [Google Scholar]
- 8.de la Monte SM, Luong T, Neely TR, Robinson D and Wands JR, Lab. Investig, 2000, 80, 1323–1335. [DOI] [PubMed] [Google Scholar]
- 9.Lovell MA and Markesbery WR, Nucleic Acids Res, 2007, 35, 7497–7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Bont R and van Larebeke N, Mutagenesis, 2004, 19, 169–185. [DOI] [PubMed] [Google Scholar]
- 11.Houtgraaf JH, Versmissen J and van der Giessen WJ, Cardiovasc. Revascularization Med, 2006, 7, 165–172. [DOI] [PubMed] [Google Scholar]
- 12.Wani AA, Gibson RE, D’Ambrosio and D’Ambrosio SM, Photochem. Photobiol, 1984, 40, 465–471. [DOI] [PubMed] [Google Scholar]
- 13.Santella RM, 1999, 733–739. [PubMed] [Google Scholar]
- 14.Braafladt S, Reipa V and Atha DH, Sci. Rep, 2016, 6, 32162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.COOKE MS, EVANS MD, Dizdaroglu M and Lunec J, 2003, 17, 1195–1214. [DOI] [PubMed] [Google Scholar]
- 16.Sinha RP and Häder D-P, Photochem. Photobiol. Sci, 2002, 1, 225–236. [DOI] [PubMed] [Google Scholar]
- 17.Cheng KC, Cahill DS, Kasai H, Nishimura S and a Loeb L, J. Biol. Chem, 1992, 267, 166–172. [PubMed] [Google Scholar]
- 18.Matsunaga T, Hieda K and Nikaido O, Photochem. Photobiol, 1991, 54, 403–410. [DOI] [PubMed] [Google Scholar]
- 19.Rastogi RP, Richa, Kumar A, Tyagi MB and Sinha RP, J. Nucleic Acids, 2010, 2010, 592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielbassa C, Roza L and Epe B, Carcinogenesis, 1997, 18, 811–816. [DOI] [PubMed] [Google Scholar]
- 21.Kvam E and Tyrrell RM, Carcinogenesis, 1997, 18, 2379–2384. [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Park HS, Lim S and Jo K, Chem. Commun, 2013, 49, 4740. [DOI] [PubMed] [Google Scholar]
- 23.Gassman NR and Holton NW, Curr. Opin. Biotechnol, 2019, 55, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Kim Y, Lim S and Jo K, Analyst, 2016, 141, 847–852. [DOI] [PubMed] [Google Scholar]
- 25.Kang Y, Lee J, Kim J, Oh Y, Kim D, Lee J, Lim S and Jo K, Analyst, 2016, 141, 4326–4331. [DOI] [PubMed] [Google Scholar]
- 26.Zirkin S, Fishman S, Sharim H, Michaeli Y, Don J and Ebenstein Y, J. Am. Chem. Soc, 2014, 136, 7771–6. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd RS, Hanawalt PC and Dodson ML, Nucleic Acids Res, 1980, 8, 5113–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boiteux S and Radicella JP, Biochimie, 81, 59–67. [DOI] [PubMed] [Google Scholar]
- 29.Jain N, Shahal T, Gabrieli T, Gilat N, Torchinsky D, Michaeli Y, Vogel V and Ebenstein Y, Epigenetics, DOI: 10.1080/15592294.2019.1638700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Orazio J, Jarrett S, Amaro-Ortiz A, Scott T, D’Orazio J, Jarrett S, Amaro-Ortiz A and Scott T, Int. J. Mol. Sci, 2013, 14, 12222–12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzpatrick TB and Sober AJ, N. Engl. J. Med, 1985, 313, 818–820. [DOI] [PubMed] [Google Scholar]
- 32.Armstrong BK and Kricker A, J. Photochem. Photobiol. B, 2001, 63, 8–18. [DOI] [PubMed] [Google Scholar]
- 33.Diffey BL, Methods, 2002, 28, 4–13. [DOI] [PubMed] [Google Scholar]
- 34.Rünger TM, J. Invest. Dermatol, 2007, 127, 2103–2105. [DOI] [PubMed] [Google Scholar]
- 35.Kalogirou SA, Pashiardis S and Pashiardi A, Renew. Energy, 2017, 111, 580–597. [Google Scholar]
- 36.Michaeli Y, Shahal T, Torchinsky D, Grunwald A, Hoch R and Ebenstein Y, Chem. Commun, 2013, 49, 8599. [DOI] [PubMed] [Google Scholar]
- 37.Nifker G, Levy-Sakin M, Berkov-Zrihen Y, Shahal T, Gabrieli T, Fridman M and Ebenstein Y, ChemBioChem, 2015, 16, 1857–1860. [DOI] [PubMed] [Google Scholar]
- 38.Gilat N, Tabachnik T, Shwartz A, Shahal T, Torchinsky D, Michaeli Y, Nifker G, Zirkin S and Ebenstein Y, Clin. Epigenetics, 2017, 9, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kafer GR, Li X, Horii T, Suetake I, Tajima S, Hatada I and Carlton PM, Cell Rep, 2016, 14, 1283–1292. [DOI] [PubMed] [Google Scholar]
- 40.Michaeli Y and Ebenstein Y, Nat. Biotechnol, 2012, 30, 762–763. [DOI] [PubMed] [Google Scholar]
- 41.Gabrieli T, Sharim H, Nifker G, Jeffet J, Shahal T, Arielly R, Levi-Sakin M, Hoch L, Arbib N, Michaeli Y and Ebenstein Y, ACS Nano, 2018, 12, 7148–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeffet J, Kobo A, Su T, Grunwald A, Green O, Nilsson AN, Eisenberg E, Ambjörnsson T, Westerlund F, Weinhold E, Shabat D, Purohit PK and Ebenstein Y, ACS Nano, 2016, 10, 9823–9830. [DOI] [PubMed] [Google Scholar]
- 43.Sharim H, Grunwald A, Gabrieli T, Michaeli Y, Margalit S, Torchinsky D, Arielly R, Nifker G, Juhasz M, Gularek F, Almalvez M, Dufault B, Sen Chandra S, Liu A, Bhattacharya S, Chen Y-W, Vilain E, Wagner KR, Pevsner J, Reifenberger J, Lam ET, Hastie AR, Cao H, Barseghyan H, Weinhold E and Ebenstein Y, Genome Res, 2019, 29, 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.