Abstract
Flaviviruses are the most medically relevant group of arboviruses causing a wide range of diseases in humans and are associated with high mortality and morbidity, as such posing a major health concern. Viruses belonging to this family can be endemic (e.g., dengue virus), but can also cause fulminant outbreaks (e.g., West Nile virus, Japanese encephalitis virus and Zika virus). Intense research efforts in the past decades uncovered shared fundamental strategies used by flaviviruses to successfully replicate in their respective hosts. However, the distinct features contributing to the specific host and tissue tropism as well as the pathological outcomes unique to each individual flavivirus are still largely elusive. The profound footprint of individual viruses on their respective hosts can be investigated using novel technologies in the field of proteomics that have rapidly developed over the last decade. An unprecedented sensitivity and throughput of mass spectrometers, combined with the development of new sample preparation and bioinformatics analysis methods, have made the systematic investigation of virus–host interactions possible. Furthermore, the ability to assess dynamic alterations in protein abundances, protein turnover rates and post-translational modifications occurring in infected cells now offer the unique possibility to unravel complex viral perturbations induced in the infected host. In this review, we discuss the most recent contributions of mass spectrometry–based proteomic approaches in flavivirus biology with a special focus on Zika virus, and their basic and translational potential and implications in understanding and characterizing host responses to arboviral infections.
Keywords: flaviviruses, Zika virus, proteomics, interactome, AP-LC-MS/MS, phosphoproteomics, Label-free Quatification, arboviruses, DENV, WNV
1. Introduction
Viruses are highly adapted intracellular pathogens that co-evolved with their hosts over millions of years. This resulted in a sophisticated adaptation that allowed viruses to engross cellular machineries for their own replication, with the benefit to encode their genetic information with high efficiency. However, this strict dependency on cellular factors comes with the obligation to replicate intracellularly and the requirement of intimate interactions between viral and cellular proteins. Moreover, most pathogens are preferentially replicating in specific organs and cause virus-associated pathologies. Technological developments in the fields of genomics and proteomics became available in recent years, allowing to systematically map interactions between viruses and their hosts in an unbiased manner. Such interactions are of particular interest especially when uncharacterized pathogens strike in a naïve population.
One such pathogen is the Zika virus, a positive stranded RNA virus belonging to the Flaviviridae family, causing the first large unexpected outbreak in Asia and French Polynesia in 2013 [1]. In 2016, a sudden raise in incidence rates of microcephaly in newborns in southern America could be linked to the ongoing Zika virus (ZIKV) outbreak in this otherwise naïve population [2,3]. In adults, ZIKV is predominantly asymptomatic or causes mild, flu-like symptoms, although recent studies report visual impairment and infertility [3,4,5]. Following vertical transmission upon maternal infection, ZIKV can cause fetal demise or Guillain-Barré syndrome, a neuro-inflammatory disease of the peripheral nervous system [6,7]. However, the limited knowledge on ZIKV pathogenesis and the sudden spread of this pathogen sparked a number of independent laboratories to characterize the virus–host interactions on a molecular level. These studies are of particular interest, since ZIKV combines many unique features of flaviviruses that are normally limited to individual species. Indeed, similarly to other members of the Flaviviridae family like Dengue (DENV), Yellow fever (YFV), Japanese encephalitis virus (JEV) and West Nile virus (WNV), ZIKV is also transmitted by mosquitoes. Furthermore, analogously to JEV, WNV and tick-borne encephalitis virus (TBEV), Zika virus infections are associated with neurological diseases, although ZIKV is the only Flaviviridae member with documented ability to cross the placental barrier and be sexually transmitted [8].
Technical advancement in mass-spectrometry analysis and the availability of replication competent epitope-tagged viruses and individual viral proteins have prompted several studies to investigate persistent modulation of protein abundances, post-translational modifications and protein–protein interactions on a global scale and with unprecedented sensitivity and quantitation [9,10,11,12,13]. Given its unique properties and the urgent need to further understand ZIKV biology, a number of laboratories started to use large-scale approaches to study how this pathogen interacts with cellular proteins. Here we review current approaches, in particular proteomic-based datasets, which have been used to identify ZIKV–host interactions.
2. Illuminating Dark Matter: Unbiased Approaches to Study Virus–Host Interactions
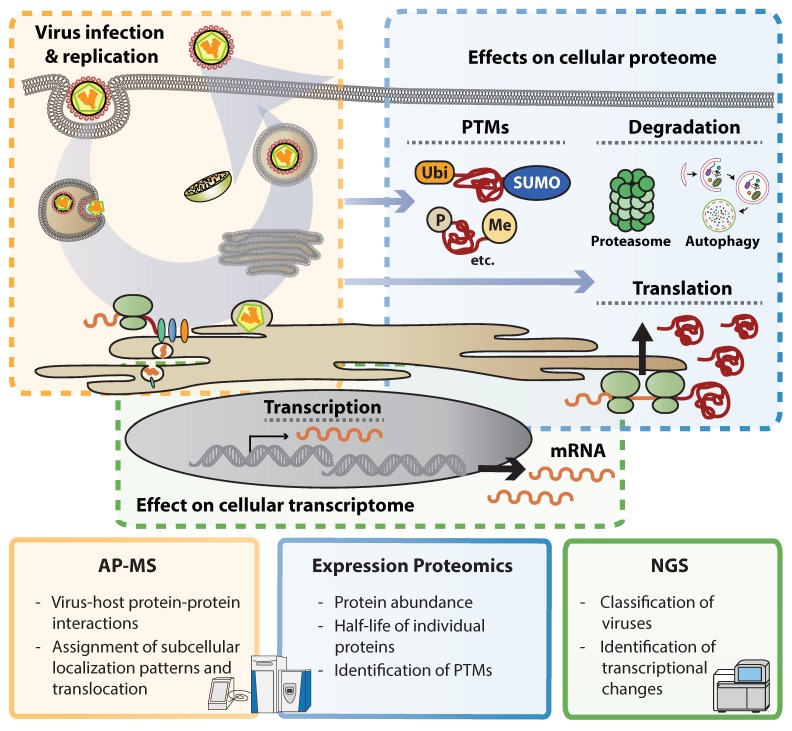
The recent technological revolution allows acquiring knowledge on biological processes at incredible speed. Several systematic and unbiased genetic approaches to identify host factors involved in flavivirus replication have been conducted to date [14,15,16,17,18,19,20,21]. For instance, next generation sequencing (NGS) became an affordable and widely used technology to unearth underrepresented sequences or estimate the abundance of RNA transcripts in complex mixtures. This made possible the identification of newly emerging viruses and virus variants. Illustrative examples in this direction are efforts during public health responses as recently shown by ZIKV and Ebola virus epidemics [22,23,24]. Furthermore, NGS can be used to study widespread transcriptional changes occurring upon virus infection, including expression of cytokines and antiviral response patterns. Deconvolution of such data using bioinformatics techniques offers a global overview of transcriptional responses and allows identifying viral activities to perturb signaling cascades. However, viruses modulate gene expression in a post-transcriptional manner by affecting mRNA processing, transport, translation, protein maturation and stability and therefore information on mRNA abundance is only partially reflecting the expression on protein level [25]. Mass spectrometry (MS)-based proteomics evolved into a sensitive and reliable technique that allows identifying proteins in complex mixtures [26]. Comparisons between mRNA and protein expression patterns in virus-infected cells revealed dramatic differences, indicating that protein abundance does not necessarily follow expression levels on transcript level [27]. Labeling proteins of a subcellular compartment (e.g., the cell surface) or combining fractionation techniques with mass spectrometry, allows assigning subcellular localization patterns to individual proteins and also monitor their translocation between compartments in a systematic manner [28,29]. MS can also identify post-translational modifications (PTMs) such as differential phosphorylation, ubiquitination or acetylation patterns. Such approaches, however, often require elaborative enrichment of modified peptides and mostly provide descriptive insights into PTMs, and functional consequences of individual modifications remain often elusive. In addition to alterations in protein expression patterns and protein quality, MS puts proteins into context. Isolation of protein complexes through affinity purification (AP) followed by MS analysis (AP-MS) gives insights into protein–protein interactions and engagement of cellular machineries. AP-MS has successfully been used to identify protein complexes and can put proteins in functional context (Figure 1).
Figure 1.
– Omics methods to study virus-induced cellular perturbations. The key advantages of Interaction proteomics (affinity purification mass spectrometry, AP-MS), expression proteomics and next generation sequencing (NGS) in studying flavivirus-induced perturbations are listed within each square. Colors indicate the respective step(s) of the viral replication cycles predominantly targeted by each method. Abbreviations: PTMs, post-translational modifications.
Development of sample preparation methodologies, highly sensitive mass analyzers as well as tailored bioinformatics processing pipelines (comprehensively summarized in [30]) have caused MS to flourish in the last years. MS is no longer a niche technology reserved to a limited number of experts but is now available for a wide range of scientists interested in biological sciences. This, however, also led to generation of datasets that are exceedingly difficult to compare and to interpret due to the complexity of the underlying analysis and the wealth of data that is generated. Here we review current literature on proteomic approaches that were employed to study flaviviruses.
3. Flavivirus Interactions with the Cellular Proteome
Some members of the Flaviviridae family are considered highly pathogenic infectious agents and knowledge on their interactions is pivotal to understand these pathogens in order to identify new forms of therapy. This is particularly true for epidemic viruses such as Zika virus, which since 2016 has left a devastating impact on large populations in southern America. In an effort to understand the pathogenicity of this virus, a number of laboratories employed systems analysis to elucidate the functions of individual proteins and the virus as a whole.
3.1. ZIKV–Host Interactions
3.1.1. Interactions of ZIKV Capsid with Host Proteins
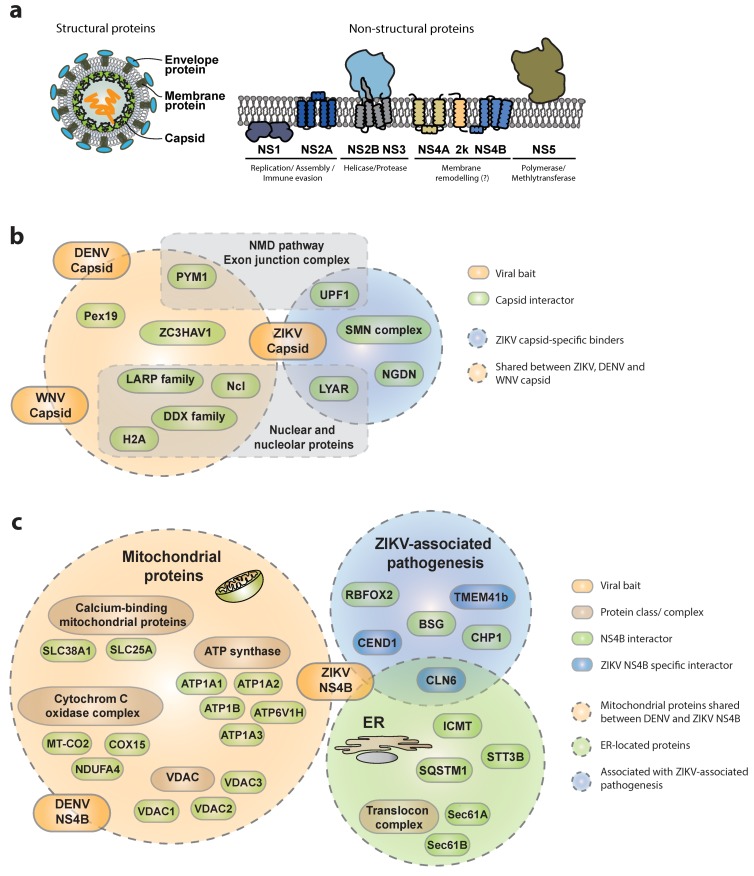
Systematic studies are of paramount importance to identify virus–host protein–protein interactions (PPIs), since they provide an unbiased view of molecular complexes and therefore insights into viral mechanisms of replication, host resources exploitation and immune evasion strategies [11,12,31,32]. Applying AP-MS in a systematic and comparative fashion can help elucidate similarities and differences in virus-specific replication strategies and even pathogenesis determinants [9,33]. A few studies recently reported combined proteomic and gene perturbation approaches integrating complementary methods to investigate the cellular interactome of each of the 10 ZIKV proteins (Figure 2a). A discrete number of previously reported bona fide interactors were consistently identified across all these studies, and appear to be shared across different flavivirus members, highlighting the relatively high degree of conservation within the genus and across diverse cellular backgrounds, thereby revealing cellular components critically required for virus replication. However, these studies also revealed specificities, reflecting the different methodologies, cellular backgrounds, enrichment strategies, gene-delivery methods and experimental designs used. For instance, using an AP-LC-MS/MS (affinity purification coupled with liquid chromatography and tandem mass spectrometry) approach in SK-N-BE2 neuroblastoma cells [34], we identified in the ZIKV capsid interactome a strong enrichment in nuclear and nucleolar-resident proteins such as nucleolin (NCL), nucleolar RNA-dependent helicases of the DDX family, core histones (H2A), as well as peroxisomal proteins including Pex19, that have been previously reported as cellular targets of Dengue (DENV) and West Nile virus (WNV) (Figure 2b) [35,36,37]. Interestingly, among the capsid-specific interactors, a completely new set of proteins associating with the ZIKV capsid was identified. Among these, a poorly characterized nucleolar protein involved in cell-growth regulation and maintenance of stem cell identity called LYAR was identified [38]. LYAR was recently implicated in viral transcription and replication of Influenza A virus (IAV) through interaction with the viral ribonucleoprotein [39], indicating a more global role of this protein in virus–host interactions. Additionally, several members of the LARP family (e.g., LARP1 and LARP7) and ZC3HAV1 (also known as ZAP) were identified as specific capsid interactors, suggesting the propensity of ZIKV capsid to interact with RNA-binding proteins. Importantly the functional relevance of some of these ZIKV capsid interactors was underlined by knock-down studies, confirming a strong reduction of viral replication upon gene depletion. In similar studies, Coyaud and collegues [40] as well as Shah et al. [41] confirmed these observations, identifying NCL, LYAR, LARP1, several members of the DDX family and ZC3HAV1, among the specific ZIKV capsid interactors using BioID- and AP-LC-MS/MS in 293T cells, respectively. Furthermore, recent work by Li et al. reported analogous enrichment for LARP1, LARP7 and ZC3HAV1 in a global proteomic survey for WNV-interacting proteins [42]. Complementary functional experiments validated a conserved role for ZC3HAV1 in restricting flaviviruses since depletion of ZC3HAV1 led to a 4- to 8-fold increase in ZIKV, DENV and WNV virus titers. Interestingly, the antiviral activity of ZC3HAV1 has been investigated for several virus families including human immunodeficiency virus (HIV) and Sindbis virus (SINV) [43,44]. The protein appears to recruit cellular RNA degradation machineries to specific ZAP-responsive elements on the viral mRNAs as well as the DCP1–DCP2 complex to initiate 5′-3′ RNA degradation of viral mRNA. Its activity has been mechanistically studied in the context of Japanese encephalitis virus, whereby ZC3HAV1 has been described to recruit the 3′-5′ degradation machinery [45]. However, the exact mechanisms of virus restriction by ZC3HAV1 remain to be elucidated.
Figure 2.
– Recently identified cellular interactors of flavivirus capsids and NS4Bs. (a) Schematic representation of flavivirus viral proteins and respective membrane topology. (b) Capsid interactors reported to bind exclusively to Zika virus (ZIKV)-C are surrounded by the light blue circle, while those within the orange circle were reported as shared interactors of ZIKV, West Nile virus (WNV) and Dengue (DENV)-C. The respective sub-cellular localization is indicated. (c) NS4B interactors reported to bind exclusively to ZIKV-NS4B are shown in blue, while those shared with DENV-NS4B are shown in green. Mitochondrial- and endoplasmic-reticulum-resident proteins are surrounded by green and orange circles, respectively. Proteins with functions associated to ZIKV-related pathologies are surrounded by the blue circle. All gene names and corresponding full protein names are listed in Table 1.
Among the other novel capsid interactors, additional work by the Ott and the Ramage groups identified the nonsense-mediated decay (NMD) pathway and the exon-junction complex (EJC) as targets of the WNV, DENV and ZIKA virus capsids [42,46]. While the antiviral activity of individual members of these complexes appears moderate in knock-down studies, further experimental evidence supports a direct interaction between the viral RNA and RBM8A (a central component of the EJC), suggesting a depletion of the cellular PYM1 pool upon recruitment by the WNV capsid to protect viral RNA from decay. Similarly, direct involvement of the NMD pathway in flavivirus is corroborated by the identification of the regulator of nonsense transcripts 1 (UPF1) among the ZIKV capsid interactors and the upregulation of several NMD substrate mRNAs upon ZIKV infection both in Huh7 and hNPC. Furthermore, depletion of UPF1 leads to a 50% increase in virus replication, supporting the notion that NMD acts as flavivirus restriction machinery that is counteracted by viral activities [46].
In addition to proteins recruited by several flavivirus capsids, the ZIKV capsid is the only one reported to specifically associate with cellular factors that are involved in neuronal development or neurological disorders [34]. These proteins include neuroguidin (NGDN), an eIF4E-interacting protein mediating CPEB-mediated translation of essential genes in early development and neuronal synaptic plasticity. Further ZIKA capsid interactors are several members of the survival motor neuron (SMN) complex [34], which is important for proper splicing, neuronal migration and differentiation. It would be interesting to assess binding specificity of other flavivirus capsid proteins in a similar neuronal background and investigate further their contribution to viral replication or pathogenesis. Such studies could pinpoint novel viral pathogenesis determinants and assist in the identification of druggable binding surfaces required for specific protein–protein interactions.
Altogether, flavivirus capsids evidently evolved evasive mechanisms through binding/sequestration of ZC3HAV1 and other RNA-binding proteins (e.g., UPF1, LARP7, NGDN). Capsid proteins are apparently exploiting their versatile subcellular localization (both nuclear and cytoplasmic) and their intrinsic hydrophobic character to simultaneously highjack and diverge regulation or expression of specific transcripts.
3.1.2. Interactions of ZIKV NS4B with the Host
Another interesting ZIKV protein that appears to have both conserved and unique cellular interactors across flaviviruses and plays a central role in virus–host adaptation, is the non-structural protein NS4B. Indeed, ZIKV-NS4B associates with subunits of the ATP synthase (ATP1A1, 1A2, 1A3, 1B and 6V1H), voltage-dependent anion selective channel proteins (VDAC1,2,3) and calcium-binding mitochondrial proteins (e.g., SLC25A, SLC38A1), as well as several components of the cytochrome C oxidase complex (COX15, MT-CO2, NDUFA4) (Figure 2c). These results largely mirror those of an earlier study on the DENV-NS4B interactome in Huh7 hepatoma cells [47], confirming ATP production, calcium homeostasis, apoptosis, and mitochondrial respiratory chain, as important targets of different flaviviruses. These interactors argue for a conserved association between flavivirus NS4Bs and mitochondria in virus-infected cells, eventually leading to a profound alteration of mitochondria morphodynamics. Since mitochondria are central to regulate antiviral immune responses, these interactions may contribute to perturbation of innate immune mechanisms through mitochondria elongation as proposed recently [47]. In addition to mitochondrial proteins, ZIKV NS4B appears to bind cellular proteins involved in protein stability and quality control in both SK-N-BE2 and HEK293T cells. For instance, STT3B, is involved in endoplasmic reticulum-associated degradation, ICMT, mediating targeting of isoprenylated proteins to the cell membrane, and the autophagosome-associated protein SQSTM1 (p62), which is in line with a general and conserved role of autophagy in flavivirus infections [34,41,48,49,50].
NS4B also appeared to associate with cellular proteins that could be associated with the whole spectrum of ZIKV-associated pathogenesis including neurodegenerative disorders and retinal degeneration (CLN6, BSG), neuronal differentiation defects (CEND1, RBFOX2) and axonal dysfunction (CHP1, TMEM41b) [34]. While these cellular factors appeared as specific binders of arthropod-borne flaviviruses in neuronal cells (such as DENV and ZIKV), a subset of proteins exhibited ZIKV specificity (TMEM41b, CEND1, CLN6), suggesting that similarly to capsid, also NS4B might have evolved partially divergent binding affinities across different flaviviruses, with potential consequences for the distinctive pathogenic outcome. Interestingly CLN6, a poorly characterized protein associated with a neurodegenerative disease with late-infantile onset, shares several cellular binding partners with NS4B, and specifically associates with mTOR. These results support reports associating ZIKV infection or NS4B overexpression to defective neurogenesis via suppression of the AKT-mTOR signaling pathway [51]. Another conserved target of multiple flavivirus NS4Bs is the translocon complex, with the Sec61A and Sec61B subunits reported to bind NS4A [41] or NS2B3 and NS4B [34]. The critical requirement of this complex for flavivirus replication both in mammalian and insect cells was further supported by the potent antiviral activity of CT8, a specific translocon inhibitor [21,52] as well as its intracellular accumulation in NS3/NS4B-positive convoluted membranes in DENV-infected cells [47].
3.1.3. Other Viral Proteins
In addition to capsid and NS4B, several novel ZIKV NS5 binders were recently identified in these proteomics surveys. Among these, multiple members of the PAF1C complex (Leo1, CDC73, CTR9 and PAF1), confirming similar observations previously made on DENV-NS5 by the Gamarnik group [53] and recently confirmed by others [54]. Importantly, while these interactions appear highly conserved among DENV, ZIKV, JEV and WNV-NS5s, a moderate and reciprocal effect on viral replication was observed upon gene silencing (2-fold increase and 2-fold decrease for ZIKV and JEV infectivity, respectively) [41,54]. Further mechanistic studies suggest that PAF1 might be recruited by NS5 to reduce expression of interferon-stimulated genes and therefore dampen immune responses [41]. Interestingly, NS5 has also been associated with modulation of alternative splicing, as reported by earlier studies on the DENV-NS5 interactome revealing an association of NS5 with the U5 snRNP particle, CD2BP2 and DDX23, providing evidence of viral interference with alternative splicing events, eventually leading to changes in the abundance of specific mRNA isoforms of known antiviral factors [53].
Furthermore, among the novel cellular targets identified in the ZIKV NS4A interactome is ANKLE2, a protein previously shown to cause autosomal recessive microcephaly in humans [41]. Interestingly, in an ANKLE2-heterozygous mutant drosophila model, expression of NS4A reduced brain size, suggesting that NS4A might induce neurotoxic effects, at least under limiting amounts of functional ANKLE2.
An important aspect is that AP-LC-MS/MS often identifies protein complexes rather than binary protein–protein interactions. This is also illustrated in an interactome study of DENV-NS1 [55] in the context of a fully replicating virus. The authors employed three different cell lines (Raji, Hela and HAP1) to exploit similarities and specificities of bona fide interactors in diverse cellular backgrounds as a selection criterion to pinpoint targets relevant for in vivo infections. This study identified a strong enrichment of the CCT complex (cytosolic chaperonin-containing T complex), and subunits of the OST (oligosaccharyl transferase) complex such as DDOST, STT3A, STT3B and RPN2, in the NS1 interactome. Previous reports identified some of these proteins as host-dependency or druggable targets in genetic [14,56] and drug-based screens [57], and the interactome study now provides potential mechanistic insights into the underlying mechanisms of anti-viral activity. However, in light of cumulative evidence on ZIKV and WNV, it is tempting to speculate that this interaction might also require additional viral binding partners (e.g., NS4A and NS4B) or a productive virus infection, since ectopic expression of NS1 does not recapitulate this interaction [13,34,40,41,42]. In this respect, mass spectrometry has also been instrumental to identify interactions between viral proteins and that could explain some of their cooperative functions. NS1 of WNV, for instance, interacts with NS4B (WNV, [58]). Likewise, DENV NS1 associates with an unprocessed viral precursor NS4A-2k-NS4B (DENV, [59]) and NS4B of DENV has been reported to bind NS3 (DENV, [60]). Proteomic analysis, in addition to identifying functional protein complexes, fosters identification of yet unreported viral gene products (e.g., NS4A-2K-NS4B). Such information is helpful to extend our view on the function of viral proteins and the mode of action of therapeutic drugs.
3.1.4. Interactions of ZIKV Viral RNA with the Host
In addition to PPI, flaviviruses were also shown to exploit their own genomic RNA (gRNA) or subgenomic derivatives, such as the subgenomic flavivirus RNA (sfRNA), to “sponge” critical RNA-binding host proteins out of the cellular pool, or to specifically inhibit or redirect their homeostatic regulatory functions [61,62,63]. In this respect, recent studies have tried to chart systematically these interactions using different MS-based approaches. For instance, using a TUX-MS-based method (thiouracil cross-linking mass-spectrometry), Viktorovskaya and colleagues reported the identification of 79 proteins specifically associated with the DENV gRNA [64]. Among these, proteins earlier involved in PPI or associated with flavivirus replication were consistently identified (e.g., PAF1 and multiple hnRNPs), as well as novel host factors previously not associated with flavivirus infections (e.g., RBMX, RNA-binding motif protein X chromosome; NONO, non-POU domain-containing octamer-binding protein; and HMCES, embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein). Importantly, depletion of RBMX and NONO by RNAi significantly reduced DENV titers, confirming their functional relevance in DENV replication. Similar studies were also performed using an alternative cross-linking approach based on the use of 5′-end biotinylated antisense oligonucleotides specific to the DENV gRNA and very stringent cut-off criteria. Interestingly this approach identified only 12 host proteins specifically associated with the gRNA and further validated as bona fide interactors in RNA-IP-WB experiments. Among these, depletion of CSDE1 (cold-shock domain-containing protein E1) and hnRNPC (heterogeneous nuclear ribonucleoproteins C1/C2) reduced ~50% of infectious titers [65].
Only one study to date reported the systematic analysis of ZIKV and DENV gRNA-associated proteins, using the ChIRP-MS (comprehensive identification of RNA-binding proteins by mass spectrometry) method. In this method, target RNA and associated proteins are retrieved after formaldehyde-based cross-linking using hybridization with antisense biotinylated oligos and streptavidin-conjugated beads, and protein complexes eluted via incubation with biotin [66]. This approach identified 464 RNA-binding proteins (RBPs) associated with DENV or ZIKV gRNAs [67], confirming association with previously reported flavivirus RBPs (e.g., YBX1, MOV10 and ADAR) and host-dependency factors (e.g., STT3B and MAGT1). Importantly, intersection of these large datasets with previously published genome-wide CRISPR/Cas9-based screens, as well as four newly performed surveys using all four DENV serotypes, identified Vigilin and RRBP1 (ribosome-binding protein 1) as novel RBPs not previously linked to flavivirus infection. Interestingly, both of these proteins promote DENV infection and gRNA stability, positively modulating translation and replication of the gRNA.
3.2. Global Proteome Expression Affected by ZIKV Infection
Several studies systematically investigated the impact of flavivirus infections on protein abundance on a global scale [68,69]. Garcez and colleagues were the first to use label-free shotgun proteomics combined with transcriptomic analysis to study the impact of a Brazilian isolate of ZIKV on human neurospheres [70]. This approach revealed 199 downregulated proteins and 259 upregulated proteins, including proteins with functions in arrest of cell cycle progression (e.g., CDK2, ERBB2) and neural differentiation (e.g., HDAC7 and NeuroD1) or previously associated with viral replication (e.g., MAP4K4, TLR4 and DDX6). Importantly, this approach also revealed an activation of the DNA repair machinery, as inferred by an upregulation of proteins such as FANCD1, BRCA1 or MRE11A. Interestingly, these observations were later complemented by the identification of several hyperphosphorylation events across the DNA damage signaling pathway (see Section 3.1), as well as recent independent reports describing induction of double-strand DNA breaks (DSBs) and activation of DNA damage response in ZIKV-infected hNPC [71].
A similar unbiased study investigating the proteome of ZIKV-infected iPSC-derived human neuronal progenitors, confirmed specific downregulation of neuronal factors upon ZIKV infection, as well as activation of general antiviral response pathways, such as upregulation of type-I interferon stimulated genes (e.g., STAT1, MX1, OAS3, IFIT1) [34]. Interestingly, this approach was also used to investigate early changes occurring in ZIKV-infected or NS4B-transduced hNPC at the onset of neuronal differentiation, revealing specific downregulation of factors involved in neuronal differentiation as well as proteins associated with neurological diseases, such as DOK3 and SUMO2, thereby revealing potential targets deregulated during neurogenesis. Importantly, in line with previous studies pointing to a role of NS4A and NS4B in inhibition of neurogenesis and deregulation of the Akt-mTOR signaling pathway [51], this study also identified shared targets deregulated both by ZIKV infection and NS4B expression (e.g., MAP2, -6, DPYSL3, -5, CNTN2).
Recently, additional reports investigated further the impact of ZIKV infection on human mesenchymal stem cells using the MudPIT method (multidimensional protein identification technology). This identified a profound downregulation of PI3K/AKT and mTOR, as well as dysregulation of several components of the phosphatidylinositol pathway [72]. Alternative studies using shotgun proteomics and TMT (tandem mass tag) labeling confirmed and expanded the observed impairment of neurogenesis and synaptogenesis previously observed in 2D- and 3D-models of ZIKV-infected NPC, also to neurons and neural stem cells [73]. Using this method to systematically compare African and Asian ZIKV isolate representatives, revealed differential responses especially in the context of neurospheres. Among these, HSPB1 emerged as one of the most downregulated proteins in neurospheres, while DCX was profoundly upregulated in NPC, confirming the existence of significant viral isolate- and cellular background-dependent specificities on proteins that are relevant for neuron-specific functions.
A unifying theme crystallizing from proteomic and transcriptomic analysis of ZIKA virus infected neuronal cells is that ZIKV infection has a profound effect on expression of proteins involved in neurogenesis or on proteins that are markers of neuronal differentiation. At this stage it is not clear whether this effect is limited to ZIKA virus or whether other viruses elicit similar effects in a neuronal background. Along these lines would be important to test whether neuropathic flaviviruses share certain binding patterns that may be etiologically linked to their pathology. Similarly, it would be of great interest to investigate the differential proteomes of diverse flaviviruses from primary biopsies of infected patients.
3.3. Phosphorylation and Other PTMs Modulated by ZIKV
Initial studies by the Cao group focused on the global protein phosphorylation status in WNV- and JEV-infected cells [74,75]. In the case of WNV, 1657 proteins were found as significantly regulated at the phosphorylation level, and gene ontology enrichment analysis identified ErbB, NFκB and mTOR signaling pathways as the strongest deregulated pathways. Furthermore, several important kinases and substrates such as RNKP, RB1 and GSK3B were hyperphosphorylated, and depletion of these proteins by RNA interference significantly reduced the level of inflammatory cytokines such as IL-6, IL-1β and TNF-α in WNV-infected cells. Similarly, analysis of the cellular phosphoproteome upon JEV infection identified over 1200 differentially modulated phosphoproteins, including AKT, GSK-3β and PKC as well as ERK and p53. Interestingly, in case of JEV, a specific enrichment of JNK1 and CK2 substrates was identified among hyperphosphorylated and hypophosporylated proteins, respectively. Importantly, pharmacological modulation of JNK1 signaling reduced JEV-induced production of inflammatory cytokines in the brain of infected mice, suggesting an important role for stress-mediated responses in JEV-mediated neurotoxicity [74].
In the case of ZIKV, only one study so far reported a time-resolved phosphoproteomic analysis of ZIKV-infected cells [34]. Similarly, to JEV and WNV, 1216 proteins were modulated upon ZIKV infection, and analysis of enriched cellular functions affected by ZIKV infection identified cellular assembly and organization, cell-cycle regulation, as well as nervous system (NS) development and neurological diseases. In line with earlier reports, also in case of ZIKV the ATM, AKT/mTOR and ERK/MAPK signaling cascades were significantly regulated, as inferred by dephosphorylation of multiple AKT1 substrates (e.g., DNMT1, TBC1D4, LARP6), mTOR targets (e.g., ANKRD17, LARP1, BAG3, EEF2K, WNK1), the central kinase RPS6KB1 and its main effector protein RPS6. Similarly, several downstream substrates of the ERK1/2 map kinases (e.g., EIF4EBP1, BAZ1B, TWIST) were significantly dephosphorylated upon ZIKV infection. In this respect, these data complement on a different level of earlier observations reporting a downregulation of the AKT/mTOR pathway in ZIKV-infected hNPC and neurospheres [51].
Furthermore, a profound upregulation of the ATM (ataxia-telangiectasia mutated)/ATR (ATM- and Rad3-related) DNA-damage pathway, was observed at multiple levels (e.g., hyperphosphorylation of several substrates of ATR, DNAPK and downstream effector proteins) including a number of proteins involved in cell-cycle regulation and DNA damage checkpoint (TOP2B, CDK1). Altogether, these effects provide possible explanations for the observed ZIKV-induced cell-cycle arrest and apoptosis [76] and p53 activation [77] and are in line with multiple other reports describing persistent activation of DNA damage repair pathways in response to ZIKV-induced mitotic abnormalities [76]. Furthermore, these effects appear to be ZIKV-specific, at least when compared to DENV infection in hNPC [71], suggesting an important role in cell death of neuronal progenitors. Phosphoproteomic analysis also supports a prominent role of Zika virus on neurogenesis. p38 MAPK and downstream targets (HSPBI and ATF7), MARCKS (one of the main PKC substrates) and DPSYL2, all of which positively modulate neurite outgrowth and brain development [78,79,80] are differentially phosphorylated in cells infected with ZIKA virus. However, at this stage only a relatively limited number of phosphorylation events can be linked to a given function in a protein. In light of the abundance of differentially phosphorylated residues that have not been reported previously, such datasets constitute a largely unexplored treasure that will reveal its enormous potential only in light of mechanistic functional studies on individual phosphorylation sites.
Another important PTM is ubiquitination, which becomes more and more accessible to proteomics analysis and has recently been studied in the context of flavivirus infections [81]. For instance, Zhang and colleagues recently reported the use of an elegant hydrolysis-resistant ubiquitin variant (HA-UbL73*) in a functional proteomic screen. This work identified several host proteins differentially mono-ubiquitylated upon DENV infection, unraveling the ER-resident protein AUP1 as differentially ubiquitin-modified in DENV-infected cells. AUP1, a lipid droplet-localized type-III membrane protein existing predominantly in the mono-ubiquitylated form, appears to be the main trigger of DENV-induced lipophagy, and the newly identified NS4A–AUP1 interaction is sufficient to trigger its acyltransferase activity.
Collectively, the analysis of PTMs in flavivirus-infected cells starts to unravel unappreciated cellular activities. Development of techniques to study PTMs as well as development in bioinformatic tools allowing better interpretation of PTM activities, hold promise to dramatically accelerate our understanding of virus–host interactions and to appreciate additional important pathways engaged during virus infections.
4. Conclusions and Future Perspectives
Large-scale approaches allowing the accurate assessment of virus–host interactions have revolutionized the way we investigate and deconvolute the complex cellular changes triggered by virus infections. While central linear pathways were the main research focus in the past, it becomes more evident that innate immune signaling, and cellular responses in general, engage many interconnected pathways in parallel. The knowledge of these pathways is paramount to holistically understand how viral pathogens interact with their hosts. Certainly more work on viral–host interactions, protein turnover as well as post-translational modifications is required to further expand our understanding of the intracellular landscape of flavivirus-infected cells. This requires well-controlled comparative studies in relevant cell culture systems of multiple members of this highly diverse arbovirus family. Similarly, rigorous validation methods by alternative approaches (e.g., co-IP-WB or proximity ligation assays), robust and transparent cut-off criteria, and functional assays on selected candidates are increasingly needed to pinpoint physiologically relevant host factors. In this respect, further efforts are required to integrate and standardize data generated by mass spectrometry based surveys in easy-to-mine databases, ideally providing critical annotations on virus isolates and cell culture systems as well as integration with other -omics data [82]. Integrative efforts to merge such information gathered at different levels into organic maps of virus-induced perturbations will be imperative to fully exploit their enormous translational potential.
Table 1.
Gene names and corresponding full protein names listed in alphabetical order.
| Gene Name | Protein Name | Gene Name | Protein Name |
|---|---|---|---|
| AKT | RAC-alpha serine/threonine-protein kinase | ICMT | protein-S-isoprenylcysteine O-methyltransferase |
| ANKLE2 | Ankyrin repeat and LEM domain-containing protein 2 | LARP | La-related protein |
| ANKRD17 | ankyrin repeat domain-containing protein 17 | LEO1 | RNA polymerase-associated protein 1 |
| ATF7 | cyclic AMP-dependent transcription factor ATF-7 | LYAR | cell growth-regulating nucleolar protein |
| AUP1 | ancient ubiquitous protein 1 | MAP4K4 | mitogen-activated protein kinase kinase kinase kinase 4 |
| BAG3 | BAG family molecular chaperone regulator 3 | MARCKS | myristoylated alanine-rich C-kinase substrate |
| BAZ1B | tyrosine-protein kinase BAZ1B | MRE11A | double-strand break repair protein MRE11 |
| BRCA1 | breast cancer type 1 susceptibility protein | MT-CO2 | cytochrome c oxidaes subunit 2 |
| CD2BP2 | CD2 antigen cytoplasmic tail-binding protein 2 | mTOR | mechanistic target of rapamycin |
| CDC73 | cell division control protein 73 | NCL | nucleolin |
| CDK1 | cyclin-dependent kinase 1 | NDUFA4 | cytochrome c oxidase subunit NDUFA4 |
| CDK2 | cyclin-dependent kinase 2 | NeuroD1 | neurogenic differentiation factor 1 |
| CEND1 | cell-cycle exit and neuronal differentiation protein 1 | NGDN | neuroguidin |
| CLN6 | ceroid-lipofuscinosis neuronal protein 6 | NONO | non-POU domain-containing octamer-binding protein |
| COX15 | cytochrome c oxidase assembly protein COX15 homolog | PAF1 | RNA polymerase II-associated factor homolog |
| CPEB | cytoplasmic polyadenylation element binding protein | PEX19 | peroxisomal biogensis factor 19 |
| CSDE1 | cold-shock domain-containing protein E1 | PYM1 | partner of Y14 and mago |
| CTR9 | RNA polymerase-associated protein CTR9 homolog | RBM8A | RNA-binding protein 8A |
| DCP1-DCP2 | mRNA-decapping enzyme subunit 1-2rbm8A | RBMX | RNA-binding motif protein X chromosome |
| DCX | neuronal migration protein doublecortin | RPN2 | dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 2 |
| DDOST | dolichyl-diphosphooligosaccharide-protein glycosyltransferase 48 kDa subunit | RRBP1 | ribosome-binding protein 1 |
| DDX | ATP-dependent RNA helicase | SMN1 | survival motor neuron protein 1 |
| DNMT1 | DNA (cytosine-5)-methyltransferase 1 | STT3A/B | dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit A/B |
| DOK3 | docking protein 3 | TBC1D4 | TBC1 domain family member 4 |
| DPYSL2 | dihydropyrimidinase-related protein 2 | TLR4 | toll-like receptor 4 |
| EEF2K | eukaryotic elongation factor 2 kinase | TMEM41b | transmembrane protein 41beta |
| ERBB2 | Receptor tyrosine-protein kinase erbB-2 | TOP2B | DNA topoisomerase 2-beta |
| FANCD1 | breast cancer type 2 susceptibility protein | TWIST | twist-related protein 1 |
| HDAC7 | histone deacetylase 7 | UPF1 | regulator of nonsense transcripts 1 |
| HMCES | embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein | VDAC1, 2, 3 | voltage-dependent anion selective channel protein 1, 2, 3 |
| hnRNPC | heterogeneous nuclear ribonucleoproteins C1/C2 | WNK1 | Serine/threonine-protein kinase WNK1 |
| HSPB1 | heat shock protein beta-1 | ZC3HAV1 | zinc finger CCCH-type antiviral protein 1 |
Funding
This research was supported by the European Research Council (ERC consolidator grant number ERC-CoG ProDAP, 817798), the Volkswagen Stiftung and the German Research Foundation (grant numbers PI 1084/2, PI 1084/3, PI 1084/4, PI 1084/5, TRR179 TP11, TRR237 A07).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Lazear H.M., Diamond M.S. Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J. Virol. 2016;90:4864–4875. doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cauchemez S., Besnard M., Bompard P., Dub T., Guillemette-Artur P., Eyrolle-Guignot D., Salje H., Van Kerkhove M.D., Abadie V., Garel C., et al. Association between Zika virus and microcephaly in French Polynesia, 2013–2015: A retrospective study. Lancet. 2016;387:2125–2132. doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ventura C.V., Maia M., Bravo-Filho V., Gois A.L., Belfort R., Jr. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet. 2016;387:228. doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- 4.Ma W., Li S., Ma S., Jia L., Zhang F., Zhang Y., Zhang J., Wong G., Zhang S., Lu X., et al. Zika Virus Causes Testis Damage and Leads to Male Infertility in Mice. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Kodati S., Palmore T.N., Spellman F.A., Cunningham D., Weistrop B., Sen H.N. Bilateral posterior uveitis associated with Zika virus infection. Lancet. 2017;389:125–126. doi: 10.1016/S0140-6736(16)32518-1. [DOI] [PubMed] [Google Scholar]
- 6.Miner J.J., Cao B., Govero J., Smith A.M., Fernandez E., Cabrera O.H., Garber C., Noll M., Klein R.S., Noguchi K.K., et al. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell. 2016;165:1081–1091. doi: 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mlakar J., Korva M., Tul N., Popovic M., Poljsak-Prijatelj M., Mraz J., Kolenc M., Resman Rus K., Vesnaver Vipotnik T., Fabjan Vodusek V., et al. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 8.Pierson T.C., Diamond M.S. The emergence of Zika virus and its new clinical syndromes. Nature. 2018;560:573–581. doi: 10.1038/s41586-018-0446-y. [DOI] [PubMed] [Google Scholar]
- 9.Pichlmair A., Kandasamy K., Alvisi G., Mulhern O., Sacco R., Habjan M., Binder M., Stefanovic A., Eberle C.A., Goncalves A., et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 10.Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F., et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 11.Batra J., Hultquist J.F., Liu D., Shtanko O., Von Dollen J., Satkamp L., Jang G.M., Luthra P., Schwarz T.M., Small G.I., et al. Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication. Cell. 2018;175:1917–1930.e13. doi: 10.1016/j.cell.2018.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis Z.H., Verschueren E., Jang G.M., Kleffman K., Johnson J.R., Park J., Von Dollen J., Maher M.C., Johnson T., Newton W., et al. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol. Cell. 2015;57:349–360. doi: 10.1016/j.molcel.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dechtawewat T., Paemanee A., Roytrakul S., Songprakhon P., Limjindaporn T., Yenchitsomanus P.T., Saitornuang S., Puttikhunt C., Kasinrerk W., Malasit P., et al. Mass spectrometric analysis of host cell proteins interacting with dengue virus nonstructural protein 1 in dengue virus-infected HepG2 cells. Biochim. Biophys. Acta. 2016;1864:1270–1280. doi: 10.1016/j.bbapap.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Marceau C.D., Puschnik A.S., Majzoub K., Ooi Y.S., Brewer S.M., Fuchs G., Swaminathan K., Mata M.A., Elias J.E., Sarnow P., et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savidis G., McDougall W.M., Meraner P., Perreira J.M., Portmann J.M., Trincucci G., John S.P., Aker A.M., Renzette N., Robbins D.R., et al. Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep. 2016;16:232–246. doi: 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Sessions O.M., Barrows N.J., Souza-Neto J.A., Robinson T.J., Hershey C.L., Rodgers M.A., Ramirez J.L., Dimopoulos G., Yang P.L., Pearson J.L., et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dukhovny A., Lamkiewicz K., Chen Q., Fricke M., Jabrane-Ferrat N., Marz M., Jung J.U., Sklan E.H. A CRISPR Activation Screen Identifies Genes That Protect against Zika Virus Infection. J. Virol. 2019;93:e00211-19. doi: 10.1128/JVI.00211-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Muffat J., Omer Javed A., Keys H.R., Lungjangwa T., Bosch I., Khan M., Virgilio M.C., Gehrke L., Sabatini D.M., et al. Genome-wide CRISPR screen for Zika virus resistance in human neural cells. Proc. Natl. Acad. Sci. USA. 2019;116:9527–9532. doi: 10.1073/pnas.1900867116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma H., Dang Y., Wu Y., Jia G., Anaya E., Zhang J., Abraham S., Choi J.G., Shi G., Qi L., et al. A CRISPR-Based Screen Identifies Genes Essential for West-Nile-Virus-Induced Cell Death. Cell Rep. 2015;12:673–683. doi: 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson R.B., Ohlson M.B., Eitson J.L., Kumar A., McDougal M.B., Boys I.N., Mar K.B., De La Cruz-Rivera P.C., Douglas C., Konopka G., et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat. Microbiol. 2018;3:1214–1223. doi: 10.1038/s41564-018-0244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R., Miner J.J., Gorman M.J., Rausch K., Ramage H., White J.P., Zuiani A., Zhang P., Fernandez E., Zhang Q., et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature. 2016;535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metsky H.C., Matranga C.B., Wohl S., Schaffner S.F., Freije C.A., Winnicki S.M., West K., Qu J., Baniecki M.L., Gladden-Young A., et al. Zika virus evolution and spread in the Americas. Nature. 2017;546:411–415. doi: 10.1038/nature22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L., Bore J.A., Koundouno R., Dudas G., Mikhail A., et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong Y.G., Shi W.F., Liu D., Qian J., Liang L., Bo X.C., Liu J., Ren H.G., Fan H., Ni M., et al. China Mobile Laboratory Testing Team in Sierra, L. Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature. 2015;524:93–96. doi: 10.1038/nature14490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edfors F., Danielsson F., Hallstrom B.M., Kall L., Lundberg E., Ponten F., Forsstrom B., Uhlen M. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. 2016;12:883. doi: 10.15252/msb.20167144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aebersold R., Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 27.Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 28.Itzhak D.N., Tyanova S., Cox J., Borner G.H. Global, quantitative and dynamic mapping of protein subcellular localization. Elife. 2016;5:e16950. doi: 10.7554/eLife.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jean Beltran P.M., Mathias R.A., Cristea I.M. A Portrait of the Human Organelle Proteome in Space and Time during Cytomegalovirus Infection. Cell Syst. 2016;3:361–373.e6. doi: 10.1016/j.cels.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinitcyn P., Rudolph J.D., Cox J. Computational Methods for Understanding Mass Spectrometry–Based Shotgun Proteomics Data. Annu. Rev. Biomed. Data Sci. 2018;1:207–234. doi: 10.1146/annurev-biodatasci-080917-013516. [DOI] [Google Scholar]
- 31.Taguwa S., Maringer K., Li X., Bernal-Rubio D., Rauch J.N., Gestwicki J.E., Andino R., Fernandez-Sesma A., Frydman J. Defining Hsp70 Subnetworks in Dengue Virus Replication Reveals Key Vulnerability in Flavivirus Infection. Cell. 2015;163:1108–1123. doi: 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M., et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 33.Rebsamen M., Kandasamy R.K., Superti-Furga G. Protein interaction networks in innate immunity. Trends Immunol. 2013;34:610–619. doi: 10.1016/j.it.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Scaturro P., Stukalov A., Haas D.A., Cortese M., Draganova K., Plaszczyca A., Bartenschlager R., Gotz M., Pichlmair A. An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature. 2018;561:253–257. doi: 10.1038/s41586-018-0484-5. [DOI] [PubMed] [Google Scholar]
- 35.Balinsky C.A., Schmeisser H., Ganesan S., Singh K., Pierson T.C., Zoon K.C. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol. 2013;87:13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colpitts T.M., Barthel S., Wang P., Fikrig E. Dengue virus capsid protein binds core histones and inhibits nucleosome formation in human liver cells. PLoS ONE. 2011;6:e24365. doi: 10.1371/journal.pone.0024365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You J., Hou S., Malik-Soni N., Xu Z., Kumar A., Rachubinski R.A., Frappier L., Hobman T.C. Flavivirus Infection Impairs Peroxisome Biogenesis and Early Antiviral Signaling. J. Virol. 2015;89:12349–12361. doi: 10.1128/JVI.01365-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Wang B., Yang A., Lu R., Wang W., Zhou Y., Shi G., Kwon S.W., Zhao Y., Jin Y. Ly-1 antibody reactive clone is an important nucleolar protein for control of self-renewal and differentiation in embryonic stem cells. Stem Cells. 2009;27:1244–1254. doi: 10.1002/stem.55. [DOI] [PubMed] [Google Scholar]
- 39.Yang C., Liu X., Gao Q., Cheng T., Xiao R., Ming F., Zhang S., Jin M., Chen H., Ma W., et al. The Nucleolar Protein LYAR Facilitates Ribonucleoprotein Assembly of Influenza a Virus. J. Virol. 2018;92:e01042-18. doi: 10.1128/JVI.01042-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coyaud E., Ranadheera C., Cheng D., Goncalves J., Dyakov B.J.A., Laurent E.M.N., St-Germain J., Pelletier L., Gingras A.C., Brumell J.H., et al. Global Interactomics Uncovers Extensive Organellar Targeting by Zika Virus. Mol. Cell Proteom. 2018;17:2242–2255. doi: 10.1074/mcp.TIR118.000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah P.S., Link N., Jang G.M., Sharp P.P., Zhu T., Swaney D.L., Johnson J.R., Von Dollen J., Ramage H.R., Satkamp L., et al. Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell. 2018;175:1931–1945.e18. doi: 10.1016/j.cell.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M., Johnson J.R., Truong B., Kim G., Weinbren N., Dittmar M., Shah P.S., Von Dollen J., Newton B.W., Jang G.M., et al. Identification of antiviral roles for the exon-junction complex and nonsense-mediated decay in flaviviral infection. Nat. Microbiol. 2019;4:985–995. doi: 10.1038/s41564-019-0375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takata M.A., Goncalves-Carneiro D., Zang T.M., Soll S.J., York A., Blanco-Melo D., Bieniasz P.D. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature. 2017;550:124–127. doi: 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M.M., Lau Z., Cheung P., Aguilar E.G., Schneider W.M., Bozzacco L., Molina H., Buehler E., Takaoka A., Rice C.M., et al. TRIM25 Enhances the Antiviral Action of Zinc-Finger Antiviral Protein (ZAP) PLoS Pathog. 2017;13:e1006145. doi: 10.1371/journal.ppat.1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu H.P., Chiu H., Yang C.F., Lee Y.L., Chiu F.L., Kuo H.C., Lin R.J., Lin Y.L. Inhibition of Japanese encephalitis virus infection by the host zinc-finger antiviral protein. PLoS Pathog. 2018;14:e1007166. doi: 10.1371/journal.ppat.1007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fontaine K.A., Leon K.E., Khalid M.M., Tomar S., Jimenez-Morales D., Dunlap M., Kaye J.A., Shah P.S., Finkbeiner S., Krogan N.J., et al. The Cellular NMD Pathway Restricts Zika Virus Infection and Is Targeted by the Viral Capsid Protein. MBio. 2018;9:e02126-18. doi: 10.1128/mBio.02126-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatel-Chaix L., Cortese M., Romero-Brey I., Bender S., Neufeldt C.J., Fischl W., Scaturro P., Schieber N., Schwab Y., Fischer B., et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host Microbe. 2016;20:342–356. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metz P., Chiramel A., Chatel-Chaix L., Alvisi G., Bankhead P., Mora-Rodriguez R., Long G., Hamacher-Brady A., Brady N.R., Bartenschlager R. Dengue Virus Inhibition of Autophagic Flux and Dependency of Viral Replication on Proteasomal Degradation of the Autophagy Receptor p62. J. Virol. 2015;89:8026–8041. doi: 10.1128/JVI.00787-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heaton N.S., Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y., Cherry S. Zika virus infection activates sting-dependent antiviral autophagy in the Drosophila brain. Autophagy. 2019;15:174–175. doi: 10.1080/15548627.2018.1528813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang Q., Luo Z., Zeng J., Chen W., Foo S.S., Lee S.A., Ge J., Wang S., Goldman S.A., Zlokovic B.V., et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell. 2016;19:663–671. doi: 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heaton N.S., Moshkina N., Fenouil R., Gardner T.J., Aguirre S., Shah P.S., Zhao N., Manganaro L., Hultquist J.F., Noel J., et al. Targeting Viral Proteostasis Limits Influenza Virus, HIV, and Dengue Virus Infection. Immunity. 2016;44:46–58. doi: 10.1016/j.immuni.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Maio F.A., Risso G., Iglesias N.G., Shah P., Pozzi B., Gebhard L.G., Mammi P., Mancini E., Yanovsky M.J., Andino R., et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog. 2016;12:e1005841. doi: 10.1371/journal.ppat.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovanich D., Saisawang C., Sittipaisankul P., Ramphan S., Kalpongnukul N., Somparn P., Pisitkun T., Smith D.R. Analysis of the Zika and Japanese Encephalitis Virus NS5 Interactomes. J. Proteome. Res. 2019;18:3203–3218. doi: 10.1021/acs.jproteome.9b00318. [DOI] [PubMed] [Google Scholar]
- 55.Hafirassou M.L., Meertens L., Umana-Diaz C., Labeau A., Dejarnac O., Bonnet-Madin L., Kummerer B.M., Delaugerre C., Roingeard P., Vidalain P.O., et al. A Global Interactome Map of the Dengue Virus NS1 Identifies Virus Restriction and Dependency Host Factors. Cell Rep. 2017;21:3900–3913. doi: 10.1016/j.celrep.2017.11.094. [DOI] [PubMed] [Google Scholar]
- 56.Lin D.L., Inoue T., Chen Y.J., Chang A., Tsai B., Tai A.W. The ER Membrane Protein Complex Promotes Biogenesis of Dengue and Zika Virus Non-structural Multi-pass Transmembrane Proteins to Support Infection. Cell Rep. 2019;27:1666–1674.e4. doi: 10.1016/j.celrep.2019.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrows N.J., Campos R.K., Powell S.T., Prasanth K.R., Schott-Lerner G., Soto-Acosta R., Galarza-Munoz G., McGrath E.L., Urrabaz-Garza R., Gao J., et al. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Youn S., Li T., McCune B.T., Edeling M.A., Fremont D.H., Cristea I.M., Diamond M.S. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J. Virol. 2012;86:7360–7371. doi: 10.1128/JVI.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plaszczyca A., Scaturro P., Neufeldt C.J., Cortese M., Cerikan B., Ferla S., Brancale A., Pichlmair A., Bartenschlager R. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Pathog. 2019;15:e1007736. doi: 10.1371/journal.ppat.1007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chatel-Chaix L., Fischl W., Scaturro P., Cortese M., Kallis S., Bartenschlager M., Fischer B., Bartenschlager R. A Combined Genetic-Proteomic Approach Identifies Residues within Dengue Virus NS4B Critical for Interaction with NS3 and Viral Replication. J. Virol. 2015;89:7170–7186. doi: 10.1128/JVI.00867-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roby J.A., Pijlman G.P., Wilusz J., Khromykh A.A. Noncoding subgenomic flavivirus RNA: Multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses. 2014;6:404–427. doi: 10.3390/v6020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chavali P.L., Stojic L., Meredith L.W., Joseph N., Nahorski M.S., Sanford T.J., Sweeney T.R., Krishna B.A., Hosmillo M., Firth A.E., et al. Neurodevelopmental protein Musashi-1 interacts with the Zika genome and promotes viral replication. Science. 2017;357:83–88. doi: 10.1126/science.aam9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charley P.A., Wilusz J. Sponging of cellular proteins by viral RNAs. Curr. Opin. Virol. 2014;9:14–18. doi: 10.1016/j.coviro.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Viktorovskaya O.V., Greco T.M., Cristea I.M., Thompson S.R. Identification of RNA Binding Proteins Associated with Dengue Virus RNA in Infected Cells Reveals Temporally Distinct Host Factor Requirements. PLoS Negl. Trop. Dis. 2016;10:e0004921. doi: 10.1371/journal.pntd.0004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phillips S.L., Soderblom E.J., Bradrick S.S., Garcia-Blanco M.A. Identification of Proteins Bound to Dengue Viral RNA in Vivo Reveals New Host Proteins Important for Virus Replication. MBio. 2016;7:e01865-e15. doi: 10.1128/mBio.01865-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ooi Y.S., Majzoub K., Flynn R.A., Mata M.A., Diep J., Li J.K., van Buuren N., Rumachik N., Johnson A.G., Puschnik A.S., et al. An RNA-centric dissection of host complexes controlling flavivirus infection. Nat. Microbiol. 2019 doi: 10.1038/s41564-019-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiu H.C., Hannemann H., Heesom K.J., Matthews D.A., Davidson A.D. High-throughput quantitative proteomic analysis of dengue virus type 2 infected A549 cells. PLoS ONE. 2014;9:e93305. doi: 10.1371/journal.pone.0093305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pastorino B., Boucomont-Chapeaublanc E., Peyrefitte C.N., Belghazi M., Fusai T., Rogier C., Tolou H.J., Almeras L. Identification of cellular proteome modifications in response to West Nile virus infection. Mol. Cell Proteom. 2009;8:1623–1637. doi: 10.1074/mcp.M800565-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcez P.P., Nascimento J.M., de Vasconcelos J.M., Madeiro da Costa R., Delvecchio R., Trindade P., Loiola E.C., Higa L.M., Cassoli J.S., Vitoria G., et al. Zika virus disrupts molecular fingerprinting of human neurospheres. Sci. Rep. 2017;7:40780. doi: 10.1038/srep40780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hammack C., Ogden S.C., Madden J.C., Jr., Medina A., Xu C., Phillips E., Son Y., Cone A., Giovinazzi S., Didier R.A., et al. Zika virus infection induces DNA damage response in human neural progenitors that enhances viral replication. J. Virol. 2019 doi: 10.1128/JVI.00638-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beys-da-Silva W.O., Rosa R.L., Santi L., Berger M., Park S.K., Campos A.R., Terraciano P., Varela A.P.M., Teixeira T.F., Roehe P.M., et al. Zika Virus Infection of Human Mesenchymal Stem Cells Promotes Differential Expression of Proteins Linked to Several Neurological Diseases. Mol. Neurobiol. 2019;56:4708–4717. doi: 10.1007/s12035-018-1417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosa-Fernandes L., Cugola F.R., Russo F.B., Kawahara R., de Melo Freire C.C., Leite P.E.C., Bassi Stern A.C., Angeli C.B., de Oliveira D.B.L., Melo S.R., et al. Zika Virus Impairs Neurogenesis and Synaptogenesis Pathways in Human Neural Stem Cells and Neurons. Front. Cell Neurosci. 2019;13:64. doi: 10.3389/fncel.2019.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye J., Zhang H., He W., Zhu B., Zhou D., Chen Z., Ashraf U., Wei Y., Liu Z., Fu Z.F., et al. Quantitative phosphoproteomic analysis identifies the critical role of JNK1 in neuroinflammation induced by Japanese encephalitis virus. Sci. Signal. 2016;9:ra98. doi: 10.1126/scisignal.aaf5132. [DOI] [PubMed] [Google Scholar]
- 75.Zhang H., Sun J., Ye J., Ashraf U., Chen Z., Zhu B., He W., Xu Q., Wei Y., Chen H., et al. Quantitative Label-Free Phosphoproteomics Reveals Differentially Regulated Protein Phosphorylation Involved in West Nile Virus-Induced Host Inflammatory Response. J. Proteome Res. 2015;14:5157–5168. doi: 10.1021/acs.jproteome.5b00424. [DOI] [PubMed] [Google Scholar]
- 76.Souza B.S., Sampaio G.L., Pereira C.S., Campos G.S., Sardi S.I., Freitas L.A., Figueira C.P., Paredes B.D., Nonaka C.K., Azevedo C.M., et al. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci. Rep. 2016;6:39775. doi: 10.1038/srep39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghouzzi V.E., Bianchi F.T., Molineris I., Mounce B.C., Berto G.E., Rak M., Lebon S., Aubry L., Tocco C., Gai M., et al. ZIKA virus elicits P53 activation and genotoxic stress in human neural progenitors similar to mutations involved in severe forms of genetic microcephaly. Cell Death Dis. 2016;7:e2440. doi: 10.1038/cddis.2016.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morooka T., Nishida E. Requirement of p38 mitogen-activated protein kinase for neuronal differentiation in PC12 cells. J. Biol. Chem. 1998;273:24285–24288. doi: 10.1074/jbc.273.38.24285. [DOI] [PubMed] [Google Scholar]
- 79.Xu X.H., Deng C.Y., Liu Y., He M., Peng J., Wang T., Yuan L., Zheng Z.S., Blackshear P.J., Luo Z.G. MARCKS regulates membrane targeting of Rab10 vesicles to promote axon development. Cell Res. 2014;24:576–594. doi: 10.1038/cr.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morimura R., Nozawa K., Tanaka H., Ohshima T. Phosphorylation of Dpsyl2 (CRMP2) and Dpsyl3 (CRMP4) is required for positioning of caudal primary motor neurons in the zebrafish spinal cord. Dev. Neurobiol. 2013;73:911–920. doi: 10.1002/dneu.22117. [DOI] [PubMed] [Google Scholar]
- 81.Zhang J., Lan Y., Li M.Y., Lamers M.M., Fusade-Boyer M., Klemm E., Thiele C., Ashour J., Sanyal S. Flaviviruses Exploit the Lipid Droplet Protein AUP1 to Trigger Lipophagy and Drive Virus Production. Cell Host Microbe. 2018;23:819–831.e5. doi: 10.1016/j.chom.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 82.Zhang B., Kuster B. Proteomics Is Not an Island: Multi-omics Integration Is the Key to Understanding Biological Systems. Mol. Cell Proteom. 2019;18(Suppl. 1):S1–S4. doi: 10.1074/mcp.E119.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]