Abstract
Leaves constitute the main photosynthetic plant organ and even though their importance is not debated, the origin and development of leaves still is. The leaf developmental network has been elucidated for angiosperms, from genes controlling leaf initiation, to leaf polarity and shape. There are four KANADI (KAN) paralogs in Arabidopsis thaliana needed for organ polarity with KAN1 and KAN2 specifying abaxial leaf identity. Yet, studies of this gene lineage outside angiosperms are required to better understand the evolutionary patterns of leaf development and the role of KAN homologs. We studied the evolution of KAN genes across vascular plants and their expression by in situ hybridization in the fern, Equisetum hyemale and the lycophyte Selaginella moellendorffii. Our results show that the expression of KAN genes in leaves is similar between ferns and angiosperms. However, the expression patterns observed in the lycophyte S. moellendorffii are significantly different compared to all other vascular plants, suggesting that the KAN function in leaf polarity is likely only conserved across ferns, gymnosperms, and angiosperms. This study indicates that mechanisms for leaf development are different in lycophytes compared to other vascular plants.
Keywords: Equisetum, ferns, in situ hybridization, lycophytes, KANADI, megaphyll, microphyll, plant evo-devo, Selaginella, telome theory
1. Introduction
Leaves are the most easily recognizable plant organ, yet surprisingly they may have evolved independently more than once [1]. Leaves are thought to have evolved between 2 and 11 times in vascular plants (lycophytes, ferns, and seed plants). Leaves in lycophytes are commonly termed microphylls while leaves in ferns and seed plants are termed megaphylls [1,2,3,4]. The fossil record indicates that leaves evolved independently in lycophytes and euphyllophytes (ferns and seed plants), and molecular genetics generally supports this idea [2,3,5,6,7,8]. The discrepancy in the number of times leaves have evolved is due to varying results mainly within ferns due to an incomplete fossil record and the paucity of developmental genetic studies in this group [1]. The most widely accepted theory for leaf evolution is the telome theory where leaves are proposed to have evolved through independent processes of branching, overtopping, planation, and webbing [9]. The evolution of webbing would result in the development of a laminar structure.
Leaves are usually thought of as the flat lateral organs in the plant, because one of the main characters that defines most leaves is their bilateral symmetry. As the leaf primordium develops, it generally forms two anatomically distinct surfaces, the adaxial and abaxial sides. Morphological diversity of leaves across land plants can be associated, in many instances, to changes in the adaxial/abaxial identity (Figure 1) [10]. Experimental evidence indicates that the specification and maintenance of bilateral symmetry during leaf development is necessary for lamina outgrowth [10,11,12,13].
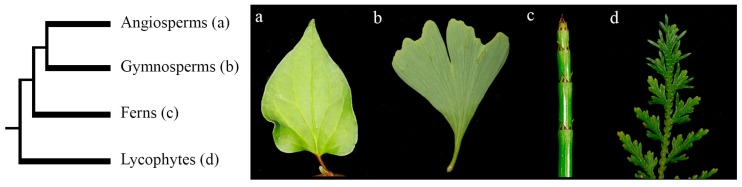
Figure 1.
Schematic representation of the evolution of vascular plants and examples of their leaves. (a) Angiosperm Houttuynia cordata single simple leaf; (b) gymnosperm Ginkgo biloba single simple leaf; (c) fern Equisetum hyemale shoot with whorls of leaves; (d) lycophyte Selaginella moellendorffii shoot with leaves.
The genetic network specifying the proper development of axes during leaf development and consequently lamina outgrowth, particularly in Arabidopsis thaliana, has been well studied and involves multiple regulatory mechanisms, from transcription factors to small RNAs and hormones [10,11,12,13]. Briefly, ASYMMETRIC LEAVES1 and 2 (AS1/AS2) [14,15,16,17], along with small RNAs, regulate Class III HD-Zips, which specify adaxial identity in A. thaliana [13,15,16,17]. While abaxial identity of the A. thaliana leaf, is specified by ETTIN/AUXIN RESPONSE FACTOR 3(ETT/ARF3), ARF4, and KANADIs (KANs) [18,19,20,21,22,23,24,25,26,27,28]. Of particular importance is the mutual antagonism of the specification of adaxial and abaxial leaf identities [29]. That is, in mutants lacking adaxial identity, the organ has abaxial identity only, and vice versa [12,13,18,19,20]. In addition, if adaxial or abaxial identity is lost then the resulting organ is more radial rather than a flat laminar structure [11,12,13].
Comparative studies to understand the specification of ab/adaxial identity have mainly focused on angiosperms [reviewed in 10], with the exception of expression studies of AS1 homologs in the lycophyte Selaginella kraussiana and the fern Osmunda regalis [6], and Class III HD-Zip expression studies in lycophytes and ferns [8,30,31]. Similar to results found in seed plants;,Class III HD-Zips have been shown to be expressed in the adaxial side of leaf primordia throughout ferns from Equisetum to leptosporangiate ferns [8]. However, expression studies of Class III HD-Zips in lycophytes, show that these genes are specifically expressed in leaf primordia, but the expression is not polar and is not maintained in older leaves [8,30,31]. These results suggest that the function of Class III HD-Zips genes in leaf adaxial identity is only conserved between ferns and seed plants but not lycophytes.
To better understand the evolution and development of bilateral symmetry in vascular plant leaves, we investigated KANADI homologs across lycophytes and ferns. KAN genes belong to the GARP family of transcription factors, characterized by the plant-specific GARP DNA binding domain. This domain is part of the helix-loop-helix distantly related to the MYB domain [27]. The GARP domain is crucial for regulating the transcription of downstream genes [19]. There are four KAN paralogs in Arabidopsis thaliana (KAN1–4), even though KAN1 and KAN2 have been described by their function in leaf polarity, all four KAN paralogs are involved in proper determination of organ polarity, including the integuments of the ovules [19,32,33,34]. Phylogenetic analyses for the KANADI gene family have been focused on angiosperm model species, including Arabidopsis thaliana, Zea mays, and Oryza sativa homologs [27]. Functional studies have also been performed only in these three model species, and indicate conserved functions in the proper determination of the abaxial leaf identity [27,28].
Available data regarding the evolution and expression of KAN genes are not sufficient to assess how these genes have functionally changed over time and their implications in leaf morphological evolution. Here we provide the first phylogenetic tree with sampling across vascular plants, with a focus on lycophytes and ferns. Furthermore, we present the expression of KANADI homologs in the developing shoots of the lycophyte Selaginella moellendorffii and the fern Equisetum hyemale, that allow us to hypothesize about the functional evolution of the KAN genes and the leaf developmental network in vascular plants.
2. Materials and Methods
2.1. KANADI Phylogenetic Analyses across Vascular Plants
We used the Basic Local Alignment Search Tool (BLAST) with the four KANADI paralogs, in nucleotides, from Arabidopsis thaliana (At5g16560, At1g32240, At4g17695, and At5g42630) as queries. We focused mainly on fern sequences publicly available in the OneKP transcriptome database (https://sites.google.com/a/ualberta.ca/onekp/) [35,36,37] with 28 sequences included; 25 sequences from gymnosperms and angiosperms available in the genome database Phytozome (https://phytozome.jgi. doe.gov/pz/portal.html) [38] and the NCBI (https://www.ncbi.nlm.nih.gov/; Table S1). Additionally, we included Ceratopteris richardii transcriptome sequences from the Ceratopteris Genome Project. Sequences were aligned with the online version of MAFFT [39] (https://mat.cbrc.jp/alignment/server/) with a gap open penalty of 3.0, an offset value of 0.5, and all the other parameters by default. Alignment was manually refined with AliView [40]. Phylogenetic relationships were inferred by Maximum Likelihood (ML) analysis using the complete nucleotide alignment, through the CIPRES Science Gateway [41] with RaxML-HPC BlackBlox [42]. The final topology was observed and edited using FigTree v.1.4.3 [43]. This analysis included, as ingroup, 53 sequences from across vascular plants, and three KANADI sequences from Physcomitrella patens were used as outgroups (Table S1).
2.2. In Situ Hybridization Expression Analyses for KANADI Homologs in Ferns and Lycophytes
Equisetum hyemale and Selaginella moellendorffii plants were grown under controlled conditions in the NYBG Nolen greenhouses. Young shoots from Equisetum hyemale, and sterile and fertile shoots from Selaginella moellendorffii were collected and immediately fixed in formaldehyde-acetic acid-ethanol and water (FAA; 3.7% formaldehyde: 5% glacial acetic acid: 50% ethanol). The material was dehydrated through an alcohol-Histo-Clear II (National Diagnostics, Atlanta, GA, United States) series and embedded in Paraplast X-tra (Fisher Healthcare, Houston, TX, United States). The samples were sectioned at 8 μm with a MICROM HM355 (Fisher Scientific, Pittsburgh, PA, United States) rotary microtome. DNA templates for RNA probe synthesis were obtained by amplification of 290–350 bp fragments. To ensure specificity, the probes were designed flanking the GARP domain (Table S2) S. moellendorffii primers were designed towards the 3’ end of the sequence (Figure S1) while E. hyemale primers were designed towards the 5’ UTR end of the sequence. Fragments were cleaned using QIAquick PCR purification Kit (Qiagen, Valencia, CA, United States). Digoxigenin labeled RNA probes were prepared using T7 RNA polymerase (Roche, Switzerland), RNAse inhibitor RNasin (New England Biolabs, Ipswich, MA, United States), and RNA labeling-mix (Roche, Switzerland) according to the manufacturer’s protocol. RNA in situ hybridization was performed according to Ambrose et al. (2000) [44].
3. Results
3.1. Evolution of KANADI in Lycophytes and Ferns
Phylogenetic analyses include representatives from all major plant lineages across vascular plants containing lycophytes, ferns, gymnosperms, and angiosperms, for an in-group of 53 sequences and three Physcomitrella homologs used as outgroups (Table S1). Angiosperm sequences are for the most part complete coding sequences as they were downloaded from genome databases, whereas some gymnosperm and fern sequences are missing 30–60 amino acids (AA) from the start codon, and others lack the stop codon. We were able to identify the GARP domain in all the sequences included in the analysis.
Our maximum likelihood (ML) analyses recovered two clades of fern KAN homologs, one sister to Arabidopsis ATS comprised only by Equisetum sequences, EdiKAN4, 6 and EhyKAN3 and the other, sister to all lycophyte sequences (Figure 2). The clade containing most fern sequences has two subclades, likely as the result of a major duplication event (Figure 2). Both subclades include sequences from several ferns such as Blechnum spicant, Ceratopteris richardii, Ceratopteris thalictroides, Cryptogramma acrostichoides, and Vittaria lineata (Figure 2). Equisetum sequences are recovered in only one of the subclades, thus it is unclear if the inferred duplication occurred before or after the evolution of Equisetaceae (Figure 2). Sister to these two fern clades are the three S. moellendorfii homologs included in this study, possibly as the result of a limited sampling among lycophytes (Figure 2).
Figure 2.
A maximum likelihood hypothesis for the evolution of KANADI genes across vascular plants. The key to the colors in the topology is shown in the upper left corner of the figure. The yellow star indicates a possible duplication event within ferns. Bootstrap (BS) values higher than 50 are shown on the corresponding branch.
3.2. Expression of KANADI in the Lycophyte Selaginella moellendorffii
To better understand the role of KANADI homologs in lycophytes, we examined the expression of these genes in the heterosporous lycophyte Selaginella moellendorffii in vegetative and reproductive tissues by in situ hybridization (Figure 3).
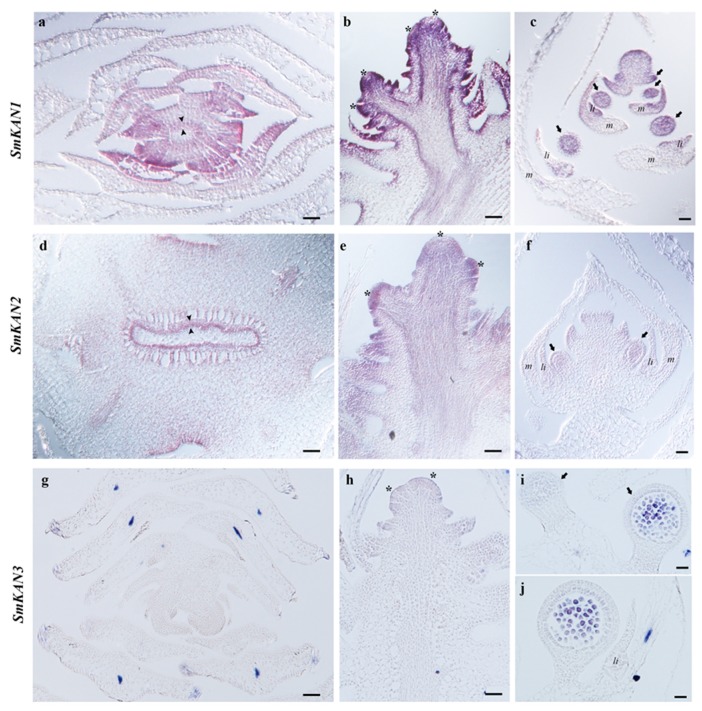
Figure 3.
SmKAN expression in Selaginella moellendorffii by in situ hybridization. (a–c) SmKAN1 expression patterns. (a) SmKAN1 expression is detected in the vasculature in a transverse section of a shoot. (b) SmKAN1 expression is detected in the emerging microphyll primordia but not the meristem of a longitudinal section through a shoot. (c) SmKAN1 expression is detected in the emerging sporangia primordia and is maintained throughout their development in a longitudinal section through a strobilus. (d–f) SmKAN2 expression patterns. (d) SmKAN2 expression is detected in the vasculature in a transverse section of shoot. (e) SmKAN2 expression is detected in the emerging microphyll primordia but not the meristem of a longitudinal section through a shoot. (f) SmKAN2 expression is detected in the emerging sporangia primordia and is maintained throughout their development in a longitudinal section through a strobilus. (g–j) SmKAN3 expression results. (g) Cross section of a shoot, SmKAN3 expression is not detected. (h) Expression of SmKAN3 is not detected in the leaf primordia. (i) SmKAN3 expression is not detected during early sporangia development but is detected late in sporocoyte proliferation. (j) SmKAN3 expression in a nearly mature sporangium prior to meiosis. Scale bars: 50 µm (a,b,e,g); 20 µm (c,d,f,h); 10 µm (i,j). Asterisk indicates apical meristem; black arrowheads indicate phloem; black arrows indicate sporangia; li = ligule; m = microphyll.
We found the expression of SmKAN1 and SmKAN2 to be very similar except that SmKAN1 appeared to be more highly expressed than SmKAN2 at all developmental stages (Figure 3). In transverse sections of the shoot axis, SmKAN1 expression is detected as a ring in the developing vasculature which likely represents the developing phloem (Figure 3a, arrowheads). Later in shoot development, the expression of SmKAN2 is still detected in the developing vasculature, specifically in the phloem that surrounds the central xylem pole (Figure 3d). In longitudinal sections of the shoot axis, SmKAN1 and SmKAN2 are detected in the earliest emerging leaf primordia and as the leaf primordia continue to grow (Figure 3b,e). SmKAN1 and SmKAN2 expression is not detected in older leaves (Figure 3a,b,e). SmKAN3 had a more discrete pattern of expression compared to SmKAN1 and SmKAN2. Notably, SmKAN3 expression is not detected during any stage of leaf development (Figure 3a,d,g). While expression of SmKAN1 and SmKAN2 during S. moellendorfii development is detected in the leaf primordia, neither of them show expression patterns polarized to one side of the developing leaf (Figure 3a,b,e,h). In addition, SmKAN1, SmKAN2, and SmKAN3 expression is not detected in the shoot apical meristem (SAM).
During development of the S. moellendorffii strobilus (fertile sporophytic axis), the expression of SmKAN1 and SmKAN2 was analogous to what was found in the vegetative shoot axis (Figure 3c,f). SmKAN1, SmKAN2, and SmKAN3 expression is not detected in the meristem of the strobilus. However, SmKAN1 and SmKAN2 expression was found in the sporangium incipient primordia on the flanks of the strobilus and this expression is maintained in the sporangia at least through the proliferation of the sporocytes. Expression of SmKAN3 is detected in sporangium development during later stages of sporocyte proliferation prior to meiosis (Figure 3i,j).
3.3. Expression of KANADI in the Fern Equisetum hyemale
To better understand the expression of KANADI homologs in ferns we assessed the expression pattern of these in the fern Equisetum hyemale (Figure 4).
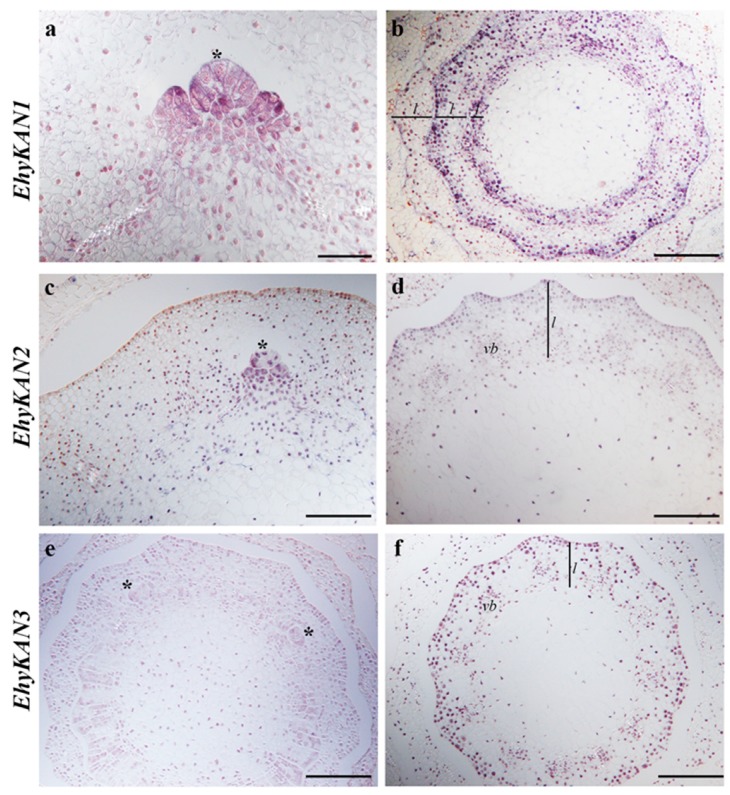
Figure 4.
EhyKAN expression in Equisetum hyemale by in situ hybridization. (a) EhyKAN1 expression is detected in the apical meristem and emerging leaf primordia of a branch in a transverse section of a shoot. (b) EhyKAN1 expression is detected throughout the leaf primordia (innermost whorl) and then becomes restricted to the abaxial side of each leaf whorl (outer two whorls shown here) in a transverse section through a shoot. (c) EhyKAN2 expression is detected in the apical meristem and emerging leaf primordia of a branch in a transverse section of a shoot. (d) EhyKAN2 becomes restricted to the abaxial side of the leaf whorl in transverse section through a shoot. (e) EhyKAN3 expression is detected in the apical meristem of a branch in a transverse section of a shoot. (f) EhyKAN3 becomes restricted to the abaxial side of the leaf whorl in transverse section through a shoot. Scale bars: 50 µm (a); 100 µm (b–f). Asterisk indicates apical meristem; lines with l indicates leaf whorl; vb = vascular bundle.
The leaves of Equisetum are morphologically distinct from the more common fern leaves that arise with circinate vernation [1]. The leaves of E. hyemale are formed in a whorl around the shoot axis (Figure 1c) and are easily seen in transverse sections of the shoot (Figure 4b,d,f). In addition, branches do not form consistently at every node of E. hyemale as in other Equisetum species, but branches do occasionally form (Figure 4a,c,e). We found the expression of all three KANADI homologs similar to each other with only an apparent difference in the level of expression between copies. EhyKAN1/2/3 are all expressed in the SAM (asterisk) and emerging leaf primordia (Figure 4a,c,e). However, EhyKAN3 showed lower levels of expression in the shoot apical meristem compared with the other two homologs. Expression of EhyKAN1, 2, and 3 is detected in vascular bundles (Figure 4b,d,f). Notably, we detected that the three homologs are expressed in the leaves early in the development and become restricted on the abaxial side as the leaves mature (Figure 4b,d,f).
4. Discussion
KANADI genes belong to the GARP family of transcription factors [45] and the GARP domain confers the DNA-binding function [45,46,47]. Our analysis focused strictly on determining the evolutionary history of KANADI homologs across vascular plants, lycophytes, ferns, and seed plants (Figure 2). By doing the alignment of KAN homologs, our results indicate that all KAN homologs have a DNA binding function, as the GARP domain is highly conserved across vascular plants. Phylogenetic analyses showing the evolutionary history of the KANADI gene lineage are scarce, most of them including only model species [24,27,28]. Therefore, we performed a BLAST search across all major plant groups (Figure 2). Even though the analysis was focused in understanding the evolution of the KAN genes in lycophytes and ferns, our results allow us to hypothesize that lycophyte and fern homologs appeared before the diversification of the traditionally known KAN1, KAN2, KAN3 and KAN4 (ATS) in seed plants. To determine at which point during the evolution of seed plants the four copies of KANADIs evolved, a more exhaustive search of homologs across seed plants will be needed. However, such duplications are most likely associated with a whole genome duplication event (WGD) [48,49,50]. We performed a search in publicly available databases for KAN homologs across ferns. Here we report a duplication event within one of the KANADI clades of ferns for the first time (Figure 2). This duplication could have been the result of the whole genome duplication predating the core leptosporangiate ferns [51]. Nevertheless, additional sequences are required in order to corroborate this hypothesis.
We examined the expression of KANADI homologs for the first time outside of model angiosperm species. We found similar expression patterns in the three KAN homologs found in Equisetum hyemale; all have polar expression, specifically in the abaxial side of developing leaves, suggesting that the three copies are redundant for leaf development (Figure 4). In order to assess differences in the expression of E. hyemale homologs, it would require looking at different tissues such as the strobili and the roots, as expression in these structures has been reported in angiosperms [52]. On the other hand, we have not detected polar expression of KANADI genes as expected in lycophytes. Significantly, the expression of SmKAN1, SmKAN2, and SmKAN3, is not detected in a polar fashion, namely the abaxial side of developing leaves (Figure 3).
Our results for the KAN gene lineage in lycophytes and ferns, together with previous analyses for other genes involved in the adaxial/abaxial leaf developmental genetic network, such as Class III HD-Zip genes [8], allow us to better understand changes of this genetic network across the evolution of vascular plants. Class III HD-Zip genes are expressed on the adaxial side of leaves from across ferns with diverse leaf morphologies [8], although KAN expression in ferns was only assessed in one species, it was found restricted to the abaxial side of the leaves (Figure 4). Therefore, our study provides further molecular genetic support that ferns share a broad leaf developmental mechanism with seed plants [1,8].
Expression patterns of an adaxial/abaxial network in lycophytes are different, as Class III HD-Zips have not been detected in the adaxial side of leaves in Selaginella moellendorffii or other lycophytes [8,30,31], and KANADI homologs were not detected in the abaxial side of the leaves (Figure 3). It would appear that the lycophyte Selaginella moellendorffii does not use the same genetic module to specify the abaxial and adaxial identities of its leaves. However, similar to what is found in angiosperms, the expression of the KANADI homologs in S. moellendorffi is found in the phloem. This may represent the ancestral function of KANADI gene lineage in vascular plants.
There is a potential to unravel the evolution of the Class III HD-Zip–KANADI leaf developmental module by studying the expression and function of these genes throughout lycophytes and ferns. The ancestral function of Class III HD-Zip was proposed to be in meristem development, as these genes were found in all vascular plants, and in flowering plants are found expressed in vasculature, meristem, and the adaxial side of lateral organs [30]. However, previous expression analyses showed that Class III HD-Zip orthologs are not expressed in the meristems of the lycophyte Selaginella moellendorffii or the meristems of the eusporangiate ferns Equisetum diffusum, Psilotum nudum, or Osmunda regalis [8,31]. However, expression of Class III HD-Zip orthologs was detected in the meristem of the leptosporangiate ferns Pilularia globulifera and Elaphoglossum peltatum [8]. Here we show that KANADI homologs are not expressed in the meristem of the lycophyte S. moellendorffii, but are expressed in the meristem of the fern Equisetum hyemale. It will be important to further study the expression of KANADI homologs in lycophytes and ferns to better understand when these genes acquired meristem expression, providing more data to untangle the evolution of the Class III HD-Zip and KANADI developmental module.
Although, we did not detect the expression of KAN homologs in the abaxial side of developing microphylls of Selaginella moellendorffii, we did find that KAN homologs are expressed in developing sporangia (Figure 3). This is similar to the expression profile previously found for Class III HD-Zips with expression in sporangia [8]. This may provide support for a modified telome theory with microphylls evolving by the progressive sterilization of sporangia [3]. This result also raises an intriguing question about the molecular genetics of microphyll development of which we still know little.
5. Conclusions
Our results suggest that the genetic network that determines abaxial identity is conserved within ferns, gymnosperms and angiosperms but not in lycophytes. However, additional studies are necessary. Identifying the precise role of KAN homologs across vascular plants will require an exhaustive evaluation of spatiotemporal expression patterns throughout ferns as well as in other lycophytes, coupled with functional analyses in diverse vascular plant species.
Acknowledgments
We thank Tynisha Smalls for excellent technical assistance, and the Nolen glass house staff for plant care. We thank the Ceratopteris Genome Project for Ceratopteris richardii sequences.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/8/9/313/s1. Table S1: List of genes included in the ML analysis. Table S2: Primers used for expression analyses in E. hyemale and S. moellendorffii.
Author Contributions
Conceptualization: B.A.A. and A.V.; methodology: B.A.A., C.Z.-C., and A.V.; investigation: B.A.A., C.Z.-C., and A.V.; data curation: C.Z.-C.; writing—original draft preparation: B.A.A. and C.Z-C.; writing—review and editing: B.A.A., C.Z.-C., and A.V.; visualization: C.Z.-C. and B.A.A.; funding acquisition: B.A.A.
Funding
This research was funded by National Science Foundation DEB-1020443 to B.A.A.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Vasco A., Moran R.C., Ambrose B.A. The evolution, morphology, and development of fern leaves. Front. Plant Sci. 2013;4:345. doi: 10.3389/fpls.2013.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bower F.O. Primitive Land Plants—Also Known as the Archegoniate. Macmillan and Co.; London, UK: 1935. [Google Scholar]
- 3.Kenrick P., Crane P.R. The Origin and Early Diversification of Land Plants: A Cladistic Study. Smithsonian Institution Press; Washington, DC, USA: 1997. [Google Scholar]
- 4.Tomescu A.M.F. Megaphylls, microphylls, and the evolution of leaf development. Trends Plant Sci. 2009;14:5–12. doi: 10.1016/j.tplants.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296:1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- 6.Harrison C.J., Corley S.B., Moylan E.C., Alexander D.L., Scotland R.W., Langdale J.A. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature. 2005;434:509–514. doi: 10.1038/nature03410. [DOI] [PubMed] [Google Scholar]
- 7.Sano R., Juárez C.M., Hass B., Sakakibara K., Ito M., Banks J.A., Hasebe M. KNOX homeobox genes potentially have similar functions in both diploid unicellular and multicellular meristems but not in haploid meristems. Evol. Dev. 2005;7:69–78. doi: 10.1111/j.1525-142X.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 8.Vasco A., Smalls T.L., Graham S.W., Cooper E.D., Wong G.K., Stevenson D.W., Moran R.C., Ambrose B.A. Challenging the paradigms of leaf evolution: Class III HD-Zips in ferns and lycophytes. New Phytol. 2016;212:745–758. doi: 10.1111/nph.14075. [DOI] [PubMed] [Google Scholar]
- 9.Zimmermann W. Main results of the ‘telome theory’. Paleobotanist. 1952;1:456–470. [Google Scholar]
- 10.Fukushima K., Hasebe M. Adaxial–abaxial polarity: The developmental basis of leaf shape diversity. Genesis. 2014;52:1–8. doi: 10.1002/dvg.22728. [DOI] [PubMed] [Google Scholar]
- 11.Waites R., Hudson A. Phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development. 1995;121:2143–2154. [Google Scholar]
- 12.McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 13.Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. Radial patterning of Arabidopsis shoots by Class III HD-Zip and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 14.Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- 15.Xu L., Xu Y., Dong A., Sun Y., Pi L., Xu Y., Huang H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- 16.Fu Y., Xu L., Xu B., Yang L., Ling Q., Wang H., Huang H. Genetic interactions between leaf polarity-controlling genes and ASYMMETRIC LEAVES1 and 2 in Arabidopsis leaf patterning. Plant Cell Physiol. 2007;48:724–735. doi: 10.1093/pcp/pcm040. [DOI] [PubMed] [Google Scholar]
- 17.Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- 18.McConnell J.R., Barton M.L. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- 19.Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- 20.Eshed Y., Baum S.F., Bowman J.L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99:199–209. doi: 10.1016/S0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- 21.Pekker I., Alvarez J.P., Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sessions A., Nemhauser J.L., McColl A., Roe J.L., Feldmann K.A., Zambryski P.C. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- 23.Sessions R.A., Zambryski P.C. Arabidopsis gynoecium structure in the wild type and in ettin mutants. Development. 1995;121:1519–1532. doi: 10.1242/dev.121.5.1519. [DOI] [PubMed] [Google Scholar]
- 24.Yan S., Yan C.J., Zeng X.H., Yang Y.C., Fang Y.W., Tian C.Y., Sun Y.W., Cheng Z.K., Gu M.H. ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant Mol. Biol. 2008;68:239–250. doi: 10.1007/s11103-008-9365-x. [DOI] [PubMed] [Google Scholar]
- 25.Kelley D.R., Arreola A., Gallagher T.L., Gasser C.S. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development. 2012;139:1105–1109. doi: 10.1242/dev.067918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eshed Y., Baum S.F., Perea J.V., Bowman J.L. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001;11:1251–1260. doi: 10.1016/S0960-9822(01)00392-X. [DOI] [PubMed] [Google Scholar]
- 27.Zhang G.H., Xu Q., Zhu X.D., Qian Q., Xue H.W. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell. 2009;21:719–735. doi: 10.1105/tpc.108.061457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Candela H., Johnston R., Gerhold A., Foster T., Hake S. The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell. 2008;20:2073–2087. doi: 10.1105/tpc.108.059709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conklin P.A., Strable J., Li S., Scanlon M.J. On the mechanisms of development in monocot and eudicot leaves. New Phytol. 2019;221:706–724. doi: 10.1111/nph.15371. [DOI] [PubMed] [Google Scholar]
- 30.Floyd S.K., Zalewski C.S., Bowman J.L. Evolution of Class III homeodomain-leucine zipper genes in streptophytes. Genetics. 2006;173:373–388. doi: 10.1534/genetics.105.054239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prigge M.J., Clark S.E. Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 2006;8:350–361. doi: 10.1111/j.1525-142X.2006.00107.x. [DOI] [PubMed] [Google Scholar]
- 32.Bowman J.L., Eshed Y., Baum S.F. Establishment of polarity in angiosperm lateral organs. Trends Genetics. 2002;18:134–141. doi: 10.1016/S0168-9525(01)02601-4. [DOI] [PubMed] [Google Scholar]
- 33.McAbee J.M., Hill T.H., Skinner D.J., Izhaki A., Hauser B.A., Meister R.J., Reddy G.V., Meyerowitz E.M., Bowman J.L., Gasser C.S. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 2006;46:522–531. doi: 10.1111/j.1365-313X.2006.02717.x. [DOI] [PubMed] [Google Scholar]
- 34.Arnault G., Vialette A., Andres Robin A., Fogliani B., Gâteblé G., Scutt C.P. Evidence for the extensive conservation of mechanisms of ovule integument development since the most recent common ancestor of living angiosperms. Front. Plant Sci. 2018;9:1352. doi: 10.3389/fpls.2018.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickett N.J., Mirarab S., Nguyen N., Warnow T., Carpenter E., Matasci N., Ayyampalayam S., Barker M.S., Burleigh J.G., Gitzendanner M.A., et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matasci N., Hung L.H., Yan Z., Carpenter E.J., Wickett N.J., Mirarab S., Nguyen N., Warnow T., Ayyampalayam S., Barker M., et al. Data access for the 1,000 Plants (1KP) project. Gigascience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Y., Wu G., Tang J., Luo R., Patterson J., Liu S., Huang W., He G., Gu S., Li S., et al. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics. 2014;30:1660–1666. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- 38.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011;40:D1178-86. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsson A. AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics. 2014;22:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller M.A., Pfeier W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010. [Google Scholar]
- 42.Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 43.Fig Tree. [(accessed on 1 July 2019)]; Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 44.Ambrose B.A., Lerner D.R., Ciceri P., Padilla C.M., Yanofsky M.F., Schmid R.J. Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell. 2000;5:569–579. doi: 10.1016/S1097-2765(00)80450-5. [DOI] [PubMed] [Google Scholar]
- 45.Riechmann J.L., Heard J., Martin G., Reuber L., Jiang C.Z., Keddle J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 46.Hosada K., Imamura A., Katoh E., Hatta T., Tachiki M., Yamada H., Mizuno T., Yamazaki T. Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis Response Regulators. Plant Cell. 2002;14:2015–2029. doi: 10.1105/tpc.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grefen C., Harter K. Plant two-component systems: Principles, functions, complexity and cross talk. Planta. 2004;219:733–742. doi: 10.1007/s00425-004-1316-4. [DOI] [PubMed] [Google Scholar]
- 48.Barker M.S., Vogel H., Schranz M.E. Paleopolyploidy in the Brassicales: Analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol. Evol. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donoghue M.T., Keshavaiah C., Swamidatta S.H., Spillane C. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol. Biol. 2011;11:47. doi: 10.1186/1471-2148-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiao Y., Wickett N.J., Ayyampalayam S., Chanderbali A.S., Landherr L., Ralph P.E., Tomsho L.P., Hu Y., Liang H., Soltis P.S., et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 51.Li F.W., Brouwer P., Carretero-Paulet L., Cheng S., deVries J., Delaux P.-M., Eily A., Koppers N., Kuo L.-Y., Li Z., et al. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants. 2018;4:460–472. doi: 10.1038/s41477-018-0188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hawker N.P., Bowman J.L. Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 2004;135:2261–2270. doi: 10.1104/pp.104.040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.