Abstract
Ochratoxin A (OTA) is a well-known, natural contaminant in foods and feeds because of its toxic effects, such as nephrotoxicity in various animals. Recent studies have revealed that Alcaligenes faecalis could generate enzymes to efficiently degrade OTA to ochratoxin α (OTα) in vitro. In an effort to obtain the OTA degrading mechanism, we purified and identified a novel degrading enzyme, N-acyl-L-amino acid amidohydrolase (AfOTase), from A. faecalis DSM 16503 via mass spectrometry. The same gene of the enzyme was also encountered in other A. faecalis strains. AfOTase belongs to peptidase family M20 and contains metal ions at the active site. In this study, recombination AfOTase was expressed and characterized in Escherichia coli. The molecular mass of recombinant rAfOTase was approximately 47.0 kDa, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The enzyme exhibited a wide temperature range (30–70 °C) and pH adaptation (4.5–9.0) and the optimal temperature and pH were 50 °C and 6.5, respectively.
Keywords: ochratoxin A, degradation, amidohydrolase, Alcaligenes faecalis, mycotoxin
1. Introduction
Ochratoxin A (OTA) is a natural, toxic, secondary metabolite produced by several fungal species of the genera Penicillium and Aspergillus [1]. OTA spreads widely in, for example, cereals, corn, grape, wine, beer, fish, pork, seasoning, oilseeds, and edible oils. In areas with a hot and humid climate, OTA contamination has a higher incidence because of the suitable conditions for the growth of toxigenic fungi. Owing to the physical and chemical stability of OTA, it is hard to be removed or degraded in agricultural commodities [2]. Thus, there may be contamination risk in each step of the agricultural supply chain, including pre-harvest, post-harvest, and during processing and storage [3,4,5].
OTA, the effects of which are mainly observed in the kidney and liver, is one of the most dangerous mycotoxins for humans and animals [6]. Nephrotoxicity, hepatotoxicity, carcinogenicity, and teratogenicity are the most relevant toxic effects [7,8]. Many researchers have suggested that OTA exposure is responsible for several human diseases, including Balkan endemic nephropathy [9]. It has also been considered that OTA has genotoxicity and immunotoxicity [10,11].
As a foodborne hazard, the presence of OTA in agricultural and sideline products has attracted great public attention and the food safety regulatory departments have recently enforced the controls of its legal limitation [12]. Therefore, to reduce the risk of human and animal exposure, several physical, chemical, and microbiological strategies were proposed to eliminate the hazard [13]. Among these strategies, biological methods using microorganisms or enzymes are more efficient, because they have high efficiency and high target specificity and are environmentally friendly, and they can preserve the nutritious value of food and feed products [14].
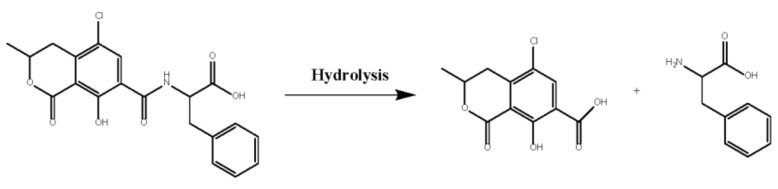
Adsorption and biodegradation are the main approaches for OTA detoxification by micro-organisms. Lactobacillus species and yeasts are the most studied microorganisms for OTA bio-sorption, which depends on the cell wall components and structure [15,16]. The biodegradation of OTA is implemented by hydrolysis, hydroxylation, lactone ring opening, and conjugation to other entities [17]. Among the final degradation products of this reaction, ochratoxin α (OTα), a product of the OTA amide bond hydrolysis reaction, is considered to be barely toxic or non-toxic (Figure 1). It is approximately 1000 times less toxic than OTA in brain cell cultures and its half-life in the blood is 10 times shorter than that of OTA (103 h) [18,19].
Figure 1.
Enzymatic degradation of ochratoxin A via hydrolysis of its amide bond using N-acyl-L-amino acid amidohydrolase from Alcaligenes faecalis.
Recent studies have discovered that the A. faecalis strain is able to efficiently degrade OTA to OTα in vitro [20]. In an effort to obtain the OTA degrading mechanism, we purified a novel degrading enzyme from A. faecalis DSM 16503 and identified it as N-acyl-L-amino acid amidohydrolase (AfOTase) via mass spectrometry (unpublished data). The same gene of the enzyme is also encountered in other A. faecalis strains. In this study, the AfOTase gene was cloned, expressed, and purified in Escherichia coli BL21 (DE3), and the effects of temperature and pH on the catalytic activity of recombinant AfOTase (rAfOTase) were characterized. The hydrolysis products of ochratoxin A by rAfOTase are β-phenylalanine and ochratoxin A (Supplementary Figure S1).
2. Results and Discussion
2.1. Cloning and Amino Acid Sequencing of The AfOTase Gene from A. Faecalis DSM 16503
The sequence of the AfOTase gene from the gDNA of A. faecalis DSM 16305 contains a complete open reading frame (ORF) (1317 bp) that employs ATG and TAG as start and stop codons, respectively. The overall G+C content of the gene is 57%. The full-length gene encodes a polypeptide of 438 amino acid residues. The primers OD-F and OD-R allowed amplification of a 1251 bp long gene without the signal peptide, which consists of 22 amino acids, predicted by SignalP 4.0. The sequence of the cloned gene was deposited in GenBank accession no. OSZ37025.1.
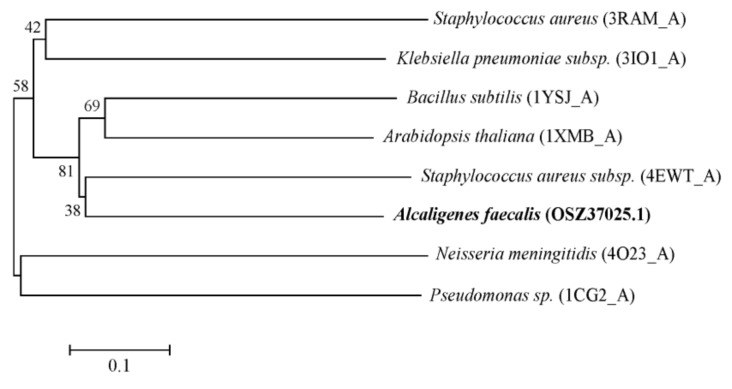
An amino acid homology analysis by BLASTP showed the highest identity (37%) with LAA-amino acid hydrolase from Arabidopsis thaliana, followed by aminohydrolase from methicillin-resistant Staphylococcus aureus (36%), Yxep protein from Bacillus subtilis (34%), aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae (25%), and Hmra from S. aureus (25%) (Figure 2). The enzyme belongs to the peptidase family M20 (clan MH) [21]. The peptidases of this clan have two catalytic zinc ions at the active site, bound by His/Asp, Asp, Glu, Asp/Glu, and His. This family comprises a peptidase dimerization domain that consists of four beta-strands and two alpha-helices which make up the dimerization surface.
Figure 2.
A phylogenetic tree was constructed based on the amino acid sequences of AfOTase by means of neighbor-joining analysis. Bootstrap values (n = 1000 replicates) are reported as percentages.
Moreover, AfOTase has low homology in the amino acid sequence with other known OTA degrading enzymes, such as OTase from A. niger (Supplementary Figure S2).
2.2. Expression and Purification of rAfOTase
AfOTase gene was successfully cloned into pET-28a (+) vector (Supplementary Figure S3), confirmed by DNA sequencing and double digestion of the recombinant vector with BamHI and HindIII. The constructed pET-28a-rAfOTase vector was transformed and efficiently expressed in E. coli BL21 (DE3) intracellularly after it was induced with 0.5 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) at 20 °C for 16 h.
The recombinant AfOTase was purified by a nitrilotriacetic acid complex of nickel (II) (Ni2+-NTA) resin affinity chromatography. The molecular mass of rAfOTase was slightly above 45.0 kDa as determined by SDS-PAGE (Supplementary Figure S4), and very close to the predicted molecular weight of the protein (~47.0 kDa).
2.3. Enzymatic Properties of rAfOTase
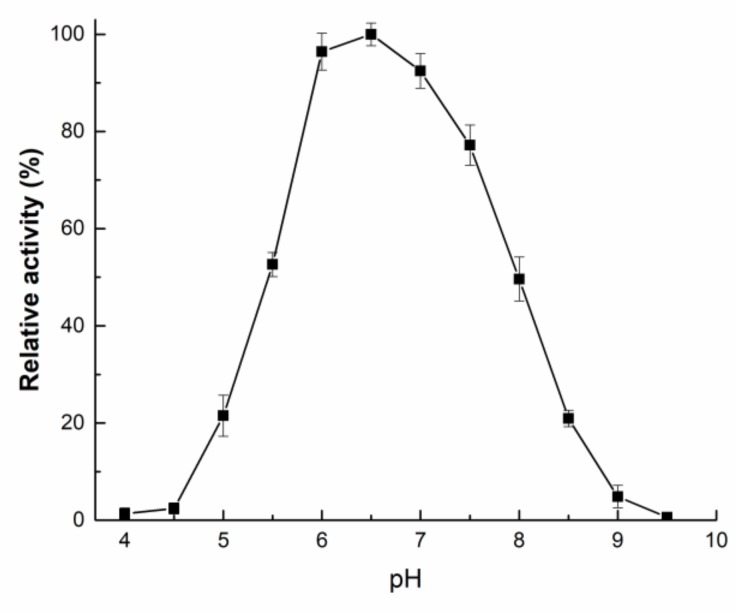
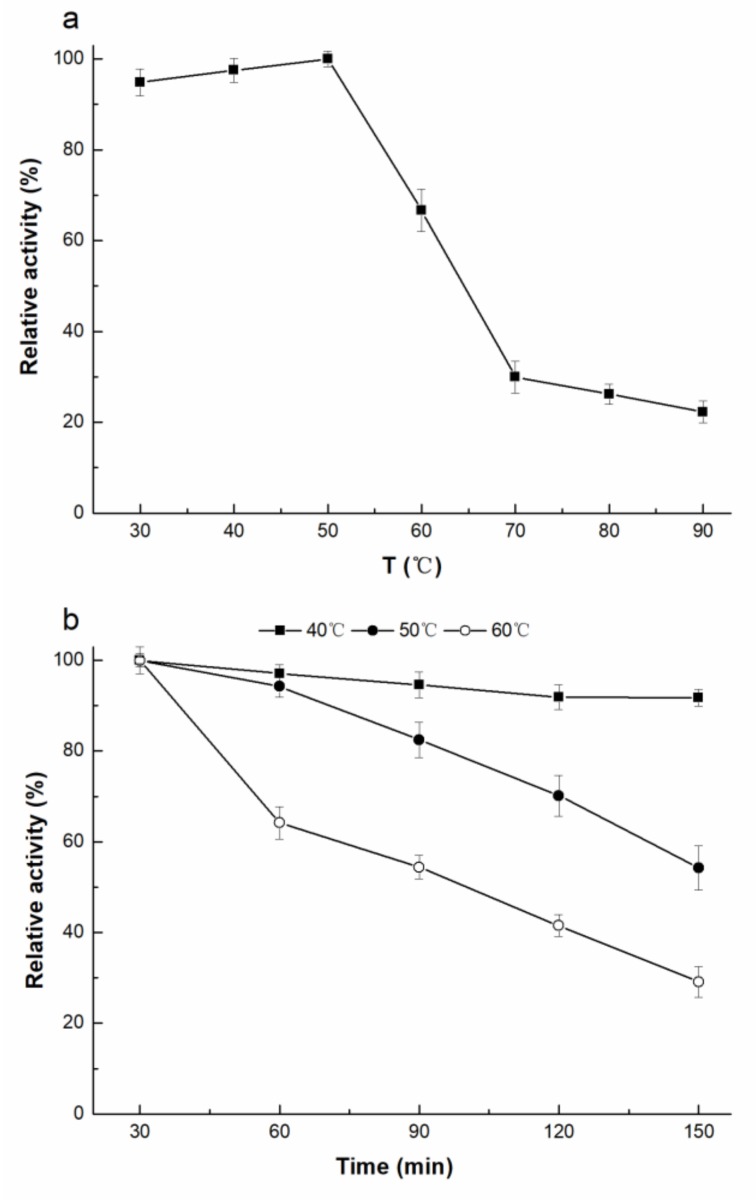
rAfOTase exhibits activity at various temperatures (30–70 °C) and pH values (4.5–9.0) (Figure 3). rAfOTase showed high levels of activity at pH 6.0 to 7.0 (> 90%), with an optimum pH of 6.5. When the pH was below 5.0 or above 8.5, the rAfOTase retained less than 20% of its optimal catalytic activity. The enzyme activity remained in varying degrees within 30–90 °C after incubation for 60 min with a substrate (Figure 4a). The optimal temperature for rAfOTase was 50 °C. rAfOTase was stable and retained no less than 90% of activity after it was incubated for 150 min at temperatures below 50 °C. After being incubated at 50 °C for 120 min, approximately 70% of activity was retained but the activity was substantially reduced at a temperature above 50 °C (Figure 4b).
Figure 3.
Effects of pH on rAfOTase catalytic activity. Reaction system consisted of 15 μg/mL rAfOTase, 20 mM buffer, and 1 μg/mL ochratoxin A.
Figure 4.
Effects of temperature on rAfOTase (a) catalytic activity and (b) stability. Reaction system consisted of 15 μg/mL rAfOTase, 20 mM buffer (pH 6.5), and 1 μg/mL ochratoxin A.
Over the last two decades, only a few enzymes involved in the OTA degrading process have been purified and characterized [22,23]. Bovine pancreas carboxypeptidase A (CPA) (EC 3.4.17.1) was reported for the first time [24]. However, CPA has a weak degrading ability to detoxify completely. Subsequently, carboxypeptidase Y (CPY) (EC 3.4.16.1) from Saccharomyces cerevisiae was found but its degrading activity is very low, with optimal activity at pH 5.6 and 37 °C [13]. Further, similar enzymes are reported in different strains [23,25] and a few commercial proteases from Aspergillus niger are reported to hydrolyze OTA to OTα. For example, protease A and pancreatin could degrade 87.3% and 43.4%, respectively, of OTA (at 1 μg/mL) after 25 h at pH 7.5 and 37 °C [26].
Remarkably, Stander, et al. [27] first reported that AmanoTM lipase from A. niger had ochratoxin-degrading activity. Subsequent research showed that the degrading enzyme from A. niger is a metalloenzyme and more efficient than CPA [26,28]. Further purification and analysis of the product revealed that the active enzyme is an amidase, named ochratoxinase (OTase), and its hydrolytic activity is approximately 600 times higher than CPA in OTA degradation at pH 7.5 after being incubated for 60 min at 37 °C [29].
The AfOTase from A. faecalis exhibited not only efficient detoxification ability but also relatively wide pH and mesophilic stability when compared to other enzymes, as described above. However, for large-scale production of rAfOTase for industrial applications, a more cost-effective expression system needs to be employed and the pH and thermal stability should be enhanced.
3. Conclusions
In conclusion, AfOTase, a novel OTA degrading enzyme from A. faecalis, was cloned and produced using an E. coli expression system. It showed commendable thermal and pH stability. These important properties of AfOTase make it a novel potential candidate for commercial applications, such as detoxifying OTA in food or feed processing. Further study of this enzyme will be focused on the meaningful value of detoxifying OTA in the food and feed industries.
4. Materials and Methods
4.1. Strain and Growth Conditions
Alcaligenes faecalis DSM 16503, Escherichia coli Top10, and E. coli BL21 (DE3) were aerobically grown at 37 °C in Luria–Bertani (LB) medium. When necessary, 100 μg/mL ampicillin or 50 μg/mL kanamycin was added to the medium as screening pressure.
4.2. DNA Isolation and Sequence Data Analysis
Genomic DNA was extracted from A. faecalis DSM 16503 as described by Lee and Taylor [30]. Plasmid DNA from E. coli was purified by using the SanPrep Column Plasmid Mini-Preps Kit (Sangon Biotech, China). The amplified DNA fragments were purified using a PCR Product Purification Kit (Sangon Biotech, Shanghai, China) and sequenced with an ABI 3730XL DNA analyzer at Sangon Biotech Co. Ltd. (Shanghai, China).
The nucleotide and protein sequences were compared with the National Center for Biotechnology Information (NCBI) nucleotide/protein database by BLASTN and BLASTP, respectively. The signal peptide sequence was analyzed by SignalP version 4.0 [31]. Multiple sequence alignments were performed with ClustalW version 2.1. A phylogenetic tree was constructed using the neighbor-joining (NJ) method in MEGA version 5.0 [32]. Confidence in the tree topology was estimated using bootstrap values based on 1000 replicates.
4.3. Cloning of AfOTase Gene
The AfOTase gene was amplified from A. faecalis DSM 16503 genomic DNA (gDNA) by PCR using LA Taq (TaKaRa, Kusatsu, Japan). Two primers, OD-F (CGCGGATCCCAAGCCAGTAATCCCATGATGG, with a BamHI site) and OD-R (CCCAAGCTTCTACGGTTTTTTGTGATCCATC, with a HindIII site) were used for PCR amplification and partial sequencing of the AfOTase gene. The gene was ligated into pET-28a (+) which was linearized by BamHI-HF and HindIII-HF (NEB). The vector pET-28a (+) with His6-tags at the N-terminus was used for recombinant expression in this study.
4.4. Expression of AfOTase in E.coli
The recombinant plasmid was transformed into E. coli BL21 (DE3). The recombinant E. coli BL21 (DE3) was first inoculated into 3 mL LB medium with 50 μg/mL kanamycin and cultured overnight at 37 °C with shaking at 220 rpm. Then 1 mL of the overnight culture was transferred into 100 mL LB medium with 50 μg/mL kanamycin and grown at 37 °C with shaking at 220 rpm until OD600 reached 0.6, then the temperature was decreased to 20 °C and IPTG was added for a final concentration of 0.5 mM. After 16 h, the cells were harvested by centrifugation (4 °C, 4000 × g, 15 min) and stored at −80 °C before purification.
4.5. Purification of Recombinant AfOTase
For every gram of induced cells pellet, 5 mL of buffer A (20 mM sodium phosphate, 0.5 M NaCl, pH 7.5), 2 μL of 250 U benzonase (BioLong, Shanghai, China), and 5 μL of 1M MgCl2 were added. The cell solution was fully dissolved by stirring and lysed (30 kpsi, 4 °C) by a high-pressure cell disrupter (Constant Systems, Northants, UK) twice, followed by centrifugation (15,000 × g, 4 °C) for 20 min to remove cell debris.
rAfOTase with His6-tags was purified by Ni2+-NTA resin using a biomolecular liquid chromatography system (ÄKTA purifier, GE Healthcare, Boston, MA, USA). The target proteins were eluted with buffer B (20 mM sodium phosphate, 0.5 M NaCl, 250 mM imidazole, pH 7.5). Proteins in the supernatant were precipitated by adding solid ammonium sulfate to 20% and 80% saturation. The fractions precipitating at 20% and 80% saturation were redissolved in 1 mL buffer A and desalted on a Sephadex G-25F column (5mL; GE Healthcare, Boston, MA, USA) with buffer C (20 mM sodium phosphate, 0.1 M NaCl, pH 7.5). The fractions with OTA degrading activity were collected and protein purity was checked by SDS-PAGE.
4.6. rAfOTase Activity Assays
rAfOTase activity assays followed the method of Dobritzsch et al. [29] with minor variations. The rAfOTase assay reaction system consisted of 5 μL sample, 240 μL phosphate buffer (pH 7.5), and 5 μL OTA stock solution (50 μg/mL). Reactions were performed for 10 to 120 min at 37 °C and stopped by adding 250 μL acetonitrile. Then, the sample was centrifuged at 12,000 × g at 4 °C for 10 min. The supernatants were filtered using 0.22 μm syringe filters (Millipore, Cork, Ireland). The concentration of OTA was determined by HPLC equipped with a fluorescence detector (FLD) and the quantitative analysis method was described in detail by Zhang et al. [20].
4.7. Biochemical Characterization of rAfOTase
The optimum pH of rAfOTase was determined by incubating the recombinant enzyme in a series of 20 mM buffers for 30 min. The pH range from 4.0 to 9.5 was used (20 mM citric acid–phosphate buffer, pH 4.0–7.0; 20 mM Tris–HCl buffer, pH 7.5–8.5; 20 mM glycine–NaOH buffer, pH 9.0–9.5). Similarly, the rAfOTase activity assays were done at different temperatures (30–90 °C) to determine the optimum temperature at pH 6.5 for 60 min. When studying the thermal stability of the enzyme, the remaining rAfOTase activity was measured after incubating the recombinant enzyme at a certain temperature for 30–150 min. Reactions were stopped as described above. The untreated enzyme was used as the control.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6651/11/9/518/s1, Figure S1. The extract ion chromatogram and MS/MS spectrum of (a1 and b1) ochratoxin A, (a2 and b2) ochratoxin α or (a3 and b3) β-phenylalanine, which the sample processed by rAfOTase; Figure S2. Multiple alignment of partial known ochratoxin A degrading enzymes; Figure S3. Plasmid map for Alcaligenes faecalis OTase expression vector pET-28a(+)-rAfOTase; Figure S4. SDS–PAGE analysis of rAfOTase; M, protein size markers; L1, rAfOTase was purified by Ni2+-NTA resin.
Author Contributions
Conceptualization, H.Z. and X.Z.; methodology, H.Z.; software, H.Z.; validation, Y.Z. and T.Y.; formal analysis, H.Z., J.W. and X.Z.; resources, H.Z.; data curation, J.W.; writing—original draft preparation, H.Z., J.W. and X.Z.; writing—review and editing, H.Z., Y.Z., T.Y., J.W. and X.Z.; supervision, J.W. and X.Z.; project administration, J.W. and X.Z.; funding acquisition, X.Z.
Funding
This research was funded by National Key Research and Development Program (2017YFC1600604) and the Natural Science Foundation of Gansu Province (1506RJZA008).
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
A novel OTA degrading enzyme, as a N-acyl-L-amino acid amidohydrolase, isolated from Alcaligenes faecalis is reported.
References
- 1.Bellí N., Ramos A.J., Coronas I., Sanchis V., Marín S. Aspergillus carbonarius growth and ochratoxin A production on a synthetic grape medium in relation to environmental factors. J. Appl. Microbiol. 2005;98:839–844. doi: 10.1111/j.1365-2672.2004.02469.x. [DOI] [PubMed] [Google Scholar]
- 2.André el Khoury A., Atoui A. Ochratoxin A: General overview and actual molecular status. Toxins. 2010;2:461–493. doi: 10.3390/toxins2040461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edite Bezerra da Rocha M., Freire F.d.C.O., Erlan Feitosa Maia F., Izabel Florindo Guedes M., Rondina D. Mycotoxins and their effects on human and animal health. Food Control. 2014;36:159–165. doi: 10.1016/j.foodcont.2013.08.021. [DOI] [Google Scholar]
- 4.Limay-Rios V., Miller J.D., Schaafsma A.W. Occurrence of penicillium verrucosum, ochratoxin A, ochratoxin B and citrinin in on-farm stored winter wheat from the canadian great lakes region. PLoS ONE. 2017;12:e0181239. doi: 10.1371/journal.pone.0181239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soto J.B., Fernándezfranzón M., Ruiz M., Juangarcía A. Presence of ochratoxin A (OTA) mycotoxin in alcoholic drinks from southern european countries: Wine and beer. J. Agr. Food Chem. 2014;62:7643–7651. doi: 10.1021/jf501737h. [DOI] [PubMed] [Google Scholar]
- 6.Pfohl-Leszkowicz A., Manderville R.A. Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol. Biol. Evol. 2007;51:61–99. doi: 10.1002/mnfr.200600137. [DOI] [PubMed] [Google Scholar]
- 7.Limonciel A., Jennings P. A review of the evidence that ochratoxin A is an nrf2 inhibitor: Implications for nephrotoxicity and renal carcinogenicity. Toxins. 2014;6:371–379. doi: 10.3390/toxins6010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heussner A.H., Bingle L.E. Comparative ochratoxin toxicity: A review of the available data. Toxins. 2015;7:4253–4282. doi: 10.3390/toxins7104253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buiklimke T.R., Wu F. Ochratoxin A and human health risk: A review of the evidence. Crit. Rev. Food Sci. 2015;55:1860–1869. doi: 10.1080/10408398.2012.724480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorrenti V., Di Giacomo C., Acquaviva R., Barbagallo I., Bognanno M., Galvano F. Toxicity of ochratoxin A and its modulation by antioxidants: A review. Toxins. 2013;5:1742–1766. doi: 10.3390/toxins5101742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfohlleszkowicz A., Manderville R.A. An update on direct genotoxicity as a molecular mechanism of ochratoxin A carcinogenicity. J. Chem. Res. in Toxicol. 2012;25:252–262. doi: 10.1021/tx200430f. [DOI] [PubMed] [Google Scholar]
- 12.NHC . Food Safety National Standard. Volume GB 2716–2017 National Health Commission; Beijing, China: 2017. Limits of mycotoxins in foods. [Google Scholar]
- 13.Abrunhosa L., Paterson R.R., Venancio A. Biodegradation of ochratoxin A for food and feed decontamination. Toxins. 2010;2:1078–1099. doi: 10.3390/toxins2051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlovsky P. Biological detoxification of fungal toxins and its use in plant breeding, feed and food production. Nat. Toxins. 1999;7:1–23. doi: 10.1002/(SICI)1522-7189(199902)7:1<1::AID-NT37>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Piotrowska M. The adsorption of ochratoxin A by lactobacillus species. Toxins. 2014;6:2826–2839. doi: 10.3390/toxins6092826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farbo M.G., Urgeghe P.P., Fiori S., Marceddu S., Jaoua S., Migheli Q. Adsorption of ochratoxin A from grape juice by yeast cells immobilised in calcium alginate beads. Int. J. Food. Microbiol. 2016;217:29–34. doi: 10.1016/j.ijfoodmicro.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Wu Q., Dohnal V., Huang L., Kuca K., Wang X., Chen G., Yuan Z. Metabolic pathways of ochratoxin A. Curr. Drug Metab. 2011;12:1–10. doi: 10.2174/138920011794520026. [DOI] [PubMed] [Google Scholar]
- 18.Li S., Marquardt R., Frohlich A., Vitti T., Crow G. Pharmacokinetics of ochratoxin A and its metabolites in rats. Toxicol. Appl. Pharm. 1997;145:82–90. doi: 10.1006/taap.1997.8155. [DOI] [PubMed] [Google Scholar]
- 19.Bruinink A., Rasonyi T., Sidler C. Differences in neurotoxic effects of ochratoxin A, ochracin and ochratoxin α in vitro. Nat. Toxins. 1998;6:173–177. doi: 10.1002/(SICI)1522-7189(199809/10)6:5<173::AID-NT10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H.H., Wang Y., Zhao C., Wang J., Zhang X.L. Biodegradation of ochratoxin A by alcaligenes faecalis isolated from soil. J. Appl. Microbiol. 2017;123:661–668. doi: 10.1111/jam.13537. [DOI] [PubMed] [Google Scholar]
- 21.Rawlings N.D., Barrett A.J. Evolutionary families of metallopeptidases. Method. Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 22.Chen W., Li C., Zhang B., Zhou Z., Shen Y., Liao X., Yang J., Wang Y., Li X., Li Y., et al. Advances in biodetoxification of ochratoxin A-A review of the past five decades. Front. Microbiol. 2018;9:1386. doi: 10.3389/fmicb.2018.01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez H., Reveron I., Doria F., Costantini A., Rivas B.D.L., Muňoz R., Garcia-Moruno E. Degradation of ochratoxin A by brevibacterium species. J. Agr. Food Chem. 2011;59:10755–10760. doi: 10.1021/jf203061p. [DOI] [PubMed] [Google Scholar]
- 24.Pitout M.J. The hydrolysis of ochratoxin A by some proteolytic enzymes. Biochem. Pharmacol. 1969;18:485–491. doi: 10.1016/0006-2952(69)90224-X. [DOI] [PubMed] [Google Scholar]
- 25.Liuzzi V.C., Fanelli F., Tristezza M., Haidukowski M., Picardi E., Manzari C., Lionetti C., Grieco F., Logrieco A.F., Thon M.R. Transcriptional analysis of acinetobacter sp. Neg1 capable of degrading ochratoxin A. Front. Microbiol. 2016;7:2162. doi: 10.3389/fmicb.2016.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abrunhosa L., Santos L., Venâncio A. Degradation of ochratoxin A by proteases and by a crude enzyme of aspergillus niger. Food Biotechnol. 2006;20:231–242. doi: 10.1080/08905430600904369. [DOI] [Google Scholar]
- 27.Stander M.A., Bornscheuer U.T., Henke E., Steyn P.S. Screening of commercial hydrolases for the degradation of ochratoxin A. J. Agr. Food Chem. 2000;48:5736–5739. doi: 10.1021/jf000413j. [DOI] [PubMed] [Google Scholar]
- 28.Abrunhosa L., Venancio A. Isolation and purification of an enzyme hydrolyzing ochratoxin A from aspergillus niger. Biotechnol. Lett. 2007;29:1909–1914. doi: 10.1007/s10529-007-9479-2. [DOI] [PubMed] [Google Scholar]
- 29.Dobritzsch D., Wang H., Schneider G., Yu S. Structural and functional characterization of ochratoxinase, a novel mycotoxin-degrading enzyme. Biochem. J. 2014;462:441–452. doi: 10.1042/BJ20140382. [DOI] [PubMed] [Google Scholar]
- 30.Lee S.B., Taylor J.W. PCR protocols: A guide to methods and applications. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. Molecular Reproduction and Development. Volume 28. Academic Press; San Diego, CA, USA: 1990. pp. 282–287. [Google Scholar]
- 31.Petersen T.N., Brunak S., Von H.G., Nielsen H. Signalp 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Boil. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.