Abstract
The diffusion of Panton–Valentine leukocidin (PVL)–positive methicillin-resistant S. aureus (MRSA) is a health problem in Algeria. The objectives of the study were to investigate the global distribution of methicillin-susceptible S. aureus (MSSA) and MRSA isolates in different ecological niches in this country. In total, 2246 samples were collected from humans, livestock, wild animals, pets, food products and the aquatic environment, from 12 Algerian provinces. A total of 312 S. aureus were detected from 2446 samples (12.7%) in the screened niches. We observed the emergence of toxinogenic S. aureus representing 41% of the isolates. Among them, we noted the diffusion of ST80-IV CA-MRSA PVL + strains isolated in human, animals, and food and genetic diversity of MSSA PVL + isolates. This study suggests an alarming dissemination of MRSA-ST80 PVL + in both human and extra-human sources in Algeria. Moreover, MSSA may become a permanent reservoir of the PVL genes necessary for human infections.
Keywords: MRSA-ST80, PVL, TSST-1, ecological niches, Algeria, one health, Staphylococcus aureus, epidemiology
1. Introduction
Staphylococcus aureus constitutes a serious public health concern due to its ability to colonize and/or infect humans and animals [1,2,3]. S. aureus is the most frequently occurring pathogen in hospitals and the second most common pathogen in outpatient settings [4]. Following the introduction of anti-staphylococcal penicillins, the emergence and diffusion of methicillin-resistant S. aureus (MRSA) rapidly appeared in hospitals [1,2]. Successively, a series of predominant clonal strains was characterized. The most important recent clones found in the world are the health-care associated MRSA (HA-MRSA) ST5 and ST239, the community-associated MRSA (CA-MRSA) USA300 and the livestock-associated MRSA (LA-MRSA) ST398. In parts of western Europe, northern Africa and the Middle East, the European ST80 CA-MRSA represents one of the main diffusing clones [1,2].
The pathogenicity of S. aureus arises from a large arsenal of virulence factors such as staphylococcal enterotoxins (SEs), leukotoxins, hemolysins, exfoliative toxin (ET), toxic shock syndrome toxin-1 (TSST-1) and Panton–Valentine leucocidin (PVL) [5]. The success of some dominant clonal MRSA strains is due to their chromosomal integration of different virulence traits acquired through mobile genetic elements (MGEs) that change the overall fitness of specific clones [6]. Thus, the acquisition of arginine-catabolic mobile element (ACME) has been associated with the spread of USA300 [7]. Moreover, the PVL, a pore-forming cytotoxic secreted toxin encoded in prophage Sa2int, has been associated with severe S. aureus pneumonia and prototypical skin lesions [8]. This toxin has been linked to CA-MRSA disease worldwide, even though some of these strains do not carry the PVL genes [9].
In Algeria, the ‘European’ ST80-IV CA-MRSA PVL + clone is widely disseminated in both community and healthcare settings and has given rise to a pandemic clone [2]. Prevalence of ST80-IV CA-MRSA among CA-MRSA isolates varied from 20.7% in Eastern Algeria [10] to 96% in Algiers [11]. However, Algeria and, more widely, Africa has the largest gaps in data on the prevalence of antimicrobial resistance in large reservoirs such as livestock, wild animals or food. While different reports have demonstrated the presence of MRSA in pets, livestock or wild terrestrial or aquatic species [2], no data are available on the prevalence and dissemination of methicillin-sensitive S. aureus (MSSA)-PVL + and MRSA-PVL + in extra-human niches, such as farm animals, pets, wild animals, environmental sources, and food products in the same geographical zone [12,13]. The aims of this study were to: (i) estimate the prevalence of S. aureus strains recovered in a large collection of samples obtained from different ecological niches distributed among 12 Algerian provinces, (ii) describe the different clones diffusing in these niches and (iii) characterize the genetic contexts of MRSA-PVL + and MSSA-PVL + strains.
2. Results
2.1. Population of the Isolates
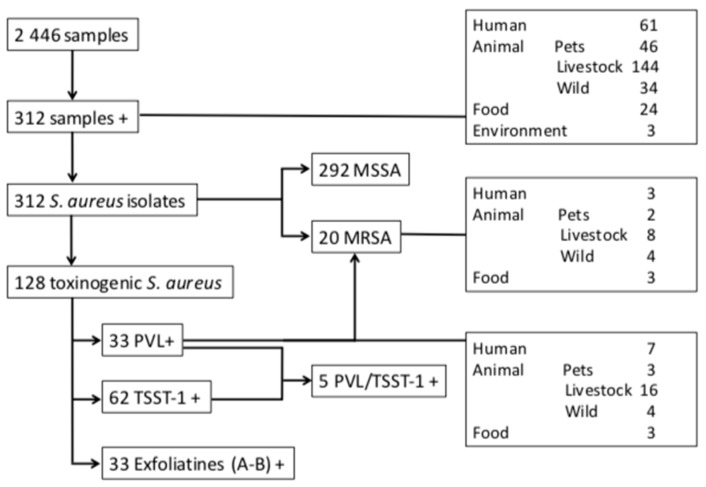
A total of 312 S. aureus isolates obtained from 312 samples were identified giving an overall prevalence of 12.7% (312/2 446) (Figure 1). The isolates were recovered from all the screened niches in the 12 provinces: humans (n = 61; 19.3%), farm animals (n = 144; 18%), pets (n = 46; 14%), wild animal (n = 34; 5.2%), food products (n = 24; 8.6%), and the aquatic environment (n = 3; 4%) (Tables S1 and S2).
Figure 1.
Distribution of the Staphylococcus aureus strains isolated from different ecological niches in Algeria.
2.2. Antimicrobial Susceptibility Profiles
The results of the antimicrobial susceptibility of the S. aureus isolates are listed in Table 1. Of the 312 S. aureus isolates tested, 20 (6.4%) were MRSA harboring mecA gene. In contrast, no mecC gene was detected. The prevalence of MRSA in the different niches was as follows (Figure 1): farm animals (n = 8/147, 5.4%), wild animals (n = 4/29, 13.7%), food of animal origin (n = 3/22, 13.6%), human (n = 3/61, 4.9%) and pets (n = 2/46, 4.3%). The strains recovered from humans and animals were isolated from nasal swabs (n = 11), feces (n = 3), oral cavity (n = 2) and rectal swab (n = 1). No MRSA strains were detected in environmental samples.
Table 1.
Resistance profiles of S. aureus strains isolated from different niches in 12 Algerian provinces*.
| Antibiotics | Origin of Isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Bovine | Ovine | Goats | Rabbits | Poultry | Pets | Wild Animals | Food Products | Aquatic Environment | Total Samples | |
| Total | 61 | 18 | 34 | 8 | 15 | 69 | 46 | 34 | 24 | 3 | 312 |
| Penicillin G | 51 (83.6) | 11 (61.1) | 19 (55.8) | 4 (50) | 8 (53.3) | 51 (73.9) | 28 (60.8) | 25 (73.5) | 11 (45.8) | 1 (33.3) | 209 (66.9) |
| Cefoxitin | 3 (4.9) | 1 (5.5) | 1 (2.9) | 0 (0) | 3 (20) | 3 (4.3) | 2 (4.3) | 5 (14.7) | 2 (8.3) | 0 (0) | 20 (6.4) |
| Erythromycin | 5 (8.1) | 0 (0) | 1 (2.9) | 0 (0) | 0 (0) | 6 (8.6) | 4 (8.6) | 2 (5.8) | 6 (25) | 0 (0) | 24 (7.6) |
| Ceftobiprole | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Clindamycin | 1 (1.6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0.3) |
| Quinupristin/Dalfopristin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Kanamycin | 4 (6.5) | 3 (16.6) | 4 (11.7) | 0 (0) | 3 (20) | 4 (5.7) | 2 (4.3) | 5 (14.7) | 4 (16.6) | 1 (33.3) | 30 (9.6) |
| Tobramycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (8.3) | 0 (0) | 2 (0.6) |
| Gentamicin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Minocycline | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1.4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0.3) |
| Ofloxacin | 2 (3.2) | 0 (0) | 0 (0) | 0 (0) | 2 (13.3) | 1 (1.4) | 1 (2.1) | 3 (8.8) | 2 (8.3) | 0 (0) | 11 (3.5) |
| Fusidic acid | 1 (1.6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1.4) | 0 (0) | 5 (14.7) | 1 (4.1) | 0 (0) | 8 (2.5) |
| Fosfomycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Rifampicin | 3 (4.9) | 1 (5.5) | 1 (2.9) | 0 (0) | 3 (20) | 3 (4.3) | 2 (4.3) | 7 (20.5) | 2 (8.3) | 0 (0) | 22 (7.0) |
| Cotrimoxazole | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
*Values indicate the number of isolates tested with the percentage of resistance in parenthesis (%).
Concerning the co-resistances, most S. aureus showed a high prevalence of resistance to penicillin (n = 209, 67%), followed by variable resistance levels to kanamycin (n = 30, 9.6%), erythromycin (n = 24, 7.7%), rifampicin (n = 22, 7%), ofloxacin (n = 11, 3.5%), fusidic acid (n = 8, 2.5%), tobramycin (n = 2, 0.6%), minocycline (n = 1, 0.3%), and clindamycin (n = 1, 0.3%). All isolates were susceptible to ceftobiprole, quinupristin/dalfopristin, gentamicin, fosfomycin and cotrimoxazole. In comparison to MSSA isolates, the 20 MRSA isolates were statistically more frequently resistant to erythromycin (30% vs. 5.4%, p = 0.001), ofloxacin (45% vs. 0.6%, p < 0.001), fusidic acid (15% vs. 0%, p < 0.001), kanamycin (100% vs. 3.7%, p < 0.001), and rifampicin (100% vs. 0.6%, p < 0.001) and harbored a MLSb phenotype (35% vs. 4.7%, p < 0.001). No statistical difference was observed between the different niches.
2.3. Importance of Toxinogenic S. Aureus
Of the 312 S. aureus isolates recovered from different sources, 128 (41.0%) were toxinogenic with 33 producing PVL (10.6%), 62 TSST-1 (19.8%), and 33 exfoliatins A and B (10.6%) (Figure 1). Among these strains, four co-harbored PVL and TSST-1, one PVL/TSST-1/EdinA, six TSST-1 and an exfoliatin, and 25 PVL + were also EtD + /EdinB + (Table S3).
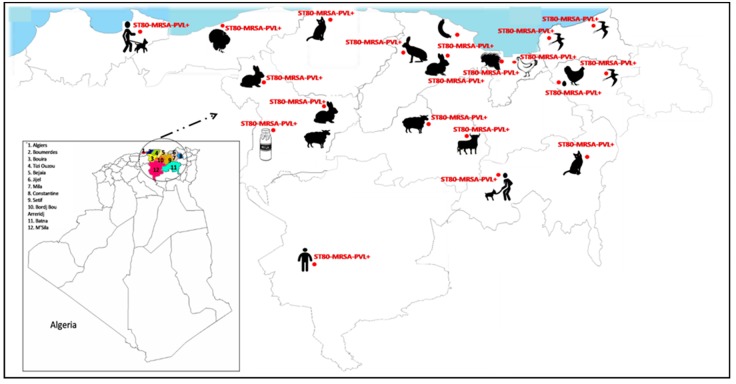
The PVL + strains were isolated from different sources including animal farms (n = 16, 48.5%), human (n = 7, 21.2%), wild animals (n = 4, 12.1%), cats (n = 3, 9.1%), and food of animal origin (n = 3, 9.1%) (Figure 1 and Figure 2).
Figure 2.
Distribution of the ST80-IV community-associated methicillin-resistant S. aureus (CA-MRSA) and the methicillin-susceptible S. aureus (MSSA) Panton–Valentine leukocidin (PVL) positive clones circulating in different ecological niches in Algeria.
The TSST-1 + strains were isolated from humans (n = 18), farm animals (n = 29), pets (n = 13), wild animals (n = 1) and environment (n = 1). Interestingly, the majority of ETs + strains (24/33, 72.7%) were detected from laying hens (5 etA + and 19 etB +) isolated in different provinces (Table S3). The others were isolated from humans (3 etA + and 1 etB +), rabbits (2 etB +), broilers (1 etB +), eggs (1 etB +) and dog (1 etB +). Finally, the 25 EtD + isolates were PVL +.
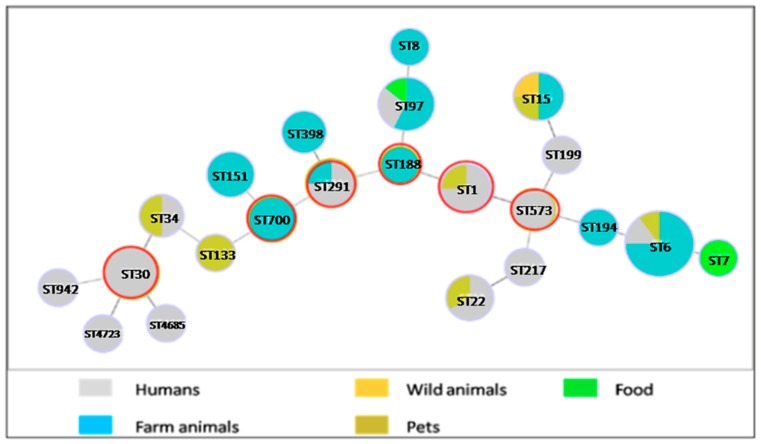
The Figure 3 shows the goeBURST analysis of the different toxinogenic MSSA-STs identified according to their origin.
Figure 3.
Population snapshot of the methicillin-sensitive Staphylococcus aureus MultiLocus Sequence Typing (MLST) dataset generated using the goeBURST algorithm, indicating the sequence types (STs) and their distribution in different sources. The dominant STs are represented by the circles with larger diameters. The founder STs are represented by a red circle.
Twenty-two STs of this study formed within six groups with ST1, ST30, ST188, ST291, ST573 and ST700 as predicted founders. The formation of four double locus variants (DLV) and seven triple locus variants (TLV) among STs was observed, while no singleton was found. ST6 was the dominant clone (28.5%) recovered among 22 STs identified in this study, distributed in farm animals, humans and pets. The majority of the human isolates were concentrated in ST30 (8.5%), and ST291 (4.2%). Most farm animal isolates were recovered in ST6 followed by ST97, ST151 and ST700. ST133 and ST7 were found in wild animal and food, respectively. Pet isolates were identified in ST15, ST1, ST22 and ST34.
2.4. Characteristics of Panton–Valentine Leukocidin (PVL) + S. aureus
Of the 33 PVL + S. aureus isolates recovered from different sources, 20 (60.6%) isolates were MRSA and 13 (39.4%) were MSSA. All the MRSA-PVL + isolates belonged to the European ST80-IV CA-MRSA clone and were isolated from humans, animals and food (Figure 1 and Figure 2). The MSSA-PVL + isolates had a more diverse origin: ST6 (n = 7), ST1 (n = 2), ST15 (n = 1), ST398 (n = 1), ST942 (n = 1), and ST97 (n = 1) (Figure 3) and were detected from pets, livestock and humans (Table 2).
Table 2.
Distribution of the main virulence and resistance genes among the S. aureus PVL + isolated from different ecological niches in Algeria.
| Virulence and Resistance Genes | MRSA-PVL + no. (%) |
MSSA-PVL + no. (%) |
p Value |
|---|---|---|---|
| Virulence Genotyping | |||
| Enterotoxins | |||
| sea | 0 (0) | 8 (61.5) | < 0.001 |
| seb | 0 (0) | 2 (15.3) | NS |
| seg | 0 (0) | 2 (15.3) | NS |
| seh | 0 (0) | 3 (23) | 0.05 |
| sei | 0 (0) | 5 (38.4) | 0.005 |
| sek | 0 (0) | 1 (7.6) | NS |
| seq | 0 (0) | 1 (7.6) | NS |
| egc clustera | 0 (0) | 0 (0) | NS |
| Other Toxins | |||
| tst | 0 (0) | 5 (38.4) | 0.005 |
| etA | 0 (0) | 0 (0) | NS |
| etB | 0 (0) | 0 (0) | NS |
| etD | 20 (100) | 5 (38.4) | 0.005 |
| edinA | 0 (0) | 1 (7.6) | NS |
| edinB | 20 (100) | 5 (38.4) | 0.005 |
| Leukocidins | |||
| lukD | 20 (100) | 12 (92.3) | NS |
| lukE | 19 (95) | 12 (92.3) | NS |
| lukX | 7 (35) | 13 (100) | < 0.001 |
| lukY | 15 (75) | 13 (100) | NS |
| Hemolysins | |||
| hla | 10 (50) | 12 (92.3) | 0.02 |
| hlb | 15 (75) | 6 (46.1) | NS |
| hld | 20 (100) | 13 (100) | NS |
| hlgA | 20 (100) | 13 (100) | NS |
| hlgv | 20 (100) | 13 (100) | NS |
| Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSRAMMs) | |||
| bbp | 20 (100) | 11 (84.6) | NS |
| clfA | 20 (100) | 13 (100) | NS |
| clfB | 20 (100) | 13 (100) | NS |
| cna | 0 (0) | 5 (38.4) | 0.005 |
| ebpS | 20 (100) | 13 (100) | NS |
| fib | 20 (100) | 12 (92.3) | NS |
| fnbA | 20 (100) | 12 (92.3) | NS |
| fnbB | 20 (100) | 13 (100) | NS |
| Capsule Components | |||
| cap5 | 0 (0) | 3 (23) | 0.05 |
| cap8 | 20 (100) | 10 (76.9) | NS |
| Intracellular Adhesion Polysaccharide | |||
| icaA | 20 (100) | 13 (100) | NS |
| icaC | 20 (100) | 13 (100) | NS |
| icaD | 20 (100) | 13 (100) | NS |
| Immune Evasion Cluster and Other | |||
| sak | 20 (100) | 10 (76.9) | NS |
| chp | 0 (0) | 3 (23) | 0.05 |
| scn | 17 (85) | 11 (84.6) | NS |
| ACME clusterb | 0 (0) | 0 (0) | NS |
| Accessory Gene Regulators | |||
| agr1 | 0 (0) | 9 (69.2) | < 0.001 |
| agr2 | 0 (0) | 1 (7.6) | NS |
| agr3 | 20 (100) | 3 (23) | < 0.001 |
| agr4 | 0 (0) | 0 (0) | NS |
| Resistance Genotyping | |||
| mecA | 20 (100) | 0 (0) | < 0.001 |
| mecC | 0 (0) | 0 (0) | NS |
| blaZ | 2 (10) | 10 (76.9) | < 0.001 |
| ermA | 0 (0) | 0 (0) | NS |
| ermC | 3 (15) | 0 (0) | NS |
| aphA3 | 20 (100) | 1 (7.6) | < 0.001 |
| sat | 20 (100) | 1 (7.6) | < 0.001 |
| fosB | 1 (5) | 3 (23) | NS |
aegc cluster corresponds to seg, sei, sem, sen and seo genes; b ACME (arginine catabolic mobile element) includes the arc genes (arcA to arcD) and the oligopeptide permease operon genes (opp-3A to opp-3E). NS, not significant (p > 0.05).
The virulence profiles of MRSA and MSSA-PVL + isolates are presented in Table 2. The MRSA-PVL + isolates had some virulence genes significantly associated with their profile compared to MSSA-PVL +. All these isolates harbored the etD gene compared to 5 MSSA-PVL + isolates (p = 0.005). As the edinB gene is on the same operon as etD, the same profile was seen for both (p = 0.005). Moreover, agr3 was significantly predominant in MRSA-PVL + isolates (p < 0.001). Conversely, the MSSA-PVL + isolates harbored significantly more enterotoxins- (sea, seh and sei), leukocidin- (lukX), hemolysin- (hla), MSCRAMM- (cna), capsule- (cap5), and an immune evasion cluster- (chp) encoding genes. While most of the hemolysins-, leukocidins- and MSCRAMMs-encoding genes were present in both MSSA- and MRSA-PVL + isolates, enterotoxins and cap5 genes were absent in MRSA-PVL + isolates. No strain harbored the ACME cluster. Finally, the regulator agr1 gene was significantly predominant in these isolates (p < 0.001).
2.5. Characteristics of the Other Toxinogenic S. aureus
The main characteristics of virulence profile of the other toxinogenic S. aureus isolates are presented in Table 3. We noted that six TSST-1 + isolates harbored etA (n = 2) and etB (n = 4) genes. The TSST-1 + isolates presented significantly more enterotoxins (seg, seh) compared to non-toxinogenic isolates (p < 0.001). These isolates were also significantly associated with the edinB gene (p < 0.001) but no etB gene was detected. The seg gene was significantly associated with the ET + isolates (p < 0.001).
Table 3.
Distribution of the main virulence genes among the toxinogenic and non toxinogenic S. aureus (excluded PVL +) isolated from different ecological niches in Algeria.
| Virulence and Resistance Genes | TSST-1 + no. (%) |
Et + no. (%) |
Non Toxinogenic MSSA no. (%) |
p valuea |
|---|---|---|---|---|
| Isolates | n = 62 | n = 33 | n = 203 | |
| Enterotoxins | ||||
| sea | 36 (58.0) | 26 (78.7) | 115 (56.6) | NS |
| seb | 7 (11.2) | 2 (6.0) | 14 (6.8) | NS |
| seg | 43 (69.3) | 30 (90.9) | 38 (18.7) | < 0.001 |
| seh | 18 (29.0) | 0 (0) | 16 (7.8) | 0.001 |
| sei | 62 (100) | 32 (96.9) | 179 (88.1) | NS |
| sek | 13 (20.9) | 2 (6.0) | 20 (9.8) | NS |
| seq | 13 (20.9) | 2 (6.0) | 16 (7.8) | NS |
| Other Toxins | ||||
| tst | 62 (100) | 6 (18.1) | 0 (0) | < 0.001 |
| etA | 2 (3.2) | 8 (24.2) | 0 (0) | < 0.001 |
| etB | 4 (6.4) | 25 (75.7) | 0 (0) | < 0.001 |
| etD | 0 (0) | 0 (0) | 0 (0) | NS |
| edinA | 0 (0) | 0 (0) | 0 (0) | NS |
| edinB | 24 (38.7) | 1 (3.0) | 18 (8.8) | < 0.001 |
| Leukocidins | ||||
| lukD | 62 (100) | 33 (100) | 203 (100) | NS |
| lukE | 62 (100) | 33 (100) | 203 (100) | NS |
| Hemolysins | ||||
| hla | 55 (88.7) | 33 (100) | 191 (94.0) | NS |
| hlb | 24 (38.7) | 1 (3.0) | 43 (21.1) | NS |
| hld | 62 (100) | 33 (100) | 203 (100) | NS |
| hlgA | 49 (79.0) | 33 (100) | 188 (92.6) | NS |
| hlgv | 60 (96.7) | 32 (96.9) | 197 (97.0) | NS |
| MSCRAMMs | ||||
| bbp | 62 (100) | 33 (100) | 203 (100) | NS |
| clfA | 62 (100) | 33 (100) | 203 (100) | NS |
| clfB | 62 (100) | 33 (100) | 203 (100) | NS |
| cna | 0 (0) | 0 (0) | 14 (6.8) | 0.006 |
| ebpS | 62 (100) | 33 (100) | 203 (100) | NS |
| fib | 55 (88.7) | 32 (96.9) | 187 (92.1) | NS |
| fnbA | 10 (16.1) | 7 (21.2) | 26 (12.8) | NS |
| fnbB | 28 (45.1) | 6 (18.1) | 62 (30.5) | NS |
| Capsule Components | ||||
| cap5 | 35 (56.5) | 20 (60.6) | 138 (68.0) | NS |
| cap8 | 27 (44.5) | 13 (39.4) | 65 (32.0) | NS |
| Intracellular Adhesion Polysaccharide | ||||
| icaA | 62 (100) | 33 (100) | 203 (100) | NS |
| icaC | 62 (100) | 33 (100) | 203 (100) | NS |
| icaD | 62 (100) | 33 (100) | 203 (100) | NS |
| Other | ||||
| ACME clusterb | 0 (0) | 0 (0) | 0 (0) | NS |
| Accessory Gene Regulators | ||||
| agr1 | 42 (67.7) | 25 (75.7) | 153 (75.3) | NS |
| agr2 | 6 (9.6) | 3 (9.0) | 31 (15.2) | NS |
| agr3 | 14 (22.5) | 2 (6.0) | 19 (9.3) | NS |
| agr4 | 0 (0) | 3 (9.0) | 0 (0) | NS |
ap value corresponds to comparison between toxinogenic and non toxinogenic S. aureus isolates using Fisher exact test. b ACME (arginine catabolic mobile element) includes the arc genes (arcA to arcD) and the oligopeptide permease operon genes (opp-3A to opp-3E). NS, not significant (p > 0.05).
3. Discussion
MRSA isolates remain a worldwide public health problem and have been reported from both hospital and non-hospital reservoirs. In Algeria, the main studies on MRSA have been published from hospital settings. They showed that this resistance mechanism was endemic, involved in a majority of infection and was mainly due to the diffusion of the ST80-IV CA-MRSA clone carrying the PVL genes among the CA-MRSA isolated: 20.7% in Eastern Algeria [10], 73.9% in Western Algeria [14], and 35.7 to 96% in different hospitals of Algiers [11,15,16]. Nasal carriage of this clone has been also described in hospital but with a low prevalence (4/159 enrolled patients) [17]. Finally, the presence of this clone was also found in animals (healthy sheep and camels) [18]. Recently, the CDC proposed the “One Health” concept to organize a worldwide strategy for expanding interdisciplinary collaborations and communications in all aspects of health care for humans, animals and the environment [19]. The authors encouraged prospective prevalence studies to better define the dissemination of the multidrug resistant bacteria, resulting in studies showing the CA-MRSA transmission between livestock and humans especially concerning the ST398 clone [1,2]. Due to the importance of this problem and the unclear and neglected situation of the epidemiology in Algeria, we sought to describe the prevalence, geographic distribution and clonal expansion of MRSA (particularly the ST80-IV CA-MRSA clone) in different ecological niches (human, food, animals, and water) in 12 Algerian provinces. The knowledge of the host specificity and geographic distribution of these bacteria are important parameters for understanding ecology, epidemiology, and diffusion of this resistance gene. Therefore, our study has established, for the first time to our knowledge, a valuable identification of the ST80-IV CA-MRSA isolates recovered from different sources and their virulence profiles.
Across a wide range of different reservoirs screened, it is interesting to note that ST80-IV CA-MRSA PVL + was the single methicillin-resistant clone found in our study, representing an overall prevalence of 6.4% of all the environments screened (except aquatic environment). This wide diffusion is of importance notably due to the presence of this clone in niches other than humans. This highlights the worrisome situation of multidrug resistance in this country. Our results also show the dynamic capacity of adaptation of the ST80-IV CA-MRSA isolates, and highlight that farm animals, companion animals, wildlife, food, etc. may become a permanent reservoir for human infections. The hospital environment is a visible threat for this country, according to the main studies, however, the community is not spared. Several practices allow the spread of hospital isolates to the community, and vice versa [20]. Therefore, even though control measures can be taken at the hospital, the eradication of this clone will be nearly impossible due to its presence in the community. Surveillance programs outside the hospital will need to be implemented to control its spread.
Although pvl genes were associated with ST80-IV CA-MRSA, our results also showed that some MSSA PVL + isolates emerged from different sources. While the distribution of MSSA PVL + clones is scarcely described in Algeria, it is important to quantify their prevalence because these strains are frequently associated with skin and soft-tissue infections in Algerian hospitals. Their potential to cause infections in humans through the food chain similarly needs attention. Six STs of MSSA-PVL + were found (ST1, ST6, ST15, ST97, ST398, and ST942), mainly from farm animals. All of these STs have been previously described by Agabou et al. in Algeria [18]. If ST15-PVL + MSSA is a very common worldwide strain reported in many countries [21,22,23], the ST6-PVL + MSSA was more unusual and has previously been identified in reports from Africa, as well as from China [22,24,25].The ST97-PVL + MSSA has been reported only in one human case from Bali [26]. The ST398 is frequently isolated from pigs [1,2,27] and the ST97 is globally linked with bovine mastitis [2,28,29]. In addition, all these types of MSSA have been reported from a variety of clinical infections [30,31,32,33,34].
One of the other main findings in our study is the high prevalence of toxinogenic MSSA strains isolated from different niches screened, in particular the TSST-1 + MSSA isolates (19.8%). This virulence factor is rare in human in Algeria [15,35] but was previously described in farm animals and food product [18,36]. Twenty-one MSSA TSST-1 + clones were distributed across different sources comprising a wide range of clones. These findings demonstrated a large diversity among strains obtained from different sources. The S. aureus pathogenicity islands containing the tst gene are classically mobilized in high frequency following infection by some bacteriophages [37]. Using the PHYLOVIZ algorithm, which provides a hypothetical pattern of descent for the analyzed isolates, we concluded that most of the PVL + and TSST-1 + MSSA isolates of this study formed different clonal complexes, suggesting a high genotypic diversity. ST6 was found to be the predominant ST representing about 23.5% of the total STs, which was recovered in farm animals, humans and pets suggesting that the strain may be transmissible across different host species. This clone had previously been reported from humans (MSSA and MRSA), feral cat (MRSA) and camel (MSSA) [18,38,39]. ST30 was only recovered from human isolates [40]. The clustered MSSA isolates included in this study demonstrated different clone variations circulating from humans, farm animal and pets.
Interestingly, S. aureus can generate genetic variations in its genome mainly by horizontal gene transfer enabling it to adapt rapidly to new ecological niches [6,7]. The introduction of new gene combinations may play an important role in the increase of the pathogenicity or the diffusion of the bacterium and in the better adaptation to new hosts [2]. Here, the presence of some genes in strains isolated from animals suggests a human origin. For example, chp, ebh, fnbA, hlb, sak, scn, sea genes have been associated with a human specificity and their absence may be a valuable indicator of S. aureus animal adaptation [41]. We observed this trend with a low prevalence of these markers in the animal isolates (16/224 for fnbA, 91/224 for sea or 25/224 for hlb). Moreover, fnbA, ebh, sak, hlb (15/20 isolates) and scn (17/20 isolates) genes were detected in ST80-IV CA-MRSA isolates, which confirms their human origin [42]. Finally, the detection of the edinB and etD genes in these MRSA isolates illustrates the dissemination of toxinogenic strains with multiple profiles (PVL +, TSST-1 +, EdinB + and EtD +) and a high pathogenicity potential relevant for human and animal health. This also demonstrates the high potential of MGEs such as phages (for lukS-PV and lukF-PV) or pathogenicity islands (for tst) to appear in a single isolate, to reach different STs (3 for 5 isolates) and to diffuse in different environments (ovine, cat, horse owner, farmer) suggesting that bacteriophages could have originally allowed the spread of these genes among lineages, as previously hypothesized [43]. The combination of major virulence factors in the same strain must be further studied. Their persistence and expansion will depend on the fitness cost for the bacteria [44]. Thus, the presence of these multiple toxinogenic genes in S. aureus isolated in different ecosystems may pose a public health risk by convergences between habitats leading to frequent contact between animals, environment and humans.
4. Conclusions
This study suggests a high prevalence and a wild diffusion of toxinogenic S. aureus isolates and especially the ST80-IV CA-MRSA clones among different niches in 12 Algerian provinces. This wide dissemination in different niches other than the clinical setting may constitute a reservoir of resistant strains, which could be responsible for transmission from humans to their cohabitants and within veterinary hospitals. This dissemination is certainly due to the excessive use of antimicrobial agents and the presence of antibiotics in different environments (animals, food, soil, waste water) as previously suggested [45], along with insufficient infection control measures, and further studies are required to elucidate the cycle of transmission of these bacteria between these different habitats and to explore the origin of MRSA isolates circulating outside hospitals. This is of importance due to the high potential of virulence of these isolates.
5. Materials and Methods
5.1. Sampling and Microbiological Procedures
The study was approved by the local Ethical Committee (Béjaia, Algeria). The approval date is 17 October 2017. The humans included in this study gave their consent. From January to July 2018, different randomly and prospectively selected niches were chosen, localized in 12 provinces in Algeria (Bejaia, Tizi Ouzou, Bouira, Jijel, Mila, Setif, Algiers, Boumerdes, Constantine, Batna, M’Sila and Bordj Bou Arreridj). This large group of samples was then randomized for testing. A total of 2446 samples were randomly collected, including samples from humans (community, n = 100; pet’ owners, n = 181 and farmers, n = 35), farm animals (n = 800), pets (n = 328), wild animals (n = 652), food products (n = 276) and water environment (n = 74) (Tables S1 and S2).
Humans and domestic animals screening was by nasal swabs, whilst wild animals and fish were screened via fresh fecal droppings and intestinal content samples, respectively. Poultry were screened for S. aureus buccal and rectal carriage. Local food products (chicken, meat, sausage, milk and eggs) were randomly obtained from different markets, stores and farms of the 12 studied provinces. Samples of aquatic environment were collected from seawater (Mediterranean Sea, n = 22), spring water (n = 16), rivers (n = 5), lakes (n = 7), dam water (n = 12) and fountains (n = 12). One hundred milliliters of milk and water samples were taken in sterile flasks.
All samples were immediately transported at + 4 °C to the Microbiological Ecology Laboratory at the University of Bejaia (Algeria) for analysis. They were processed within the day after sampling. The samples were cultured in trypticase soy broth (TSB) (Fluka, St Louis, MO, USA) supplemented with colistin (10 mg/L), aztreonam (10 mg/L) and a mphotericin B (2 mg/L) and incubated for 18 h at 37 °C. For food products, 25 g of each food item or 25 mL of milk were cultured in 225 mL of TSB supplemented with the same antibiotics as described above and incubated for 18 h at 37 °C. Following incubation, a 200 μL aliquot was plated onto mannitol salt agar (Fluka, St Louis, MO, USA) incubated for 24 and 48 h at 37 °C. For water, the procedure of isolation consisted on inoculation of 200 μL aliquot of 1:10 diluted samples on mannitol salt agar.
Presumptive isolates were sub-cultured on TSA and identified as S. aureus by the Vitek MS® system (bioMérieux, Marcy l’Etoile, France).
5.2. Antibiotic Susceptibility Testing
Susceptibility to 14 antimicrobial agents (penicillin G, cefoxitin, erythromycin, clindamycin, quinupristin/dalfopristin, kanamycin, tobramycin, gentamicin, minocycline, ofloxacin, fusidic acid, fosfomycin, rifampicin and cotrimoxazole) was tested by the disk diffusion method on Mueller Hinton agar (BioRad, Marnes La Coquette, France) according to recommendations of EUCAST 2018 [46]. A cefoxitin disk was used to screen the MRSA isolates. To confirm the presence/absence of mecA and mecC genes, we performed PCR as previously described [47].
5.3. DNA Arrays Procedures
All the S. aureus strains isolated during the study were then analyzed by Alere Staphy Type DNA microarray according to protocols and procedures previously detailed [38,48]. This array simultaneously detects 333 S. aureus target sequences, including species markers, antimicrobial resistance and virulence-associated genes, and SCCmec-associated genes and typing markers, allowing isolates to be assigned to MultiLocus Sequence Typing (MLST) sequence types (STs), clonal complexes (CCs) and SCCmec types. Primer and probe sequences have been previously published [48]. Raw data were interpreted as “positive”, “negative” or “ambiguous” using a previously described algorithm [48]. The affiliation of isolates to CCs or STs as defined by MLST was determined by an automated comparison of hybridization profiles with a collection of reference strains previously characterized [49].
5.4. MultiLocus Sequence Typing (MLST)
To confirm array results, MLST analysis was performed on the toxinogenic MSSA isolates as previously described [50]. Seven housekeeping genes (arc, aroE, glpF, gmk, pta, tpi and yqi) were sequenced to determine allelic profile. Strains were assigned to a ST using the MLST database [51].
5.5. goeBURST Analysis of Methicillin-Susceptible S. Aureus (MSSA) Distribution
In order to determine the distribution of STs of toxinogenic MSSA strains isolated from different sources, MLST dataset was performed using goeBURST software [52] implemented by the PHYLOVIZ program [53] available at http://phyloviz.net. Related ST is inserted in the software to provide an unrooted tree-based representation of the relationship of the founding genotype, reflected in the appearance of STs differing only in one housekeeping gene sequence from the founder genotype-single locus variants (SLVs). Further diversification of those SLVs results in the appearance of variations of the original genotype with more than one difference in the allelic profile: double locus variants (DLVs), triple locus variants (TLVs) and so on.
5.6. Statistical Analysis
Data are displayed as frequencies (effective, %) and pairwise comparisons between the resistance profile of MSSA and MRSA isolates, MRSA-PVL + and MSSA-PVL + isolates and toxinogenic and non-toxinogenic isolates were assessed by a chi-square test or Fisher’s exact two-tailed test (when n < 5) using the statistical software GraphPad Instat Prism vers.6.04 (GraphPad Software Inc., San Diego, CA, USA. 2014). Statistical significance was set at a p ≤ 0.05.
Acknowledgments
We thank Sarah Kabani for her editing assistance. A.P., A.S., C.D.-R. and J.-P.L. belong to the FHU INCh (Federation Hospitalo Universitaire Infections Chroniques), Aviesan. We thank the Nîmes University hospital for its structural, human and financial support through the award obtained by our team during the internal call for tenders « Thématiques phares ».
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6651/11/9/500/s1: Table S1: Numbers and distribution of samples collected from different ecological niches in 12 Algerian provinces, Table S2: Numbers and distribution of samples in the different ecological niches, Table S3: Characteristics of MSSA strains isolated from different niches in Algeria.
Author Contributions
conceptualization, J.-P.L., A.M., A.T. and A.S.; methodology, A.M., A.T., J.-P.L. and A.Y.M.; software, A.Y.M.; validation, J.-P.L., A.M., A.T. and A.S.; formal analysis, A.M.; investigation, A.M., A.P., K.Z. and A.T.; resources, A.P., K.Z., and C.D.-R.; writing—original draft preparation, A.M. and J.-P.L.; writing—review and editing, A.T., A.P., K.Z., A.Y.M., C.D.-R. and A.S.; visualization, A.T., A.P. and C.D.-R.; supervision, J.-P.L. and A.S.; project administration, A.M., A.T. and J.-P.L.; funding acquisition, A.T., A.S. and J.-P.L.
Funding
This research was funded by Institut National de la Santé et de la Recherche Médicale: U1047; Campus France: 17MDU980; Campus France: 73-PROFAS B+.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Key Contribution
This article demonstrates the wild dissemination of methicillin-resistant Staphylococcus aureus ST80 carrying the Panton–Valentine leukocidin genes in different niches (human, animals and environment) in Algeria.
References
- 1.Turner N.A., Sharma-Kuinkel B.K., Maskarinec S.A., Eichenberger E.M., Shah P.P., Carugati M., Holland T.L., Fowler V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Lancet Infect. Dis. 2019;17:203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakhundi S., Zhang K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018;31:e00020-18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newsom S.W. Ogston’s coccus. J. Hosp. Infect. 2008;70:369–372. doi: 10.1016/j.jhin.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Grundmann H., Aires-de-Sousa M., Boyce J., Tiemersma E. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 5.Ayeni F.A., Ruppitsch W., Allerberger F. Molecular characterization of clonal lineage and staphylococcal toxin genes from S. aureus in Southern Nigeria. Peer J. 2018;6:e5204. doi: 10.7717/peerj.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Planet P.J., Narechania A., Chen L., Mathema B., Boundy S., Archer G., Kreiswirth B. Architecture of a species phylogenomics of Staphylococcus aureus. Trends Microbiol. 2017;25:153–166. doi: 10.1016/j.tim.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Uhlemann A.C., Dordel J., Knox J.R., Raven K.E., Parkhill J., Holden M.T., Peacock S.J., Lowy F.D. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc. Natl. Acad. Sci. USA. 2014;111:6738–6743. doi: 10.1073/pnas.1401006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saeed K., Gould I., Esposito S., Ahmad-Saeed N., Ahmed S.S., Alp E., Bal A.M., Bassetti M., Bonnet E., Chan M., et al. Panton-Valentine leukocidin-positive Staphylococcus aureus: A position statement from the International Society of Chemotherapy. Int. J. Antimicrob. Agents. 2018;51:16–25. doi: 10.1016/j.ijantimicag.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Shallcross L.J., Fragaszy E., Johnson A.M., Hayward A.C. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: A systematic review and meta-analysis. Lancet Infect. Dis. 2013;13:43–54. doi: 10.1016/S1473-3099(12)70238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alioua M.A., Labid A., Amoura K., Bertine M., Gacemi-Kirane D., Dekhil M. Emergence of the European ST80 clone of community-associated methicillin-resistant Staphylococcus aureus as a cause of healthcare-associated infections in Eastern Algeria. Med. Mal. Infect. 2014;44:180–183. doi: 10.1016/j.medmal.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Djoudi F., Bonura C., Benallaoua S., Touati A., Touati D., Aleo A., Cala C., Fasciana T., Mammina C. Panton-Valentine leukocidin positive sequence type 80 methicillin-resistant Staphylococcus aureus carrying a staphylococcal cassette chromosome mec type IVc is dominant in neonates and children in an Algiers hospital. New Microbiol. 2013;36:49–55. [PubMed] [Google Scholar]
- 12.Irek E.O., Amupitan A.A., Obadare T.O., Aboderin A.O. A systemic review of healthcare-associated infections in Africa: An antimicrobial resistance perspective. Afr. J. Lab. Med. 2018;7:796. doi: 10.4102/ajlm.v7i2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dweba C.C., Zishiri O.T., El Zowalaty M.E. Methicillin-resistant Staphylococcus aureus: Livestock-associated, antimicrobial, and heavy metal resistance. Infect. Drug Resist. 2018;11:2497–2509. doi: 10.2147/IDR.S175967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bekkhoucha S.N., Cady A., Gautier P., Itim F., Donnio P.Y. A portrait of Staphylococcus aureus from the other side of the Mediterranean Sea: Molecular characteristics of isolates from Western Algeria. Eur. J. Clin. Microbiol. Infect. Dis. 2009;28:553–555. doi: 10.1007/s10096-008-0660-x. [DOI] [PubMed] [Google Scholar]
- 15.Antri K., Rouzic N., Dauwalder O., Boubekri I., Bes M., Lina G., Vandenesch F., Tazir M., Ramdani-Bouguessa N., Etienne J. High prevalence of methicillin-resistant Staphylococcus aureus clone ST80-IV in hospital and community settings in Algiers. Clin. Microbiol. Infect. 2011;17:526–532. doi: 10.1111/j.1469-0691.2010.03273.x. [DOI] [PubMed] [Google Scholar]
- 16.Basset P., Amhis W., Blanc D.S. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus in an Algerian hospital. J. Infect. Dev. Ctries. 2015;9:206–209. doi: 10.3855/jidc.4620. [DOI] [PubMed] [Google Scholar]
- 17.Djoudi F., Benallaoua S., Aleo A., Touati A., Challal M., Bonura C., Mammina C. Descriptive epidemiology of nasal carriage of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus among patients admitted to two healthcare facilities in Algeria. Microb. Drug Resist. 2015;21:218–223. doi: 10.1089/mdr.2014.0156. [DOI] [PubMed] [Google Scholar]
- 18.Agabou A., Ouchenane Z., Ngba Essebe C., Khemissi S., Chehboub M.T.E., Chehboub I.B., Sotto A., Dunyach-Remy C., Lavigne J.P. Emergence of nasal carriage of ST80 and ST152 PVL + Staphylococcus aureus isolates from livestock in Algeria. Toxins (Basel) 2017;9:303. doi: 10.3390/toxins9100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.One Health. [(accessed on 31 July 2019)]; Available online: https://www.cdc.gov/onehealth/index.html.
- 20.Coll F., Harrison E.M., Toleman M.S., Reuter S., Raven K.E., Blane B., Palmer B., Kappeler A.R.M., Brown N.M., Török M.E., et al. Longitudinal genomic surveillance of MRSA in the UK reveals transmission patterns in hospitals and the community. Sci. Transl. Med. 2017;9:eaak9745. doi: 10.1126/scitranslmed.aak9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolo J., Miragaia M., Turlej-Rogacka A., Empel J., Bouchami O., Faria N.A., Tavares A., Hryniewicz W., Fluit A.C., de Lencastre H., et al. High genetic diversity among community-associated Staphylococcus aureus in Europe: Results from a multicenter study. PLoS ONE. 2012;7:e34768. doi: 10.1371/journal.pone.0034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gharsa H., Ben Slama K., Gómez-Sanz E., Lozano C., Zarazaga M., Messadi L., Boudabous A., Torres C. Molecular characterization of Staphylococcus aureus from nasal samples of healthy farm animals and pets in Tunisia. Vector Borne Zoonotic Dis. 2015;15:109–115. doi: 10.1089/vbz.2014.1655. [DOI] [PubMed] [Google Scholar]
- 23.Orchein R.C., Hundstad D.A., Fritz S.A., Loughman J.A., Mitchell K., Storch E.K., Gaudreault M., Sellenriek P.L., Armstrong J.R., Mardis E.R., et al. Contribution of gentetically restricted, mthcillin-susceptible strains to the ongoing epidemic of community-acquired Staphylococcus aureus infections. Clin. Infect. Dis. 2009;49:536–542. doi: 10.1086/600881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oosthuysen W.F., Orth H., Lombard C.J., Sinha B., Wasserman E. Population structure analyses of Staphylococcus aureus at Tygerberg hospital, South Africa, reveals a diverse population, a high prevalence of Panton-Valentine leukocidin genes, and unitque local methicillin-resistant S. aureus clone. Clin. Microbiol. Infect. 2014;20:652–659. doi: 10.1111/1469-0691.12452. [DOI] [PubMed] [Google Scholar]
- 25.Li G., Wu S., Luo W., Su Y., Luan Y., Wang X. Staphylococcus aureus ST6-t701 isolates from food-posoning outbreaks (2006–2013) in Xi’an, China. Foodborne Pathog. Dis. 2015;12:203–206. doi: 10.1089/fpd.2014.1850. [DOI] [PubMed] [Google Scholar]
- 26.Santosaningsih D., Santoso S., Setijowati N., Rasyid H.A., Budayanti N.S., Suata K., Widhyatmoko D.B., Purwono P.B., Kuntaman K., Damayanti D., et al. Prevalence and characterisation of Staphylococcus aureus causing community-acquired skin and soft tissue infections on Java and Bali, Indonesia. Trop. Med. Int. Health. 2018;23:34–44. doi: 10.1111/tmi.13000. [DOI] [PubMed] [Google Scholar]
- 27.Pirolo M., Gioffrè A., Visaggio D., Gherardi M., Pavia G., Samele P., Ciambrone L., Di Natale R., Spatari G., Casalinuovo F., et al. Prevalence, molecular epidemiology, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus from swine in southern Italy. BMC Microbiol. 2019;19:51. doi: 10.1186/s12866-019-1422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cormican P., Keane O.M. Complete genome sequences of Sequence Type 71 (ST71) and ST97 Staphylococcus aureus isolates from bovine milk. Microbiol. Resour. Announc. 2018;7:e00954-18. doi: 10.1128/MRA.00954-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panahi M., Saei H.D. Genetic diversity and methicillin resistance of Staphylococcus aureus originating from buffaloes with mastitis in Iran. Comp. Immunol. Microbiol. Infect. Dis. 2019;62:19–24. doi: 10.1016/j.cimid.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Donkor E.S., Jamrozy D., Mills R.O., Dankwah T., Amoo P.K., Egyir B., Badoe E.V., Twasam J., Bentley S.D. A genomic infection control study for Staphylococcus aureus in two Ghanaian hospitals. Infect. Drug Resist. 2018;11:1757–1765. doi: 10.2147/IDR.S167639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundmann H., Aanensen D.M., van den Wijngaard C.C., Spratt B.G., Harmsen D., Friedrich A.W. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: A molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozano C., Marí A., Aspiroz C., Gómez-Sanz E., Ceballos S., Fortuño B., Barcenilla F., Jover-Sáenz A., Torres C. Nasal carriage of coagulase positive staphylococci in patients of a primary-healthcare-center: Genetic lineages and resistance and virulence genes. Enferm. Infecc. Microbiol. Clin. 2015;33:391–396. doi: 10.1016/j.eimc.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Suhaili Z., Lean S.S., Mohamad N.M., Rachman A.R., Desa M.N., Yeo C.C. Draft genome sequence of Staphylococcus aureus KT/312045, an ST1-MSSA PVL positive isolated from pus sample in East Coast Malaysia. Genom. Data. 2016;9:111–112. doi: 10.1016/j.gdata.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valentin-Domelier A.S., Girard M., Bertrand X., Violette J., François P., Donnio P.Y., Talon D., Quentin R., Schrenzel J., van der Mee-Marquet N., et al. Methicillin-susceptible ST398 Staphylococcus aureus responsible for bloodstream infections: An emerging human-adapted subclone? PLoS ONE. 2011;6:e28369. doi: 10.1371/journal.pone.0028369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramdani-Bouguessa N., Bes M., Meugnier H., Forey F., Reverdy M.E., Lina G., Vandenesch F., Tazir M., Etienne J. Detection of methicillin-resistant Staphylococcus aureus strains resistant to multiple antibiotics and carrying the Panton-Valentine Leukocidin genes in an Algiers hospital. Antimicrob. Agents Chemother. 2006;50:1083–1085. doi: 10.1128/AAC.50.3.1083-1085.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaalal W., Chaalal N., Bourafa N., Kihal M., Diene S.M., Rolain J.M. Characterization of Staphylococcus aureus isolated from food products in Western Algeria. Foodborne Pathog. Dis. 2018;15:353–360. doi: 10.1089/fpd.2017.2339. [DOI] [PubMed] [Google Scholar]
- 37.Malachowa N., DeLeo F.R. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010;67:3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monecke S., Coombs G., Shore A.C., Coleman D.C., Akpaka P., Borg M., Chow H., Ip M., Jatzwauk L., Jonas D., et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011;6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bierowiec K., Ploneczka-Janeczko K., Rypula K. Prevalence and risk factors of colonisation with Staphylococcus aureus in healthy pet cats kept in the city households. Biomed. Res. Int. 2016;2016:3070524. doi: 10.1155/2016/3070524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papadimitriou-Olivgeris M., Drougka E., Fligou F., Dodou V., Kolonitsiou F., Filos K.S., Anastassiou E.D., Petinaki E., Marangos M., Spiliopoulou I. Spread of Tst-positive Staphylococcus aureus strains belonging to ST30 clone among patients and healthcare workers in two intensive care units. Toxins (Basel) 2017;9:270. doi: 10.3390/toxins9090270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sung J.M., Lloyd D.H., Lindsay J.A. Staphylococcus aureus host specificity: Comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology. 2008;154:1949–1959. doi: 10.1099/mic.0.2007/015289-0. [DOI] [PubMed] [Google Scholar]
- 42.Asanin J., Misic D., Aksentijevic K., Tambur Z., Rakonjac B., Kovacevic I., Spergser J., Loncaric I. Genetic profiling and comparison of human and animal methicillin-resistant Staphylococcus aureus (MRSA) isolates from Serbia. Antibiotics (Basel) 2019;16:26. doi: 10.3390/antibiotics8010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarraud S., Mougel C., Thioulouse J., Lina G., Meugnier H., Forey F., Nesme X., Etienne J., Vandenesch F. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamrozy D., Coll F., Matther A.E., Harris S.R., Harrison E.M., MacGowan A., Karas A., Elston T., Estée Török M., Parkhill J., et al. Evolution of mobile genetic element composition in an epidemic methicillin-resistant Staphylococcus aureus: Temporal changes correlated with frequent loss and gain events. BMC Genom. 2017;18:684. doi: 10.1186/s12864-017-4065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karakonstantis S., Kalemaki D. Antimicrobial overuse and misuse in the community in Greece and link to antimicrobial resistance using methicillin-resistant S. aureus as an example. J. Infect. Public Health. 2019;12:460–464. doi: 10.1016/j.jiph.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 46.European Committee on Antimicrobial Susceptibility Testing. [(accessed on 31 July 2019)]; Available online: https://www.sfm-microbiologie.org/wp-content/uploads/2018/12/CASFMV2_SEPTEMBRE2018.pdf.
- 47.Sahebnasagh R., Saderi H., Owlia P. The prevalence of resistance to methicillin in Staphylococcus aureus strains isolated from patients by PCR method for detection of mecA and nuc genes. Iran. J. Public Health. 2014;43:84–92. [PMC free article] [PubMed] [Google Scholar]
- 48.Monecke S., Jatzwauk L., Weber S., Slickers P., Ehricht R. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin. Microbiol. Infect. 2008;14:534–545. doi: 10.1111/j.1469-0691.2008.01986.x. [DOI] [PubMed] [Google Scholar]
- 49.Monecke S., Slickers P., Ehricht R. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 2008;53:237–251. doi: 10.1111/j.1574-695X.2008.00426.x. [DOI] [PubMed] [Google Scholar]
- 50.Enright M.C., Day N.P., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staphylococcus Aureus MLST Databases. [(accessed on 31 July 2019)]; Available online: http://www.mlst.net/
- 52.Francisco A.P., Bugalho M., Ramirez M., Carriço J.A. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 2009;10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Francisco A.P., Vaz C., Monteiro P.T., Melo-Cristino J., Ramirez M., Carriço J.A. PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinform. 2012;13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.