Abstract
Peanuts are widely consumed in many local dishes in southeast Asian countries, especially in Malaysia which is one of the major peanut-importing countries in this region. Therefore, Aspergillus spp. and aflatoxin contamination in peanuts during storage are becoming major concerns due to the tropical weather in this region that favours the growth of aflatoxigenic fungi. The present study thus aimed to molecularly identify and characterise the Aspergillus section Flavi isolated from imported peanuts in Malaysia. The internal transcribed spacer (ITS) and β-tubulin sequences were used to confirm the species and determine the phylogenetic relationship among the isolates, while aflatoxin biosynthesis genes (aflR, aflP (omtA), aflD (nor-1), aflM (ver-1), and pksA) were targeted in a multiplex PCR to determine the toxigenic potential. A total of 76 and one isolates were confirmed as A. flavus and A. tamarii, respectively. The Maximum Likelihood (ML) phylogenetic tree resolved the species into two different clades in which all A. flavus (both aflatoxigenic and non-aflatoxigenic) were grouped in the same clade and A. tamarii was grouped in a different clade. The aflatoxin biosynthesis genes were detected in all aflatoxigenic A. flavus while the non-aflatoxigenic A. flavus failed to amplify at least one of the genes. The results indicated that both aflatoxigenic and non-aflatoxigenic A. flavus could survive in imported peanuts and, thus, appropriate storage conditions preferably with low temperature should be considered to avoid the re-emergence of aflatoxigenic A. flavus and the subsequent aflatoxin production in peanuts during storage.
Keywords: peanuts, aflatoxins, Aspergillus flavus, aflatoxin biosynthesis gene
1. Introduction
Aspergillus section Flavi is one of the most important sections in the genus Aspergillus as the majority of the species in this section are able to produce aflatoxins, of which aflatoxin B1 (AFB1) is a carcinogenic compound that can cause acute and chronic diseases related to aflatoxin poisoning [1]. Acute exposure of aflatoxin may lead to death as reported in Kenya in 2004 [2], while chronic exposure may lead to liver cancer [3]. AFB1 has been classified as a Group 1 carcinogen by the International Agency of Cancer Research [4]. According to [5], Aspergillus section Flavi could be separated into two groups based on their impact on food and human health. The first group includes the main aflatoxigenic species such as A. flavus, A. parasiticus and A. nomius, while the second group comprises the non-aflatoxin-producing species such as A. tamarii, A. oryzae and A. sojae. These are the main important species found in crops—especially in nuts, spices, cereals and also fermented product such as meju [6,7,8,9,10]. These species are closely related to each other in terms of morphology and phylogeny [5,11]. However, A. flavus is reported to be more diverse in terms of morphological characters and toxigenic potential [12,13].
Molecular and phylogenetic analyses are commonly used to validate the morphological identification of Aspergillus section Flavi. The DNA sequence of the conserved regions in fungi, especially in the genus Aspergillus such as the ITS regions, β-tubulin and calmodulin, could be used to differentiate the closely related species of Aspergillus section Flavi such as A. parasiticus, A. oryzae, A. minisclerotigenes, A. parvisclerotigenus and A. arachidicola [14,15].
The occurrence of aflatoxin and Aspergillus spp. in raw peanuts (imported) and peanut-based products marketed in Malaysia have been documented since 1980s [16] and the contamination has been increasing along the supply chain, especially at the level of manufacturers and retailers as reported by [17]. The authors [17] then reported on the presence of aflatoxigenic and non-aflatoxigenic A. flavus and one isolate of A. nomius from the same peanut samples based on the morphological and chemical (extrolites) characteristics [18]. Based on the findings, some of the aflatoxigenic A. flavus were able to produce both aflatoxin B and G group aflatoxins which are the common features in other species in section Flavi such as A. parasiticus, A. arachidicola and A. minisclerotigenes [19]. In a previous study, A. tamarii and A. nomius were misidentified as A. flavus due to the phenotypic resemblance as reported by [20]. In this regard, the misidentification of aflatoxigenic and non-aflatoxigenic A. flavus could have occurred due to these similarities and thus requires additional molecular data to support the results [18], which is presented and discuss in this paper.
Even though the Aspergillus flavus and A. parasiticus agar (AFPA) medium was specifically formulated by [21] to isolate and enumerate A. flavus and A. parasiticus from food samples, recent studies have reported that other Aspergillus section Flavi species were also capable of producing aspergillic acid, which could react with the ferric citrate in the medium to produce orange colour reverse [14,22,23]. Therefore, gene sequencing and phylogenetic analysis were required to resolve and confirm the identification of Aspergillus section Flavi species. The ITS region has been widely used as the ‘barcode’ for fungal identification [24]. However, depending on a single gene identification is not always accurate due to the intra- and inter-species variation of the Aspergillus spp. The analysis of DNA sequences from two or more genes are thus deemed more accurate and reliable. Therefore, β-tubulin was used as the secondary identification marker in the present study to validate the species identification [25].
The aflatoxin-producing ability of A. flavus in the previous study [18] was found to be inconsistent since a large number of strains were found to be non-aflatoxigenic. Other than the environmental conditions, the ability of A. flavus to produce aflatoxin is highly determined by the genetic variation of the strains. It could be due to any disruption in the aflatoxin biosynthesis genes or belong to other species that do not produce aflatoxin. The aflatoxin biosynthesis gene cluster has been sequenced and extensively studied in order to understand the mechanism and biosynthesis pathway of aflatoxin in aflatoxigenic fungi [26]. The presence of these genes is required by the Aspergillus spp. to produce aflatoxin, and any changes therein might cause disruption in the biosynthetic pathway. The precise identification of Aspergillus section Flavi is therefore important to determine the risk of aflatoxin contamination as results in the previous study [17,18] indicated a high occurrence of these species in raw peanuts (imported) and peanut-based products marketed in Malaysia.
Therefore, the objectives of the present study were to molecularly confirm the identity of A. flavus and A. nomius from the previous study [18], to determine the phylogenetic relationships among the Aspergillus section Flavi strains, and to detect the presence of aflatoxin biosynthesis genes in those strains.
2. Results
2.1. PCR Amplification and Basic Local Alignment Search Tool (BLAST) Search
The PCR amplifications of the ITS region and β-tubulin genes for all strains were positive, generating products of ~600 bp and ~595 bp, respectively. Based on the BLAST search against the GenBank database, both ITS and β-tubulin genes gave a similar result for all 77 Aspergillus section Flavi strains in this study (Table 1). Results from the ITS and β-tubulin gene sequencing are in line with the previous identification of A. flavus except for A52R. The BLAST results showed that a total of 76 strains were identified as A. flavus/A. oryzae with 99 to 100% similarity, while A52R, which was previously identified as A. nomius, was re-identified as A. tamarii based on the ITS and β-tubulin gene sequences. The DNA sequences analysis confirmed the absence of A. parasiticus in raw peanuts and peanut-based products tested in the previous study [18]. A total of 37 out of 46 (92.5%) aflatoxigenic A. flavus (Chemotype I, II, and V) were isolated from raw peanut kernels in which 57 and 35% of the them were imported from India and China, respectively (Table 1).
Table 1.
GenBank accession numbers of the Aspergillus section Flavi strains used.
| Isolate No. | Species | Stakeholder | * Source | ** Chemotype | GenBank Accession Number | |
|---|---|---|---|---|---|---|
| ITS | β-Tubulin | |||||
| A8R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095114 | MN148806 |
| A34R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095115 | MN148807 |
| A35R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095116 | MN148808 |
| A42R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095117 | MN148809 |
| A45R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095118 | MN148810 |
| A46R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095119 | MN148811 |
| A47R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095120 | MN148812 |
| A50R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095121 | MN148813 |
| A53R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095122 | MN148814 |
| A54R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095123 | MN148815 |
| A55R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095124 | MN148816 |
| A57R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095125 | MN148817 |
| A58R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095126 | MN148818 |
| A59R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095127 | MN148819 |
| A60R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095128 | MN148820 |
| A61R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095129 | MN148821 |
| A63R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095130 | MN148822 |
| A68R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095131 | MN148823 |
| A74R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095132 | MN148824 |
| A81R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095133 | MN148825 |
| A90R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095134 | MN148826 |
| A91R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095135 | MN148827 |
| A92R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095136 | MN148828 |
| A95R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095137 | MN148829 |
| A96R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095138 | MN148830 |
| A116P | A. flavus | Retailer | Peanut-based product (roasted peanut) | I | MN095139 | MN148831 |
| A87R | A. flavus | Manufacturer | Raw peanut kernel (China) | II | MN095140 | MN148832 |
| A88R | A. flavus | Manufacturer | Raw peanut kernel (China) | II | MN095141 | MN148833 |
| A5R | A. flavus | Importer | Raw peanut kernel (China) | III | MN095142 | MN148834 |
| A24R | A. flavus | Importer | Raw peanut kernel (China) | III | MN095143 | MN148835 |
| A40R | A. flavus | Manufacturer | Raw peanut kernel (India) | III | MN095144 | MN148836 |
| A43R | A. flavus | Manufacturer | Raw peanut kernel (China) | III | MN095145 | MN148837 |
| A48R | A. flavus | Manufacturer | Raw peanut kernel (China) | III | MN095146 | MN148838 |
| A49R | A. flavus | Manufacturer | Raw peanut kernel (China) | III | MN095147 | MN148839 |
| A71R | A. flavus | Manufacturer | Raw peanut kernel (India) | III | MN095148 | MN148840 |
| A73R | A. flavus | Manufacturer | Raw peanut kernel (India) | III | MN095149 | MN148841 |
| A77R | A. flavus | Importer | Raw peanut in shell (Indonesia) | III | MN095150 | MN148842 |
| A78R | A. flavus | Manufacturer | Raw peanut kernel (China) | III | MN095151 | MN148843 |
| A94R | A. flavus | Retailer | Raw peanut kernel (India) | III | MN095152 | MN148844 |
| A97R | A. flavus | Retailer | Raw peanut kernel (India) | III | MN095153 | MN148845 |
| A108P | A. flavus | Manufacturer | Peanut-based product (roasted peanut) | III | MN095154 | MN148846 |
| A109P | A. flavus | Retailer | Peanut-based product (peanut candy) | III | MN095155 | MN148847 |
| A118P | A. flavus | Retailer | Peanut-based product (roasted peanut) | III | MN095156 | MN148848 |
| A9R | A. flavus | Retailer | Raw peanut kernel (China) | IV | MN095157 | MN148849 |
| A12R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095158 | MN148850 |
| A13R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095159 | MN148851 |
| A14R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095160 | MN148852 |
| A16R | A. flavus | Importer | Raw peanut in shell (Vietnam) | IV | MN095161 | MN148853 |
| A19R | A. flavus | Importer | Raw peanut in shell (China) | IV | MN095162 | MN148854 |
| A20R | A. flavus | Importer | Raw peanut in shell (China) | IV | MN095163 | MN148855 |
| A21R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095164 | MN148856 |
| A22R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095165 | MN148857 |
| A23R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095166 | MN148858 |
| A26R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095167 | MN148859 |
| A27R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095168 | MN148860 |
| A67R | A. flavus | Manufacturer | Raw peanut kernel (India) | IV | MN095169 | MN148861 |
| A75R | A. flavus | Importer | Raw peanut in shell (Vietnam) | IV | MN095170 | MN148862 |
| A76R | A. flavus | Importer | Raw peanut in shell (Vietnam) | IV | MN095171 | MN148863 |
| A98P | A. flavus | Manufacturer | Peanut-based product (peanut snack) | IV | MN095172 | MN148864 |
| A104P | A. flavus | Manufacturer | Peanut-based product (peanut sauce) | IV | MN095173 | MN148865 |
| A111P | A. flavus | Retailer | Peanut-based product (peanut snack) | IV | MN095174 | MN148866 |
| A114P | A. flavus | Retailer | Peanut-based product (roasted peanut) | IV | MN095175 | MN148867 |
| A115P | A. flavus | Retailer | Peanut-based product (roasted peanut) | IV | MN095176 | MN148868 |
| A122R | A. flavus | Retailer | Raw peanut kernel (India) | IV | MN095177 | MN148869 |
| A123R | A. flavus | Retailer | Raw peanut kernel (India) | IV | MN095178 | MN148870 |
| A1R | A. flavus | Importer | Raw peanut kernel (India) | V | MN095179 | MN148871 |
| A15R | A. flavus | Importer | Raw peanut kernel (China) | V | MN095180 | MN148872 |
| A25R | A. flavus | Importer | Raw peanut kernel (China) | V | MN095181 | MN148873 |
| A29R | A. flavus | Importer | Raw peanut in shell (China) | V | MN095182 | MN148874 |
| A41R | A. flavus | Manufacturer | Raw peanut kernel (India) | V | MN095183 | MN148875 |
| A44R | A. flavus | Manufacturer | Raw peanut kernel (India) | V | MN095184 | MN148876 |
| A69R | A. flavus | Manufacturer | Raw peanut kernel (India) | V | MN095185 | MN148877 |
| A80R | A. flavus | Manufacturer | Raw peanut kernel (China) | V | MN095186 | MN148878 |
| A82R | A. flavus | Manufacturer | Raw peanut kernel (China) | V | MN095187 | MN148879 |
| A102P | A. flavus | Manufacturer | Peanut-based product (peanut cookies) | V | MN095188 | MN148880 |
| A107P | A. flavus | Manufacturer | Peanut-based product (peanut cookies) | V | MN095189 | MN148881 |
| A52R | A. tamarii | Manufacturer | Raw peanut in shell (China) | VI | MN095190 | MN148882 |
| A. flavus NRRL 3357 | n.a | n.a | MF966967 | M38265 | ||
| A. oryzae CBS 100925 | n.a | n.a | KJ175432 | EF203138 | ||
| A. minisclerotigenes NRRL 29000 | n.a | n.a | KY937929 | KY924668 | ||
| A. minisclerotigenes CBS 117620 | n.a | n.a | JF422073 | EF203150 | ||
| A. parvisclerotigenus CBS 121.62 | n.a | n.a | EF409240 | EF203130 | ||
| A. parasiticus CBS 100926 | n.a | n.a | KJ175437 | KJ175497 | ||
| A. parasiticus CBS 100308 | n.a | n.a | KJ175436 | KJ175496 | ||
| A. nomius NRRL 25393 | n.a | n.a | AF027864 | AF255068 | ||
| A. nomius NRRL 13137 | n.a | n.a | AF027860 | AF255067 | ||
| A. tamarii CBS 121599 | n.a | n.a | KJ175443 | KJ175501 | ||
| A. tamarii CBS 118098 | n.a | n.a | KJ175442 | KJ175500 | ||
| A. arachidicola CBS 117610 | n.a | n.a | EF409241 | EF203158 | ||
| A. arachidicola CBS 117614 | n.a | n.a | KY937923 | KY924665 | ||
| A. niger CBS 113.46 | n.a | n.a | FJ629351 | FJ629302 | ||
* Source: Type of peanuts (County of origin or the type of peanut-based products); n.a: not applicable; ** Chemotype I (AFB, CPA), Chemotype II (AFB), Chemotype III (CPA), Chemotype IV (none), Chemotype V (AFB, AFG, CPA), Chemotype VI (AFB and AFG) [18].
2.2. Phylogenetic Analysis
The Maximum Likelihood (ML) tree was constructed based on the ITS, β-tubulin and combined sequences to describe the phylogenetic relationships among the Aspergillus section Flavi strains as shown in Figure 1, Figure 2 and Figure 3, respectively. The individual ML tree for ITS sequences does not clearly separate different species into different clades as shown in Figure 1. The reference strains of A. flavus, A. oryzae, A. parvisclerotigenus and A. minisclerotigenes were grouped together in the same clade. This result demonstrated that ITS alone is not enough to resolve the closely related species of Aspergillus section Flavi. However, β-tubulin and the combined ITS and β-tubulin sequences showed better separation of each species into different clades and were supported with medium to high bootstrap values ranging from 60–99%.
Figure 1.
Maximum likelihood tree showing the phylogenetic relationships among the Aspergillus section Flavi strains based on the ITS sequences. *A. flavus includes all strains in Chemotypes I—V as listed in Table 1. Values on branches are the bootstrap values.
Figure 2.
Maximum likelihood tree showing the phylogenetic relationships among the Aspergillus section Flavi strains based on the β-tubulin sequences. *A. flavus includes all strains in Chemotypes I—V as listed in Table 1. Values on branches are the bootstrap values.
Figure 3.
Maximum likelihood tree showing the phylogenetic relationships among Aspergillus section Flavi strains based on the combined ITS and β-tubulin sequences. *A. flavus includes all strains in Chemotypes I—V as listed in Table 1. Values on branches are the bootstrap values.
The ML tree for β-tubulin, as shown in Figure 2, grouped 76 strains in the present study in the same clade with the reference strains A. flavus NRRL 3357 and A. oryzae CBS 100925. Both reference strains could not be separated due to the high genetic similarity in both species [27]. However, the identities of the strains in this group were confirmed as A. flavus as they originated from peanuts and the majority of them showed the ability to produce aflatoxins (Table 1). In contrast, A. oryzae does not produce aflatoxin, and it has never been reported in peanuts [28]. The species is mainly used in koji fermentation for traditional fermented food in Japan. On the other hand, one isolate of A. tamarii was consistently grouped together with the reference strains of A. tamarii CBS 121599 and CBS 118098.
The ML tree of the combined dataset (Figure 3) shows similar tree topology with the individual β-tubulin. All strains were grouped together with the reference strains A. flavus NRRL 3357 and A. oryzae CBS 100925 except for A52R which was grouped together with A. tamarii CBS 121599 and A. tamarii CBS 113.46. The outgroup A. niger CBS 113.46 formed a separate clade.
Generally, the A. flavus strains in the present study were clustered in the same clade and not according to the source of isolation. A. flavus strains isolated from raw peanuts and peanut-based products collected from different stakeholders did not show any genetic variation as they were consistently grouped in the same clade. Furthermore, the aflatoxigenic and non-aflatoxigenic A. flavus did not form a separate clade, since ITS and β-tubulin genes were mainly used for identification purposes, and they were not involved in the biosynthesis of aflatoxin.
2.3. Detection of Aflatoxin Biosynthesis Genes in Aspergillus Section Flavi Strains
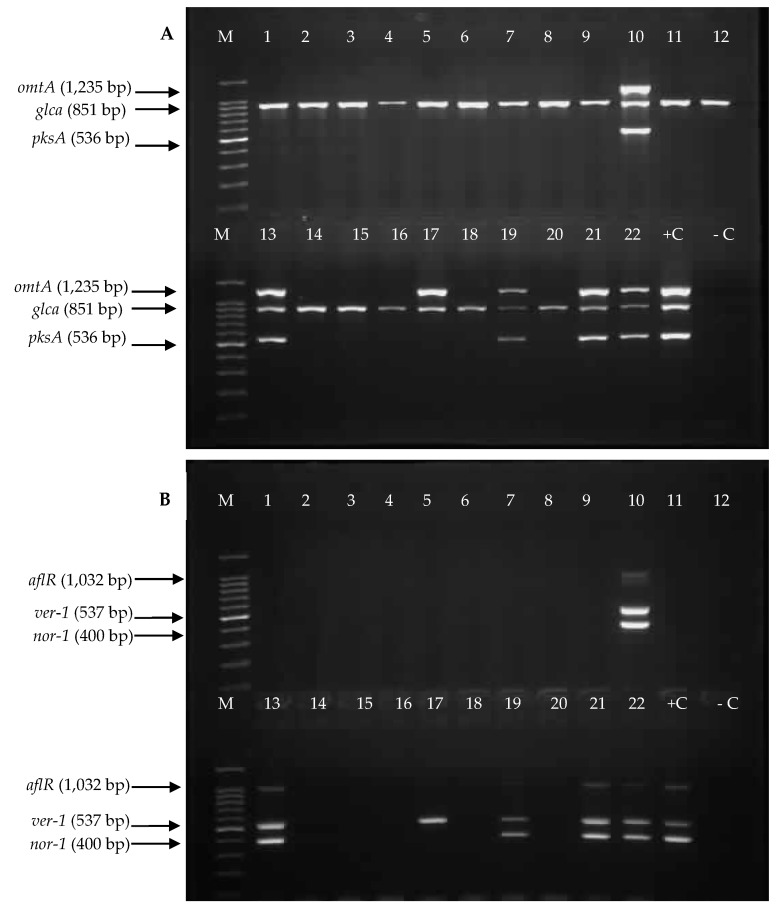
The PCR method was used to amplify the targeted aflatoxin biosynthesis genes aflR, aflP (omtA), aflD (nor-1), aflM (ver-1), pksA, and one sugar utilisation gene, glcA. The glcA gene, which is located adjacent to the 3′ end of aflatoxin biosynthesis gene cluster, was used as a positive marker for A. flavus, as this gene is consistently present in this species regardless of toxigenic potentials [29,30]. Figure 4A,B show the representative amplification patterns of the reference strain A. flavus NRRL 3357 and the aflatoxigenic A. flavus isolates from Chemotype V in Multiplex PCR set 1 and 2, respectively. All the targeted genes were successfully amplified and corresponded to the sizes of their PCR products. The results support the ability of these strains to produce aflatoxin.
Figure 4.
Amplification of (A) Multiplex PCR set 1: omtA, glca, and pksA and (B) Multiplex PCR set 2: aflR, ver-1 and nor-1 genes in the representative aflatoxigenic A. flavus (Chemotype V). M: 100-bp DNA ladder; Lane 1: A1R; Lane 2: A15R; Lane 3: A25R; Lane 4: A29R; Lane 5: A41R; Lane 6: A44R; Lane 7: A69R; Lane 8: A80R; Lane 9: A82R, Lane 10: A102R; Lane 11: A107R and Lane 12: +C Positive control (A. flavus NRRL 3357).
In contrast, the representative non-aflatoxigenic A. flavus strains from Chemotype IV failed to amplify almost all the genes required for aflatoxin biosynthesis as shown by the results of Multiplex PCR set 1 and set 2 in Figure 5A,B, respectively. The amplification patterns of the aflatoxin biosynthesis genes in all A. flavus strains and A. tamarii in the present study are summarised in Table 2. At least one gene was missing as depicted by the amplification patterns that caused the strains to fail to produce aflatoxin except for A23R, A67R, A122R and A123R in Chemotype IV. The majority (59%) of the non-aflatoxigenic strains in Chemotype IV that failed to amplify all the genes originated from raw peanut samples collected from the importers. However, all non-aflatoxigenic A. flavus in Chemotype III showed a complete amplification of all genes except for A40R, A43R and A48R. As a comparison, the glcA gene was amplified in all strains, as this gene is not involved in the aflatoxin biosynthetic pathway. In addition, the complete amplification pattern in reference strain A. flavus NRRL 3357 confirmed that the absence of these genes in the non-aflatoxigenic A. flavus isolates was not caused by any technical error.
Figure 5.
Representative amplification of (A) multiplex PCR set 1: omtA, glca, and pksA and (B) Multiplex PCR set 2: aflR, ver-1 and nor-1 genes in the representative non-aflatoxigenic A. flavus (Chemotype IV). M: 100-bp DNA ladder; +C: Positive control (A. flavus NRRL 3357); -C: Negative control (without DNA template); Lane 1: A9R; Lane 2: A12R; Lane 3: A13R; Lane 4: A14R; Lane 5: A16R; Lane 6: A19R; Lane 7: A20R; Lane 8: A21R; Lane 9: A22R; Lane 10: A23R; Lane 11: A26R; Lane 12: A27R; Lane 13: A67R; Lane 14; A75R; Lane 15: A76R; Lane 16: A98R; Lane 17: A104P; Lane 18: A111P; Lane 19: A114P; Lane 20: A115P; Lane 21: A122R; Lane 22: A123R.
Table 2.
Amplification pattern of aflatoxin biosynthesis and sugar utilisation genes in Aspergillus section Flavi strains.
| No. | Strain | * Chemotype | Aflatoxin Biosynthesis Gene | |||||
|---|---|---|---|---|---|---|---|---|
| aflR | aflP (omtA) | aflD (nor-1) | aflM (ver-1) | pksA | glcA | |||
| 1 | A8R | I | + | + | + | + | + | + |
| 2 | A34R | I | + | + | + | + | + | + |
| 3 | A35R | I | + | + | + | + | + | + |
| 4 | A42R | I | + | + | + | + | + | + |
| 5 | A45R | I | + | + | + | + | + | + |
| 6 | A46R | I | + | + | + | + | + | + |
| 7 | A47R | I | + | + | + | + | + | + |
| 8 | A50R | I | + | + | + | + | + | + |
| 9 | A53R | I | + | + | + | + | + | + |
| 10 | A54R | I | + | + | + | + | + | + |
| 11 | A55R | I | + | + | + | + | + | + |
| 12 | A57R | I | + | + | + | + | + | + |
| 13 | A58R | I | + | + | + | + | + | + |
| 14 | A59R | I | + | + | + | + | + | + |
| 15 | A60R | I | + | + | + | + | + | + |
| 16 | A61R | I | + | + | + | + | + | + |
| 17 | A63R | I | + | + | + | + | + | + |
| 18 | A68R | I | + | + | + | + | + | + |
| 19 | A74R | I | + | + | + | + | + | + |
| 20 | A81R | I | + | + | + | + | + | + |
| 21 | A90R | I | + | + | + | + | + | + |
| 22 | A91R | I | + | + | + | + | + | + |
| 23 | A92R | I | + | + | + | + | + | + |
| 24 | A95R | I | + | + | + | + | + | + |
| 25 | A96R | I | + | + | + | + | + | + |
| 26 | A116P | I | + | + | + | + | + | + |
| 27 | A87R | II | + | + | + | + | + | + |
| 28 | A88R | II | + | + | + | + | + | + |
| 29 | A5R | III | + | + | + | + | + | + |
| 30 | A24R | III | + | + | + | + | + | + |
| 31 | A40R | III | + | + | + | − | + | + |
| 32 | A43R | III | + | + | + | − | + | + |
| 33 | A48R | III | + | + | + | − | + | + |
| 34 | A49R | III | + | + | + | + | + | + |
| 35 | A71R | III | + | + | + | + | + | + |
| 36 | A73R | III | + | + | + | + | + | + |
| 37 | A77R | III | + | + | + | + | + | + |
| 38 | A78R | III | + | + | + | + | + | + |
| 39 | A94R | III | + | + | + | + | + | + |
| 40 | A97R | III | + | + | + | + | + | + |
| 41 | A108P | III | + | − | + | + | + | + |
| 42 | A109P | III | + | + | + | + | + | + |
| 43 | A118P | III | + | + | + | + | + | + |
| 44 | A9R | IV | − | − | − | − | − | + |
| 45 | A12R | IV | − | − | − | − | − | + |
| 46 | A13R | IV | − | − | − | − | − | + |
| 47 | A14R | IV | − | − | − | − | − | + |
| 48 | A16R | IV | − | − | − | − | − | + |
| 49 | A19R | IV | − | − | − | − | − | + |
| 50 | A20R | IV | − | − | − | − | − | + |
| 51 | A21R | IV | − | − | − | − | − | + |
| 52 | A22R | IV | − | − | − | − | − | + |
| 53 | A23R | IV | + | + | + | + | + | + |
| 54 | A26R | IV | − | − | − | − | − | + |
| 55 | A27R | IV | − | − | − | − | − | + |
| 56 | A67R | IV | + | + | + | + | + | + |
| 57 | A75R | IV | − | − | − | − | − | + |
| 58 | A76R | IV | − | − | − | − | − | + |
| 59 | A98P | IV | − | − | − | − | − | + |
| 60 | A104P | IV | − | + | − | + | − | + |
| 61 | A111P | IV | − | − | − | − | − | + |
| 62 | A114P | IV | − | + | + | + | + | + |
| 63 | A115P | IV | − | − | − | − | − | + |
| 64 | A122R | IV | + | + | + | + | + | + |
| 65 | A123R | IV | + | + | + | + | + | + |
| 66 | A1R | V | + | + | + | + | + | + |
| 67 | A15R | V | + | + | + | + | + | + |
| 68 | A25R | V | + | + | + | + | + | + |
| 69 | A29R | V | + | + | + | + | + | + |
| 70 | A41R | V | + | + | + | + | + | + |
| 71 | A44R | V | + | + | + | + | + | + |
| 72 | A69R | V | + | + | + | + | + | + |
| 73 | A80R | V | + | + | + | + | + | + |
| 74 | A82R | V | + | + | + | + | + | + |
| 75 | A102P | V | + | + | + | + | + | + |
| 76 | A107P | V | + | + | + | + | + | + |
| 77 | A52R | VI | + | + | + | + | + | + |
+ present; − absent; * Chemotype I (AFB, CPA), Chemotype II (AFB), Chemotype III (CPA), Chemotype IV (none), Chemotype V (AFB, AFG, CPA), Chemotype VI (AFG) [18].
3. Discussion
A comparison of both ITS and β-tubulin sequences with fungal sequences deposited in the GenBank showed a high similar percentage for A. flavus and A. oryzae. The strong phylogenetic relationship between A. flavus and A. oryzae has been explained by many researchers and they concluded that A. oryzae is actually the domesticated species of A. flavus through years of selection under artificial production environments [27,31,32,33]. A. oryzae has been widely used for commercial application such as the starter culture for koji fermentation in the production of traditional fermented foods such as soy sauce, sake and shochu [34], and it has earned the Generally Regarded as Safe (GRAS) status due to its long history of safe use in the food fermentation industry. Payne et al. [27] who studied the whole genome comparison of A. flavus and A. oryzae revealed that these fungi are very similar in the genome size and number of predicted genes. However, due to the economics and food safety issues, A. oryzae continues to be classified as a separate species from A. flavus even though it has been proven to be genetically similar to A. flavus [27,31].
A. oryzae is not a plant pathogen and it has never been reported to contaminate peanuts in the field [35,36,37]. It is believed that A. oryzae rarely survives in the field due to the low production of sclerotia, which could be detrimental to its survival [31,33]. According to [19], no aflatoxin production has been recorded from this species. Besides, a study on the comparative chemistry of A. flavus and A. oryzae also revealed that the latter species does not produce CPA [38]. Therefore, A. flavus was confirmed as the main aflatoxigenic and non-aflatoxigenic strains detected in raw peanuts and peanut-based products in the present study. Interestingly, A. parasiticus was absent in the present study even though it was reported as one of the main aflatoxin producers in peanuts by previous researchers [6,35,37].
Peanuts were dried to achieve a moisture content of <9% during post-harvest, and this condition is maintained throughout the shipping period to the importing countries to prevent fungal proliferation. However, the ability of A. flavus to produce sclerotia, which is a compact mass of hardened mycelium that contains food reserves, helps them to survive in the extreme environmental conditions until favourable growth conditions return [39,40].
In the previous study [18], one strain (A52R) has been morphologically identified as A. nomius due to the production of AFB, AFG, aspergillic acid and limited growth on CZ agar at 42 °C. However, molecular analysis based on the sequences of ITS and β-tubulin region revealed the identity as A. tamarii. It was found to be an unusual observation of A. tamarii since it was able to produce aflatoxins, which is contradictory to its typical characteristics. According to [15], A. tamarii does not produce aflatoxins, and it has been used in the food industry for the production of soy sauce and various enzymes such as amylases, proteases and xylanolytic enzymes since a long time ago.
However, an isolated case was reported for the first time by [41], in which several strains of A. tamarii isolated from a tea field were found to produce aflatoxin and CPA. The strains were also reported to produce sclerotia and exhibited dark olive to olive brown colour on CZ agar. A strain was then re-examined for the morphology, mycotoxin production, and the sequences of ITS, β-tubulin and calmodulin gene. Based on these results, a new species named A. pseudotamarii was given to replace the previous identification of A. tamarii [42]. The characteristics described by [41] are in line with our observation on strain A52R except for the production of CPA. However, the molecular identification based on ITS and β-tubulin were not in agreement with [42], in which the identity of A52R in the present study remains as A. tamarii instead of A. pseudotamarii. Another study by [14] also reported the presence of A. tamarii in peanuts from Argentina, but no aflatoxin was produced by this isolate.
The misidentification of the closely related species in Aspergillus section Flavi has been reported previously [20]. The authors reported on the misidentification of A. nomius and A. tamarii as A. flavus. According to the authors, this occurred due to the lack of expertise in mycological identification. The similar morphological characteristic of those three species observed on Sabouraud Dextrose Agar (yellow colour) led to the identification of A. flavus. However, the sequencing of β-tubulin and calmodulin gene finally and unambiguously identified the species as A. nomius and A. tamarii. Their finding was also supported by the metabolic fingerprinting, in which A. flavus, A. tamarii and A. nomius were separated into three clusters based on the UHPLC-MS analysis.
A. arachidicola and A. minisclerotigenes, which were first isolated from the Argentinean peanuts, are known as the closely related species to A. parasiticus and A. flavus, respectively, and they were also reported to produce AFB, AFG and aspergillic acid [14]. However, none of these species were recorded from peanut samples in the present study even though some strains exhibited similar morphological and chemical characteristics as reported by the author. This indicated that the geographical area might be one of the factors that determine the type of Aspergillus spp. that colonise peanuts in fields. In the present study, the raw peanut samples marketed in Malaysia were mostly imported from other countries such as India, China and Vietnam. None of them were from Argentina.
Based on the phylogenetic analysis, both aflatoxigenic and non-aflatoxigenic A. flavus could not be differentiated based on the sequences of ITS and β-tubulin sequences. They were grouped in the same clade except for A. tamarii that formed a separate clade. The current results are supported by a previous study on A. flavus population from maize [43] and chestnut [22] in Italy, which reported that A. flavus was the main species responsible for aflatoxin contamination, and both aflatoxigenic and non-aflatoxigenic strains were also grouped in the same clade.
Molecular analysis on the aflatoxin biosynthesis gene cluster has proven to be most useful to differentiate the aflatoxigenic and non-aflatoxigenic strains of A. flavus. In recent decades, aflatoxin biosynthesis genes have been targeted for the detection of aflatoxigenic fungi in food samples, as the presence of these genes is compulsory for the synthesis of aflatoxin [9,43,44,45]. According to [43], the variability in the aflatoxin gene cluster that exists in the A. flavus population is useful in order to understand the risk of aflatoxin contamination as well as the selection of biocontrol agents.
Two sets of multiplex PCRs were used in the present study to detect the presence of aflatoxin biosynthesis genes that code for proteins involved in the aflatoxin biosynthetic pathway at the early stage (aflD and pksA), middle stage (aflM), and the late stage (aflP), and in the regulatory gene (aflR) that plays an important role in controlling structural gene expressions [46]. All genes were successfully amplified in the aflatoxigenic A. flavus strains (Chemotypes I, II, V), while the non-aflatoxigenic strains failed to amplify at least one of the targeted genes except for a few strains (Chemotype IV). The present findings are in agreement with previous studies [9,44,45,47]. According to [48], the non-aflatoxigenic fungi have varyious amplification patterns. This was further supported by [43] who successfully grouped the non-aflatoxigenic strains into four different amplification patterns.
In contrast, A. flavus strains in Chemotype III were unable to produce aflatoxin even though all the genes were present. Similar findings were also demonstrated by previous researchers [43,48]. The authors suggested that other genes involved in the aflatoxin biosynthesis (which was not tested in the present study) might be lacking or carry some deletions. Chang et al. [29] studied the deletions of a part or the entire aflatoxin biosynthesis gene cluster in non-aflatoxigenic A. flavus and suggested that small deletions or mutations in the related genes such as those involved in the signalling pathway or with a regulatory role might have inactivated the aflatoxin biosynthesis pathway of these strains. Besides, the expression of these genes is crucial in determining their ability to produce aflatoxin, as the protein (enzymes) coded by these genes is needed to catalyse the conversion of each aflatoxin precursors. However, gene expression varied among the A. flavus strains depending on the physiological and environmental conditions [46,49]. A study by [46] demonstrated a significant difference between aflD gene expression at three water activity (aw) levels in which a higher expression was observed at 0.90 aw as compared to 0.95 aw, and no expression occurred at 0.85 aw. aflD gene expression was also reported as a reliable marker to differentiate between aflatoxigenic and non-aflatoxigenic A. flavus [50]. Besides, the authors suggested to grow the non-aflatoxigenic A. flavus strain on the natural food matrix in order to confirm their aflatoxigenic potential.
According to [48], simple mutations (substitution of some bases) could lead to the formation of non-functional products. For example, the aflR gene is a regulatory gene and plays an important role in regulating the activity of other structural genes such as aflP (omtA), aflD (nor-1) and aflM (ver-1), and any mutations occurring in the gene will produce a non-functional AFLR gene product that fails to regulate the expression of the structural gene. As a result, no aflatoxin will be produced. The aflR gene is also present in some strains of A. oryzae and A. sojae despite having no record of aflatoxin production [28]. However, the sequences of the amplified aflR gene, which was named A. oryzae-type aflR, showed a consistent variation and can be distinguished from A. flavus. It was postulated that this change might affect the DNA-binding capacity of the AFLR protein and disrupt the aflatoxin biosynthesis.
In the present study, A. flavus was the predominant species from section Flavi that was found in raw peanut kernel samples collected from all stakeholders along the supply chain. The findings are in line with a previous study by [51], who reported the predominance of A. flavus in peanuts from the Busia and Homa bay districts of Western Kenya. Another study by [52] also reported that A. flavus was the dominant species found in peanuts during storage. A. flavus was able to survive even after the peanuts had been dried prior to storage to reach the moisture content level of less than 11% before they were packed for export. The occurrence of A. flavus in the imported peanuts as reported in the present study has proven the survival of its conidia or sclerotia in dried peanut kernels. In contrast, A. parasiticus is more dominant in soils from the peanut field as reported by [53], and this might explain the absence of A. parasiticus in the present study.
The surveillance and enforcement conducted on the imported raw peanuts by the authorities are only focusing on the aflatoxin level but not the aflatoxigenic fungi that are responsible for aflatoxin production. Thus, the presence of aflatoxin in peanuts at any points along the supply chain, mainly with the manufacturers and retailers, could be due to contamination during storage. The favourable storage conditions are the main cause for the conidia or sclerotia from the aflatoxigenic A. flavus to germinate, grow and subsequently produce aflatoxins [54,55]. Moreover, A. flavus was also reported in peanut-based products, which demonstrated its ability to invade processed food [8,37]. Thus, the presence of aflatoxin in peanut-based products could be explained by the accumulation and carryover of aflatoxin from raw peanuts or post-contamination of A. flavus in the product itself, especially during storage.
4. Conclusions
Molecular analyses on the DNA sequences of ITS and β-tubulin genes have confirmed that A. flavus was the only species in section Flavi that contaminated raw peanuts and peanut-based products in this study except for one isolate of A. tamarii. The phylogenetic analysis grouped all A. flavus strains from Chemotypes I–V in the same clade, and A. tamarii in a separate clade. In addition, the aflatoxigenic and non-aflatoxigenic A. flavus have been described based on the molecular analysis of the aflatoxin biosynthesis genes, aflR, aflP (omtA), aflD (nor-1), aflM (ver-1) and pksA, in which the results are in line with the aflatoxin production that was described in the previous study [18] except for A. flavus strains in Chemotype III. The non-aflatoxigenic A. flavus showed varying amplification patterns, which are related to the inability of these isolates to produce aflatoxin.
5. Materials and Methods
5.1. Fungal Isolates
A total of 77 out of 128 aflatoxigenic and non-aflatoxigenic Aspergillus section Flavi strains (morphologically identified as A. flavus and A. nomius) isolated from raw peanuts and peanut-based products from the previous study [18] were used for molecular identification and characterisation in the present study. The source of isolation, chemotype groups, and the GenBank accession number are listed in Table 1. The strains used in this study have been characterised previously using a morphological and chemical approach in which all strains were grouped into five different chemotype profiles depending on the production of B- and G-group aflatoxins, aspergillic acid, and cyclopiazonic acid (CPA). All strains consistently produced aspergillic acid, which was indicated by the orange colour on the reverse of AFPA media. However, the production of aflatoxins and CPA varied and were classified into six different chemotype profiles: Chemotype I (AFB and CPA), Chemotype II (AFB), Chemotype III (CPA), Chemotype IV (none), Chemotype V (AFB, AFG and CPA), and Chemotype VI (AFG). A reference culture of A. flavus (NRRL 3357) was used as a positive control. Fungal isolates were sub-cultured on PDA slant and incubated at 30 °C for seven days to enhance the growth and sporulation before they were refrigerated at 4 °C for further use.
5.2. Molecular Identification of Aspergillus Section Flavi
5.2.1. Genomic DNA Extraction
Fungal mycelia for genomic DNA extraction were prepared by inoculating the fungal conidia in in a 150-mL Erlenmeyer flask containing 50 mL Potato Dextrose Broth (PDB) for seven days with shaking at 150 rpm and 30 °C. The mycelia were then filtered using sterile filter paper No. 1 (Whatman, Maidstone, England) and dried under laminar flow. The dried mycelia were subsequently ground to fine powder in liquid nitrogen using a mortar and pestle. The powdered mycelia were weighed and transferred into a 1.5-mL microcentrifuge tube. The genomic DNA extraction was performed by using the DNeasy Plant MiniKit (QIAGEN, Hilden, Germany), following the manufacturer’s instructions, and the purified DNA was kept at −20 °C until further use.
5.2.2. PCR Amplification and Sequencing of ITS and β-Tubulin Genes
The primer pairs used for the ITS region and β-tubulin gene are listed in Table 2. The amplification reaction was carried out in a 25 µL reaction containing 1.0 µL of template DNA (~100 ng), 12.5 µL of EnonoTaq Plus Green 2× Master Mix (Lucigen, Middleton, WI, USA), 2.5 µL of each primer (1.0 µM) (MyTACG Bioscience Enterprise, Selangor, Malaysia) and 6.5 µL of sterile dH2O from Elga PureLab Water Purification System (Elga LabWater, High Wycombe, UK). The Master Mix contains the following materials: 0.1 units/µL of EconoTaq DNA Polymerase, Reaction Buffer (pH 9.0), 400 µM dATP, 400 dGTP, 400 dCTP, 400 dATP, 3 mM MgCl2, and blue and yellow tracking dyes. The negative control was prepared by using sterile dH2O to replace the fungal DNA template. The PCR amplification was performed by using a Veriti Thermal Cycler machine (Applied Biosystems, Waltham, MA, USA). A PCR program for each primer was optimised by using gradient PCR. The optimised condition was as follows: an initial step at 95 °C for one minute, followed by 35 cycles of denaturation at 95 °C for one minute, annealing (at 55 °C for ITS and 61 °C for β-tubulin) for one minute, extension at 72 °C for one minute, and final extension at 72 °C for five minutes. Next, 5 µL of PCR product was loaded in the well and a 100-bp DNA ladder (GeneDireX, Taiwan) was used as a comparison to estimate the size of the PCR product. Gel electrophoresis was conducted by using 1.5% agarose gel (1st Base, Selangor, Malaysia) stained with 0.01% ethidium bromide (Vivantis Technologies, Selangor, Malaysia), and run for 30 min (100 V, 400 mA) using 1× Tris Borate-Ethylenediaminetetraacetic acid, EDTA (TBE) Buffer (1st Base, Selangor, Malaysia). The gel was visualised under UV light and captured using a gel documentation system (SynGene, Cambridge, UK). The PCR products were sent for DNA purification and sequencing to a local service provider (MyTACG Bioscience Enterprise, Selangor, Malaysia).
5.2.3. Sequence Alignment and Species Identification
Following sequencing, consensus sequences were obtained by aligning and editing the forward and reverse sequences using ClustalW in Molecular Evolution and Genetic Analysis (MEGA 7) 2016 software [56]. The consensus sequences were then used to compare with the existing sequences in the GenBank database (http://www.ncbi.nlm.nih.gov) using the Basic Local Alignment Search Tool (BLAST). The identity of isolates was determined by the closest matches between the query and existing sequence from the BLAST search and presented as percentage match of similarity (from 99% to 100%).
5.3. Phylogenetic Analysis
Multiple sequence alignment and phylogenetic analysis was performed using MEGA 7 2016 software [56]. The Maximum Likelihood (ML) method was used on individual and combined ITS and β-tubulin sequences to construct the phylogenetic tree. The ex-type for eight species of Aspergillus section Flavi as listed in Table 1 were downloaded from the GenBank and included in the phylogenetic analysis for comparison with the current isolates. A. niger CBS 113.56 was used as the outgroup. A model test was run to determine the best substitution DNA models with the lowest Akaike Information Criterion (AIC) scores. Tamura 3-parameter model was used to construct the ML tree and the tree reliability was estimated using the bootstrap method with 1000 replicates. Gaps and missing data were treated as complete deletion and excluded from the analysis. A total of 430, 389, and 819 nucleotide characters in the final dataset of individual ITS, β-tubulin, and combined sequences were used in constructing the ML tree respectively.
5.4. PCR Amplification and Detection of Aflatoxin Biosynthesis Genes
Five genes, namely the aflR, aflP (omtA), aflD (nor-1), aflM (ver-1) and pksA genes, from the aflatoxin biosynthesis cluster and one sugar utilisation gene (glcA), as listed previously in Table 3, were amplified using two sets of multiplex PCRs as shown in Table 4. A gradient PCR was used to optimise the annealing temperature from 60–70 °C.
Table 3.
List of primers used for DNA sequencing, aflatoxin biosynthesis genes and sugar utilisation gene detection.
| Target Gene | Primer | Primer Sequences | Size (bp) | References |
|---|---|---|---|---|
| ITS region | ITS 1 | 5′-TCC GTA GGT GAA CCT GCG G-3′ | 600 | [57,58] |
| ITS 4 | 5′-TCC TCC GCT TAT TGA TAT GC-3′ | |||
| β-tubulin | Bt2a | 5′- GGT AAC CAA ATC GGT GCT GCT TTC-3′ | 495 | [57] |
| Bt2b | 5′-ACC CTC AGT GTA GTG ACC CTT GGC-3′ | |||
| aflR | aflr1 | 5′-TAT CTC CCC CCG GGC ATC TCC CGG-3′ | 1032 | [48,59] |
| aflr2 | 5′-CCG TCA GAC AGC CAC TGG ACA CGG-3′ | |||
| aflP (omtA) | omt1 | 5′-GGC CCG GTT CCT TGG CTC CTA AGC-3′ | 1024 | [59] |
| omt2 | 5′-CGC CCC AGT GAG ACC CTT CCT CG-3′ | |||
| aflD (nor-1) | nor1 | 5′-ACC GCT ACG CCG GCA CTC TCG GCA C-3′ | 400 | [48] |
| nor2 | 5′-GTT GGC CGC CAG CTT CGA CAC TCC G-3′ | |||
| aflM (ver-1) | ver1 | 5′-GCC GCA GGC CGC GGA GAA AGT GGT-3′ | 537 | [48] |
| ver2 | 5′-GGG GAT ATA CTC CCG CGA CAC AGC C-3′ | |||
| pksA | pksa1 | 5′-GCT GGG ATT CTG CAT GGG TT-3′ | 536 | [30] |
| pksa2 | 5′-CAG TTG CTC CCA AGG AGT GGT-3′ | |||
| glcA | glca1 | 5′-GTA CGA TGC AAA TGG CGT CC-3′ | 851 | [60] |
| glca2 | 5′-GAA GCT CTG TGT CGT TGG GA-3′ |
Table 4.
Multiplex PCR condition.
| Set | Primers | PCR Reaction Condition | Cycle |
|---|---|---|---|
| 1 | omt1/omt2, pksa1/pksa2, glca1/glca2 | Initial denaturation: 95 °C, 1 min Denaturation: 95 °C, 1 min Annealing: 61 °C, 1 min Extension: 72 °C, 1 min Final extension: 72 °C, 5 min |
1 30 30 30 1 |
| 2 | aflr1/aflr2, ver1/ver2, nor1/nor2 | Initial denaturation: 95 °C, 1 min Denaturation: 95 °C, 1 min Annealing: 67 °C, 1 min Extension: 72 °C, 1 min Final extension: 72 °C, 5 min |
1 30 30 30 1 |
Acknowledgments
The authors would like to acknowledge the contribution of the Ministry of Education Malaysia for the HICOE research grant (6369114) and research facility at the Institute of Tropical Agriculture and Food Security, and for sponsoring the first author for her PhD studies under the Academic Staff Training Scheme through Universiti Sains Malaysia.
Author Contributions
Conceptualization, M.N., S.J., M.A.R.N.-K. and C.K.C.; Formal analysis, M.N.; Funding acquisition, S.J. and M.A.R.N.-K.; Investigation, M.N. and A.H.F.; Methodology, M.N., A.H.F., S.J., M.A.R.N.-K. and S.R.; Project administration, S.J. and M.A.R.N.-K.; Resources, S.J., M.A.R.N.-K. and S.R.; Supervision, S.J., M.A.R.N.-K., S.R. and C.K.C.; Writing—original draft, M.N.; Writing—review and editing, S.J., M.A.R.N.-K., S.R. and N.I.P.S.
Funding
This research was funded by Universiti Putra Malaysia, UPM/800-3/3/1/GPB/2018/9658100.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
This study provides data on the occurrence of aflatoxigenic and non-aflatoxigenic A. flavus in imported peanuts, and the findings suggest an urgent intervention and implementation of control strategies for the fungal growth and aflatoxin accumulation in imported peanuts at the storage facilities.
References
- 1.Gnonlonfin G.J.B., Hell K., Adjovi Y., Fandohan P., Koudande D.O., Mensah G.A., Sanni A., Brimer L. A review on aflatoxin contamination and its implications in the developing world: A sub-Saharan African perspective. Crit. Rev. Food Sci. Nutr. 2013;53:349–365. doi: 10.1080/10408398.2010.535718. [DOI] [PubMed] [Google Scholar]
- 2.Lewis L., Onsongo M., Njapau H., Schurz-Rogers H., Luber G., Kieszak S., Nyamongo J., Backer L., Dahiye A., Misore A., et al. Aflatoxin contamination of commercial maize products during an outbreak of acute aflatoxicoses in Eastern and Central Kenya. Environ. Health Perspect. 2005;113:1763–1767. doi: 10.1289/ehp.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y., Wu F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Perspect. 2010;118:818–824. doi: 10.1289/ehp.0901388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. IARC Monographs on the Evaluation of Carcinogenic Risk of Chemicals to Humans. Volume 56. International Agency for Research on Cancer; Lyon, France: 1993. IARC Aflatoxins; pp. 245–395. [Google Scholar]
- 5.Godet M., Munaut F. Molecular strategy for identification in Aspergillus section Flavi. FEMS Microbiol. Lett. 2010;304:157–168. doi: 10.1111/j.1574-6968.2009.01890.x. [DOI] [PubMed] [Google Scholar]
- 6.Nyirahakizimana H., Mwamburi L., Wakhisi J., Mutegi C.K., Christie M.E., Wagacha J.M. Occurrence of Aspergillus species and aflatoxin contamination in raw and roasted peanuts from formal and informal markets in Eldoret and Kericho, Kenya. Adv. Microbiol. 2013;03:333–342. doi: 10.4236/aim.2013.34047. [DOI] [Google Scholar]
- 7.Ozbey F., Kabak B. Natural co-occurrence of aflatoxins and ochratoxin A in spices. Food Control. 2012;28:354–361. doi: 10.1016/j.foodcont.2012.05.039. [DOI] [Google Scholar]
- 8.Reddy K.R.N., Farhana N.I., Salleh B. Occurrence of Aspergillus spp. and aflatoxin B1 in Malaysian foods used for human consumption. J. Food Sci. 2011;76:99–104. doi: 10.1111/j.1750-3841.2011.02133.x. [DOI] [PubMed] [Google Scholar]
- 9.Kim D.M., Chung S.H., Chun H.S. Multiplex PCR assay for the detection of aflatoxigenic and non-aflatoxigenic fungi in meju, a Korean fermented soybean food starter. Food Microbiol. 2011;28:1402–1408. doi: 10.1016/j.fm.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 10.Kim K.M., Lim J., Lee J.J., Hurh B.S., Lee I. Characterization of aspergillus sojae isolated from meju, korean traditional fermented soybean brick. J. Microbiol. Biotechnol. 2017;27:251–261. doi: 10.4014/jmb.1610.10013. [DOI] [PubMed] [Google Scholar]
- 11.Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.1080/15572536.2008.11832477. [DOI] [PubMed] [Google Scholar]
- 12.Pildain M.B., Vaamonde G., Cabral D. Analysis of population structure of Aspergillus flavus from peanut based on vegetative compatibility, geographic origin, mycotoxin and sclerotia production. Int. J. Food Microbiol. 2004;93:31–40. doi: 10.1016/j.ijfoodmicro.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Reis T.A., Baquião A.C., Atayde D.D., Grabarz F., Corrêa B. Characterization of Aspergillus section Flavi isolated from organic Brazil nuts using a polyphasic approach. Food Microbiol. 2014;42:34–39. doi: 10.1016/j.fm.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Pildain B., Frisvad J.C., Vaamonde G., Cabral D., Varga J., Samson R.A. Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int. J. Syst. Evol. Microbiol. 2008;58:725–735. doi: 10.1099/ijs.0.65123-0. [DOI] [PubMed] [Google Scholar]
- 15.Varga J., Frisvad J.C., Samson R.A. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud. Mycol. 2011;69:57–80. doi: 10.3114/sim.2011.69.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afsah-Hejri L., Jinap S., Hajeb P., Radu S., Shakibazadeh S. A review on mycotoxins in food and feed: Malaysia case study. Compr. Rev. Food Sci. Food Saf. 2013;12:629–651. doi: 10.1111/1541-4337.12029. [DOI] [PubMed] [Google Scholar]
- 17.Norlia M., Nor-Khaizura M.A., Selamat J., Abu Bakar F., Radu S., Chin C.K. Evaluation of aflatoxins and Aspergillus sp. contamination in raw peanuts and peanut-based products along the supply chain in Malaysia. Food Addit. Contam. Part A. 2018;23:1–16. doi: 10.1080/19440049.2018.1488276. [DOI] [PubMed] [Google Scholar]
- 18.Norlia M., Jinap S., Nor-khaizura M.A.R., Son R., Chin C.K. Sardjono Polyphasic approach to the identification and characterization of aflatoxigenic strains of Aspergillus section Flavi isolated from peanuts and peanut-based products marketed in Malaysia. Int. J. Food Microbiol. 2018;282:9–15. doi: 10.1016/j.ijfoodmicro.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 19.Frisvad J.C., Hubka V., Ezekiel C.N., Hong S.B., Nováková A., Chen A.J., Arzanlou M., Larsen T.O., Sklenář F., Mahakarnchanakul W., et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019;93:1–63. doi: 10.1016/j.simyco.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tam E.W.T., Chen J.H.K., Lau E.C.L., Ngan A.H.Y., Fung K.S.C., Lee K.C., Lam C.W., Yuen K.Y., Lau S.K.P., Woo P.C.Y. Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: Characterization by internal transcribed spacer, β-tubulin, and calmodulin gene sequencing, metabolic fingerprinting, and matrix-assisted laser des. J. Clin. Microbiol. 2014;52:1153–1160. doi: 10.1128/JCM.03258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitt J.I., Hocking A.D., Glenn D.R. An improved medium for the detection fo Aspergillus flavus and A. parasiticus. J. Appl. Bacteriol. 1983;54:109–114. doi: 10.1111/j.1365-2672.1983.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 22.Prencipe S., Siciliano I., Contessa C., Botta R., Garibaldi A., Gullino M.L., Spadaro D. Characterization of Aspergillus section Flavi isolated from fresh chestnuts and along the chestnut flour process. Food Microbiol. 2018;69:159–169. doi: 10.1016/j.fm.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Samson R.A., Hong S.B., Frisvad J.C. Old and new concepts of species differentiation in Aspergillus. Med. Mycol. 2006;44:133–148. doi: 10.1080/13693780600913224. [DOI] [PubMed] [Google Scholar]
- 24.Bellemain E., Carlsen T., Brochmann C., Coissac E., Taberlet P., Kauserud H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010;10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samson R.A., Visagie C.M., Houbraken J., Hong S.-B., Hubka V., Klaassen C.H.W., Perrones G., Seifert K.A., Susca A., Tanney J.B., et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J., Chang P., Ehrlich K.C., Cary J.W., Bhatnagar D., Cleveland T.E., Payne G.A., Linz J.E., Woloshuk C.P., Bennett W., et al. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payne G.A., Nierman W.C., Wortman J.R., Pritchard B.L., Brown D., Dean R.A., Bhatnagar D., Cleveland T.E., Machida M., Yu J. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 2006;44:9–11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- 28.Lee C.Z., Liou G.Y., Yuan G.F. Comparison of the aflR gene sequences of strains in Aspergillus section Flavi. Microbiology. 2006;152:161–170. doi: 10.1099/mic.0.27618-0. [DOI] [PubMed] [Google Scholar]
- 29.Chang P.K., Horn B.W., Dorner J.W. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 2005;42:914–923. doi: 10.1016/j.fgb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Yin Y., Lou T., Yan L., Michailides T.J., Ma Z. Molecular characterization of toxigenic and atoxigenic Aspergillus flavus isolates, collected from peanut fields in China. J. Appl. Microbiol. 2009;107:1857–1865. doi: 10.1111/j.1365-2672.2009.04356.x. [DOI] [PubMed] [Google Scholar]
- 31.Chang P.K., Ehrlich K.C. What does genetic diversity of Aspergillus flavus tell us about Aspergillus oryzae? Int. J. Food Microbiol. 2010;138:189–199. doi: 10.1016/j.ijfoodmicro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 32.Geiser D.M., Pitt J.I., Taylor J.W. Cryptic Speciation and Recombination in the Aflatoxin-producing Fungus Aspergillus flavus. Proc. Natl. Acad. Sci. USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiser D.M., Dorner J.W., Horn B.W., Taylor J.W. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genet. Biol. 2000;31:169–179. doi: 10.1006/fgbi.2000.1215. [DOI] [PubMed] [Google Scholar]
- 34.Machida M., Yamada O., Gomi K. Genomics of Aspergillus oryzae: Learning from the history of koji mold and exploration of its future. DNA Res. 2008;15:173–183. doi: 10.1093/dnares/dsn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martins L.M., Sant’Ana A.S., Fungaro M.H.P., Silva J.J., Nascimento M. da S. do; Frisvad, J.C.; Taniwaki, M.H. The biodiversity of Aspergillus section Flavi and aflatoxins in the Brazilian peanut production chain. Food Res. Int. 2017;94:101–107. doi: 10.1016/j.foodres.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Passone M.A., Rosso L.C., Ciancio A., Etcheverry M. Detection and quantification of Aspergillus section Flavi spp. in stored peanuts by real-time PCR of nor-1 gene, and effects of storage conditions on aflatoxin production. Int. J. Food Microbiol. 2010;138:276–281. doi: 10.1016/j.ijfoodmicro.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Wagacha J.M., Mutegi C., Karanja L., Kimani J., Christie M.E. Fungal species isolated from peanuts in major Kenyan markets: Emphasis on Aspergillus section Flavi. Crop Prot. 2013;52:1–9. doi: 10.1016/j.cropro.2013.05.004. [DOI] [Google Scholar]
- 38.Rank C., Klejnstrup M.L., Petersen L.M., Kildgaard S., Frisvad J.C., Held Gotfredsen C., Ostenfeld Larsen T. Comparative chemistry of Aspergillus oryzae (RIB40) and A. flavus (NRRL 3357) Metabolites. 2012;2:39–56. doi: 10.3390/metabo2010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wicklow D.T., Wilson D.M., Nelsen T.C. Survival of Aspergillus flavus sclerotia and conidia buried in soil in Illinois or Georgia. Phytopathology. 1993;83:1141–1147. doi: 10.1094/Phyto-83-1141. [DOI] [Google Scholar]
- 40.Horn B., Sorensen R., Lamb M., Sobolev V., Olarte R., Worthington C., Carbone I. Sexual reproduction in Aspergillus flavus sclerotia naturally produced in corn. Phytopathology. 2014;104:75–85. doi: 10.1094/PHYTO-05-13-0129-R. [DOI] [PubMed] [Google Scholar]
- 41.Goto T., Wicklow D.T., Ito Y. Aflatoxin and cyclopiazonic acid production by a sclerotium-producing Aspergillus tamarii strain. Appl. Environ. Microbiol. 1996;62:4036–4038. doi: 10.1128/aem.62.11.4036-4038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito Y., Peterson S.W., Wicklow D.T., Goto T. Aspergillus pseudotamarii, a new aflatoxin producing species in Aspergillus section Flavi. Mycol. Res. 2001;105:233–239. doi: 10.1017/S0953756200003385. [DOI] [Google Scholar]
- 43.Gallo A., Stea G., Battilani P., Logrieco A.F., Perrone G. Molecular characterization of an Aspergillus flavus population isolated from maize during the first outbreak of aflatoxin contamination in Italy. Phytopathol. Mediterr. 2012;51:198–206. [Google Scholar]
- 44.Erami M., Hashemi S., Pourbakhsh S., Shahsavandi S., Mohammadi S., Shooshtari A., Jahanshiri Z. Application of PCR on detection of aflatoxinogenic fungi. Arch. Razi Inst. 2007;62:95–100. [Google Scholar]
- 45.Davari E., Mohsenzadeh M., Mohammadi G., Rezaeian-Doloei R. Characterization of aflatoxigenic Aspergillus flavus and A. parasiticus strain isolates from animal feedstuffs in northeastern Iran. Iran. J. Vet. Res. 2015;16:150–155. [PMC free article] [PubMed] [Google Scholar]
- 46.Abdel-Hadi A., Carter D., Magan N. Temporal monitoring of the nor-1 (aflD) gene of Aspergillus flavus in relation to aflatoxin B1 production during storage of peanuts under different water activity levels. J. Appl. Microbiol. 2010;109:1914–1922. doi: 10.1111/j.1365-2672.2010.04820.x. [DOI] [PubMed] [Google Scholar]
- 47.Rodrigues P., Venâncio A., Kozakiewicz Z., Lima N. A polyphasic approach to the identification of aflatoxigenic and non-aflatoxigenic strains of Aspergillus Section Flavi isolated from Portuguese almonds. Int. J. Food Microbiol. 2009;129:187–193. doi: 10.1016/j.ijfoodmicro.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 48.Criseo G., Bagnara A., Bisignano G. Differentiation of aflatoxin-producing and non-producing strains of Aspergillus favus group. Lett. Appl. Microbiol. 2001;33:291–295. doi: 10.1046/j.1472-765X.2001.00998.x. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt-Heydt M., Abdel-Hadi A., Magan N., Geisen R. Complex regulation of the aflatoxin biosynthesis gene cluster of Aspergillus flavus in relation to various combinations of water activity and temperature. Int. J. Food Microbiol. 2009;135:231–237. doi: 10.1016/j.ijfoodmicro.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 50.Abdel-Hadi A., Carter D., Magan N. Discrimination between aflatoxigenic and non-aflatoxigenic strains of Aspergillus section Flavi group contaminationg Egyption peanuts using molecular and analystical techniques. World Mycotoxin J. 2011;4:69–77. doi: 10.3920/WMJ2010.1223. [DOI] [Google Scholar]
- 51.Mutegi C.K., Ngugi H.K., Hendriks S.L., Jones R.B. Factors associated with the incidence of Aspergillus section Flavi and aflatoxin contamination of peanuts in the Busia and Homa bay districts of western Kenya. Plant Pathol. 2012;61:1143–1153. doi: 10.1111/j.1365-3059.2012.02597.x. [DOI] [Google Scholar]
- 52.Zorzete P., Baquião A.C., Atayde D.D., Reis T.A., Gonçalez E., Corrêa B. Mycobiota, aflatoxins and cyclopiazonic acid in stored peanut cultivars. Food Res. Int. 2013;52:380–386. doi: 10.1016/j.foodres.2013.03.029. [DOI] [Google Scholar]
- 53.Kachapulula P.W., Akello J., Bandyopadhyay R., Cotty P.J. Aspergillus section Flavi community structure in Zambia influences aflatoxin contamination of maize and groundnut. Int. J. Food Microbiol. 2017;261:49–56. doi: 10.1016/j.ijfoodmicro.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torres A.M., Barros G.G., Palacios S.A., Chulze S.N., Battilani P. Review on pre- and post-harvest management of peanuts to minimize aflatoxin contamination. Food Res. Int. 2014;62:11–19. doi: 10.1016/j.foodres.2014.02.023. [DOI] [Google Scholar]
- 55.Wagacha J.M., Mutegi C.K., Christie M.E., Karanja L.W., Kimani J. Changes in fungal population and aflatoxin levels and assessment of major aflatoxin types in stored peanuts (Arachis hypogaea Linnaeus) J. Food Res. 2013;2:10–23. doi: 10.5539/jfr.v2n5p10. [DOI] [Google Scholar]
- 56.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glass N.L., Donalson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White T.J., Bruns T., Lee S., Taylor J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols. A Guide to Methods and Applications. Academic Press; New York, NY, USA: 1990. pp. 315–322. [Google Scholar]
- 59.Shapira R., Paster N., Eyal O., Menasherov M., Mett A., Salomon R. Detection of aflatoxigenic molds in grains by PCR. Appl. Environ. Microbiol. 1996;62:3270–3273. doi: 10.1128/aem.62.9.3270-3273.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siti-Aminah I. Master’s Thesis. Universiti Putra Malaysia; Selangor, Malaysia: 2017. Screening of Toxigenic and Atoxigenic Aspergillus flavus Isolates Collected from Corn Fields in UPM, Serdang. [Google Scholar]