Abstract
Staphylococcus aureus is an important nosocomial pathogen and its multidrug resistant strains, particularly methicillin-resistant S. aureus (MRSA), poses a serious threat to public health due to its limited therapeutic options. The increasing MRSA resistance towards vancomycin, which is the current drug of last resort, gives a great challenge to the treatment and management of MRSA infections. While vancomycin resistance among Malaysian MRSA isolates has yet to be documented, a case of vancomycin resistant S. aureus has been reported in our neighboring country, Indonesia. In this review, we present the antimicrobial resistance profiles of S. aureus clinical isolates in Malaysia with data obtained from the Malaysian National Surveillance on Antimicrobial Resistance (NSAR) reports as well as various peer-reviewed published records spanning a period of nearly three decades (1990–2017). We also review the clonal types and characteristics of Malaysian S. aureus isolates, where hospital-associated (HA) MRSA isolates tend to carry staphylococcal cassette chromosome mec (SCCmec) type III and were of sequence type (ST)239, whereas community-associated (CA) isolates are mostly SCCmec type IV/V and ST30. More comprehensive surveillance data that include molecular epidemiological data would enable further in-depth understanding of Malaysian S. aureus isolates.
Keywords: Antimicrobial resistance, community-associated (CA), hospital-associated (HA), Staphylococcus aureus, macrolide-lincosamide-streptogramin B (MLSB), Malaysian clinical isolates, methicillin-resistance S. aureus (MRSA), methicillin-susceptible S. aureus (MSSA), staphylococcal cassette chromosome mec (SCCmec) type, sequence types (STs)
1. Introduction
Staphylococcus aureus is a frequently encountered Gram-positive nosocomial pathogen that is associated with a wide array of diseases, ranging from simple skin infection to more serious and potentially life-threatening infections such as infective endocarditis and toxic shock syndrome [1,2]. Its ability to rapidly develop and acquire antimicrobial resistance has led to the emergence of multidrug resistant strains such as methicillin-resistant S. aureus (MRSA) [1]. The MRSA strains have become endemic in most hospitals worldwide, including in Asia [3], and are a treatment challenge to physicians.
In their 2014 global report, the World Health Organization (WHO) listed MRSA as one of the seven pathogens of international concern and that has been associated with a high number of mortality and septic shock cases compared to MSSA [1]. In Malaysia, the national prevalence rate of MRSA among S. aureus clinical isolates ranged from 17.2% to 28.1%, with the rates recorded at 18% and 19.8% for the years 2016 and 2017, respectively [4,5]. At present, vancomycin remains the drug of last resort for the treatment of MRSA infections [6]. However, reports regarding MRSA strains that had developed resistance to vancomycin have emerged in many parts of the world, with the first of such strains reported in the United States almost two decades ago [7]. Nevertheless, the occurrence of such strains, known as vancomycin-resistant S. aureus (VRSA), is not as abundant as MRSA. To date, in the United States alone, 14 VRSA strains have been reported, of which only one is community-associated (CA) while the rest are hospital-associated (HA) strains [8]. While VRSA is still considered as a rare bacterium that has yet to achieve endemic status like MRSA, the possibility of VRSA isolates becoming widely disseminated must not be ignored, as it could cause serious implications for public health. There has been no published report of vancomycin resistance among Malaysian MRSA isolates so far. However, a VRSA strain with a vancomycin MIC value of ≥ 32 µg/mL has been isolated from a patient in the neighboring country, Indonesia [9]. Thus, we believe VRSA strains could also be detected in Malaysia sooner or later.
The purpose of this review is to provide a general overview of the antimicrobial resistance trends of clinical S. aureus isolates in Malaysia obtained from various published reports of individual studies as well as the Malaysian National Surveillance on Antimicrobial Resistance (NSAR) annual reports. The molecular mechanism of antibiotic resistance and other characteristics of the S. aureus isolates, such as their sequence types (STs) and other molecular epidemiological markers, are also presented.
2. Antibiotic Susceptibility Profiles
Malaysian NSAR is an active antimicrobial surveillance program which is published online annually and encompasses various bacterial pathogens including S. aureus. NSAR is under the Malaysian One Health Antimicrobial Resistance (MyOHAR) program, which is hosted by the Institute for Medical Research (IMR) starting from year 2003 (with the exception of year 2006) and involves clinical isolates tested by hospital microbiology laboratories throughout Malaysia (https://www.imr.gov.my/MyOHAR/index.php/site/archive_rpt). The antimicrobials tested for S. aureus in the NSAR reports vary from year to year but usually include erythromycin, gentamicin, co-trimoxazole, rifampicin, fusidic acid, and clindamycin, besides many others. Most Malaysian studies, as well as NSAR data, tend to focus more on MRSA isolates rather than methicillin-susceptible S. aureus (MSSA) isolates, thus data concerning antimicrobial resistance among Malaysian MSSA isolates are rather limited. In the guidelines published by the Clinical and Laboratories Standard Institute (CLSI), a total of 43 antibiotics from 16 antimicrobial classes were listed for S. aureus (Table 1) [10]. The European Centre for Disease Prevention and Control (ECDC) in conjunction with the Centers for Disease Control and Prevention (CDC) of the United States also published a list of antimicrobials recommended for susceptibility testing of S. aureus isolates, which included 22 antibiotics from 17 antimicrobial classes, including three agents which were not listed in the CLSI guideline, namely fusidic acid, tigecycline, and fosfomycin [11]. Nevertheless, the guideline published by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) covered as many as 16 antimicrobial classes encompassing 38 antimicrobial agents, and includes the three agents that are not covered by CLSI [12].
Table 1.
List of antimicrobials recommended by the joint commission of the European Centre for Disease Prevention and Control (ECDC) and the United States Centers for Disease Control and Prevention (CDC) for antimicrobial susceptibility testing of S. aureus [11]. The availability of standard breakpoints for the antimicrobials from CLSI or EUCAST and the antimicrobials reported by the Malaysian National Surveillance on Antimicrobial Resistance (NSAR) reports are indicated.
| Antimicrobial Category | Antimicrobial Agent | CLSI Breakpoint | EUCAST Breakpoint | Reported by Malaysian NSAR |
|---|---|---|---|---|
| Aminoglycosides | Gentamicin | Yes | Yes | 2005–2017 |
| Ansamycins | Rifampin/rifampicin | Yes | Yes | 2005–2017 |
| Anti-MRSA cephalosporins | Ceftaroline | Yes | Yes | No |
| Anti-staphylococcal β-lactams (or cephamycins) | Oxacillin (or cefoxitin) | Yes | Yes | 2003–2005 |
| Fluoroquinolones | Ciprofloxacin | Yes | Yes | 2007–2014 |
| Moxifloxacin | Yes | Yes | No | |
| Folate pathway inhibitors | Trimethoprim-sulfamethoxazole | Yes | Yes | 2005–2017 |
| Fucidanes | Fusidic acid | No | Yes | 2005–2017 |
| Glycopeptides | Vancomycin | Yes | Yes | 2003–2017 |
| Teicoplanin | Yes | Yes | 2007–2008 | |
| Telavancin | Yes | Yes | No | |
| Glycylcyclines | Tigecycline | No | Yes | No |
| Lincosamides | Clindamycin | Yes | Yes | 2005–2017 |
| Lipopeptides | Daptomycin | Yes | Yes | No |
| Macrolides | Erythromycin | Yes | Yes | 2005–2017 |
| Oxazolidinones | Linezolid | Yes | Yes | 2009–2017 |
| Phenicols | Chloramphenicol | Yes | Yes | 2005–2014 |
| Phosphonic acids | Fosfomycin | No | Yes | No |
| Streptogramins | Quinupristin-dalfopristin | Yes | Yes | 2011 |
| Tetracyclines | Tetracycline | Yes | Yes | 2009–2014 |
| Doxycycline | Yes | Yes | No | |
| Minocycline | Yes | Yes | No |
CLSI, Clinical and Laboratories Standard Institute; EUCAST, European Committee on Antimicrobial Susceptibility Testing.
Thus far, a total of 11 individual studies and 12 NSAR reports regarding the antimicrobial resistance profiles of clinical S. aureus in Malaysia were documented from 1990 to 2017. The details regarding the framework of data from these NSAR reports as well as individual studies are shown in Table 2.
Table 2.
Reported clinical Staphylococcus aureus studies in Malaysia.
| Study (Period) | No. of S. aureus (n) or Percentage (%) of MRSA | Study Site(s) or No. of Study Site(s) (n) | Reference |
|---|---|---|---|
| Cheong et al., 1994 (1990–1991) |
2586 35.5% (905) MRSA 539 MRSA included in study |
HKL | [13] |
| Norazah et al., 2001 (1997–1999) |
400 MRSA | Various, collected from ten hospitals throughout Malaysia | [14] |
| Al-Talib et al., 2010 (2002–2007) |
8200 1979 MRSA |
HUSM | [15] |
| Thong et al., 2009 (2003–2004 and 2007) |
66 MRSA | N/A | [16] |
| Neela et al., 2008 (2006–2007) |
32 MRSA | N/A | [17] |
| Lim et al., 2013 (2003 and 2008) |
162 MRSA | UMMC | [18] |
| Al-Talib et al., 2015 (2008) |
34 MRSA, 124 MSSA | HUSM | [19] |
| Noordin et al., 2016 (2009) |
318 MRSA | UKMMC | [20] |
| Ho et al., 2017 (2011–2012) |
175 MRSA | HRPB, KPJ and GP | [21] |
| Sit et al., 2018 (2013) |
67 MRSA | UMMC | [22] |
| Che Hamzah et al., 2019 (2016–2017) |
90 MRSA, 109 MSSA | HSNZ | [23] |
| NSAR (2003–2005) |
N/A | N/A | [24] |
| NSAR (2006) ** |
12,370 [25] 31.5% MRSA [26] |
10 GH [25] | [25,26] |
| NSAR (2007) |
13,548 [25] 28% MRSA |
12 GH [25,26] | [25,26] |
| NSAR (2008) |
23,176 26% MRSA |
13 GH | [25] |
| NSAR (2009) |
20,053 21% MRSA |
16 GH | [27] |
| NSAR (2010) |
20,007 22.2% MRSA |
16 GH | [28] |
| NSAR (2011) |
31,140 19.2% MRSA |
36 hospitals* (35 GH and 1 UH) | [29] |
| NSAR (2012) |
32,611 17.3% MRSA |
37 hospitals* (35 GH and 2 UH) | [30] |
| NSAR (2013) |
34,492 17.7% MRSA |
38 hospitals* (36 GH and 2 UH) | [31] |
| NSAR (2014) |
37,341 17.2% MRSA |
39 hospitals* (37 GH and 2 UH) | [32] |
| NSAR (2015) |
37,416 19.3% MRSA |
39 hospitals* (37 GH and 2 UH) | [33] |
| NSAR (2016) |
37,207 18% MRSA |
39 hospitals* (37 GH and 2 UH) | [4] |
| NSAR (2017) |
39,447 19% MRSA |
39 hospitals* (37 GH and 2 UH) | [5] |
* Government hospitals (GH) and university hospitals (UH) distributed in all the 13 states in Malaysia; ** No antibiotic susceptibility profile available, the national MRSA rate and number of hospitals involved were obtained from the 2007 and 2008 NSAR reports [25,26]; MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; NSAR, National Surveillance of Antibiotic Resistance; HKL, Hospital Kuala Lumpur; Hospital HUSM, Hospital Universiti Sains Malaysia; UMMC, University of Malaya Medical Centre; UKMMC, Universiti Kebangsaan Malaysia Medical Centre; HRPB, Hospital Raja Permaisuri Bainun; KPJ, KPJ Ipoh Specialist Hospital; GP, Gribbles Pathology Ipoh; HSNZ, Hospital Sultanah Nur Zahirah; GH, government hospitals; UH, university hospitals; N/A, not available.
2.1. β-Lactams (Penicillin, Oxacilin, Cefoxitin, and Cefoperazone)
Resistance towards β-lactam antimicrobials among MRSA isolates in Malaysia has always been high due to the nature of MRSA itself, where the presence of the mecA gene that encodes the penicillin binding protein 2a (PBP 2a) confers resistance to β-lactams through a mechanism in which the drug binding affinity is affected [34]. An early report involving 539 MRSA isolates from Hospital Kuala Lumpur (HKL) between 1990 and 1991 recorded full resistance to penicillin [13]. Thirty two MRSA isolates obtained between 2006 and 2007 from an unnamed tertiary hospital were also fully resistant to penicillin, oxacillin, and cefoxitin [17]. Similarly, complete resistance was also reported for oxacillin and penicillin in 66 MRSA isolates that were collected from an unspecified teaching hospital in 2003, 2004, and 2007 [16] and 1979 MRSA isolates from Hospital Universiti Sains Malaysia (HUSM), obtained from 2002 until 2007 [15]. Similar observations were also recorded among 175 MRSA isolates collected between 2011 and 2012 from Hospital Raja Permaisuri Bainun (HRPB), KPJ Ipoh Specialist Hospital (KPJ), and Gribbles Pathology Ipoh (GP) (located at Ipoh, the capital city of Perak state on the west coast of peninsular Malaysia [21]), as well as 67 MRSA strains obtained from adult patients (above 16 years old) with bacteremia in University of Malaya Medical Centre (UMMC) in 2013 [22]. Ninety MRSA isolates were obtained between 2016 and 2017 from Hospital Sultanah Nur Zahirah (HSNZ), located at Kuala Terengganu, the state capital of Terengganu on the east coast of peninsular Malaysia also showed full resistance to penicillin, oxacillin and cefoxitin [23].
Cefoperazone is rarely used in Malaysia and thus there is a scarcity of data reporting the rates of cefoperazone resistance among Malaysian MRSA isolates. To the best of our knowledge, our recent study on isolates from HSNZ is the only report of cefoperazone resistance among Malaysian MRSA isolates, with the prevalence of resistance recorded at 86.7% for the 2016–2017 isolates [23].
Reports describing β-lactam resistance among Malaysian MSSA isolates are even scarcer. An early study reported an approximately 90% prevalence of resistance to penicillin and full susceptibility to oxacillin among 124 MSSA isolates from HUSM between March to August 2008 [19], while our recent study reported 84.4% and 5.5% prevalence of resistance to penicillin and oxacillin among 109 HSNZ MSSA isolates obtained in 2016 and 2017 [23].
2.2. Aminoglycosides (Gentamicin, Amikacin, Netilmicin, and Kanamycin)
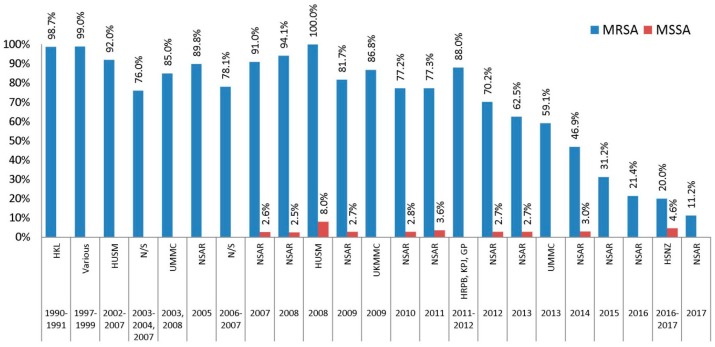
Gentamicin resistance among Malaysian MRSA isolates was generally high in the earlier years of the NSAR reports but showed a declining trend towards recent years (Figure 1). In general, MSSA isolates tend to be much more susceptible to gentamicin as compared to MRSA isolates. Among the antibiotics in the aminoglycoside group, only gentamicin was frequently reported by NSAR, while amikacin resistance was only reported in 2007 and 2008, whereas netilmicin and kanamycin resistance were never reported. In the proposed CDC-ECDC criteria for S. aureus, only gentamicin is recommended to be tested for the aminoglycoside group [11].
Figure 1.
The prevalence of gentamicin resistance among Malaysian S. aureus isolates, 1990–2017. Data from the National Surveillance of Antibiotic Resistance (NSAR) reports, 2003–2005 [24], 2007 [26], 2008 [25], 2009 [27], 2010 [28], 2011 [29], 2012 [30], 2013 [31], 2014 [32], 2015 [33], 2016 [4], and 2017 [5]; Hospital Kuala Lumpur (HKL) between 1990 and 1991 [13]; Various, collected from ten hospitals throughout Malaysia between 1997 and 1999 [14]; Hospital Universiti Sains Malaysia (HUSM) between 2002 and 2007 [15] and in 2008 [19]; not specified (N/S) between 2003 and 2004 and in 2007 [16], and between 2006 and 2007 [17]; University of Malaya Medical Centre (UMMC) in 2003 and 2008 [18], and in 2013 [22]; Universiti Kebangsaan Malaysia Medical Centre (UKMMC) in 2009 [20]; Hospital Raja Permaisuri Bainun (HRPB), KPJ Ipoh Specialist Hospital (KPJ) and Gribbles Pathology Ipoh (GP) between 2011 and 2012 [21]; and Hospital Sultanah Nur Zahirah (HSNZ) between 2016 and 2017 [23].
Isolates obtained in the 1990s showed very high prevalence of gentamicin resistance (>98%) (Figure 1). The MRSA isolates collected between 2003 and 2009 in various studies as well as the NSAR reports showed a fluctuating trend of resistance but the prevalence remained above 70%. The first NSAR report in 2005 showed a national gentamicin resistance rate of 78.1% and this increased to more than 90% in 2007 and 2008 before dropping to around 80% in 2009. Individual studies of isolates obtained within that period showed contrasting results. The prevalence of gentamicin resistance in 66 MRSA isolates collected from an unnamed teaching hospital in 2003, 2004, and 2007 was 76% [16]. In another study, 32 isolates collected in 2006 and 2007 from a likewise undetermined hospital showed a similar resistance rate of 78.1% [17]. Isolates from HUSM, which is located in the north-eastern state of Kelantan in Peninsular Malaysia, appeared to display high prevalence of gentamicin resistance in the 2000s. The 1979 HUSM isolates collected from 2002 to 2007 showed a gentamicin resistance rate of 92.0% [15] and a 100% gentamicin resistance rate for 34 isolates obtained in 2008 [19]. However, no further reports of S. aureus isolates from HUSM were available in the 2010s. From 2010 onwards, data obtained from the NSAR reports showed a steady decline in the national prevalence of gentamicin resistance among Malaysian MRSA isolates. Between 2010 and 2012, gentamicin resistance was recorded in the range of 70% to 77% [28,29,30] and decreased to 62.5% and 46.9% in 2013 and 2014, respectively [31,32]. The latest NSAR report showed that gentamicin resistance among MRSA isolates has dropped further to only 11.2% in 2017 [5] (Figure 1).
The prevalence of gentamicin resistance among MSSA isolates were much lower, with NSAR reporting rates of below 3%, except for the year 2011 and 2014 where the resistance was recorded at 3.6% and 3.0%, respectively [29,32]. The HUSM study of 124 MSSA isolates obtained in 2008 indicated a gentamicin resistance rate of 8.0% [19]. However, NSAR did not mention the prevalence of antimicrobial resistance for MSSA isolates in their reports from 2015 onwards.
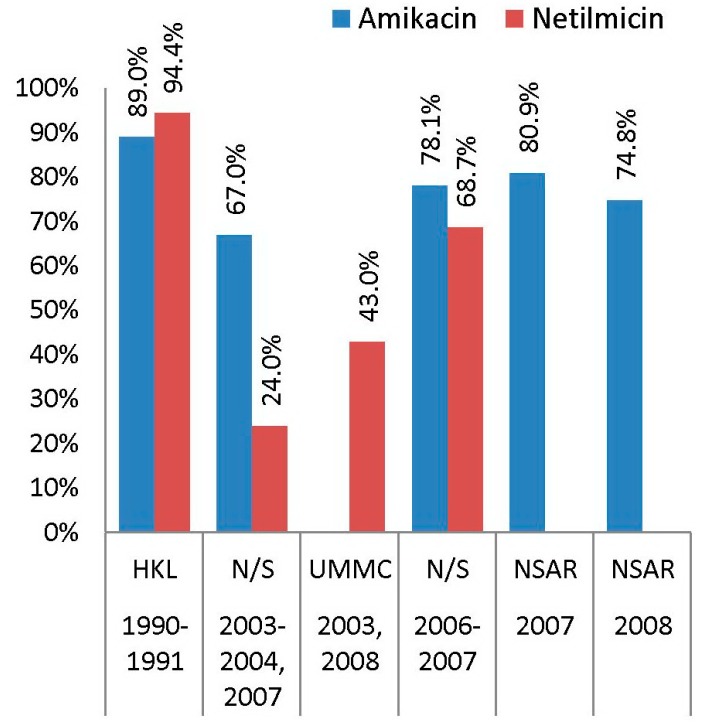
Data on amikacin and netilmicin resistance among Malaysian S. aureus isolates were only available for MRSA isolates. However, data from two NSAR reports in 2007 and 2008 included the amikacin resistance rate for all S. aureus isolates (both MRSA and MSSA), which were 78.0% and 65.0%, respectively [25,26]. Amikacin resistance among MRSA isolates is generally in the upper range, with resistance rates mostly reported above 70% (Figure 2). However, one study on 66 MRSA isolates from a local teaching hospital in 2003, 2004, and 2007 reported amikacin resistance at a slightly lower range of 67.0% [16], whereas our more recent study involving 90 MRSA isolates obtained from HSNZ in 2016 and 2017 recorded an even lower rate, which was 12.2% [23].
Figure 2.
The prevalence of amikacin and netilmicin resistance among Malaysian MRSA isolates, 1990–2008. Data from the NSAR reports; HKL between 1990 and 1991 [13]; not specified (N/S) between 2003 and 2004 and in 2007 [16], and between 2006 and 2007 [17]; and UMMC in 2003 and 2008 [18].
Netilmicin resistance among MRSA isolates had a more fluctuating pattern (Figure 2), with the lowest prevalence of resistance, reported at 24.0% by Thong et al., for isolates obtained in 2003, 2004, and 2007 [16], while the highest prevalence of resistance was reported among 539 HKL MRSA isolates collected in 1990 and 1991, which was 94.4% [13]. Neela et al. also tested their 32 MRSA isolates collected in 2006 and 2007 for susceptibilities towards amikacin, netilmicin, and kanamycin, besides gentamicin. The aminoglycoside resistance rates for their MRSA isolates were similar (both to gentamicin and amikacin at 78.1% and kanamycin at 75.0%), with netilmicin the lowest at 68.7% [17].
2.3. Macrolides (Erythromycin)
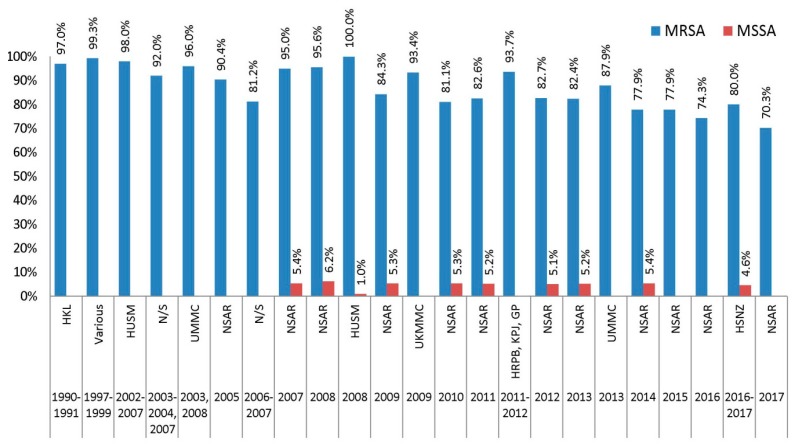
Almost all Malaysian MRSA isolates in the 1990s and early 2000s were erythromycin resistant with resistance rates of >90% (Figure 3). The first NSAR report in 2005 showed a national prevalence of 90.5% in 2005 and this increased to 95% in 2007 and 2008. Other studies of isolates obtained around this period also indicated very high prevalence of erythromycin resistance. Only a study on 32 MRSA isolates collected in 2006 and 2007 from an undisclosed tertiary hospital revealed a lower erythromycin resistance rate of 84.3% [17]. However, the NSAR report in 2009 showed that the prevalence of erythromycin resistance among the Malaysian MRSA isolates had decreased to 84.3% [27] and remained around the 80% level until 2013. The national resistance rates for erythromycin decreased further to 70.3% in the latest NSAR report of 2017 [5].
Figure 3.
The prevalence of erythromycin resistance among Malaysian S. aureus isolates, 1990–2017. Data from the NSAR reports; HKL between 1990 and 1991 [13]; Various, collected from ten hospitals throughout Malaysia between 1997 and 1999 [14]; HUSM between 2002 and 2007 [15] and in 2008 [19]; not specified (N/S) between 2003 and 2004 and in 2007 [16], and between 2006 and 2007 [17]; UMMC in 2003 and 2008 [18], and in 2013 [22]; UKMMC in 2009 [20]; HRPB, KPJ and GP between 2011 and 2012 [21]; and HSNZ between 2016 and 2017 [23].
As for MSSA isolates, data from NSAR were only available from 2007 until 2014, where the resistance rates stayed consistent at around 5% to 6% (Figure 3). Only two independent studies reported erythromycin resistance among MSSA isolates, where the rate was much lower compared to the national rates with only approximately 1.0% among 124 MSSA isolates collected in HUSM in 2008 [19], whereas the HSNZ MSSA isolates recorded a slightly higher rate at 4.6%.
2.4. Lincosamides (Clindamycin)
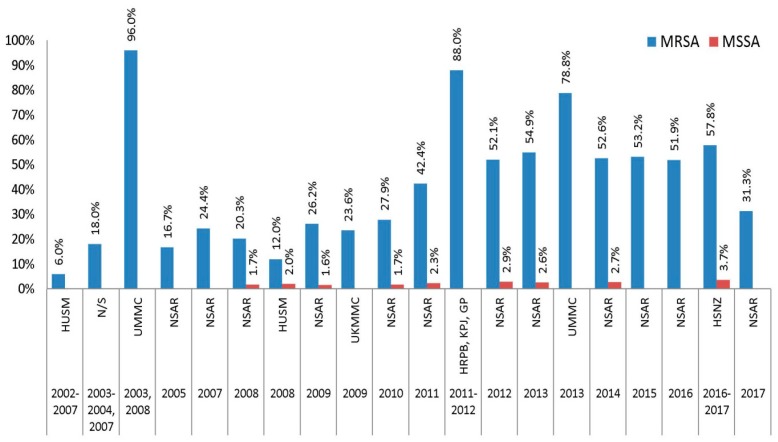
From 2002 until 2010, data from the NSAR reports as well as various studies showed that the prevalence of clindamycin resistance among Malaysian MRSA isolates is mostly low, with resistance rates hardly reaching 30% (Figure 4). The lowest prevalence of resistance was recorded at 6.0% in 1979 HUSM MRSA isolates collected from 2002 to 2007 [15]. A subsequent study of 34 HUSM MRSA isolates collected in 2008 showed an increase in the prevalence of clindamycin resistance to 12% [19]. However, a study of 162 MRSA isolates from UMMC collected in 2003 and 2008 yielded an extremely high prevalence of clindamycin resistance at 96.0% [18]. The NSAR reports from that time period showed a prevalence of between 20%–30%. Nevertheless, in 2011 there was an increase in the national clindamycin resistance rate to 42.4% and this increased further to 52.1% in 2012. One study reported a very high resistance rate of 88.0% among MRSA isolates obtained from HRPB, KPJ, and GP in Ipoh [21]. The national clindamycin resistance rate maintained around the 50–55% level from 2013 to 2016 until it suddenly decreased by nearly 20% to 31.3% in the 2017 NSAR report [5]. During this time period, one study reported a resistance rate of 78.0% among isolates obtained from UMMC in 2013, although this could be attributed to the lower number of samples studied, which was 67 [22].
Figure 4.
The prevalence of clindamycin resistance in Malaysian S. aureus isolates, 2002–2017. Data from the NSAR reports; HUSM between 2002 and 2007 [15] and in 2008 [19]; not specified (N/S) between 2003 and 2004 and in 2007 [16], and between 2006 and 2007 [17]; UMMC in 2003 and 2008 [18], and in 2013 [22]; UKMMC in 2009 [20]; HRPB, KPJ and GP between 2011 and 2012 [21]; and HSNZ between 2016 and 2017 [23].
Data concerning clindamycin resistance among national MSSA isolates were only available from 2008 until 2014, where the resistance rates remained low throughout the years, i.e., below 3.0%, with the exception for our recent study of 109 HSNZ MSSA isolates from 2016 and 2017, where the prevalence of clindamycin resistance was slightly higher at 3.7% [23] (Figure 4).
2.5. Streptogramins (Quinupristin-Dalfopristin)
There is scarce data on the prevalence of quinupristin-dalfopristin resistance among Malaysian S. aureus isolates. Neela et al. reported that all 32 MRSA isolates from a tertiary hospital in 2006 and 2007 were fully susceptible to quinupristin-dalfopristin. We also recorded a similar observation in the MRSA and MSSA isolates obtained from HSNZ in 2016 and 2017 [23]. The only available data from NSAR regarding resistance to this antimicrobial was in year 2011, in which resistance was recorded as 3.9% for all S. aureus isolates (i.e., both MRSA and MSSA) [29].
2.6. Macrolide-Lincosamide-Streptogramin B (MLSB) Phenotypes
As a result of overlapping binding sites for macrolides, lincosamides, and streptogramin B, resistance involving ribosomal modification will affect the binding affinity of all three drugs. This is the situation observed in macrolide-lincosamide-streptogramin B (MLSB) resistance, where the resistance mechanism lies in the production of an enzyme called erythromycin ribosome methylase, encoded by the erm gene, which causes ribosomal modification by catalyzing the dimethylation of nucleotide 2058A [35]. The MLSB resistance can be categorized as constitutive (cMLSB), inducible (iMLSB), or macrolides streptogramin (MS) phenotype. Out of these three, the iMLSB phenotype is of utmost importance in the clinical settings. In this phenotype, the isolate appears to be erythromycin resistant and clindamycin susceptible in normal disc diffusion assays but the use of clindamycin as antimicrobial therapy in patients that were infected with iMLSB strains has been associated with treatment failure [36]. The iMLSB (or inducible clindamycin resistance) phenotype could only be detected through double-disc diffusion or D-zone test where erythromycin (15 µg) and clindamycin (2 µg) discs were placed 15 mm apart and iMLSB isolates would appear resistant to erythromycin and have a clindamycin zone of ≥21 mm with a D-shaped zone [10].
There was limited data available for the prevalence of inducible clindamycin resistance among Malaysian S. aureus isolates. Earlier studies reported a very high prevalence of the iMLSB phenotype. Neela et al. reported an iMLSB prevalence of 96.1% among 32 MRSA isolates obtained in 2006 and 2007 [17]. Similarly, Lim et al. also found an iMLSB prevalence of 96% among their 156 erythromycin-resistant MRSA isolates collected from UMMC in 2003 and 2008 [18]. Our more recent study however documented a much lower iMLSB prevalence among HSNZ isolates obtained in 2016 and 2017, which was 46.7% among 90 MRSA isolates and 1.8% among 109 MSSA isolates [23].
2.7. Tetracyclines (Tetracycline, Doxycycline, and Minocycline)
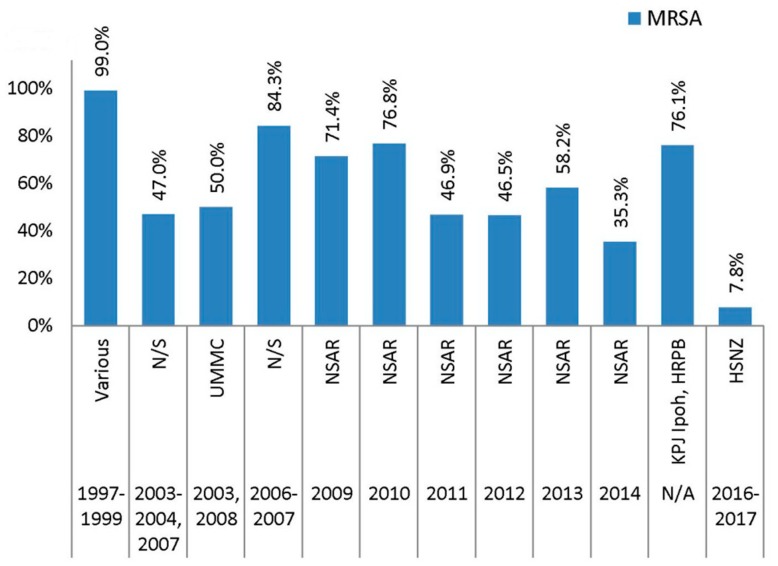
The CDC-ECDC proposed guidelines listed three agents in the tetracycline class of antimicrobials, namely tetracycline, doxycycline, and minocycline, for S. aureus susceptibility testing [11]. NSAR data for the Malaysian MRSA isolates only covered the prevalence of tetracycline resistance and only for the period of 2009–2014. Focusing just on the NSAR data, the prevalence of tetracycline resistance in the Malaysian MRSA isolates was at a downward trend with the highest prevalence at 76.8% in 2010 and the lowest at 35.3% in 2014. An early report of 400 MRSA isolates obtained from ten hospital laboratories throughout Malaysia between 1997 and 1999 [14] (Figure 5) showed nearly all isolates (99.0%) were resistant to tetracycline. From 2003 to 2008, data from two separate studies showed that the prevalence of tetracycline resistance was around 50% [16,18], but another study on MRSA isolates obtained in 2006 and 2007 showed a higher prevalence of 84.3% [17]. A recent study documented a similarly high rate of tetracycline resistance (76.1%) among 117 MRSA isolates obtained from HRPB and KPJ in Ipoh [37]. However, the study did not mention the year of the sample collection. The lowest tetracycline resistance was reported at 7.8%, involving 90 MRSA isolates obtained from HSNZ in 2016 and 2017 [23]. Thus, over a period of nearly 30 years, the prevalence of tetracycline resistance appeared to decrease in MRSA isolates from Malaysia.
Figure 5.
The prevalence of tetracycline resistance among Malaysian MRSA isolates, 1997–2017. Data from the NSAR reports; Various, collected from ten hospitals throughout Malaysia between 1997 and 1999 [14]; not specified (N/S) between 2003 and 2004 and in 2007 [16], and between 2006 and 2007 [17]; UMMC in 2003 and 2008 [18]; KPJ and HRPB with an unspecified year of collection [37]; and HSNZ between 2016 and 2017 [23].
So far, only one study has reported on the prevalence of minocycline resistance among 32 MRSA isolates obtained in 2006 and 2007 and with a rate of 65.6% [17], while doxycycline resistance has yet to be documented.
2.8. Fluoroquinolones (Ciprofloxacin, Moxifloxacin, Norfloxacin, and Ofloxacin)
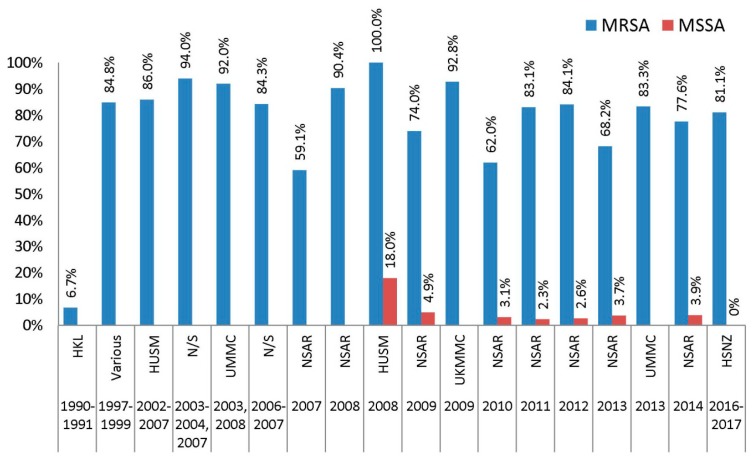
The earliest report on ciprofloxacin resistance in Malaysian MRSA isolates was on 539 MRSA isolates from HKL that were obtained in 1990 and 1991 [13], whereby a very low prevalence of 6.7% was reported (Figure 6). A subsequent report on isolates obtained from ten different hospitals in 1997–1999 showed a dramatically higher prevalence of 84.8% [14]. MRSA isolates from various studies obtained in 2002–2009 showed a consistently high prevalence of ciprofloxacin resistance (>85%) [14,15,16,22,23]. However, the first NSAR report indicated a national resistance rate of 59.1% for 2007, but this rate increased substantially to 90.4% the following year. The national ciprofloxacin resistance rates fluctuated between 62–84% from 2009 until the final NSAR report for ciprofloxacin resistance on 2014. Our recent report on MRSA isolates from HSNZ in 2016–2017 still indicated a high prevalence of ciprofloxacin resistance, at 81.1% [23].
Figure 6.
The prevalence of ciprofloxacin resistance among Malaysian S. aureus isolates, 1990–2017. Data from the NSAR reports; HKL between 1990 and 1991 [13]; Various, collected from ten hospitals throughout Malaysia between 1997 and 1999 [14]; HUSM between 2002 and 2007 [15] and in 2008 [19]; not specified (N/S) between 2003 and 2004 and in 2007 [16], and between 2006 and 2007 [17]; UMMC in 2003 and 2008 [18], and 2013 [22]; UKMMC in 2009 [20]; and HSNZ in 2016 and 2017 [23].
MSSA isolates were mostly susceptible to ciprofloxacin, with NSAR reports from 2009–2014 indicating resistance rates of between 2–5%. However, Al-Talib et al. found a higher rate of ciprofloxacin resistance (18.0%) among their 124 HUSM MSSA isolates collected in 2008 [19], whereas our recent data on HSNZ isolates in 2016 and 2017 showed that all MSSA isolates were susceptible to ciprofloxacin [23].
Data on resistance towards other fluoroquinolone antimicrobials among Malaysian S. aureus isolates were scarce (Figure 6). An early study reported a 3% prevalence of resistance towards moxifloxacin among 32 MRSA isolates which were obtained from nine major hospitals between 1997 and 1999 [38]. However in this study, the authors only chose to test isolates that were both resistant to fusidic acid and rifampicin, hence the low number of isolates may not truly reflect the prevalence of moxifloxacin resistance among MRSA isolates for that particular period [38]. Neela et al. tested their 32 MRSA isolates obtained in 2006 and 2007 against two other fluoroquinolone antimicrobials, which were norfloxacin and ofloxacin, and found the prevalence of resistance towards both agents was at 78.1% and 53.1%, respectively [17].
2.9. Folate Pathway Inhibitors (Co-Trimoxazole)
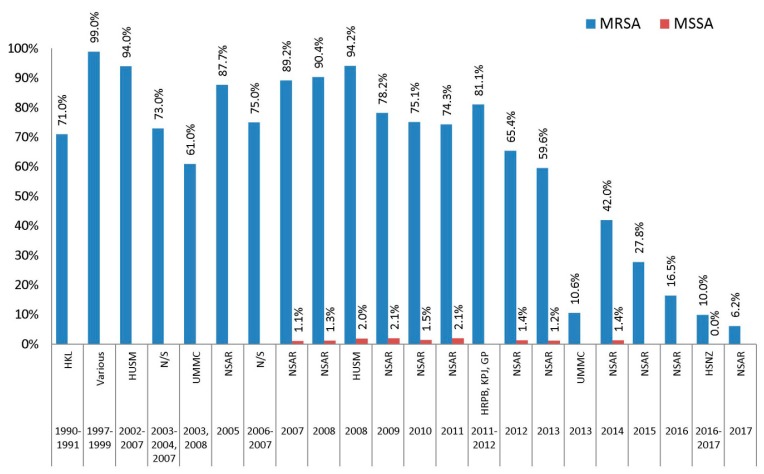
There was generally a high prevalence (>70%) of co-trimoxazole resistance in the Malaysian MRSA isolates from the 1990s to 2000s, with fluctuations in between (Figure 7). The NSAR reports for 2005–2008 documented a resistance prevalence of close to 90%, but which gradually declined to around 70% by 2011. Studies in HUSM supported this as MRSA isolates from 2002–2007 showed prevalence of co-trimoxazole resistance at 94.0% [15] and 94.2% for isolates from 2008 [19]. Studies from other healthcare settings, however, showed a lower prevalence of co-trimoxazole, with the lowest rate at 61.0% in UMMC isolates collected in 2003 and 2008 [18]. Interestingly, the national prevalence of co-trimoxazole resistance started to noticeably decline from 2012 onwards, with rates of 65.4% in 2012 to a very low level of 6.2% in 2017. Studies from individual hospitals showed support for this trend with MRSA isolates obtained from UMMC in 2013–2014 indicating a prevalence of 10.6% [22], whereas our recent study of HSNZ MRSA isolates from 2016–2017 also showed a similar prevalence of 10.0% [23]. The reason for this drastic decline in co-trimoxazole resistance in the Malaysian MRSA isolates is not known as we do not have data for the use of co-trimoxazole and other antibiotics in the treatment of MRSA and other bacterial infections in the Malaysian hospitals. Most Malaysian MSSA isolates were still very much susceptible to co-trimoxazole, where data recorded from 2007 until 2014 showed the resistance rates lingering around 1.1% to 2.1% (Figure 7).
Figure 7.
Prevalence of co-trimoxazole resistance in Malaysian S. aureus isolates, 1990–2017. Data from the NSAR reports; HKL between 1990 and 1991 [13]; various, collected from ten hospitals throughout Malaysia between 1997 and 1999 [14]; HUSM between 2002 and 2007 [15] and in 2008 [19]; not specified (N/S) between 2003 and 2004 and in 2007 [16], and between 2006 and 2007 [17]; UMMC in 2003 and 2008 [18], and in 2013 [22]; UKMMC in 2009 [20]; HRPB, KPJ and GP between 2011 and 2012 [21]; and HSNZ between 2016 and 2017 [23].
2.10. Phenicols (Chloramphenicol)
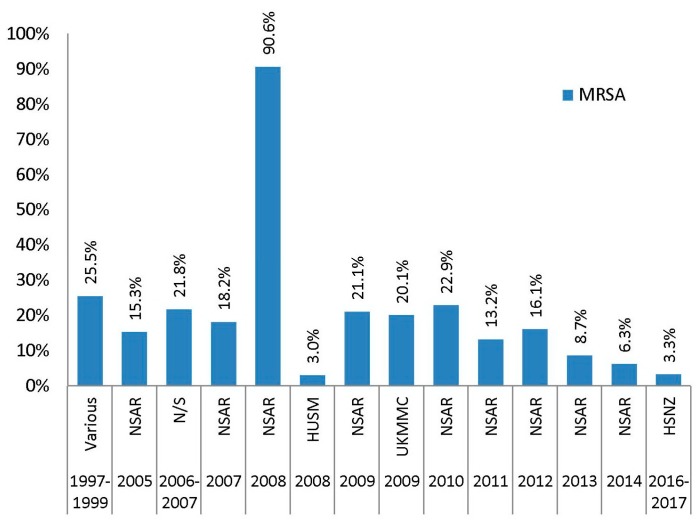
Malaysian MRSA isolates were moderately resistant to chloramphenicol. In most NSAR reports as well as individual studies, the prevalence of chloramphenicol resistance was observed below 30% (Figure 8). The NSAR report for 2008, however, documented a great increase in chloramphenicol resistance among MRSA isolates, with a documented rate of 90.6% [25]. This was the highest prevalence ever reported and was markedly different from the rest of the NSAR reports and studies, prompting doubt as to the accuracy of the 2008 NSAR data regarding chloramphenicol resistance. Data from 2009 until 2010 showed a prevalence of resistance in the range of 20% to 23%, but which gradually declined in 2012 (16.1%) until the final NSAR report for MRSA chloramphenicol resistance in 2014 (6.3%). Our recent report on HSNZ isolates from 2016–2017 supported this decline, where we observed prevalence of chloramphenicol resistance was only at 3.3% [23]. Data regarding chloramphenicol resistance among Malaysian MSSA isolates were not covered by the NSAR reports. Our recent study on 109 HSNZ MSSA isolates indicated their full susceptibility towards chloramphenicol [23].
Figure 8.
The prevalence of chloramphenicol resistances in Malaysian MRSA isolates, 1997–2017. Data from the NSAR reports; various, collected from ten hospitals throughout Malaysia between 1997 and 1999 [14]; not specified (N/S) between 2006 and 2007 [17]; HUSM in 2008 [19]; UKMMC in 2009 [20]; and HSNZ in 2016 and 2017 [23].
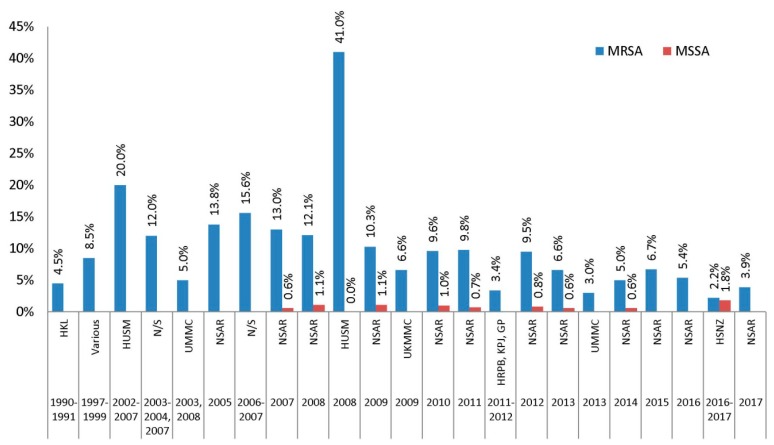
2.11. Ansamycins (Rifampin)
The prevalence of rifampin resistance in Malaysian MRSA isolates is generally low, mostly around 10–15% in the 2000s and dropping to below 10% in the 2010s, with the latest NSAR report of 2017 showing a prevalence of only 3.9% (Figure 9). However, the prevalence of rifampin resistance in MRSA isolates from two studies carried out in HUSM were much higher than the national average. HUSM isolates from 2002 to 2007 showed a resistance rate of 20% [15] and this increased considerably to 41% for isolates obtained in 2008 [19].
Figure 9.
The prevalence of rifampin resistance in Malaysian S. aureus isolates, 1990–2017. Data from the NSAR reports; HKL between 1990 and 1991 [13]; various, collected from ten hospitals throughout Malaysia between 1997 and 1999 [14]; HUSM between 2002 and 2007 [15] and in 2008 [19]; not specified (N/S) between 2003 and 2004 and in 2007 [16], and between 2006 and 2007 [17]; UMMC in 2003 and 2008 [18], and in 2013 [22]; UKMMC in 2009 [20]; HRPB, KPJ and GP between 2011 and 2012 [21]; and HSNZ between 2016 and 2017 [23].
As for MSSA isolates, the prevalence of rifampin resistance was very low, as observed from data available from 2007 to 2014, with resistance rates rarely reaching 1% (Figure 9). We reported a slightly higher prevalence of rifampin resistance (1.8%) among the HSNZ MSSA isolates from 2016–2017 [23].
2.12. Glycopeptides (Vancomycin and Teicoplanin)
Resistance to vancomycin can exist in three forms depending on the minimum inhibition concentration (MIC) values. Isolates showing intermediate resistance towards vancomycin (vancomycin-intermediate S. aureus, VISA) have MIC values of 4–8 µg/mL, while those with complete resistance have MIC values of ≥16 µg/mL [10]. Heterogeneous VISA (hVISA) isolates, on the other hand, have susceptible MIC values (≤2 µg/mL), but a proportion of the population also have MIC values in the intermediate range [39].
In the earliest NSAR reports, which were from years 2003 to 2005, the vancomycin resistance rate was recorded at 0.1% for three consecutive years [24]. This was the rate recorded among all S. aureus isolates, including MRSA. In 2005, NSAR reported a similar resistance rate, but exclusively for MRSA isolates [24]. Not much information could be obtained from these early reports. The numbers as well as the name of participating hospitals were not listed and further information regarding these vancomycin-resistant isolates such as the origin, source of the isolates, and how the vancomycin resistance was tested (whether by disc diffusion method or MIC value) were also not available. The validity of these results could also not be confirmed as there were no peer-reviewed published reports regarding these supposedly vancomycin-resistant isolates. However, in the following years, there have been no reports of vancomycin-resistant isolates in any of the NSAR reports as well as in any individual studies.
While complete resistance towards vancomycin has not been reported in Malaysia in published journals, several studies have documented the presence of VISA and hVISA isolates. The hVISA isolates were mostly detected among isolates collected in 2009. Seven out of 320 MRSA isolates collected from HKL in 2009 were hVISA isolates [40], while two out of 43 MRSA isolates collected during the same year from Hospital Selayang were also confirmed as hVISA. A report of the emergence of a VISA isolate in Malaysia was just made recently, where the isolate was obtained from a female patient who was first admitted for leptospirosis in an unnamed referral hospital. The isolate showed a vancomycin MIC value of 4 µg/mL and was obtained from the blood sample of the patient who had previously received a total of 31 days of intravenous vancomycin therapy during her hospital stay [41].
Teicoplanin resistances were only reported in the 2007 and 2008 NSAR, where the rate was recorded at 0.2% for MRSA isolates in 2007 [26]. In 2008, the rate was recorded at 0.1% for MRSA isolates and 0.2% for MSSA isolates [25]. However, it should be noted that these rates were marked as unconfirmed in the NSAR reports. In a study involving 162 MRSA isolates obtained from UMMC in 2003 and 2008, teicoplanin resistance was recorded at 1%, which was just slightly higher than the national rate [18]. In another study, 32 MRSA isolates collected from an unnamed tertiary hospital from 2006 to 2007 showed full susceptibility towards teicoplanin [17]. A similar observation was also reported among 124 MSSA and 34 MRSA isolates obtained in 2008 from HUSM, where all the isolates showed full sensitivity towards teicoplanin [19]. Teicoplanin-resistant isolates were also not detected among the 318 MRSA isolates collected in Universiti Kebangsaan Malaysia Medical Centre (UKMMC) in 2009 [20].
2.13. Oxazolidinones (Linezolid)
The prevalence of linezolid resistance in Malaysian S. aureus isolates was very low, with most NSAR data and other individual studies reporting resistance rates below 1.0% (Figure 10). However, NSAR reports for 2010 and 2015 documented higher linezolid resistance rates with 7.7% for MRSA and 3.3% for MSSA isolates in 2010, and 3.1% for MRSA isolates in 2015 (MSSA data was unavailable for 2015). The latest NSAR report for 2017 showed a linezolid resistance rate of 0.9% for MRSA.
Figure 10.
The prevalence of linezolid resistance in Malaysian S. aureus isolates, 2003–2017. Data from the NSAR; UMMC in 2003 and 2008 [18]; not specified (N/S) between 2006 and 2007 [17]; HUSM in 2008 [19]; HRPB, KPJ and GP between 2011 and 2012 [21]; and HSNZ in 2016 and 2017 [23].
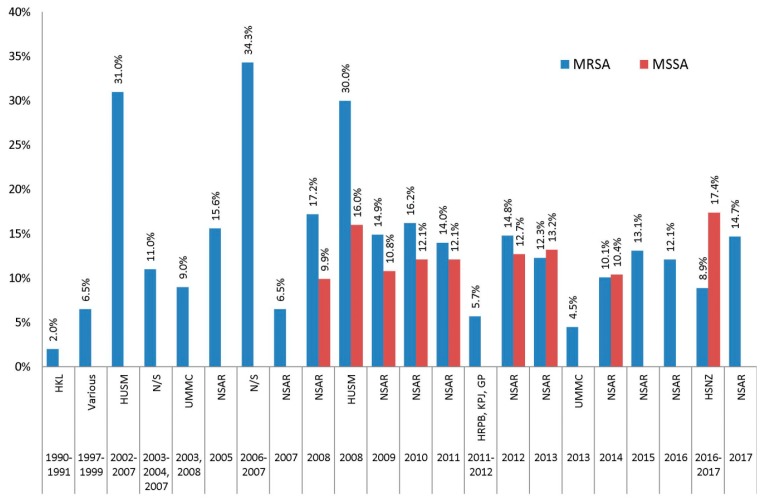
2.14. Fucidanes (Fusidic Acid)
The NSAR reports indicated that the prevalence of fusidic acid resistance in Malaysian MRSA isolates remained relatively consistent between 10–16% from the first NSAR report in 2005 (15.6%) to the latest report in 2017 (14.7%), apart from 2007 when NSAR reported a prevalence of only 6.5% (Figure 11). Individual studies, particularly from HUSM, showed much higher prevalence of fusidic acid resistance in their MRSA isolates, with 31.0% for isolates collected from 2002–2007 [15] and 30.0% for isolates obtained in 2008 [19]. MRSA isolates obtained from an unspecified healthcare centre in 2006–2007 also showed higher prevalence of fusidic acid resistance at 34.3% [17]. From 2011 onwards, fusidic acid resistance isolates were reported in three independent studies, with HSNZ isolates showing higher resistance rate among MSSA isolates (17.4%) compared to MRSA isolates (8.9%) [21,22,23].
Figure 11.
The prevalence of fusidic acid resistance in Malaysian S. aureus isolates, 1990–2017. Data from the NSAR reports; HKL between 1990 and 1991 [13]; various, collected from ten hospitals throughout Malaysia between 1997 and 1999 [14]; HUSM between 2002 and 2007 [15] and in 2008 [19]; not specified (N/S) between 2003 and 2004 and in 2007 [16], and between 2006 and 2007 [17]; UMMC in 2003 and 2008 [18], and in 2013 [22]; UKMMC in 2009 [20]; HRPB, KPJ and GP between 2011 and 2012 [21]; and HSNZ between 2016 and 2017 [23].
2.15. Glycylcyclines (Tigecycline)
There was very limited data concerning tigecycline resistance among Malaysian S. aureus isolates. NSAR did not report the prevalence of tigecycline resistance and individual studies were also very scarce. One such study was by Al-Talib et al., who found full tigecycline susceptibility among the HUSM MRSA and MSSA isolates obtained in 2008 [19]. Isolates collected approximately a year later (2009–2010) in HKL showed a higher rate of resistance to tigecycline among MRSA isolates upon determination of MIC values, which was 26.7% [42]. Both studies involved a small number of MRSA isolates (34 isolates in the first study and 30 isolates in the latter) and yet there was a difference in the resistance rates between the two studies. A more recent study, however, documented a much lower prevalence of tigecycline resistance at 5.6% among 90 HSNZ MRSA isolates in 2016 and 2017 [23]. Tigecycline is a useful alternative for the treatment of vancomycin-resistant isolates. Therefore, the rise in tigecycline resistance should not be taken lightly and overuse of this antibiotic should be avoided.
2.16. Lipopeptides (Daptomycin)
Daptomycin is a relatively new antimicrobial, having been approved for clinical use in 2003. Reports on daptomycin resistance among Malaysian S. aureus isolates remain very scarce. The national resistance rate for daptomycin was not available in any of the NSAR reports. To the best of our knowledge, there has only been one study involving 30 MSSA and 30 MRSA isolates collected from HKL during 2009 and 2010, where a 100% susceptibility rate for daptomycin was recorded when tested using E-test strips to obtain the MIC values [42].
2.17. Overview of Staphylococcus aureus Antimicrobial Resistance Trends in Malaysia
Generally, the resistance rates reported by NSAR seemed to be lower than that of individual hospitals, possibly due to the pooling effect. NSAR reported the compilation of antibiotic susceptibility data from hospital microbiology laboratories mainly of government hospitals and university hospitals from all the 13 states in Malaysia. Hence, the sample size of each NSAR report was frequently above 10,000 isolates, which is more than 10 times higher than the sample sizes of individual reports from selected hospitals (which were well below a thousand).
Amikacin and netilmicin, two older line of drugs that were previously used were stopped for empirical treatment of MRSA. According to the second edition of the Malaysian National Antibiotic Guideline 2014, drugs to be used for treating MRSA infections include clindamycin, erythromycin, vancomycin, rifampicin, tetracycline, linezolid, and gentamicin [43]. The second edition of the National Antibiotic Guideline 2014 is in line with the Protocol on Antimicrobial Stewardship (AMS) Program in healthcare facilities launched at the end of 2014 [44]. Overall, the prevalence of resistance to some of these drugs seems to be reduced as the positive outcome of the implementation of the AMS program [45]. The five core strategies of AMS are as follows: (i) Formulation of an AMS team in each hospital, health district office and health clinics; (ii) surveillance and feedback mechanism on specific antimicrobial consumption; (iii) implementation of prospective audit and feedback according to local needs; (iv) formalize regular antimicrobial rounds by AMS team especially in state and specialist hospitals; and (v) establishment of formulary restriction and a pre-authorization/approval system [46].
The decreasing resistance to the older line of drugs observed in this review for Malaysia has also been described in the United States and China [47,48]. Possible explanations for this scenario include a more rational use of antibiotics and a shift in antimicrobial selection pressures [47,48]. In Malaysia, vancomycin or linezolid are the drug of choice to treat MRSA infections, whereas cloxacillin is the recommended antimicrobial therapy for MSSA infections, particularly for adult patients in the intensive care units [49]. These could thus result in less usage of older antimicrobials and therefore a shift in antibiotic pressures, ultimately leading to decrease resistance to the older drugs. Some authors also suggested the changing molecular epidemiology of S. aureus clones circulating in the hospital as a possible reason for the shift in antimicrobial resistance patterns [50,51].
3. Molecular Mechanisms of Antimicrobial Resistance
Data regarding the molecular mechanisms of antimicrobial resistance in Malaysia were limited. There were two studies describing the prevalence of tetracycline resistance genes among Malaysian MRSA strains, where tetM and tetK were found to be the most prevalent among tetracycline-resistant MRSA isolates. Ong et al. (2017) found that tetM was the predominant gene, with 97.8% prevalence compared to 42.7% for tetK, among 89 tetracycline-resistant MRSA isolates and also with a 90.0% prevalence of tetM among 10 MRSA isolates that were intermediately-resistant [37]. Similarly, in another study, tetM was also found to be the more predominant tetracycline-resistant gene, with a prevalence of 49.0% compared to tetK (21.0%), among 188 MRSA isolates [52]. The higher prevalence of tetM was postulated to be due to the stable nature of Tn916, the tetM-containing conjugative transposon [37,53], whereas tetK is usually carried in a small plasmid (pT181) and therefore can only affect a smaller number of hosts compared to Tn916, resulting in a lower prevalence [37,54].
The prevalence of MLSB resistance genes among clinical MRSA isolates has only been described in one clinical study. Lim et al. (2012) reported a prevalence of 84.0% for ermA, 21.0% for ermC, and only 2.0% for msrA among 188 MRSA hospital isolates [52]. In a community study, 49 S. aureus isolates obtained from 200 healthy undergraduate students in 2012 and 2013 yielded eight erythromycin-resistant isolates, of which six were found to carry the msrA gene and one isolate harbored ermC [55].
In S. aureus, the multidrug efflux pump NorA (encoded by the norA gene) provides a low level resistance to fluoroquinolone antibiotics [56,57], whereas the efflux pump MdeA (encoded by mdeA gene) can confer resistance to fusidic acid [58]. The prevalence of these efflux genes in 16 MRSA/MSSA isolates from HUKM was reported, whereby the norA gene was detected in 13 isolates whereas the mdeA gene was detected in 15 isolates [59].
4. Clonal Types and Characteristics of Malaysian S. aureus Isolates
MRSA isolates can be differentiated into two categories depending on the origin of the isolates, i.e., hospital-associated MRSA (HA-MRSA) and community-associated MRSA (CA-MRSA). As the name implies, HA-MRSA is typically found in the hospital settings whereas CA-MRSA isolates originate from the community. The CDC defines CA-MRSA as an isolate obtained within 48 hours after hospitalization with no previous history of hospitalization, surgery, residence in long-term care facility, and dialysis within the last 12 months, presence of percutaneous device or indwelling catheter, and previous MRSA infection or colonization. Isolates that do not fulfill the above definition are considered HA-MRSA [60]. There are several distinguishing features between the two types of MRSA. While HA-MRSA generally affects people with chronic diseases, hospitalized patients and the elderly, CA-MRSA usually affects young and healthy individuals [61]. Clinically, most CA-MRSA infections involve the skin and soft tissue, while HA-MRSA has a wider spectrum of disease [62]. Genetically, MRSA strains are commonly grouped into different clones based on their sequence types (STs) as well as staphylococcal cassette chromosome mec (SCCmec) types. Generally, HA-MRSA tend to carry SCCmec types I, II, and III while CA-MRSA tend to carry SCCmec types IV or V [63]. Identifying MRSA clones enables researchers to carry out surveillance and outbreak investigations. Some of the most well-known MRSA clones include the Brazillian clone (ST239-III), the Iberian clone (ST247-IA), the Berlin clone (ST45-IV), the New York/Japan clone (ST5-II), epidemic MRSA (EMRSA)-15 (ST22-IV-PVL negative), and EMRSA-16 (ST36-II) [64,65,66].
Various studies found that the predominant HA-MRSA clone circulating in Malaysian hospitals is ST239 belonging to clonal complex (CC) 8 and spa type t037, with the most common SCCmec type being type III or its variant IIIA. Ghaznavi-Rad et al. reported the majority of MRSA strains (92.8%) from HKL were ST239 belonging to CC8 for isolates collected from 2007–2008 [67]. These findings were similar to another study published in 2010, where 83.3% of their MRSA isolates were also found to be ST239 belonging to CC8 for isolates collected from 2006–2007 [68].
Other STs that have been documented in Malaysia include ST1, ST6, ST7, ST22, ST30 ST188, ST772, ST1178, and ST1283 [18,67,68,69,70,71] (Table 3). The emergence of ST22 is of particular interest as this strain mostly circulates in Europe and belongs to the EMRSA-15 [72]. ST22 is also a strain that is capable of quickly spreading and therefore able to replace the other strains [73]. The emergence of this strain means that it has spread to Malaysia and could replace the currently dominating ST239. In Singapore, MRSA ST239 containing SCCmec type III was the dominant clone in the mid-1980s, but was replaced by ST22-SCCmec type IV isolates beginning in the early 2000s [72]. However, from 2007 onwards, the genetic diversity of circulating ST239 clones showed an increase along with the emergence of a subclone of ST239, which was attributed to the multiple acquisition of the Arginine Catabolic Mobile Element (ACME) that encode genes associated with enhanced skin colonization [72].
Table 3.
Clonal characteristics of Staphylococcus aureus isolates in Malaysia.
| A. Community- and Hospital-Associated Staphylococcus aureus Isolates in Malaysia | |||||
| Year | MRSA/MSSA | SCCmec (%) | Sequence Type (ST) | Origin | Reference |
| 2003, 2004 | MRSA | III (78.8) IV (18.2) Untypable (3.0) |
N/A | N/A | [16] |
| 2003, 2006, 2007 | MRSA | IV IVE |
ST6, ST30 ST22, ST1178, ST1179 |
HA, CA | [74] |
| 2003, 2008 | MRSA | III (90.0) IV (9.0) V (1.0) |
(ST239, ST772, ST6, ST22, ST1178) * | HA, CA | [18] |
| 2003, 2004, 2007, 2008 | MRSA | NA | ST5, ST6, ST20, ST22, ST80, ST239 | NA | [75] |
| 2006–2007 | MRSA | III (41.7) IIIA (52.8) V (5.5) |
ST239 ST239 ST1, ST772 |
HA, CA | [68] |
| 2006–2008 | MRSA | III (96.8) IV (3.2) |
NA ST30, ST22, ST45, ST80, ST101, ST188, ST1284, ST1285, ST1286, ST1287, ST1288 |
HA, CA | [69] |
| 2007–2008 | MRSA | III (20.0) IIIA (72.8) V (5.7) IVh (1.5) |
(ST239, ST1, ST7, ST22, ST188, ST1283) * | NA | [67] |
| 2008 | MSSA | - | ST1, ST3, ST5, ST8, ST9, ST12, ST15, ST18, ST20, ST25, ST45, ST80, ST88, ST97, ST121, ST152, ST188, ST231, ST239, ST427, ST508, ST769, ST833, ST1050, ST1153 | HA, CA | [76] |
| 2008–2010 | MRSA | III (91.4) IV (2.9) V (5.7) |
(ST239, ST772) * | N/A | [77] |
| MSSA | - | ST1, ST7, ST30, ST239, ST508, ST779, ST1179, ST1659 | |||
| 2009 | MRSA | II (0.4) III (94.5) IV (3.4) V (1.7) |
ST239 ST239 ST30, ST1178 ST772 |
N/A | [78] |
| 2009 | MRSA | III (72.0) IV (2.5) V (1.3) II (0.3) Untypable (9.7) Novel (14.2) |
N/A | NA | [20] |
| 2010 | MRSA | III (78.5) IV (21.5) |
N/A | NA | [79] |
| 2011–2012 | MRSA | III (81.1) IV (12.6) II (0.6) V (5.7%) |
N/A | NA | [21] |
| 2011–2012 | MRSA | II (0.9) III (66.5) IV (28.2) V (3.3) Untypable (0.9) |
NA ST239 ST1, ST6, ST22, ST1137 ST5, ST772 ST239, ST508 |
HA, CA | [80] |
| 2013 | MRSA | III (55.2) I (1.5) IV (29.9) V (11.9) Untypable (1.5) |
ST239 ST152 ST6, ST22, ST30, ST1179 ST1, ST45, ST772, ST951 ST5 |
HA, CA | [22] |
| 2015–2017 | MRSA | IVa (35.0) IVc (2.7) V (2.7) II (2.7) Untypable (57.0) |
N/A | CA | [81] |
| B. Livestock-Associated Staphylococcus aureus Isolates in Malaysia | |||||
| Year | Livestock | Prevalence (%) | SCCmec (%)/Sequence Type (ST) | Origin | Reference |
| N/A | Raw chicken meat | S. aureus (24.0) | N/A | LA | [82] |
| N/A | Cat and dog | MRSA (11.7) | N/A | LA | [83] |
| N/A | Cat and dog | MRSA (1.9) | N/A | LA | [84] |
| N/A | Chicken | MRSA (18.0) | N/A | LA | [85] |
| N/A | Piglet | MRSA (2.4) | N/A | LA | [86] |
| 2007–2008 | Cat and dog | MRSA (8.0) | N/A | LA | [87] |
| N/A | Horse | S. aureus (44.0) | N/A | LA | [88] |
| 2010 | Chicken | S. aureus (1.0) | ST692 | LA | [89] |
| 2014 | Cat and dog | MRSA in cat (7.7) MRSA in dog (11.7) |
N/A | LA | [90] |
| N/A | Pig | MRSA (1.4) | V (100.0)/ST9 | LA | [91] |
| N/A | Pig |
S. aureus (24.6) MRSA (0.8) |
N/A | LA | [92] |
* Sequence type not correlated with SCCmec type; ST, sequence type; MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; CA, community-associated; HA, hospital-associated; LA, livestock-associated; N/A, not available.
Generally, SCCmec type III or its variant IIIA is the predominant type in Asian countries like Singapore, Indonesia, and Thailand [93]. This is also true for Malaysia, as demonstrated by Ghaznavi-Rad et al., where they documented 20% of SCCmec type III and 72.8% of SCCmec type IIIA in their isolates [67]. Other studies corroborating this finding include Neela et al., with 41.6% of type III and 52.7% of type IIIA [68], and Mustafa et al., who reported 78.5% of type III in their isolates collected from Hospital Tengku Ampuan Afzan [79]. Two other studies in 2009 and 2011 also have similar results with the former study and reported 90% of type III and the latter detected the same type in 96.8% (608/628) MRSA isolates [18,69]. Likewise, more recent studies from UMMC of MRSA isolates obtained from 2011 to 2013 indicated the majority were ST239-SCCmec type III, followed by ST22-SCCmec type IV [22,80], a situation that was similar to that in Singapore before 2007 [72]. Another paper that reported on MRSA isolates obtained in the Malaysian state of Perak in 2011 and 2012 also showed the predominance of SCCmec type III, but with the emergence of SCCmec type IV among 175 non-repetitive isolates [21]. Although the predominant clone in Malaysia appeared to be ST239 with SCCmec type III, Samat et al. made an interesting discovery of the first ST239 containing an SCCmec type II [78]. The strain, which was isolated from a skin swab in an orthopedic clinic, was the first of its kind to be reported.
The prevalence rate of CA-MRSA is much lower compared to HA-MRSA. The first CA-MRSA isolate in Malaysia was reported by Shamsudin et al. when they screened 100 healthy students for nasal carriage. Out of the 100 nasal cultures, 26 were positive for S. aureus, of which three were identified as CA-MRSA based on their SCCmec types, which were IVa and V [94]. In clinical settings, Sam et al. found two CA-MRSA isolates when performing an analysis of cases from 2002 to 2007 [74], while Rashid et al. discovered five isolates throughout 2007 [95] and Ahmad et al. documented nine isolates from a total of 628 MRSA strains collected from 2006–2008 [69].
One feature attributable to CA-MRSA infections is that they usually involve the skin and soft tissues. Several studies in Malaysia supported this theory as their CA-MRSA isolates were obtained from patients with skin and soft tissue infections [74,95]. Chan et al. presented a case report of necrotizing fasciitis of the left lower limb in a healthy 20 year old male due to CA-MRSA, which started out as a soft tissue infection [96].
Aside from the type of infection, another characteristic found to be associated with CA-MRSA is the possession of Panton-Valentine leucocidin (PVL), which is a pore-forming toxin. Isolates of CA-MRSA in Malaysia were mostly positive for PVL [69,74,94,95]. However, there were also reports of PVL-negative strains detected in these isolates [69,94].
The dominant CA-MRSA strains in Malaysia were found to mostly belong to ST30. The two CA-MRSA samples from Sam et al. were of ST6 and ST30 [74]. Ahmad et al. found eight ST30 strains from nine CA-MRSA isolates, while the remaining one isolate was ST80 [69]. One of the three isolates from Shamsudin et al. was ST80, while the other two were not found to match any type in the multilocus sequence typing (MLST) database [94].
While HA-MRSA generally harbors SCCmec type I, II, and III, CA-MRSA is often associated with type IV and V [63]. Shamsudin et al. reported two isolates with SCCmec type V and one with type IVA [94], while Rashid et al. also found four isolates with type IV [95]. However, two studies found isolates that did not meet the definition of CA-MRSA and yet harbored SCCmec types related to CA-MRSA. Sam et al. described seven nosocomial isolates carrying SCCmec type IV, with three of them even testing positive for PVL, and four strains belong to ST6 and ST30 [74]. Similarly, Ahmad et al. detected SCCmec type IV in 11 nosocomial samples, with three being PVL-positive. Two samples were ST30 and five had no match in the database [69].
Studies reporting on the clonal characteristics of MSSA isolates were limited in number. Even so, two studies reporting on the STs of Malaysian MSSA isolates described an interesting finding, in which MSSA appeared to be very genetically diverse. In one study, a group of researchers screened 268 MSSA isolates obtained from both the clinical and community settings in 2008 where they managed to discover as many as 26 different types of STs [76] (Table 3). Similarly, in another study involving 35 MRSA and 21 MSSA isolates collected between 2008 and 2010, the MSSA isolates also showed more diverse STs compared to MRSA isolates where eight STs were detected among MSSA isolates as compared to only two STs among MRSA isolates [77].
In recent years, sequencing of the full genome of pathogens via whole-genome sequencing (WGS) has revolutionized the global epidemiological investigation in hospitals and communities including for S. aureus [97,98]. However, until now, no large-scale WGS of S. aureus for epidemiological investigations from Malaysia has been published, with only limited reports of one or several isolates. In Malaysia, a brief WGS report was found for a HA-MRSA ST239 clone isolated from a patient with septicemia. This isolate was resistant to oxacillin, ampicillin, cefuroxime, ceftriaxone, gentamicin, erythromycin, ciprofloxacin, and co-trimoxazole [99]. Hashim et al. (2018) also conducted WGS for a hospital-acquired VISA isolate with a vancomycin MIC of 4 µg/mL from a patient who was admitted for leptospiral infection and received an intravenous course of vancomycin for 31 days prior to the detection of the VISA isolate [41]. They reported mutations in the graR, graS, walR, and vraR genes [41], but without further detailed genome analyses of the VISA isolate. Other WGS studies included one for a SCCmec V-ST772-PVL positive MRSA obtained from a blood sample of a newborn [70], a ST1-PVL positive MSSA obtained from pus [71], and three SCCmec III-ST239 and one SCCmec IV-ST1178 MRSA isolates [100]. All of these are brief reports without detailed analyses of the genome characteristics and phylogeny.
The importance of S. aureus as a pathogen not only lies in human populations, but also in animals and from the environment. This is fundamental for the One Health approach, which recognizes that human, animal, and environmental health are all interlinked [101]. The antibiotics used in livestock animals belong to the same group of those used for humans and the issue of antimicrobial resistance in livestock-associated MRSA (LA-MRSA) should be treated with equal concern as per a One Health problem. Irrational antibiotic use in animals has been shown to contribute to the rise of antimicrobial resistance where these resistant strains were also capable of affecting humans [102]. For instance, LA-MRSA has been shown to be capable of causing human infections [103]. In Malaysia, several studies have described the varying prevalence of S. aureus in different types of animals (Table 3), with the highest prevalence for S. aureus colonization at 44.0% (22/50) from the nares of horses [88] and the highest prevalence for MRSA was 18.0% (9/50) from the feathers of chickens [85]. One matter of concern is that some of these studies demonstrated high level of resistance among the S. aureus isolates. For example, one MRSA strain isolated from a cat was shown to have a high level of oxacillin resistance with an MIC of 256 µg/mL [87]. In another study, 13 MSSA isolates from horses were found to be multidrug resistant, with resistances to three to 10 types of antimicrobials [88]. This indicates the importance of continuous S. aureus surveillance among animals, as well as proper antimicrobial use, to minimize their contribution to increasing prevalence of antimicrobial resistance.
Thus far, data on antimicrobial usage in livestock in Malaysia is very limited. Neela et al. (2009) reported the detection of a novel ST9-spa type t4358-MRSA strain from pigs as well as pig handlers [91]. LA-MRSA ST9 is the primary sequence type in Asia, but this clone harbored different types of SCCmec in different countries, as follows: Type V in Malaysia, types IV and V in Taiwan, types II and IVb in China, and types IV, IVb, or V in Hong Kong [104]. More surveillance efforts are clearly needed to monitor the emergence of LA-MRSA in farm animals and animal handlers, as well as animal products, in view of their potential impact on human health.
5. Conclusions
Our review of the prevalence of antimicrobial resistance in Malaysian S. aureus isolates obtained from the annual NSAR reports as well as various peer-reviewed published papers indicated an intriguing trend whereby the prevalence of resistance for the older line of antibiotics such as gentamicin, tetracycline and, to a lesser extent, erythromycin, has decreased over the past three decades. Chloramphenicol and co-trimoxazole also showed decreasing prevalence of resistance, whereas ciprofloxacin and rifampin retained almost the same level of prevalence despite fluctuations over the years. Only clindamycin showed a gradually increasing trend in the prevalence of resistance, although there was a nearly 20% decrease when comparing the prevalence in 2016 with 2017. So far, no Malaysian S. aureus isolates have been reported to be fully vancomycin resistant in peer-reviewed journals, although hVISA and VISA isolates have been reported. Some of the data from NSAR suggested an error in reporting (such as the chloramphenicol resistance data from 2008, which showed a massive ~80% increase in prevalence for that year only for the prevalence to decrease again to its pre-2008 levels the following years), but we were unable to assess the quality assurance or the validity of the data. Good quality surveillance data is essential in the global fight against the spread of antimicrobial resistance, hence the data and analysis provided by NSAR should be vastly improved. Molecular epidemiology data for S. aureus in Malaysia is still scarce and from the few published papers, the predominant HA-MRSA clone in Malaysia appeared to be ST239 with SCCmec type III whereas CA-MRSA isolates were mostly ST30 SCCmec type IV/V. As for LA-MRSA, the clones that have been reported so far were ST9 in pigs and ST692 in chickens. A comprehensive surveillance program which includes molecular epidemiological data should be established, which will enable us to have an in-depth understanding of the S. aureus clones that are circulating in the various healthcare institutions in Malaysia and to assess the extent of the spread of antimicrobial resistance among the S. aureus isolates. In our opinion, a more advanced and standardized tool such as WGS should be implemented in the government hospitals to obtain more comprehensive and better-quality surveillance data for Malaysia in order to further combat S. aureus infections in this country. This is feasible to be achieved one day if the current constraints, mainly due to the lack of financial and infrastructure support, can be overcome.
Abbreviations
| ACME | Arginine Catabolic Mobile Element |
| CA | community-associated |
| CC | clonal complex |
| CDC | Centers for Disease Prevention and Control |
| CLSI | Clinical and Laboratories Standard Institute |
| cMLSB | constitutive macrolide-lincosamide-streptogramin B |
| ECDC | European Centre for Disease Prevention and Control |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| GP | Gribbles Pathology Ipoh |
| HA | hospital-associated |
| HKL | Hospital Kuala Lumpur |
| HRPB | Hospital Raja Permaisuri Bainun |
| HSNZ | Hospital Sultanah Nur Zahirah |
| hVISA | heterogeneous vancomycin-intermediate S. aureus |
| HUSM | Hospital Universiti Sains Malaysia |
| iMLSB | inducible macrolide-lincosamide-streptogramin B |
| IMR | Institute for Medical Research |
| KPJ | KPJ Ipoh Specialist Hospital |
| LA | livestock-associated |
| MIC | minimum inhibition concentration |
| MLSB | macrolide-lincosamide-streptogramin B |
| MRSA | methicillin-resistance S. aureus |
| MS | macrolides streptogramin |
| MSSA | methicillin-susceptible S. aureus |
| MLST | multilocus sequence typing |
| MyOHAR | Malaysian One Health Antimicrobial Resistance |
| NSAR | National Surveillance on Antimicrobial Resistance |
| PBP 2a | penicillin binding protein 2a |
| PVL | panton-valentine leucocidin |
| SCCmec | staphylococcal cassette chromosome mec |
| ST | sequence type |
| UKMMC | Universiti Kebangsaan Malaysia Medical Centre |
| UMMC | University of Malaya Medical Centre |
| VISA | vancomycin-intermediate S. aureus |
| VRSA | vancomycin-resistant S. aureus |
| WGS | whole genome sequencing |
| WHO | World Health Organization |
Author Contributions
Data curation, formal analysis, figures and table preparation, writing-original draft preparation, and editing the manuscript, A.M.C.H.; supervision, conceptualization, formal analysis, table preparation, writing-review, and editing the manuscript, C.H.C., C.C.Y.; review and editing the manuscript, S.M.P. and K.H.C.
Funding
This study was funded by Fundamental Research Grant Scheme (grant number: FRGS/1/2019/SKK11/UNISZA/02/1) and Research Acculturation Grant Scheme (grant number: RAGS/1/2015/SKK0/UNISZA/02/1) from the Ministry of Education Malaysia.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.World Health Organization [(accessed on 20 June 2019)];Antimicrobial Resistance: Global Report on Surveillance. 2014 Available online: https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf?sequence=1.
- 2.Lowy F.D. Staphylococcus Aureus Infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Chen C.J., Huang Y.C. New Epidemiology of Staphylococcus Aureus Infection in Asia. Clin. Microbiol. Infect. 2014;20:605–623. doi: 10.1111/1469-0691.12705. [DOI] [PubMed] [Google Scholar]
- 4.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2016 Available online: https://www.imr.gov.my/images/uploads/NSAR/NSAR_2016/NSAR_report_2016.pdf.
- 5.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2017 Available online: https://www.imr.gov.my/images/uploads/NSAR/NSAR_2017/NSAR_report_2017-edited-31.1.2019.pdf.
- 6.Rodvold K.A., McConeghy K.W. Methicillin-Resistant Staphylococcus Aureus Therapy: Past, Present and Future. Clin. Infect. Dis. 2014;58:20–27. doi: 10.1093/cid/cit614. [DOI] [PubMed] [Google Scholar]
- 7.CDC Staphylococcus Aureus Resistant to Vancomycin-United States, 2002. MMWR. Morb. Mortal. Wkly. Rep. 2002;51:565–567. [PubMed] [Google Scholar]
- 8.Walters M.S., Eggers P., Albrecht V., Travis T., Lonsway D., Hovan G., Taylor D., Rasheed K., Limbago B., Kallen A. Notes from the Field: Vancomycin-Resistant Staphylococcus Aureus—Delaware, 2015. Morb. Mortal. Wkly. Rep. 2015;64:1056. doi: 10.15585/mmwr.mm6437a6. [DOI] [PubMed] [Google Scholar]
- 9.Ramazoni M., Siregar M.L., Jamil K.F. Vancomycin-Resistant Staphylococcus Aureus (VRSA) in Hepatic Cirrhosis Patient: A Case Report. IOP Conf. Ser. Earth Environ. Sci. 2018;25:012096. doi: 10.1088/1755-1315/125/1/012096. [DOI] [Google Scholar]
- 10.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. [Google Scholar]
- 11.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 12.The European Committee on Antimicrobial Susceptibility Testing [(accessed on 20 June 2019)];Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2018 Version 8.1. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf.
- 13.Cheong I., Tan S.C., Wong Y.H., Zainudin B.M., Rahman M.Z. Methicillin-Resistant Staphylococcus Aureus (MRSA) in a Malaysian Hospital. Med. J. Malays. 1994;49:24–28. [PubMed] [Google Scholar]
- 14.Norazah A., Koh Y.T., Ghani Kamel A., Alias R., Lim V.K.E. Mupirocin Resistance among Malaysian Isolates of Methicillin-Resistant Staphylococcus Aureus. Int. J. Antimicrob. Agents. 2001;17:411–414. doi: 10.1016/S0924-8579(01)00314-4. [DOI] [PubMed] [Google Scholar]
- 15.Al-Talib H., Chan Y.Y., Al-Jashamy K., Hasan H. Methicillin-Resistant Staphylococcus Aureus Nosocomial Infection Trends in Hospital Universiti Sains Malaysia during 2002–2007. Ann. Saudi Med. 2010;30:358–363. doi: 10.4103/0256-4947.67077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thong K.L., Junnie J., Liew F.Y., Yusof M.Y., Hanifah Y.A. Antibiograms and Molecular Subtypes of Methicillin-Resistant Staphylococcus Aureus in Local Teaching Hospital, Malaysia. J. Microbiol. Biotechnol. 2009;19:1265–1270. [PubMed] [Google Scholar]
- 17.Neela V., Sasikumar M., Ghaznavi G.R., Zamberi S., Mariana S. In Vitro Activities of 28 Antimicrobial Agents against Methicillin-Resistant Staphylococcus Aureus (MRSA) from a Clinical Setting in Malaysia. Southeast Asian J. Trop. Med. Public Health. 2008;39:885–892. [PubMed] [Google Scholar]
- 18.Lim K.T., Hanifah Y.A., Mohd Yusof M.Y., Ito T., Thong K.L. Comparison of Methicillin-Resistant Staphylococcus Aureus Strains Isolated in 2003 and 2008 with an Emergence of Multidrug Resistant ST22: SCCmec IV Clone in a Tertiary Hospital, Malaysia. J. Microbiol. Immunol. Infect. 2013;46:224–233. doi: 10.1016/j.jmii.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Al-Talib H., Al-Khateeb A., Hassan H. Antimicrobial Resistance of Staphylococcus Aureus Isolates in Malaysian Tertiary Hospital. Int. Med. J. 2015;22:1–3. [Google Scholar]
- 20.Noordin A., Sapri H.F., Mohamad Sani N.A., Leong S.K., Tan X.E., Tan T.L., Mohamad Zin N., Neoh H., Hussin S. Antimicrobial Resistance Profiling and Molecular Typing of Methicillin-Resistant Staphylococcus Aureus Isolated from a Malaysian Teaching Hospital. J. Med. Microbiol. 2016;65:1476–1481. doi: 10.1099/jmm.0.000387. [DOI] [PubMed] [Google Scholar]
- 21.Ho W.Y., Choo Q.C., Chew C.H. Predominance of Three Closely Related Methicillin-Resistant Staphylococcus Aureus Clones Carrying a Unique CcrC-Positive SCC Mec Type III and the Emergence of Spa T304 and T690 SCC Mec Type IV Pvl + MRSA Isolates in Kinta Valley, Malaysia. Microb. Drug Resist. 2017;23:215–223. doi: 10.1089/mdr.2015.0250. [DOI] [PubMed] [Google Scholar]
- 22.Sit P.S., Teh C.S.J., Idris N., Ponnampalavanar S. Methicillin-Resistant Staphylococcus Aureus (MRSA) Bacteremia: Correlations between Clinical, Phenotypic, Genotypic Characteristics and Mortality in a Tertiary Teaching Hospital in Malaysia. Infect. Genet. Evol. 2018;59:132–141. doi: 10.1016/j.meegid.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 23.Che Hamzah A.M., Yeo C.C., Puah S.M., Chua K.H., Rahman N.I., Abdullah F.H., Othman N., Chew C.H. Tigecycline and Inducible Clindamycin Resistance in Clinical Isolates of Methicillin-Resistant Staphylococcus Aureus from Terengganu, Malaysia. J. Med. Microbiol. 2019 doi: 10.1099/jmm.0.000993. [DOI] [PubMed] [Google Scholar]
- 24.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance 2003–2005. Available online: https://www.imr.gov.my/images/uploads/NSAR%20Archives/NSAR%20data%202002-2005.pdf.
- 25.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2008 Available online: https://www.imr.gov.my/images/uploads/NSAR%20Archives/Summary%20of%20antibiotic%20resistance%202008.pdf.
- 26.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2007 Available online: https://www.imr.gov.my/images/uploads/NSAR%20Archives/Summary%20of%20Antibiotic%20Resistance%202007.pdf.
- 27.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2009 Available online: https://www.imr.gov.my/images/uploads/NSAR%20Archives/Summary%20of%20antibiotic%20resistance%202009.pdf.
- 28.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2010 Available online: https://www.imr.gov.my/images/uploads/NSAR%20Archives/National%20Surveillance%20of%20Antibiotic%20Resistance%20report%202010.pdf.
- 29.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2011 Available online: https://www.imr.gov.my/images/uploads/NSAR%20Archives/Summary%20of%20antibiotic%20resistance%202011.pdf.
- 30.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2012 Available online: https://www.imr.gov.my/images/uploads/NSAR/Summary_of_antibiotic_resistance_2012_for_website.pdf.
- 31.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2013 Available online: https://www.imr.gov.my/images/uploads/NSAR/NSAR_2013/Summary_of_NSAR_2013_report.pdf.
- 32.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2014 Available online: https://www.imr.gov.my/images/uploads/NSAR/NSAR_2014/Summary_of_Antibiotic_Resistance_2014_report.pdf.
- 33.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Surveillance of Antimicrobial Resistance. 2015 Available online: https://www.imr.gov.my/images/uploads/NSAR/NSAR_2015/edited_251616_NSAR_Antibiotic_Resistance_Surveillance_data_2015.pdf.
- 34.Wielders C.L.C., Fluit A.C., Brisse S., Verhoef J., Schmitz F.J. MecA Gene Is Widely Disseminated in Staphylococcus Aureus Population. J. Clin. Microbiol. 2002;40:3970–3975. doi: 10.1128/JCM.40.11.3970-3975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leclercq R. Mechanisms of Resistance to Macrolides and Lincosamides: Nature of the Resistance Elements and Their Clinical Implications. Clin. Infect. Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 36.Drinkovic D., Fuller E.R., Shore K.P., Holland D.J., Ellis-Pegler R. Clindamycin Treatment of Staphylococcus Aureus Expressing Inducible Clindamycin Resistance. J. Antimicrob. Chemother. 2001;48:315–316. doi: 10.1093/jac/48.2.315. [DOI] [PubMed] [Google Scholar]
- 37.Ong M.H.L., Ho W.Y., Ng W.W., Chew C.H. High Prevalence of TetM as Compared to TetK amongst Methicillin-Resistant Staphylococcus Aureus (MRSA) Isolates from Hospitals in Perak, Malaysia. Jundishapur J. Microbiol. 2017;10:e13935. [Google Scholar]
- 38.Norazah A., Lim V.K.E., Rohani M.Y., Kamel A.G.M. In-Vitro Activity of Quinupristin/Dalfopristin, Levofloxacin and Moxifloxacin against Fusidic Acid and Rifampicin-Resistant Strains of Methicillin-Resistant Staphylococcus Aureus (MRSA) from Malaysian Hospitals. Med. J. Malays. 2005;60:411–415. [PubMed] [Google Scholar]
- 39.Hiramatsu K. Vancomycin-Resistant Staphylococcus Aureus: A New Model of Antibiotic Resistance. Lancet Infect. Dis. 2001;1:147–155. doi: 10.1016/S1473-3099(01)00091-3. [DOI] [PubMed] [Google Scholar]
- 40.Ramli S.R., Neoh H.M., Aziz M.N., Hussin S. Screening and Detection of Heterogenous Vancomycin Intermediate Staphylococcus Aureus in Hospital Kuala Lumpur Malaysia, Using the Glycopeptide Resistance Detection Etest and Population Analysis Profiling. Infect. Dis. Rep. 2012;4:e20. doi: 10.4081/idr.2012.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashim R., Hamzah H.H., Dahalan N.A., Amran F., Ahmad N., Hazwani N.Z., Baharudin S., Zainal S., Raj A.S.S. Vancomycin-Intermediate Staphylococcus Aureus in Malaysia: A Case Study. J. Clin. Case Reports. 2018;8:1178. [Google Scholar]
- 42.Atshan S.S., Nor Shamsudin M., Lung L.T.T., Sekawi Z., Pei Pei C., Karunanidhi A., Jeevajothi Nathan J., Mateg Ali A., Ghaznavi-Rad E., Abduljaleel S.A., et al. Genotypically Different Clones of Staphylococcus Aureus Are Diverse in the Antimicrobial Susceptibility Patterns and Biofilm Formations. Biomed Res. Int. 2013;2013:515712. doi: 10.1155/2013/515712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ministry of Health Malaysia [(accessed on 20 June 2019)];National Antibiotic Guideline. 2014 Available online: https://www.pharmacy.gov.my/v2/sites/default/files/document-upload/national-antibiotic-guideline-2014-full-versionjun2015_1.pdf.
- 44.Ministry of Health Malaysia [(accessed on 20 June 2019)];Protocol on Antimicrobial Stewardship Program in Healthcare Facilities. 2014 Available online: https://www.pharmacy.gov.my/v2/sites/default/files/document-upload/protocol-antimicrobial-stewardship.pdf.
- 45.Sing D.Y.F., Boo Y.L., Mukhlis R., Chin P.W., Hoo F.K. Antimicrobial Stewardship Program in a Malaysian District Hospital: First Year Experience. Pakistan J. Med. Sci. 2016;32:999–1004. doi: 10.12669/pjms.324.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerald N.M.J. [(accessed on 20 June 2019)];Malaysian Action Plan on Antimicrobial Resistance (MyAP-AMR) 2017–2021. 2018 Available online: http://www.moh.gov.my/moh/resources/Penerbitan/Garis%20Panduan/Garis%20panduan%20Umum%20(Awam)/National_Action_Plan_-_FINAL_29_june.pdf.
- 47.Kanjilal S., Sater M.R.A., Thayer M., Lagoudas G.K., Kim S., Blainey P.C., Grad Y.H. Trends in Antibiotic Susceptibility in Staphylococcus Aureus in Boston, Massachusetts, from 2000 to 2014. J. Clin. Microbiol. 2018;56:e01160-17. doi: 10.1128/JCM.01160-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen K., Huang Y., Song Q., Wu C., Chen X., Zeng L. Drug-Resistance Dynamics of Staphylococcus Aureus between 2008 and 2014 at a Tertiary Teaching Hospital, Jiangxi Province, China. BMC Infect. Dis. 2017;17:97. doi: 10.1186/s12879-016-2172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malaysian Society of Intensive Care [(accessed on 20 June 2019)];Guide to Antimicrobial Therapy in the Adult ICU. 2017 Available online: http://www.msic.org.my/download/AntibioticGuidelines.pdf.
- 50.Amorim M.L., Faria N.A., Oliveira D.C., Vasconcelos C., Cabeda J.C., Mendes A.C., Calado E., Castro A.P., Ramos M.H., Amorim J.M., et al. Changes in the Clonal Nature and Antibiotic Resistance Profiles of Methicillin-Resistant Staphylococcus Aureus Isolates Associated with Spread of the EMRSA-15 Clone in a Tertiary Care Portuguese Hospital. J. Clin. Microbiol. 2007;45:2881–2888. doi: 10.1128/JCM.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai Y., Liu J., Guo W., Meng H., Huang Q., He L., Gao Q., Lv H., Liu Y., Wang Y., et al. Decreasing Methicillin-Resistant Staphylococcus Aureus (MRSA) Infections Is Attributable to the Disappearance of Predominant MRSA ST239 Clones, Shanghai, 2008-2017. Emerg. Microbes Infect. 2019;8:471–478. doi: 10.1080/22221751.2019.1595161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim K.T., Hanifah Y.A., Yusof M.Y.M., Thong K.L. ErmA, ErmC, TetM and TetK Are Essential for Erythromycin and Tetracycline Resistance among Methicillin-Resistant Staphylococcus Aureus Strains Isolated from a Tertiary Hospital in Malaysia. Indian J. Med. Microbiol. 2012;30:203–207. doi: 10.4103/0255-0857.96693. [DOI] [PubMed] [Google Scholar]
- 53.Naglich J.G., Andrews R.E. Introduction of the Streptococcus Faecalis Transposon Tn916 into Bacillus Thuringiensis Subsp. Israelensis. Plasmid. 1988;19:84–93. doi: 10.1016/0147-619X(88)90047-9. [DOI] [PubMed] [Google Scholar]
- 54.Villedieu A., Diaz-Torres M.L., Hunt N., McNab R., Spratt D.A., Wilson M., Mullany P. Prevalence of Tetracycline Resistance Genes in Oral Bacteria. Antimicrob. Agents Chemother. 2003;47:878–882. doi: 10.1128/AAC.47.3.878-882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suhaili Z., Rafee P.A., Mat Azis N., Yeo C.C., Nordin S.A., Abdul Rahim A.R., Al-Obaidi M.M.J., Mohd Desa M.N. Characterization of Resistance to Selected Antibiotics and Panton-Valentine Leukocidin-Positive Staphylococcus Aureus in a Healthy Student Population at a Malaysian University. Germs. 2018;8:21–30. doi: 10.18683/germs.2018.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costa S.S., Viveiros M., Amaral L., Couto I. Multidrug Efflux Pumps in Staphylococcus Aureus: An Update. Open Microbiol. J. 2013;7:59–71. doi: 10.2174/1874285801307010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng X., Sun F., Ji Q., Liang H., Missiakas D., Lan L., He C. Expression of Multidrug Resistance Efflux Pump Gene NorA Is Iron Responsive in Staphylococcus Aureus. J. Bacteriol. 2012;194:1753–1762. doi: 10.1128/JB.06582-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang J., O’Toole P.W., Shen W., Amrine-Madsen H., Jiang X., Lobo N., Palmer L.M., Voelker L., Fan F., Gwynn M.N., et al. Novel Chromosomally Encoded Multidrug Efflux Transporter MdeA in Staphylococcus Aureus. Antimicrob. Agents Chemother. 2004;48:909–917. doi: 10.1128/AAC.48.3.909-917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saiful A.J., Mastura M., Zarizal S., Mazurah M.I., Shuhaimi M., Ali A.M. Efflux Genes and Active Efflux Activity Detection in Malaysian Clinical Isolates of Methicillin-Resistant Staphylococcus Aureus (MRSA) J. Basic Microbiol. 2008;48:245–251. doi: 10.1002/jobm.200700387. [DOI] [PubMed] [Google Scholar]
- 60.CDC . Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Methicillin-Resistant Staphylococcus Aureus. Centers for Disease Control and Prevention; Atlanta, GA, USA: 2015. [Google Scholar]
- 61.Nathwani D., Morgan M., Masterton R.G., Dryden M., Cookson B.D., French G., Lewis D. Guidelines for UK Practice for the Diagnosis and Management of Methicillin-Resistant Staphylococcus Aureus (MRSA) Infections Presenting in the Community. J. Antimicrob. Chemother. 2008;61:976–994. doi: 10.1093/jac/dkn096. [DOI] [PubMed] [Google Scholar]
- 62.Stryjewski M.E., Chambers H.F. Skin and Soft-tissue Infections Caused by Community-acquired Methicillin-resistant Staphylococcus Aureus. Clin. Infect. Dis. 2008;46:S368–S377. doi: 10.1086/533593. [DOI] [PubMed] [Google Scholar]
- 63.Kilic A., Li H., Stratton C.W., Tang Y.-W. Antimicrobial Susceptibility Patterns and Staphylococcal Cassette Chromosome Mec Types of, as Well as Panton-Valentine Leukocidin Occurrence among, Methicillin-Resistant Staphylococcus Aureus Isolates from Children and Adults in Middle Tennessee. J. Clin. Microbiol. 2006;44:4436–4440. doi: 10.1128/JCM.01546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Enright M.C., Robinson D.A., Randle G., Feil E.J., Grundmann H., Spratt B.G. The Evolutionary History of Methicillin-Resistant Staphylococcus Aureus (MRSA) Proc. Natl. Acad. Sci. USA. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oliveira D.C., Tomasz A., de Lencastre H. The Evolution of Pandemic Clones of Methicillin-Resistant Staphylococcus Aureus: Identification of Two Ancestral Genetic Backgrounds and the Associated Mec Elements. Microb. Drug Resist. 2001;7:349–361. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 66.Oliveira D.C., Tomasz A., de Lencastre H. Secrets of Success of a Human Pathogen: Molecular Evolution of Pandemic Clones of Meticillin-Resistant Staphylococcus Aureus. Lancet Infect. Dis. 2002;2:180–189. doi: 10.1016/S1473-3099(02)00227-X. [DOI] [PubMed] [Google Scholar]
- 67.Ghaznavi-Rad E., Shamsudin M.N., Sekawi Z., Khoon L.Y., Aziz M.N., Hamat R.A., Othman N., Chong P.P., Van Belkum A., Ghasemzadeh-Moghaddam H., et al. Predominance and Emergence of Clones of Hospital-Acquired Methicillin-Resistant Staphylococcus Aureus in Malaysia. J. Clin. Microbiol. 2010;48:867–872. doi: 10.1128/JCM.01112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neela V., Ghasemzadeh Moghaddam H., Van Belkum A., Horst-Kreft D., Mariana N.S., Ghaznavi Rad E. First Report on Methicillin-Resistant Staphylococcus Aureus of Spa Type T037, Sequence Type 239, SCCmec Type III/IIIA in Malaysia. Eur. J. Clin. Microbiol. Infect. Dis. 2010;29:115–117. doi: 10.1007/s10096-009-0813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmad N., Ruzan I.N., Ghani M.K.A., Hussin A., Nawi S., Aziz M.N., Maning N., Eow V.L.K. Characteristics of Community- and Hospital-Acquired Meticillin-Resistant Staphylococcus Aureus Strains Carrying SCCmec Type IV Isolated in Malaysia. J. Med. Microbiol. 2009;58:1213–1218. doi: 10.1099/jmm.0.011353-0. [DOI] [PubMed] [Google Scholar]
- 70.Suhaili Z., Lean S.S., Yahya A., Mohd Desa M.N., Ali A.M., Yeo C.C. Draft Genome Sequence of Methicillin-Resistant Staphylococcus Aureus KT/Y21, a Sequence Type 772 (ST772) Strain Isolated from a Pediatric Blood Sample in Terengganu, Malaysia. Genome Announc. 2014;2:e00271–e14. doi: 10.1128/genomeA.00271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suhaili Z., Lean S.S., Mohamad N.M., Abdul Rachman A.R., Mohd Desa M.N., Yeo C.C. Draft Genome Sequence of Staphylococcus Aureus KT/312045, an ST1-MSSA PVL Positive Isolated from Pus Sample in East Coast Malaysia. Genom. Data. 2016;9:111–112. doi: 10.1016/j.gdata.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsu L.Y., Harris S.R., Chlebowicz M.A., Lindsay J.A., Koh T.H., Krishnan P., Tan T.Y., Hon P.Y., Grubb W.B., Bentley S.D., et al. Evolutionary Dynamics of Methicillin-Resistant Staphylococcus Aureus within a Healthcare System. Genome Biol. 2015;16:81. doi: 10.1186/s13059-015-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knight G.M., Budd E.L., Whitney L., Thornley A., Al-Ghusein H., Planche T., Lindsay J.A. Shift in Dominant Hospital-Associated Methicillin-Resistant Staphylococcus Aureus (HA-MRSA) Clones over Time. J. Antimicrob. Chemother. 2012;67:2514–2522. doi: 10.1093/jac/dks245. [DOI] [PubMed] [Google Scholar]
- 74.Sam I.C., Kahar-Bador M., Chan Y.F., Loong S.K., Mohd Nor Ghazali F. Multisensitive Community-Acquired Methicillin-Resistant Staphylococcus Aureus Infections in Malaysia. Diagn. Microbiol. Infect. Dis. 2008;62:437–439. doi: 10.1016/j.diagmicrobio.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 75.Lim K.T., Hanifah Y.A., Mohd Yusof M.Y., Goering R.V., Thong K.L. Temporal Changes in the Genotypes of Methicillin-Resistant Staphylococcus Aureus Strains Isolated from a Tertiary Malaysian Hospital Based on MLST, Spa, and Mec-Associated Dru Typing. Diagn. Microbiol. Infect. Dis. 2012;74:106–112. doi: 10.1016/j.diagmicrobio.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 76.Ghasemzadeh-Moghaddam H., Ghaznavi-Rad E., Sekawi Z., Yun-Khoon L., Aziz M.N., Hamat R.A., Melles D.C., van Belkum A., Shamsudin M.N., Neela V. Methicillin-Susceptible Staphylococcus Aureus from Clinical and Community Sources Are Genetically Diverse. Int. J. Med. Microbiol. 2011;301:347–353. doi: 10.1016/j.ijmm.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 77.Lim K.T., Yeo C.C., Suhaili Z., Thong K.L. Comparison of Methicillin-Resistant and Methicillin-Sensitive Staphylococcus Aureus Strains Isolated from a Tertiary Hospital in Terengganu, Malaysia. Jpn. J. Infect. Dis. 2012;65:502–509. doi: 10.7883/yoken.65.502. [DOI] [PubMed] [Google Scholar]
- 78.Samat Muttaqillah N.A., Hussin S., Neoh H.M., Noordin A., Ding C.H., Wahab A.A., Rahman M.M. Clonal Diversity of Methicillin-Resistant Staphylococcus Aureus in UKM Medical Centre: Characterisation by Multilocus Sequence Typing of Different SCCmec Type Representatives. Sains Malays. 2015;44:1315–1323. doi: 10.17576/jsm-2015-4409-14. [DOI] [Google Scholar]
- 79.Mustafa M.I., Alarosi N.A., Amjad N. Staphylococcal Cassette Chroromosome Mec (SCCmec) Typing of Methicillin-Resistant Staphylococcus Aureus (MRSA) Isolates from Patients Attending Tengku Ampuan Afzan Hospital (HTAA) in Kuantan, Pahang, Malaysia. Int. J. Infect. Dis. 2012;16:e426. doi: 10.1016/j.ijid.2012.05.594. [DOI] [Google Scholar]
- 80.Sit P.S., Teh C.S.J., Idris N., Sam I.C., Syed Omar S.F., Sulaiman H., Thong K.L., Kamarulzaman A., Ponnampalavanar S. Prevalence of Methicillin-Resistant Staphylococcus Aureus (MRSA) Infection and the Molecular Characteristics of MRSA Bacteraemia over a Two-Year Period in a Tertiary Teaching Hospital in Malaysia. BMC Infect. Dis. 2017;17:274. doi: 10.1186/s12879-017-2384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amit L.N., Yung C.V.L., Moy F.S., John D.V. Molecular Characteristics of Infection and Colonization Isolates of Community-Acquired Methicillin-Resistant Staphylococcus Aureus (CA-MRSA) Trop. Biomed. 2018;35:442–452. [PubMed] [Google Scholar]
- 82.Aklilu E., Ab Manah H.A., Abu Daud N. Presence of Antimicrobial Resistant Staphylococcus Aureus in Chicken Meat and Its Potential Public Health Implications. Malays. J. Microbiol. 2016;12:418–422. [Google Scholar]
- 83.Saleha A.A., Shamzarina M., Fauziah O., Zunita Z. Methicillin-Resistant Staphylococcus Aureus (MRSA) Isolated from Pet Animals and Pet and Non-Pet Owners. J. Vet. Malays. 2006;18:13–16. [Google Scholar]
- 84.Ng W.S.J., Zunita Z., Goh Y.M., Raha A.R. Occurrence of Methicillin-Resistant Staphylococcus Aureus (MRSA) in Cats and Dogs; Proceedings of the 17th Veterinary Association Malaysia Congress; Kuala Lumpur, Malaysia. 27–30 July 2005; pp. 43–45. [Google Scholar]
- 85.Abdulkadir M.M., Zunita Z., Goh Y.M., Saleha A.A., Son R. Occurrence of Methicillin-Resistant Staphylococcus Aureus (MRSA) in Chickens; Proceedings of the 19th Veterinary Association Malaysia Congress; Kuala Lumpur, Malaysia. 3–5 August 2007; pp. 182–185. [Google Scholar]
- 86.Khairina A.K., Zunita Z., Ooi P.T. Methicillin-Resistant Staphylococcus in Pigs. Emerg. Infect. Dis. 2005;11:1965. [Google Scholar]
- 87.Aklilu E., Zunita Z., Hassan L., Chen H.C. Phenotypic and Genotypic Characterization of Methicillin-Resistant Staphylococcus Aureus (MRSA) Isolated from Dogs and Cats at University Veterinary Hospital, Universiti Putra Malaysia. Trop. Biomed. 2010;27:483–492. [PubMed] [Google Scholar]
- 88.Zunita Z., Bashir A., Hafizal A. Occurrence of Multidrug Resistant Staphylococcus Aureus in Horses in Malaysia. Vet. World. 2008;1:165–167. [Google Scholar]
- 89.Neela V., Ghaznavi-Rad E., Ghasemzadeh-Moghaddama H., Nor Shamsudin M., van Belkum A., Karunanidhi A. Frequency of Methicillin Resistant Staphylococcus Aureus in the Noses of Malaysian Chicken Farmers and Their Chicken. Iran. J. Vet. Res. 2013;14:226–231. [Google Scholar]
- 90.Kanagarajah R.R., Lee D.C.W., Lee D.Z.F., Yusoff K., Paramasivam S.J., Low W.Y., Jeevaratnam K., Lim S.H.E. Antibiotic Profiling of Methicillin Resistant Staphylococcus Aureus (MRSA) Isolates in Stray Canines and Felines. Cogent Biol. 2017;3 doi: 10.1080/23312025.2017.1412280. [DOI] [Google Scholar]
- 91.Neela V., Mohd Zafrul A., Mariana N.S., van Belkum A., Liew Y.K., Rad E.G. Prevalence of ST9 Methicillin-Resistant Staphylococcus Aureus among Pigs and Pig Handlers in Malaysia. J. Clin. Microbiol. 2009;47:4138–4140. doi: 10.1128/JCM.01363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khalid K.A., Zakaria Z., Toung O.P., McOrist S. Low Levels of Meticillin-Resistant Staphylococcus Aureus in Pigs in Malaysia. Vet. Rec. 2009;164:626–627. doi: 10.1136/vr.164.20.626. [DOI] [PubMed] [Google Scholar]
- 93.Chongtrakool P., Ito T., Ma X.X., Kondo Y., Trakulsomboon S., Tiensasitorn C., Jamklang M., Chavalit T., Song J.H., Hiramatsu K. Staphylococcal Cassette Chromosome mec (SCCmec) Typing of Methicillin-Resistant Staphylococcus aureus Strains Isolated in 11 Asian Countries: A Proposal for a New Nomenclature for SCCmec Elements. Antimicrob. Agents Chemother. 2006;50:1001–1012. doi: 10.1128/AAC.50.3.1001-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nor Shamsudin M., Sekawi Z., van Belkum A., Neela V. First Community-Acquired Methicillin-Resistant Staphylococcus Aureus in Malaysia. J. Med. Microbiol. 2008;57:1180–1181. doi: 10.1099/jmm.0.47844-0. [DOI] [PubMed] [Google Scholar]
- 95.Rashid Z.Z., Bahari N., Othman A., Jaafar R., Mohamed N.A., Jabbari I., Sulong A., Hashim R., Ahmad N. Community-Acquired Methicillin-Resistant Staphylococcus Aureus in a Malaysian Tertiary Centre. Southeast Asian J. Trop. Med. Public Health. 2013;44:104–108. [PubMed] [Google Scholar]
- 96.Chan C., Merican A., Nawar A., Hanifah Y.A., Thong K.L. Necrotising Fasciitis of the Lower Limb Caused by Community-Acquired Methicillin-Resistant Staphylococcus Aureus. Malaysian Orthop. J. 2010;4:36–38. doi: 10.5704/MOJ.1011.010. [DOI] [Google Scholar]
- 97.Cunningham S.A., Chia N., Jeraldo P.R., Quest D.J., Johnson J.A., Boxrud D.J., Taylor A.J., Chen J., Jenkins G.D., Drucker T.M., et al. Comparison of Whole-Genome Sequencing Methods for Analysis of Three Methicillin-Resistant Staphylococcus Aureus Outbreaks. J. Clin. Microbiol. 2017;55:1946–1953. doi: 10.1128/JCM.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Toleman M.S., Watkins E.R., Williams T., Blane B., Sadler B., Harrison E.M., Coll F., Parkhill J., Nazareth B., Brown N.M., et al. Investigation of a Cluster of Sequence Type 22 Methicillin-Resistant Staphylococcus Aureus Transmission in a Community Setting. Clin. Infect. Dis. 2017;65:2069–2077. doi: 10.1093/cid/cix539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee L., Teh L., Zainuddin Z., Salleh M. The Genome Sequence of a Type ST239 Methicillin-Resistant Staphylococcus Aureus Isolate from a Malaysian Hospital. Stand. Genomic. Sci. 2014;9:933–939. doi: 10.4056/sigs.3887716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Neoh H.M., Mohamed-Hussein Z.A., Tan X.E., B Raja Abd Rahman R.M.F., Hussin S., Mohamad Zin N., Jamal R. Draft Genome Sequences of Four Nosocomial Methicillin-Resistant Staphylococcus Aureus (MRSA) Strains (PPUKM-261-2009, PPUKM-332-2009, PPUKM-377-2009, and PPUKM-775-2009) Representative of Dominant MRSA Pulsotypes Circulating in a Malaysian University Teaching Hospital. Genome Announc. 2013;1:e00103-12. doi: 10.1128/genomeA.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.World Health Organization [(accessed on 20 June 2019)];Global Action Plan on Antimicrobial Resistance. 2015 Available online: http://www.wpro.who.int/entity/drug_resistance/resources/global_action_plan_eng.pdf.
- 102.Mattar C., Ore A.S., Fagerberg S.K., Ramachandran R., Tun W., Wiley E., Chapman H.J. One Health and Antimicrobial Resistance. World Med. J. 2016;62:108–111. [Google Scholar]
- 103.Cuny C., Wieler L.H., Witte W. Livestock-Associated MRSA: The Impact on Humans. Antibiotics. 2015;4:521–543. doi: 10.3390/antibiotics4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Butaye P., Argudín M.A., Smith T.C. Livestock-Associated MRSA and Its Current Evolution. Curr. Clin. Microbiol. Rep. 2016;3:19–31. doi: 10.1007/s40588-016-0031-9. [DOI] [Google Scholar]