Abstract
Background
Circular RNAs (circRNAs), a newly discovered type of endogenous noncoding RNA, have been proposed to mediate the progression of diverse types of tumors. Systematic studies of circRNAs have just begun, and the physiological roles of circRNAs remain largely unknown. Here, we focused on elucidating the potential role and molecular mechanism of circular forkhead box O3 (circFOXO3) in glioblastoma (GBM) progression.
Methods
First, we analyzed circFOXO3 alterations in GBM and noncancerous tissues through real-time quantitative reverse transcription PCR (qRT-PCR). Next, we used loss- and gain-of-function approaches to evaluate the effect of circFOXO3 on GBM cell proliferation and invasion. Mechanistically, fluorescent in situ hybridization, RNA pull-down, dual luciferase reporter, and RNA immunoprecipitation assays were performed to confirm the interaction between circFOXO3 and miR-138-5p/miR-432-5p in GBM. An animal model was used to verify the in vitro experimental findings.
Results
CircFOXO3 expression was significantly higher in GBM tissues than in noncancerous tissues. GBM cell proliferation and invasion were reduced by circFOXO3 knockdown and enhanced by circFOXO3 overexpression. Further biochemical analysis showed that circFOXO3 exerted its pro-tumorigenic activity by acting as a competing endogenous RNA (ceRNA) to increase expression of nuclear factor of activated T cells 5 (NFAT5) via sponging both miR-138-5p and miR-432-5p. Notably, tumor inhibition by circFOXO3 downregulation could be reversed by miR-138-5p/miR-432-5p inhibitors in GBM cells. Moreover, GBM cells with lower circFOXO3 expression developed less aggressive tumors in vivo.
Conclusions
Our data demonstrate that circFOXO3 can exert regulatory functions in GBM and that ceRNA-mediated microRNA sequestration might be a potential strategy for GBM therapy.
Keywords: circFOXO3, ceRNA, glioblastoma, miR-138-5p/miR-432-5p, NFAT5
Key Points.
CircFOXO3 is aberrantly upregulated in GBM tissues.
CircFOXO3 can function as a miR-138-5p and miR-432-5p sponge to regulate NFAT5 expression.
CircFOXO3 is a new factor and potential therapeutic target in GBM.
Importance of the Study.
GBM is one of the most aggressive brain tumors in the central nervous system and has a high incidence of recurrence. The standard treatment for GBM is limited to surgery, irradiation, and chemotherapy. Here we provide the first evidence that circFOXO3 is overexpressed in GBM tissues and sponges specific microRNAs (miR-138-5p and miR-432-5p) to regulate NFAT5 expression through a ceRNA mechanism, thus promoting tumorigenesis in vitro and in vivo. MiR-138-5p/miR-432-5p were aberrantly downregulated and act as tumor suppressor genes by targeting NFAT5 in GBM. Notably, the tumor inhibition caused by circFOXO3 downregulation could be reversed by miR-138/432-5p inhibitors in GBM cells. Targeting circFOXO3/miR-138/432-5p signaling appears to be a promising treatment for GBM.
Glioblastoma (GBM) is the most frequently occurring lethal central nervous system neoplasm.1 Largely because of the propensity of GBM to infiltrate adjacent normal brain tissues, the median survival of GBM patients is merely 12 months. As a result of such efficient invasive activity, these tumors are unresectable and prone to reoccur. Therefore, the mechanism underlying GBM cell invasion has received a great deal of attention. The 2016 World Health Organization (WHO) GBM classification emphasizes the importance of elucidating the molecular signature of GBM. Isocitrate dehydrogenase 1 and 2 (IDH1/2) mutations, 1p/19q codeletion, H3F3A or HIST1H3B/C K27M(H3-K27M) mutations, and C11orf95–RELA fusions have been introduced as determining criteria for GBM, which could assist in diagnosis and therapy selection.2 This cancer type has gradually become a socioeconomic problem, and clarifying the molecular mechanism and treatment targets for GBM is of paramount importance.
Circular RNAs (circRNAs) are characterized by a covalently closed continuous structure with neither a 5′ cap nor a 3′ polyadenylated tail, which differs from linear RNAs; thus, these molecules represent a novel class of noncoding RNAs. Due to their unique structure, circRNAs are highly stable, predominantly distributed in the cytoplasm, and conserved across species.3 There is accumulating evidence that circRNAs are aberrantly expressed in cancer and involved in tumor progression, including proliferation, survival, and motility.3,4 Many studies have indicated that circRNAs could function mainly as miRNA sponges; they communicate with and regulate each other by competitive binding to miRNA response elements (MREs) to further regulate miRNA-targeted gene expression,5 bind proteins,6 encode proteins,7 and promote nuclear translocation.8 These findings indicate that ectopically expressed circRNAs are important in tumorigenesis and cancer progression. Although preliminary studies on circRNA function in GBM have been conducted,7,9,10 the clinical significance and underlying mechanisms of circRNAs in GBM remain poorly defined.
CircFOXO3 (hsa_circ_0006404) was reported to be downregulated in breast cancer11 and non–small cell lung cancer12 and to act as a powerful tumor suppressor by sponging certain miRNAs targeting the parental transcript FOXO3.13 Nevertheless, William et al reported that circFOXO3 was upregulated in heart samples from aged patients.14 However, the role of circFOXO3 in GBM has not been reported. More importantly, whether other mechanisms through which circFOXO3 regulates tumor progression exist remains unknown.
The biological functions of miRNAs, which are endogenous, nonprotein-coding, single-stranded RNAs of 19 to 25 nucleotides, have been extensively studied. MiRNAs have been shown to play several roles in tumor progression. MiR-138-5p and miR-432-5p exert tumor suppressor functions in different tumors. For example, miR-138-5p is always downregulated in cancer.15,16 There are very few reports in the literature concerning miR-432-5p; it has been shown to mediate cyclin-dependent kinase (CDK)4 and 6 inhibitor resistance in breast tumors17 and is highly overexpressed in senescent fibroblasts.18 The possible mechanism needs to be further investigated. Importantly, the expression and underlying mechanisms of miR-138-5p and miR-432-5p in GBM progression have not been fully clarified.
Nuclear factor of activated T cells 5 (NFAT5) is a transcription factor discovered as part of DNA binding in the renal medulla.19 Recent studies have suggested that NFAT5 is dysregulated in tumors and involved in tumor progression, including in melanoma,20 hepatocellular carcinoma,21 renal carcinoma,22 and breast cancer.23 Moreover, Yu et al demonstrated that NFAT5 promotes GBM cell-driven angiogenesis by reducing the expression and secretion of epidermal growth factor–like domain 7.24 However, the potential role of NFAT5 in GBM invasiveness and its upstream regulatory mechanisms currently remain unclear.
In the present study, we used real-time quantitative reverse transcription PCR (qRT-PCR) to first identify the expression of circFOXO3 in GBM tissues and cells. Subsequent studies clarified the functional effect and underlying molecular mechanisms of circFOXO3 in GBM progression through a series of in vitro and in vivo assays. The aim was to provide a potential therapeutic target in patients with GBM.
Materials and Methods
Human Tissue Specimens
Forty-eight glioma tissues (WHO grade II: n = 16, WHO grade III: n = 10, WHO grade IV: n = 22) and 10 normal brain tissues were obtained at the Department of Neurosurgery of the Affiliated Wuxi No. 2 People’s Hospital of Nanjing Medical University between 2013 and 2018. Patient diagnoses were independently re-reviewed by 2 pathologists and classified according to WHO criteria. All specimens were obtained with written informed consent from the patients. Tumor volumes were measured using preoperative contrast-enhanced T1-weighted MRI scans that were routinely acquired on the day of or prior to surgery. The area of contrast enhancement including central necrosis was calculated for each axial section, and the tumor volume was quantified semi-automatically based on the sum of axial areas by OsiriX software. All of these samples were obtained at the initial diagnosis. The study was approved by the ethics committee of the Affiliated Wuxi No. 2 People’s Hospital of Nanjing Medical University.
RNA Sample Treatment with RNase R and PCR
Total RNA was isolated with TRIzol (Life Technologies) according to the manufacturer’s instructions. RNase R treatment was carried out for 15 min at 37°C using 3 U/mg RNase R (Epicentre, cat. #RNR07250). The detailed steps of the qRT-PCR, IDH1/2 mutation, and O6-methylguanine DNA methyltransferase (MGMT) methylation PCR analyses are provided in the Supplementary Material. All primers are listed in Supplementary Table 1.
Cell culture, western blotting, and construction of cells with stable knockdown (KD) or overexpression (OE) of circFOXO3 assays were performed as we previously described.25 Recombinant lentivirus and negative control (NC) lentivirus were purchased from Hanyin Co. (Shanghai, China). The details are described in the Supplementary Methods.
RNA Fluorescence In Situ Hybridization
CircFOXO3 probes were designed and synthesized by RiboBio. The probe signals were detected with a fluorescence in situ hybridization (FISH) kit (RiboBio) according to the manufacturer’s instructions.
RNA Pull-Down Assay
Biotinylated miR-138-5p/miR-432-5p or its mutant were transfected into T98G cells with circFOXO3 OE. Then cells were harvested, lysed, sonicated, and incubated with C-1 magnetic beads at 4°C for 3 hours. After washing with wash buffer, the RNA mix bound to the beads was eluted and extracted with TRIzol (Invitrogen) for qRT-PCR.
RNA Immunoprecipitation Assay
RNA immunoprecipitation (RIP) assay was carried out using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. Antibodies against argonaute 2 (AGO2) (cat. #ab5072) and immunoglobulin G were purchased from Abcam for the RIP assays. Purified RNAs were extracted and analyzed by qRT-PCR.
Cell counting kit-8, transwell migration, luciferase reporter, and immunohistochemistry (IHC) assays were performed as previously described.9,25 The details were described in Supplementary Methods.
Colony Formation and Wound Healing Assay
For colony formation assay, GBM cells with or without circFOXO3-KD/OE were examined; for wound healing assay, GBM cells were seeded in 6-well plates and allowed to reach confluence. The details are in the Supplementary Methods.
Animal Experiments
Animal experiments were performed as we previously described.25 The details are in the Supplementary Methods. The animal study was approved by the Institutional Animal Care and Use Committee of the Affiliated Wuxi No. 2 People’s Hospital of Nanjing Medical University.
Statistical Analysis
The experimental data are presented as the mean ± SD from at least 3 replicates. Student’s two-tailed unpaired t-test was used to determine differences between 2 groups, and one-way ANOVA was performed to test differences among at least 3 groups. The chi-square test was applied to determine the association of circFOXO3 levels with clinicopathological features. The Kaplan–Meier method was used to estimate survival curves for the mice. Correlations were measured by Pearson correlation analysis. All statistical analyses were performed using SPSS software for Windows v17.0. Significance was defined as P < 0.05.
Results
High CircFOXO3 Expression in Human GBM Tissue
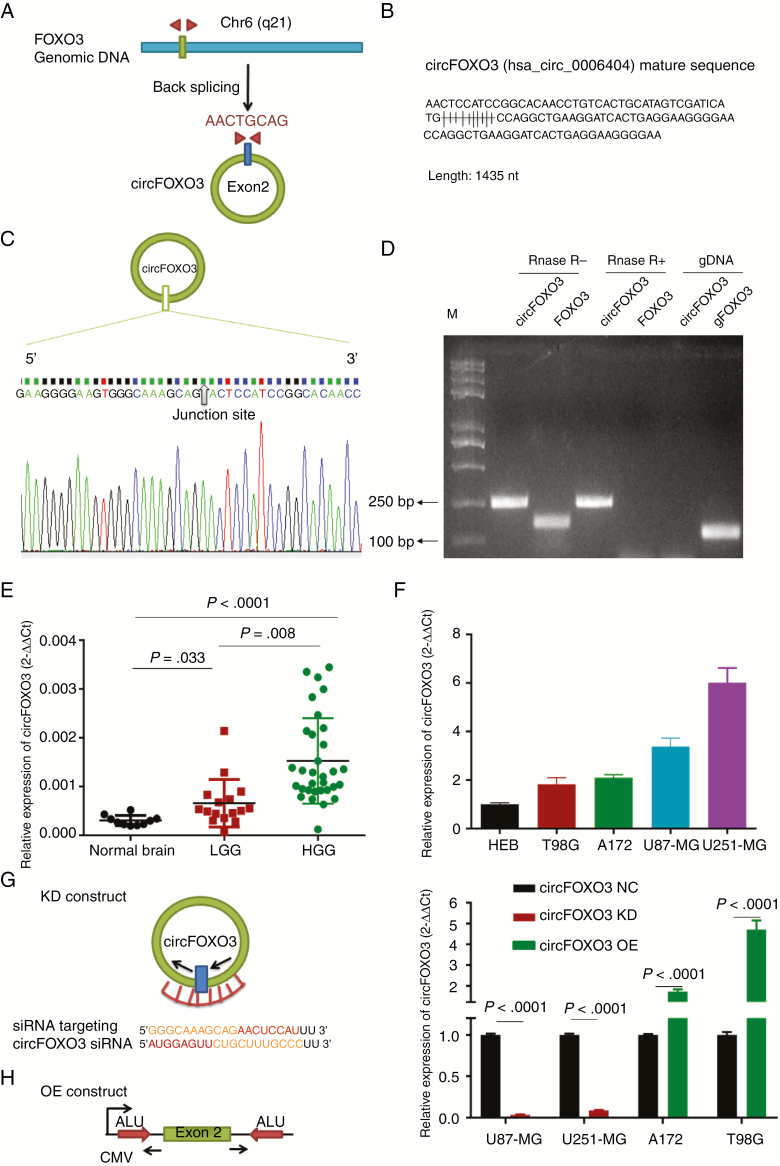
CircFOXO3 contains one exon that ultimately creates a transcript of 1435 nucleotides by back-splicing (Fig. 1A, B). CircFOXO3 cDNA approximately 100 bp upstream and downstream from the junction site was amplified in GBM cells using divergent primers and analyzed by Sanger sequencing. The results confirmed the circFOXO3 junction (Fig. 1C). The divergent primers detected circRNAs in cDNA with or without RNase R treatment, demonstrating that the circRNAs were truly circular and could not amplify any product from genomic DNA. The convergent primers amplified PCR products from linear FOXO3 mRNA, but these products disappeared after RNase R treatment (Fig. 1D).
Fig. 1.
Identification and expression of circFOXO3 in GBM tissues and cells. (A) The structure and part of the sequence of the junction of circFOXO3 are provided, and divergent (red) primers were designed to amplify the back-splicing products. (B) Part of the mature circFOXO3 sequence. (C) Sanger sequencing after PCR using the indicated divergent flanking primers confirmed the “head-to-tail” splicing of circFOXO3. (D) Total RNA from cells with or without RNase R treatment was subjected to PCR. (E) CircFOXO3 transcript levels were significantly higher in HGG than in LGG (LGG vs normal controls: P = 0.033, HGG vs LGG, P = 0.008; one-way, P < 0.0001). (F) Quantitative RT-PCR analysis of circFOXO3 in GBM cells and HEBs. (G) A sketch of the short hairpin circFOXO3 vector structure. (H) A sketch of the circFOXO3 overexpression vector structure. ALU: Complementary ALU pairs were identified as at least one plus strand and one minus strand ALU element on opposite sides of the back-splice, which was required for the formation of circRNA; CMV: a common promoter in vectors. (I) Quantitative RT-PCR analysis of circFOXO3 expression in GBM cells. A multiple comparisons test adjusted P-value of <0.05 was considered statistically significant. Error bars represent the mean ± SD.
To investigate whether circFOXO3 expression varied in GBM, we detected circFOXO3 expression in 48 gliomas and 10 normal controls. As shown in Fig. 1E, circFOXO3 was significantly higher in high-grade glioma (HGG) than in low-grade glioma (LGG) (LGG vs normal controls: P = 0.033, HGG vs LGG, P = 0.008; one-way, P < 0.0001). For a detailed association analysis of circFOXO3 with clinicopathological characteristics and molecular markers, see Supplementary Tables 2 and 3. We analyzed IDH1/2 mutation expression and MGMT methylation status in 48 gliomas. Consistent with previous reports,26–28 the analysis revealed that 68.75% of WHO grade II, 60% of WHO grade III, and 4.5% of WHO grade IV samples were IDH1/2 mutations and that 40.9% of GBMs (n = 9) exhibited MGMT methylation (Supplementary Fig. 1 and Supplementary Table 2). Based on the median relative circFOXO3 expression in glioma, the patients were separated into low and high expression groups (0.001061 ± 0.0001233, n = 48). Briefly, circFOXO3 expression was significantly associated with tumor size (P = 0.009), histologic grade (P = 0.014), wild-type IDH expression (P = 0.035), and MGMT methylation status (P = 0.00017) (Supplementary Table 3).
Altogether, these data suggest that increased circFOXO3 expression might be critically involved in GBM progression.
CircFOXO3 Promotes GBM Tumorigenesis and Invasion In Vitro
We determined circFOXO3 expression levels in 4 GBM cells (U87-MG, U251-MG, A172, and T98G) and human normal glial cells (HEBs). Compared with HEBs, GBM cells showed significant upregulation of circFOXO3, with high expression in U251-MG and U87-MG and low expression in A172 and T98G. Therefore, U251-MG and U87-MG were selected for circFOXO3 KD, and A172 and T98G were selected for circFOXO3 OE (Fig. 1F).
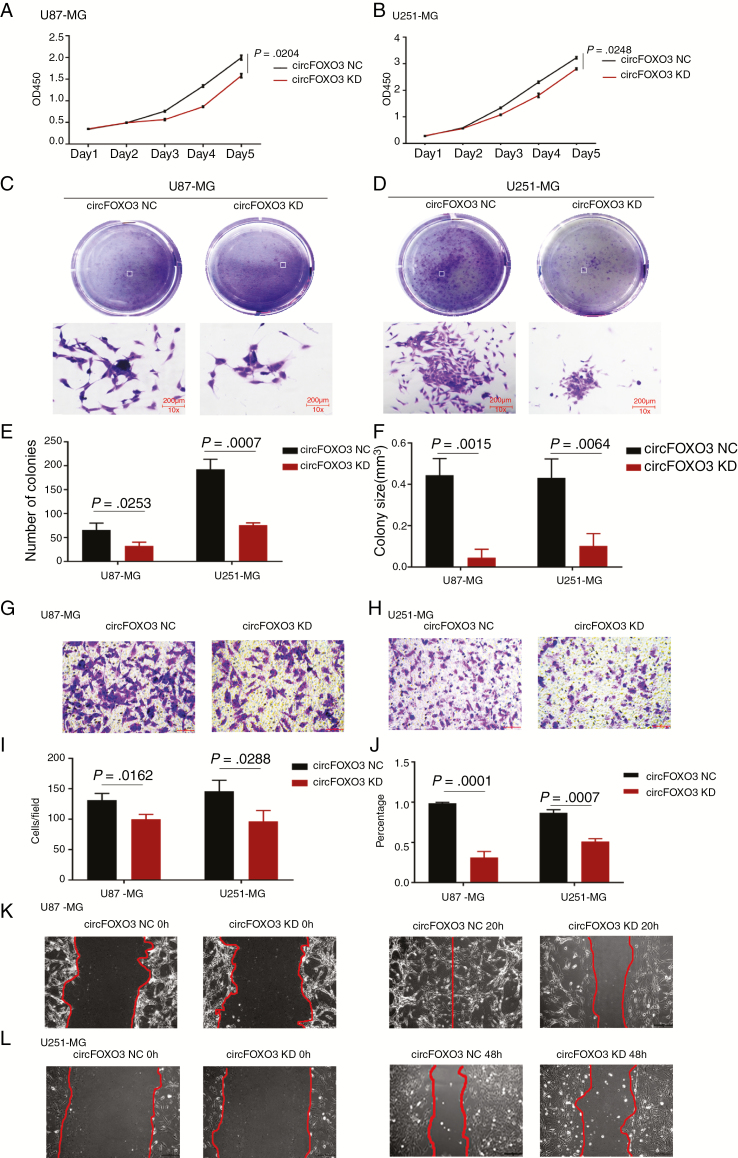
To explore the function of circFOXO3 in GBM cells, we effectively knocked down circFOXO3 in U87-MG and U251-MG (Fig. 1G and 1I). We also successfully constructed a circFOXO3 expression lentivirus and overexpressed circFOXO3 in A172 and T98G (Fig. 1H, I). CCK-8 and colony formation assays were carried out to explore the effects of circFOXO3 on GBM cell proliferation. The CCK-8 assays showed that circFOXO3 KD significantly inhibited U87-MG and U251-MG cell proliferation (Fig. 2A, B). In addition, colony formation assays demonstrated that circFOXO3 KD reduced the colony numbers (Fig. 2C–E) and size (Fig. 2F). Moreover, circFOXO3 OE enhanced the proliferative ability of A172 and T98G (Supplementary Fig. 2A, B), significantly increased the number of colonies (Supplementary Fig. 2C–E), and notably increased the size of each colony (Supplementary Fig. 2F).
Fig. 2.
CircFOXO3 KD suppressed GBM cell proliferation, colony formation, invasion, and migration. (A, B) CCK-8 assays were performed using U87-MG and U251-MG with or without circFOXO3-KD. (C–F) Colony formation assays were performed using U87-MG and U251-MG with or without circFOXO3 KD (magnification: 100×; scale bar = 200 μm). Colony number (E) and size (F) were calculated. (G–I) Transwell analyses of U87-MG and U251-MG with or without circFOXO3 KD were performed. Representative staining images are presented (magnification: 200×; scale bar = 100 μm). (J–L) Wound healing assays were conducted on U87-MG and U251-MG with or without circFOXO3 KD (magnification: 100×; scale bar = 200 μm). A multiple comparisons test adjusted P-value of <0.05 was considered statistically significant. Error bars represent the mean ± SD.
Then, we used transwell and wound healing assays to analyze the effects of circFOXO3 on GBM cell invasion. Transwell assays showed that cell invasion was obviously attenuated in circFOXO3 KD cells (Fig. 2G–I), and the number of invaded A172 and T98G cells increased after circFOXO3 OE (Supplementary Fig. 2G–I). Moreover, wound healing assays showed that wound closure in U87-MG and U251-MG with circFOXO3 KD was slower than that in controls (Fig. 2J–L), but wound closure was much faster after circFOXO3 expression upregulation in A172 and T98G (Supplementary Fig. 2J–L). Collectively, these findings provide evidence that circFOXO3 KD can inhibit GBM cell proliferation and invasion in vitro and that increased circFOXO3 levels are crucial for promoting GBM cell tumorigenesis and invasion.
CircFOXO3 Functions as a Sponge for MiR-138-5p and MiR-432-5p in GBM Cells
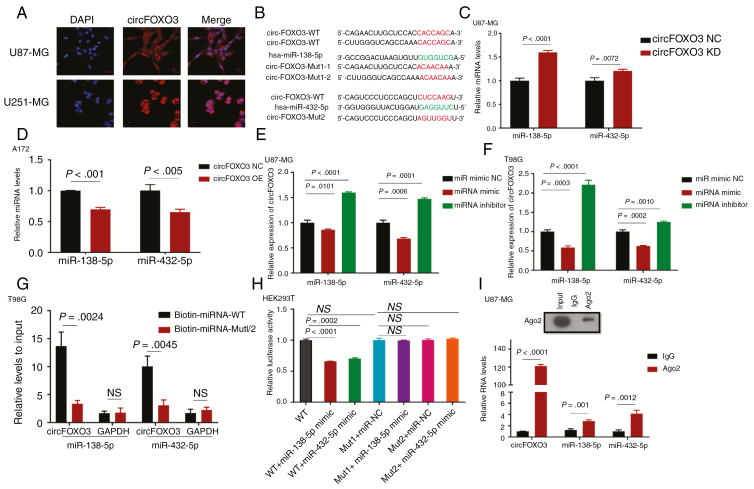
CircRNAs located predominantly in the cytoplasm usually serve as miRNA sponges to regulate expression and activity. Thus, we first searched the circRNADb database (http://202.195.183.4:8000/circrnadb/circRNADb.php) and found that no protein features were predicted for circFOXO3. Second, FISH analysis showed that circFOXO3 was abundant and located in the cytoplasm (Fig. 3A). Based on the above findings we explored whether circFOXO3 binds to miRNAs.
Fig. 3.
CircFOXO3 may function as a miR-138-5p and miR-432-5p sponge. (A) CircFOXO3 was detected by FISH in U87-MG and U251-MG. Nuclei were stained with 4′,6′-diamidino-2-phenylindole. Scale bar = 20 μm. (B) A schematic drawing showing the putative miR-138-5p/miR-432-5p binding sites with respect to circFOXO3. (C, D) MiR-138-5p/miR-432-5p expression increased upon circFOXO3 KD and decreased after circFOXO3 OE. (E, F) CircFOXO3 expression decreased upon ectopic miR-138-5p/miR-432-5p expression and increased after miR-138-5p/miR-432-5p inhibition. (G) Biotinylated wild-type/mutant miR-138-5p/miR-432-5p were transfected into T98G cells overexpressing circFOXO3. After streptavidin capture, circFOXO3 expression was detected by qRT-PCR. (H) Luciferase reporter assays were reported in HEK293T cotransfected with reporter plasmid (or the corresponding mutant reporter) and the indicated miRNAs. (I) RIP experiments were performed using an anti-AGO2 antibody and U87-MG cells. A multiple comparisons test adjusted P-value of <0.05 was considered statistically significant. Error bars represent the mean ± SD.
Because various algorithms use different methods to score miRNA targets, we screened and analyzed 2 miRNAs (miR-138-5p and miR-432-5p) identified in different public databases (RegRNA, starBase, miRDB, and CircInteractome) and in previous reports12,13; the expression of these miRNAs was downregulated in glioma, and they had more than one binding site for specific miRNAs related to circFOXO3. In searching for miRNAs that bind to circFOXO3, we found binding sites for miR-138-5p/miR-432-5p in circFOXO3 (Fig. 3B). Subsequently, we investigated the correlation between circFOXO3 and miR-138-5p/miR-432-5p in GBM cells. MiR-138-5p and miR-432-5p expression was upregulated after circFOXO3 KD and downregulated after circFOXO3 OE (Fig. 3C, D). Moreover, circFOXO3 levels were significantly decreased after the overexpression of miR-138-5p/miR-432-5p via mimics and increased after the inhibition of miR-138-5p/miR-432-5p (Fig. 3E, F; Supplementary Fig. 3).
To validate the direct binding between circFOXO3 and miR-138-5p/miR-432-5p, we conducted biotin-coupled miRNA capture and luciferase activity assay. Biotin-labeled miR-138-5p/miR-432-5p and its mutant mimics were designed to pull down circFOXO3 in T98G cells overexpressing circFOXO3. We found obvious enrichment of circFOXO3 in wild-type miR-138-5p/miR-432-5p compared with mutant (Fig. 3G). Additionally, HEK293T cells were cotransfected with miR-138-5p/miR-432-5p mimics and luciferase reporters. Compared with the controls, miR-138-5p/miR-432-5p transfection reduced luciferase reporter activity (Fig. 3H). We then mutated the predicted binding sites for miR-138-5p/miR-432-5p and found no difference in luciferase reporter activity between miR-138-5p/miR-432-5p mimics and the control (Fig. 3H).
Previous studies have demonstrated that the binding of circRNAs to miRNAs is mediated by AGO2. Thus, we performed an anti-AGO2 RIP assay in U87-MG and showed that circFOXO3 and miR-138-5p/miR-432-5p were enriched in AGO2 (Fig. 3I).
These results indicated that circFOXO3 can function as a sponge for miR-138-5p/miR-432-5p.
MiR-138-5p/miR-432-5p Are Downregulated and Act as Tumor Suppressor Genes by Targeting NFAT5 in GBM Cells
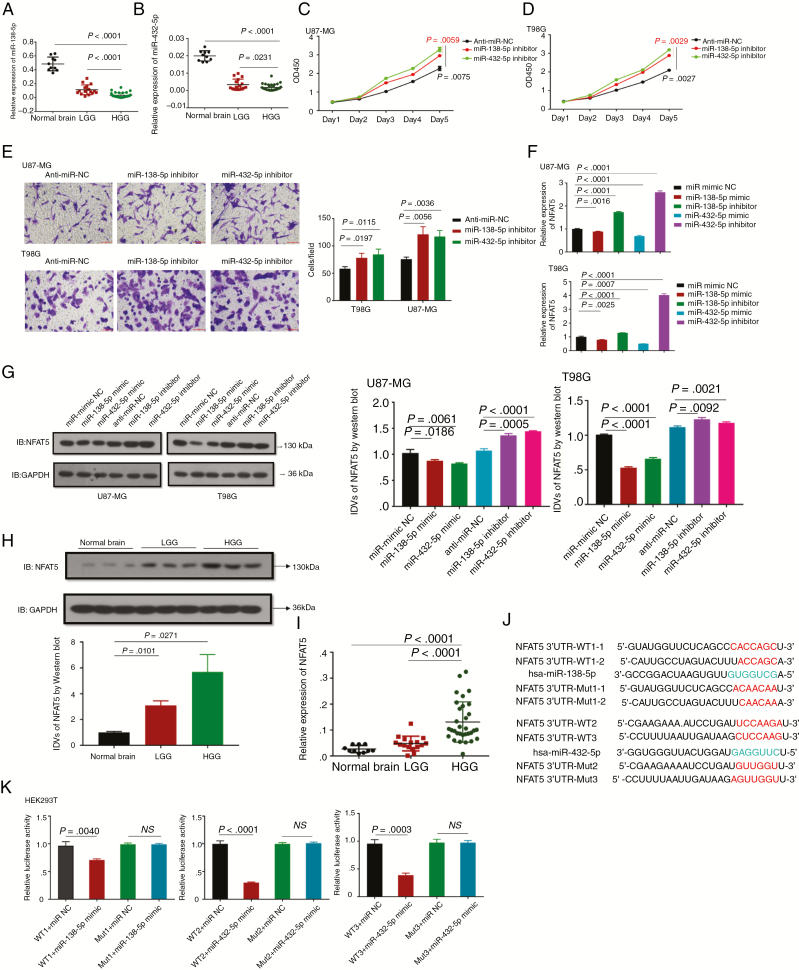
Considering the interaction between circFOXO3 and miR-138-5p/miR-432-5p, we next assessed the expression and function of miR-138-5p/miR-432-5p in GBM. As shown in Fig. 4A and B, miR-138-5p/miR-432-5p were found to be significantly lower in HGG than in normal controls. Furthermore, CCK-8 assays revealed that decreased expression of miR-138-5p/miR-432-5p significantly promoted cell viability (Fig. 4C, D). Additionally, transwell assays indicated that the invasive ability was obviously higher in miR-138-5p/miR-432-5p inhibitor-transfected cells than in NC-transfected cells (Fig. 4E).
Fig. 4.
MiR-138-5p/miR-432-5p suppress cell proliferation and invasion by targeting NFAT5 in GBM cells. (A, B) The relative expression levels of miR-138-5p (A) and miR-432-5p (B) were determined by qRT-PCR in 48 gliomas (WHO grade II: n = 16, WHO grade III: n = 10, WHO grade IV: n = 22) and 10 normal controls. (C, D) CCK-8 assays of U87-MG and T98G transfected with anti-miR NC, miR-138-5p or miR-432-5p inhibitor were performed. At the indicated time points, the number of cells per well was measured according to the absorbance (450 nm). (E) Transwell analysis of U87-MG and T98G transfected with anti-miR NC, miR-138-5p or miR-432-5p inhibitor was performed. Representative images are presented (magnification: 200×; scale bar = 100 μm). (F, G) The relative NFAT5 mRNA and protein expression levels in U87-MG and T98G transfected with different vectors were analyzed by qRT-PCR (F) and western blotting (G). (H) Western blot analysis of NFAT5 was performed in fresh glioma tissues (LGG: n = 12; HGG: n = 12) and normal controls (n = 5). Images of representative expression patterns are presented. (I) The relative NFAT5 expression level was analyzed by qRT-PCR in 48 gliomas (WHO grade II: n = 16, WHO grade III: n = 10, WHO grade IV: n = 22) and 10 normal controls. (J) A schematic drawing shows the putative miR-138-5p/miR-432-5p binding sites with respect to NFAT5. (K) Luciferase activity assays were performed in wild-type and mutant HEK293T cells transfected with different vectors. A multiple comparisons test adjusted P-value of <0.05 was considered statistically significant. Error bars represent the mean ± SD.
According to previous reports,24,29,30 certain GBM oncogenes are targets of miR-138-5p/miR-432-5p. Thus, 14 oncogenes among the miR-138-5p/miR-432-5p targets were selected using the mirDIP database (http://ophid.utoronto.ca/mirDIP/) (Supplementary Table 4). Among these oncogenes, 3 (CDK6, NFAT5, and SP1) are closely involved in tumor progression. Thus, we altered miR-138-5p/miR-432-5p expression and detected the levels of the respective targets (Fig. 4F, G and Supplementary Fig. 4). Western blot analysis revealed that NFAT5 expression was downregulated in cells overexpressing miR-138-5p or miR-432-5p but increased in cells transfected with a miR-138-5p or miR-432-5p inhibitor (Fig. 4G). Moreover, high NFAT5 mRNA and protein levels were observed in GBM (Fig. 4H, I). As predicted, NFAT5 was shown to have binding sites for miR-138-5p/miR-432-5p (Fig. 4J). Then, wild-type and mutant 3′ untranslated region sequences of NFAT5 were cloned to construct reporter plasmids and mutant vectors, respectively. Cotransfection with miR-138-5p/miR-432-5p mimics and reporter plasmids strongly reduced luciferase activity. Conversely, cotransfection with miR-138-5p/miR-432-5p mimics and mutated vectors had no obvious effect on luciferase activity (Fig. 4K). Consequently, the findings confirmed that NFAT5 is a direct target of miR-138-5p/miR-432-5p.
Altogether, these data indicated that miR-138-5p/miR-432-5p promoted the proliferation and invasion of GBM cells by targeting NFAT5.
CircFOXO3 Modulates MiR-138-5p and MiR-432-5p Targets
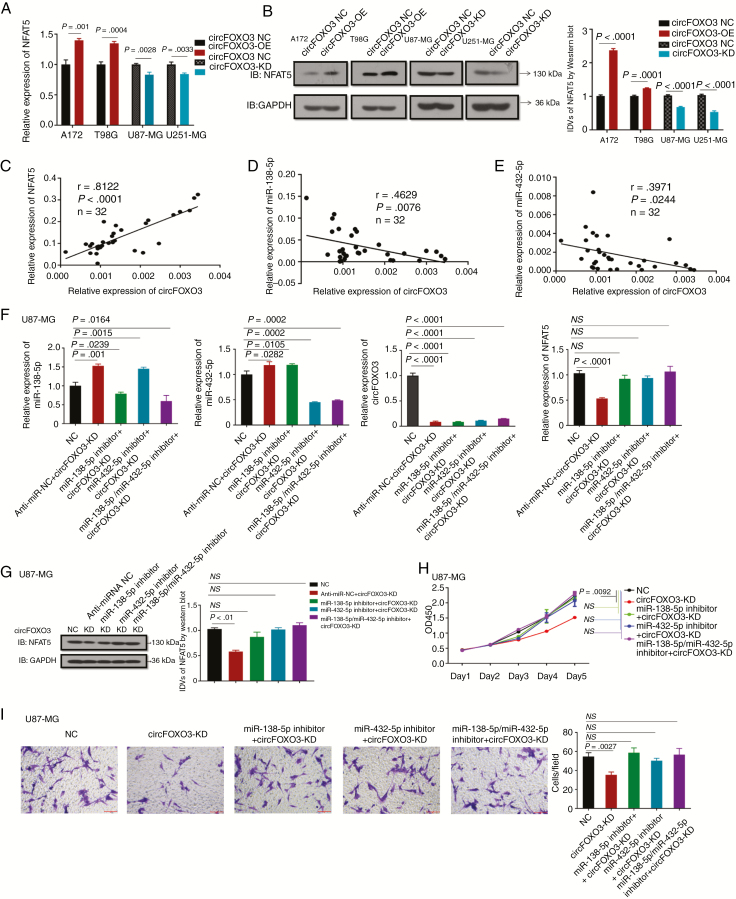
Next, we measured the expression of these targets after circFOXO3 OE or KD. The results showed that NFAT5 was the most strikingly upregulated when circFOXO3 was overexpressed (Fig. 5A, B and Supplementary Fig. 5).
Fig. 5.
CircFOXO3 modulates the expression of NFAT5, an endogenous target of miR-138-5p and miR-432-5p. (A, B) The relative NFAT5 mRNA and protein expression levels in cells with or without circFOXO3-OE/KD were analyzed by qRT-PCR (A) and western blotting (B). (C) CircFOXO3 was positively correlated with the expression of NFAT5 (r = 0.8122, P < 0.0001) in HGGs. (D, E) CircFOXO3 was inversely correlated with the expression of miR-138-5p (D) (r = −0.4629, P = 0.0076) and miR-432-5p (E) (r = −0.3971, P = 0.0244) in HGGs. (F, G) Quantitative RT-PCR (F) and western blotting (G) analyses revealed NFAT5 mRNA and protein levels in U87-MG transfected with different vectors. (H) CCK-8 assays were performed after cells were cotransfected with different vectors. (I) Transwell assays were conducted after cells were cotransfected with different vectors (magnification: 200×; scale bar = 100 μm). A multiple comparisons test adjusted P-value of <0.05 was considered statistically significant. Error bars represent the mean ± SD.
Given that NFAT5 shares MREs with circFOXO3, we conducted a correlation analysis and rescue assays to investigate whether circFOXO3 exerts its oncogenic effect by modulating NFAT5 expression via sponging miR-138-5p/miR-432-5p. Correlation analysis demonstrated a moderate positive correlation between circFOXO3 and NFAT5 (r = 0.8122, P < 0.0001) (Fig. 5C). In addition, a moderate negative correlation was observed between circFOXO3 and miR-138-5p (r = −0.4629, P = 0.0076) or miR-432-5p (r = −0.3971, P = 0.0244) (Fig. 5D, E).
Furthermore, rescue experiments were performed by cotransfecting circFOXO3 KD and miR-138-5p and/or miR-432-5p inhibitors in U87-MG. Quantitative RT-PCR and western blot assays indicated that NFAT5 mRNA and protein levels were partly increased in U87-MG cells cotransfected with circFOXO3 KD and miR-138-5p/miR-432-5p inhibitor compared with those in the circFOXO3 KD group (Fig. 5F, G). Also, the proliferation ability of U87-MG cells cotransfected with circFOXO3 KD and miR-138-5p/miR-432-5p inhibitor was increased compared with those in the circFOXO3 KD group (Fig. 5H). These findings suggest that inhibition of miR-138-5p/miR-432-5p could partly restore the proliferation suppression induced by circFOXO3 KD. Similarly, circFOXO3 KD could also partly attenuate the downregulation of invasion promotion mediated by miR-138-5p/miR-432-5p in U87-MG (Fig. 5I).
Altogether, these data demonstrated that circFOXO3 promotes GBM progression by eliminating miR-138-5p/miR-432-5p, which target NFAT5.
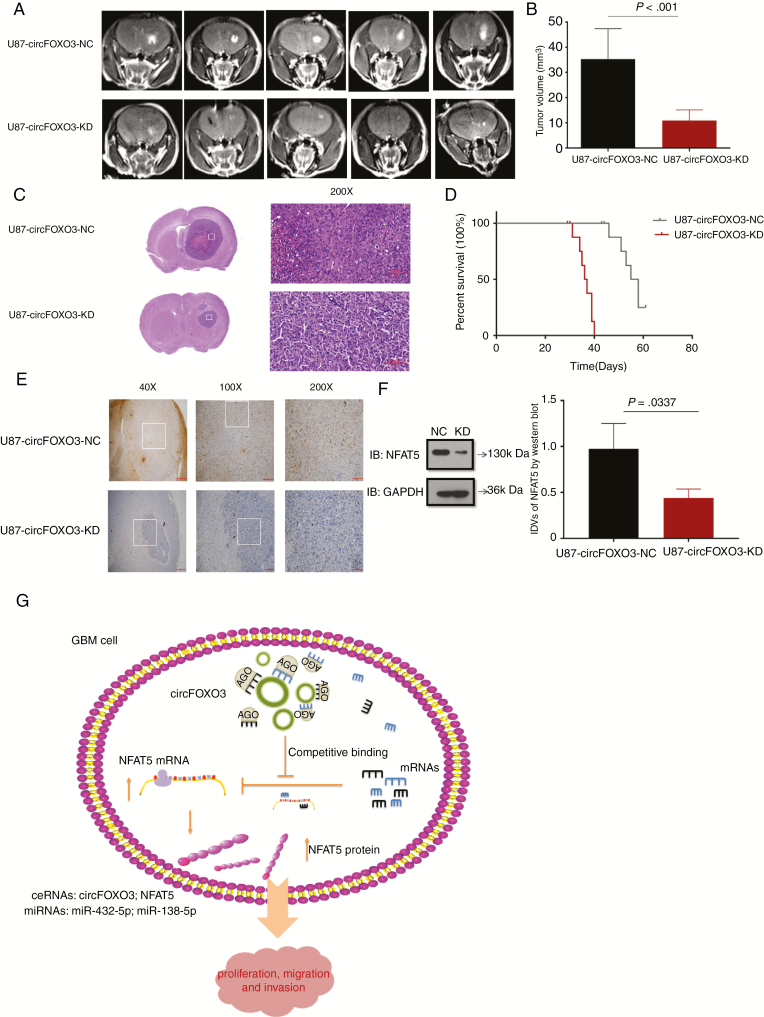
Suppression of CircFOXO3 Inhibits Xenograft Growth In Vivo
To determine the in vivo effect of circFOXO3 on GBM progression, we injected circFOXO3 KD or circFOXO3 NC cells into the corpus striatum of anesthetized nude mice. After implantation, a few animals started to show signs of morbidity, and mice were assessed by MRI to confirm intracranial tumor formation (Fig. 6A). CircFOXO3 suppression significantly reduced tumor growth and invasion (Fig. 6A–C). Tumor volumes were also decreased by approximately 2-fold in the circFOXO3 KD group (P = 0.001; Fig. 6B). Strikingly, mice implanted with circFOXO3 KD cells had a median survival of 56.5 days, whereas 100% of the controls died within 37 days (Fig. 6D). Consistently, NFAT5 expression was decreased in the circFOXO3 KD group (Fig. 6E, F). Altogether, these results suggest that circFOXO3 is critical for GBM progression in vivo.
Fig. 6.
CircFOXO3 KD inhibits GBM progression in vivo. (A) Representative MR images of xenograft GBM tumors orthotopically inoculated with cells with NC or circFOXO3 KD on 3 weeks post-implantation. (B)Tumor volumes were calculated for each group (n = 8 per group). (C) Representative hematoxylin and eosin images of each group are shown (magnification: 200×; scale bar = 100 μm). (D) The comparative survival of mice bearing NC or circFOXO3 KD tumors was determined. The time of death was recorded as days after U87-MG implantation. (E) Representative immunohistochemistry images of NFAT5 in tumors collected from each group (magnification: 40×, 100×, 200×; scale bar = 500 μm, 200 μm, 100 μm). (F) Western blot analysis of NFAT5 in tumor collected from each group was performed. A multiple comparisons test adjusted P-value of <0.05 was considered statistically significant. The error bars represent the mean ± SD. (G) Model for the mechanism of circFOXO3 in GBM. CircFOXO3 sponges miR-138-5p/miR-432-5p through MREs and thus acts as a ceRNA to regulate NFAT5 expression and modulate tumor cell function in vitro and in vivo.
Discussion
Accumulating evidence has confirmed that circRNAs play important roles in tumor pathology and might be used as diagnostic and therapeutic targets. For instance, a decrease in circMTO1 (circular mitochondrial transfer RNA translation optimization 1) in hepatocellular carcinoma may be a prognostic indicator of poor patient survival.5 The growth and migration of hepatocellular carcinoma cells are inhibited by cSMARCA5, which could be a potential therapeutic target.31 Another study indicated that ectopic expression of circFOXO3 could suppress tumor growth and cancer cell proliferation and survival.13 However, the expression, function, and molecular mechanism of circFOXO3 in GBM remain unknown.
Here, we found that circFOXO3 was aberrantly upregulated in GBM tissues compared with normal controls. High circFOXO3 expression was significantly associated with tumor size, clinical stage, wild-type IDH expression, and MGMT methylation status. Recently, it was reported that IDH mutations occur in a subset of GBM patients and in the majority of WHO grades II and III diffuse gliomas.32 Additionally, IDH mutations predict greater sensitivity to temozolomide.33 MGMT methylation is a well-established confounding prognostic marker for GBM patients and predicts the response of GBM patients to alkylating agents.28,34 The findings reported herein indicate that circFOXO3 plays a vital role in GBM progression. At a functional level, we found that circFOXO3 KD inhibited GBM cell proliferation and invasion. In contrast, circFOXO3 OE enhanced GBM cell proliferation and invasion in vitro. These data suggest that circFOXO3 acts as an oncogene in GBM.
RNAs can communicate with each other as ceRNAs through competitively shared miRNAs.35 In accordance with previous reports,36–38 we presented the ceRNA mechanism in a series of thorough experiments. The cytoplasmic accumulation of circFOXO3 in GBM cells illustrated by FISH indicated that circFOXO3 may function through posttranscriptional regulation. Next, bioinformatics analysis identified several miRNAs that might interact with circFOXO3. Interestingly, both miR-138-5p and miR-432-5p could bind and be regulated by circFOXO3. In addition, RNA pull-down, luciferase assays, and RIP showed that circFOXO3 interacted with miR-138-5p/miR-432-5p, which provided evidence that circFOXO3 competes with miR-138-5p/miR-432-5p in GBM cells. In loss-of-function experiments, a miR-138-5p or miR-432-5p inhibitor reduced the number of invading cells upon circFOXO3 KD. Moreover, the effects of circFOXO3 KD on cell proliferation could be reversed by a miR-138-5p/miR-432-5p inhibitor. Therefore, circFOXO3 may exert its physiological functions via sponging both miR-138-5p and miR-432-5p. To date, there has been little information regarding the role of miR-138-5p and miR-432-5p in GBM. The expression levels of miR-138-5p and miR-432-5p were decreased and negatively associated with circFOXO3 expression in GBM. Additionally, CCK-8 and transwell assays suggested that miR-138-5p and miR-432-5p may participate in GBM tumorigenesis.
Recent researches have pointed to an important role for NFAT5 in tumor progression.20–24 In this study, we report for the first time that NFAT5 is directly targeted by miR-138-5p and miR-432-5p. CircFOXO3 indirectly regulated NFAT5 expression via sequence matching with both miR-138-5p and miR-432-5p. Moreover, the effects of circFOXO3 downregulation on NFAT5 expression could be reversed by a miR-138-5p/miR-432-5p inhibitor in GBM cells. Additionally, we found that NFAT5 was upregulated and positively associated with circFOXO3 expression in GBM. This finding is in accordance with previous reports indicating that NFAT5 promotes tumor progression.22–24 Therefore, circFOXO3 may regulate the proliferation and invasion of GBM cells by modulating NFAT5 expression. However, the opposite expression pattern of circFOXO3 was also reported in breast cancer11 and non–small cell lung cancer.12 Du et al found that circFOXO3 suppresses cell proliferation and cell cycle progression by binding to CDK2 and p21, resulting in the formation of a ternary complex. Additionally, circFOXO3 was shown to interact with other cell cycle–associated proteins, including CDK6, p16, and p27 and to facilitate p53 ubiquitination and degradation by binding to both p53 and murine double minute 2.6,39 Furthermore, circFOXO3 was shown to be highly expressed in heart samples from aged patients and to interact with inhibitor of DNA binding 1 protein, E2F1, focal adhesion kinase, and hypoxia-inducible factor 1α.14 These studies indicate that the circFOXO3 tertiary conformation differs in various diseases, resulting in the formation of different ternary complexes. Here we observed different trends in circFOXO3 expression, and CDK6 expression was not correlated with circFOXO3 in GBM cells, suggesting that the previously reported roles of circFOXO3 in other tumors are not ubiquitous. This discrepancy is complicated and might be caused by different tertiary structures of circFOXO3 in GBM, which awaits further investigation.
In this study, we provide the first evidence that circFOXO3 is overexpressed in GBM tissues and functions as a miR-138-5p/miR-432-5p sponge to regulate NFAT5 expression through a ceRNA mechanism, thus promoting tumorigenesis in vitro and in vivo (Fig. 6G). We demonstrated that circFOXO3 is a new factor and potential therapeutic target in GBM. Our findings emphasize the significance of the interaction between circRNAs and miRNAs in tumorigenesis.
Funding
This study was partially supported by grants from General Programs of Nanjing Medical University (2017NJMU172), Pudong New Area health system key specialist construction support (PWZzk2017-16), Shanghai Pudong New Area health system leading talent training plan (PWRl2017-03), Pudong New Area science and technology development fund, special funds for people’s livelihood, medical and health projects (PKJ2016-Y45).
Conflict of interest statement. The authors declare no potential conflicts of interest.
Authorship statement.
Conception and design: S.Z., K.L., Z.M., Q.W., X.L., Y.Q. Collection and assembly of data: S.Z., K.L., Z.M., Q.W., X.L., Y.Q. Data analysis and interpretation: All authors. Manuscript writing: All authors. Final approval of manuscript: All authors. Accountable for all aspect of the study: All authors.
Supplementary Material
References
- 1. Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015; 17(Suppl 4):iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 3. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. [DOI] [PubMed] [Google Scholar]
- 4. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019; 176(4):869–881 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. [DOI] [PubMed] [Google Scholar]
- 6. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang Q, Du WW, Wu N, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He Q, Zhao L, Liu Y, et al. circ-SHKBP1 regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol Ther Nucleic Acids. 2018;10:331–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang R, Zhang S, Chen X, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu WY. Roles of the circular RNA circ-Foxo3 in breast cancer progression. Cell Cycle. 2017;16(7):589–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y, Zhao H, Zhang L. Identification of the tumor‑ suppressive function of circular RNA FOXO3 in non‑ small cell lung cancer through sponging miR-155. Mol Med Rep. 2018;17(6):7692–7700. [Google Scholar]
- 13. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35(30):3919–3931. [DOI] [PubMed] [Google Scholar]
- 14. Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–1412. [DOI] [PubMed] [Google Scholar]
- 15. Yang R, Liu M, Liang H, et al. miR-138-5p contributes to cell proliferation and invasion by targeting survivin in bladder cancer cells. Mol Cancer. 2016;15(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pan X, Chen Y, Shen Y, Tantai J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019;10(6):429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cornell L, Wander SA, Visal T, Wagle N, Shapiro GI. MicroRNA-mediated suppression of the TGF-beta pathway confers transmissible and reversible CDK4/6 inhibitor resistance. Cell Rep. 2019;26(10):2667–2680 e2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dhahbi JM, Atamna H, Boffelli D, Magis W, Spindler SR, Martin DI. Deep sequencing reveals novel microRNAs and regulation of microRNA expression during cell senescence. PLoS One. 2011;6(5):e20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berry MR, Mathews RJ, Ferdinand JR, et al. Renal sodium gradient orchestrates a dynamic antibacterial defense zone. Cell. 2017;170(5):860–874 e819. [DOI] [PubMed] [Google Scholar]
- 20. Levy C, Khaled M, Iliopoulos D, et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell. 2010;40(5):841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee JH, Suh JH, Choi SY, et al. Tonicity-responsive enhancer-binding protein promotes hepatocellular carcinogenesis, recurrence and metastasis. Gut. 2019;68(2):347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Küper C, Beck FX, Neuhofer W. NFAT5-mediated expression of S100A4 contributes to proliferation and migration of renal carcinoma cells. Front Physiol. 2014;5:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jauliac S, López-Rodriguez C, Shaw LM, Brown LF, Rao A, Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4(7):540–544. [DOI] [PubMed] [Google Scholar]
- 24. Yu H, Zheng J, Liu X, et al. Transcription factor NFAT5 promotes glioblastoma cell-driven angiogenesis via SBF2-AS1/miR-338-3p-mediated EGFL7 expression change. Front Mol Neurosci. 2017;10:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lan J, Guo P, Lin Y, et al. Role of glycosyltransferase PomGnT1 in glioblastoma progression. Neuro Oncol. 2015;17(2):211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 29. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Li F, Yang YT, et al. IGFBP2 promotes vasculogenic mimicry formation via regulating CD144 and MMP2 expression in glioma. Oncogene. 2019;38(11):1815–1831. [DOI] [PubMed] [Google Scholar]
- 31. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. [DOI] [PubMed] [Google Scholar]
- 32. Leu S, von Felten S, Frank S, et al. IDH/MGMT-driven molecular classification of low-grade glioma is a strong predictor for long-term survival. Neuro Oncol. 2013;15(4):469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75(17):1560–1566. [DOI] [PubMed] [Google Scholar]
- 34. Vassella E, Vajtai I, Bandi N, Arnold M, Kocher V, Mariani L. Primer extension based quantitative polymerase chain reaction reveals consistent differences in the methylation status of the MGMT promoter in diffusely infiltrating gliomas (WHO grade II-IV) of adults. J Neurooncol. 2011;104(1):293–303. [DOI] [PubMed] [Google Scholar]
- 35. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hall IF, Climent M, Quintavalle M, et al. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ Res. 2019;124(4):498–510. [DOI] [PubMed] [Google Scholar]
- 37. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019. [DOI] [PubMed] [Google Scholar]
- 39. Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.