1. Introduction
Personalized or precision treatments are targeted therapies that are particularly effective or have reduced harms for a subgroup of patients within a given condition [3]. This type of targeted therapy has considerable potential benefits, including reduced exposure of patients and study participants to treatments from which they are not likely to benefit and reduced financial resources spent on treatments that are ineffective or potentially harmful for certain individuals. Personalized treatment may be potentially beneficial for chronic pain conditions, in which the percentages of patients who meaningfully improve after initiation of efficacious pharmacologic therapies has been modest [19]. However, the ability of a particular patient characteristic to predict treatment response must be demonstrated in clinical trials before such predictors can be implemented in clinical practice.
Significant advances have occurred in developing a foundation for personalized pain treatment [10, 22]. Phenotypic or genotypic characteristics (e.g., predictive biomarkers [15]) that potentially identify groups of patients that are more likely to respond to a particular treatment have been suggested in multiple studies. For example, quantitative sensory testing (QST) can be used to group patients with similar sensory phenotypes within neuropathic pain conditions and has been shown to predict response to oxcarbazepine. [1, 5, 26]. Patients with specific genetic mutations in receptors that are targeted by certain drugs (e.g., sodium channels [2]) may respond more robustly to those drugs. Psychological characteristics associated with negative affect and pain catastrophizing have been associated with worse outcomes in uncontrolled, prospective studies of single treatments and in studies that qualitatively compare subgroup differences in treatment effects (active vs. placebo) groups [9, 27, 28]. Other potential phenotypic predictors include pain qualities (e.g., neuropathic vs. nociceptive) and sleep quality [10]. The evidence derived from uncontrolled studies is very difficult to interpret because it is possible that the observation of better outcomes in a subgroup might reflect an enhanced placebo effect in that subgroup. For example, increased brain connectivity identified using functional magnetic resonance imaging (fMRI) has been shown to predict placebo response in recent studies [17, 18, 21]. Also, increased variability in baseline pain ratings was shown to predict placebo response in retrospective analyses of data from multiple randomized clinical trials [12, 16].
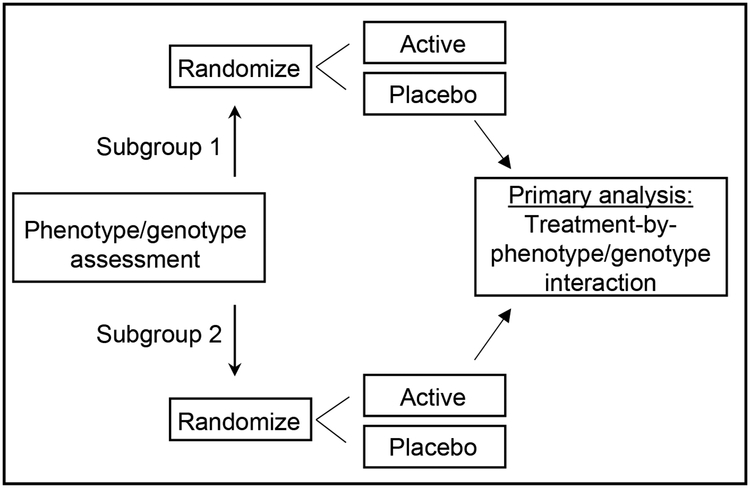
Much of the current literature regarding potential predictors of treatment response, including the studies referenced above, is based on secondary or retrospective analyses of randomized clinical trial (RCT) data that demonstrate a treatment effect in only 1 subgroup [7, 10]. However, the finding that a treatment effect (vs. placebo) yields a significant result in 1 subgroup but not the other is not sufficient to demonstrate that the treatment is more efficacious in 1 subgroup than the other. Concluding that the treatment effect in 1 subgroup is larger than that in another subgroup because the effect is what some authors term “more significant” (i.e., associated with a smaller p-value) in 1 subgroup (e.g., p = 0.001 vs. p=0.045) is also not appropriate [15]. In such a case, differences in statistical significance could be simply due to differences in subgroup sample sizes rather than the magnitude of the treatment effect. Additionally, even if the difference in statistical significance is partially due to differences in the magnitudes of the treatment effect, it does not follow that the magnitudes of the treatment effects for the 2 subgroups are significantly different from each another [15]. In order to demonstrate rigorously the ability of a phenotype or genotype to predict response to active treatment, a RCT with a pre-specified primary analysis that tests the significance of the difference between the effect sizes in the 2 subgroups must be conducted. This is accomplished by testing what is termed the treatment-by-phenotype/genotype interaction. [11, 25] (Figure 1). Demant et al. [5] recently reported such a trial in which the primary analysis was a test of significance of the treatment-by-“irritable nociceptor” phenotype interaction. The trial successfully demonstrated that an “irritable nociceptor” phenotype in patients with peripheral neuropathic pain detected using QST predicted greater response to oxcarbazepine vs. placebo compared with patients who did not have the irritable nociceptor phenotype. Specifically, the estimated standardized effect size (SES) (i.e., (meanactive – meanplacebo)/pooled standard deviation) was 0.73 in the irritable nociceptor group and 0.38 in the non-irritable nociceptor group, and the treatment-by-phenotype interaction was statistically significant (p = 0.047). However, a second trial with a similar design and primary analysis that compared the effects of lidocaine on neuropathic pain between patients with and without the “irritable nociceptor” phenotype did not successfully demonstrate a treatment-by-phenotype interaction [4]. The remainder of this article will discuss sample size determination for prospective randomized clnical trials designed specifically to evaluate treatment-by-phenotype/genotype interactions and the interpretation of such trials. These methods apply to all randomized clinical trials, including those that test pharmacologic, psychological, physical, surgical, and device interventions. Of course, those with the most rigorous blinding possible for the particular intervention will produce the highest quality data.
Figure 1.
Illustrative treatment-by-phenotype/genotype interaction trial schema.
2. Sample size determination for testing interactions
A standard sample size calculation for a parallel group RCT that is designed to compare the means of two groups for a normally distributed outcome variable requires assumptions for the type I error probability (alpha or significance level), power, the minimum group difference in means (treatment effect) that one would like to detect, and the variance of the outcome variable in each group. If the variance is the same in each group, as is often assumed, then one only needs to specify alpha, power, and the ratio of the treatment effect to the standard deviation of the outcome variable (i.e., the SES). For a parallel group RCT that is designed to compare the treatment effects between 2 subgroups (i.e., to evaluate whether the treatment-by-phenotype/genotype interaction is equal to zero), the sample size calculation requires assumptions for alpha, power, the percentages of the randomized sample that will be in each subgroup, and the difference in SES between the 2 phenotype/genotype subgroups that one would like to detect. This assumes that the variance of the outcome variable is the same in each treatment group/subgroup combination, which simplifies the presentation below.
Methods are presented below for determining the sample sizes required for parallel group RCTs and crossover trials that are designed to compare the treatment effects between 2 subgroups (i.e., to evaluate whether the treatment-by-phenotype/genotype interaction is equal to zero).
2.1. Parallel group RCTs
Let μij be the mean outcome in treatment group j (0 = placebo, 1 = treatment) for Subgroup i (i = 1, 2). Let Δ = (μ21 − μ20) – (μ11 − μ10) be the subgroup difference in the treatment effect, where μ21 − μ20 is the treatment effect in Subgroup 2 and μ11 − μ10 is the treatment effect in Subgroup 1. It is of interest to test the null hypothesis that Δ = 0 (i.e., that the treatment effects are the same in each subgroup). Note that the means μij will be estimated by the sample means and the subgroup difference in the treatment effect will be estimated by . Assuming equal variances (σ2) in each treatment group-subgroup combination and that there is equal allocation of treatment and placebo within each subgroup, then it is easy to show that the variance of is equal to
where n1 and n2 are the sample sizes for Subgroups 1 and 2, respectively. A formula for the approximate sample size required to provide 100(1 − β)% power to detect a subgroup difference in treatment effect of size Δ, using a t-test and a two-tailed significance level α, is
where r = n1/n2 is the ratio of the subgroup sample sizes and Zυ is the upper 100υth percentile of the standard normal distribution (e.g., when α = 0.05 and power = 0.80, Z0.025 = 1.96 and Z0.20 = 0.842). The total sample size required is N = n1 + n2.
Alternatively, if one has access to software to compute sample size for a two-group comparison of means (using the t-distribution), the required input would be the significance level (α), desired power 100(1 – β)%, subgroup difference in treatment effect (Δ), ratio of the subgroup sample sizes (r), and variance (4σ2). Note here that it is necessary to multiply the variance of the outcome variable by 4 to obtain the required sample size.
2.2. Crossover trials
In a crossover trial, each participant receives both the active treatment and placebo in random order. Let μdi be the mean treatment – placebo difference (treatment effect) among the ni participants in Subgroup i, i = 1, 2. Then Δ = μd2 − μd1 is the subgroup difference in treatment effect, estimated by Let be the variance of the within-participant treatment – placebo difference in outcome, assumed to be the same for each subgroup. Then it is easy to show that the variance of is equal to
A formula for the approximate sample size required to provide 100(1 – β)% power to detect a subgroup difference in treatment effect of size Δ, using a t-test and a two-tailed significance level α, is
The total sample size required is N = n1 + n2.
As in the case of a parallel group trial, if one has access to software to compute sample size for a two-group comparison of means (using the t-distribution), the required input would be the significance level (α), desired power (1 – β), subgroup difference in treatment effect (Δ), ratio of the subgroup sample sizes (r), and variance It is sometimes the case that there is no information available concerning the variance of the within-participant treatment – placebo difference in outcome, but there is information on the variance of the outcome measured at a single time point (σ2). Assuming that the variance is the same for each treatment condition, the two quantities are related by the formula
where ρ is the correlation between the outcomes for the treatment and placebo conditions. If information concerning σ2 is all that is available, one would have to make an educated guess regarding the value of ρ to compute the required sample size.
2.3. Results of sample size calculations
Table 1 provides the total sample sizes required to provide either 80% or 90% power to detect specified subgroup differences in treatment effect, using a t-test and a two-tailed significance level of 5%, for various subgroup allocations. The subgroup differences in treatment effect are expressed in terms of the subgroup difference in standardized effect size (SES) Δ/σ. Table 2 provides the sample sizes required to provide either 80% or 90% power to detect specified subgroup differences in treatment effect, using a t-test and a two-tailed significance level of 5%, for various subgroup allocations and values of the correlation ρ. The subgroup differences in treatment effect are again expressed in terms of the subgroup difference in SES. As is usual practice, the above calculations assume that there are no period effects and no treatment-by-period interaction (e.g., no carry-over effects). For both the parallel group and crossover studies, the calculations were performed using R, Version 3.5.1.
Table 1:
Total sample sizes necessary to yield 80% or 90% power to detect a treatment-by-subgroup interaction in parallel group studies
| SES | 80% Power | 90% Power | ||||
|---|---|---|---|---|---|---|
| Subgroup Allocation | ||||||
| 50:50 | 34:66 | 25:75 | 50:50 | 34:66 | 25:75 | |
| Sample Size | ||||||
| 1.0 | 126 | 142 | 170 | 170 | 189 | 226 |
| 0.9 | 157 | 174 | 209 | 209 | 233 | 277 |
| 0.8 | 198 | 220 | 262 | 265 | 295 | 352 |
| 0.7 | 258 | 286 | 342 | 345 | 383 | 458 |
| 0.6 | 350 | 389 | 466 | 469 | 522 | 625 |
| 0.5 | 505 | 561 | 671 | 674 | 751 | 898 |
| 0.4 | 786 | 876 | 1049 | 1053 | 1172 | 1402 |
| 0.3 | 1397 | 1557 | 1861 | 1869 | 2083 | 2493 |
SES = Difference in standardized effect size between subgroups
Table 2:
Total sample sizes necessary to yield 80% or 90% power to detect a treatment-by-subgroup interaction in crossover studies.
| 80% Power | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SES | Subgroup Allocation | ||||||||||||||
| 50:50 | 34:66 | 25:75 | |||||||||||||
| Correlation† | |||||||||||||||
| 0.8 | 0.6 | 0.5 | 0.4 | 0.2 | 0.8 | 0.6 | 0.5 | 0.4 | 0.2 | 0.8 | 0.6 | 0.5 | 0.4 | 0.2 | |
| Sample Size | |||||||||||||||
| 1.0 | 15 | 27 | 34 | 39 | 54 | 17 | 31 | 37 | 44 | 58 | 19 | 35 | 44 | 53 | 70 |
| 0.9 | 18 | 34 | 42 | 50 | 64 | 20 | 37 | 46 | 55 | 72 | 22 | 43 | 54 | 64 | 86 |
| 0.8 | 22 | 42 | 51 | 62 | 82 | 26 | 46 | 58 | 68 | 90 | 29 | 54 | 67 | 82 | 107 |
| 0.7 | 27 | 54 | 67 | 79 | 106 | 31 | 60 | 74 | 88 | 117 | 37 | 70 | 87 | 106 | 139 |
| 0.6 | 38 | 71 | 90 | 107 | 142 | 41 | 80 | 99 | 120 | 158 | 50 | 94 | 118 | 142 | 188 |
| 0.5 | 54 | 103 | 127 | 154 | 203 | 58 | 114 | 143 | 170 | 226 | 70 | 136 | 171 | 203 | 270 |
| 0.4 | 82 | 159 | 199 | 238 | 315 | 90 | 178 | 221 | 264 | 352 | 107 | 211 | 263 | 316 | 422 |
| 0.3 | 142 | 282 | 351 | 422 | 562 | 158 | 314 | 390 | 469 | 624 | 188 | 374 | 467 | 561 | 747 |
| 90% Power | |||||||||||||||
| SES | Subgroup Allocation | ||||||||||||||
| 50:50 | 34:66 | 25:75 | |||||||||||||
| Correlation† | |||||||||||||||
| 0.8 | 0.6 | 0.5 | 0.4 | 0.2 | 0.8 | 0.6 | 0.5 | 0.4 | 0.2 | 0.8 | 0.6 | 0.5 | 0.4 | 0.2 | |
| Sample Size | |||||||||||||||
| 1.0 | 19 | 35 | 45 | 54 | 70 | 22 | 40 | 49 | 58 | 78 | 26 | 46 | 59 | 70 | 91 |
| 0.9 | 23 | 43 | 54 | 66 | 86 | 26 | 49 | 61 | 73 | 95 | 30 | 59 | 70 | 86 | 114 |
| 0.8 | 30 | 55 | 67 | 82 | 107 | 31 | 61 | 75 | 90 | 120 | 38 | 73 | 91 | 107 | 142 |
| 0.7 | 38 | 71 | 87 | 106 | 139 | 40 | 78 | 98 | 117 | 155 | 48 | 94 | 117 | 139 | 187 |
| 0.6 | 50 | 95 | 119 | 143 | 190 | 55 | 107 | 132 | 158 | 211 | 65 | 126 | 158 | 190 | 251 |
| 0.5 | 70 | 138 | 171 | 203 | 271 | 78 | 152 | 190 | 227 | 302 | 91 | 182 | 227 | 270 | 362 |
| 0.4 | 107 | 214 | 266 | 318 | 423 | 120 | 237 | 296 | 354 | 470 | 142 | 283 | 353 | 422 | 563 |
| 0.3 | 190 | 375 | 470 | 563 | 750 | 211 | 419 | 523 | 627 | 834 | 251 | 501 | 626 | 750 | 998 |
SES = Difference in standardized effect size between subgroups
Correlation between the within-subject outcomes (treatment and placebo conditions)
In both Tables 1 and 2, if an investigator assumes that the SES will be 0.5 in one subgroup and 0.2 in the other subgroup, the sample size for the 0.3 difference in SES (i.e., the treatment-by-subgroup interaction) displayed in the table should be selected. Table 1 expands on the sample size considerations discussed by Vollert et al. [26], who estimate the numbers of patients that need to be screened to identify a sufficient number of patients for adequately powered clinical trials aimed at assessing treatment effects in patients with only a single sensory phenotype. The sample sizes required to have sufficient power to detect a treatment-by-subgroup interaction are typically much higher than that required to detect an overall effect of the treatment. For example, in a parallel group trial, the total sample size required to detect an overall SES of 0.3 with 80% power and a significance level of 5% is approximately 350 patients. If the subgroups are of equal size (50:50 allocation) and the SESs in the subgroups are 0.1 and 0.5, respectively (i.e., an SES of 0.3 for the overall treatment effect), the total sample size required to demonstrate that these subgroups have significantly different treatment effects (i.e., the treatment-by-subgroup interaction, subgroup difference in SES of 0.4) is 786.
Given that larger sample sizes are required to test an interaction, when appropriate and feasible, cross-over designs can be used to reduce the necessary sample sizes. Note that use of the cross-over design can decrease the sample size requirements compared to the parallel group design to varying degrees depending on the assumption regarding the within-patient correlation in outcomes (Tables 1 and 2).
3. Implications and Discussion
In addition to genotypes and phenotypes inherent to patients, the sample size considerations presented here are also applicable to trials aimed at evaluating potentially modifying factors that are assigned at randomization. For example, it has been suggested that the assay sensitivity of clinical trials can be improved by implementing various research design modifications, such as improving the accuracy of pain reporting through patient training programs [6, 20]. However, in order to adequately demonstrate the effects of such efforts, they must be evaluated by testing a treatment-by-training program interaction. The sample size requirements presented in Tables 1 and 2 are also applicable to such trials aimed at evaluating factors that are assigned at randomization. For example, in a clinical trial in which patients are randomized to receive active or placebo treatment and within each treament group are randomized to receive pain rating training or no training, an analysis of the treatment-by-training group interaction will test whether the training increased the effect size of the treatment. These types of trials are necessary to determine the effectiveness of various training methods or other clinical trial design modifications intended to increase assay sensitivity [8, 14, 24]. Such data would be especially important if the methods were expensive to implement or burdensome to patients.
A significant treatment-by-phenotype/genotype interaction indicates that the response to the active treatment relative to placebo is significantly different between groups. In order to understand the potential mechanisms underlying this difference, it can be helpful to examine the magnitude of change over time in each treatment group (i.e., active and placebo) separately in each subgroup and to consider the biological plausability of a differential response to active treatment. This is especially important given the subjective nature of pain ratings and the potential for substantial placebo effects. For example, retrospective analyses suggest that patients with excessively variable baseline pain ratings in RCTs may demonstrate larger placebo responses than those with lower variability in baseline pain ratings, but this difference in response does not seem to occur to the same degree in people receiving the active treatment [12, 16]. It is biologically plausible that patients whose pain ratings are more influenced by their external environment than others will have more variable pain ratings and also be more susceptible to the placebo influences of a clinical trial [23]. Such an interpretation, however, presumes that this placebo-induced response is not added to the true effect of the treatment in this subgroup, but instead largely replaces it. Another example is given by the Demant et al. [5] trial, in which the difference between subgroups in the change over time in subjects receiving oxcarbazepine was large, whereas subjects receiving placebo exhibited almost no change over time in either subgroup. It is also biologically plausible that patients with irritable nociceptors would respond more favorably to oxcarbazepine than those without irritable nociceptors.
4. Conclusions
Continued efforts to develop personalized pain treatments have the potential to accelerate the identification of novel therapies with greater efficacy or safety, or both, for certain subgroups. Proper design of clinical trials with pre-specification of a primary analysis that tests the significance of a treatment-by-phenotype/genotype interaction is necessary to demonstrate the differential efficacy of treatments. Appropriate sample size estimation is needed to ensure that such trials have adequate power to detect minimally important subgroup differences in efficacy. These practices will ensure that personalized pain treatment strategies can be identified in clinical trials as rigorously and efficiently as possible.
Conflicts of Interest statement
Financial support for this project was provided by the ACTTION public-private partnership, which has received research contracts, grants, or other revenue from the FDA, multiple pharmaceutical and device companies, philanthropy, and other sources. The authors have no conflicts of interest related to this work to disclose.
References
- 1.Baron R, Maier C, Attal N, Binder A, Bouhassira D, Cruccu G, Finnerup NB, Haanpaa M, Hansson P, Hullemann P, Jensen TS, Freynhagen R, Kennedy JD, Magerl W, Mainka T, Reimer M, Rice AS, Segerdahl M, Serra J, Sindrup S, Sommer C, Tolle T, Vollert J, Treede RD. Peripheral neuropathic pain: a mechanism-related organizing principle based on sensory profiles. Pain 2017;158:261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brouwer BA, Merkies IS, Gerrits MM, Waxman SG, Hoeijmakers JG, Faber CG. Painful neuropathies: the emerging role of sodium channelopathies. J Peripher Nerv Syst 2014;19:53–65. [DOI] [PubMed] [Google Scholar]
- 3.Collins FS and Varmus H. A new initiatve on precision medicine. NEJM. 2015;26:793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demant DT, Lund K, Finnerup NB, Vollert J, Maier C, Segerdahl MS, Jensen TS, Sindrup SH. Pain relief with lidocaine 5% patch in localized peripheral neuroapthic pain in relation to pain phenitype: a randomised, dobule-blind, and placebo-controlled, phenotype panel study. Pain 2015;156:2234–44. [DOI] [PubMed] [Google Scholar]
- 5.Demant DT, Lund K, Vollert J, Maier C, Segerdahl M, Finnerup NB, Jensen TS, Sindrup SH. The effect of oxcarbazepine in peripheral neuropathic pain depends on pain phenotype: A randomised, double-blind, placebo-controlled phenotype-stratified study. Pain 2014;155:2263–73. [DOI] [PubMed] [Google Scholar]
- 6.Dworkin RH, Burke LB, Gewandter JS, Smith SM. Reliability is Necessary but Far From Sufficient: How Might the Validity of Pain Ratings be Improved? Clin J Pain 2015;31:599–602. [DOI] [PubMed] [Google Scholar]
- 7.Dworkin RH, Edwards RR. Phenotypes and treatment response: it’s difficult to make predictions, especially about the future. Pain 2017;158:187–189. [DOI] [PubMed] [Google Scholar]
- 8.Dworkin RH, Turk DC, Peirce-Sandner S, Burke LB, Farrar JT, Gilron I, Jensen MP, Katz NP, Raja SN, Rappaport BA, Rowbotham MC, Backonja MM, Baron R, Bellamy N, Bhagwagar Z, Costello A, Cowan P, Fang WC, Hertz S, Jay GW, Junor R, Kerns RD, Kerwin R, Kopecky EA, Lissin D, Malamut R, Markman JD, Mcdermott MP, Munera C, Porter L, Rauschkolb C, Rice AS, Sampaio C, Skljarevski V, Sommerville K, Stacey BR, Steigerwald I, Tobias J, Trentacosti AM, Wasan AD, Wells GA, Williams J, Witter J, Ziegler D. Considerations for improving assay sensitivity in chronic pain clinical trials: IMMPACT recommendations. Pain 2012;153:1148–58. [DOI] [PubMed] [Google Scholar]
- 9.Edwards RR, Cahalan C, Mensing G, Smith M, Haythornthwaite JA. Pain, catastrophizing, and depression in the rheumatic diseases. Nat Rev Rheumatol 2011;7:216–24. [DOI] [PubMed] [Google Scholar]
- 10.Edwards RR, Dworkin RH, Turk DC, Angst MS, Dionne R, Freeman R, Hansson P, Haroutounian S, Arendt-Nielsen L, Attal N, Baron R, Brell J, Bujanover S, Burke LB, Carr D, Chappell AS, Cowan P, Etropolski M, Fillingim RB, Gewandter JS, Katz NP, Kopecky EA, Markman JD, Nomikos G, Porter L, Rappaport BA, Rice AS, Scavone JM, Scholz J, Simon LS, Smith SM, Tobias J, Tockarshewsky T, Veasley C, Versavel M, Wasan AD, Wen W, Yarnitsky D. Patient phenotyping in clinical trials of chronic pain treatments: IMMPACT recommendations. Pain 2016;157:1851–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Medicines Agency. Guideline on the investigation of subgroups in confirmatory clinical trials. 2014; Accessed 9/25/2018 [http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2014/02/WC500160523.pdf].
- 12.Farrar JT, Troxel AB, Haynes K, Gilron I, Kerns RD, Katz NP, Rappaport BA, Rowbotham MC, Tierney AM, Turk DC, Dworkin RH. Effect of variability in the 7-day baseline pain diary on the assay sensitivity of neuropathic pain randomized clinical trials: an ACTTION study. Pain 2014;155:1622–31. [DOI] [PubMed] [Google Scholar]
- 13.FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource. Accessed 9/25/2018 [https://www.ncbi.nlm.nih.gov/books/NBK326791/]. [PubMed]
- 14.Finnerup NB, Haroutounian S, Baron R, Dworkin RH, Gilron I, Haanpaa M, Jensen TS, Kamerman PR, Mcnicol E, Moore A, Raja SN, Andersen NT, Sena ES, Smith BH, Rice ASC, Attal N. Neuropathic pain clinical trials: factors associated with decreases in estimated drug efficacy. Pain 2018;Epub Ahead of Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelman A, Stern H. The difference between “significant” and “not significant” is not itself statistically significant. The American Statistician 2006;60:328–331. [Google Scholar]
- 16.Harris RE, Williams DA, Mclean SA, Sen A, Hufford M, Gendreau RM, Gracely RH, Clauw DJ. Characterization and consequences of pain variability in individuals with fibromyalgia. Arthritis Rheum 2005;52:3670–4. [DOI] [PubMed] [Google Scholar]
- 17.Hashmi JA, Baria AT, Baliki MN, Huang L, Schnitzer TJ, Apkarian AV. Brain networks predicting placebo analgesia in a clinical trial for chronic back pain. Pain 2012;153:2393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Ma S, Mu J, Chen T, Xu Q, Dun W, Tian J, Zhang M. Integration of white matter network is associated with interindividual differences in psychologically mediated placebo response in migraine patients. Hum Brain Mapp 2017;38:5250–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore A, Derry S, Eccleston C, Kalso E. Expect analgesic failure; pursue analgesic success. BMJ 2013;346:f2690. [DOI] [PubMed] [Google Scholar]
- 20.Smith SM, Amtmann D, Askew RL, Gewandter JS, Hunsinger M, Jensen MP, Mcdermott MP, Patel KV, Williams M, Bacci ED, Burke LB, Chambers CT, Cooper SA, Cowan P, Desjardins P, Etropolski M, Farrar JT, Gilron I, Huang IZ, Katz M, Kerns RD, Kopecky EA, Rappaport BA, Resnick M, Strand V, Vanhove GF, Veasley C, Versavel M, Wasan AD, Turk DC, Dworkin RH. Pain intensity rating training: results from an exploratory study of the ACTTION PROTECCT system. Pain 2016;157:1056–64. [DOI] [PubMed] [Google Scholar]
- 21.Tetreault P, Mansour A, Vachon-Presseau E, Schnitzer TJ, Apkarian AV, Baliki MN. Brain Connectivity Predicts Placebo Response across Chronic Pain Clinical Trials. PLoS Biol 2016;14:e1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Themistocleous AC, Crombez G, Baskozos G, Bennett DL. Using stratified medicine to understand, diagnose, and treat neuropathic pain. Pain 2018;159 Suppl 1:S31–S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treister R, Honigman L, Lawal OD, Lanier R, Katz NP. A deeper look at pain variability and its relationship with the placebo response: Results from a randomized, double-blind, placebo-controlled clinical trial of naproxen in osteoarthritis of the knee. Pain 2018;Under Review. [DOI] [PubMed] [Google Scholar]
- 24.Treister R, Lawal OD, Shecter JD, Khurana N, Bothmer J, Field M, Harte SE, Kruger GH, Katz NP. Accurate pain reporting training diminishes the placebo response: Results from a randomised, double-blind, crossover trial. PLoS One 2018;13:e0197844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.U.S. Department of Health and Human Services Food and Drug Administration. Guidance for industry: Enrichment strategies for cliinical trials to support approval of human drugs and biological products. 2012.
- 26.Vollert J, Maier C, Attal N, Bennett DLH, Bouhassira D, Enax-Krumova EK, Finnerup NB, Freynhagen R, Gierthmuhlen J, Haanpaa M, Hansson P, Hullemann P, Jensen TS, Magerl W, Ramirez JD, Rice ASC, Schuh-Hofer S, Segerdahl M, Serra J, Shillo PR, Sindrup S, Tesfaye S, Themistocleous AC, Tolle TR, Treede RD, Baron R. Stratifying patients with peripheral neuropathic pain based on sensory profiles: algorithm and sample size recommendations. Pain 2017;158:1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wasan AD, Davar G, Jamison R. The association between negative affect and opioid analgesia in patients with discogenic low back pain. Pain 2005;117:450–61. [DOI] [PubMed] [Google Scholar]
- 28.Wasan AD, Jamison RN, Pham L, Tipirneni N, Nedeljkovic SS, Katz JN. Psychopathology predicts the outcome of medial branch blocks with corticosteroid for chronic axial low back or cervical pain: a prospective cohort study. BMC Musculoskelet Disord 2009;10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]