Abstract
Hepatocellular carcinoma (HCC) accounts for >700,000 deaths worldwide, largely related to poor rates of diagnosis. Our previous work identified glycoproteins with increased levels of fucosylation in HCC. Plate-based assays to measure this change were compromised by increased levels of heterophilic antibodies with glycan lacking terminal galactose residues, which allowed for increased binding to the lectins used in these assays. To address this issue, we developed a multi-step protein A/G incubation and filtration method to remove the contaminating signal. However, this method was time consuming and expensive so alternative methods were desired. Herein, we describe a simple method relying on PEG precipitation that allows for the removal of IgG and IgM but retention of glycoproteins of interest. This method was tested on three sample sets, two internal and one external. PEG depletion of heterophilic IgG and IgM reduced in the coefficient of variation as observed with the protein A/G filtration method from 26.82% to 7.50% and allowed for the measurement of fucosylated protein. This method allowed for the measurement of fucosylated kininogen, which could serve as a biomarker of HCC. In conclusion, a new and simple method for the depletion of heterophilic IgG and IgM was developed and allowed for the analysis of fucosylated kininogen in patients with liver disease.
Keywords: biomarker, glycosylation, hepatocellular carcinoma, lectin, liver cance
1.0. Introduction:
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide and the incidence in the United States (USA) is increasing [1–3]. The progression of liver disease into liver cancer can be monitored with serum levels of alpha-fetoprotein (AFP). However, AFP’s limited sensitivity and specificity has reduced its reliability as a primary screening tool for HCC[4], and more sensitive biomarkers for HCC are desired.
Using fucose-specific lectins, we previously identified more than 50 glycoproteins that contained increased fucosylation in HCC[5] as compared to those with liver disease but without HCC, and have used these in plate-based assays to detect HCC in the background of cirrhosis[6–8]. However, the plate-based assays were hampered by the presence of heterophilic antibodies (human antibodies that can interact with assay antibodies) that themselves had altered glycan[9]. This alteration in glycosylation occurred with liver fibrosis and cirrhosis, the background from which most HCC develops[3,10]. To deal with this issue, we have utilized several methods to deplete heterophilic immunoglobulins from serum. Unfortunately, all of these methods required expensive protein A/G mixtures and several time consuming filtration steps limiting their translation and clinical use [9,11]. In the current study, we have developed a simple and rapid method for the removal of these contaminants before plate-based lectin analysis. This method was used in three independent sample sets to validate the method and the performance of fucosylated kininogen as a biomarker of HCC. Fucosylated kininogen is of interest as we have shown in prior studies that this marker can combine with AFP and other clinical factors to achieve AUC of >0.90 in the differentiation of HCC from cirrhosis [12]. The potential use of this method as a diagnostic tool for the detection of liver cancer is discussed.
2.0. Material and Methods.
2.1. Patient Samples and Ethics Statements
Three sets of serum samples were utilized for this study. The first was from the University of Texas Southwestern (UTSW) Medical Center. Serum samples were obtained via a study protocol approved by the UTSW Institutional Review Board. In all cases, written informed consent was obtained from each subject. Diagnosis of cirrhosis was based on liver histology or clinical, laboratory and imaging evidence of hepatic decompensation or portal hypertension (15). Each non-HCC patient had a normal ultrasound; if serum AFP was elevated, a CT or MRI showed no liver mass. For HCC patients, the diagnosis of HCC was made by histopathology or imaging (magnetic resonance imaging [MRI] or computed tomography [CT]) showing a vascular enhancing mass with delayed washout) (5). Tumor staging was determined using the Milan Criteria[13]. Demographic and clinical information were obtained, and a blood sample was collected from each subject. A 20-ml blood sample was drawn from each subject, spun, aliquoted, and serum stored at –80°C until testing. Blood samples for HCC patients were drawn prior to initiation of HCC treatment. Patient details are provided in Supplementary Table S1.
The second set was obtained from the University Michigan under a study protocol approved by the University Michigan Review Board and written informed consent was obtained from each subject. Patients details regarding samples these are found in our previous publication[14].
The third patient set was from the Fudan University Shanghai Cancer Center. As before these samples sets were collected under study protocols approved by the Institutional Review Board and written informed consent was obtained from each subject. Detailed information regarding patients from the Fudan University Shanghai Cancer Center is found in Table 1.
Table 1.
Patient characteristics for validation study.
| Disease Diagnosis | HCC1 | Cirrhosis2 | p Value3 |
|---|---|---|---|
| Number | 75 | 75 | |
| Etiology% (HBV/HCV/other) 4 | 100/0/10 | 100/0/0 | |
| Age (mean, SD) 5 | 54.1 (11.7) | 47.8 (11.9) | 0.0005 |
| AFP ng/mL (mean, SD) 6 | 842.7 (1291) | 5.2 (9.2) | <0.0001 |
| AST IU/mL (mean, SD) 7 | 50.9 (65.4) | 30.5 (21.6) | 0.0003 |
| ALK IU/mL (mean, SD) 8 | 126.2(113.7) | 69.2 21.1) | <0.0001 |
| Gender(M:F)% 9 | 80/20 | 48/52 | <0.0001 |
| Tumor Stage (1/2/3/4/unk)%10 | 3/24/25/8/40 | N/A |
Disease diagnosis was determined by MRI (HCC) or by liver biopsy (in the case of cirrhosis). All HCC patients had cancer in the background of cirrhosis.
P value comparing two groups. Patient’s characteristics were analyzed through the use of ChiSquare test, Fisher’s exact test or Welch’s approximate t test as appropriate. All test were two-sided, and p<0.05 was considered significant.
For Etiology: HBV, hepatitis B virus; HCV, hepatitis C virus; other, liver disease consisting of cryptogenic liver disease or alcohol induced liver disease.
Mean age of groups.
Mean level of AFP.
Mean level of aspartate aminotransferase (AST) with the standard deviation indicated (in U/L).
Mean level of alkaline phosphatase with the range indicated (in U/L).
Gender as a % male or female.
Tumor staging information. Unknown indicates patients with unknown tumor stage.
2.2. Lectin Fluorescence-Linked Immunosorbent Assay (FLISA)
The basic design of the commonly used lectin FLISA is described elsewhere[6]. Liver fibrosis (and cirrhosis) is associated with increased levels of heterophilic immunoglobulins and significantly, an alteration in the glycosylation of these molecules[15–17]. This change in glycosylation increases their reactivity to fucose binding lectins. Two methods to deal with these lectin reactive heterophilic antibodies were used. For the protein A/G method[15–17], a Pall Omega Nanosep 100K spin filter (Pall Corporation, #OD100C35) is washed twice with 300 µl of X PBS to prepare the filter. Subsequently, 150 µl of Pierce Protein A/G Plus™ (Pierce Chemicals, # 20423/4) is added to the filter, spun at 14,000g for 5 minutes and the Protein A/G beads re-washed twice in 300 µl 1X PBS. In a separate tube, 12.5 μL of serum is diluted to a final volume of 125 μL in 1X PBS and transfered to the washed Pierce Protein A/G Plus™ in the centrifugal filter. The filter lid is wrapped with parafilm to ensure no leaks and vortexed. Tubes are placed on a shaker for 2 hours at 37°C. Samples are spun at 10,000 g for 5 minutes and flow through is collected and used for subsequent analysis. In the final PEG method, 20 µl of serum is incubated with 20 µl of 40% polyethylene glycol (PEG)-8000 for 30 minutes at room temperature with shaking and the samples centrifuged at 14,000g for 30 minutes at 4°C. The supernatant, free of all immunoglobulins, contains fucosylated kininogen (as well as many other fucosylated proteins of interest such as fucosylated A1AT and fetuin-A) and is used for subsequent analysis.
Two assays for the measurement of fucosylated kininogen were performed. Our plate based assays that utilized detection of bound biotinylated lectin using IRDyelabelled streptavidin and visualized using the LI-COR Odyssey imaging system [8,14,7,18,19,15,12] and a second method using more conventional horse radish peroxidase-labelled streptavidin where development occurs with 3,3’,5,5’-tetramethylbenzidine (TMB) chromogenic substrate. In this case, fucosylated protein standards (66%, 55%, 44%, 33%, 22% 11% 7% and 0%) are used to generate the calibration curve and assign %fucosylation values for each sample. In all cases, fucosylation is detected using a recombinant Aleuria aurantia lectin (AAL) that is modified at site 3 (N224Q). Information on this lectin is found elsewhere[20–22].
2.3. Immunoblotting
Specific proteins or human serum[23] depleted as above were resolved via SDS-PAGE and either stained with colloidal Coomassie brilliant blue (Colloidal Blue Staining Kit, Thermo Fisher) or transferred PVDF membranes for immunoblot analysis. IgM, IgG, or kininogen was detected using polyclonal antibodies (Abcam, Cambridge, MA and AbBiotec, San Diego, CA, GenScript, Piscataway, NJ). Bound antibody was visualized using IRDye® 800-conjugated anti-mouse-800, IRDye®-conjugated anti-mouse-antibody, IRDye® 800-conjugated anti-goat antibody.
2.4. Statistical analysis
Descriptive statistics for patient groups were compared by scatter plots that included the outliers. All values were reported as mean values ±SD unless otherwise stated. For Lectin-FLISA data, as the signal intensity varied following sample processing, signal intensity was normalized using z-score transformation[24] as it facilitates statistical power for group comparisons. We applied t-test, Mann-Whitney, Chi-square test or Fisher’s exact test for statistical comparison based on data type and data distribution appropriately. We also built ROC curves and using Area under ROC curve(AUROC) to identify the discriminant ability of the different FLISAs methods.
The details on the development of the Kininogen algorithm is found in our recent publication [12] but the output equation is as follows: P=1/ (1+exp(−[−17.7221+(0.1646*age)+ (3.9453*male)+(2.4343*logAFP)+ (1.3748*Kininogen) + (0.0239*ALK)+ (−0.0222*ALT)])).
In all cases, a P output value of 0.5 was used as a fixed cutoff and patients were classified as being HCC positive when p>=0.5, otherwise they were classified as cirrhotic (p<0.5). The basic methods that were used to develop the different algorithms are described in more detail in our recent publication[25,12].
3.0. Results.
We have previously identified heterophilic IgG and IgM as contaminants in a plate based lectin assay[15]. Our initial method to remove these factors involved depletion using Pierce Protein A/G Plus™ agarose and a 100K spin filter[15]. This method was complicated, costly and not suitable for high throughput clinical use. To that end a new method was developed that utilized polyethylene glycol-8000 (PEG-8000) for the depletion of IgM and IgG with no loss of protein of interest. Supplementary Figures S1-S2 present a coomassie brilliant blue stained 15% SDS-PAGE gel from a mixture of 2 µg of IgG and 2 µg IgM following incubation with a final concentration of 3–14% PEG8000. As Supplementary Figures S1-S2 show, treatment with a final concentration of 14% PEG was required to precipitate IgM and IgG when only these two proteins were present. More complex protein mixtures such as serum required higher levels of PEG. Figure 1 shows the impact of 16–23% PEG-8000 on IgM and IgG and one protein of interest, kininogen in human serum[12]. In this experiment, 10 µl of human serum was brought to final concentrations of 16–23% PEG-8000 (lanes 4–11) and the depletion of proteins from the supernatant determined by western blotting. In Figure 1A, an anti-IgM immunoblot following PEG-8000 treatment is shown. Figure 1B is the same membrane following incubation with anti-IgG antibodies and Figure 1C is the same membrane after stripping and incubation with anti-kininogen antibodies. The control of IgG alone (Lane 2 of Figure 1), IgM alone (Lane 3) and kininogen alone (lane 12) was run on the gel directly, without any treatment or filtration. As this figure shows, IgM (lane 3, panel A), IgG (lane 3, panel B) and kininogen (lane 12, panel C) reactive bands are observed. PEG8000 concentrations of 16% or higher were required to pellet (remove) IgM from the serum while concentrations of >17% were required for the removal of IgG (Figure 1B). Kininogen levels remained constant in the supernatant in all concentrations (Figure 1C). It is noted that PEG treatment resulted in a mobility shift as a result of excess PEG in the sample (data not shown).
Figure 1: Methods for the removal of IgM and IgG via PEG-8000 precipitation.
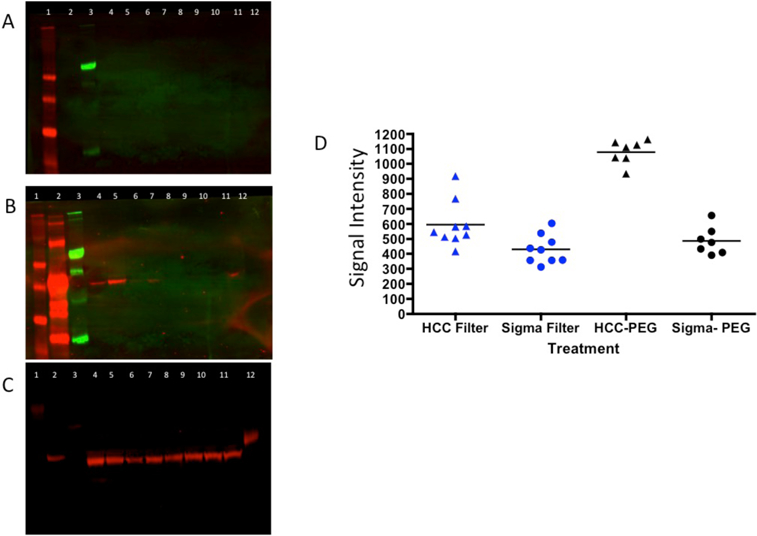
A) Anti-IgM immunoblot following PEG treatment of samples to remove IgM and IgG. B) Immunoblot of the same samples with anti-IgG or (C) anti-kininogen antibodies. D) Scatter plots of a HCC sample or control sample run multiple times for determination of the coefficient of variation. For A–C: Lane 21, markers. Lane 2, IgG alone. Lane 3, IgM alone. Lane 4, 10 µl of human serum treated with 16% Peg-8000. Lanes 5–11 are 17–23% PEG concentrations. Lane 12, kininogen alone (2 µg). Molecule weight markers are shown on left of gels.
The robustness of this assay was determined by comparison of the coefficient of variation between the two methods. To that end, we utilized the PEG method and the Protein A/G-filter method on the same HCC and healthy control serum multiple times to compare the reproducibility of two methods. As Figure 1D shows, the PEG method of sample preparation on the HCC serum results in much smaller variation than that of the Protein A/G-filter method. We derived the coefficient of variation (CV) to measure and compare the two variations (Supplementary Table S2). The CV of the protein A/G filter method on the HCC serum was 25.82% while the CV of the PEG method on HCC serum was 7.49% (p=0.0096, asymptotic test). For the healthy control serum, there was no significant difference (p=0.6722) between the two methods. This is the result of low signal from these patients and the lack of lectin reactive heterophilic antibodies in these individuals[9].
After validating the ability of this method to remove IgG and IgM from human cirrhotic serum (Supplementary Figures S3-S4), we applied this procedure to the analysis of fucosylated kininogen in two internal patient cohorts. The first consisted of 32 cirrhotic patients and 23 patients with early stage HCC in the background of cirrhosis, all of which had HCV associated liver disease. The second set consisted of 20 cirrhotic and 20 HCC patients also in the background of cirrhosis and also with HCV associated liver disease. Clinical information on the first set is found in Supplementary Table S1 and the second set is found in [14]. As Figure 2 shows, in the first set of patient samples without any serum processing, there was no statistical difference in the level of fucosylated kininogen between the HCC and cirrhotic patients(p=0.6056). The AUROC of this assay was 0.5421 (Figure 2D). In contrast, when samples were first processed using the Protein A/G Plus −100K spin filter serum processing method[15], it decreased the lectin signal observed in the cirrhotic samples (from IgG and IgM) and resulted in a significant difference between HCC and cirrhotic patients (p=0.0163) with an AUROC of 0.690. Similarly, processing of samples with PEG-8000 significantly increased the difference between HCC and cirrhotic patients (p=0.0035) and resulted in an AUROC of 0.730. Importantly, every sample that was positive by the Protein A/G Plus depletion−100K spin filter method was shown also to be positive by the PEG-8000 method.
Figure 2. Impact of the PEG mediated depletion method on Lectin-FLISA for fucosylated kininogen:
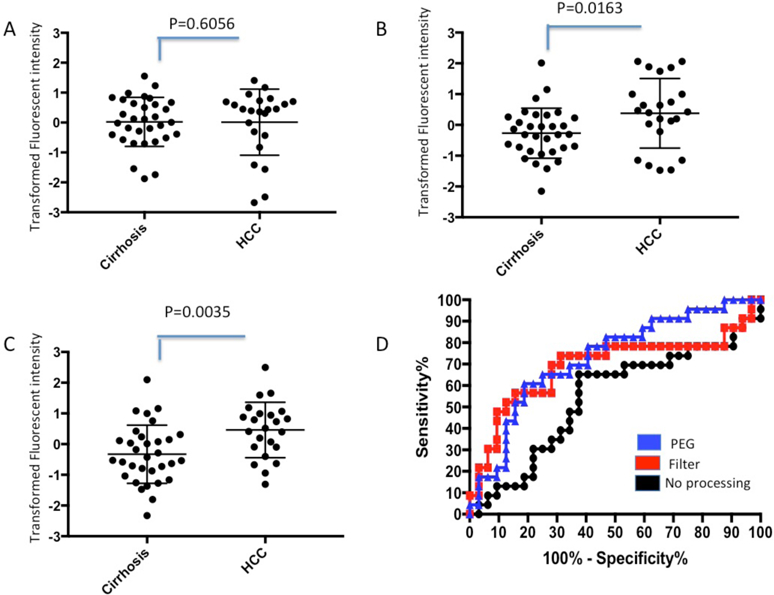
A) Scatter plot of kininogen FLISA without any treatment in 55 samples (32 cirrhotic patients and 23 cirrhotic plus HCC patients). Y-axis are Z-transformed raw intensities from the Li-Core Odyssey Scanner. B) Scatter plot of kininogen FLISA after depletion of IgM and IgG using Protein A/G and 100 K spin filter filtration. C). Scatter plot of kininogen FLISA after PEG-8000 treatment. For Panels A–C, the p value is indicated. The mean and standard deviation of the mean is indicated. D) ROC curves for data in Panels A–C.
A similar result was achieved in second set of 20 HCC and 20 cirrhotic patients. As supplementary Figure S5 shows, in this set, when using no processing procedure before the Lectin- FLISAs, there was no statistical significant difference in fucosylated kininogen levels between HCC and cirrhotic patients (p=0.5877), with an AUROC=0.551. Sample processing with the Protein A/G Plus −100K spin filter method increased the AUROC to 0.665. Similarly, the PEG processing method resulted in a significant difference between the HCC and cirrhotic patients (p=0.0143) with an AUROC 0.725 (Supplementary Figure 5D).
The PEG method in conjunction with the new fucosylated kininogen assay was tested in a third independent set at the clinical laboratory of the Fudan University Shanghai Cancer Center. This study was performed using a plate-based kit developed by a commercial entity, Glycotest, Inc, to determine % fucosylation values for kininogen. This set consisted of 75 patients with hepatitis B virus (HBV) associated chronic liver disease and 75 patients with HCC in the background of HBV associated chronic liver disease. Details on these patients are found in Table 1. In addition, we analysed 10 serum samples from healthy subjects (normal). Using the PEG-based serum processing method, the lectin ELISA for kininogen gave a mean value of 14.1% fucose (SD: 4.9%) in the cirrhotic samples and 26.1% (SD: 11.7%) in the HCC samples (Figure 3A). Serum from healthy patients was 9.8% (SD: 3.6). HCC values were statistically different then either the cirrhotic or healthy (p<0.0001). There was also statistical difference between the healthy individuals and those with cirrhosis (p=0.0018). The AUROC (HCC versus cirrhosis) of fucosylated kininogen (Figure 3C) was 0.840 which is similar to what has been observed in other studies[14]. Forty-five of the HCC patients had early stage HCC (Stage 1 or 2 UNOS) and fucosylated kininogen had a an average % fucosylated kininogen value of 24.65 (SD: 12.2). Again there was statistical difference between the healthy individuals and those with cirrhosis (P<0.0001).
Figure 3. Analysis of the PEG depletion method in an independent sample set.
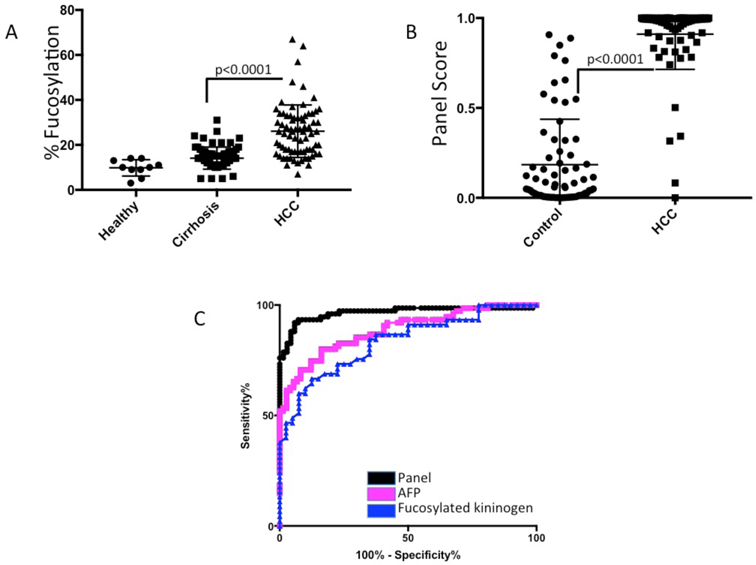
A) Scatter plot of kininogen lectin ELISA following PEG-8000 depletion in 10 healthy patients, 75 patients with HCC and 75 patients with HBV associated chronic liver disease (detailed in Table 1). B) Scatter plot of the kininogen panel scores in 75 patients with HCC and 75 patients with liver cirrhosis C) ROC curves for fucosylated kininogen, AFP or the kininogen panel.
We have recently described a kininogen based algorithm for the detection of HCC[12], and thus were interested to see how this panel worked in this cohort. This equation, which consists of age, gender, logAFP, alkaline phosphatase (ALK), aspartate transaminase (AST) and fucosylated kininogen, was used to differentiate HCC from the control group. The output of the logistic regression equation is a number between 0 and 1 and as figure 3B shows, a significant difference in the panel score was obtained between the control and HCC samples (p<0.0001). The AUROC of the Panel (Figure 3C) was 0.970, which is similar to what has been observed in other studies[12]. If only those early stage HCC patients were examined the AUROC was 0.972. AFP in this set had an AUROC of 0.88.
4.0. Discussion.
In our previous analysis of fucosylated glycoforms, we used proteomics to identify the nonspecific lectin reactive factors present in patient samples and identified IgM and IgG as the major contaminants[15]. A method for depletion of these interfering components was developed that involved incubation of sample with one specific source of Protein A/G agarose (Protein A/G Plus, Pierce Chemicals) followed by separation of the unbound material with a 100K spin filter enabled analysis of specific proteins of interest. However, this method required significant effort and time in sample preparation, including a 2-hour incubation before analysis. In addition, the use of Protein A/G Plus agarose limited the amount of serum that could be used. To that end, another method to remove heterophilic IgG and IgM was developed. The PEG-8000 method described here dramatically reduced complexity and time. This method involves the simple addition of PEG-8000 (20% final concentration) to serum and incubation of the mixture on a shaker for 30 minutes prior to centrifugation. Any volume of serum can be used and as PEG8000 is inexpensive, the overall assay cost is very low. In addition, as Figure 1D showed, the use of the PEG precipitation method reduced variability leading to a more robust assay.
The molecular basis for PEG mediated precipitation of proteins is not fully understood, but it thought to involve the relative hydrophobicity of the protein as well as the size of the protein, with larger proteins being precipitated by lower concentrations of PEG[26]. However, there is still great ambiguity regarding the nature by which PEG precipitates proteins[27].
We have used this method for the analysis of one specific fucosylated protein, fucosylated kininogen. It is noted that fucosylated kininogen alone is not a superior biomarker as compared to the current marker, AFP but consistently has AUC of 0.7 to 0.84[17,12]. However, in our recent work we have shown this protein complements AFP alone, and importantly, AFP based algorithms [6,28]. That is, we have recently developed an AFP based algorithm that acts as a biomarker “base” and validated this in over 3,000 patients[29]. Further work building upon this identified fucosylated kininogen as the best partner for this base algorithm[12]. In that previous study, the fucosylated kininogen algorithm resulted in AUROC curves of >0.95 and in the work here, in an independent sample set, the same exact algorithm results in an AUROC of 0.97, highlighting the robustness of the diagnostic algorithm.
In conclusion, having identified the material confounding our lectin-assays, we were able to develop a workflow that allowed for the lectin based assay of a specific protein in a timely and cost effective manner. Testing of this marker in an independent patient set in a clinical laboratory confirmed the ability of this marker and assay platform to differentiate HCC from cirrhosis with a high degree of accuracy. Current work is focusing on examining other markers and incorporation of these markers into existing biomarker combinations and algorithms.
Supplementary Material
Acknowledgements:
Funding: This work was supported by the National Cancer Institute [grant numbers R01 CA120206 and U01 CA168856.
Abbreviations:
- HCC
Hepatocellular carcinoma
- PEG
poly-ethylene glycol
- AUROC
area under receiver operator curve
- AUC
area under curve
- AAL
Aleuria aurantia lectin
- AFP
alpha-fetoprotein
Reference:
- 1.Block TM, Mehta AS, Fimmel CJ, Jordan R: Molecular viral oncology of hepatocellular carcinoma. Oncogene 22(33), 5093–5107 (2003). doi: 10.1038/sj.onc.1206557 [DOI] [PubMed] [Google Scholar]
- 2.Singal AG, El-Serag HB: Hepatocellular Carcinoma From Epidemiology to Prevention: Translating Knowledge into Practice. Clin Gastroenterol Hepatol 13(12), 2140–2151 (2015). doi: 10.1016/j.cgh.2015.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atiq O, Tiro J, Yopp AC, Muffler A, Marrero JA, Parikh ND, Murphy C, McCallister K, Singal AG: HCC Surveillance: Striving for a Better Balance of Benefits and Harms. Hepatology (Baltimore, Md (2017) doi: 10.1002/hep.29286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherman M: Hepatocellular carcinoma: epidemiology, risk factors, and screening. Semin Liver Dis 25(2), 143–154 (2005). doi: 10.1055/s-2005-871194 [DOI] [PubMed] [Google Scholar]
- 5.Comunale MA, Lowman M, Long RE, Krakover J, Philip R, Seeholzer S, Evans AA, Hann HWL, Block TM, Mehta AS: Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. Journal of Proteome Research 6(5), 308–315 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, Block TM, Mehta AS: Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 18(6), 1914–1921 (2009). doi: 10.1158/1055-9965.EPI-08-0980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, Long RE, Junaidi O, Bisceglie AM, Block TM, Mehta AS: Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. J Proteome Res 8(2), 595–602 (2009). doi: 10.1021/pr800752c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comunale MA, Rodemich-Betesh L, Hafner J, Wang M, Norton P, Di Bisceglie AM, Block T, Mehta A: Linkage specific fucosylation of alpha-1antitrypsin in liver cirrhosis and cancer patients: implications for a biomarker of hepatocellular carcinoma. PLoS One 5(8), e12419 (2010). doi: 10.1371/journal.pone.0012419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehta AS, Long RE, Comunale MA, Wang M, Rodemich L, Krakover J, Philip R, Marrero JA, Dwek RA, Block TM: Increased levels of galactosedeficient anti-Gal immunoglobulin G in the sera of hepatitis C virus-infected individuals with fibrosis and cirrhosis. Journal of virology 82(3), 1259–1270 (2008). doi: 10.1128/JVI.01600-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singal AG, Marrero JA, Yopp A: Screening process failures for hepatocellular carcinoma. J Natl Compr Canc Netw 12(3), 375–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balagopal A, Philp FH, Astemborski J, Block TM, Mehta A, Long R, Kirk GD, Mehta SH, Cox AL, Thomas DL, Ray SC: Human immunodeficiency virus-related microbial translocation and progression of hepatitis C. Gastroenterology 135(1), 226–233 (2008). doi:S0016-5085(08)004587 [pii] 10.1053/j.gastro.2008.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Sanda M, Comunale MA, Herrera H, Swindell C, Kono Y, Singal AG, Marrero J, Block T, Goldman R, Mehta A: Changes in the Glycosylation of Kininogen and the Development of a Kininogen-Based Algorithm for the Early Detection of HCC. Cancer Epidemiol Biomarkers Prev 26(5), 795–803 (2017). doi: 10.1158/1055-9965.EPI-16-0974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazzaferro V, Regalia E, Doci R, Andreola S, Pulvirenti A, Bozzetti F, Montalto F, Ammatuna M, Morabito A, Gennari L: Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med 334(11), 693–699 (1996). doi: 10.1056/NEJM199603143341104 [DOI] [PubMed] [Google Scholar]
- 14.Comunale MA, Wang M, Anbarasan N, Betesh L, Karabudak A, Moritz E, Devarajan K, Marrero J, Block TM, Mehta A: Total serum glycan analysis is superior to lectin-FLISA for the early detection of hepatocellular carcinoma. Proteomics. Clinical applications 7, 690–700 (2013). doi: 10.1002/prca.201200125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, Comunale MA, Herrera H, Betesh L, Kono Y, Mehta A: Identification of IgM as a contaminant in lectin-FLISA assays for HCC detection. Biochem Biophys Res Commun 476(3), 140–145 (2016). doi: 10.1016/j.bbrc.2016.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comunale MA, Wang M, Anbarasan N, Betesh L, Karabudak A, Moritz E, Devarajan K, Marrero J, Block TM, Mehta A: Total serum glycan analysis is superior to lectin-FLISA for the early detection of hepatocellular carcinoma. Proteomics Clin Appl 7(9–10), 690–700 (2013). doi: 10.1002/prca.201200125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, Block TM, Mehta AS: Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev 18(6), 1914–1921 (2009). doi: 10.1158/1055-9965.EPI-08-0980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comunale MA, Wang M, Rodemich-Betesh L, Hafner J, Lamontagne A, Klein A, Marrero J, Di Bisceglie AM, Gish R, Block T, Mehta A: Novel changes in glycosylation of serum Apo-J in patients with hepatocellular carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 20(6), 1222–1229 (2011). doi: 10.1158/1055-9965.EPI10-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamontagne A, Long RE, Comunale MA, Hafner J, Rodemich-Betesh L, Wang M, Marrero J, Di Bisceglie AM, Block T, Mehta A: Altered functionality of anti-bacterial antibodies in patients with chronic hepatitis C virus infection. PLoS ONE 8(6), e64992 (2013). doi: 10.1371/journal.pone.0064992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norton P, Comunale MA, Herrera H, Wang M, Houser J, Wimmerova M, Romano PR, Mehta A: Development and application of a novel recombinant Aleuria aurantia lectin with enhanced core fucose binding for identification of glycoprotein biomarkers of hepatocellular carcinoma. Proteomics 16(24), 3126–3136 (2016). doi: 10.1002/pmic.201600064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houser J, Kozmon S, Mishra D, Mishra SK, Romano PR, Wimmerova M, Koca J: Influence of Trp flipping on carbohydrate binding in lectins. An example on Aleuria aurantia lectin AAL. PLoS One 12(12), e0189375 (2017). doi: 10.1371/journal.pone.0189375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romano PR, Mackay A, Vong M, DeSa J, Lamontagne A, Comunale MA, Hafner J, Block T, Lec R, Mehta A: Development of recombinant Aleuria aurantia lectins with altered binding specificities to fucosylated glycans. Biochem Biophys Res Commun 414(1), 84–89 (2011). doi: 10.1016/j.bbrc.2011.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, Long RE, Junaidi O, Bisceglie AM, Block TM, Mehta AS: Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. Journal of Proteome Research 8(2), 595–602 (2009). doi: 10.1021/pr800752c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J, Pei J, Kamber M: Data mining: concepts and techniques Elsevier, (2011) [Google Scholar]
- 25.Wang XW, Zhang FX, Yang F, Ding ZF, Agarwal N, Guo ZK, Mehta JL: Effects of linagliptin and liraglutide on glucose- and angiotensin II-induced collagen formation and cytoskeleton degradation in cardiac fibroblasts in vitro. Acta Pharmacol Sin (2016). doi: 10.1038/aps.2016.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atha DH, Ingham KC: Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J Biol Chem 256(23), 1210812117 (1981). [PubMed] [Google Scholar]
- 27.Sim SL, He T, Tscheliessnig A, Mueller M, Tan RB, Jungbauer A: Protein precipitation by polyethylene glycol: a generalized model based on hydrodynamic radius. J Biotechnol 157(2), 315–319 (2012). doi: 10.1016/j.jbiotec.2011.09.028 [DOI] [PubMed] [Google Scholar]
- 28.Wang M, Sanda M, Comunale MA, Herrera H, Swindell C, Kono Y, Singal AG, Marrero JA, Block T, Goldman R, Mehta A: Changes in the glycosylation of kininogen and the development of a kininogen based algorithm for the early detection of HCC. Cancer Epidemiol Biomarkers Prev (2017). doi: 10.1158/1055-9965.EPI-16-0974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, Devarajan K, Singal AG, Marrero JA, Dai J, Feng Z, Rinaudo JA, Srivastava S, Evans A, Hann HW, Lai Y, Yang H, Block TM, Mehta A: The Doylestown Algorithm: A Test to Improve the Performance of AFP in the Detection of Hepatocellular Carcinoma. Cancer Prev Res (Phila) 9(2), 172–179 (2016). doi: 10.1158/1940-6207.CAPR-15-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


