Short abstract
The liver plays a central role in the innate immune response to endotoxemia. While previous studies have demonstrated lobe-specific transcriptional responses to various insults, whether this is true in response to endotoxemia is unknown. We sought to assess whether there were significant intra- and inter-lobe differences in the murine hepatic innate immune transcriptional response to endotoxemia. Adult male ICR mice were exposed to i.p. LPS (5 mg/kg, 30 min, 60 min, 5 h) and primary (Tnf, Cxcl1, Nfkbia, Tnfiap3) and secondary (Il6, Nos2) innate immune response gene expression was assessed in the left medial, right medial, left lateral, and right lateral lobes, and the papillary and caudate processes. The expression of all innate immune response genes increased following i.p. LPS challenge. When tested at the early time points (30 and 60 min), the left medial lobe and caudate process consistently demonstrated the highest induction of gene expression. Most inter-lobe differences were attenuated at later time points (5 h). To improve reproducibility of the study of endotoxemia induced by i.p. LPS challenge, inclusion of appropriate methodological details regarding collection of hepatic tissue should be included when reporting scientific results in published manuscripts.
Keywords: Endotoxemia, innate immune response, primary response genes, secondary response genes, liver
Introduction
Despite its limitations, the murine model of endotoxemia is an accepted and validated approach for the study of the innate immune response to pro-inflammatory stimuli. The benefits of this model include technical ease and reproducible inflammatory response.1 Increasingly, the liver is viewed as an “immunological organ” that is central to the innate immune response.2–4 This is particularly relevant in the study of endotoxemia, as the hepatic macrophage (Kupffer cell) plays an important role in clearing LPS from the systemic circulation.5–16 Thus, it is not surprising that LPS-induced hepatic expression of primary and secondary response genes is often included in studies of the innate immune response to endotoxemia.
While the hepatic innate immune response to endotoxemia is of interest, whether this is lobe-specific is unknown. There is biologic plausibility that there would be lobe-specific responses to i.p. LPS challenge. It is well recognized that hepatic anatomy and blood flow likely dictate sources of variation in hepatic gene expression.17 It is well established that i.p. injections are considered a “parenteral administration,” as absorption occurs through the mesenteric vessels, ultimately draining into the portal system.18 Previous studies have shown that i.p. injected drugs rapidly accumulate in the liver, before significant elevation in systemic concentrations.19 Furthermore, other models of hepatic injury, including ischemia-reperfusion and acetaminophen, show lobe-specific effects.20,21 Interestingly, we could find no reports on the hepatic lobe-specific transcriptional response to i.p. LPS exposure, nor could we find methodologic details in previously published manuscripts that would support a standardized approach for testing hepatic gene expression. If previously unrecognized differences did exist, this could lead to variability in results and interpretation. A better understanding of these differences would lead to consistency in study design and reporting that would stand to affect a large body of literature.
Therefore, we hypothesized that, after an i.p. LPS challenge, primary response gene expression would be significantly different between hepatic lobes. In this study, we exposed adult male mice to 30 min, 60 min, or 5 h of an i.p. LPS challenge, and evaluated gene expression in the left medial, right medial, left lateral, and right lateral lobes, and the papillary and caudate processes of the caudate lobe. Importantly, i.p. LPS challenge significantly increased the expression of all primary response genes tested (TNF-α, CXCL1, IκBα, A20). Next, we tested gene expression across lobes and assessed whether expression was significantly different across lobes, and whether the variation in expression from three randomly obtained samples obtained from each lobe would be similar across lobes. Importantly, gene expression was significantly different between lobes, and the variation in the three samples taken from individual lobes was similar between lobes. Next, we compared level of induction between lobes across the first 5 h of endotoxemia. Of note, at the earliest time point (30 min), the left medial lobe consistently demonstrated the highest induction of gene expression, most frequently statistically higher than induction in the left lateral lobe. By 60 min, expression in the left medial lobe and the caudate process were consistently higher than the other lobes, and were frequently higher than the right medial lobe and the papillary process. Most, but not all, differences in gene expression tended to attenuate at later time points (5 h). These results justify a standardized approach in the collection of hepatic tissue and mRNA/protein after LPS exposure, as well as the inclusion of appropriate methodological details when reporting scientific results in published manuscripts.
Material and methods
Murine model of endotoxemia
Adult (8–10 wk, male) Institute of Cancer Research (ICR) mice were exposed to LPS (Sigma L2630, 5 mg/kg, i.p.). All procedures were approved by the Institutional Animal Care and Use Committee at the University of Colorado (Aurora, CO), and care and handling of the animals was in accord with the National Institutes of Health guidelines for ethical animal treatment.
Collection of hepatic tissue
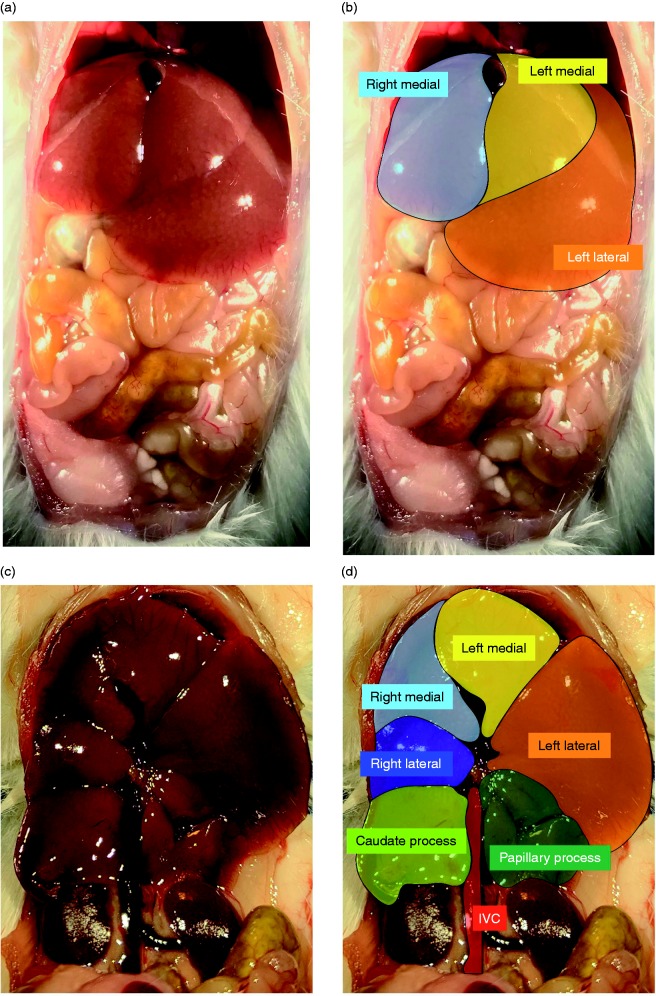
Mice were sacrificed at 30 min, 60 min, and 5 h of exposure with a fatal dose of pentobarbital sodium. During dissection, the hepatic lobes were identified as previously described: hepatic left medial lobe, left lateral lobe, right medial lobe, right lateral lobe, and the caudate process and papillary process of the caudate lobe (Figure 1).22 Lobes were collected in the following order: right medial lobe, left medial lobe, left lateral lobe, right lateral lobe, and the caudate process and papillary process of the caudate lobe, and immediately flash-frozen in liquid nitrogen. The time from opening the abdomen to the collection of the last lobe took under 1 min and was performed by a single individual (CJW). Tissues were stored at –80°C.
Figure 1.
Murine liver lobes as assessed in the current study, as adapted from Fiebig.22 (a) Ventral view of a male ICR mouse liver with corresponding color coding (b) showing the right medial lobe (light blue), left medial lobe (yellow) and left lateral lobe (orange). (c) Ventral view of the male ICR mouse liver with the stomach, small, and large intestine removed and corresponding color coding (d) right medial lobe (light blue), left medial lobe (yellow), right lateral lobe (dark blue), left lateral lobe (orange), caudate process (light green), and papillary process (dark green).
Isolation of mRNA, cDNA synthesis, and analysis of relative mRNA levels by RT-qPCR
Frozen tissue was placed in RLT buffer (Qiagen) and tissue was homogenized using the Bullet Blender (NextAdvance). Hepatic mRNA was collected from homogenized tissue using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Initially, tissue RNA was assessed for purity and concentration using the NanoDrop (ThermoFisher Scientific), and cDNA synthesized using the Verso cDNA synthesis Kit (ThermoFisher Scientific). Relative mRNA levels were evaluated by quantitative real-time PCR using exon spanning primers (Table 1), TaqMan gene expression and StepOnePlus Real-Time PCR System (Applied Biosystems). Relative quantitation was performed via normalization to the endogenous control 18S using the cycle threshold (ΔΔCt) method.
Table 1.
Primers used for PRC analysis of gene expression.
| Target | Assay ID |
|---|---|
| Tnf | Mm00443258_m1 |
| Cxcl1 | Mm04207460_m1 |
| Nfkbia | Mm00477798_m1 |
| Tnfaip3 | Mm00437121_m1 |
| Il6 | Mm00446190_m1 |
| Nos2 | Mm00440502_m1 |
Isolation of protein and Western blot analysis
Frozen hepatic tissue was homogenized using the Bullet Blender (NextAdvance) and hepatic whole cell lysates were collected in T-PER (ThermoFisher Scientific). Lysates were electrophoresed on a 4–12% polyacrylamide gel (Invitrogen) and proteins were transferred to an Immobilon membrane (Millipore) and blotted with Abs (A20, Cell Signaling, #5630; GAPDH, Cell Signaling, #5174). Blots were imaged using the LiCor Odyssey imaging system and densitometric analysis was performed using ImageStudio (LiCor).
Statistical analysis
First, we tested the null hypothesis that no difference existed in hepatic gene expression between control and LPS challenge. Thus, the means of samples taken from the left lateral lobe of three separate animals were tested by Student’s t-test. This experiment was repeated in triplicate. Next, we tested the null hypothesis that no difference existed in hepatic gene expression between liver lobes following 30 min of LPS challenge. Thus, the means of three samples taken from the same lobe from the same animal were tested by ANOVA without multiple comparisons and equality of group variances assessed using the Brown-Forsythe test. Based on these results, we tested the null hypothesis that no difference existed in hepatic gene expression between liver lobes following 30 min, 60 min, and 5 h of LPS challenge. Thus, the means of samples taken from the separate lobes from three separate animals were tested by Kruskal-Wallis test. This experiment was repeated in triplicate. Statistical significance was defined as P < 0.05, and all statistical analysis was performed using Prism (GraphPad Software, Inc.).
Results
Intraperitoneal LPS challenge significantly increases hepatic expression of primary and secondary response genes
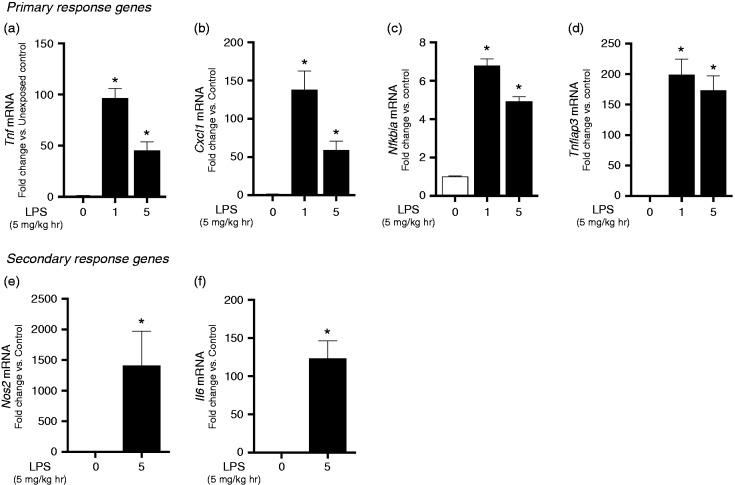
First, we sought to determine the hepatic expression of known primary and secondary response genes in adult male mice challenged with an exposure to i.p. LPS (5 mg/kg). For this study, we evaluated the expression of four, well characterized, primary innate immune response genes: Tnf (TNF-α), Cxcl1, Nfkbia (IκBα), and Tnfaip3 (A20). We chose these genes as they have CpG island promoters and are SWI/SNF-independent, thus facilitating “promiscuous induction” of expression following LPS exposure.23 For this initial assessment, samples were taken from the left lateral lobe of the liver following exposure. We observed significant induction of Tnf, Cxcl1, Nfkbia, and Tnfaip3 (A20) at both 60 min and 5 h following exposure (Figure 2a–d).
Figure 2.
Intraperitoneal LPS challenge induces hepatic primary and secondary response gene expression in adult male mice. LPS i.p. challenge (5 mg/kg, 1–5 h) significantly increases hepatic left lateral lobe mRNA expression of the primary response genes (a) Tnf (b) Cxcl1 (c) Nfkbia (d) Tnfaip3 and secondary response genes (e) Nos2 and (F) Il6. Values are means ± SE (n = 9/time point). * P < 0.05 vs. unexposed controls.
Next, we assessed the level at which LPS induced hepatic expression of the secondary response genes Il6 and Nos2. As these are secondary response genes dependent on new protein synthesis for activation, expression was assessed at the 5 h time point. Exposure to i.p. LPS challenge significantly increased hepatic Nos2 and Il6 expression (Figure 2e and f).
Based on these results, we concluded that i.p. LPS exposure significantly increased the expression of both primary and secondary response innate immune genes in the left lateral lobe of the murine liver.
Variance in repeated samples taken from the same lobe is not different between lobes, while expression of primary response genes is significantly different between lobes
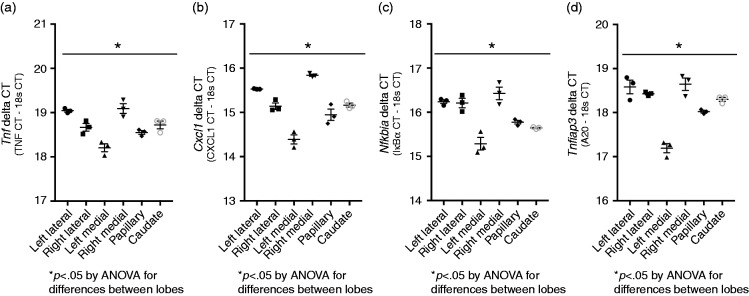
Prior to testing whether expression varied between lobes, we sought to demonstrate that the variance in expression in repeated samples from the same lobe would be similar across lobes. Thus, following tissue collection, we isolated mRNA from three separate tissue sections from each of the six liver lobes/regions assessed in this study (left lateral lobe, right lateral lobe, left medial lobe, right medial lobe, papillary process, caudate process). The expression of Tnf, Cxcl1, Nfkbia, and Tnfaip3 (A20) was tested in each of the three samples, and the variance in expression tested between groups. As expected, there was variance in expression between the repeated samples taken from the individual lobes; however, statistical testing (Brown-Forsythe test) demonstrated that the variance was not different between lobes (Figure 3a–d). Furthermore, ANOVA demonstrated that expression, as assessed by the delta CT (CT of gene of interest minus 18 s), was significantly different between lobes. Given these findings, we sought to formally test the hypothesis that after an i.p. LPS challenge, primary response gene expression would be significantly different between hepatic lobes.
Figure 3.
LPS-induced primary response gene expression varies significantly by lobe while intra-lobe variance does not. ΔCT values (CT of gene of interest minus CT of 18s) of (a) Tnf, (b) Cxcl1, (c) Nfkbia, and (d) Tnfaip3 from three separate samples taken from each lobe of a single adult male ICR mouse exposed to i.p. LPS challenge (5 mg/kg, 1–5 h). Each point represents a single value, error bars represent mean with SE. *P < 0.05 for differences between lobes by one-way ANOVA.
LPS-induced hepatic expression of primary innate immune response genes varies by lobe and changes over time
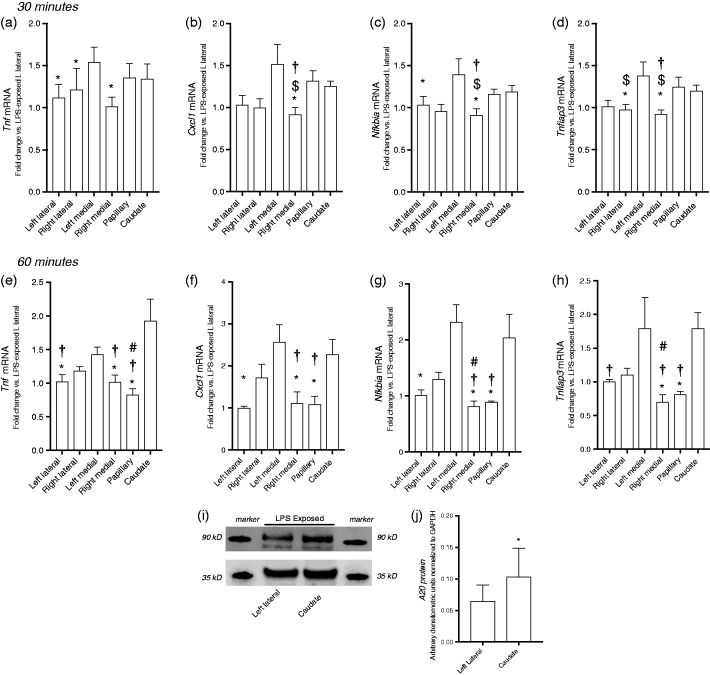
Having demonstrated that i.p. LPS challenge induced expression of primary response innate immune genes in the left lateral lobe, we compared the expression of these same genes in the other lobes to each other, normalizing expression to the left lateral lobe. Of note, systemic LPS exposure significantly increased expression of all genes tested in all liver lobes tested. However, our results showed that the level of induction was unique across lobes. At 30 min of exposure, expression of Tnf, Cxcl1, Nfkbia, and Tnfaip3 (A20) was consistently highest in the left medial lobe (Figure 4a–d). For every gene tested, LPS-induced expression levels in the left medial lobe were significantly higher than at least one other lobe (denoted by * in Figure 4a–d). In contrast, expression levels were consistently lowest in the right medial lobe. Expression of these selected primary response genes in the right medial lobe is significantly lower than the expression in at least one other lobe (vs. left medial; Tnf; Figure 4a) and up to three lobes (vs. left medial; papillary, caudate, Cxcl1, Nfkibia, Tnfiap3; Figure 4b, c, and d).
Figure 4.
LPS-induced primary response gene expression varies significantly by lobe at 30 and 60 min of exposure. LPS-induced fold change of primary response genes relative to left lateral lobe. Expression following 30 min of exposure of (a) Tnf, (b) Cxcl1, (c) Nfkbia, and (d) Tnfaip3 and following 60 min of exposure of (e) Tnf, (f) Cxcl1, (g) Nfkbia, and (h) Tnfaip3. Values are means ± SE (n = 9/time point). *P < 0.05 vs. left medial lobe; †P < 0.05 vs. caudate process; $P < 0.05 vs. papillary process; #P < 0.05 vs. right lateral lobe; % P < 0.05 vs. right medial lobe; by Kruskal-Wallis test. (i) Representative Western blot showing LPS-induced A20 protein expression in the left lateral lobe and caudate process with GAPDH as loading control. (j) Densitometric analysis of A20 in hepatic lysate following LPS exposure. *P < 0.05 vs left lateral, by Wilcoxon matched-pairs signed rank test. Values shown as means ± SEM; n = 5–6/timepoint.
Interestingly, while gene expression remained significantly different between lobes at 60 min of exposure, the pattern of these differences had changed. Expression levels remained highest in the left medial lobe for three (Cxcl1, Nfkibia, Tnfiap3; Figure 4f, g, and h) of the four genes tested. For every gene tested, LPS-induced expression levels in the left medial lobe were significantly higher than two to three other lobes (denoted by * in Figure 4a–d). Furthermore, at this later time point, expression in the caudate process had increased to a point where it was significantly higher (denoted by † in Figure 4e–h) than at two (vs. right medial and papillary; Cxcl1 and Nfkbia; Figure 4f and g) or three (vs. left lateral, right medial and papillary process; Tnf and Tnfaip3, Figure 4e and h) other regions. Expression levels were lowest in the right medial lobe and papillary process, with expression being significantly lower than at least one and up to three other lobes for every gene tested (Figure 4e–h). Furthermore, we queried whether the observed differences in transcription would lead to differences in protein expression. Having observed differences in A20 expression between the caudate process and the left lateral lobe, these regions were assessed for differences in A20 protein expression. We found that following systemic LPS exposure, A20 expression was significantly higher in the caudate process when compared with the left lateral lobe (Figure 4i and j).
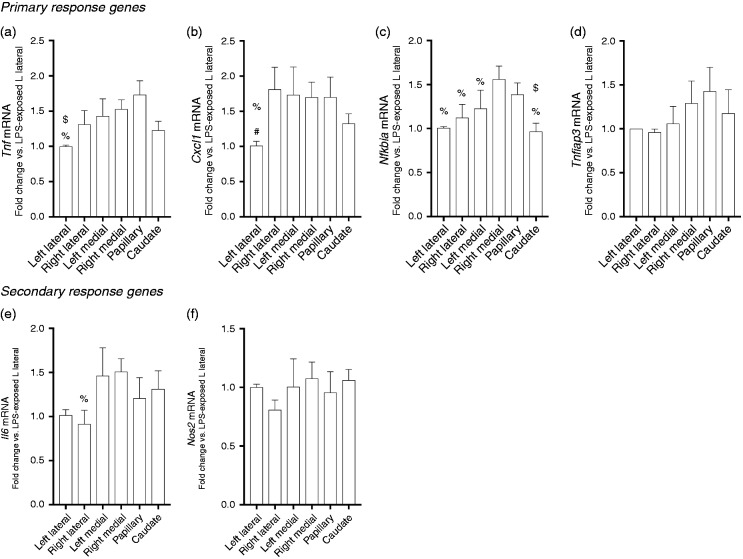
By 5 h of exposure, differences in primary response gene expression by lobe persisted, but were attenuated compared with earlier time points. Specifically, expression of Tnfiap3 was not different between lobes (Figure 5d, expression of both Tnf and Cxcl1 were lowest in the left lateral lobe (Figure 5a and b), while expression of Nfkbia was highest in the right medial lobe and the papillary process, with right medial lobe expression being significantly higher than the other four lobes, and papillary process expression being higher than just the caudate process (Figure 5c).
Figure 5.
LPS-induced primary response gene expression varies significantly by lobe at 5 h of exposure. LPS-induced fold change of primary and secondary response genes relative to left lateral lobe. Expression following 5 h of exposure of the primary response genes (a) Tnf, (b) Cxcl1, (c) Nfkbia, and (d) Tnfaip3 and secondary response genes (e) Nos2 and (f) Il6. Values are means ± SE (n = 9/time point). *P < 0.05 vs. left medial lobe; †P < 0.05 vs. caudate process; $P < 0.05 vs. papillary process; #P < 0.05 vs. right lateral lobe; %P < 0.05 vs. right medial lobe; by Kruskal-Wallis test.
LPS-induced hepatic expression of secondary innate immune response genes demonstrates little variability by lobe
Next, we assessed the expression of secondary innate immune response genes at 5 h of LPS exposure. Secondary response genes are expressed in response to prior signaling events and their expression is dependent upon new protein synthesis.24 The expression of IL6 was different only between the right medial and right lateral lobe (Figure 5e). There were no differences in Nos2 expression between the six separate hepatic lobes (Figure 5f).
Discussion
In this study, we found that following exposure to i.p. LPS challenge, the hepatic expression of primary innate immune response genes was lobe-specific and variable over time. Importantly, i.p. LPS challenge induced primary innate immune response gene expression at all times assessed in the current study. Additionally, we demonstrated that the variance between multiple samples taken from the same lobe was similar between lobes. However, at early time points following exposure (30–60 min), the left medial lobe and caudate process consistently demonstrated the highest level of primary response gene expression. In contrast, the right medial lobe and papillary process most consistently demonstrated the lowest levels of induction. Differences between lobes were generally attenuated at a later time point (5 h) of exposure. Finally, we found very little evidence for lobe-specific expression of secondary innate immune response genes at this later time point.
These results are interesting because they reveal nuances in the hepatic transcriptional response to i.p. LPS challenge. Specifically, at early time points following exposure, there are significant differences in lobe-specific hepatic expression of primary response cytokines. For the current study, we investigated the expression innate immune response genes with CpG island promoters and whose expression is SWI/SNF-independent.23 Due to this configuration, TLR4 signaling induces gene expression rapidly without the need for nucleosome remodeling or other factors needed for promoter remodeling necessary for transcription. Given the promiscuous induction of these genes downstream of TLR4 signaling, we reasoned that assessing their expression would clearly reveal any lobe-specific differences in expression if they in fact existed. Based on these results, it is clear that in the early stages of endotoxemia induced by i.p. LPS, there are significant differences in lobe-specific expression of these primary response genes. Whether there are physiologic implications of these early differences is unclear. However, the practical implications of these findings are significant. This study demonstrates the need for future studies assessing the hepatic response to i.p. LPS challenge must be consistent in how hepatic tissue is collected and assessed. Importantly, while our results showed that LPS exposure significantly increased expression of all genes tested in all liver lobes, the level of induction was unique across lobes. Thus, it likely does not matter what lobe is chosen to study, as long as there is consistency in both study design and in reporting methodologic approach and results.
We could not find any previous reports investigating the lobe-specific expression of primary innate immune response genes following i.p. LPS challenge. However, other groups have shown that there are lobe-specific responses to various exposures and injuries. Previous studies have shown “functional heterogeneity among individual liver lobes in the absence or presence of environmental factors.”17 Specifically, lobe-specific responses have been reported in rodent models of ischemia/reperfusion,20 Helicobacter hepaticus infection,25 furan-mediated hepatotoxicity,26 and bile duct ligation-induced obstructive cholestasis.27 Furthermore, lobe-specific differences in hepatic response to i.p. administered toxins. These observations have been made in the development of injury and hepatocarcinogenesis in rats exposed to i.p. carbon tetrachloride or diethylnitrosamine (DEN),28,29 as well as in hepatic distribution of acetaminophen following i.p. exposure.21 Of note, our findings are consistent with previous reports demonstrating that at baseline, hepatic gene expression exhibits very little intra-organ variance.30,31 Our findings add to a growing body of literature that advocates for consistent experimental design, with full disclosure of methods and tissue collection in studies of the hepatic response to various insults, including i.p. LPS challenge.
Perhaps more interesting are the multiple possible mechanisms underlying our results. Previous authors have hypothesized that asymmetric portal blood flow during fetal development32 programs specific liver lobes for unique responses to various stimuli.17 It is possible that these mechanisms underlie the significant differences in the LPS-induced expression of primary innate immune response genes observed in our study. Alternatively, unequal delivery of LPS to specific liver lobes via the portal circulation following peritoneal absorption could explain the differences seen in the current study. The i.p. injection is “considered a parenteral route of administration”,18 as absorption occurs mainly through the mesenteric vessels and delivered directly to the portal circulation.19 Portal streamlining of blood to the liver is one potential explanation for the observed differences.33,34 Of note, the murine portal venous system has recently been delineated in fine detail using µCT.35 Although there was some variation, the majority of the mice shared the same portal vein anatomy. From the common portal vein, the first branch (designated as the common right portal vein) feeds the caudate process of the caudate lobe and the right lateral lobe. The next branch feeds the papillary process of the caudate lobe. The portal vein then bifurcates, with the left branch feeding the left medial and left lateral lobes and the final right branch feeding the right medial lobe. Beyond these anatomical details, nothing is known about how vein caliber, turbulent flow, and branching affects portal flow. However, based on those findings, it is possible to speculate on the contribution of portal delivery of LPS to the specific lobes and how this might affect gene expression. For example, the last branch of the portal blood supply to the liver is delivered to the right medial lobe, where induction is consistently lowest. Whether this explains in part the differences seen between lobes remains to be tested. Furthermore, whether the LPS-induced expression of vasoactive innate immune factors (Edn1, endothelin 1; Nos2, inducible NO synthase) affects portal blood flow to specific lobes, and, ultimately, LPS delivery, is unknown. Additionally, it is well recognized that the liver sinusoidal endothelial cells (LSEC) play an important role in eliminating blood-borne LPS.11,12,36,37 In fact, recent evidence suggests that the LSEC is more efficient than the hepatic macrophage in clearing LPS following i.v. administration.36 Whether lobe-specific differential clearance of LPS by LSEC contributes to our findings remains to be determined. Finally, more significant differences in gene expression were found in the early time points following exposure. At later time points, for the genes tested, differences between lobes appear to attenuate. The mechanisms underlying this finding are unknown; however, we believe that the attenuation in differences at later time points may reflect a more systemic process occurring as the animal is progressively affected by endotoxemia. In contrast, the early difference likely reflects regional differences due to programming or LPS delivery via the portal flow.
There are multiple limitations to the current study. We investigated expression of a limited number of genes, specifically primary response genes with CpG island promoters and whose expression is SWI/SNF-independent and secondary response genes.23 It is very likely that other genes with other transcriptional requirements (e.g., IRF3 dependent) would show different patterns of expression and lobe-specific differences. Additionally, we tested only three time points of exposure: 30 min, 60 min, and 5 h. While our findings revealed a dynamic system with multiple changes, a more thorough time course would likely reveal even more insights into lobe-specific changes. Finally, we performed gross assessments of hepatic gene expression. No cell type specific assessment of LPS-induced gene expression was performed. This is of particular relevance because previous studies have demonstrated that i.p. LPS challenge increases hepatic activated macrophage number.37 Whether these increases are lobe-specific is unknown. It is reasonable to hypothesize that a lobe-specific change in the number of hepatic macrophages would contribute to differences in innate immune gene expression following i.p. LPS challenge. These studies remain to be done.
Another limitation to the current report is the use of only one route of LPS administration was used. Systemic endotoxemia can be induced by direct i.v. administration. Previous studies have shown that the liver plays a central role in clearing LPS after i.v. administration.5–13,38–41 However, whether there would be lobe-specific differences LPS-induced expression of innate immune response genes after i.v. exposure is unknown. There are important differences in hepatic distribution of LPS following i.v. and i.p. administration.12 It is possible that after systemic i.v. exposure, no lobe-specific differences in gene expression would be observed. However, given the current findings, the most controlled approach would include being consistent with hepatic collection and assessment of gene expression. Additionally, for the current study, we used ICR mice. It is well known that there are strain and interspecies differences in endotoxin sensitivities.7,42–44 Whether our findings in ICR mice would be blunted, or exaggerated, in different murine strains or other species is unknown. Finally, how these data apply to humans is unknown. The murine liver is lobulated, while the human liver is not. In the human, the portal vein divides the liver into left and right perfusion areas.45 Making direct comparisons between this perfusion pattern, and the murine portal anatomy described above is difficult. Finally, the hepatic artery contribution to lobar perfusion increases, and the portal contribution decreases, as the animal increases in size. It is estimated that the portal contribution is 25% lower in men than in mice.45 While important to note these differences, the fact remains that during the study of murine endotoxemia significant lobe-specific differences exist that must be accounted for during study design and sample collection.
In this study, we hypothesized that the different hepatic lobes would demonstrate significant differences in primary response gene expression following i.p. LPS challenge. We found significant lobe-specific differences in LPS-induced primary immune response gene expression that were variable over the first 5 h of exposure. Importantly, there was significant induction of expression of all genes tested [primary response genes: Tnf (TNF-α), Cxcl1, Nfkbia (IκBα), Tnfaip3 (A20); secondary response genes: Nos2 (iNOS) and Il6)] in all lobes. We conclude that, unless a standardized approach is taken to tissue collection in these types of studies, and similar studies using i.p. routes of administration, any perceived differences in gene expression may result simply from variation across lobes. Our results justify a consistent approach in the collection of hepatic tissue and mRNA/protein extraction after i.p. administration of drugs, toxins and inflammatory stimuli. Furthermore, this rigorous approach should be made clear through the inclusion of appropriate methodological details when reporting scientific results in published manuscripts.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Institutes of Health (NIH) grant R01HL132941 to CJW.
References
- 1.Stortz JA, Raymond SL, Mira JC, et al. Murine models of sepsis and trauma: can we bridge the gap? ILAR J 2017; 58: 90–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenne CN, Kubes P. Immune surveillance by the liver. Nat Immunol 2013; 14: 996–1006. [DOI] [PubMed] [Google Scholar]
- 3.Gao B, Jeong WI, Tian Z. Liver: An organ with predominant innate immunity. Hepatology 2008; 47: 729–736. [DOI] [PubMed] [Google Scholar]
- 4.Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology 2006; 43: S54–62. [DOI] [PubMed] [Google Scholar]
- 5.Mathison JC, Ulevitch RJ. The clearance, tissue distribution, and cellular localization of intravenously injected lipopolysaccharide in rabbits. J Immunol 1979; 123: 2133–2143. [PubMed] [Google Scholar]
- 6.Praaning-van Dalen DP, Brouwer A, Knook DL. Clearance capacity of rat liver Kupffer, endothelial, and parenchymal cells. Gastroenterology 1981; 81: 1036–1044. [PubMed] [Google Scholar]
- 7.McCuskey RS, McCuskey PA, Urbaschek R, et al. Species differences in Kupffer cells and endotoxin sensitivity. Infect Immun 1984; 45: 278–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freudenberg N, Piotraschke J, Galanos C, et al. The role of macrophages in the uptake of endotoxin by the mouse liver. Virchows Arch B Cell Pathol Incl Mol Pathol 1992; 61: 343–349. [DOI] [PubMed] [Google Scholar]
- 9.Ge Y, Ezzell RM, Tompkins RG, et al. Cellular distribution of endotoxin after injection of chemically purified lipopolysaccharide differs from that after injection of live bacteria. J Infect Dis 1994; 169: 95–104. [DOI] [PubMed] [Google Scholar]
- 10.Nakao A, Taki S, Yasui M, et al. The fate of intravenously injected endotoxin in normal rats and in rats with liver failure. Hepatology 1994; 19: 1251–1256. [PubMed] [Google Scholar]
- 11.Takeuchi M, Nakashima Y, Miura Y, et al. The localization of lipopolysaccharide in an endotoxemic rat liver and its relation to sinusoidal thrombogenesis: light and electron microscopic studies. Pathol Res Pract 1994; 190: 1123–1133. [DOI] [PubMed] [Google Scholar]
- 12.Yasui M, Nakao A, Yuuki T, et al. Immunohistochemical detection of endotoxin in endotoxemic rats. Hepatogastroenterology 1995; 42: 683–690. [PubMed] [Google Scholar]
- 13.Ge Y, Ezzell RM, Clark BD, et al. Relationship of tissue and cellular interleukin-1 and lipopolysaccharide after endotoxemia and bacteremia. J Infect Dis 1997; 176: 1313–1321. [DOI] [PubMed] [Google Scholar]
- 14.Shao B, Lu M, Katz SC, et al. A host lipase detoxifies bacterial lipopolysaccharides in the liver and spleen. J Biol Chem 2007; 282: 13726–13735. [DOI] [PubMed] [Google Scholar]
- 15.Shao B, Munford RS, Kitchens R, et al. Hepatic uptake and deacylation of the LPS in bloodborne LPS-lipoprotein complexes. Innate Immun 2012; 18: 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng M, Scott MJ, Loughran P, et al. Lipopolysaccharide clearance, bacterial clearance, and systemic inflammatory responses are regulated by cell type-specific functions of TLR4 during sepsis. J Immunol 2013; 190: 5152–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corton JC, Bushel PR, Fostel J, et al. Sources of variance in baseline gene expression in the rodent liver. Mutat Res 2012; 746: 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner PV, Brabb T, Pekow C, et al. Administration of substances to laboratory animals: routes of administration and factors to consider. J Am Assoc Lab Anim Sci 2011; 50: 600–613. [PMC free article] [PubMed] [Google Scholar]
- 19.Lukas G, Brindle SD, Greengard P. The route of absorption of intraperitoneally administered compounds. J Pharmacol Exp Ther 1971; 178: 562–564. [PubMed] [Google Scholar]
- 20.Palladini G, Ferrigno A, Rizzo V, et al. Lobe-specific heterogeneity and matrix metalloproteinase activation after ischemia/reperfusion injury in rat livers. Toxicol Pathol 2012; 40: 722–730. [DOI] [PubMed] [Google Scholar]
- 21.Irwin RD, Parker JS, Lobenhofer EK, et al. Transcriptional profiling of the left and median liver lobes of male f344/n rats following exposure to acetaminophen. Toxicol Pathol 2005; 33: 111–117. [DOI] [PubMed] [Google Scholar]
- 22.Fiebig T, Boll H, Figueiredo G, et al. Three-dimensional in vivo imaging of the murine liver: a micro-computed tomography-based anatomical study. PLoS One 2012; 7: e31179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez-Carrozzi VR, Braas D, Bhatt DM, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 2009; 138: 114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fowler T, Sen R, Roy AL. Regulation of primary response genes. Mol Cell 2011; 44: 348–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh R, Leuratti C, Josyula S, et al. Lobe-specific increases in malondialdehyde DNA adduct formation in the livers of mice following infection with Helicobacter hepaticus. Carcinogenesis 2001; 22: 1281–1287. [DOI] [PubMed] [Google Scholar]
- 26.Hamadeh HK, Jayadev S, Gaillard ET, et al. Integration of clinical and gene expression endpoints to explore furan-mediated hepatotoxicity. Mutat Res 2004; 549: 169–183. [DOI] [PubMed] [Google Scholar]
- 27.Ferrigno A, Palladini G, Bianchi A, et al. Lobe-specific heterogeneity in asymmetric dimethylarginine and matrix metalloproteinase levels in a rat model of obstructive cholestasis. Biomed Res Int 2014: 327537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson FC, Boucheron JA, Dyroff MC, et al. Biochemical and morphologic studies of heterogeneous lobe responses in hepatocarcinogenesis. Carcinogenesis 1986; 7: 247–251. [DOI] [PubMed] [Google Scholar]
- 29.Lawson TA, Pound AW. The different susceptibility of rat liver lobes to carbon tetrachloride and dimethylnitrosamine. Br J Exp Pathol 1974; 55: 583–588. [PMC free article] [PubMed] [Google Scholar]
- 30.Vedell PT, Svenson KL, Churchill GA. Stochastic variation of transcript abundance in C57BL/6J mice. BMC Genomics 2011; 12: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard CC, Hsu L, Delrow J, et al. Project normal: defining normal variance in mouse gene expression. Proc Natl Acad Sci USA 2001; 98: 13266–13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Germain A, Garrido A, Canas P, et al. Differences in the lipid peroxidative status, cytochrome P-450 content and microsomal oxygen consumption between right and left lobes of the liver in fetal sheep. Comparison with maternal liver. Biochem Int 1987; 15: 571–577. [PubMed] [Google Scholar]
- 33.Daniel GB, DeNovo RC, Sharp DS, et al. Portal streamlining as a cause of nonuniform hepatic distribution of sodium pertechnetate during per-rectal portal scintigraphy in the dog. Vet Radiol Ultrasound 2004; 45: 78–84. [DOI] [PubMed] [Google Scholar]
- 34.Duchen LW. The effects of deprivation of portal blood on the liver and its influence on carbon tetrachloride liver injury in the rat. Br J Exp Pathol 1961; 42: 247–252. [PMC free article] [PubMed] [Google Scholar]
- 35.Sanger C, Schenk A, Schwen LO, et al. Intrahepatic vascular anatomy in rats and mice—variations and surgical implications. PLoS One 2015; 10: e0141798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao Z, Mates JM, Cheplowitz AM, et al. Blood-borne lipopolysaccharide is rapidly eliminated by liver sinusoidal endothelial cells via high-density lipoprotein. J Immunol 2016; 197: 2390–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen LC, Gordon RE, Laskin JD, et al. Role of TLR-4 in liver macrophage and endothelial cell responsiveness during acute endotoxemia. Exp Mol Pathol 2007; 83: 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hopf U, Ramadori G, Moller B, et al. Hepatocellular clearance function of bacterial lipopolysaccharides and free lipid A in mice with endotoxic shock. Am J Emerg Med 1984; 2: 13–19. [DOI] [PubMed] [Google Scholar]
- 39.Musson RA, Morrison DC, Ulevitch RJ. Distribution of endotoxin (lipopolysaccharide) in the tissues of lipopolysaccharide-responsive and -unresponsive mice. Infect Immun 1978; 21: 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braude AI, Carey FJ, Zalesky M. Studies with radioactive endotoxin. II. Correlation of physiologic effects with distribution of radioactivity in rabbits injected with radioactive sodium chromate. J Clin Invest 1955; 34: 858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noyes HE, McInturf CR, Blahuta GJ. Studies on distribution of Escherichia coli endotoxin in mice. Proc Soc Exp Biol Med 1959; 100: 65–68. [DOI] [PubMed] [Google Scholar]
- 42.Hoshino K, Takeuchi O, Kawai T, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the LPS gene product. J Immunol 1999; 162: 3749–3752. [PubMed] [Google Scholar]
- 43.Vogel SN, Moore RN, Sipe JD, et al. BCG-induced enhancement of endotoxin sensitivity in C3H/HeJ mice. I. In vivo studies. J Immunol 1980; 124: 2004–2009. [PubMed] [Google Scholar]
- 44.Fink MP. Animal models of sepsis. Virulence 2014; 5: 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kruepunga N, Hakvoort TBM, Hikspoors J, et al. Anatomy of rodent and human livers: What are the differences? Biochim Biophys Acta Mol Basis Dis 2018; pii: S0925-4439(18)30189-3. [DOI] [PubMed]